Significance
The identification and subsequent grafting of a unique peptide binding site within the Fab domain offers a unique means of adding functionality to monoclonal antibodies through a noncovalent interaction including improved pretargeted imaging, alternative payload delivery, and cross-linking of mAbs on cell surfaces to enhance their therapeutic potential.
Keywords: protein engineering, molecular recognition, cancer
Abstract
Capitalizing on their extraordinary specificity, monoclonal antibodies (mAbs) have become one of the most reengineered classes of biological molecules. A major goal in many of these engineering efforts is to add new functionality to the parental mAb, including the addition of cytotoxins and imaging agents for medical applications. Herein, we present a unique peptide-binding site within the central cavity of the fragment antigen binding framework region of the chimeric, anti-epidermal growth factor receptor mAb cetuximab. We demonstrate through diffraction methods, biophysical studies, and sequence analysis that this peptide, a meditope, has moderate affinity for the Fab, is specific to cetuximab (i.e., does not bind to human IgGs), and has no significant effect on antigen binding. We further demonstrate by diffraction studies and biophysical methods that the meditope binding site can be grafted onto the anti-human epidermal growth factor receptor 2 mAb trastuzumab, and that the antigen binding affinity of the grafted trastuzumab is indistinguishable from the parental mAb. Finally, we demonstrate a bivalent meditope variant binds specifically and stably to antigen-bearing cells only in the presence of the meditope-enabled mAbs. Collectively, this finding and the subsequent characterization and engineering efforts indicate that this unique interface could serve as a noncovalent “linker” for any meditope-enabled mAb with applications in multiple mAb-based technologies including diagnostics, imaging, and therapeutic delivery.
Monoclonal antibodies (mAbs) are indispensable tools in research laboratories and have become a central component in modern medicine. Thousands of antibodies are routinely used in research to detect and/or label specific proteins in a variety of settings. In recent years, dozens of mAbs that block signaling pathways, sequester growth factors, and/or induce an immune response have been successfully implemented in the clinic to treat cancer and other diseases, with hundreds still in active development (1, 2). Antibodies are being reengineered to best capitalize on their extraordinary ligand specificity to add new functionalities for a broad range of applications, mostly for clinical use, such as antibody-drug conjugates (ADCs), antibody-directed enzyme prodrug therapies (ADEPT), immune system engagement [e.g., Fc modifications (3), chemokine fusion, bispecific T-cell engagers (4)] and disease imaging (e.g., immunoPET and pretargeted radionuclide imaging; refs. 5 and 6).
Invariably, these engineering efforts have been achieved either through posttranslational chemical modifications or manipulation of the gene sequence (7–9). Many of the chemical modifications lead to undesirable consequences that are detrimental to therapeutic and imaging applications (10). For example, to create ADCs to target cytotoxins to disease sites with the mAb, the chemical conjugation of the toxin (typically involves lysines, reduced cysteines, or sugars on the mAb) produces a heterogeneous mixture, which can adversely affect the specificity and stability of the mAb and alter its biodistribution (11). Reducing the size of the mAb to Fab fragments [e.g., single-chain Fab variable domain (scFvs)] to facilitate tumor penetration and enhance imaging reduces the valency and, thus, affects both the affinity and the tissue specificity compared with the original mAb. In other cases, as part of a pretargeted imaging protocol to enhance detection, mAbs conjugated with streptavidin or multiple Fabs/scFvs stitched together through a scaffold (e.g., “lock-and-dock”; ref. 12) can be immunogenic (13), unstable in serum (14), and technically difficult and expensive to produce, especially at scales necessary for clinical use (15, 16). Although significant advances in antibody engineering have been achieved, practical challenges remain, many requiring alternative approaches.
Herein, we present a unique interaction that has the potential to overcome some of the difficulties in mAb engineering. We identified a binding site for a small peptide in the center of the Fab cavity of cetuximab via diffraction studies. Because of the position of the binding interface, we have named the peptide “meditope” (medius, middle; topos, place). We demonstrated that the meditope does not interfere with antigen binding. Based on structural and biophysical analysis, we identified key residues within the meditope binding site of the Fab and observed that the combination of these residues is absent in other clinically relevant mAbs, including a humanized mAb framework. To confirm the structural model and determine whether we could enable meditope binding in other mAbs, we grafted selected residues onto trastuzumab, a humanized anti-HER2 mAb, and demonstrated by diffraction and biophysical studies that the meditope binds to this meditope-enabled trastuzumab in a similar manner and affinity compared with cetuximab. Finally, we created a bivalent analog of the meditope to demonstrate that this interaction can be used to specifically target antigen-bearing cells in the presence of a meditope-enabled mAb that binds to the specific antigen. Overall, these results establish the basis for an alternative mAb platform that offers unique opportunities to improve mAb-based delivery systems for cancer and other diseases.
Results
Identification of the Meditope Binding Site on Cetuximab Fab Framework.
In our efforts to create tumor activated antibodies (17), we cocrystallized two peptides, CQFDLSTRRLKC (cQFD) and CQYNLSSRALKC (cQYN), individually with the Fab domain of cetuximab. These peptides were originally identified by phage display (18) and thought to mimic the tumor epitope EGFR domain III. The initial diffraction data indicated that the unit cell dimensions of each cocrystal were the same as that of the apo cetuximab Fab [Protein Data Bank (PDB) ID code 1YY8; ref. 19]. Because the complementarity determining region (CDR) loops in the published structure make extensive lattice contacts, the data suggested that the peptides were either absent or bound to a different region of the Fab. To rule out the latter case, we solved both structures by molecular replacement. After the first round of refinement, we observed a continuous stretch of unmodeled electron density that was consistent with the cyclic peptides not at the CDRs, but within the cavity formed by the Fab light and heavy chains (Fig. 1 A and B). We built the peptides into the electron density and the R and RFree dropped accordingly. Detailed description of crystallization and structural refinement procedures is found in SI Appendix, and diffraction and refinement statistics of the final models are presented in SI Appendix, Table S1.
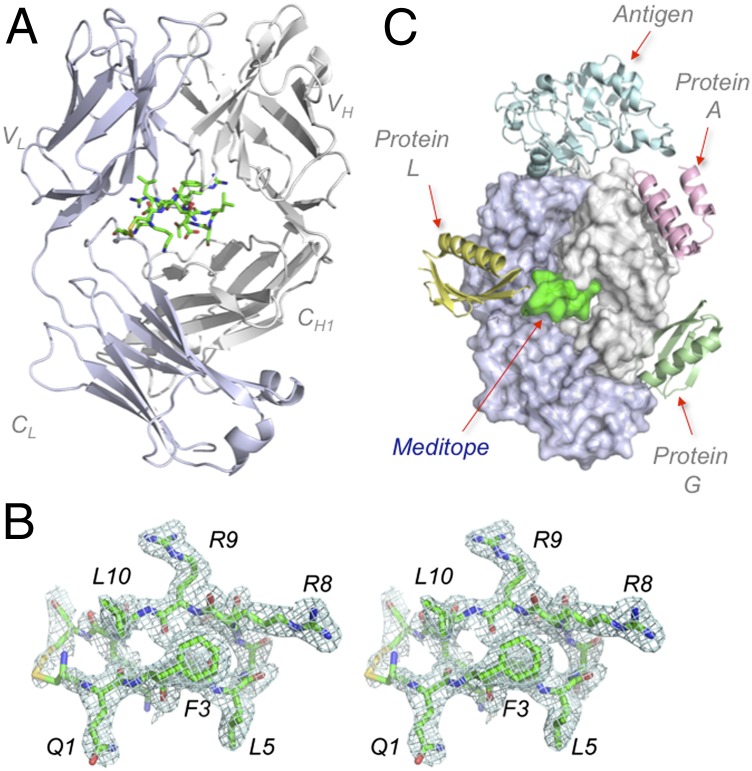
Fig. 1.
Meditope binds to cetuximab Fab framework. (A) Ribbon representation of cetuximab Fab (light chain, blue-white; heavy chain, white) with a stick representation of the cyclic CQFDLSTRRLKC meditope (green). (B) Electron density maps of cQFD meditope, in stereo and contoured at 1σ. (C) Superposition of the cetuximab-EGFR (1YY9) structure to Fab framework ligands: protein L (1HEZ), protein A (1DEE), protein G (1QKZ), and cQFD meditope (4GW1).
This study shows that a molecule can specifically dock to this cavity in the Fab framework, despite being apparent in any surface rendered Fab structures. Contrary to known Fab binding proteins such as proteins A (20), L (21), or G (22), which all bind to only a single domain of the heavy or light chain of the Fab (e.g., protein L binds only to the kappa variable light chain domain), the meditope–Fab interface is distributed among all four domains of the Fab and distinct from the solvent-exposed binding sites of protein A, G, or L (Fig. 1C).
Binding Studies Support Structural Model.
To ensure that this unanticipated binding site is not an artifact of crystallization, we used surface plasmon resonance (SPR) and determined the dissociation constant (Kd) between the cQFD meditope and cetuximab Fab to be 430 (± 30) nM and for the cQYN meditope and cetuximab Fab to be 3.5 (± 0.1) μM (Fig. 2A). Of note, these Kd values are similar to those reported for protein A or L binding to human Fab (23, 24). In addition, the buried surface area of the meditope-Fab interface, ∼850 Å, is similar to the that of protein A, G, and L. (23) (SI Appendix, Table S2).
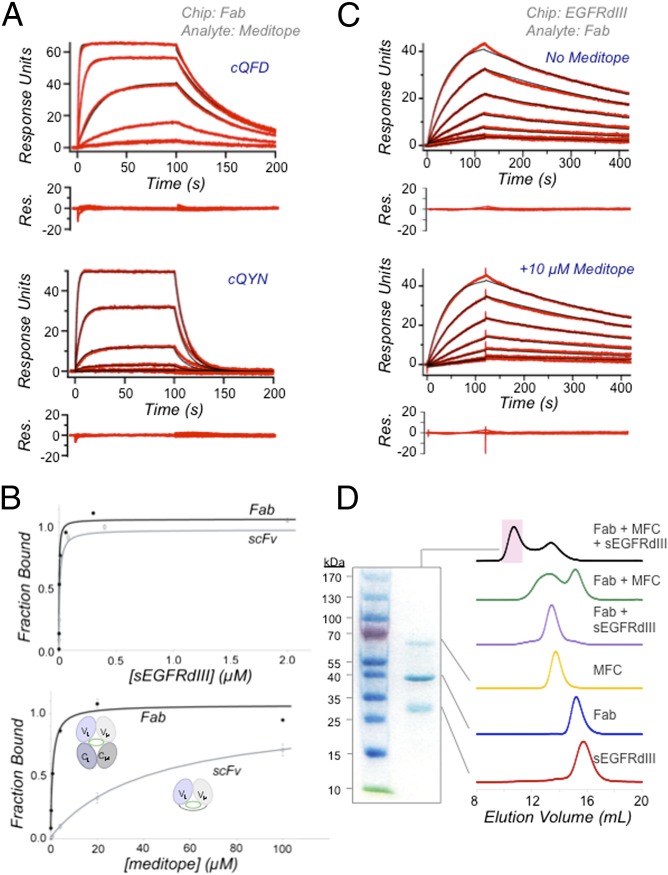
Fig. 2.
Biophysical characterization of meditope–Fab interaction. (A) SPR measurements of immobilized cetuximab Fab with 20, 4, 0.8, 0.16, and 0.032 μM cQFD meditope and 100, 20, 4, 0.8, and 0.16 μM cQYN meditope passing over the chip. (B) SPR saturation experiments using immobilized cetuximab Fab and scFv and passing EGFRdIII (Upper) or cQFD meditope (Lower). Also, Inset is a schematic representation of the scFv and the Fab. (C) SPR measurements of immobilized EGFRdIII with 10, 5, 2.5, 1.3, 0.63, and 0.31 nM cetuximab Fab passed over in the absence (Upper) or presence of 10 μM cQFD meditope (Lower). (D) SEC of cetuximab Fab, EGFRdIII, MFC, Fab–EGFRdIII and Fab–MFC complexes and an admixture of all three. Each trace has been normalized to one. Shown on Left is a nonreducing SDS/PAGE of the new peak (shaded pink on SEC chromatogram) formed from the admixture.
As an additional confirmation of the structural model, we identified key residues on the meditopes that enabled the interaction with the Fab based on the superposition of the cQFD-Fab and cQYN-Fab structures. We observed that the hydrophobic side chains of Phe/Tyr3, Leu5, and Leu10 of both meditopes were positioned nearly identically, suggesting that they make important contacts to the Fab (SI Appendix, Fig. S1). The importance of these residues is confirmed by SPR experiments showing alanine mutations at Phe3, Leu5, or Leu10 of the cQFD meditope resulted in a 5- to 140-fold decrease in affinity (SI Appendix, Table S3). We also observed that the guanidinium group of Arg8 of cQFD forms a hydrogen bond to the backbone carbonyl of Gln111 of the Fab heavy chain. Accordingly, Arg8Ala substitution decreased the affinity by 325-fold (SI Appendix, Table S3). The extended conformation of Arg8 is sterically blocked by the hydroxyl group of Tyr3 of the cQYN meditope, which causes a backbone rotation and eliminates the Arg8–Gln111 interaction (SI Appendix, Fig. S1). The loss of this interaction partially accounts for the nearly 10-fold difference in the binding affinity between the two meditopes (Fig. 2A and SI Appendix, Table S3) Thus, although the point mutations agree with the structural model, there are other subtle differences (e.g., electrostatics) between the two peptides and their interactions with the Fab that would require further study.
To provide further evidence that the meditopes do not bind to the CDRs of cetuximab, we measured the affinity of the meditope to a cetuximab single-chain Fab variable construct (scFv). Specifically, the scFv is generated by linking the variable domains of the light and heavy chains through a 19-residue linker (VL-SGSTSGSGKPGSGEGSTKG-VH), which keeps the CDRs intact (see schematic in Fig. 2B) but causes a partial loss of the meditope cavity because of the removal of the constant domains of the light and heavy chains. Thus, if the meditope binds to the CDR loop region, it should bind to the scFv and the Fab with similar binding affinity; however, if the meditope binds to the cavity, a significantly decreased affinity to the scFv should be observed. We first demonstrated by SPR that EGFR domain III (EGFRdIII), the antigen of cetuximab, readily saturated both immobilized Fab and scFv, indicating that the CDRs of the scFv are intact and functional. Next, we demonstrated that the meditope saturated the Fab as expected, but did not saturate the scFv at concentrations up to 100 μM because of the partial loss of the meditope binding in the scFv construct, indicating that the meditope does not bind to the CDRs, consistent with the crystal structure (Fig. 2B).
Meditope Occupancy Does Not Interfere with Antigen Binding.
Multiple studies indicate that the interaction between a Fab (or mAb) and protein A, G, or L does not affect antigen binding. However, because the meditope interacts with all four domains of the Fab (Fig. 1C), it is possible that meditope binding may affect the orientation and/or the conformational dynamics of the Fab and, thus, interfere with antigen binding (25).
To test whether meditope binding affects the affinity of cetuximab for its antigen, we measured the affinity and kinetics of cetuximab binding to EGFR in the presence and absence of meditope by SPR. We tethered EGFRdIII to an SPR chip and first determined, in the absence of meditope, the Kd of the Fab–EGFR interaction to be 0.76 nM, similar to the reported value (19). Next, we added a saturating concentration of 10 μM of the cQFD meditope (∼20-fold the meditope–Fab binding constant) to cetuximab Fab and all buffers (thus holding the meditope concentration constant throughout the experiment) and determined the Kd of the Fab and EGFRdIII to be 0.82 nM (Fig. 2C), approximately the same value as the Fab–EGFRdIII interaction in the absence of the meditope. Had meditope binding strongly affected antigen binding of cetuximab (either facilitating or interfering with antigen binding), we would expect a significant difference in the kinetics (26); instead, we observed the association and dissociation kinetics of cetuximab-EGFR are approximately the same both in cases: the association constants were 2.5 × 106 M−1⋅s−1 in the presence and 2.6 × 106 M−1⋅s−1 in the absence of the meditope; the dissociation constants were 2.1 × 10−3 s−1 in the presence and 2.0 × 10−3 s−1 in the absence of the meditope.
Furthermore, we demonstrate by size exclusion chromatography (SEC) that cetuximab Fab, EGFRdIII, and meditope form a complex. To facilitate monitoring, we increased the mass of the cQFD meditope by fusing to the N terminus of an Fc domain via a polyglycine-serine linker (denoted “MFC” hereafter). We added this construct to a stoichiometric complex of EGFRdIII and cetuximab Fab and observed a new peak, which eluted much earlier than the Fab–EGFRdIII complex, consistent with an increase in the hydrodynamic mass (Fig. 2D). SDS/PAGE of this new peak showed the presence of all three proteins.
Taken together, the SPR and SEC experiments indicate that the meditope does not significantly interfere with antigen recognition.
Meditope Binding Site Is Unique to Cetuximab.
To understand how the original phage display experiment (18) identified a unique peptide binding site distinct of the CDRs, we sought sequence differences between cetuximab and the decoy mAb CH14.18 (also a human-murine chimeric mAb) used in the experiment to filter out false positives. Such differences provide additional information concerning the specificity of the interaction from the point of view of the Fab. Because there is no experimental structure of CH14.18, we aligned the sequences of cetuximab and CH14.18 light and heavy chains (CH14.18 sequences 2 and 5 found in patent US 7169904). In addition, we aligned these sequences with: rituximab, another murine-human chimeric mAb; trastuzumab, a humanized anti-HER2 mAb; and M425, a murine anti-EGFR mAb (Fig. 3A). Although the CDR loops are expectedly different, there are several significant differences between cetuximab and the other mAbs at the meditope binding site, the most apparent being the cetuximab light chain residues Thr40, Asn41, and Asp85 (Fig. 3A). Asp85 (Val in CH14.18) makes hydrogen bonds to the guanidinium group of Arg9 and the backbone amide of Leu10 in the meditope (Fig. 3B); Thr40 (Gly in CH14.18) also makes a hydrogen bond to the guanidinium group of Arg9 (Fig. 3B). Likewise, the amide nitrogen of Asn41 (Pro in CH14.18) makes a hydrogen bond to the amide carbonyl of Thr7 in the meditope (Fig. 3B), and Thr40 averts the steric interference posed by the proline ring in CH14.18. Other sequence differences such as the loop defined by residues 39–44 in the heavy chain of cetuximab also are likely to influence meditope binding (SI Appendix, Fig. S2). These differences may account for the selection of the meditopes in the phage display experiment.
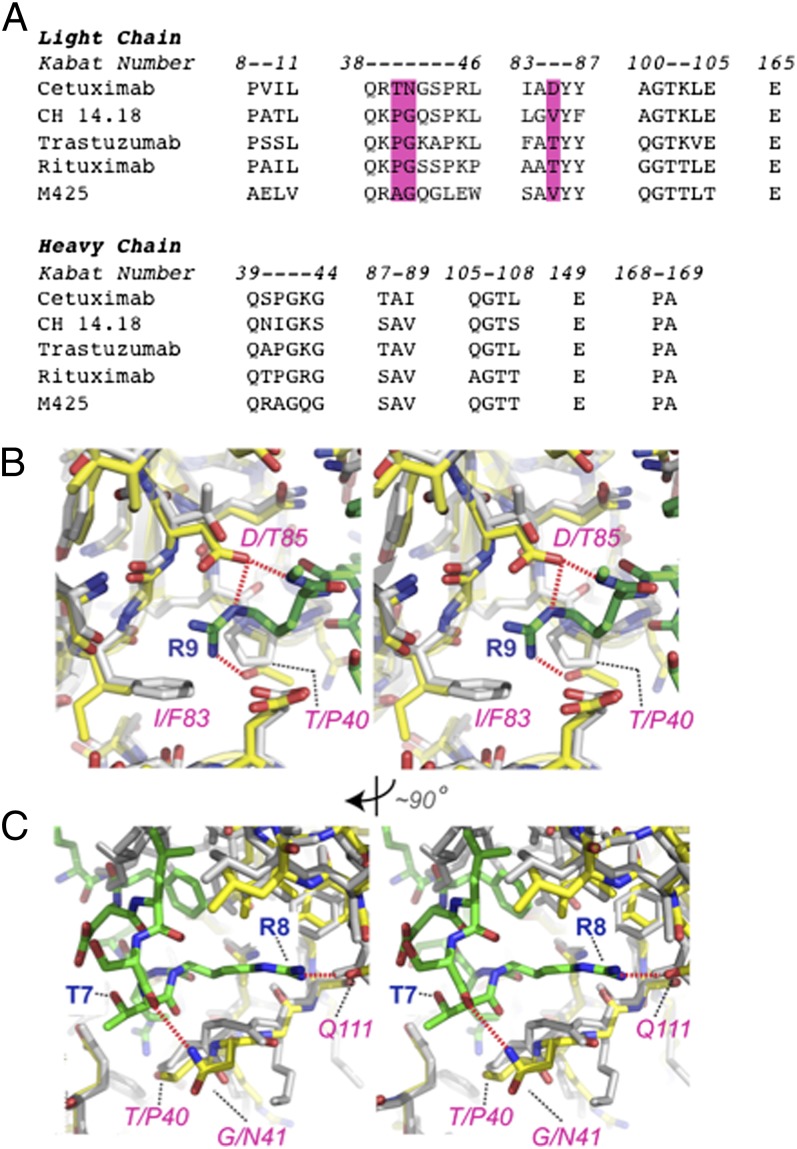
Fig. 3.
Critical framework residues that confer meditope binding. (A) Alignment of cetuximab, CH14.18, trastuzumab, rituximab, and M425 light and heavy chain residues within 5 Å of the meditope-binding site. Magenta bars denote the combination of residues unique to cetuximab in Kabat notation. (B) A stereoview highlighting Arg9 of the cQFD meditope (green) and its occupation of a distinct pocket in cetuximab (yellow). Trastuzumab Fab (1N8Z; gray carbons) is superimposed onto cetuximab. Red dotted lines indicate salt bridge from Asp85 to the meditope Arg9 and Leu10. (C) Rotated by ∼90°, the hydrogen bond between Asn41 of cetuximab Fab to Thr7 of the meditope is shown. Also shown is the extended side chain of Arg8 of the meditope making a backbone hydrogen bond to Gln111 in the heavy chain.
Based on these observations, we performed a BLAST search by using cetuximab in an attempt to find other mAbs containing this combination of residues. Although occasionally a hit from this query contained an individual substitution, the specific combination of Thr40, Asn41, and Asp85 was absent in the top 1,000 hits including human mAbs (alignment S1; Methods). In support of these observations, SPR measurements showed that a synthetic cQFD meditope coupled to the SPR chip bound cetuximab but not to rituximab or trastuzumab (SI Appendix, Fig. S3). We could not test CH14.18 because it is not commercially available. Collectively, these observations indicate that the meditope site is rare, if not all together absent, in human or murine mAbs.
Grafting of the Meditope Binding Site onto Trastuzumab.
To better understand the specificity of the interaction and to test whether this interaction could be expanded to other mAbs, we attempted to graft the meditope binding site of cetuximab onto a human mAb framework. To do so, we aligned the sequences of cetuximab and trastuzumab and mapped the sequence identity onto the atomic model of trastuzumab (PDB ID code 1N8Z; ref. 27). Based on this mapping and the superposition of cQFD-cetuximab and trastuzumab structures, we identified 13 residues that differ between cetuximab and trastuzumab and whose side chains either make direct contact with the meditope or may indirectly affect meditope binding (Fig. 4A). They are as follows: Thr40, Asn41, and Asp85 as detailed above, Arg39 and Arg45 in the light chain that coordinates a nonbonded phosphate group and may potentially stabilize the loop containing Thr40 and Asn41 in the light chain, Val9 and Ile10 that participates in forming a shallow hydrophobic surface near the side chain of Leu10 in the meditope, and Gly42, Ser43, Ile83, and Ala100 of the light chain as well as Ser40 and Ile89 in the heavy chain. We mutated the equivalent positions in the trastuzumab sequence and produced and purified the protein. Using SPR, we observed that the cQFD meditope bound to the grafted trastuzumab Fab with similar affinity as cetuximab (Kd = 1.2 μM) (Fig. 4B). We also observed that the meditope-enabled trastuzumab Fab bound to soluble HER2 with similar affinity as the Fab isolated from the commercial source, indicating that the mutations have no deleterious effect on antigen binding (SI Appendix, Fig. S4A). Finally, we demonstrated by SEC that the meditope-enabled trastuzumab, sHER2, and meditope–Fc coeluted (SI Appendix, Fig. S4B). Hereafter, we will refer to the meditope-enabled mAbs as memAbs.
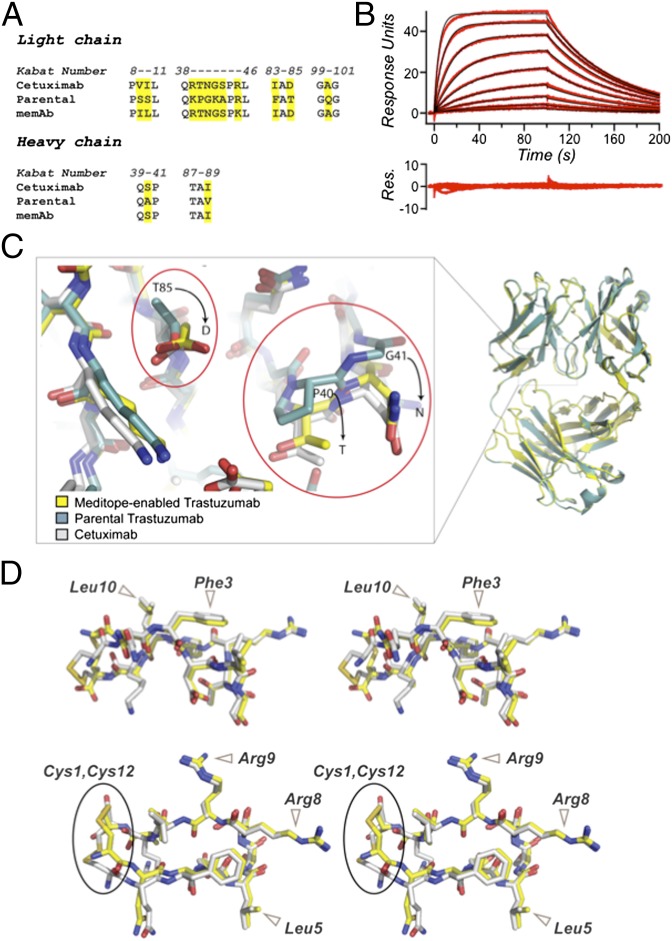
Fig. 4.
Meditope-enabling of trastuzumab. (A) Sequence alignment of light and heavy chains of parental and memAb trastuzumab showing residues, in Kabat notation, mutated to confer meditope binding. (B) SPR sensogram showing the binding of 16, 8, 4, 2, 1, 0.5, 0.25, and 0.063 µM cQFD meditope to immobilized memAb trastuzumab ligand. (C) Superposition of memAb (yellow carbons) and parental trastuzumab (cyan carbons) with expanded view of critical mutations in meditope-enable trastuzumab. Cetuximab (white carbons) is included for comparison. (D). Stereoviews of the superposition of the meditope bound to memAb trastuzumab (yellow carbons) and cetuximab (white carbons). Phe3 and Leu10 are highlighted in Upper; Leu5, Arg8, and Arg-9 in Lower. The orientation of the disulfide bond between the terminal cysteines, however, is slightly different in the memAb trastuzumab.
To further confirm that the point mutations did not significantly perturb the overall structure of the Fab, we crystallized the Fab of the trastuzumab memAb with and without the cQFD meditope. We obtained well-diffracting crystals in the presence of protein A and protein L to aid crystallization. The structures of the apo-memAb trastuzumab and the meditope-containing complex were solved at 1.95 and 2.0 Å, respectively (SI Appendix, Table S1). The parental trastuzumab Fab bound to protein A and protein L was also crystallized and solved to 2.08 Å resolution (SI Appendix, Table S1). Superposition of either the apo-parental trastuzumab Fab or HER2-bound parental trastuzumab Fab (27) onto apo-memAb trastuzumab revealed only minor changes in the overall structure (rmsd = 0.22 Å and 0.56 Å over 433 and 431 Cα atoms, respectively). Superposition of the apo- and meditope-liganded memAb trastuzumab showed that meditope occupancy stabilizes the heavy chain loop (residues 39–44) in an open conformation, similar to what is observed for cetuximab and accounts in part for the slightly higher rmsd. More importantly, critical residues (Thr40, Asn41, and Asp85) identified by the sequence mapping in the apo- and meditope-liganded memAb were essentially in the same positions as their counterparts in cetuximab and formed similar interactions with the meditope (Fig. 4C). Critical side chain rotamers of the meditope were also essentially the same as they were in the cQFD-cetuximab structure (Fig. 4D). As with cetuximab, we did not observe significant alterations in the CDR loops (Fig. 4C).
Taken together, the binding data and structural information demonstrate that it is possible to graft the meditope binding site onto a human Fab framework without significantly affecting antigen binding. The successful grafting of the meditope site further validates the initial cetuximab-meditope model obtained through diffraction data and the specificity of the interaction.
Engineering a Bivalent, High-Affinity Meditope.
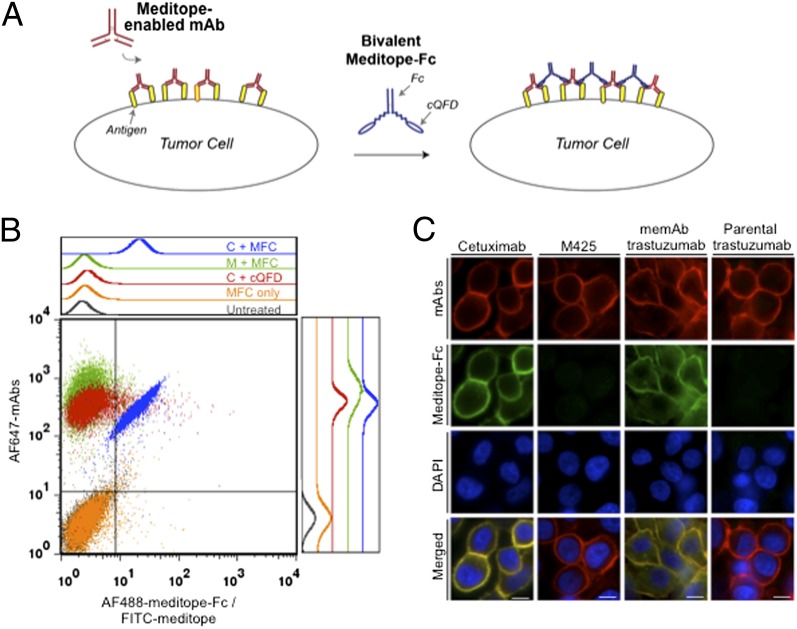
Although the meditope–memAb interaction is specific and that the meditope did not interfere with antigen binding, the moderate affinity of the meditopes preclude their potential use in antibody pretargeted imaging and/or drug delivery through a monovalent interaction. Fortunately, specificity and affinity are commonly gained through avidity/multivalency (28). Thus, we tested whether the aforementioned bivalent meditope–Fc construct would significantly enhance the apparent affinity of the meditope to cell-associated cetuximab or cell-associated trastuzumab memAb (Fig. 5A). We observed that the MFC bound to MDA-MB-468 cells (an EGFR-overexpressing breast cancer cell line) pretreated with cetuximab and remained bound despite an extensive washing procedure (essentially an ∼106-fold dilution of the bivalent meditope). The meditope–Fc construct did not bind to untreated cells or cells pretreated with M425, a murine anti-EGFR mAb that does not have the meditope binding site (Fig. 5B) (17, 29). We also demonstrated that an FITC-labeled, monomeric cQFD meditope bound to cetuximab pretreated MDA-MB-468 cells, but its signal, as measured by FACS, is reduced to background levels after a single wash (∼100-fold dilution and consistent with its binding affinity) (SI Appendix, Fig. S5).
Fig. 5.
Specific binding of meditope–Fc to memAb-labeled cells. (A) A schematic demonstrating the proposed enhanced affinity of the bivalent meditope to antigen overexpressing cells that are pretreated with a meditope-enabled mAb. (B) FACS analysis of AF488-labeled meditope–Fc (MFC) binding to untreated, AF647-labeled cetuximab-treated (C) or M425-treated (M) MDA-MB-468 cells. Monomeric meditope (cQFD) did not show appreciable binding. (C) Wide-field fluorescent microscopy showing meditope–Fc colocalization with cetuximab on MDA-MB-468 cells, but not to M425 pretreated cells (left two columns). Likewise, meditope–Fc colocalized with memAb trastuzumab, but not to parental trastuzumab pretreated SKBR3 cells. (Scale bars: 10 μm.)
The stable and specific interaction of the meditope–Fc with cetuximab is also clearly seen by their colocalization on the surface of MDA-MB-468 cells (Fig. 5C). These in vitro studies indicate that the avidity gained by creating a bivalent meditope enabled its selective binding to cells that express pathological concentrations of EGFR only in the presence of cetuximab.
Finally, we tested the memAb trastuzumab on HER2-overexpressing SKBR3 breast cancer cells. By either FACS or fluorescent microcopy, the grafted trastuzumab bound to the cells at similar levels as the parental trastuzumab. FACS analysis showed that the trastuzumab memAb and the parental trastuzumab we produced had the same range of affinity to cell surface HER2 as the commercial trastuzumab (SI Appendix, Fig. S6). As with cetuximab, we used microscopy to show that the meditope–Fc colocalized with memAb trastuzumab on SKBR3 cells (Fig. 5C). The same specific binding was recapitulated. Most importantly, the memAb trastuzumab and the parental control only differ at the grafted residues, thus proving that the specificity originated entirely from the meditope-enabling point mutations.
Discussion
Our structural investigations have led us to identify and characterize a unique peptide interface within the Fab framework of cetuximab. We demonstrated that the interaction does not perturb the ability of the mAb to bind to its cognate antigen, and that the site could be grafted onto a human IgG framework, exemplified by our meditope-enabled trastuzumab, with similar specificity and without compromise to antigen recognition. We also demonstrated the feasibility of enhancing the meditope binding affinity through avidity, and showed that a bivalent meditope is specific and binds with high affinity, but only in the presence of the cognate memAb, to cells overexpressing the antigen.
Beyond the description of a unique interaction in one of the most intensely studied protein families, we suggest that this interaction has practical utility and can potentially overcome hurdles facing mAb-based technologies. Specifically, many of the advantages sought in reengineered mAb and Fab fragments often give rise to new issues. A common problem is that these nonnative reagents expose new surfaces, which can be antigenic. Also, there are significant concerns with stability and the scalability in the manufacturing of such molecules (30). We note that cetuximab is already in the clinic and, thus, stable and produced at scale. We also note that the unique combination of residues that line the meditope binding site is distant in sequence (i.e., unlikely to be a peptide T-cell epitope) and located within a deep cavity (i.e., unlikely to be a surface exposed B-cell epitope) (Figs. 1C and 3B). Consequently, mAbs containing the meditope binding site should not be immunogenic.
As demonstrated by the bivalent meditope–Fc’s stable interaction with memAb-treated cells, we envision that the meditope–memAb interaction will have applications in pretargeted imaging and drug delivery methods (5, 6). We also envision that the meditope–memAb interaction can be used to derive synergistic tumor inhibition (17, 31, 32) as observed in the trastuzumab-pertuzumab combination (33). The meditope–Fc could be viewed as a second mAb, but with the flexibility of being able to pair up with any memAb, hence potentially reducing the cost of identifying and developing a second therapeutic mAb. It is also possible to fine tune the properties of a multivalent meditope (e.g., through valency and linker geometry) to further enhance specificity and synergistic effects.
By using the meditope binding site as a specific point of conjugation, we propose that issues of heterogeneity and conjugation efficiency that typically affect the development of effective ADCs can be avoided. A toxin-bearing meditope can direct a reactive chemical to the Fab cavity, favoring specificity and unit stoichiometry.
Although the moderate affinity of the cQFD and cQYN meditopes may be advantageous when considering a multivalent approach (28), we suggest that it is possible to incorporate different unnatural amino acids, different cyclization strategies, and use different screening methods to not only improve the affinity but also address potential pharmacokinetics, pharmacodynamics, and toxicity issues. In addition, as we have demonstrated by the meditope enabling of trastuzumab, it is equally possible to tailor the meditope binding site to accommodate changes in the meditope.
In summary, this unanticipated interaction between the meditope and a nonantigen binding surface of the Fab of cetuximab, together with the demonstrated feasibility in grafting this peptide-binding surface on to other mAbs, have the potential to offer opportunities to improve mAb-based technologies in general.
Methods
Materials.
Meditopes were synthesized by the Drug Discovery and Structural Biology Core at the City of Hope and by CS Bio except for the cQFD meditope and its alanine point mutants used in the SPR experiment presented in SI Appendix, Table S3, which were produced bacterially by encoding the peptides at the C terminus of SMT (34) with the corresponding codon changes generated by site-directed mutagenesis. The peptides were oxidized by dialysis into buffer without DTT, purified by SEC to obtain monomers and released from the SMT tag by ubiquitin-like protease (Ulp1) before analysis. Cetuximab, rituximab, and trastuzumab were purchased from the City of Hope pharmacy. The genes for the light and heavy chains of memAb and parental trastuzumab were synthesized by DNA2.0, cotransfected, and selected in NS0 cells. Each Fab was generated through the digestion of the IgG with immobilized papain (Pierce) and purified by reverse purification with protein A (GE Healthcare) and SEC on a HiLoad 16/600 pg Superdex 75 column (GE Healthcare). scFv of cetuximab was expressed in Sf9 cells and purified as described (35). The gene for the meditope–Fc was constructed by DNA2.0 (cQFD meditope connected to human IgG gamma heavy chain 3; accession no. AAW65947) residues 22–241 via a 37-residue linker, cloned into pAcGP67A vector (BD Biosciences), and produced in Sf9 cells. The protein was purified by using protein A and SEC. His-EGFRdIII (residues 309–512) and His-sHER2 (residues 1–630) were expressed in Sf9 cells and purified by using standard methods. The sequences of Fab binding domains of protein A (N-terminal Ser-Tyr, 1804–1855) and protein L (N-terminal Ser, residues 820–880, C-terminal Gly) (20, 21) were obtained from the crystal structure Fasta files, synthesized by Genscript, and purified in a similar manner as the cQFD mutants.
Crystallization and Diffraction.
Please see SI Appendix, SI Methods or the PDB entries for crystallization, structure determination, and refinement details. The structures have been deposited in the PDB as: 4GW1 (cetuximab with cQFD), 4GW5 (cetuximab with cQYN), 4HKZ (parental trastuzumab with protein A and L fragments), 4HJG (memAb trastuzumab with protein A and L fragments), and 4IOI (memAb trastuzumab with cQFD and protein A and L fragments).
Analytical SEC.
Ten micromolar cetuximab Fab, EGFRdIII, and 5 μM meditope–Fc were used for the individual runs and for the binary complexes. The complex of the three components was formed with stoichiometric complex of the Fab and EGFR added to the meditope–Fc. The proteins were mixed and incubated at room temperature for 20 min and applied to a Superdex 200 10/300 column (GE Healthcare) at 4 °C. Similar procedures were used with the parental or meditope-enabled trastuzumab IgGs, sHER2 and meditope–Fc.
BLAST Search.
Using the nonredundant database and filtering for Homo sapiens, we used the cetuximab and trastuzumab sequences as search sequences and aligned the top 1,000 sequences returned from each. A multisequence alignment of light chain residues 26–89, starting with cetuximab (1YY8_A), is presented as Alignment S1 with Thr40, Asn41, and Asp85 highlighted in magenta.
SPR Binding Experiments.
All SPR experiments were performed on a GE Biacore T100 instrument at 20 °C, and analysis was performed by using Biacore T100 Evaluation software version 2.0.1. Ligands were amine coupled to CM5 chips at low densities suitable for kinetics. Concentrations of analytes were prepared in HBS-EP+ buffer (10 mM Hepes at pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.05% vol/vol surfactant P20), which was used as a running buffer in all experiments. For saturation analysis, cQFD meditope or EGFRdIII affinities to immobilized cetuximab scFv or Fab were assessed by equilibrium methods at 20 °C and fit to the equation: RU = (RMAX • [L])/([L] • Kd) + ROFFSET.
Supplementary Material
Acknowledgments
We thank Dr. Stephen Gillies for providing us the CH14.18 sequences. J.M.D. acknowledges the Foerderer Scholar Fellowship Award, the Dubbs Scholar Fellowship Award, the Measey MD/PhD Student Fellowship, and Ruth L. Kirschstein National Research Service Award T32 DK07705; and support from the Alicia and John Kruger Gift, the Nesvig Foundation, and National Cancer Institute (NCI) Grant R21 CA135216. C.Z. acknowledges support from the Gastrointestinal Cancers Program Pilot Grant at City of Hope. The project described was also supported by NCI Grant P30 CA033572.
Footnotes
Conflict of interest statement: D.A.H. and J.C.W. have founded Meditope Biosciences, a company based on some of the observations presented here.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4GW1, 4GW5, 4HKZ, 4HJG, and 4IOI).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307309110/-/DCSupplemental.
References
- 1.Weiner LM, Murray JC, Shuptrine CW. Antibody-based immunotherapy of cancer. Cell. 2012;148(6):1081–1084. doi: 10.1016/j.cell.2012.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat Rev Cancer. 2012;12(4):278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 3.Desjarlais JR, Lazar GA. Modulation of antibody effector function. Exp Cell Res. 2011;317(9):1278–1285. doi: 10.1016/j.yexcr.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 4.Wolf E, Hofmeister R, Kufer P, Schlereth B, Baeuerle PA. BiTEs: Bispecific antibody constructs with unique anti-tumor activity. Drug Discov Today. 2005;10(18):1237–1244. doi: 10.1016/S1359-6446(05)03554-3. [DOI] [PubMed] [Google Scholar]
- 5.Goldenberg DM, et al. Pretargeted molecular imaging and radioimmunotherapy. Theranostics. 2012;2(5):523–540. doi: 10.7150/thno.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pagel JM, et al. Comparison of a tetravalent single-chain antibody-streptavidin fusion protein and an antibody-streptavidin chemical conjugate for pretargeted anti-CD20 radioimmunotherapy of B-cell lymphomas. Blood. 2006;108(1):328–336. doi: 10.1182/blood-2005-11-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yazaki PJ, et al. Biodistribution and tumor imaging of an anti-CEA single-chain antibody-albumin fusion protein. Nucl Med Biol. 2008;35(2):151–158. doi: 10.1016/j.nucmedbio.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orcutt KD, et al. Engineering an antibody with picomolar affinity to DOTA chelates of multiple radionuclides for pretargeted radioimmunotherapy and imaging. Nucl Med Biol. 2011;38(2):223–233. doi: 10.1016/j.nucmedbio.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Junutula JR, et al. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat Biotechnol. 2008;26(8):925–932. doi: 10.1038/nbt.1480. [DOI] [PubMed] [Google Scholar]
- 10.Nelson AL, Dhimolea E, Reichert JM. Development trends for human monoclonal antibody therapeutics. Nat Rev Drug Discov. 2010;9(10):767–774. doi: 10.1038/nrd3229. [DOI] [PubMed] [Google Scholar]
- 11.Wakankar AA, et al. Physicochemical stability of the antibody-drug conjugate Trastuzumab-DM1: Changes due to modification and conjugation processes. Bioconjug Chem. 2010;21(9):1588–1595. doi: 10.1021/bc900434c. [DOI] [PubMed] [Google Scholar]
- 12.Sharkey RM, et al. Signal amplification in molecular imaging by pretargeting a multivalent, bispecific antibody. Nat Med. 2005;11(11):1250–1255. doi: 10.1038/nm1322. [DOI] [PubMed] [Google Scholar]
- 13.Onda M, et al. Recombinant immunotoxin against B-cell malignancies with no immunogenicity in mice by removal of B-cell epitopes. Proc Natl Acad Sci USA. 2011;108(14):5742–5747. doi: 10.1073/pnas.1102746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuesta AM, et al. Improved stability of multivalent antibodies containing the human collagen XV trimerization domain. MAbs. 2012;4(2):226–232. doi: 10.4161/mabs.4.2.19140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ducry L. Challenges in the development and manufacturing of antibody-drug conjugates. Methods Mol Biol. 2012;899:489–497. doi: 10.1007/978-1-61779-921-1_29. [DOI] [PubMed] [Google Scholar]
- 16.Ducry L, Stump B. Antibody-drug conjugates: Linking cytotoxic payloads to monoclonal antibodies. Bioconjug Chem. 2010;21(1):5–13. doi: 10.1021/bc9002019. [DOI] [PubMed] [Google Scholar]
- 17.Kamat V, et al. Enhanced EGFR inhibition and distinct epitope recognition by EGFR antagonistic mAbs C225 and 425. Cancer Biol Ther. 2008;7(5):726–733. doi: 10.4161/cbt.7.5.6097. [DOI] [PubMed] [Google Scholar]
- 18.Riemer AB, et al. Vaccination with cetuximab mimotopes and biological properties of induced anti-epidermal growth factor receptor antibodies. J Natl Cancer Inst. 2005;97(22):1663–1670. doi: 10.1093/jnci/dji373. [DOI] [PubMed] [Google Scholar]
- 19.Li S, et al. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell. 2005;7(4):301–311. doi: 10.1016/j.ccr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Graille M, et al. Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: Structural basis for recognition of B-cell receptors and superantigen activity. Proc Natl Acad Sci USA. 2000;97(10):5399–5404. doi: 10.1073/pnas.97.10.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graille M, et al. Complex between Peptostreptococcus magnus protein L and a human antibody reveals structural convergence in the interaction modes of Fab binding proteins. Structure. 2001;9(8):679–687. doi: 10.1016/s0969-2126(01)00630-x. [DOI] [PubMed] [Google Scholar]
- 22.Derrick JP, Feavers I, Maiden MC. Use of streptococcal protein G in obtaining crystals of an antibody Fab fragment in complex with a meningococcal antigen. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 1):314–316. doi: 10.1107/S0907444998010269. [DOI] [PubMed] [Google Scholar]
- 23.Graille M, et al. Evidence for plasticity and structural mimicry at the immunoglobulin light chain-protein L interface. J Biol Chem. 2002;277(49):47500–47506. doi: 10.1074/jbc.M206105200. [DOI] [PubMed] [Google Scholar]
- 24.Young WW, Jr, Tamura Y, Wolock DM, Fox JW. Staphylococcal protein A binding to the Fab fragments of mouse monoclonal antibodies. J Immunol. 1984;133(6):3163–3166. [PubMed] [Google Scholar]
- 25.Torres M, Fernandez-Fuentes N, Fiser A, Casadevall A. Exchanging murine and human immunoglobulin constant chains affects the kinetics and thermodynamics of antigen binding and chimeric antibody autoreactivity. PLoS ONE. 2007;2(12):e1310. doi: 10.1371/journal.pone.0001310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rouhana J, et al. Kinetics of interaction between ADP-ribosylation factor-1 (Arf1) and the Sec7 domain of Arno guanine nucleotide exchange factor, modulation by allosteric factors, and the uncompetitive inhibitor brefeldin A. J Biol Chem. 2013;288(7):4659–4672. doi: 10.1074/jbc.M112.391748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho HS, et al. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421(6924):756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 28.Mammen M, Choi S, Whitesides G. Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitor. Angew Chem Int Ed. 1998;37:2755. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 29.Rodeck U, et al. Tumor growth modulation by a monoclonal antibody to the epidermal growth factor receptor: Immunologically mediated and effector cell-independent effects. Cancer Res. 1987;47(14):3692–3696. [PubMed] [Google Scholar]
- 30.Nelson AL. Antibody fragments: Hope and hype. MAbs. 2010;2(1):77–83. doi: 10.4161/mabs.2.1.10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koefoed K, et al. Rational identification of an optimal antibody mixture for targeting the epidermal growth factor receptor. MAbs. 2011;3(6):584–595. doi: 10.4161/mabs.3.6.17955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spangler JB, et al. Combination antibody treatment down-regulates epidermal growth factor receptor by inhibiting endosomal recycling. Proc Natl Acad Sci USA. 2010;107(30):13252–13257. doi: 10.1073/pnas.0913476107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baselga J, et al. CLEOPATRA Study Group Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med. 2012;366(2):109–119. doi: 10.1056/NEJMoa1113216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5(5):865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 35.Donaldson JM, Kari C, Fragoso RC, Rodeck U, Williams JC. Design and development of masked therapeutic antibodies to limit off-target effects: Application to anti-EGFR antibodies. Cancer Biol Ther. 2009;8(22):2147–2152. doi: 10.4161/cbt.8.22.9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.