Significance
Metallocofactors enable enzymes to catalyze difficult reactions that would otherwise not be possible, such as the reduction of oxygen. Nature utilizes a number of different metals, and it is crucial that proteins bind the correct metals to execute their function. Nonetheless, the principles that govern metal specificity in proteins remain poorly understood. Here we use an enzyme that forms a heterodinuclear Mn/Fe cofactor with the same protein ligands in both metal-coordinating positions to study how proteins can differentiate between two such similar metals. We show that the protein is intrinsically capable of site-specific metal discrimination. Surprisingly, specificity is achieved in a stepwise process involving not only fundamental affinity differences, but also chemical maturation upon reaction with molecular oxygen.
Keywords: protein metallation, di-metal carboxylate protein, ferritin superfamily, X-ray crystallography, EPR spectroscopy
Abstract
Although metallocofactors are ubiquitous in enzyme catalysis, how metal binding specificity arises remains poorly understood, especially in the case of metals with similar primary ligand preferences such as manganese and iron. The biochemical selection of manganese over iron presents a particularly intricate problem because manganese is generally present in cells at a lower concentration than iron, while also having a lower predicted complex stability according to the Irving–Williams series (MnII < FeII < NiII < CoII < CuII > ZnII). Here we show that a heterodinuclear Mn/Fe cofactor with the same primary protein ligands in both metal sites self-assembles from MnII and FeII in vitro, thus diverging from the Irving–Williams series without requiring auxiliary factors such as metallochaperones. Crystallographic, spectroscopic, and computational data demonstrate that one of the two metal sites preferentially binds FeII over MnII as expected, whereas the other site is nonspecific, binding equal amounts of both metals in the absence of oxygen. Oxygen exposure results in further accumulation of the Mn/Fe cofactor, indicating that cofactor assembly is at least a two-step process governed by both the intrinsic metal specificity of the protein scaffold and additional effects exerted during oxygen binding or activation. We further show that the mixed-metal cofactor catalyzes a two-electron oxidation of the protein scaffold, yielding a tyrosine–valine ether cross-link. Theoretical modeling of the reaction by density functional theory suggests a multistep mechanism including a valyl radical intermediate.
Half of all enzymes are estimated to contain metallocofactors (1). An important subset uses transition metal ions to perform key redox reactions such as oxygen activation. The diiron cofactor of the ferritin-like superfamily of proteins is particularly versatile (2). While ferritin itself simply oxidizes and sequesters iron (3), in other family members the diiron center acts as a transient one- or two-electron oxidant. In the R2 subunits of class I ribonucleotide reductases (RNRs) it generates a redox-active tyrosyl radical (4, 5), whereas in the bacterial multicomponent monooxygenases (BMMs) it catalyzes the hydroxylation of a variety of hydrocarbons (6). For four decades it was assumed that all ferritin superfamily proteins contained diiron cofactors. However, in recent years new subfamilies harboring either a dimanganese or heterodinuclear Mn/Fe cofactor have been documented (7–14). The Mn/Fe cofactor was discovered in class Ic RNR R2 subunits, where its MnIV/FeIII state functionally replaces the diiron-tyrosyl radical cofactor of class Ia R2s (9, 10). After a long controversy, class Ib R2 proteins were shown to use a dimanganese cofactor in the same scaffold (7, 8). These recent developments highlight the complexity of correctly identifying the metals that make up native metallocofactors. While the metal preferences of some primary coordination motifs are well known and distinct, others are more promiscuous and less well understood. Manganese and iron are two of the most important and versatile metals for biological redox chemistry. Their primary ligand preferences are very similar, and their binding sites in enzymes often appear virtually identical (15). However, their redox potentials differ greatly, and correct discrimination between them is therefore paramount for redox-active enzymes (16). Metal specificity is commonly discussed in terms of the intracellular availability of metal ions and the Irving–Williams series of metal complex stabilities (MnII < FeII < NiII < CoII < CuII > ZnII) (17). Manganese is generally present in cells at a lower concentration than iron (18) and also has a lower predicted complex stability. The biochemical selection of manganese over iron thus presents an intricate problem. Metallochaperones are required for correct metallation of some metalloproteins (19–22), but to date no chaperones have been identified for manganese, and it is unclear whether they are required for diiron clusters (16). Protein-folding location or general metal status can also control metallation (16, 23). However, these mechanisms cannot be used for proteins with mixed-metal cofactors.
The heterodinuclear Mn/Fe cofactor recently discovered in a protein from Mycobacterium tuberculosis provides an ideal model system to study manganese/iron metallation. The protein was found to belong to a novel group of R2-like proteins, denoted R2-like ligand-binding oxidases (R2lox) (11, 24). These proteins are particularly interesting because the primary protein-derived metal-binding ligands are identical in both metal sites, and also identical to the closely related diiron-binding BMMs. To verify that the mixed-metal cofactor is a common feature of the R2lox group, here we characterize a homolog from the thermophilic bacterium Geobacillus kaustophilus (GkR2loxI). Using crystallography, spectroscopy, and quantum chemical calculations, we show that site-specific metal discrimination is inherent to the protein structure and diverges from the Irving–Williams series without requiring auxiliary factors in vitro. The mixed-metal cofactor activates oxygen and catalyzes the formation of an ether cross-link in the protein scaffold, demonstrating the chemical potential of this cofactor.
Results
A Stable Heterodinuclear MnIII/FeIII Cofactor Obtained Under Aerobic Conditions.
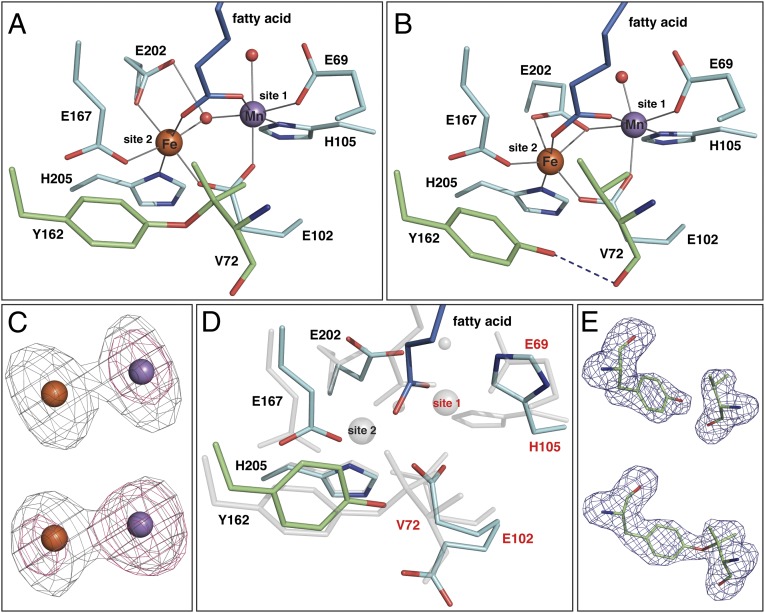
GkR2loxI was produced in Escherichia coli and subsequently reconstituted with MnII and FeII in the presence of air. Its crystal structure, solved at 1.9 Å resolution (Table S1), is very similar to the previously reported structure of its homolog from M. tuberculosis (MtR2lox) (11). The active site architectures of both proteins are virtually identical: The metal ions are coordinated by two histidine and four glutamate residues and bridged by a µ-oxo/hydroxo ligand (Fig. 1A and Figs. S1A and S2). A water molecule is bound at the open coordination site of the metal ion in site 1 (the N-terminal metal-binding site). When purified from E. coli, both GkR2loxI and MtR2lox contain a long-chain fatty acid bound in a hydrophobic tunnel extending from the active site toward the protein surface (Fig. 1A and Fig. S3) (11). The carboxyl head group coordinates both metal ions so that all six coordination sites of both metals are occupied.
Fig. 1.
Crystal structures of GkR2loxI. (A) The oxidized Mn/Fe-bound active site at 1.9 Å resolution. (B) The reduced Mn/Fe-bound active site at 1.9 Å resolution. (C) Anomalous difference density at the Fe (gray) and Mn (pink) edges from apoprotein crystals soaked with MnII and FeII in the absence (Upper) or presence (Lower) of oxygen for 3 h, contoured at four electrons per cubic angstrom. At the Fe edge, both Fe and Mn display an anomalous signal. (D) Superposition of the active sites in the metal-free state (at 2.3 Å resolution, colored as in A) and the oxidized Mn/Fe-bound state (transparent gray). Site 2 is preformed before metal binding, whereas site 1 is disordered, with E102 adopting two alternative conformations, and a stretch of four residues including E69 and V72 being invisible in the electron density. (E) mFo-DFc omit electron density for residues Y162 and V72 in apoprotein crystals soaked with MnII and FeII in the absence (Upper) or presence (Lower) of oxygen for 1 h, contoured at 3.0 σ.
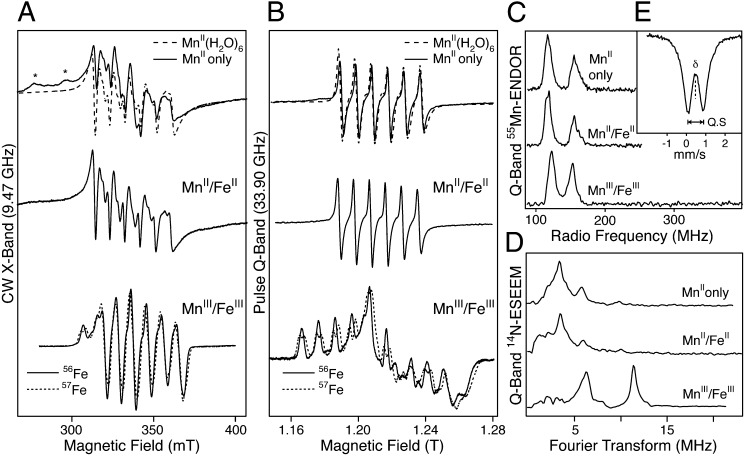
EPR spectroscopy confirms that both manganese and iron comprise the aerobically reconstituted metallocofactor. A structured EPR signal centered at g ≈2 is obtained both at X- and Q-band microwave frequencies (Fig. 2 A and B). The hyperfine splitting observed in continuous wave (CW) EPR and commensurate 55Mn-electron nuclear double resonance (ENDOR) spectra is a fingerprint for a Mn complex (≈10 mT/250 MHz) (Fig. 2 A–C and SI Results). The additional line splitting observed for the 57Fe-labeled cofactor demonstrates that the EPR signal arises from a complex containing both Mn and Fe (Fig. 2 A and B). 14N-electron spin echo envelope modulation (ESEEM) measurements at Q-band frequencies resolve at least one nitrogen interaction with the metal ions, indicating that the metals are ligated by protein-derived nitrogen ligands, presumably histidine residues, in agreement with the crystal structure (Fig. 2D). The oxidation state of the cofactor was determined by a combination of EPR and Mössbauer spectroscopy. The EPR signal, which represents a S = 1/2 species, comes about because the unpaired electrons of the two metal ions antiferromagnetically couple. This coupling has the effect of “pairing” all but one of the electrons on the Fe ion with a corresponding electron of opposite spin on the Mn ion, which leads to a ground electronic state with a single unpaired electron (i.e., S = 1/2). Zero-field Mössbauer measurements show that the Fe ion is in the +III oxidation state (δ = 0.47 mm/s) with local high spin (five unpaired electrons, d5 ion) (Fig. 2E) (25). Hence, the Mn ion must carry four unpaired electrons (d4 ion), and therefore its oxidation state must also be +III (SI Results and ref. 26). The small quadrupole splitting obtained in the Mössbauer measurements (ΔEQ = 0.83 mm/s) suggests that the two metal ions are bridged by a µ-hydroxo ligand (25), which is supported by spin Hamiltonian simulations of the EPR spectra (Fig. S4 and Table S2) (26). A large “transfer of hyperfine anisotropy” is observed, requiring the exchange interaction between the two metal ions to be small (J < |20| cm−1). Similar small exchange interactions have previously been observed in μ-hydroxo bridged FeIII/FeIII (25) and MnII/MnIII (26) model complexes. The inferred intrinsic hyperfine anisotropy for the 55Mn nucleus is consistent with a tetragonally elongated octahedral MnIII ion, as seen in the crystal structure. It is emphasized that only one type of MnIII/FeIII cofactor is observed, requiring selective metallation of the R2lox scaffold (for a full discussion see SI Results).
Fig. 2.
EPR spectra of GkR2loxI. Labels refer to protein loaded with only Mn (MnII only), 1:1 Mn:Fe under anaerobic conditions (MnII/FeII), and 1:1 Mn to natural abundance Fe (56Fe) or 57Fe under aerobic conditions (MnIII/FeIII). (A) CW X-band EPR spectra. The EPR spectrum of MnII(H2O)6 is overlaid with the Mn-only spectrum for comparison. Asterisks indicate interfering signals from the EPR cavity. (B) Echo-detected, pseudomodulated Q-band EPR spectra. (C) Q-band 55Mn-ENDOR spectra. (D) Q-band 14N-ESEEM spectra, showing the absolute value of the Fourier transform of the time domain measurements. (E) Zero-field Mössbauer spectrum of the MnIII/57FeIII cofactor. The dashed line denotes the fit to the data with an isomer shift of δ = 0.47 mm/s and quadrupole splitting ΔEQ = 0.83 mm/s.
The in Vitro Reconstituted Mn/Fe Cofactor Activates Oxygen.
When the metal cofactor is reconstituted in the absence of oxygen, the bridging hydroxo ligand is absent, and instead E202 bridges the metal ions (Fig. 1B and Fig. S1B). In contrast to the aerobically reconstituted cofactor, the EPR and 55Mn-ENDOR spectra of this state show only a MnII ion with a typical nuclear hyperfine coupling of ≈10 mT/250 MHz, which also has a nitrogen ligand, as shown using ESEEM (Fig. 2 A–D). The enhanced magnetic relaxation of the MnII signal suggests that the MnII weakly interacts with an unobserved paramagnetic center nearby, i.e., a high-spin FeII. This weak interaction excludes a direct link between the two metal ions such as a bridging oxo/hydroxo ligand, in accordance with the crystal structure. Oxidation of this sample by exposure to air results in the immediate appearance of the characteristic EPR spectrum of the MnIII/FeIII cofactor, further indicating that iron is bound within the active site of the protein in the reduced state, and also demonstrating that the in vitro reconstituted Mn/Fe cofactor is capable of activating oxygen.
The Two Metal-Binding Sites Exhibit Differential Metal Specificity.
To investigate the intrinsic metal binding properties of GkR2loxI, crystals of metal-free protein were soaked with a large excess of MnII and FeII in equal concentrations. The influence of oxygen on cofactor assembly was assessed by performing these experiments in air-saturated buffer or under reducing anoxic conditions (in the presence of 0.5% sodium dithionite). Crystallographic quantification of the site-specific metal content by anomalous dispersion reveals that the two sites have intrinsically different affinities for either metal. In the absence of oxygen, site 2 preferentially binds Fe (on the order of 4:1 Fe:Mn), as expected based on the Irving–Williams series. In contrast, site 1 binds equal amounts of Mn and Fe (Table 1 and Fig. 1C and Fig. S5). The low probability of simultaneous Mn binding in both metal sites is also observed using EPR. GkR2loxI loaded only with Mn displays a typical MnII signal in both EPR and 55Mn-ENDOR spectra (Fig. 2 A–C). The absence of any further structure, particularly in the Q-band EPR spectrum, requires that the MnII ion is isolated within the protein, i.e., that it occupies only one site. As ESEEM measurements resolve the same nitrogen interaction as seen for the reduced cofactor, it is expected that the MnII ion is bound to the same protein site (Fig. 2D). It is noted, however, that although the EPR/ESEEM/ENDOR data of both Mn/Fe and Mn-only loaded samples suggest MnII only binds to site 1, these data do not preclude FeII binding at site 1. Thus, these results are consistent with the selective binding of FeII in site 2 and the nonselective (1:1 Mn:Fe) loading of site 1 seen in the crystallographic data.
Table 1.
Relative amounts of Mn and Fe in the two metal sites obtained when reconstituting the cofactor in the absence or presence of oxygen
| Soaking duration | Site 1 |
Site 2 |
||
| Mn | Fe | Mn | Fe | |
| Anoxic conditions | ||||
| 1 h | 0.46 | 0.54 | 0.24 | 0.76 |
| 0.49 | 0.51 | 0.17 | 0.83 | |
| 3 h | 0.57 | 0.43 | 0.21 | 0.79 |
| 0.49 | 0.51 | 0.20 | 0.80 | |
| Aerobic conditions | ||||
| 1 h | 0.78 | 0.22 | 0.27 | 0.73 |
| 0.86 | 0.14 | 0.35 | 0.65 | |
| 3 h | 1.10 | −0.10 | 0.30 | 0.70 |
| 1.01 | −0.01 | 0.31 | 0.69 | |
| 6 h | 0.99 | 0.01 | 0.45 | 0.55 |
| 24 h | 0.87 | 0.13 | 0.26 | 0.74 |
Metal-free GkR2loxI crystals were soaked with an excess of MnII and FeII in equal concentrations under reducing anoxic (in the presence of 0.5% sodium dithionite) or aerobic conditions (in air-saturated buffer). The relative amounts are derived from the integrated intensity of the anomalous difference density peaks at the Mn and Fe absorption edges. For 1-h and 3-h soaking duration, data from two crystals each were analyzed. The values obtained indicate that the error margin is in the range of 10% (SI Materials and Methods).
Metal Specificity Seems to Be Dictated Predominantly by the First Ligand Sphere.
The primary protein-derived metal-binding ligands are identical in both metal sites. Nevertheless, computational modeling of the active site indicates that the structure of the first coordination sphere directly influences metal selection. Theoretical estimates for the relative energies were made by comparing the total calculated energy of the reduced active site with different combinations of bound MII/MII (M = Mn, Fe) using density functional theory (DFT). In this analysis, the calculated energy for the metallocofactor must be added to the corresponding pair of solvated MII ions for each different metal permutation to allow the set combinations to be compared (Table 2). These calculations reproduce the experimentally observed trends. For GkR2loxI, formation of the FeII/FeII and the MnII/FeII site is predicted to be isoenergetic within the error of the calculation, whereas formation of the FeII/MnII or the MnII/MnII site is less favorable. The same calculations were performed for the E. coli class Ia R2 protein, which uses a diiron cofactor. In contrast to R2lox, the N-terminal metal-coordinating carboxylate of class Ia R2 proteins is an aspartate, and the coordination geometry in the reduced active site of the E. coli class Ia R2 (PDB ID code 1XIK) (27) is different from that of R2lox (Fig. S6C). In the class Ia R2 active site, a homodinuclear FeII/FeII center is calculated to be favored over all other permutations, again in agreement with its experimentally observed metal preference. However, it is not possible to attribute different metal preferences to particular coordination geometries, as becomes evident when comparing the reduced active site architectures of R2 and R2-like proteins with different metal specificities (Fig. S6). In this context it is interesting to note that DFT calculations performed using the first ligand sphere can nonetheless reproduce the experimentally observed metal preferences.
Table 2.
Calculated relative energies for the binding of different combinations of MnII and FeII to the reduced structures of GkR2loxI and class Ia E. coli R2 (EcR2)
| Protein | Protein metal site | Solvated ions | Relative energy, kcal/mol |
| GkR2loxI | FeII/FeII | MnII, MnII | 0.0 |
| MnII/FeII | MnII, FeII | 1.3 | |
| FeII/MnII | MnII, FeII | 3.7 | |
| MnII/MnII | FeII, FeII | 6.9 | |
| Class Ia EcR2 | FeII/FeII | MnII, MnII | 0.0 |
| MnII/FeII | MnII, FeII | 4.3 | |
| FeII/MnII | MnII, FeII | 3.2 | |
| MnII/MnII | FeII, FeII | 7.2 |
The relative energy values are obtained by combining the DFT energy of the active site with a particular combination of the two metal ions with the DFT energy of the remaining two metal ions coordinated by six water molecules in a continuum water solvent. All energy values are relative to the system with the lowest energy (the FeII/FeII-bound protein), which is set to zero. Positive values represent a higher relative energy and thus a less favorable assembly.
The Heterodinuclear Cofactor Is Assembled More Efficiently in the Presence of Oxygen.
Under aerobic conditions, site 2 displays the same preference for Fe as under anoxic conditions within experimental error, whereas site 1 now binds virtually only Mn (Table 1 and Fig. 1C and Fig. S5). This enrichment of Mn in site 1 in the presence of oxygen must be caused by an additional mechanism other than the affinity differences observed under anoxic conditions. Assembly of the heterodinuclear cofactor therefore seems to be achieved by at least a two-step process involving the intrinsic metal specificity of the protein scaffold and additional effects exerted during oxygen binding or activation. It should be noted that while in crystals full Mn/Fe loading was achieved, in solution the Mn/Fe cofactor occupies 0.5 protein monomers under ideal loading conditions, as estimated by spin quantification of the EPR signal. The systematically lower concentration of the Mn/Fe cofactor seen in solution likely stems from the relatively low metal ion concentrations used during protein loading to avoid protein precipitation (SI Materials and Methods).
The Mn/Fe Cofactor Catalyzes Formation of an Ether Cross-Link in the Protein Scaffold.
Once activated, the Mn/Fe cofactor of R2lox goes on to perform a unique chemical reaction. In the aerobically reconstituted Mn/Fe-bound state, GkR2loxI displays a covalent ether cross-link close to the active site that was also observed in MtR2lox, formed between the phenolic oxygen of Y162 and the Cβ atom of V72 (Fig. 1A and Fig. S2B) (11). Because these two residues are conserved in the R2lox family (24), the cross-link is likely to be a common feature of R2lox proteins. The chemical nature of the cross-link was verified by mass spectrometry (Fig. S7). It is not present before metal binding: When the protein is produced in metal-free form, four residues including the first metal ligand E69 and the cross-link residue V72 are disordered and invisible in the electron density, while Y162 is ordered (Fig. 1D). Upon metal binding under anoxic conditions all active site residues become ordered, bringing V72 into close proximity to Y162 (Fig. 1B). However, no electron density connecting Y162 and V72 is observed (Fig. 1E), and the hydroxyl group of Y162 instead forms a hydrogen bond to the carbonyl oxygen of V72. In contrast, the cross-link is clearly present in apoprotein crystals soaked with Mn and Fe under aerobic conditions (Fig. 1E). Hence, the cross-link is generated by the metal cofactor upon oxygen activation. Formally, the amino acid side chains are oxidized by two electrons with the concomitant loss of two protons. This type of reaction has been explicitly documented for the diiron (6), but not the Mn/Fe cofactor.
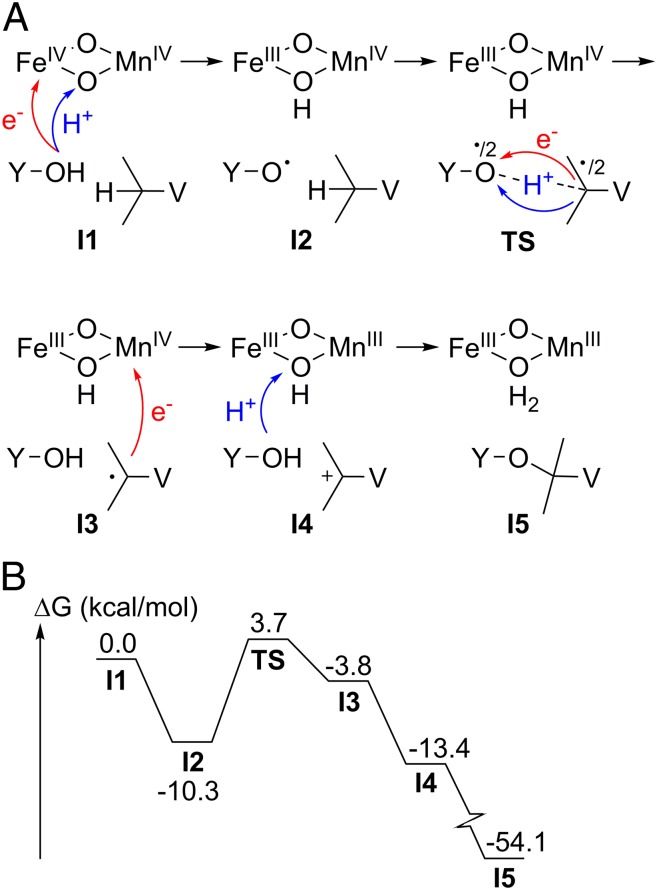
The mechanism of cross-link formation was investigated using DFT, assuming an initial bis-µ-oxo Mn/Fe cofactor that lacks the fatty acid ligand, poised in the MnIV/FeIV oxidation state (Fig. 3A). This state, formed by reductive cleavage of molecular oxygen, is observed in the closely related class Ic R2 heterodinuclear Mn/Fe center (28, 29). Oxygen activation by the Mn/Fe cofactor has previously been computationally described (30). The first step of cross-link synthesis is analogous to tyrosyl radical formation in class Ia diiron R2s (5): A Y162 radical is formed with the concomitant reduction of FeIV to FeIII. The phenolic proton is transferred to the proximal µ-oxo-bridge of the cofactor via a chain of two water molecules. Direct cross-link formation from the tyrosyl radical is not possible. The radical is transferred from Y162 to V72, forming a transient valyl radical. This is the rate-limiting step of the reaction, with a reasonably small barrier of 14 kcal/mol (Fig. 3B). A subsequent second electron transfer to the cofactor yields a tertiary valine carbocation with the concomitant reduction of MnIV to MnIII. The cross-link is then formed upon a nucleophilic attack of the Y162 phenolic oxygen on the V72 carbocation. Tyrosyl radical formation from the MnIV/FeIV state is exergonic, leading to a more stable state than the valyl radical. There are no structural properties indicating that the tyrosyl radical state can be avoided, and direct formation of the valyl radical is therefore unlikely.
Fig. 3.
Density functional theory study of the mechanism of tyrosine–valine ether cross-link formation. (A) Reaction scheme for the proposed mechanism. (B) Energy profile with key intermediates and rate-limiting transition state.
The hydrophobic tunnel extending from the active site toward the protein surface, and the fatty acid ligand observed in it (Fig. 1A and Fig. S3) (11), suggest that the natural substrates of R2lox proteins could be long-chain hydrocarbons. Our calculations indicate that from the valine carbocation intermediate of cross-link formation a desaturation of V72 could in principle occur, implying that the substrate reaction catalyzed by R2lox proteins might be the desaturation of a hydrocarbon (Fig. S8).
Discussion
The data presented here show that the heterodinuclear Mn/Fe cofactor of R2lox can assemble spontaneously in vitro, and that the protein scaffold itself is capable of site-specifically discriminating between manganese and iron. This observation suggests that a metallochaperone may not be required in vivo. It should be noted that metal partitioning into the two sites was more distinct in MtR2lox that was purified in metal-bound form from a heterologous expression system (11) than in GkR2loxI reconstituted in vitro. Although this observation also argues against a requirement for dedicated metal delivery systems, it does indicate that additional factors such as the intracellular availability of metal ions play a role in metal selection as well.
The active site model used for the metal binding energy calculations only contained the first-shell metal ligands in their reduced state conformation and the outer-sphere phenylalanine corresponding to the radical-bearing tyrosine in class Ia R2, but nevertheless reproduced the experimentally observed metal distribution in the reduced state. Thus, it seems that if outer-sphere effects play a role in determining metal specificity, a significant part of these effects is likely exerted indirectly by influencing the structure and/or dynamics of the first ligand sphere. Although the protein-derived ligands in the first sphere of the two metal-binding sites are identical, subtle differences are clearly used by the protein to tune metal specificity. The electronic differences between Mn and Fe, resulting in a minor Jahn–Teller effect for FeII but not MnII, could potentially be used by the protein to achieve differences in specificity. Certain structural differences are observed between the two sites, for example a water ligand in site 1, a more distorted coordination sphere in site 2, and differences in coordination mode by one of the bridging carboxylates (E202, Fig. 1B). However, from analysis of the structures presented in this study as well as structures of related diiron and dimanganese proteins, it is not possible to pinpoint certain active site residues or geometries as being responsible for the observed metal distribution (Fig. S6). It is likely a combination of several factors that modulate metal binding. Nevertheless, our data in combination with theoretical calculations show that structural differences in the local complex do influence metal selection. Although the specific differences leading to selectivity in the reduced state can presently not be defined, the fact that metal binding can be computationally reproduced is noteworthy, because it indicates the extent of the metal coordination sphere needed to yield these properties.
Interestingly, the Fe-specific site 2 is preformed in the apoprotein, whereas the initially nonselective site 1 is disordered (Fig. 1D). It seems that in the hetero-metallated R2lox scaffold site 2 must be filled with Fe before Mn binds in site 1, because only a very low percentage of Mn-bound protein was obtained when GkR2loxI was incubated with only Mn. Thus, cooperative effects together with an intrinsically low affinity in site 1 might dominate metal loading of this site and contribute to nonselectivity. In contrast, in the E. coli class Ia diiron R2, both metal sites are preformed in the apoprotein (31), and while site 2 has a higher affinity for Fe than site 1, Fe binding is not cooperative (32, 33).
Following initial metal (+II) binding, the heterodinuclear cofactor is enriched through a process involving oxygen. The experimentally observed equal filling of site 1 with MnII or FeII (Table 1) as well as the small relative energy differences between the Mn/Fe and Fe/Fe cofactor (Table 2) indicate that metal ions in site 1 can exchange as long as the two metal sites are reduced. DFT calculations suggest that oxygen activation, i.e., formation of the IV/IV state, the presumed active state in R2lox, is significantly faster with the heterodinuclear Mn/Fe center than with the Fe/Fe center (30). Thus, we hypothesize that oxygen activation may function as a selectivity filter by predominantly “trapping” the Mn/Fe cofactor in the oxidized state. We cannot exclude the possibility that the oxidation of FeII in aerobic solution and concomitant precipitation of FeIII contributes to the observed enrichment of Mn in site 1. However, metal ions bound in the active site do not exchange after oxygen activation (32), and it is expected that metal (+II) binding and activation are fast processes that take place before oxidation of FeII begins to have a significant effect on the MnII/FeII ratio in solution. This hypothesis would also predict that the kinetics of metal exchange in relation to the kinetics of oxygen activation would influence the resulting metal loading of the protein, in line with the observation that in reconstitution experiments performed in solution where no or only a very small excess of metal ions over protein was used it was not possible to achieve 100% loading of the heterodinuclear cofactor. Although our hypothesis remains to be verified, the total metal specificity of this system seems to be the result of a stepwise process including enrichment of a particular state. Our data therefore imply that complementary mechanisms other than the direct metal affinity, such as differences in chemical properties, can be used to achieve particular metallation states.
Diiron proteins have long been known to be capable of catalyzing two-electron oxidations from highly oxidized metal cofactor intermediates (6). Here we have shown that the Mn/Fe cofactor can also perform such reactions. While currently available data do not allow the conclusion that R2lox proteins indeed use a heterodinuclear Mn/Fe cofactor in vivo, this seems likely in light of the intrinsic metal specificity of the protein observed. Although the redox potential of the MnIV/FeIV state is likely too low to oxidize small hydrocarbons such as methane, the most prominent substrate of the diiron cofactor of BMMs, it is high enough to oxidize other substrates, as in the cross-link–forming reaction described here. Computational studies also suggest that it is capable of oxidizing larger exergonically bound substrates, and indicate that the MnIV/FeIV state is more stable than the FeIV/FeIV state (34). In line with these calculations, in the Chlamydia trachomatis class Ic R2 protein the MnIV/FeIV state has been experimentally observed, whereas it has not been possible to trap the FeIV/FeIV state (28, 35–39). The rationale for using the more complex heterodinuclear cofactor to perform functions that a diiron cofactor is also capable of might therefore be its greater stability. While BMMs have evolved into complex multicomponent proteins that can stabilize the FeIV/FeIV state and use it to perform two-electron oxidations (40–46), it seems that in R2lox proteins the simple one-subunit scaffold has been maintained and the cofactor exchanged to achieve a IV/IV state that is stable enough to perform catalysis. The potential for two-electron oxidation chemistry extends the known versatility of the Mn/Fe cofactor (9, 10) to rival that of its well-known diiron counterpart (2, 4, 6). Further development of such catalysts may be important for a large variety of academic and industrial applications.
This study shows that protein scaffolds can directly break the Irving–Williams series of metal complex stabilities to achieve site-specific metal discrimination, and reveals additional criteria that need to be considered when assessing metallation in biological systems. The fact that metal preferences can be computationally reproduced suggests that density functional theory may be used to predict protein metallation.
Materials and Methods
Detailed descriptions are given in SI Materials and Methods.
Protein Production, Purification, and Reconstitution of the Metallocofactor.
A construct encoding full-length GkR2loxI (accession number yp_148624) was expressed as an N-terminally His-tagged protein in metal-free form in E. coli and purified by heat denaturation of contaminating proteins and nickel chelate affinity chromatography. To reconstitute the Mn/Fe cofactor, apoprotein was incubated with a twofold molar excess each of (NH4)2Fe(SO4)2 and MnCl2 under aerobic or anaerobic conditions. Excess metal ions were removed by desalting.
Crystallography.
Apoprotein was crystallized by vapor diffusion at 22 °C in 12.5–32.5% (wt/vol) PEG 1500 and 100 mM Hepes- Na (pH 7.0–7.5). To reconstitute the Mn/Fe cofactor, crystals were soaked in 5 mM each (NH4)2Fe(SO4)2 and MnCl2 in the presence of 0.5% sodium dithionite or in air-saturated buffer. Data were collected at beamlines PX14.1/BESSY, ID23-2/ESRF, and X06SA/SLS. The structure of oxidized Mn/Fe-bound GkR2loxI was determined by single-wavelength anomalous dispersion phasing from a Fe peak wavelength dataset. The structures of GkR2loxI in the reduced Mn/Fe-bound and the metal-free state were solved using the oxidized state structure not containing any ligands as a starting model. Data and refinement statistics are given in Table S1.
EPR Spectroscopy.
EPR spectra were measured at 4.8 K using a Bruker ELEXSYS 500 (X-band) and E580 (Q-band) spectrometer. Experimental settings used were as reported in ref. 26 and in SI Results.
DFT Calculations.
DFT calculations were performed on active site cluster models with the B3LYP and B3LYP* functionals. The reported values include solvation, zero point, and dispersion corrections. The active site models include the metals, first-shell residues, and the second-shell residues relevant to the investigated mechanisms. The computational details are described in ref. 30.
Note Added in Proof.
A recent study by Dassama et al. (47) on assembly of the Mn/Fe cofactor in the Chlamydia trachomatis class Ic R2 protein found that site 1 specifically binds MnII under anaerobic conditions. This observation was made following a sequential metal loading scheme, thus making it difficult to directly compare to the present study. However, it indicates that there may be similarities as well as differences between cofactor assembly in the class Ic R2 and R2lox systems, perhaps relating to the divergent chemistry these two systems perform.
Supplementary Material
Acknowledgments
We thank Margareta Blomberg, Manfred Weiss, Torbjörn Astlind, and Karin Skaar for discussions and technical assistance; the PHENIX team, especially Jeff Headd, for advice on structure refinements; and the staff at beamlines PX14.1/BESSY, ID23-2/ESRF, and X06SA/SLS for assistance. Financial support was provided by the Swedish Research Council (M.H., A.G., P.E.M.S., and J.L.), the Swedish Foundation for Strategic Research and the Knut and Alice Wallenberg Foundation (M.H.), the Max Planck Gesellschaft (N.C. and W.L.), the Alexander von Humboldt Foundation (H.S.S.), and BioStruct-X for synchrotron travel expenses.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4HR0, 4HR4, and 4HR5).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304368110/-/DCSupplemental.
References
- 1.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460(7257):823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 2.Nordlund P, Eklund H. Di-iron-carboxylate proteins. Curr Opin Struct Biol. 1995;5(6):758–766. doi: 10.1016/0959-440x(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 3.Theil EC, Matzapetakis M, Liu X. Ferritins: Iron/oxygen biominerals in protein nanocages. J Biol Inorg Chem. 2006;11(7):803–810. doi: 10.1007/s00775-006-0125-6. [DOI] [PubMed] [Google Scholar]
- 4.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 5.Stubbe J. Di-iron-tyrosyl radical ribonucleotide reductases. Curr Opin Chem Biol. 2003;7(2):183–188. doi: 10.1016/s1367-5931(03)00025-5. [DOI] [PubMed] [Google Scholar]
- 6.Sazinsky MH, Lippard SJ. Correlating structure with function in bacterial multicomponent monooxygenases and related diiron proteins. Acc Chem Res. 2006;39(8):558–566. doi: 10.1021/ar030204v. [DOI] [PubMed] [Google Scholar]
- 7.Cotruvo JA, Jr, Stubbe J. An active dimanganese(III)-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Biochemistry. 2010;49(6):1297–1309. doi: 10.1021/bi902106n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox N, et al. A tyrosyl-dimanganese coupled spin system is the native metalloradical cofactor of the R2F subunit of the ribonucleotide reductase of Corynebacterium ammoniagenes. J Am Chem Soc. 2010;132(32):11197–11213. doi: 10.1021/ja1036995. [DOI] [PubMed] [Google Scholar]
- 9.Jiang W, et al. A manganese(IV)/iron(III) cofactor in Chlamydia trachomatis ribonucleotide reductase. Science. 2007;316(5828):1188–1191. doi: 10.1126/science.1141179. [DOI] [PubMed] [Google Scholar]
- 10.Voevodskaya N, Lendzian F, Ehrenberg A, Gräslund A. High catalytic activity achieved with a mixed manganese-iron site in protein R2 of Chlamydia ribonucleotide reductase. FEBS Lett. 2007;581(18):3351–3355. doi: 10.1016/j.febslet.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 11.Andersson CS, Högbom M. A Mycobacterium tuberculosis ligand-binding Mn/Fe protein reveals a new cofactor in a remodeled R2-protein scaffold. Proc Natl Acad Sci USA. 2009;106(14):5633–5638. doi: 10.1073/pnas.0812971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersson CS, et al. The manganese ion of the heterodinuclear Mn/Fe cofactor in Chlamydia trachomatis ribonucleotide reductase R2c is located at metal position 1. J Am Chem Soc. 2012;134(1):123–125. doi: 10.1021/ja209678x. [DOI] [PubMed] [Google Scholar]
- 13. Dassama LM, Boal AK, Krebs C, Rosenzweig AC, Bollinger JM (2012) Evidence that the beta subunit of Chlamydia trachomatis ribonucleotide reductase is active with the manganese ion of its manganese(IV)/iron(III) cofactor in site 1. J Am Chem Soc 134(5):2520–2523. [DOI] [PMC free article] [PubMed]
- 14.Tomter AB, et al. Ribonucleotide reductase class I with different radical generating clusters. Coord Chem Rev. 2013;257(1):3–26. [Google Scholar]
- 15.Tabares LC, Gätjens J, Un S. Understanding the influence of the protein environment on the Mn(II) centers in superoxide dismutases using high-field electron paramagnetic resonance. Biochim Biophys Acta. 2010;1804(2):308–317. doi: 10.1016/j.bbapap.2009.09.027. [DOI] [PubMed] [Google Scholar]
- 16.Cotruvo JA, Jr, Stubbe J. Metallation and mismetallation of iron and manganese proteins in vitro and in vivo: the class I ribonucleotide reductases as a case study. Metallomics. 2012;4(10):1020–1036. doi: 10.1039/c2mt20142a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irving H, Williams RJP. The stability of transition-metal complexes. J Chem Soc. 1953;(Oct):3192–3210. [Google Scholar]
- 18.Outten CE, O’Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292(5526):2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 19.Pufahl RA, et al. Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science. 1997;278(5339):853–856. doi: 10.1126/science.278.5339.853. [DOI] [PubMed] [Google Scholar]
- 20.Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O’Halloran TV. Undetectable intracellular free copper: The requirement of a copper chaperone for superoxide dismutase. Science. 1999;284(5415):805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 21.Maier RJ, Benoit SL, Seshadri S. Nickel-binding and accessory proteins facilitating Ni-enzyme maturation in Helicobacter pylori. Biometals. 2007;20(3-4):655–664. doi: 10.1007/s10534-006-9061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamoto S, Van Petegem F, Patrauchan MA, Eltis LD. AnhE, a metallochaperone involved in the maturation of a cobalt-dependent nitrile hydratase. J Biol Chem. 2010;285(33):25126–25133. doi: 10.1074/jbc.M110.109223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tottey S, et al. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature. 2008;455(7216):1138–1142. doi: 10.1038/nature07340. [DOI] [PubMed] [Google Scholar]
- 24.Högbom M. The manganese/iron-carboxylate proteins: What is what, where are they, and what can the sequences tell us? J Biol Inorg Chem. 2010;15(3):339–349. doi: 10.1007/s00775-009-0606-5. [DOI] [PubMed] [Google Scholar]
- 25.Kurtz DM. Oxo-bridged and hydroxo-bridged diiron complexes - a chemical perspective on a biological unit. Chem Rev. 1990;90(4):585–606. [Google Scholar]
- 26.Cox N, et al. Electronic structure of a weakly antiferromagnetically coupled Mn(II)Mn(III) model relevant to manganese proteins: A combined EPR, 55Mn-ENDOR, and DFT study. Inorg Chem. 2011;50(17):8238–8251. doi: 10.1021/ic200767e. [DOI] [PubMed] [Google Scholar]
- 27.Logan DT, et al. Crystal structure of reduced protein R2 of ribonucleotide reductase: The structural basis for oxygen activation at a dinuclear iron site. Structure. 1996;4(9):1053–1064. doi: 10.1016/s0969-2126(96)00112-8. [DOI] [PubMed] [Google Scholar]
- 28.Jiang W, Hoffart LM, Krebs C, Bollinger JM., Jr A manganese(IV)/iron(IV) intermediate in assembly of the manganese(IV)/iron(III) cofactor of Chlamydia trachomatis ribonucleotide reductase. Biochemistry. 2007;46(30):8709–8716. doi: 10.1021/bi700906g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han WG, Giammona DA, Bashford D, Noodleman L. Density functional theory analysis of structure, energetics, and spectroscopy for the Mn-Fe active site of Chlamydia trachomatis ribonucleotide reductase in four oxidation states. Inorg Chem. 2010;49(16):7266–7281. doi: 10.1021/ic902051t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roos K, Siegbahn PEM. Oxygen cleavage with manganese and iron in ribonucleotide reductase from Chlamydia trachomatis. J Biol Inorg Chem. 2011;16(4):553–565. doi: 10.1007/s00775-011-0755-1. [DOI] [PubMed] [Google Scholar]
- 31.Aberg A, Nordlund P, Eklund H. Unusual clustering of carboxyl side chains in the core of iron-free ribonucleotide reductase. Nature. 1993;361(6409):276–278. doi: 10.1038/361276a0. [DOI] [PubMed] [Google Scholar]
- 32.Bollinger JM, et al. Differential iron(II) affinity of the sites of the diiron cluster in protein R2 of Escherichia coli ribonucleotide reductase: Tracking the individual sites through the O-2 activation sequence. J Am Chem Soc. 1997;119(25):5976–5977. [Google Scholar]
- 33.Yang YS, Baldwin J, Ley BA, Bollinger JM, Solomon EI. Spectroscopic and electronic structure description of the reduced binuclear non-heme iron active site in ribonucleotide reductase from E. coli: Comparison to reduced Delta(9) desaturase and electronic structure contributions to differences in O-2 reactivity. J Am Chem Soc. 2000;122(35):8495–8510. [Google Scholar]
- 34.Roos K, Siegbahn PEM. A comparison of two-electron chemistry performed by the manganese and iron heterodimer and homodimers. J Biol Inorg Chem. 2012;17(3):363–373. doi: 10.1007/s00775-011-0858-8. [DOI] [PubMed] [Google Scholar]
- 35.Högbom M, et al. The radical site in chlamydial ribonucleotide reductase defines a new R2 subclass. Science. 2004;305(5681):245–248. doi: 10.1126/science.1098419. [DOI] [PubMed] [Google Scholar]
- 36.Voevodskaya N, Lendzian F, Gräslund A. A stable FeIII-FeIV replacement of tyrosyl radical in a class I ribonucleotide reductase. Biochem Biophys Res Commun. 2005;330(4):1213–1216. doi: 10.1016/j.bbrc.2005.03.104. [DOI] [PubMed] [Google Scholar]
- 37.Voevodskaya N, et al. Chlamydial ribonucleotide reductase: Tyrosyl radical function in catalysis replaced by the FeIII-FeIV cluster. Proc Natl Acad Sci USA. 2006;103(26):9850–9854. doi: 10.1073/pnas.0600603103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voevodskaya N, et al. Structure of the high-valent FeIIIFeIV state in ribonucleotide reductase (RNR) of Chlamydia trachomatis—combined EPR, 57Fe-, 1H-ENDOR and X-ray studies. Biochim Biophys Acta. 2007;1774(10):1254–1263. doi: 10.1016/j.bbapap.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Jiang W, et al. Branched activation- and catalysis-specific pathways for electron relay to the manganese/iron cofactor in ribonucleotide reductase from Chlamydia trachomatis. Biochemistry. 2008;47(33):8477–8484. doi: 10.1021/bi800881m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SK, Nesheim JC, Lipscomb JD. Transient intermediates of the methane monooxygenase catalytic cycle. J Biol Chem. 1993;268(29):21569–21577. [PubMed] [Google Scholar]
- 41.Lee SK, Fox BG, Froland WA, Lipscomb JD, Munck E. A transient intermediate of the methane monooxygenase catalytic cycle containing an Fe(Iv)Fe(Iv) cluster. J Am Chem Soc. 1993;115(14):6450–6451. [Google Scholar]
- 42.Liu KE, et al. Spectroscopic detection of intermediates in the reaction of dioxygen with the reduced methane monooxygenase hydroxylase from Methylococcus-capsulatus (Bath) J Am Chem Soc. 1994;116(16):7465–7466. [Google Scholar]
- 43.Liu KE, et al. Kinetic and spectroscopic characterization of intermediates and component interactions in reactions of methane monooxygenase from Methylococcus-capsulatus (Bath) J Am Chem Soc. 1995;117(41):10174–10185. [Google Scholar]
- 44.Valentine AM, Stahl SS, Lippard SJ. Mechanistic studies of the reaction of reduced methane monooxygenase hydroxylase with dioxygen and substrates. J Am Chem Soc. 1999;121(16):3876–3887. [Google Scholar]
- 45.Brazeau BJ, Lipscomb JD. Kinetics and activation thermodynamics of methane monooxygenase compound Q formation and reaction with substrates. Biochemistry. 2000;39(44):13503–13515. doi: 10.1021/bi001473l. [DOI] [PubMed] [Google Scholar]
- 46.Lundin D, Poole AM, Sjöberg BM, Högbom M. Use of structural phylogenetic networks for classification of the ferritin-like superfamily. J Biol Chem. 2012;287(24):20565–20575. doi: 10.1074/jbc.M112.367458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dassama LM, Krebs C, Bollinger JM, Jr., Rosenzweig AC, Boal AK. Structural basis for assembly of the Mn(IV)/Fe(III) cofactor in the class Ic ribonucleotide reductase from Chlamydia trachomatis. Biochemistry. 2013;52(37):6424–6436. doi: 10.1021/bi400819x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.