Significance
Shigella sonnei is a globally emerging agent of bacterial dysentery. Here, we use genomics to examine the microevolution of S. sonnei in Vietnam. We show that S. sonnei was introduced into Vietnam in the early 1980s, where it continued to evolve, spreading geographically to establish localized founder populations. The population in Ho Chi Minh City has undergone several localized clonal replacement events, during which a small number of microevolutionary changes have risen to dominance. These changes, induced by horizontal gene transfer and substitution mutations, confer high-level antimicrobial resistance and the ability to kill other gut bacteria. This work provides a unique, high-resolution insight into the microevolution of a pioneering human pathogen during its establishment in a new host population.
Keywords: enteric disease, drug resistance, phylogeography, genomics
Abstract
Shigella sonnei is a human-adapted pathogen that is emerging globally as the dominant agent of bacterial dysentery. To investigate local establishment, we sequenced the genomes of 263 Vietnamese S. sonnei isolated over 15 y. Our data show that S. sonnei was introduced into Vietnam in the 1980s and has undergone localized clonal expansion, punctuated by genomic fixation events through periodic selective sweeps. We uncover geographical spread, spatially restricted frontier populations, and convergent evolution through local gene pool sampling. This work provides a unique, high-resolution insight into the microevolution of a pioneering human pathogen during its establishment in a new host population.
The bacterium Shigella sonnei is a human-adapted bacterial pathogen that accounts for approximately one-sixth of the global dysentery burden of >160 million infections and 1 million deaths annually (1, 2). We have recently shown that S. sonnei emerged in Europe ∼500 y ago and spread intercontinentally in the last few decades to establish new and locally evolving populations in countries where it is now considered endemic (3). Most of these newly disseminated S. sonnei populations belonged to a single, globally distributed, multidrug-resistant (MDR) clade of S. sonnei lineage III, which we refer to as Global III. Members of this clade are biotype g and carry a class II integron insertion within the chromosome bearing MDR genes (3). Recent shifts have been reported in the dominant agents of bacterial dysentery, with S. sonnei replacing Shigella flexneri in Vietnam, Thailand, Malaysia, China, and several other countries undergoing economic development (4–8). Here, we have sequenced the genomes of >250 S. sonnei isolated in Vietnam over a 15-y period to investigate the microevolution of this pathogen during its establishment in a naïve human population.
Results
Introduction of S. sonnei into Vietnam.
We sequenced the genomes of 244 S. sonnei isolated in Vietnam between 1995 and 2010 and examined their position within the global S. sonnei phylogeny, which already included 19 Vietnamese isolates (3) (SI Appendix, Fig. S1 and Table S1). All but two of the 263 sequenced Vietnamese S. sonnei genomes belonged to S. sonnei Global III, and 90% of all Vietnamese isolates (n = 237) formed a single clonal group comprising isolates sourced exclusively from Vietnam and which we refer to as the VN clone (Fig. 1). A comparison of isolation dates with root-to-tip distances (SI Appendix, Fig. S2) showed that point mutations have accumulated within the Vietnamese S. sonnei population with clock-like uniformity, at a rate of ∼3.6 SNPs per chromosome per year [Pearson correlation between date of isolation and maximum-likelihood (ML) branch lengths = 0.92; P < 0.0001].
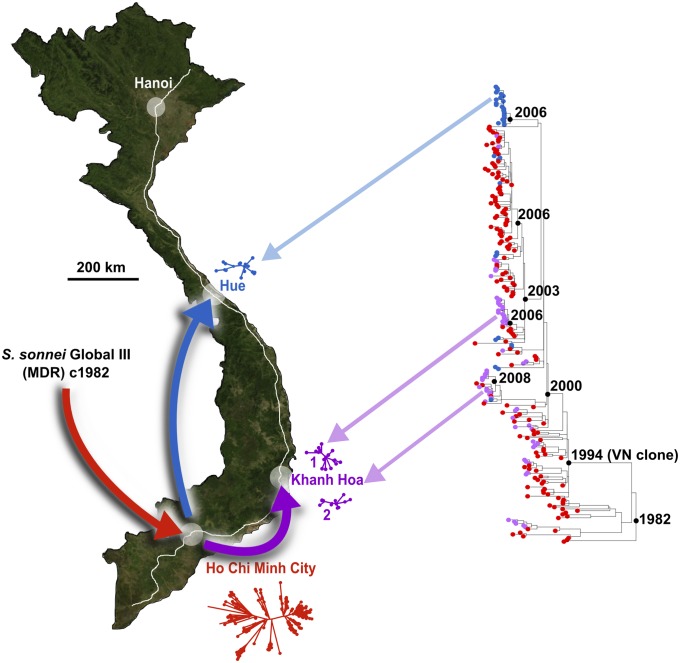
Fig. 1.
Tracking S. sonnei after entering and establishing successive founder populations across Vietnam. North-orientated map of Vietnam shows the location of the study sites in Ho Chi Minh City (HCMC), Khanh Hoa (KH), and Hue; proposed pattern of geographical spread is indicated with arrows, and National Highway 1 is shown in white. Maximum-likelihood (ML) phylogeny for the Vietnamese S. sonnei is shown to the right, with divergence dates for major clones labeled (black internal nodes). Colors indicate location of isolation: red, HCMC; blue, Hue; and purple, KH. Where distinct locally evolving populations were detected, the local phylogenies are represented next to the city; blue and purple arrows indicate where these localized Hue and KH phylogenies fit within the larger tree.
We performed a Bayesian analysis, using BEAST (9), to estimate the mutation rate and divergence dates. The estimated median clock rate was 8.5 × 10−7 substitutions⋅base−1⋅y−1 [95% highest posterior density (HPD); 7.6 × 10−7 to 9.5 × 10−7], which is faster than the mean rate across the global S. sonnei phylogeny and consistent with our previous observation that lineage III has a faster mutation rate than other lineages (3). This analysis demonstrated that all of the Vietnamese S. sonnei included in this study could be traced back to a single common ancestor that emerged in Vietnam in 1982 (95% HPD 1978–1986), shortly after the country’s postwar reunification in 1975 (Fig. 1). This finding could reflect a novel introduction of S. sonnei into Vietnam, or it could also be explained by a historical bottleneck leading to replacement of the previously circulating S. sonnei population. Consistent with the former, Vietnamese S. sonnei form a subgroup of the Global III clade (SI Appendix, Fig. S1), which we have previously shown to have spread out of Europe only since the 1970s, after the acquisition of chromosomally encoded MDR (resistance to trimethoprim, streptothricin, and streptomycin) (3). Therefore, it is most probable that the emergence of S. sonnei in the post-reunification era represents a novel introduction of S. sonnei into Vietnam during a recent global dissemination.
Phylogeography of S. sonnei in Vietnam.
Our dataset contains genome sequences for 167 S. sonnei isolated in Ho Chi Minh City (HCMC; close to the Mekong Delta region in the south), 60 from Khanh Hoa province (KH), and 36 from the central province of Hue (Fig. 1), offering an opportunity to explore the phylogeographical spread of S. sonnei within Vietnam. Notably, these provinces account for a substantial proportion of the reported dysentery burden in Vietnam (10). Although Hue and KH isolates were present in multiple subgroups within the phylogeny, the majority of isolates from these locations clustered into localized subclones (Fig. 1). Twenty-two (61%) isolates from Hue formed an independent subclone (Hue1), and 27 (45%) from KH belonged to one of two further subclones (KH1, 11 isolates, and KH2, 16 isolates; Fig. 1). These geographically constrained subclones represent local clonal expansions of S. sonnei within these provinces, indicating the recent establishment of local S. sonnei populations responsible for ongoing pathogen transmission in these locations (Fig. 1). The most recent common ancestors of the KH and Hue clones were closely related to HCMC isolates (Fig. 1), suggesting that the local populations of S. sonnei in KH and Hue are likely derived from HCMC strains. It is apparent that multiple KH and Hue S. sonnei isolates, clustering outside the geographically localized clonal groups, are distributed throughout the phylogeny (Fig. 1), suggesting that S. sonnei has been periodically transferred from HCMC to KH and Hue (16 and 9 independent events, respectively). However, these transfer events have only rarely been successful in forming new local populations (∼10% establishment rate; 2 of 16 occasions in KH and 1 of 9 occasions in Hue).
Evolutionary Dynamics of S. sonnei in HCMC.
Having determined that S. sonnei has only recently become established in Vietnam, we used the genomic sequences to investigate further the evolutionary dynamics of this pioneering pathogen within HCMC. We have previously reported that the proportion of dysentery cases in HCMC attributable to S. sonnei rose from 20% in 1995–1996 to 75% in 2005–2007 (4), and in 2009–2010 we found the rate to be 99% (Fig. 2A). The genome sequence data reveal that this change was not due to a general increase in transmission of the circulating S. sonnei population but, rather, coincides with a series of four population bottlenecks that have occurred within the locally evolving source population every 3–6 y, beginning in the mid-1990s (Fig. 2B). During each of these bottlenecks, a subset of accumulating mutations has become fixed in the local population, whereas others have been permanently lost (SI Appendix, Fig. S3). This process has resulted in an overall decline in observed genetic diversity within the HCMC S. sonnei population over time (SI Appendix, Fig. S4), which could be attributable to clonal selection or genetic drift. Because these bottlenecks coincide with increasing infection rates and the accumulation of antimicrobial resistance, we suggest that they represent selective sweeps, indicating that competition and selection are driving the local emergence of increasingly successful S. sonnei clones in HCMC.
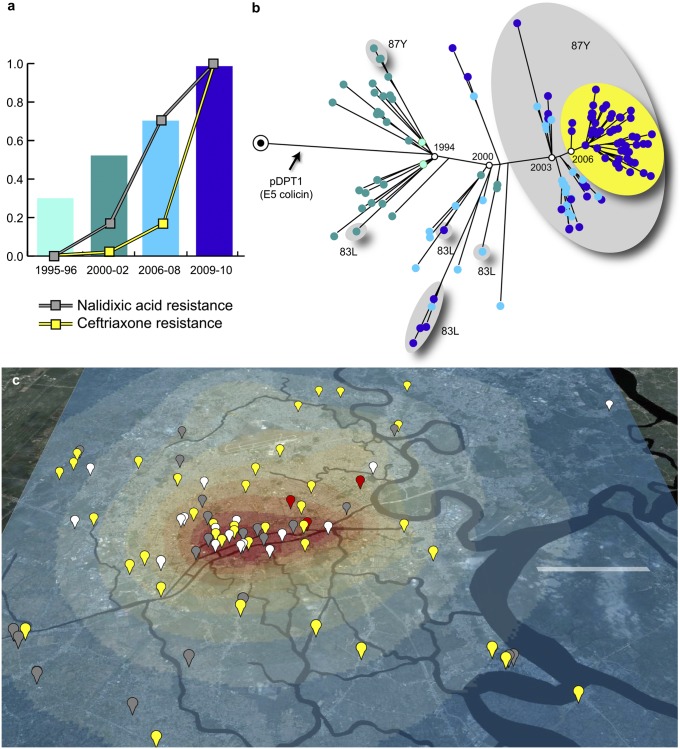
Fig. 2.
The dynamics of S. sonnei microevolution in HCMC. (A) Bars indicate the frequency of S. sonnei isolated in HCMC as a proportion of total culture-confirmed Shigellosis cases; line plots indicate the proportion of S. sonnei resistant to nalidixic acid and ceftriaxone. (B) Phylogenetic tree for HCMC S. sonnei isolates, showing estimated dates for major sweeps, acquisition of pDPT1 colicin plasmid, and fixation of key nalidixic resistance mutations (gray groups) and pKHSB1 plasmid conferring resistance to third-generation cephalosporins (yellow group). (C) North-orientated map of central HCMC, with highlighted waterways, showing the GPS locations of 53 S. sonnei patient residences identified between 2009 and 2010 (gray, sweep 3/gyrA-87Y; yellow, sweep 4/gyrA-87Y/CTX-M-15; white, presweep 3). Heat map indicates the spatial distribution of 1,419 diarrheal disease episodes in HCMC; colors of the heat map indicate the probability of all-cause diarrhea per 0.02 km2, ranging from 2 × 10−6 (dark blue) up to 1.67 × 10−4 (red). (Scale bar: 5 km.)
Global positioning system (GPS) data—available for patient residences for 53 S. sonnei isolated in HCMC between 2009 and 2010 (SI Appendix, Table S1)—were used to investigate the spatial distribution of S. sonnei infections within HCMC. All S. sonnei isolated during this period were closely related members of the VN clone with >50% derived from the most recent sweep (Fig. 2 B and C). Although there was no significant association between genetic distance and geographical distance among these isolates (P = 0.3; Mantel test)—indicating that S. sonnei circulates widely throughout the city—we found that most of the genetic diversity of S. sonnei in HCMC clustered within a southwestern region of the city (Fig. 2C). This region represents a likely infection hotspot or hub for S. sonnei transmission and evolution, contributing to disease transmission in other districts of HCMC (Fig. 2C). Some of the HCMC S. sonnei isolates were collected during a large study of diarrheal infections in children, conducted at three HCMC hospitals (n = 1,419 cases). There was no significant difference between the spatial distributions of S. sonnei infections and other diarrheal episodes attributed to all other pathogens (P > 0.05; Bernoulli model), indicating that the pattern of S. sonnei transmission is typical of overall diarrheal pathogen transmission patterns in HCMC.
Microevolution of S. sonnei in Vietnam.
To understand the extent of microevolution within the HCMC S. sonnei population, we investigated the distribution of chromosomal SNPs and the presence of horizontally acquired DNA sequences among the Vietnamese S. sonnei. Given the fundamental role of the S. sonnei invasion plasmid—encoding the O antigen and the type III secretion system required for host cell invasion—for virulence (11, 12), we determined the sequence of pINV B to determine whether plasmid-related functions were under continuing purifying or adaptive selection. Although notoriously unstable on laboratory media, pINV B was present in 66% (186) of the sequenced S. sonnei isolates collected in Vietnam. In all instances, the phylogeny of the plasmid matched the host chromosome, showing no evidence of pINV B transfer between S. sonnei hosts; rather, we surmise that the invasion plasmid is evolving in parallel with the host chromosome. We identified four SNPs that became fixed in the Vietnamese S. sonnei population during the first evolutionary bottleneck (two intergenic, one synonymous, and one nonsynonymous in a coding sequence of unknown function) and one SNP, a nonsynonymous change in invasion plasmid gene (ipg)D, which became fixed during the second bottleneck. The ipgD gene encodes an effector protein that plays a role in cellular invasion (13) and impairment of T-cell immune function (14); hence, this nonsynonymous SNP may be under selection. We detected no fixation of plasmid SNPs during the third or fourth bottlenecks.
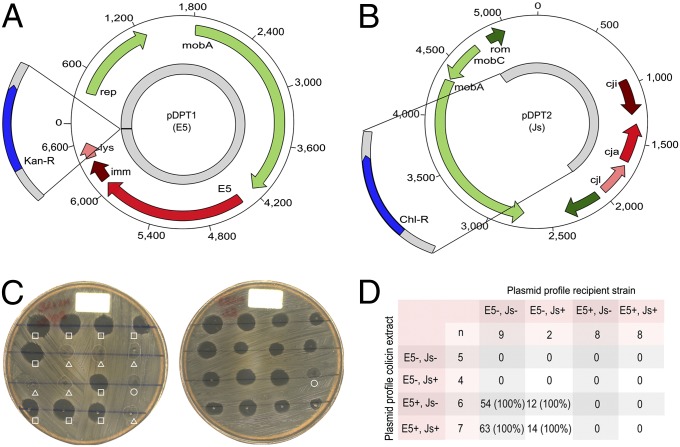
We identified >20 additional plasmids, 2 of which became fixed in the Vietnam population (Fig. 2B). The first was a 7-kbp plasmid (pDPT1) encoding an E5-type colicin and an associated immunity gene (15) (Fig. 3A). This colicin exhibited potent bactericidal activity against nonimmune Shigella and Escherichia coli (SI Appendix) and became fixed in the first sweep in ∼1994 (Fig. 3C). The second was a 100-kbp IncI1 plasmid (pKHSB1) encoding a CTX-M-15 extended spectrum beta-lactamase (ESBL) gene that became fixed in the most recent sweep in ∼2006 (Fig. 2B and SI Appendix, Figs. S5 and S6). This finding, together with evidence of novel prophage acquisition (SI Appendix, Fig. S5), indicates that the recently established Vietnamese S. sonnei population has access to, and is regularly sampling from, an extensive accessory gene pool. Taxonomic analysis of the accessory genes (SI Appendix, Table S2) indicated that they were derived from other gut-dwelling bacteria, including Escherichia/Shigella (54%), Salmonella (9%), and Yersinia (7%), consistent with other studies (16, 17). Although most of the horizontally acquired DNA was strain-specific and not fixed within subsequent lineages, the data show that sampling from the gene pool occasionally provides advantageous functions that became fixed in the evolving population. Most notable was the acquisition of the pDPT1 colicin plasmid, coinciding with the first genetic bottleneck in the mid-1990s (Figs. 2B and 3), after which the contribution of S. sonnei infections to dysentery in HCMC increased dramatically (Fig. 2A).
Fig. 3.
Colicin plasmids in the VN S. sonnei clone. (A) Schematic representation of the 6,826-bp E5 colicin-encoding plasmid pDPT1 orientated from the hypothesized origin of replication. ORFs are strand orientated and colored according to function as follows: green, plasmid replication and mobility; red, colicin activity; brown, colicin immunity; pink, colicin lysis. The gray region highlights the area of the plasmid used to create plasmid pDPT3 containing an inserted kanamycin-resistance gene (blue). (B) Schematic representation of the 5,262-bp Js colicin encoding plasmid pDPT2 orientated from the hypothesized origin of replication. ORFs are strand-orientated and colored as above. The gray region highlights the area of the plasmid used to create plasmid pDPT4 and containing an inserted chloramphenicol-resistance gene (blue). (C) Example of colicin extracts growth inhibition experiments. (Left) Lawn of S. sonnei MS128 (E5−/Js−) challenged with colicin extracts from 15 different E5+ isolates (squares), Js+ isolates (triangles), and PBS (circle). (Right) Lawn of S. sonnei MS83 (E5−/Js−) challenged with colicin extracts from 15 different E5+/Js+ isolates and PBS (circle). (D) Tabulated summary of >600 colicin extract growth inhibition experiments with 27 different bacterial isolates.
Selective Pressures Acting on the Vietnamese S. sonnei Population.
We found no general patterns of selection within the 1,388 chromosomal SNPs detected across the VN clone. Rather, mutations were randomly distributed around the chromosome (SI Appendix, Figs. S3 and S7), and nonsynonymous mutations were detected at a slightly lower rate than synonymous mutations (chromosome-wide dN/dS = 0.7; P < 1 × 10−9 against null hypothesis of dN/dS = 1 using χ2 test). dN/dS is most appropriately applied to assess interspecies variation rather than microevolution within species (18); however, this genome-wide dN/dS value is similar to that observed in other enteric pathogens, including Salmonella enterica (19, 20), and may result from a combination of purifying selection, diversifying selection, and genetic drift across different parts of the genome. A limited number of chromosomal genes displayed evidence of convergent evolution (defined as independent mutations in phylogenetically distinct backgrounds resulting in changes to identical or neighboring amino acids) or diversifying selection (a higher rate of nonsynonymous mutations over synonymous mutations; dN/dS > 1.5). Nearly all such genes were associated with stress response (e.g., oxidative stress, nutrient starvation, heavy metal stress) or antimicrobial susceptibility (SI Appendix, Table S3), indicating that the most plausible selective pressures acting on the S. sonnei population in HCMC were environmental stresses and antimicrobial exposure. For example, mutations in codons 83 or 87 of the DNA gyrase, gyrA, are associated with quinolone resistance and reduced susceptibility to fluoroquinolones (21). We detected these gyrA mutations in 157 isolates: 31 possessed a S83L mutation, 3 had an A87G mutation, and 105 had an A87Y mutation. The phylogenetic positioning of these isolates indicates that these mutations have arisen independently on at least six occasions within the HCMC population (Fig. 2B). Notably, the A87Y mutation became fixed during the third sweep (Fig. 2B), accounting for the high prevalence of resistance to nalidixic acid (NAL; Fig. 2A) and elevated minimum inhibitory concentrations (MICs) to ofloxacin (OFX) currently observed in all S. sonnei from HCMC (≥0.38 μg/mL in all 2009–2010 isolates).
The VN clone was almost certainly MDR at the time of its introduction into Vietnam (3) and has since accumulated additional resistances to quinolones (in the form of gyrA mutations; fixed since ∼2004) and third-generation cephalosporins (via acquisition of a CTX-M–bearing plasmid; sustained since ∼2006) in HCMC. Convergent evolution, inducing the same antimicrobial resistance phenotypes, has occurred in KH, where an entirely unrelated plasmid (pEG356) bearing a CTX-M-14 gene was identified (SI Appendix). Plasmid pEG356 was detected in multiple isolates of both KH clones (SI Appendix, Fig. S5) (22), indicative of local plasmid transfer between S. sonnei clones cocirculating within the province. Principal components analysis of the accessory gene content (i.e., presence/absence of noncore genes among the 263 S. sonnei isolates) showed that variation in accessory gene content among the Vietnamese S. sonnei genomes was strongly associated with geographical location (SI Appendix, Fig. S8). The distribution of accessory genes among the phylogenetic tree (SI Appendix, Fig. S5A) indicates some phylogenetic clustering of accessory genes (mainly for the HCMC ESBL plasmid pKHSB1), but also many cases where the same accessory genes are distributed across different phylogenetic lineages (including the KH ESBL plasmid pEG356, which is present in both KH clones). This finding suggests that the S. sonnei populations in HCMC, Hue, and KH differ in terms of the local gene pools to which they have access, further supporting the conclusion that localized S. sonnei populations have become established in Hue and KH and are now evolving separately from the HCMC population.
Discussion
The expansion of S. sonnei within Vietnam reflects the successful establishment of a local pathogen population within the country, which has continued to evolve and has since become responsible for almost the entire national bacterial dysentery disease burden (4). These findings provide an explanation for the majority of S. sonnei in Vietnam being MDR (4)—because they are all descendants of the MDR Global III clade—and predict that a similar pattern of local microevolution will be mirrored in other countries in which MDR Global III S. sonnei arrived some decades previously. Specifically, we hypothesize that fluoroquinolone-resistant and cephalosporin-resistant MDR S. sonnei will emerge in other locations, not by international spread of the Vietnam clone but via local microevolutionary processes within locally evolving members of the MDR Global III clade, including substitution mutations in gyrA (as we have observed, these mutations arose on multiple independent occasions in Vietnam) and the accumulation of resistance determinants acquired from other local microbial populations (as we have observed, multiple distinct CTX-M plasmids acquired in a location-specific way in different localized subpopulations of S. sonnei in Vietnam). Our data also indicate that the replacement of S. flexneri by S. sonnei in Vietnam coincided with the expansion of a single successful clone of S. sonnei rather than a generalized increase in the prevalence of diverse circulating S. sonnei strains. Further studies are required to determine whether similar replacement events that have been observed in other countries are also associated with clonal expansion.
Our observations show that S. sonnei probably entered Vietnam in HCMC, where we have described the sequential evolution and acquisition of drug-resistance determinants. We identified a spatial hotspot in HCMC in which S. sonnei infections were concentrated. This region is no more densely populated than other areas of the city, but it coincides with a canal system that may be linked to transmission. This finding is consistent with a previous spatial analysis of Shigellosis in the Vietnamese city of Nha Trang, in which the risk of S. sonnei infection was associated with proximity to the river (23). We have also shown that S. sonnei has since established new founder populations in other parts of Vietnam provinces, which are linked phylogenetically to those originating in HCMC, in which localized microevolution has continued through the very recent accumulation of novel SNPs and sampling of local gene pools.
One limitation of this study is the lack of isolates for comparison from Hanoi, Vietnam’s capital and northern center. However, Hanoi has the lowest burden of dysentery in the country (10), and it is, therefore, unlikely that this city is a major contributor to the dissemination of S. sonnei throughout Vietnam. Furthermore, the majority of the phylogenetic diversity captured within the Vietnamese S. sonnei was observed among the HCMC isolates. These data implicate HCMC as the probable source of the S. sonnei populations in the other locations and a likely location for the initial founder population of the VN clone following its original introduction into Vietnam. Because dysentery is seldom diagnosed through microbiological culture in Vietnam, our isolate collection is necessarily incomplete, yet it does include three of the provinces with the greatest dysentery burden (10). Future prospective collection of S. sonnei across the country may provide further resolution regarding the early origins of this pathogen in Vietnam; however, the temporal spread of our collection (>15 y) has been crucial in enabling us to estimate mutation rates and observe microevolutionary dynamics. In founder populations like those observed here, which are expected to be small, genetic drift can be a major driver of evolution and can allow potentially deleterious mutations to become fixed. We identified several deletion events within S. sonnei chromosomes from HCMC, KH, and Hue. However, in the HCMC population, these deletions were each restricted to one or two isolates, and we could not identify any that that became fixed in newly arising subclones. In contrast, the KH and Hue populations each harbored chromosomal deletion mutations that became fixed within the local clones. This finding suggests that genetic drift has greater impact in the KH and Hue populations, consistent with our hypothesis that these populations are smaller and more recently established than the HCMC population, from which they are likely derived. This finding also supports the notion that the bottlenecks observed within the HCMC population may be selective events rather than genetic drift.
In addition to geographical and temporal features of S. sonnei evolution, our data provide insight into the specific selective pressures faced by S. sonnei during its establishment as an endemic pathogen in a naïve human population in Vietnam. We detected selection for a plasmid-borne colicin system. This plasmid was intimately associated with the first major genetic bottleneck, because all S. sonnei isolated since 1996 bore this plasmid and formed a single clonal group. This finding is in contrast to the diversity present in the 1980s and early 1990s, which was replaced by this colicin-positive clone. We therefore hypothesize that the acquisition of this colicin plasmid is likely to have been a critical step in the establishment of S. sonnei as a significant endemic pathogen in HCMC. Our experiments demonstrated the colicin’s functionality, and we suggest that this plasmid enhances the ability of its S. sonnei host to establish infections by outcompeting gut microbiota in vivo. A recent study demonstrated an in vivo fitness advantage conferred by a plasmid-borne colicin system, transferred between Salmonella and E. coli resident in the murine gut during a burst of horizontal gene transfer associated with a period of gut inflammation (17). It is, therefore, likely that the large-scale gene acquisition we identified among S. sonnei in this study, which appears to be derived from other enteric bacteria, occurs within the human gut and may be facilitated by inflammation associated with S. sonnei infection.
We also identified diversifying selection in stress-response genes and strong selection for the accumulation of antimicrobial resistances. The ESBL encoding genes CTX-M-14 and CTX-M-15 has been reported in S. sonnei previously in geographical locations outside Vietnam (24–26), yet the associated plasmids and clones have not been described. Based on our observations of convergent evolution of ESBL-positive S. sonnei in different Vietnamese provinces—which indicates strong selective pressure for this phenotype—we predict that ESBL-expressing S. sonnei will emerge in other locales with a history of high antimicrobial use, via parallel acquisition of locally circulating ESBL plasmids rather than widespread dissemination of Vietnamese or other ESBL-containing S. sonnei clones. Antimicrobials are not universally used to treat Shigella infections in Vietnam. However, the acquisition of these ESBL-encoding plasmids inducing resistance to third-generation cephalosporins, within a highly quinolone-resistant, MDR population, is concerning and demonstrates the speed with which extensive drug resistance can accumulate within populations of enteric bacteria after escalating antimicrobial use, even if they are not first-line agents for that bacterium. Quinolone resistance has been increasing among Shigella populations globally, with the sharpest upsurges reported in Asia and Africa (27). All classes of antimicrobials are widely available without prescription in Vietnam, and we have shown previously that resistance to, and coselection against, third-generation cephalosporins and fluoroquinolones is exceptionally common in Gram-negative members of the commensal gut microbiota in healthy children and adults resident in HCMC (28). Hence, we propose that antimicrobial resistance in S. sonnei is not only a strategy for survival and onward transmission during antimicrobial therapy but may be analogous to the effect of the colicin, providing both (i) a selective advantage over antimicrobial-sensitive bacterial competitors, and (ii) a mechanism by which to maintain competitiveness in the presence of other, similarly antimicrobial-resistant organisms, which are very common within the human gut microbiota in Vietnam. These observations not only predict a fundamental role for human gut microbiota in regulating susceptibility, duration, and recovery for S. sonnei infections, but also highlight S. sonnei as a sentinel organism for monitoring the human enteric bacterial pan-genome and changing trends in antimicrobial-resistance strategies of human enteric bacterial pathogens.
Materials and Methods
Ethics.
The studies providing the data and bacterial isolates for this investigation were approved by the scientific and ethical committees of the Hospital for Tropical Diseases in HCMC, all other participants’ hospitals, and the Oxford Tropical Research Ethics Committee (OXTREC) in the United Kingdom. All parents of the subject children were required to provide written informed consent for the collection of samples and subsequent analyses, except when samples were collected as part of routine care.
Accessions.
The S. sonnei short-read sequence data reported in this work are available in the European Read Archive (accession nos. ERP000182 and ERP000631). Annotated assemblies for novel plasmids are available under accession nos. HF565446 (pDPT1), HF565445 (pDPT2), and HF572032 (pKHSB1).
Spatial Analyses.
The latitude and longitude coordinates of patients’ homes with diarrheal infections enrolled in the study conducted between May 2009 and April 2010 in HCMC were collected by using a handheld GPS device (Garmin). The distribution of isolates around HCMC and the overlaying of the phylogenetic tree was performed by using the GenGIS software package, based on the household GPS coordinates of each patient and the whole-genome ML phylogeny of their corresponding S. sonnei isolates. Association of spatial distances between patient homes and ML phylogenetic distances between their corresponding isolates was performed via Mantel test, implemented in the ade4 package for R. The reported P value is based on 100,000 permutations of the distance matrices. A kernel density estimation of the absolute density of S. sonnei cases in central HCMC was performed to generate a smoothed map of the distribution of S. sonnei by using CrimeStat software (Version 3.3; www.icpsr.umich.edu/CrimeStat/). A normal distribution interpolation method was used with an adaptive bandwidth and minimum sample size of 10 points per square kilometer. The output was visualized by using Quantum GIS (Version 1.7.3; www.qgis.org). A Bernoulli model, in SaTScan software (Version 9.1.1; www.satscan.org/), was used to examine the spatial clusters of S. sonnei cases, by using all non-S. sonnei diarrheal cases to represent the background distribution of the total diarrheal population. The upper limit for cluster detection was specified as 10% of the study population. The significance of the detected clusters was assessed by a likelihood ratio test, with a P value obtained by 999 Monte Carlo simulations generated under the null hypothesis of a random spatiotemporal distribution.
Study Sites.
The primary location providing S. sonnei isolates for this study was the pediatric gastrointestinal infections ward at the Hospital for Tropical Diseases in HCMC in southern Vietnam (136 S. sonnei isolates). Secondary locations were the pediatric gastrointestinal departments at Children’s Hospital 1 (22 S. sonnei isolates) and Children’s Hospital 2 (10 S. sonnei isolates) in HCMC, Hue Central Hospital in Hue (36 S. sonnei isolates), and Khanh Hoa General Hospital in Nha Trang (41 S. sonnei isolates). Also included in the analysis were 19 previously sequenced isolates (3) originating from Khanh Hoa General Hospital (2). Details of additional studies contributing data and bacterial isolates are given in SI Appendix.
Bacteriology.
Stool samples were collected from hospitalized patients and cultured on the day of sampling at the microbiology laboratories. Stool samples were cultured overnight in selenite F broth (Oxoid) and plated onto MacConkey and xylose lysine desoxycholate agar (Oxoid) at 37 °C. Non-lactose-fermenting colonies were subcultured on nutrient agar and identified by using API20E (Biomerieux). Serologic identification was performed by slide agglutination with polyvalent somatic (O) antigen grouping sera, followed by testing with available monovalent antisera for specific serotype identification according to the manufacturer's recommendations (Denka Seiken). All organisms were stored in 20% glycerol at −70 °C or by freeze drying in a 20% powdered milk solution and stored at room temperature.
Antimicrobial susceptibility testing was performed for all S. sonnei against ampicillin, chloramphenicol, trimethoprim–sulfamethoxazole, tetracycline, NAL, OFX, and ceftriaxone (CRO). Testing was performed initially by the disk diffusion method (Oxoid) and latterly by MICs by E test, according to the manufacturer's recommendations (AB Biodisk). Antimicrobial testing was performed for all isolates on Mueller–Hinton agar at the Hospital for Tropical Diseases microbiology laboratory; data were interpreted according to the Clinical and Laboratory Standards Institute guidelines (29). Strains that were identified as resistant to CRO by disk diffusion were further subjected to the combination disk method to confirm ESBL production. The combination disk method uses discs containing only cefotaxime (CTX) (30 µg) and ceftazidime (30 µg) and both antimicrobials combined with clavulanic acid (10 µg). ESBL-producing strains were identified as those with a >5-mm increase in zone with the single antimicrobial compared with the combined antimicrobial.
DNA Sequencing.
DNA was extracted from all S. sonnei isolates (SI Appendix, Table S1) by using the Wizard Genomic DNA Extraction Kit (Promega). The quality and concentration of the DNA were assessed by using the Quant-IT Kit (Invitrogen) before DNA sequencing. Index-tagged paired-end Illumina sequencing libraries were prepared by using 1 of 96 unique indexing tags as described (30). These were combined into pools of uniquely tagged libraries and sequenced on the Illumina Genome Analyzer GAII or HiSeq according to manufacturer’s protocols to generate tagged 54- to 100-bp paired-end reads.
Read Alignment and SNP Detection.
SNP analysis was performed as described (3). Briefly, reads were mapped to the S. sonnei reference genome (strain Ss046 chromosome, NC_007384, and pINV B plasmid, NC_007385) by using BWA (31), and SNPs were identified with SAMTools (32). Average read depths are given in SI Appendix, Table S1. SNPs called in phage regions or repetitive sequences were excluded as in ref. 3 (10.2% and 37% of bases in the reference chromosome and plasmid, respectively; gray in SI Appendix, Fig. S3), resulting in a final set of 12,311 chromosomal and 95 plasmid SNPs (SI Appendix). The allele at each locus in each isolate was determined by reference to the consensus base in that genome (using SAMTools pileup and removing low-confidence alleles with consensus base quality ≤20, read depth ≤5, or a heterozygous base call).
Phylogenetic Analyses.
Chromosomal SNP alleles were concatenated for each strain to generate a multiple alignment of all SNPs (where high-confidence base calls could not be determined, the allele was recorded as a gap character). For ML analysis, RAxML (33) was run 10 times by using the generalized time-reversible model with a Γ distribution to model site-specific rate variation (i.e., the GTR+Γ substitution model; GTRGAMMA in RAxML). One thousand bootstrap pseudoreplicate analyses were performed to assess support for the ML phylogeny. The final result (Fig. 1 and SI Appendix, Fig. S1) is the tree with the highest likelihood across all 10 runs, with ML estimates of branch length and confidence in major bipartitions calculated by using the bootstrap values across all runs. This phylogeny was rooted by using E. coli and Shigella outgroups as in ref. 3. Root-to-tip branches were extracted from the ML tree by using the program TreeStat (http://tree.bio.ed.ac.uk/software/treestat/). The relationship between root-to-tip distances and year of isolation was analyzed by using linear regression. For BEAST analysis (Version 1.6) (9), we used the GTR+Γ substitution model and defined tip dates as the year of isolation, as in prior analysis of a global collection of S. sonnei (3). We used a Bayesian skyline demographic model and either a strict molecular clock or a relaxed clock (uncorrelated lognormal distribution). Each analysis was replicated by using five chains of 100 million generations each to ensure convergence, with samples taken every 1,000 Markov chain Monte Carlo (MCMC) generations. Parameters were estimated after combining replicate analyses, totaling 450 million MCMC generations postburnin, with all reported parameter estimates [i.e., medians and 95% highest posterior densities (HPDs)] calculated by using the program Tracer (Version 1.5). Consistent with the prior global S. sonnei analysis (3), Bayesian skyline plots indicated a constant population size through time, the relaxed clock models provided better fit to the data (Bayes factor = 15, using the harmonic mean estimator of the marginal likelihood), and the SD of inferred substitution rates across branches was 0.40 [95% HPD = 0.29–0.52], providing additional support for a relaxed molecular clock. Therefore, all parameter estimates quoted are from analyses using the relaxed clock and Bayesian skyline models.
Gene Content Analysis.
Read sets were assembled by using the de novo short read assembler Velvet and Velvet Optimizer (34). Contigs of <100 bp in size were excluded from further analysis. We used an iterative mapping approach as described in ref. 3 to generate a pan-genome for the Vietnamese S. sonnei [using the nucmer algorithm in MUMmer (35)] and annotation via RAST (36). S. sonnei read sets were then aligned to the pan-genome by using BWA (31), from which the coverage (percent of bases covered) of each gene in each isolate was calculated. Details of plasmid assembly and resistance gene identification are provided in SI Appendix. Taxonomic investigation of accessory genes was performed with MG-RAST (Version 3.2; http://metagenomics.anl.gov) (37). A total of 1.2 Mbp of accessory sequence was analyzed, in which 1,141 protein features were detected. The taxonomic distribution of these protein features at genus level, based on closest matches in the M5 nonredundant protein database, is shown in SI Appendix, Table S3.
Supplementary Material
Acknowledgments
We thank Ms. Song Chau and others members of the microbiology laboratory at Oxford University Clinical Research Unit (OUCRU). The sequencing work was supported by Wellcome Trust Grant 098051. The work in Vietnam was supported by the Wellcome Trust, through core funding to OUCRU and the Vietnamese Initiative for Zoonotic Infections hospital disease surveillance consortium (with thanks to Ho Dang Trung Nghia and Tran My Phuc) and by the Li Ka Shing Foundation of China. S.B. is supported by a Royal Society Sir Henry Dale Fellowship and the Wellcome Trust. K.E.H. was supported by National Health and Medical Research Council (Australia) Fellowship 628930. D.W.K. was supported by Basic Science Research Program through the National Research Foundation of Korea, funded by Ministry of Education, Science and Technology Grant 2012R1A2A2A01009741.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The S. sonnei short-read sequence data and annotated assemblies for plasmids reported in this paper have been deposited in the European Read Archive [accession nos. ERP000182 and ERP000631 (short-read sequence data), HF565446 (pDPT1), HF565445 (pDPT2), and HF572032 (pKHSB1)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308632110/-/DCSupplemental.
References
- 1.Kotloff KL, et al. Global burden of Shigella infections: Implications for vaccine development and implementation of control strategies. Bull World Health Organ. 1999;77(8):651–666. [PMC free article] [PubMed] [Google Scholar]
- 2.von Seidlein L, et al. A multicentre study of Shigella diarrhoea in six Asian countries: Disease burden, clinical manifestations, and microbiology. PLoS Med. 2006;3(9):e353. doi: 10.1371/journal.pmed.0030353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holt KE, et al. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat Genet. 2012;44(9):1056–1059. doi: 10.1038/ng.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vinh H, et al. A changing picture of shigellosis in southern Vietnam: Shifting species dominance, antimicrobial susceptibility and clinical presentation. BMC Infect Dis. 2009;9:204. doi: 10.1186/1471-2334-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bangtrakulnonth A, et al. Shigella from humans in Thailand during 1993 to 2006: Spatial-time trends in species and serotype distribution. Foodborne Pathog Dis. 2008;5(6):773–784. doi: 10.1089/fpd.2008.0109. [DOI] [PubMed] [Google Scholar]
- 6.Banga Singh KK, Ojha SC, Deris ZZ, Rahman RA. A 9-year study of shigellosis in Northeast Malaysia: Antimicrobial susceptibility and shifting species dominance. Z Gesundh wiss. 2011;19(3):231–236. doi: 10.1007/s10389-010-0384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boehme C, et al. [Comparison of Shigella susceptibility to commonly used antimicrobials in the Temuco Regional Hospital, Chile 1990 - 2001] Rev Med Chil. 2002;130(9):1021–1026. [PubMed] [Google Scholar]
- 8.Qu F, et al. Genotypes and antimicrobial profiles of Shigella sonnei isolates from diarrheal patients circulating in Beijing between 2002 and 2007. Diagn Microbiol Infect Dis. 2012;74(2):166–170. doi: 10.1016/j.diagmicrobio.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly-Hope LA, et al. Geographical distribution and risk factors associated with enteric diseases in Vietnam. Am J Trop Med Hyg. 2007;76(4):706–712. [PubMed] [Google Scholar]
- 11.Sansonetti PJ, Kopecko DJ, Formal SB. Shigella sonnei plasmids: Evidence that a large plasmid is necessary for virulence. Infect Immun. 1981;34(1):75–83. doi: 10.1128/iai.34.1.75-83.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shepherd JG, Wang L, Reeves PR. Comparison of O-antigen gene clusters of Escherichia coli (Shigella) sonnei and Plesiomonas shigelloides O17: Sonnei gained its current plasmid-borne O-antigen genes from P. shigelloides in a recent event. Infect Immun. 2000;68(10):6056–6061. doi: 10.1128/iai.68.10.6056-6061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niebuhr K, et al. Conversion of PtdIns(4,5)P(2) into PtdIns(5)P by the S.flexneri effector IpgD reorganizes host cell morphology. EMBO J. 2002;21(19):5069–5078. doi: 10.1093/emboj/cdf522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konradt C, et al. The Shigella flexneri type three secretion system effector IpgD inhibits T cell migration by manipulating host phosphoinositide metabolism. Cell Host Microbe. 2011;9(4):263–272. doi: 10.1016/j.chom.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Lau PC, Condie JA. Nucleotide sequences from the colicin E5, E6 and E9 operons: Presence of a degenerate transposon-like structure in the ColE9-J plasmid. Mol Gen Genet. 1989;217(2-3):269–277. doi: 10.1007/BF02464892. [DOI] [PubMed] [Google Scholar]
- 16.Karberg KA, Olsen GJ, Davis JJ. Similarity of genes horizontally acquired by Escherichia coli and Salmonella enterica is evidence of a supraspecies pangenome. Proc Natl Acad Sci USA. 2011;108(50):20154–20159. doi: 10.1073/pnas.1109451108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stecher B, et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc Natl Acad Sci USA. 2012;109(4):1269–1274. doi: 10.1073/pnas.1113246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kryazhimskiy S, Plotkin JB. The population genetics of dN/dS. PLoS Genet. 2008;4(12):e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holt KE, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet. 2008;40(8):987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Z, et al. Neutral genomic microevolution of a recently emerged pathogen, Salmonella enterica serovar Agona. PLoS Genet. 2013;9(4):e1003471. doi: 10.1371/journal.pgen.1003471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopkins KL, Davies RH, Threlfall EJ. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: Recent developments. Int J Antimicrob Agents. 2005;25(5):358–373. doi: 10.1016/j.ijantimicag.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen NT, et al. The sudden dominance of blaCTX-M harbouring plasmids in Shigella spp. Circulating in Southern Vietnam. PLoS Negl Trop Dis. 2010;4(6):e702. doi: 10.1371/journal.pntd.0000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim DR, et al. Geographic analysis of shigellosis in Vietnam. Health Place. 2008;14(4):755–767. doi: 10.1016/j.healthplace.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Zhang R, et al. Serotypes and extended-spectrum β-lactamase types of clinical isolates of Shigella spp. from the Zhejiang province of China. Diagn Microbiol Infect Dis. 2011;69(1):98–104. doi: 10.1016/j.diagmicrobio.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 25.Tajbakhsh M, et al. Antimicrobial-resistant Shigella infections from Iran: An overlooked problem? J Antimicrob Chemother. 2012;67(5):1128–1133. doi: 10.1093/jac/dks023. [DOI] [PubMed] [Google Scholar]
- 26.Sabra AH, et al. Molecular characterization of ESBL-producing Shigella sonnei isolates from patients with bacilliary dysentery in Lebanon. J Infect Dev Ctries. 2009;3(4):300–305. doi: 10.3855/jidc.128. [DOI] [PubMed] [Google Scholar]
- 27.Gu B, et al. Comparison of the prevalence and changing resistance to nalidixic acid and ciprofloxacin of Shigella between Europe-America and Asia-Africa from 1998 to 2009. Int J Antimicrob Agents. 2012;40(1):9–17. doi: 10.1016/j.ijantimicag.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Le TM, et al. High prevalence of plasmid-mediated quinolone resistance determinants in commensal members of the Enterobacteriaceae in Ho Chi Minh City, Vietnam. J Med Microbiol. 2009;58(Pt 12):1585–1592. doi: 10.1099/jmm.0.010033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard. Wayne, PA: Clinical and Laboratory Standards Institute; 2012. 11th Ed. [Google Scholar]
- 30.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327(5964):469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 34.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aziz RK, et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer F, et al. The metagenomics RAST server: A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.