Significance
Plants rely on many different types of photoreceptors to control light responses, but the molecular mechanisms underlying the coaction of different photoreceptors that regulate the same light response remains unclear. This study demonstrates a mechanism of the photoreceptor coaction. In Arabidopsis, the blue-light receptor cryptochrome 2 interacts with and activates the transcription factor CRYPTOCHROME-INTERACTING basic helix–loop–helix 1 (CIB1). It was found that CIB1 protein is degraded in the absence of blue light and that the light–oxygen–voltage-domain F-box blue-light receptor ZEITLUPE mediates blue-light suppression of CIB1 degradation.
Keywords: protein degradation, photomorphogenesis, gene expression
Abstract
Plants possess multiple photoreceptors to mediate light regulation of growth and development, but it is not well understood how different photoreceptors coordinate their actions to jointly regulate developmental responses, such as flowering time. In Arabidopsis, the photoexcited cryptochrome 2 interacts with the transcription factor CRYPTOCHROME-INTERACTING basic helix–loop–helix 1 (CIB1) to activate transcription and floral initiation. We show that the CIB1 protein expression is regulated by blue light; CIB1 is highly expressed in plants exposed to blue light, but levels of the CIB1 protein decreases in the absence of blue light. We demonstrate that CIB1 is degraded by the 26S proteasome and that blue light suppresses CIB1 degradation. Surprisingly, although cryptochrome 2 physically interacts with CIB1 in response to blue light, it is not the photoreceptor mediating blue-light suppression of CIB1 degradation. Instead, two of the three light–oxygen–voltage (LOV)-domain photoreceptors, ZEITLUPE and LOV KELCH PROTEIN 2, but not FLAVIN-BINDING KELCH REPEAT 1, are required for the function and blue-light suppression of degradation of CIB1. These results support the hypothesis that the evolutionarily unrelated blue-light receptors, cryptochrome and LOV-domain F-box proteins, mediate blue-light regulation of the same transcription factor by distinct mechanisms.
Cryptochromes are the photolyase-related blue-light receptors that regulate photoresponses and/or the circadian clock in all major evolutionary lineages (1–4). The Arabidopsis genome encodes two cryptochromes, cryptochrome 1 (CRY1) and CRY2, which mediate blue-light suppression of hypocotyl elongation and photoperiodic control of flowering time (5, 6). Arabidopsis CRY2 is a nuclear protein that regulates flowering time by at least two different mechanisms (4). The photoexcited CRY2 physically interacts with the basic helix–loop–helix (bHLH) transcription factor CRYPTOCHROME-INTERACTING bHLH 1 (CIB1) to activate transcription of the flowering integrator gene FLOWERING LOCUS T (FT) (7–9). CRY2 also physically interacts with SUPPRESSOR OF PHYTOCHROME A 1 (SPA1) in response to blue light to suppress the E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), leading to the accumulation of the CONSTANS (CO) protein and activation of the transcription of the FT gene (10–12).
In addition to CRY2, other photoreceptors—such as phytochrome A (phyA) and phytochrome B and the light–oxygen–voltage (LOV)-domain F-box proteins FLAVIN-BINDING KELCH REPEAT 1 (FKF1), ZEITLUPE (ZTL), and LOV KELCH PROTEIN 2 (LKP2)—also regulate expression of the CO and FT genes to affect flowering time in response to photoperiod. For example, phytochromes interact with PHYTOCHROME-INTERACTING FACTOR 3 and 4, which has been reported to regulate FT mRNA expression by clock-dependent or -independent mechanisms (13, 14). FKF1 mediates blue-light–dependent degradation of CDF1 and stabilization of the CO protein to facilitate transcription of FT (15, 16). ZTL acts as the substrate-binding subunit of the SCFZTL E3 ubiquitin ligase that regulates protein abundance of the key circadian oscillator components, TIMING of CAB EXPRESSION 1 (TOC1) and PSEUDO-RESPONSE REGULATOR 5 (PRR5), to affect expression of a number of flowering-time genes, including FT (16–18).
Different photoreceptors are known to “coact” by direct or indirect molecular interactions to regulate expression of genes that govern the same developmental process, which presumably confer evolutionary advantages by coordinating different photoreceptor signaling “pathways” for the appropriate control of light responses. For example, both cryptochromes and phytochromes regulate activity of the COP1/SPA1 E3 ligase complex to affect seedling deetiolation responses (4, 19–22). Cryptochrome has also been reported to physically interact with the LOV-domain photoreceptor ZTL (also known as ADO1) to regulate the circadian clock (23). However, exactly how different photoreceptors coordinate a light response remains unclear. We report here a mechanism underlying the photoreceptor coaction. We show that the CRY2-intercting CIB1 protein accumulates only in the presence of blue light, whereas it is degraded by the 26S proteasome in the absence of blue light. Surprisingly, although CRY2 is the photoreceptor that physically interacts with CIB1 to regulate its activity in response to blue light, CRY2 is not the photoreceptor mediating blue-light regulation of CIB1 protein stability. Instead, it is the LOV-domain photoreceptor ZTL and its close homolog LKP2 that mediate blue-light suppression of CIB1 degradation. These results support a hypothesis that cryptochrome and LOV-domain photoreceptors mediate blue-light regulation of CIB1 via distinct mechanisms.
Results
Blue Light Regulates CIB1 Protein Expression.
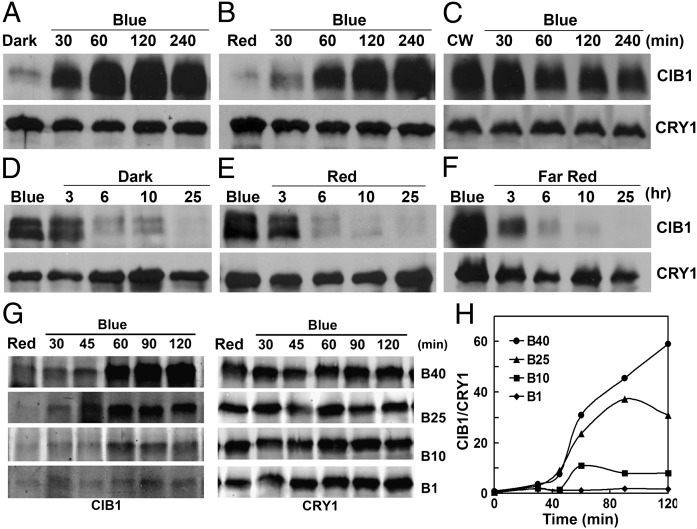
We previously reported that CRY2 interacts with CIB1 to activate its transcriptional activation activity (7). To better understand the role of CIB1 in the light regulation of plant development, we investigated whether light affects CIB1 protein expression. Because none of the antibodies we prepared against CIB1 recognizes the endogenous CIB1 proteins in plants, we used transgenic plants constitutively expressing epitope-tagged CIB1 (35S:Myc–CIB1) to analyze the CIB1 protein expression. In the first experiment, we grew transgenic plants expressing 35S:Myc–CIB1 in continuous white light (CW) for 3 wk, transferred those plants to either darkness or red light for 16 h, and then exposed the plants to blue light for various times and analyzed the level of CIB1 protein expression. The results of this experiment show that little CIB1 protein was detected in plants pretreated with darkness or red light, but the levels of the CIB1 protein increased markedly (>10-fold) within 1 h of blue-light treatment (Fig. 1 A–C and Fig. S1A). In the second experiment, we treated white-light–grown plants with blue light for 16 h, then transferred the plants to darkness, red light, or far-red light for various time periods and analyzed CIB1 protein. The results of this experiment demonstrate that the CIB1 protein is highly expressed in plants pretreated with blue light, but the levels of the CIB1 protein decreased markedly (>10-fold) within ∼6 h after plants were transferred from blue light to darkness, red light, or far-red light (Fig. 1 D–F and Fig. S1A). The blue-light regulation of CIB1 protein expression was observed not only in 3-wk-old adult plants (Fig. 1 and Fig. S1A), but also in 8-d-old young seedlings (Fig. S2 A and C), indicating that blue-light regulation of CIB1 expression is independent of developmental control. The blue-light regulation of CIB1 protein expression was also observed in transgenic plants expressing epitope-tagged CIB1 under control of the CIB1 native promoter (Fig. S2 B and D). We next examined whether light regulation of CIB1 protein expression is responsive to the photon dosage of blue light. In this experiment, transgenic plants expressing the 35S::Myc–CIB1 transgene were grown in red light for 3 wk and transferred to blue light of difference fluence rates for 30–120 min, and samples were collected for quantitative (Fig. 1 G and H) or semiquantitative immunoblot analyses (Fig. S1 B and C). The results of these experiments demonstrated that the level of the CIB1 protein increased in response to higher fluence rates of blue light. Together, our results establish that the level of the CIB1 protein expression is positively regulated by blue light in a wavelength-specific and photon density-dependent manner.
Fig. 1.
The CRY2-interacting bHLH protein CIB1 is degraded in the absence of blue light. Immunoblots show the expression of the CIB1 protein in transgenic plants expressing the 35S::Myc–CIB1 transgene. Samples were fractionated by 10% SDS/PAGE, blotted, probed with the anti-Myc antibody, stripped, and reprobed with the anti-CRY1 antibody as the loading controls. (A–C) Plants were grown in CW for 3 wk, transferred to dark (A) or red light (20 μmol⋅m−2⋅s−1) (B) or remained in CW (C) for 16 h, and then transferred to blue light (35 μmol⋅m−2⋅s−1) for the indicated time before sample collection. (D–F) Three-week-old plants were grown in CW, transferred to continuous blue light (blue, 35 μmol m−2 s−1) for 16 h, and then transferred to dark (D), red light (20 μmol⋅m−2⋅s−1) (E), or far-red light (5 μmol⋅m−2⋅s−1) (F), respectively, for the indicated time before sample collection. (G and H) Results of a fluence rate response showing the CIB1 protein expression changes in response to blue light. Plants were grown in continuous red light for 3 wk and then transferred to blue light of indicated fluence rate (1–40 μmol⋅m−2⋅s−1) and time indicated before sample collection. (H) The immunoblots shown in G were analyzed by the quantitative Odyssey analysis.
CIB1 Protein Is Degraded in the Absence of Blue Light.
Blue-light regulation of the CIB1 protein expressed from the 35S promoter-driven transgene suggests that CIB1 protein expression is regulated by a posttranscriptional mechanism. To examine this hypothesis, we analyzed light responsiveness of mRNA expression of the endogenous CIB1 gene and the 35S::Myc–CIB1 transgene in response to blue light (Figs. S3 and S4). In both 8-d-old seedlings and 3-wk-old plants, the mRNA expression of the endogenous CIB1 gene appeared to decrease slightly in the first 2 h of blue-light treatment and then increased slightly afterward (Fig. S3 A and B). In contrast, the mRNA expression of the 35S::Myc–CIB1 transgene showed little change in response to blue light (Fig. S3B). The mRNA expression of the endogenous CIB1 gene is not affected by developmental stages under the condition tested, and it is also not affected by mutations of the CRY1 and CRY2 genes (Figs. S3C and S4). Treatment of seedlings with the transcription inhibitor cordycepin, which reduced the overall protein expression as expected, failed to prevent the increase of the CIB1 protein in response to blue light (Fig. S5). These results suggest that the CIB1 protein expression is regulated by blue light primarily at the protein level, although a modest light effect on the CIB1 mRNA expression may also contribute to the overall expression of the endogenous CIB1 gene.
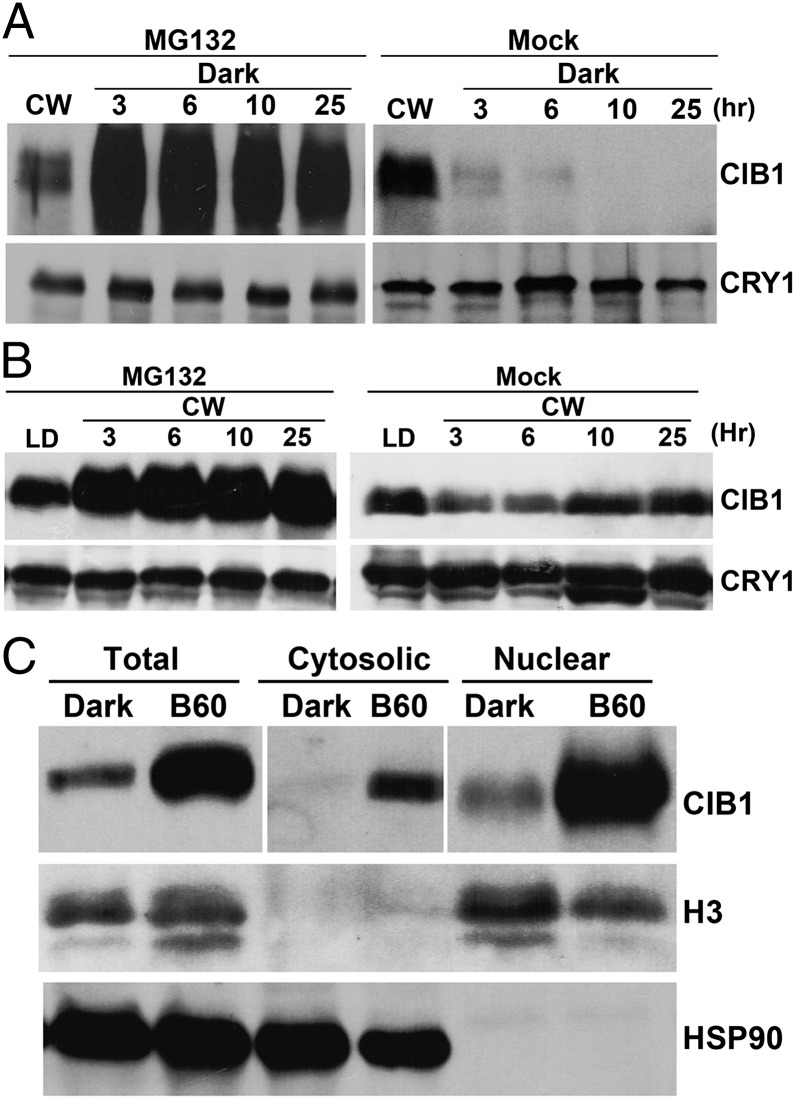
Given that light-dependent and ubiquitin/26S-proteasome–dependent proteolysis is a common mechanism regulating light signaling proteins (22), we examined blue-light effects on the CIB1 protein expression in the presence or absence of the 26S proteasome inhibitor MG132. In the first experiment, tissue samples were harvested and incubated in MG132 or mock control in darkness for up to 25 h and analyzed by immunoblots. As expected, the abundance of the CIB1 protein decreased markedly in darkness in the absence of MG132, and the CIB1 protein became barely detectable within 3 h under this condition (Fig. 2A, Mock). However, in the presence of MG132, the abundance of the CIB1 protein did not decrease in darkness for up to 25 h (Fig. 2A, MG132), suggesting that the decrease of CIB1 protein expression in the absence of blue light is due to proteolysis of CIB1 by the 26S proteasome. In the second experiment, tissue samples of plants grown in long-day (LD) photoperiods were incubated in MG132 or the mock control under CW before protein analysis. Under this condition, the level of the CIB1 protein decreased slightly in the absence of MG132 (Fig. 2B, Mock), but increased in the presence of proteasome inhibitor (Fig. 2B, MG132). This result is also consistent with the 26S-proteasome–dependent regulation of the CIB1 protein. The level of the CIB1 protein showed a modest increase in the presence of MG132 in both experiments, suggesting a continuous synthesis and turnover of the CIB1 protein in both light and dark conditions. We also analyzed the relative distribution of CIB1 protein in the nucleus and cytosol in response to blue light. Results of this experiment confirm that the CIB1 protein is mostly localized in the nucleus, and its nuclear localization is not significantly affected by blue light (Fig. 2C). Together, these experiments establish that the CIB1 protein is degraded by the 26S proteasome in the absence of blue light and that blue light suppresses CIB1 protein degradation.
Fig. 2.
The CIB1 protein is degraded in the absence of blue light by the 26S proteasome. (A and B) Immunoblot showing the inhibition of CIB1 degradation by the proteasome inhibitor MG132. Plants were grown in CW for 3 wk, and leaves were excised and incubated with MG132 (50 µmol/L) or mock solution (0.1% DMSO) in darkness (A) or white light (B) for the indicated time before sample collection. (C) Immunoblot showing the CIB1 protein in cytosolic and nuclear fractions. LD [16-h light/8-h dark (16hL/8hD)]-grown plants were transferred to dark for 16 h and then transferred to blue light (35 μmol⋅m−2⋅s−1) for 60 min. Total protein, cytosolic protein, and nuclear proteins were extracted, fractionated by 10% SDS/PAGE, blotted, and probed by the anti-Myc (CIB1), anti-histone H3 (nuclear marker), and anti-HSP90 (cytosol marker) antibodies.
Cryptochromes, phyA, Phototropins, and COP1 Are Not Required for the Blue-Light Regulation of CIB1 Protein Expression.
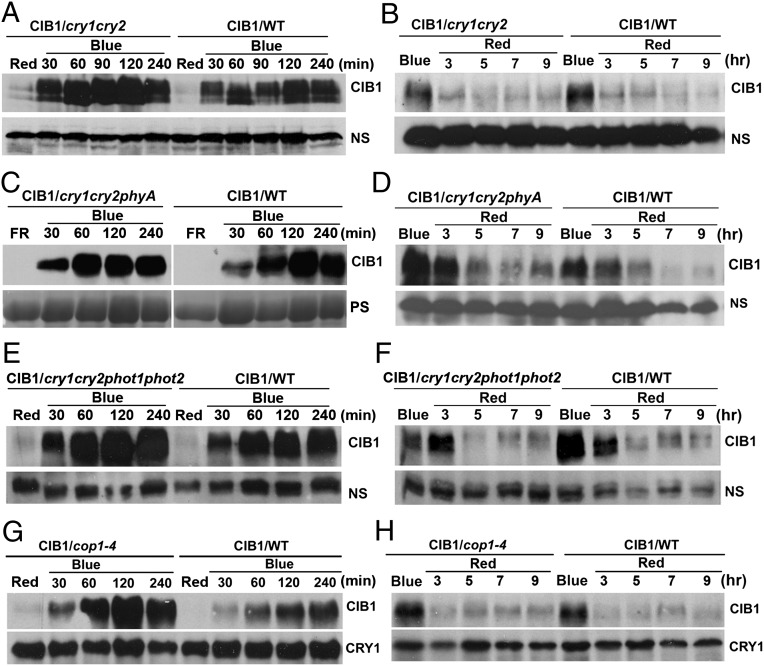
We next investigated which photoreceptor(s) mediate blue-light promotion of the CIB1 protein accumulation by examining the CIB1 protein expression in mutants impaired in various photoreceptors known to sense blue light (Fig. 3). To our surprise, although CIB1 is a cryptochrome-signaling protein that physically interacts with CRY2 in response to blue light (7–9), the cry1cry2 mutant showed no discernable defect in the blue-light regulation of CIB1 protein expression (Fig. 3 A and B). The CIB1 protein levels increased in response to blue light and decreased in the absence of blue light in both the wild-type and the cry1cry2 mutant (Fig. 3 A and B). Therefore, neither CRY1 nor CRY2 is the photoreceptor mediating blue-light suppression of CIB1 degradation. A direct involvement of two other types of photoreceptors known to sense blue light, phototropins, and phyA was also ruled out because CIB1 protein expression exhibited normal light responses in the multiple-photoreceptor mutants cry1cry2phyA and cry1cry2photphot2 (Fig. 3 C and F). We also examined a possible involvement of COP1, which is a E3 ubiquitin ligase catalyzing ubiquitination and proteolysis of many light-signaling proteins (22). However, our results indicate that COP1 is not required for CIB1 degradation in the absence of blue light, because CIB1 showed normal degradation in the cop1 mutant in the absence of blue light (Fig. 3 G and H).
Fig. 3.
Lack of effect of CRY, phyA, and COP1 on CIB1 protein expression. (A, C, E, and G) Transgenic plants expressing the 35S::Myc–CIB1 transgene in wild-type (WT) or the indicated mutant backgrounds (cry1cry2, cry1cry2phyA, cry1cry2phot1phot2, or cop1) were grown in LD (16hL/8hD) for 3 wk and exposed to red light (20 μmol⋅m−2⋅s−1) for 16 h (A, E, and G) or exposed to far-red light (5 μmol⋅m−2⋅s−1) for 16 h (C) and then transferred to blue light (35 μmol⋅m−2⋅s−1) for the indicated time before sample harvest. (B, D, F, and H) Alternatively, the 3-wk-old plants were exposed to blue light (35 μmol⋅m−2⋅s−1) for 16 h and then transferred to red light (20 μmol⋅m−2⋅s−1) for the indicated time. Samples were fractionated by 10% SDS/PAGE, blotted, and probed by the anti-Myc antibody (CIB1). CRY1 or nonspecific bands (NS) are shown as the loading controls. Because of uncontrolled exposure times of ECL of different immunoblots, results of different blots are not directly comparable.
ZTL and LKP2 Are Required for the CIB1 Protein Expression.
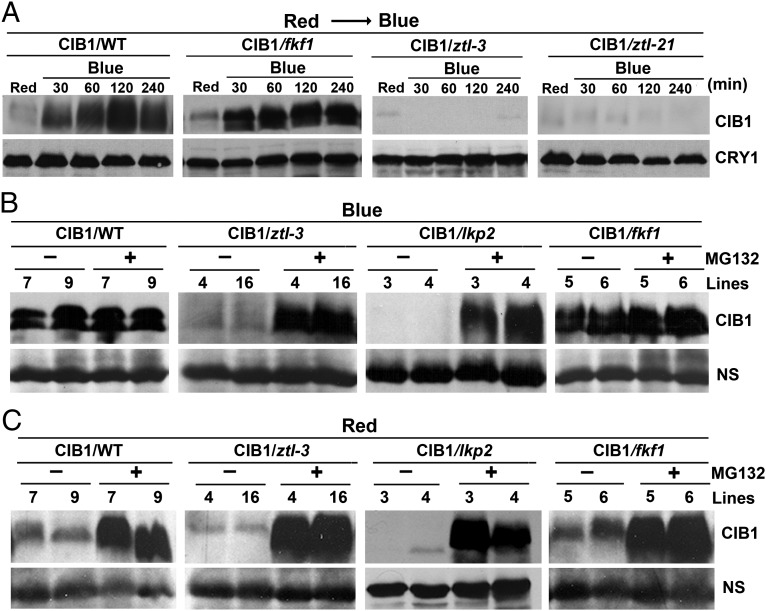
We then tested whether any of the three LOV-domain F-box proteins—ZTL, LKP2, and FKF1—may be required for the blue-light regulation of CIB1 protein expression. These related flavoproteins have been shown to act as blue-light receptors regulating the circadian clock and flowering time in Arabidopsis (16). Among the three proteins, ZTL and LKP2 are more closely related, and they have been shown to regulate expression of the same target proteins, such as TOC1 and PRR5 (17, 18, 24–26). We prepared transgenic plants expressing the 35S::Myc–CIB1 transgene in the ztl-3 mutant that showed no detectable ZTL protein expression, as well as in ztl-21, fkf1, and lkp2 mutant backgrounds (Figs. S6 and S7). We selected transgenic lines in different mutant backgrounds that expressed similar levels of mRNA of the 35S::Myc–CIB1 transgene (Figs. S6 and S7) and analyzed the level of the Myc-tagged CIB1 protein by immunoblots (Fig. 4). In the first experiment, we grew transgenic plants in LD photoperiods for 3 wk, transferred the plants to red light for 16 h to allow CIB1 degradation, then transferred the plants to blue light to facilitate reaccumulation of the CIB1 protein, and collected samples at different time after blue-light treatment for immunoblot analyses (Fig. 4A). As expected, little CIB1 protein was detected in plants treated with red light in all genetic backgrounds tested. In plants exposed to blue light, the CIB1 protein expression increased rapidly and reached maximum level of expression within 1 h in the wild type (CIB1/WT) and the fkf1 mutant background (CIB1/fkf1). In contrast, little CIB1 protein was detected in two different ztl mutant alleles (CIB1/ztl-3 and CIB1/ztl-21) exposed to blue light for as long as 4 h (Fig. 4A). This result demonstrates that ZTL, but not FKF1, is required for the blue-light–dependent CIB1 protein accumulation.
Fig. 4.
ZTL and LKP2, but not FKF1, are required for the accumulation of CIB1 protein in response to blue light. (A) Immunoblot showing the lack of blue-light–dependent CIB1 accumulation in two different ztl mutant alleles. The transgenic plants expressing the 35S::Myc–CIB1 transgene in the wild-type (CIB1/WT) and ztl-3 (CIB1/ztl-3) and ztl-21 (CIB1/ztl-21) mutant backgrounds were grown in LD (16hL/8hD) for 3 wk, transferred to continuous red light (20 µmol⋅m−2⋅s−1) for 16 h, and then transferred to blue light (35 µmol⋅m−2⋅s−1) for the indicated time before sample collection. Immunoblot was probed with the anti-Myc antibody, stripped, and reprobed with the anti-CRY1 antibody as the loading control. (B) Immunoblots showing levels of the CIB1 protein in different genetic backgrounds treated with blue light in the absence or presence of the proteasome inhibitor MG132. The transgenic plants expressing the 35S::Myc–CIB1 transgene in the wild-type (CIB1/WT) or ztl (CIB1/ztl-3), lkp2 (CIB1/lkp2), or fkf1 (CIB1/fkf1) mutants were grown in LD (16hL/8hD) for 3 wk and transferred to blue light (35 µmol⋅m−2⋅s−1) for 16 h. Leaves were excised, incubated in MG132 (50 µmol/L) or mock solution (0.1% DMSO) in blue light for 3 h, and the samples were analyzed by immunoblot probed with the anti-Myc antibody. A nonspecific band (NS) is included as the loading control. Two independent transgenic lines of each genotype were tested and shown (Lines). (C) Immunoblots showing levels of the CIB1 protein in different genetic backgrounds treated with red light in the absence or presence of the proteasome inhibitor MG132. The transgenic plants expressing the 35S::Myc–CIB1 transgene in wild-type (WT) or ztl-3, lkp2, or fkf1 mutants were grown in LD for 3 wk and transferred to red light (20 µmol⋅m−2⋅s−1) for 16 h. Leaves were excised, incubated in MG132 (50 µmol/L) or mock solution (0.1% DMSO) under red light for 3 h, and analyzed by immunoblot probed with the anti-Myc antibody. A nonspecific band (NS) is included as the loading control. Two independent transgenic lines of each genotype were tested and shown (Lines).
To examine how ZTL affects CIB1 expression and whether its close homolog LKP2 is also involved in the blue-light regulation of CIB1, we performed a second experiment. In this experiment, two independent transgenic lines expressing the 35S::Myc–CIB1 transgene in each of the ztl, fkf1, and lkp2 mutant backgrounds were grown in LD photoperiod for 3 wk; plants were transferred to blue light for 16 h; the leaf tissues were excised and incubated in the presence or absence of the proteasome inhibitor MG132 under blue light for 3 h, and the levels of the CIB1 protein were examined by immunoblot (Fig. 4B). As shown in Fig. 4B, similar levels of the CIB1 protein are detected in the wild-type and fkf1 mutant plants incubated under blue light, regardless of the proteasome inhibitor MG132 (Fig. 4B). The lack of effect of MG132 on CIB1 protein expression in these two genotypes pretreated with blue light suggests that CIB1 degradation was completely suppressed after the prolonged (16 h) blue-light treatment such that the additional inhibition of the 26S proteasome by MG132 caused no further reduction of the proteasome-dependent CIB1 degradation or further increase of the level of CIB1 (Fig. 4B). In contrast, little CIB1 protein was detected in the ztl mutant seedlings after the same 16-h blue-light treatment (Fig. 4B). However, the CIB1 protein was detected in the ztl mutant tissues treated with MG132. The levels of CIB1 accumulated in the ztl mutant treated with blue light and MG132 were comparable to that in the wild-type plants treated with blue light in the absence of MG132 (Fig. 4B). This observation demonstrates that the lack of CIB1 protein expression in the ztl mutant was primarily due to an impairment of blue-light inhibition of CIB1 degradation. Similarly, little CIB1 was detected in the lkp2 mutant unless tissues were incubated in MG132, indicating that LKP2 is also required for CIB1 protein expression in response to blue light. To further test this hypothesis, we did a third experiment, in which two independent lines of each of the photoreceptor mutant backgrounds were treated in red light in the absence or presence of MG132 (Fig. 4C). In the absence of MG132, little CIB1 protein was detected in all plants exposed to red light, although slightly more CIB1 protein was detected in the wild-type and fkf1 background. In the presence of GM132, similarly high levels of CIB1 was detected in all genetic backgrounds (Fig. 4C), confirming that the lack of the CIB1 expression in the ztl and lkp2 mutant is due to excessive degradation of the CIB1 protein. It is interesting that ZTL or LKP2 is necessary but not sufficient for the CIB1 protein expression, suggesting that the ZTL–LKP2 heterodimer may mediate blue-light suppression of CIB1 degradation. This proposition is consistent with the observation that ZTL interacts with LKP2 (27).
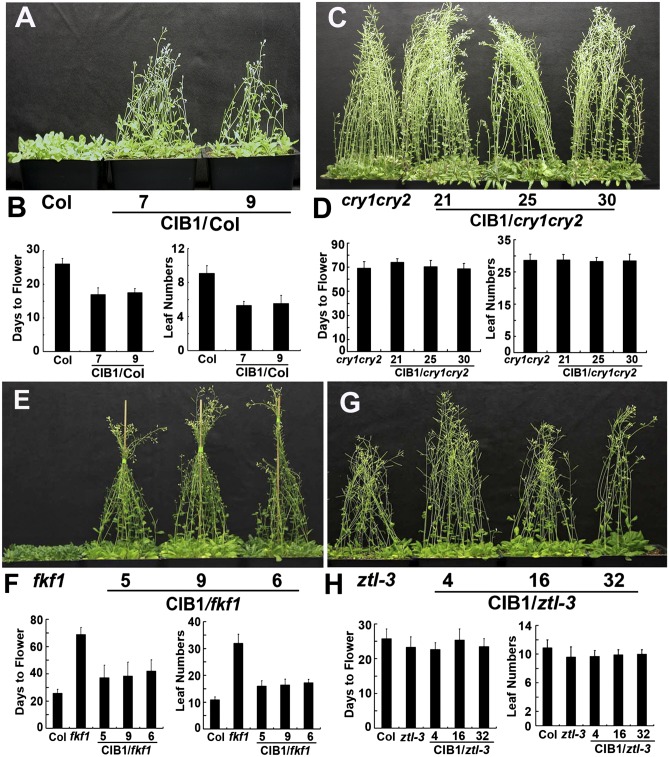
We reasoned that if ZTL and LKP2 are both required for CIB1 protein accumulation, mutation of either gene should suppress CIB1-dependent acceleration of flowering (7). To investigate this proposition, we examined the flowering time of transgenic plants expressing the 35S::Myc–CIB1 transgene in the wild-type or different photoreceptor mutant backgrounds. As reported (7), overexpression of CIB1 accelerated flowering in the wild-type, but not in the cry1cry2 mutant, background (Fig. 5 A–D). Consistent with the observation that FKF1 is not required for the CIB1 protein expression (Fig. 4A), overexpression of CIB1 in the fkf1 mutant background resulted in accelerated flowering similar to that of the wild-type background (Fig. 5 K and L), suggesting that the function of CIB1 is not dependent on FKF1 (Fig. 5 E and F). Transgenic plants expressing the 35S::Myc–CIB1 transgene in the ztl or lkp2 mutant backgrounds showed same flowering time as the respective parents (Fig. 5 G and H and Fig. S8), demonstrating that the function of CIB1 is dependent on not only CRY2 but also on ZTL and LKP2. Although transgenic plants expressing the 35S::Myc–CIB1 transgene in the ztl or lkp2 mutant accumulated high levels of the CIB1 mRNA (Figs. S6B and S7A), no CIB1 protein was detected in the ztl or lkp2 mutant backgrounds grown under normal white light conditions (Figs. S6C and S7B). This result further confirms that ZTL and LKP2 are required specifically for the CIB1 protein expression. Therefore, although the function of CIB1 is dependent on both CRY2 and ZTL/LKP2, the underlying mechanism is different: CRY2 mediates blue-light activation of the CIB1 activity whereas ZTL/LKP2 mediates blue-light stimulation of CIB1 protein expression.
Fig. 5.
The CIB1 activity promoting floral initiation is dependent on cryptochrome, ZTL, and LKP2, but independent from FKF1. (A, C, E, and G) Images of transgenic plants overexpressing CIB1 in the indicated genetic backgrounds and the respective parents. Plants were grown in LD photoperiods. (B, D, F, and H) The time to flowering, the number of rosette leaves at the time of flowering of the respective genotypes, and the SDs (n > 20) are shown.
Discussion
Plants evolve with multiple photoreceptors that function by interacting with photoreceptor-specific signaling proteins. However, the results of our previous and present studies of the function and regulation of the CRY2-signaling protein CIB1 demonstrate that different photoreceptors can also regulate the same transcription factor by distinct mechanisms. We have previously reported that CRY2 physically interacts with the transcription factor CIB1 in response to blue light to activate its transcriptional activation activity (7). We show in the present study that the CIB1 protein accumulates only in blue light, whereas it is degraded by the 26S proteasome in the absence of blue light. We further demonstrated that the LOV-domain proteins ZTL and LKP2 act as the photoreceptors mediating blue-light–dependent expression of CIB1. These results support a hypothesis that CIB1 is ubiquitinated by an unknown E3 ubiquitin ligase and degraded in the absence of blue light; ZTL (and LKP2) mediates blue-light suppression of the expression or activity of the E3 ubiquitin ligase or other proteins required for CIB1 ubiquitination and degradation (Fig. 6). We recently showed that CIB1 acts redundantly with its related proteins, CIB2, CIB4, and CIB5, to promote floral initiation and that these CIB1-related proteins are similarly degraded in the absence of blue light and stabilized in blue light in a ZTL-dependent manner (28). The ZTL/LKP2-mediated blue-light regulation of multiple CRY2-interacting CIB proteins provides a molecular mechanism to coordinate the functions of these two different types of photoreceptors in the control of plant development.
Fig. 6.
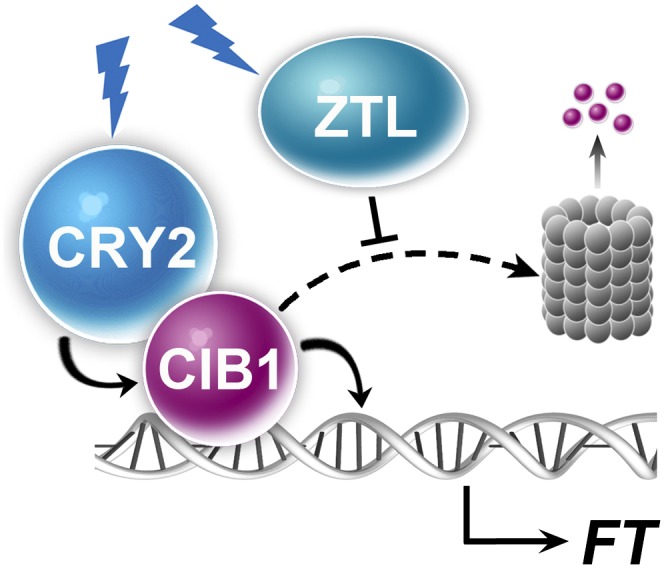
A hypothetical model depicting CRY2- and ZTL-mediated blue-light regulation of CIB1. The model hypothesizes that in response to blue light, CRY2 interacts with CIB1 to activate the activity of CIB1 promoting transcription of the FT gene, whereas ZTL suppresses CIB1 degradation by the 26S proteasome (barrel).
The effects of ZTL on the flowering-time control of plants are complex and cannot be explained simply by the ZTL-dependent stabilization of CIB proteins. Although some ztl mutant alleles, such as the missense ztl-1 allele in C24 background and missense ztl-25 allele in Ws background, showed delayed flowering phenotype; other ztl mutant alleles, such as the missense allele ztl-21 in Ws background, do not show delayed flowering (29). An apparently null ztl mutant allele, ztl-3 (T-DNA insertion mutation in Col background), showed normal flowering time in LD photoperiods but accelerated flowering in short-day photoperiods (29). Moreover, the ztllkp2 double mutant showed accelerated flowering, whereas transgenic plants overexpressing ZTL exhibited delayed flowering (30, 31). These results are consistent with previous and recent reports of ZTL acting to both stabilize GIGANTEA (GI) and facilitate its cytosolic retention (32, 33). Nuclear GI promotes flowering (34), and GI nuclear levels are higher in ztl mutants than in WT, whereas overexpressed ZTL sequesters GI in the cytosol (32). In addition to GI and CIB1, ZTL regulates other proteins that play important roles in the control of the circadian clock and photoperiodic flowering, such as PRR5 and TOC1 (18, 35, 36). Further studies are needed to elucidate both the individual and the combined effects of ZTL and ZTL-regulated proteins, including CIB1, in the control of flowering time.
Materials and Methods
Plant Materials.
Except where indicated, the Columbia accession of Arabidopsis was used. The ztl-3 (31), ztl21 (29), fkf1 (37), cry1cry2 (38), cry1cry2phyA (39), cry1cry2phot1phot2 (40), and cop1-4 (41) mutants have been described. Transgenic Arabidopsis lines were prepared by floral dip transformation method (42, 43). Phenotypes of transgenic plants were verified in at least three independent transgenic lines. The binary plasmids encoding the 35S::Myc–CIB1 and PCIB1::Myc–CIB1 were prepared by conventional and/or GATEWAY methods. PCIB1 represents the CIB1 promoter (−1,004 to +75 nt).
Gene Expression Analyses.
A mouse monoclonal anti-Myc antibody 4A6 (Millipore; no. 05-724; 1:4,000 dilution for immunoblot and 1:100 for immunostain) was used to detect Myc–CIB1 fusion protein. Immunoblots were quantified either by manually scanning the ECL (enhanced chemiluminescence) luminography films and analyzing the digitized signal by ImageJ (44) or by the Odyssey Image System, according to the manufacturer’s instructions (LI-COR). For Odyssey analysis, anti-Myc and -CRY1 antibodies were used to detect Myc–CIB1 and CRY1 proteins, IRDye 680RD goat anti-mouse (LI-COR, no. 926-68070) and IRDye 680RD goat anti-rabbit (LI-COR, no. 926-68071) were used as secondary antibodies. All antibody dilutions were made in casein blocking solution (LI-COR). Attempts to prepare anti-CIB1 antibodies, by using both peptide antigens or proteins expressed in Escherichia coli, were unsuccessful in raising antisera that recognize plant CIB1, although they recognize the proteins expressed in E. coli. Nuclear and cytoplasmic fractionation were prepared as described (45, 46).
Total RNAs were isolated by using the Illustra RNAspin Mini kit (GE Healthcare). cDNA was synthesized from 1 μg of total RNA by using SuperScript first-strand cDNA synthesis system (Invitrogen). Platinum SYBR Green qPCR Supermix-UDG (Invitrogen) or SYBR Premix Ex Tag (Takara) was used for quantitative PCR reaction, using the MX3000 System (Stratagene). The level of ACTIN mRNA expression (At3g18780; Table S1) was used as the internal control.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants GM56265 (to C.L.), GM23167 (to E.M.T.), GM079712 (to T.I.), and GM093285 (to D.E.S.); National Natural Science Foundation of China Grant 31270285 (to H.L.); the Hundred Talents Program of the Chinese Academy of Sciences (to H.L.); a Ministry of Agriculture Transgenic Research Grant (to Institute of Crop Sciences–Chinese Academy of Agricultural Sciences); University of California, Los Angeles (UCLA) Faculty Research Grants (to C.L.); and Sol Leshin Ben Gurion University–UCLA Academic Cooperation Programs (to C.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308987110/-/DCSupplemental.
References
- 1.Cashmore AR. Cryptochromes: Enabling plants and animals to determine circadian time. Cell. 2003;114(5):537–543. [PubMed] [Google Scholar]
- 2.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103(6):2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 3.Chaves I, et al. The cryptochromes: Blue light photoreceptors in plants and animals. Annu Rev Plant Biol. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- 4.Liu H, Liu B, Zhao C, Pepper M, Lin C. The action mechanisms of plant cryptochromes. Trends Plant Sci. 2011;16(12):684–691. doi: 10.1016/j.tplants.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366(6451):162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- 6.Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279(5355):1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- 7.Liu H, et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322(5907):1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- 8.Kennedy MJ, et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7(12):973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Idevall-Hagren O, Dickson EJ, Hille B, Toomre DK, De Camilli P. Optogenetic control of phosphoinositide metabolism. Proc Natl Acad Sci USA. 2012;109(35):E2316–E2323. doi: 10.1073/pnas.1211305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang S, et al. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 2008;27(8):1277–1288. doi: 10.1038/emboj.2008.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu LJ, et al. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell. 2008;20(2):292–306. doi: 10.1105/tpc.107.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21(10):841–847. [DOI] [PMC free article] [PubMed]
- 13. Oda A, Fujiwara S, Kamada H, Coupland G, Mizoguchi T (2004) Antisense suppression of the Arabidopsis PIF3 gene does not affect circadian rhythms but causes early flowering and increases FT expression. FEBS Lett 557(1–3):259–264. [DOI] [PubMed]
- 14.Kumar SV, et al. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature. 2012;484(7393):242–245. doi: 10.1038/nature10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science. 2012;336(6084):1045–1049. doi: 10.1126/science.1219644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito S, Song YH, Imaizumi T. LOV domain-containing F-box proteins: Light-dependent protein degradation modules in Arabidopsis. Mol Plant. 2012;5(3):573–582. doi: 10.1093/mp/sss013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Más P, Kim WY, Somers DE, Kay SA. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003;426(6966):567–570. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- 18.Kiba T, Henriques R, Sakakibara H, Chua NH. Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell. 2007;19(8):2516–2530. doi: 10.1105/tpc.107.053033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmad M, Jarillo JA, Smirnova O, Cashmore AR. The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell. 1998;1(7):939–948. doi: 10.1016/s1097-2765(00)80094-5. [DOI] [PubMed] [Google Scholar]
- 20.Más P, Devlin PF, Panda S, Kay SA. Functional interaction of phytochrome B and cryptochrome 2. Nature. 2000;408(6809):207–211. doi: 10.1038/35041583. [DOI] [PubMed] [Google Scholar]
- 21.Yang HQ, Tang RH, Cashmore AR. The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell. 2001;13(12):2573–2587. doi: 10.1105/tpc.010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17(10):584–593. doi: 10.1016/j.tplants.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Jarillo JA, et al. An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature. 2001;410(6827):487–490. doi: 10.1038/35068589. [DOI] [PubMed] [Google Scholar]
- 24.Somers DE, Schultz TF, Milnamow M, Kay SA. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell. 2000;101(3):319–329. doi: 10.1016/s0092-8674(00)80841-7. [DOI] [PubMed] [Google Scholar]
- 25.Baudry A, et al. F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell. 2010;22(3):606–622. doi: 10.1105/tpc.109.072843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Fujiwara S, Somers DE. PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 2010;29(11):1903–1915. doi: 10.1038/emboj.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yasuhara M, et al. Identification of ASK and clock-associated proteins as molecular partners of LKP2 (LOV kelch protein 2) in Arabidopsis. J Exp Bot. 2004;55(405):2015–2027. doi: 10.1093/jxb/erh226. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Li X, Li K, Liu H, Lin C. Plos Genetics. 2013. Multiple CIBs form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kevei E, et al. Forward genetic analysis of the circadian clock separates the multiple functions of ZEITLUPE. Plant Physiol. 2006;140(3):933–945. doi: 10.1104/pp.105.074864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takase T, et al. (2011) LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowering under non-inductive conditions, dependent on FLAVIN-BINDING KELCH REPEAT F-BOX1. Plant J 67(4):608–621. [DOI] [PubMed]
- 31.Somers DE, Kim WY, Geng R. The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell. 2004;16(3):769–782. doi: 10.1105/tpc.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J, Geng R, Gallenstein RA, Somers DE. The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development. 2013;140(19):4060–4069. doi: 10.1242/dev.096651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim WY, et al. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449(7160):356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- 34.Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318(5848):261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamichi N, et al. The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant Cell Physiol. 2005;46(4):609–619. doi: 10.1093/pcp/pci061. [DOI] [PubMed] [Google Scholar]
- 36.Sato E, Nakamichi N, Yamashino T, Mizuno T. Aberrant expression of the Arabidopsis circadian-regulated APRR5 gene belonging to the APRR1/TOC1 quintet results in early flowering and hypersensitiveness to light in early photomorphogenesis. Plant Cell Physiol. 2002;43(11):1374–1385. doi: 10.1093/pcp/pcf166. [DOI] [PubMed] [Google Scholar]
- 37.Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell. 2000;101(3):331–340. doi: 10.1016/s0092-8674(00)80842-9. [DOI] [PubMed] [Google Scholar]
- 38.Mockler TC, Guo H, Yang H, Duong H, Lin C. Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development. 1999;126(10):2073–2082. doi: 10.1242/dev.126.10.2073. [DOI] [PubMed] [Google Scholar]
- 39.Mockler T, et al. Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc Natl Acad Sci USA. 2003;100(4):2140–2145. doi: 10.1073/pnas.0437826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao J, Zhang YC, Sang Y, Li QH, Yang HQ. A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc Natl Acad Sci USA. 2005;102(34):12270–12275. doi: 10.1073/pnas.0501011102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McNellis TW, et al. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell. 1994;6(4):487–500. doi: 10.1105/tpc.6.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 43.Weigel D, et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122(4):1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu X, et al. Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell. 2007;19(10):3146–3156. doi: 10.1105/tpc.107.053017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Folta KM, Kaufman LS. Isolation of Arabidopsis nuclei and measurement of gene transcription rates using nuclear run-on assays. Nat Protoc. 2006;1(6):3094–3100. doi: 10.1038/nprot.2006.471. [DOI] [PubMed] [Google Scholar]
- 46.Després C, DeLong C, Glaze S, Liu E, Fobert PR. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell. 2000;12(2):279–290. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.