Abstract
Understanding the molecular basis of many cardiac diseases has been hampered by the lack of appropriate in vitro cell culture models that accurately reflect the human disease phenotypes. In the past few years, remarkable advances in stem cell biology have made possible this long-standing ambition—the generation of human and even patient-specific cellular models of diseases. Combined with other novel technologies in the fields of human genetics, tissue engineering, and gene-targeted manipulation, disease modeling with pluripotent stem cells has the promise to influence modern cardiovascular medicine on several fronts: molecular understanding of pathological mechanisms, early diagnosis, drug development, and effective treatment.
Strategies for generating human cardiac cells from genetically modified hESCs or patient-specific iPSCs have recently been developed. These differentiated cardiac cells can be used in disease modeling and drug screening.
Much of our current knowledge on the molecular pathways that lead to human cardiovascular disorders has come from heterologous expression systems and genetic animal models, in particular, mouse models. However, considerable differences exist between the human and mouse genomes, with many genetic modulators being human specific. In addition, species-specific electrophysiological properties of cardiac myocytes resulting from the finely balanced gating characteristics of many distinct ionic currents lead to considerable functional species differences between human and nonhuman hearts. For example, resting heart rate is tenfold higher in mice than in humans and physiological responses to exercise and arrhythmias are different. Therefore, the necessity to generate human in vitro models of cardiac disorders that accurately reflect the human disease phenotypes has become evident. The availability of such models is also crucial for the discovery and development of therapeutics. Primary human cardiac cells and especially disease-bearing ones are difficult to obtain and propagate in culture for extended periods of time. Recent advances in pluripotent stem cell biology now make it possible to generate an unlimited number of human cardiac cells in vitro from both healthy individuals and from patients with cardiac abnormalities (Fig. 1). Pluripotent stem cells (PSCs) retain the capacity to indefinite self-renewal and differentiation into cells and tissue derivatives from all three germ layers. These properties make PSCs valuable for studying early developmental biology, disease modeling, drug discovery, toxicology testing, and for regenerative medicine. Since the first derivation of pluripotent human embryonic stem cells (hESCs) from normal embryos in 1998 (Thomson et al. 1998), the potential of these cells to be used to model genetic and more complex cardiac diseases and for high-throughput pharmacological screenings has been anticipated. However, initial difficulties in genetically modifying hESCs, ethical and legislative issues associated with destroying surplus embryos during derivation and the limitation of being able to obtain disease-specific hESC lines only for rare single-gene disorders discerned by preimplantation diagnostics restricted their use and no hESC lines modeling a cardiac disease have been published to date.
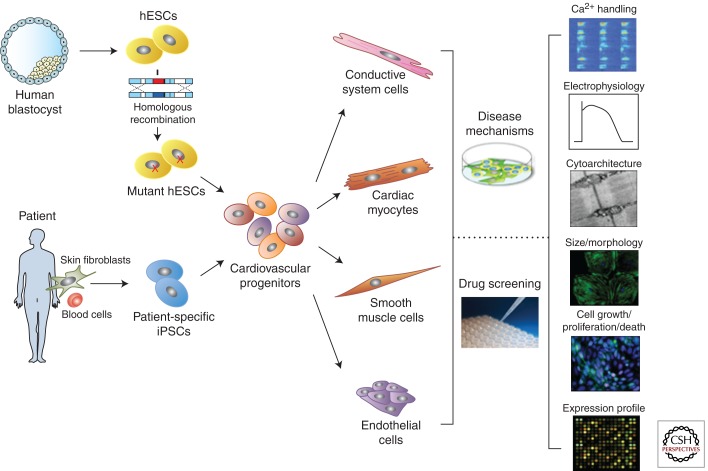
Figure 1.
Strategies for obtaining human disease-specific cardiac cells and their use in disease modeling and drug screening. Human embryonic stem cells (hESCs) can be derived from human normal blastocysts and then genetically targeted to introduce a disease-associated mutation by homologous recombination. Alternatively, patient-specific induced pluripotent stem cells (iPSCs) can be derived directly from patient somatic cells (e.g., skin fibroblasts or blood cells) by different reprogramming methods. Either type of disease-specific pluripotent stem cells can be differentiated in vitro into all kinds of cardiac cells (cardiac myocytes, cells of the conductive system, smooth muscle cells, and endothelial cells) through a cardiovascular progenitor population. Differentiated cardiac cells can then be used in disease modeling to understand the molecular mechanisms underlying disease phenotypes and in drug screening to determine the effects of candidate drugs or new compounds and identify target pathways. Examples of different cellular readouts are presented.
The revolutionary discovery of induced pluripotent stem cells (iPSCs), generated by reprogramming somatic cells into an embryonic pluripotent state by the forced expression of a defined set of transcription factors (Takahashi and Yamanaka 2006; Takahashi et al. 2007; Yu et al. 2007; Park et al. 2008), introduced a potentially transformative tool to enable in vitro cardiac disease modeling and inform drug discovery. The first disease-specific iPSCs of a pathology affecting the heart were derived from patients with LEOPARD syndrome (Carvajal-Vergara et al. 2010), an autosomal-dominant developmental disorder with pleomorphic effects on several tissues and organ systems, with hypertrophic cardiomyopathy being often the cause of mortality. Since then, a growing number of patient-specific iPSC models of genetically inherited cardiovascular diseases have been generated (Table 1). Critical for the generation of hPSC-based models of disease are (1) selecting an appropriate disease target, (2) directing the differentiation of hPSCs into phenotype-relevant cell populations, and (3) identifying disease-relevant phenotypes.
Table 1.
Cardiac diseases for which induced pluripotent stem cell models have been generated
| Disease | Affected gene and gene function | Mutation | Phenotype in hiPSC-derived cardiac cells | Cellular assays | Control iPSC lines | Patient iPSC lines | Rescue and drug testing | References |
|---|---|---|---|---|---|---|---|---|
| Cardiochannelopathies | ||||||||
| LQT1 | KCNQ1, pore-forming α subunit of the channel generating the slow component of the delayed rectifier potassium current IKs | c.569G > A; p.R190Q | Prolonged APD, impaired AP rate adaptation, EADs upon β-adrenergic stress, decreased IKs, aberrant subcellular localization of KCNQ1 owing to trafficking defect of the channel | Whole-cell patch clamp, immunohisto-chemistry | Two iPSC lines: unrelated 32- and 36- yr-old women | Two iPSC lines: 8-yr-old son and 42-yr-old father | Propanolol | Moretti et al. 2010b |
| c.1893delC; p.P631fs/33 | Prolonged FPD, EADs upon β-adrenergic stress, impaired IKs, trafficking defect of KCNQ1 to the membrane | Whole-cell patch clamp, MEA, immunohistochemistry | Two iPSC lines: unrelated individuals | One iPSC line: 13-yr-old boy | Propanolol | Egashira et al. 2012 | ||
| LQT2 | KCNH2, pore-forming α subunit of the channel generating the rapid component of the delayed rectifier potassium current IKr | c.1841C > T; p.A614V | Prolonged APD and FPD owing to reduced IKr, arrhythmia | Whole-cell patch clamp, MEA | One iPSC line: unrelated individual | One iPSC line: 28-yr-old woman | Nifedipine, Pinacidil, Ranolazine | Itzhaki et al. 2011a |
| c.1681G > A; p.A561T | Prolonged APD and FPD, arrhythmia under β-adrenergic stress, reduced IKr | Whole-cell patch clamp, MEA | One iPCS line: generated from ESC-derived fibroblasts; one ESC line | Two iPSC lines: daughter and mother | Propranolol, Nicorandil, PD-118057 | Matsa et al. 2011 | ||
| c.526C > T; p.R176W | Prolonged APD and FPD, reduced IKr, arrhythmia under β-adrenergic stress | Whole-cell patch clamp, MEA | Four iPSC lines: unrelated individuals; one ESC line | Two iPSC lines: 61-yr-old man and his sister | Erythromycin, Sotalol, Cisapride | Lahti et al. 2012 | ||
| LQT3, Brugada syndrome | SCN5A, pore-forming α subunit of the cardiac Na+ channel | c.5387_5389insTGA; p.1795insD | Increased APD, decreased INa+ | Whole-cell patch clamp | One iPSC line: unrelated 51-yr-old woman | One iPSC line: 47-yr-old man | N/A | Davis et al. 2012 |
| Timothy syndrome (LQT8) | CACNA1C, pore-forming α subunit of L-type Ca2+ channel | c.1216G > A; p.G406R | Irregular contraction, increased Ca2+ influx, prolonged APD, irregular electrical activity, reduced voltage-dependent inactivation of L-type current, arrhythmias, abnormal Ca2+ transients | Whole-cell patch clamp, Ca2+ imaging | Two iPSC lines: unrelated individuals | Two iPCS lines | Roscovitine | Yazawa et al. 2011 |
| CPVT1 | RYR2, Ca2+ release channel of the SR | c.1217C > T; p.S406 L | Increased diastolic Ca2+, reduced SR-Ca2+, increased frequency and duration of Ca2+ sparks, arrhythmia and DADs | Whole-cell patch clamp, Ca2+ imaging, immuno-histochemistry | One iPSC line: unrelated 32-yr-old woman | One iPSC line: 24-yr-old woman | Dantrolene | Jung et al. 2012 |
| c.7447T > A; p.F2483I | DADs upon catecholaminergic stimulus, higher amplitude and longer duration of Ca2+ sparks | Whole-cell patch clamp, MEA, Ca2+ imaging | Two iPSC lines: unrelated individuals; three ESC lines | One iPSC line: 46-yr-old woman | N/A | Fatima et al. 2011 | ||
| c.12056T > G; p.M4109R | DADs, irregular Ca2+ transients, store-overload-induced Ca2+ release | Whole-cell patch clamp, MEA, Ca2+ imaging | One iPSC line: unrelated individual | One iPSC line: 30-yr-old woman | Flecainide, Thapsigargin | Itzhaki et al. 2012 | ||
| CPVT2 | CASQ2, SR Ca2+-binding protein | c.1183G > C; p.D307H | DADs, oscillatory arrhythmic prepotentials, after-contractions, increased diastolic Ca2+, less organized myofibrils, enlarged SR cisternae | Whole-cell patch clamp, Ca2+ imaging, TEM, immunohisto-chemistry | Three iPSC lines: unrelated 25-yr-old man, 23- and 42-yr-old woman | Two iPSC lines: 12-yr-old boy and 30-yr-old woman | N/A | Novak et al. 2012 |
| Cardiomyopathies | ||||||||
| DCM | TNTT2, cardiac troponin T, a sarcomeric protein | p.R173W | Abnormal distribution of sarcomeric protein α-actinin, increased sarcomeric disorganization upon ionotropic and mechanical stress, impaired Ca2+ handling, decreased contractility | TEM, immuno-histochemistry, whole-cell patch clamp, Ca2+ imaging, atomic force microscopy | Three iPSC lines: father, son, and grandson | Four iPSC lines: mother, two sons, and grandson | SERCA2a overexpression, metoprolol, norepinephrine | Sun et al. 2012 |
| HCM | MYH7, β-myosin heavy chain, a sarcomeric protein | c.1988G > A; p.R663H | Hypertrophic cardiomyocytes: cellular enlargement and multinucleation, increased myofibril content and sarcomere disorganization, ANF expression, elevation of β-myosin/α-myosin ratio, calcineurin activation, NFAT nuclear translocation, contractile arrhythmia and DADs, Ca2+ cycling dysfunction and elevated [Ca2+]i | Immunohisto-chemistry, whole-cell patch clamp, MEA, Ca2+ imaging | Five iPSC lines: 55-yr-old husband and 20-, 16-, 14-, and 13-yr-old children | Five iPSC lines: 53-yr-old mother and 21-, 18-, 14-, and 10-yr-old children | Propranolol, Verapamil, Nifedipin, Diltiazem, Lidocaine, Mexiletine, Ranolazine | Lan et al. 2013 |
| LEOPARD syndrome | PTPN11, a protein tyrosine phosphatase SHP2 | c.1403C > T; p.T468M | Hypertrophic cardiomyocytes: increase in size, increased sarcomeric organization, preferential nuclear localization of NFATC4, increase of phosphorylated proteins MEK1, EGFR | Cardiomyocyte cell size determination, immunohisto-chemistry, phosphoproteomics | Two iPSC lines: unrelated individual and unaffected brother of the female patient; one ESC line | Two iPSC lines: 25-yr-old woman and 34-yr-old man | N/A | Carvajal-Vergara et al. 2010 |
| ARVC | PKP2, plakophilin 2, a desmosomal protein | c.1841T > C; p.L614P | Decreased expression of desmosomal proteins PKP2, PG, increased lipid droplets upon adipogenic stimulus | Immunohisto-chemistry, TEM, Oil Red O staining | One iPCS line: unrelated 32-yr-old man | One iPSC line: 30-yr-old man | N/A | Ma et al. 2013 |
| c.2484C > T; r2483_2489del c.2013delC; p.R672fsX683 |
Abnormal PG nuclear translocation and decreased β-catenin activity, lipogenesis and apoptosis upon adipogenic stimulus, Ca2+-handling defects | Immunohisto-chemistry, Nile Red staining, functional assays of FAO and glycolysis, Ca2+ imaging | Two iPCS lines: unrelated female and male fetuses; one ESC line |
Two iPSC lines: unrelated female and male individuals | Wild-type PKP2 overexpression | Kim et al. 2013 | ||
| Vascular diseases | ||||||||
| Pompe’s disease | GAA, acid α glucosidase, a lysosomal glycogen-degrading enzyme | c.1935C > A/1935C > A; 1935C > A/2040 + 1G > T | Increased glycogen levels, deteriorating mitochondria, autophagosome-like structures, reduced cellular respiration | Immunohistoche-mistry, electron microscopy, O2 consumption rate measurement | One iPSC line: unrelated individual, autologous patient’s iPSC lines with inducible GAA transgene; two hESC lines | One iPSC line per mutation | GAA transgene, rhGAA; 3-methyladenin L-carnitine | Huang et al. 2011 |
| Hutchinson-Gilford progeria syndrome | LMNA, lamin A | c.1824C > T; p.G608G | Premature senescence of SMC associated with vascular aging: reduced telomere length, compromised cell proliferation owing to accumulation of progerin (slice variant of lamin A) | Immunohistochemistry, cell proliferation assay, telomere length analysis | Two iPSC lines: parents of the patients | Two iPSC lines | shRNA targeting progerin | Zhang et al. 2011a |
| SVAS/WBS | ELN, elastin, monomeric protein tropoelastin, which is secreted, cross-linked, and organized into ELN polymer | SVAS: GTAT insertion in exon 9; WBS: 1.4 MB microdeletion at 7q11.23 including ELN gene | SMCs: Decreased organization of SM α-actin filament bundles, increased proliferation, increased migration to PDGF | Immunohisto-chemistry, migration and proliferation assays | Two iPCS lines: unrelated 12- and 1-yr-old boys | Two iPSC lines: 39-yr-old man with SVAS and 1-yr-old boy with WBS | Recombinant elastin, overexpression of constitutive RhoA | Ge et al. 2012 |
ANF, atrial natriuretic factor; APD, action potential duration; CPVT, catecholaminergic polymorphic ventricular tachycardia; DADs, delayed after-depolarizations; DCM, dilated cardiomyopathy; EADs, early after-depolarizations; EGFR, epidermal growth factor receptor; FAO, fatty acid oxidation; FPD, field potential duration; HCM, hypertrophic cardiomyopathy; LQT, long-QT syndrome; MEA, multielectrode array; MEK1, mitogen-activated protein kinase kinase 1; NFATC4, T-cell transcription factor NFAT3; PG, plakoglobin; SERCA2a, sarco/endoplasmic reticulum Ca2+-ATPase; SMC, smooth muscle cell; SR, sarcoplasmic reticulum; SVAS/WBS, supraventricular arrhythmias/Williams syndrome; TEM, transmission electron microscopy.
In this review, we discuss recent pluripotent stem cell developments in the cardiac field, with special emphasis on disease modeling, and address current challenges and promises of using PSCs in modeling cardiovascular disorders.
GENERATION OF HPSC DERIVATIVES FOR THE STUDY OF CARDIOVASCULAR DISEASE
In Vitro Differentiation of hPSCs into Multipotent Cardiovascular Progenitors and Their Differentiated Derivatives: Recent Progress and Current Limitations
The ability to generate, expand, and purify functional cardiac cells is critical to developing pluripotent stem cell models of human cardiovascular disease. During the past decade, substantial progress has been made to understand the regulation of cardiac differentiation from hPSCs and to develop strategies to efficiently and reliably direct hPSCs into the cardiovascular lineages (Laflamme et al. 2007; Yang et al. 2008; Kattman et al. 2011; Burridge et al. 2012). In vitro differentiation of hPSCs into cardiovascular cells is a multistep process that involves initial epithelial to mesenchymal transition, mesodermal, and subsequently cardiogenic specification and differentiation followed by functional maturation (Fig. 2). This temporally controlled process is tightly regulated by critical developmental signals, epigenetic programs, and extracellular microenvironments. Early cardiac specification and differentiation of hPSCs is modulated by four major signaling pathways: BMP, TGF-β/activin/nodal, WNT/β-catenin, and FGF, with highly specific temporal windows for effectiveness. During this phase, multipotent cardiovascular progenitors arise, which then further enter alternative differentiation pathways to generate cardiomyocytes, vascular endothelial, or smooth muscle cells (Fig. 2) (Yang et al. 2008; Bu et al. 2009). Factors including coculture with END-2 stromal cells (Mummery et al. 2003) and ascorbic acid, which enhances collagen synthesis and promotes proliferation of cardiac progenitors (Cao et al. 2012), have been shown to support generation of differentiated cardiovascular cells, highlighting the unique role of microenvironmental and cellular signaling during hPSC differentiation.
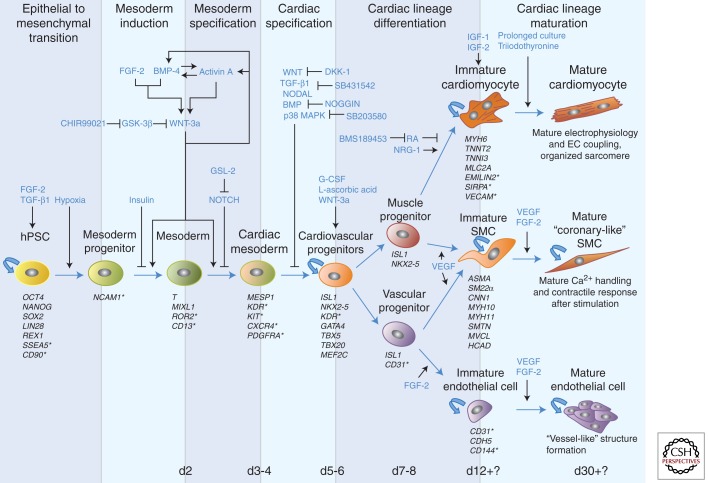
Figure 2.
Pathways and factors involved in cardiac differentiation of human pluripotent stem cells in vitro. Differentiation of human pluripotent stem cells (hPSCs) into the cardiovascular lineages (cardiomyocytes, smooth muscle cells, and endothelial cells) is a multistep process that involves initial epithelial to mesenchymal transition, mesoderm induction and specification, cardiac specification and differentiation, and functional maturation. Each of these steps is temporally controlled by specific signaling pathways and factors, and each cellular intermediate is characterized by the expression of different markers; surface markers are marked with an asterisk.
Depending on the form of cardiovascular disorder (congenital or adult) to be modeled, multipotent cardiovascular progenitor populations or terminally differentiated cardiac cells may be more relevant. In both cases, robust protocols to purify the desired cell type (as discussed below) and to reproducibly achieve a functional maturation state are essential. Recent work on hESC-derived ISL-1+ cardiovascular precursors has shown that the use of Wnt-3a-secreting mouse embryonic fibroblast feeder cells allows for the expanded growth of the cells, while still maintaining ISL-1 expression and the capability to differentiate into all major cardiac lineages (Bu et al. 2009; Moretti et al. 2010a). In particular, for cardiomyocytes, issues regarding the level of cellular maturation and the subtype heterogeneity that result from most of the available hPSC differentiation protocols need to be considered for reliable disease modeling.
Maturation of hPSC-Derived Cardiomyocytes
Although structural and electrophysiological maturation of hPSC-derived cardiomyocytes improves in a time-dependent manner (Lundy et al. 2013), these cells remain largely immature compared with their adult counterparts and resemble fetal stages of development. They lack a well-developed transverse tubule system (Lieu et al. 2009) and show automaticity (spontaneous contractions) and irregular membrane potentials, with action potential (AP) up-stroke velocity and amplitude similar to 16-wk-old embryonic hearts (Mummery et al. 2003). Moreover, fetal transcriptional and functional profiles have been reported of cardiomyocytes generated from both hESCs (Cao et al. 2008) and hiPSCs (Gupta et al. 2010). Conflicting data exist regarding the maturity of Ca2+ handling and sarcoplasmic reticulum development in hPSC-derived cardiomyocytes, although there is evidence that at least basic components of the Ca2+ cycling machinery are functional as well as excitation-contraction coupling (Satin et al. 2008; Itzhaki et al. 2011b; Jung et al. 2012). The lack of complete maturity of hPSC-derived cardiomyocytes may reduce the suitability for drug testing and limit disease modeling when the disease-causing mutation affects a gene that is not expressed until later in development or if the disease occurs postnatally and has long latency of onset.
Heterogeneity of hPSC-Derived Cardiomyocytes
Electrophysiological and gene expression analysis show that PSCs generate all three major subtypes of cardiomyocytes, showing atrial-, ventricular-, or nodal-like phenotypes. Common hPSC differentiation methodologies create a mixture of all these cell types and this might represent a limitation in modeling cardiac diseases that affect a specific cardiomyocyte type, especially if the AP is not the readout assay of the cellular phenotype, or in high-throughput drug screening. Significant efforts have been made to understand cardiac subtype specification and to reduce heterogeneity of hPSC-derived cardiomyocytes. Recent studies have shown that inhibition of NRG-1b/ERBB signaling enhances the proportion of nodal-like cells (Zhu et al. 2010) and that retinoid signals enhance atrial versus ventricular specification during cardiac hESC differentiation (Zhang et al. 2011b).
Interline Variability in Cardiac Differentiation of hPSCs
It has been well documented that a diversity in cardiac differentiation capacity between hPSC lines exists (Kattman et al. 2011). Many factors are likely to contribute to this line-to-line variation, including the conditions used to establish and maintain the lines, the levels of expression of endogenous growth factors, and, specifically for hiPSCs, the efficiency of and the methodology used for reprogramming. A recognized source of variability in the differentiation potential of hiPSC lines is also the cell type of origin. Recent data suggest that hiPSCs harbor residual DNA methylation signatures characteristic of their somatic tissue of origin, which favors differentiation along lineages related to the donor cell, while confining alternative cell fates (Kim et al. 2010). The retention of epigenetic memory of cell origin was initially thought to be restricted to low passage hiPSCs, but it has now been documented in later passages as well (Ohi et al. 2011). This issue becomes more complex given that hPSCs progressively acquire epigenetic heterogeneity after prolonged culture, which affects subsequent differentiation (Tanasijevic et al. 2009), and that different passages of the same cell line may have diverse cardiac potentials (Paige et al. 2010).
In Vitro Differentiation of hPSCs into Lineage-Specific Vascular Smooth Muscle and Endothelial Cells
Although most electrophysiological disorders and cardiomyopathies directly affect cardiomyocytes, noncardiomyocyte cell types are key to understanding other cardiovascular diseases. For example, additional cell lineages of the heart, such as vascular endothelial and smooth muscle cells or fibroblasts, are relevant to congenital heart diseases (CHD) or disorders affecting the outflow tract vessels (e.g., aortic aneurysm) and the coronary system (e.g., coronary artery disease). Lineage tracing studies have shown that vascular smooth muscle cells (SMCs) in different heart vessels have distinct embryological origin. The basal aortic root is derived from secondary heart field (Waldo et al. 2005), whereas the ascending aorta and the arch are neural crest derived (Jiang et al. 2000); descending aortic SMCs originate from paraxial (somitic) mesoderm (Wasteson et al. 2008); and coronary SMCs arise from proepicardial organ, which is of lateral plate mesoderm origin (Mikawa and Gourdie 1996; see also Brade et al. 2013). Vascular SMCs originating from different types of embryonic progenitors show lineage-specific differences in growth, gene expression, and functional properties (Majesky 2007), supporting the notion that vascular SMC lineage diversity may contribute to site-specific patterns of vascular disease in adults. Disease modeling with the appropriate origin-specific SMCs generated from patient-specific iPSCs is therefore vital for accurate assessment and therapeutic discovery. In this regard, a recent study has described an in vitro strategy for generating large numbers of lineage-specific vascular SMCs from hPSCs using chemically defined protocols to direct neuroectoderm, paraxial mesoderm, and lateral plate mesoderm progenitors to a smooth muscle fate (Cheung et al. 2012).
Similarly to SMCs, endothelial cells with arterial and venous identity develop distinct gene and protein expression profiles that depend on the embryological origins and on microenvironmental cues during organogenesis and result in functional heterogeneity (Cleaver and Melton 2003). Although the embryological origins of the endothelial cells of different heart vessels are still controversial (see Brade et al. 2013), development of robust protocols to generate hPSC-derived lineage-specific arterial and venous endothelial cells with organ-specific properties will be necessary to develop faithful hPSC-based models of specific cardiovascular disorders.
CARDIAC DISEASE MODELING WITH PSCs
The key to modeling any disease with hPSCs is the identification of a disease-relevant cellular pathology with robust phenotypic assays. Early-onset diseases that have a strong genetic component and affect a highly defined cell or tissue type are the first choice.
Gene Targeting in hPSCs: hESC Models of Cardiac Disease
Although the genetic basis of many cardiovascular diseases is incompletely understood, most of them are now appreciated to arise from genetic variants. Several chromosomal abnormalities, rare DNA variants, or common DNA variants have been associated with congenital heart disease in newborns (Bruneau 2008), and many mutations in genes encoding cardiac ion channels and sarcomeric proteins have been identified in several electrophysiological cardiac disorders (channelopathies) and cardiomyopathies (Jacoby and McKenna 2012; Martin et al. 2012). With the advent of new techniques for efficient genetic manipulation through homologous recombination, introduction of targeted mutations into hESCs is becoming a very attractive tool for generating hESC-based models of monogenic cardiac diseases. Owing to the difficulty in cloning hESCs from single cells and the low efficiency of introducing DNA-targeting plasmids using conditions well established for mouse ESCs, such as electroporation, only very few hESC lines have been targeted using classical homologous recombination protocols (Urbach et al. 2004; Irion et al. 2007; Bu et al. 2010). Zinc-finger nucleases (ZNFs) or transcription activator-like effector nucleases (TALENs), which introduce double-strand breaks at specific sites in the genome, have been shown to greatly improve the efficiency of homologous recombination when used in combination with traditional gene-targeting constructs (Wood et al. 2011). Using this approach, several genes have been targeted in hESC lines (Hockemeyer et al. 2009, 2011; Zou et al. 2009; Collin and Lako 2011), and also chromosome deletions and translocations have been successfully engineered in human cells (Brunet et al. 2009; Lee et al. 2010). At the time of this writing, no published studies have reported models of cardiovascular disease obtained by genetic modification of hESCs. However, compared with models based on patient-specific hiPSCs, genetic targeting of hESCs to introduce the disease variant offers a built-in control, because the parental hESC line is genetically matched with the recombinant hESC line except for the introduced disease mutation.
Patient-Specific hiPSC-Based Models of Cardiac Diseases
Since the first report describing iPSCs, rapid progress has been made in reprogramming various somatic cell types using many variants of the original cocktail of reprogramming genes, including chromatin-modifying small molecules, and numerous methods for delivering the reprogramming factors to the cell, including nonintegrating genomic approaches (Gonzalez et al. 2011). It is now possible to obtain easily accessible somatic cells from patients (e.g., from skin biopsies, hair follicles, or whole blood) and derive “factor-free” hiPSC lines that are genetically matched to the patients. Whereas the initial reports argued convincingly that hiPSCs were functionally equivalent to hESCs, a more refined analysis of how hiPSCs behave in vitro, coupled with genome-wide genetic and epigenetic analysis, has revealed numerous subtle but substantial molecular differences, probably owing to technical limitations inherent to reprogramming (Robinton and Daley 2012).
Advantages of hiPSC over hESC and Considerations
Patient-specific hiPSCs offer few advantages over recombinant hESCs for the generation of disease models: (1) they allow the investigators to probe genotype–phenotype relationships for conditions with a monogenic and, increasingly, with a complex basis, even when the genetic variant linked to the disease is difficult to generate with homologous recombination (e.g., a chromosomal deletion or translocation) or is unknown; and (2) they constitute the genetic background of the original patient, which may prove crucial as phenotypic variations may entail interactions between a polymorphism and modifier loci.
An important consideration in using hiPSC models for the study of any human disease is the identification of appropriate control lines, because differences in genetic background between well-matched control donors (for gender, age, and ethnicity) and patient-derived hiPSC lines could confound the analysis of the disease phenotype, in particular when the genetic variant under study is not associated with a large phenotypic effect. One way to circumvent this problem would be to rely on not just one control hiPSC line but on a panel of control lines derived from genetically diverse subjects. The major drawback of this approach is the increased cost and labor associated with the generation and maintenance of multiple control cell lines and with the performance of multiple control experiments with cells derived from the different lines. Another way to limit the genetic variance between patient and control cells would be the derivation of control hiPSC lines from a healthy sibling who is unaffected by the disease. When a monogenic disease is investigated, the ultimate control hiPSC line could be constructed by correcting the disease-causing mutation in the patient hiPSC by a gene-targeting approach. Furthermore, such an experiment could unequivocally prove that the mutation under consideration is the sole cause of the phenotypic differences between patient and control cells.
Existing Patient-Specific hiPSC Models of Cardiovascular Diseases
Soon after the first demonstration that patient-specific hiPSC-derived cardiomyocytes could recapitulate aspects of the cardiac phenotype seen in patients affected by LEOPARD syndrome (Carvajal-Vergara et al. 2010), several following studies validated the utility of hiPSCs for modeling cardiovascular disorders and drug testing (Table 1). By now, many patient-specific hiPSC-based models of diseases affecting cardiomyocytes have been reported, including channelopathies related to malfunctions of voltage-activated ion channels generating the cardiac AP (long-QT syndrome) and channels controlling the intracellular calcium levels (catecholaminergic polymorphic ventricular tachycardia [CPVT]), as well as cardiomyopathy linked to impaired cardiac muscle contractility (hypertrophic and dilated cardiomyopathy and arrhythmogenic right ventricular cardiomyopythy [ARVC]). Also for few vascular disorders, such as Hutchinson-Gilford progeria and supravalvular aortic stenosis syndromes, patient hiPSC-derived SMCs have been proved to model disease traits in vitro.
All of the so-far-available patient-specific hiPSC models of cardiovascular diseases have some common characteristics: the genetic bases of the disorders are quite well understood, with mutations in single genes being the underlying cause (monogenic diseases); the disease phenotypes develop in a paradigmatically cell-autonomous manner in single cardiomyocytes or SMCs; the pathologies have developmental or early-onset; differentiation protocols directing hiPSCs toward the cells of interest affected by the disease are available; and the phenotypic traits of the disease can be directly assessed in vitro by means of specific cellular assays (Table 1). However, most of the cardiovascular disorders in adults and CHD in newborns are multifactorial or polygenic and involve multiple interconnected tissue and cell types. Moreover, many cardiac maladies have late adult onset. The challenges now are to develop disease-modeling strategies for studying such complex cardiovascular disorders and to use hiPSC disease models to gain new insights into the molecular mechanisms leading to the disease to generate new therapeutic interventions.
Future Challenges and Considerations
Modeling of Complex Cardiovascular Disorders
During reprogramming of somatic cells to pluripotency, epigenetic histories are erased to a large extent (Ohi et al. 2011; Onder and Daley 2012; Hewitt and Garlick 2013) and standard tissue culture conditions normalize all environmental differences. We direct interested readers on epigenetics during reprogramming to recent comprehensive reviews (Onder and Daley 2012; Hewitt and Garlick 2013; Papp and Plath 2013). Of the genetic, environmental, and epigenetic variations that precipitate the disease in the patient, only the genetic variation is likely to be faithfully maintained in hiPSCs. Thus, the value of patient-specific hiPSC-based modeling for complex and multifactorial disorders is likely to be directly proportional to the disease heritability. In the case of complex cardiovascular diseases that show significant heritability but no Mendelian inheritance pattern, the genetic disease predisposition would still be present in the hiPSCs and their derivatives. In the past few years, genome-wide association studies (GWAS) have linked genetic variants with specific complex cardiac diseases, such as coronary artery disease (CAD) and myocardial infarction (MI). At least 33 loci have been identified to be associated with CAD and MI (reviewed by Kathiresan and Srivastava 2012), but it has been difficult to confirm the impact of these gene variants through experimental functional assessment in animal disease models. In all of these GWAS studies, a locus on chromosome 9p21 has emerged as most strongly associated with the clinical phenotypes, which has been suggested to represent a novel risk pathway involving vascular tissue (Helgadottir et al. 2008). The challenge is now to move from statistical association based on genetic markers to identification and functional characterization of individual risk-associated genes, gene variants, biological pathways, and proteins, and to elucidation of their roles in the resulting vascular pathology. Generation of hiPSC cellular models from patients with risk-offering DNA variants and protective DNA variants in the GWAS-identified 9p21 locus, in combination with gene expression profiling and gene silencing or overexpression approaches, as well as specific differentiation to all diverse vascular cell lineages and eventually generation of vessel-organoid systems, would facilitate such functional characterization and eventually help identification of new therapeutics. Also CHDs, occurring in approximately 1% of newborns, are now known to have genetic causes in some patients, with several of the genes affected being key players in cardiac development, such as members of the T-box family, GATA4, NKX2-5, HAND1, and NOTCH genes (Bruneau 2008; Fahed et al. 2013). A common feature of most inherited and sporadic CHDs is that the same disorder can arise from mutations in a variety of genes, and mutations in one gene can give rise to a variety of CHDs. The identification of distinct populations of cardiovascular precursors forming specific compartments of the heart hints at a new approach to understanding CHDs, not as a defect in a specific gene or transcription factor, but rather as a defect in the lineage decisions of a defined subset of cardiac precursors. In this manner, CHDs might be associated with alterations in the formation, expansion, and differentiation of embryonic cardiovascular progenitor cells, which in turn form essential components of the heart, such as the atria, ventricles, outflow tract, coronary arteries, and conduction system (see Brade et al. 2013). The ability to obtain distinct populations of embryonic cardiovascular progenitors from hPSCs and further differentiate them into specific cardiac muscle or vascular cell types (as discussed above) renders hPSCs particularly valuable for generation of human cellular models of CHDs. However, efficient methods to purify the desired subset of cardiovascular progenitors and their differentiated derivatives, as well as tissue-engineering approaches to mimic developmental cues in three-dimensional organoid systems are mandatory. If ethically possible, transplantation of cardiovascular progenitors from disease-specific hPSCs (either patient-specific iPSCs or genetically modified hESCs) into mouse embryos will be extremely informative for dissecting disease pathways and mechanisms that may appear only in an in vivo setting (discussed below).
Purification and Lineage Marking of Cardiovascular Progenitors and Cardiomyocyte Subtypes
Recently, many research efforts have been made in trying to identify cell-specific surface antigens that could allow antibody-based purification and tracking of specific cardiovascular progenitor populations and differentiated cardiomyocytes subtypes arising from hPSCs. The receptor tyrosine kinases KDR (FLK1/VEGFR2) in combination with PDGFRA or the stage-specific embryonic antigen 1 (SSEA-1) have been used to isolate cardiovascular progenitor populations after treatment of hPSCs with specific cardiac morphogens (Yang et al. 2008; Blin et al. 2010; Kattman et al. 2011). The recent identification of markers expressed specifically on cardiomyocytes, including EMILIN2 (Van Hoof et al. 2010; Dubois et al. 2011), SIRPA (Dubois et al. 2011; Elliott et al. 2011), and VCAM (Elliott et al. 2011; Uosaki et al. 2011), has made it possible to isolate highly enriched populations of these cells from hPSCs by FACS or magnetic bead sorting. Despite these previous findings, the quest for appropriate surface antigens specific for distinct hPSC-derived early cardiac progenitors (FHF and SHF progenitors) or cardiomyocyte subtypes (ventricular, atrial, and conductive system cells) still remains a relevant issue; indeed, in some cell types, highly specific surface markers may not even exist.
The modification of hPSCs by introduction of ectopic reporters or by targeting developmentally important loci is becoming a very useful tool, not only for understanding cardiac lineage specification and differentiation, but also for cell-type-specific lineage marking. Selectable markers, such as a fluorescent marker or an antibiotic resistance gene, under the control of lineage-specific promoters allow for purification of precise cell populations by either FACS or selection with antibiotics. In addition, fluorescent transgenic or knockin reporters may facilitate the tracking of diseased cells in cocultures/organoid systems and in chimeric animals after grafting or transplantation.
Using this approach, engineered hESC lines have been generated to fluorescently mark SHF ISL-1+ cardiovascular progenitors and their progenies (Bu et al. 2009) or NKX2-5+ cardiac precursors and deriving cardiomyocytes (Elliott et al. 2011). Similarly, the human α-myosin heavy-chain (α-MHC) promoter coupled to a bicistronic reporter (GFP and puromycin) has been used to achieve high purity of cardiomyocytes (>90%) from differentiating hESCs (Anderson et al. 2007). Marking and purification of distinct subtypes of hPSC-derived cardiomyocytes would be extremely important for disease modeling of several cardiac disorders affecting specifically atrial, ventricular, or conductive system cells or for subtype-specific cellular investigation of disease phenotypes. Identification of ventricular cardiomyocytes can be obtained by delivery of a selectable human myosin light chain-2v (MLC2v) enhancer driving EGFP expression via lentiviral transgene (Huber et al. 2007). Generation of reporters for the other cardiomyocyte subtypes has been hampered by the fact that, owing to the differences in gene expression profiles between hPSC-derived cardiomyocytes and their adult counterparts, classical markers for adult atrial myocytes—such as the atrial isoform of MLC2 (MLC2a), and adult cells of the conductive system, such as the hyperpolarization-activated cyclic nucleotide-gated potassium channel 4 (HCN4) or contactin-2 (CNTN2)—are not very specific for these lineages in early hPSC-derived cardiomyocytes. Identification of promoter elements that could be used for specific hPSC-derived cardiomyocyte-type selection or, alternately, strategies to improve their maturity level are current topics of extensive research.
Cardiac Tissue Engineering and In Vivo Cardiac Disease Modeling
The development of cell culture organoids from PSCs is a valuable approach to assess both cellular differentiation potential and interactions between different cell types of the same genetic background and may be essential for modeling complex disorders with noncell autonomous phenotype, such as CHDs. Cardiac muscle engineering holds the promise to engineer a cardiogenic niche in vitro with the opportunity to study and modify potential niche components—such as extracellular matrix, growth factors, physical stimuli, and cell–cell contacts—under defined conditions. This, in combination with genetic cell engineering, should provide a deeper understanding of processes governing cardiovascular progenitor differentiation, as well as improve cardiomyocyte maturation and assess the role of potential disease modifiers in affecting disease phenotypes of complex cardiovascular disorders. Since the first description 15 years ago, engineered heart tissue (EHT) is evolving rapidly and addition of mechanical loading and electrical stimulation, higher O2 content, and various medium supplements and/or miniaturization have led to improved 3D cardiac tissue structure, maturation of myocytes, reduced extracellular matrix content, and development of near-physiological contractile force (Eschenhagen et al. 2012). However, even with all of these improvements, full manifestation of adult cardiomyocyte phenotypes has not been shown thus far and the major application of hESC/hiPSC-derived EHT constructs has been drug testing (Schaaf et al. 2011).
Although cardiac tissue engineering will be extremely useful for modeling complex cardiovascular disorders, it is necessary to integrate in vivo systems with PSC biology. In most cases, such as in the CAD, MI, and CHD examples previously mentioned, investigators will need to continue using both human PSCs and mouse models to dissect polygenic traits. The advantages of using human cells are amplified by exploring interactions with in vivo environments. A fundamental way to test these interactions is through the transplantation of cardiovascular progenitors (for CHD) or differentiated derivatives (for CAD and MI) from patient-specific hiPSCs into mouse models, in which the cell-autonomous contribution of the hiPSC derivatives to the disease can be tested after physiologically relevant integration in vivo.
PSCs and Personalized Medicine
Aside from disease modeling, PSCs have an exciting and novel application in the field of “personalized medicine” that is worth mentioning. When combined with massively parallel DNA sequencing (whole exome and whole genome sequencing) and high-throughput pharmacological screenings, patient-specific hiPSCs that carry all disease-relevant genetic alterations represent valuable platforms for providing insights into factors that predispose individuals to develop overt symptoms and for revealing drug sensitivities. The influence of the genetic background on the manifestation of cardiovascular disorders is still poorly understood and substantial discrepancies remain in estimating the heritability of numerous quantitative traits related to these diseases. Moreover, it is hard in the clinic to foresee which individual receiving a particular drug will benefit or suffer toxic side effects. A panel of diseased PSC lines whose genomes are fully characterized might allow in vitro analyses predicting both drug effectiveness and toxicity at an individual level against a specific genetic background, as well as identifying biomarkers for the development of diagnostic assays. This will be beneficial for the improvement of preventive public health strategies and could open new landscapes for more effective treatment (“personalized medicine”) and reduced complications.
DIRECT REPROGRAMMING OF SOMATIC CELLS TO THE CARDIAC LINEAGE AS A TOOL TO GENERATE PATIENT-SPECIFIC MODELS OF CARDIOVASCULAR DISEASE
The original publication of iPSC reprogramming has inspired researchers to attempt manipulations of cellular identity in new and unexpected directions. Ectopic transcription factor expression is now being investigated as a tool to perform direct conversion, or “transdifferentiation,” of one differentiated cell type to another. Direct lineage reprogramming of mouse and human fibroblasts into cardiomyocytes, without reversion to a cardiac progenitor cell state, has been achieved by exogenous expression of Gata4, Mef2c, and Tbx5, all key factors involved in cardiogenesis (Ieda et al. 2010; Nam et al. 2013). “Induced cardiomyocytes” (iCMs) appear similar to neonatal cardiomyocytes in global gene expression profile and electrophysiologically, and contract spontaneously (Ieda et al. 2010). In alternative, cardiomyocytes have been generated by partially reprogramming fibroblasts with the Yamanaka factors and controlling cell fate by cardiogenic media supplementation and inhibition of JAK/STAT-driven pluripotency induction (Efe et al. 2011). Direct and partial cardiac reprogramming, which do not involve a progenitor intermediate, may have the advantage that future refinement and characterization of the reprogramming process could allow to obtain more mature cell types for cardiac disease modeling. The inability to expand mature, transdifferentiated cells is currently a rate-limiting factor of this approach, but future improvements in efficiency of reprogramming may overcome this hurdle. A recent study has also reported that a unique combination of transcription factors, ETS2 and MESP1, is sufficient to reprogram human dermal fibroblasts into cardiac progenitors (Islas et al. 2012). It remains to be seen whether ETS2/MESP1-transdifferentiated cells can be efficiently expanded and further maturated into terminal differentiated cardiovascular cells. If this will be possible, they would represent an optimal system to generate unlimited numbers of mature healthy and diseased cardiomyocytes, without the need of pluripotent cell reprogramming.
CONCLUDING REMARKS
The past few years have witnessed remarkable advances in stem cell biology and human genetics, and we have arrived at an era in which patient-specific cell and tissue models are now practical. We have outlined how hPSCs and their derivatives (cardiovascular progenitors or differentiated cells) have been and can be applied for the modeling of cardiovascular disorders of genetic origin. The key challenge now is to implement these models in an efficient manner to study complex cardiovascular diseases and to gain new insights into the disease-causing molecular mechanisms to generate new therapeutic interventions. It will also become important to be able to recapitulate environmental and epigenetic influences with which the relevant genetic and nongenetic factors collude to faithfully reproduce disease phenotypes in vitro. Emerging solutions involve several areas of biomedical research, from tissue engineering to genetic manipulation (Fig. 3), and are likely to be used for producing new molecular understanding of a wide range of cardiovascular diseases.
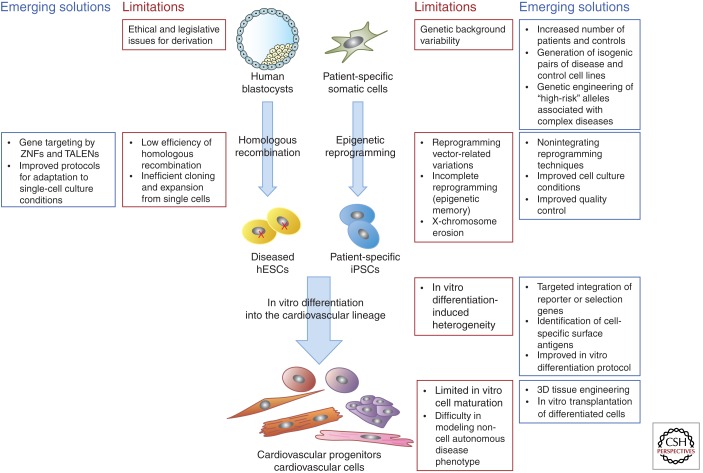
Figure 3.
The limitations of pluripotent stem cells in disease modeling and emerging solutions for identifying disease-related phenotypes.
ACKNOWLEDGMENTS
K.-L.L. thanks the German Research Foundation and the European Research Council for their ongoing support of research in the Laugwitz laboratory. A.M. and D.S. are supported by grants from the German Research Foundation and the German Ministry for Education and Research. K.-L.L. and A.M. also acknowledge the Munich Heart Alliance, a member of the German Centre for Cardiovascular Research.
Footnotes
Editors: Margaret Buckingham, Christine L. Mummery, and Kenneth R. Chien
Additional Perspectives on The Biology of Heart Disease available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Anderson D, Self T, Mellor IR, Goh G, Hill SJ, Denning C 2007. Transgenic enrichment of cardiomyocytes from human embryonic stem cells. Mol Therapy 15: 2027–2036 [DOI] [PubMed] [Google Scholar]
- Blin G, Nury D, Stefanovic S, Neri T, Guillevic O, Brinon B, Bellamy V, Rucker-Martin C, Barbry P, Bel A, et al. 2010. A purified population of multipotent cardiovascular progenitors derived from primate pluripotent stem cells engrafts in postmyocardial infarcted nonhuman primates. J Clin Invest 120: 1125–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Brade T, Pane LS, Moretti A, Chien KR, Laugwitz KL 2013. Embryonic heart progenitors and cardiogenesis. Cold Spring Harb Perspect Med 10.1101/cshperspect.a013847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG 2008. The developmental genetics of congenital heart disease. Nature 451: 943–948 [DOI] [PubMed] [Google Scholar]
- Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, Jasin M 2009. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci 106: 10620–10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, Roberts DJ, Huang PL, Domian IJ, Chien KR 2009. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature 460: 113–117 [DOI] [PubMed] [Google Scholar]
- Bu L, Gao X, Jiang X, Chien KR, Wang Z 2010. Targeted conditional gene knockout in human embryonic stem cells. Cell Res 20: 379–382 [DOI] [PubMed] [Google Scholar]
- Burridge PW, Keller G, Gold JD, Wu JC 2012. Production of de novo cardiomyocytes: Human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell 10: 16–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, Wagner RA, Wilson KD, Xie X, Fu JD, Drukker M, Lee A, Li RA, Gambhir SS, Weissman IL, et al. 2008. Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS ONE 3: e3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao N, Liu Z, Chen Z, Wang J, Chen T, Zhao X, Ma Y, Qin L, Kang J, Wei B, et al. 2012. Ascorbic acid enhances the cardiac differentiation of induced pluripotent stem cells through promoting the proliferation of cardiac progenitor cells. Cell Res 22: 219–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal-Vergara X, Sevilla A, D’Souza SL, Ang YS, Schaniel C, Lee DF, Yang L, Kaplan AD, Adler ED, Rozov R, et al. 2010. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature 465: 808–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C, Bernardo AS, Trotter MW, Pedersen RA, Sinha S 2012. Generation of human vascular smooth muscle subtypes provides insight into embryological origin-dependent disease susceptibility. Nat Biotechnol 30: 165–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver O, Melton DA 2003. Endothelial signaling during development. Nat Med 9: 661–668 [DOI] [PubMed] [Google Scholar]
- Collin J, Lako M 2011. Concise review: Putting a finger on stem cell biology: Zinc finger nuclease-driven targeted genetic editing in human pluripotent stem cells. Stem Cells 29: 1021–1033 [DOI] [PubMed] [Google Scholar]
- Davis RP, Casini S, van den Berg CW, Hoekstra M, Remme CA, Dambrot C, Salvatori D, Oostwaard DW, Wilde AA, Bezzina CR, et al. 2012. Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation 125: 3079–3091 [DOI] [PubMed] [Google Scholar]
- Dubois NC, Craft AM, Sharma P, Elliott DA, Stanley EG, Elefanty AG, Gramolini A, Keller G 2011. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat Biotechnol 29: 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S 2011. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol 13: 215–222 [DOI] [PubMed] [Google Scholar]
- Egashira T, Yuasa S, Suzuki T, Aizawa Y, Yamakawa H, Matsuhashi T, Ohno Y, Tohyama S, Okata S, Seki T, et al. 2012. Disease characterization using LQTS-specific induced pluripotent stem cells. Cardiovasc Res 95: 419–429 [DOI] [PubMed] [Google Scholar]
- Elliott DA, Braam SR, Koutsis K, Ng ES, Jenny R, Lagerqvist EL, Biben C, Hatzistavrou T, Hirst CE, Yu QC, et al. 2011. NKX2–5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods 8: 1037–1040 [DOI] [PubMed] [Google Scholar]
- Eschenhagen T, Eder A, Vollert I, Hansen A 2012. Physiological aspects of cardiac tissue engineering. Am J Physiol Heart Circ Physiol 303: H133–H143 [DOI] [PubMed] [Google Scholar]
- Fahed AC, Gelb BD, Seidman JG, Seidman CE 2013. Genetics of congenital heart disease: The glass half empty. Circ Res 112: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima A, Xu G, Shao K, Papadopoulos S, Lehmann M, Arnaiz-Cot JJ, Rosa AO, Nguemo F, Matzkies M, Dittmann S, et al. 2011. In vitro modeling of ryanodine receptor 2 dysfunction using human induced pluripotent stem cells. Cell Physiol Biochem 28: 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Ren Y, Bartulos O, Lee MY, Yue Z, Kim KY, Li W, Amos PJ, Bozkulak EC, Iyer A, et al. 2012. Modeling supravalvular aortic stenosis syndrome with human induced pluripotent stem cells. Circulation 126: 1695–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F, Boue S, Izpisua Belmonte JC 2011. Methods for making induced pluripotent stem cells: Reprogramming a la carte. Nat Rev Genet 12: 231–242 [DOI] [PubMed] [Google Scholar]
- Gupta MK, Illich DJ, Gaarz A, Matzkies M, Nguemo F, Pfannkuche K, Liang H, Classen S, Reppel M, Schultze JL, et al. 2010. Global transcriptional profiles of beating clusters derived from human induced pluripotent stem cells and embryonic stem cells are highly similar. BMC Dev Biol 10: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgadottir A, Thorleifsson G, Magnusson KP, Gretarsdottir S, Steinthorsdottir V, Manolescu A, Jones GT, Rinkel GJ, Blankensteijn JD, Ronkainen A, et al. 2008. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet 40: 217–224 [DOI] [PubMed] [Google Scholar]
- Hewitt KJ, Garlick JA 2013. Cellular reprogramming to reset epigenetic signatures. Mol Aspects Med 34: 841–848 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. 2009. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 27: 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. 2011. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29: 731–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HP, Chen PH, Hwu WL, Chuang CY, Chien YH, Stone L, Chien CL, Li LT, Chiang SC, Chen HF, et al. 2011. Human Pompe disease-induced pluripotent stem cells for pathogenesis modeling, drug testing and disease marker identification. Hum Mol Genet 20: 4851–4864 [DOI] [PubMed] [Google Scholar]
- Huber I, Itzhaki I, Caspi O, Arbel G, Tzukerman M, Gepstein A, Habib M, Yankelson L, Kehat I, Gepstein L 2007. Identification and selection of cardiomyocytes during human embryonic stem cell differentiation. FASEB J 21: 2551–2563 [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D 2010. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142: 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion S, Luche H, Gadue P, Fehling HJ, Kennedy M, Keller G 2007. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat Biotechnol 25: 1477–1482 [DOI] [PubMed] [Google Scholar]
- Islas JF, Liu Y, Weng KC, Robertson MJ, Zhang S, Prejusa A, Harger J, Tikhomirova D, Chopra M, Iyer D, et al. 2012. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci 109: 13016–13021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, et al. 2011a. Modelling the long QT syndrome with induced pluripotent stem cells. Nature 471: 225–229 [DOI] [PubMed] [Google Scholar]
- Itzhaki I, Rapoport S, Huber I, Mizrahi I, Zwi-Dantsis L, Arbel G, Schiller J, Gepstein L 2011b. Calcium handling in human induced pluripotent stem cell derived cardiomyocytes. PLoS ONE 6: e18037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki I, Maizels L, Huber I, Gepstein A, Arbel G, Caspi O, Miller L, Belhassen B, Nof E, Glikson M, et al. 2012. Modeling of catecholaminergic polymorphic ventricular tachycardia with patient-specific human-induced pluripotent stem cells. J Am Coll Cardiol 60: 990–1000 [DOI] [PubMed] [Google Scholar]
- Jacoby D, McKenna WJ 2012. Genetics of inherited cardiomyopathy. Eur Heart J 33: 296–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM 2000. Fate of the mammalian cardiac neural crest. Development 127: 1607–1616 [DOI] [PubMed] [Google Scholar]
- Jung CB, Moretti A, Mederos y Schnitzler M, Iop L, Storch U, Bellin M, Dorn T, Ruppenthal S, Pfeiffer S, Goedel A, et al. 2012. Dantrolene rescues arrhythmogenic RYR2 defect in a patient-specific stem cell model of catecholaminergic polymorphic ventricular tachycardia. EMBO Mol Med 4: 180–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Srivastava D 2012. Genetics of human cardiovascular disease. Cell 148: 1242–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G 2011. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8: 228–240 [DOI] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. 2010. Epigenetic memory in induced pluripotent stem cells. Nature 467: 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S, Kan NG, Forcales S, Puri PL, Leone TC, et al. 2013. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature 494: 105–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, et al. 2007. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol 25: 1015–1024 [DOI] [PubMed] [Google Scholar]
- Lahti AL, Kujala VJ, Chapman H, Koivisto AP, Pekkanen-Mattila M, Kerkela E, Hyttinen J, Kontula K, Swan H, Conklin BR, et al. 2012. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis Model Mech 5: 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, Han L, Yen M, Wang Y, Sun N, et al. 2013. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell 12: 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Kim E, Kim JS 2010. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res 20: 81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieu DK, Liu J, Siu CW, McNerney GP, Tse HF, Abu-Khalil A, Huser T, Li RA 2009. Absence of transverse tubules contributes to non-uniform Ca2+ wavefronts in mouse and human embryonic stem cell-derived cardiomyocytes. Stem Cells Dev 18: 1493–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundy SD, Zhu WZ, Regnier M, Laflamme M 2013. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev 22: 1991–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Wei H, Lu J, Ho S, Zhang G, Sun X, Oh Y, Tan SH, Ng ML, Shim W, et al. 2013. Generation of patient-specific induced pluripotent stem cell-derived cardiomyocytes as a cellular model of arrhythmogenic right ventricular cardiomyopathy. Eur Heart J 34: 1122–1133 [DOI] [PubMed] [Google Scholar]
- Majesky MW 2007. Developmental basis of vascular smooth muscle diversity. Arterioscler Thromb Vasc Biol 27: 1248–1258 [DOI] [PubMed] [Google Scholar]
- Martin CA, Matthews GD, Huang CL 2012. Sudden cardiac death and inherited channelopathy: The basic electrophysiology of the myocyte and myocardium in ion channel disease. Heart 98: 536–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsa E, Rajamohan D, Dick E, Young L, Mellor I, Staniforth A, Denning C 2011. Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation. Eur Heart J 32: 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa T, Gourdie RG 1996. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol 174: 221–232 [DOI] [PubMed] [Google Scholar]
- Moretti A, Bellin M, Jung CB, Thies TM, Takashima Y, Bernshausen A, Schiemann M, Fischer S, Moosmang S, Smith AG, et al. 2010a. Mouse and human induced pluripotent stem cells as a source for multipotent Isl1+ cardiovascular progenitors. FASEB J 24: 700–711 [DOI] [PubMed] [Google Scholar]
- Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flugel L, Dorn T, Goedel A, Hohnke C, Hofmann F, et al. 2010b. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med 363: 1397–1409 [DOI] [PubMed] [Google Scholar]
- Mummery C, Ward-van Oostwaard D, Doevendans P, Spijker R, van den Brink S, Hassink R, van der Heyden M, Opthof T, Pera M, de la Riviere AB, et al. 2003. Differentiation of human embryonic stem cells to cardiomyocytes: Role of coculture with visceral endoderm-like cells. Circulation 107: 2733–2740 [DOI] [PubMed] [Google Scholar]
- Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R, et al. 2013. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci 110: 5588–5593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak A, Barad L, Zeevi-Levin N, Shick R, Shtrichman R, Lorber A, Itskovitz-Eldor J, Binah O 2012. Cardiomyocytes generated from CPVTD307H patients are arrhythmogenic in response to β-adrenergic stimulation. J Cell Mol Med 16: 468–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, Qi Z, Downey SL, Manos PD, Rossi DJ, et al. 2011. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol 13: 541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder TT, Daley GQ 2012. New lessons learned from disease modeling with induced pluripotent stem cells. Curr Opin Genet Dev 22: 500–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige SL, Osugi T, Afanasiev OK, Pabon L, Reinecke H, Murry CE 2010. Endogenous Wnt/β-catenin signaling is required for cardiac differentiation in human embryonic stem cells. PLoS ONE 5: e11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Plath K 2013. Epigenetics of reprogramming to induced pluripotency. Cell 152: 1324–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ 2008. Disease-specific induced pluripotent stem cells. Cell 134: 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinton DA, Daley GQ 2012. The promise of induced pluripotent stem cells in research and therapy. Nature 481: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satin J, Itzhaki I, Rapoport S, Schroder EA, Izu L, Arbel G, Beyar R, Balke CW, Schiller J, Gepstein L 2008. Calcium handling in human embryonic stem cell-derived cardiomyocytes. Stem Cells 26: 1961–1972 [DOI] [PubMed] [Google Scholar]
- Schaaf S, Shibamiya A, Mewe M, Eder A, Stohr A, Hirt MN, Rau T, Zimmermann WH, Conradi L, Eschenhagen T, et al. 2011. Human engineered heart tissue as a versatile tool in basic research and preclinical toxicology. PLoS ONE 6: e26397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, et al. 2012. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med 4: 130ra147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- Tanasijevic B, Dai B, Ezashi T, Livingston K, Roberts RM, Rasmussen TP 2009. Progressive accumulation of epigenetic heterogeneity during human ES cell culture. Epigenetics 4: 330–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM 1998. Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147 [DOI] [PubMed] [Google Scholar]
- Uosaki H, Fukushima H, Takeuchi A, Matsuoka S, Nakatsuji N, Yamanaka S, Yamashita JK 2011. Efficient and scalable purification of cardiomyocytes from human embryonic and induced pluripotent stem cells by VCAM1 surface expression. PLoS ONE 6: e23657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach A, Schuldiner M, Benvenisty N 2004. Modeling for Lesch-Nyhan disease by gene targeting in human embryonic stem cells. Stem Cells 22: 635–641 [DOI] [PubMed] [Google Scholar]
- Van Hoof D, Dormeyer W, Braam SR, Passier R, Monshouwer-Kloots J, Ward-van Oostwaard D, Heck AJ, Krijgsveld J, Mummery CL 2010. Identification of cell surface proteins for antibody-based selection of human embryonic stem cell-derived cardiomyocytes. J Proteome Res 9: 1610–1618 [DOI] [PubMed] [Google Scholar]
- Waldo KL, Hutson MR, Ward CC, Zdanowicz M, Stadt HA, Kumiski D, Abu-Issa R, Kirby ML 2005. Secondary heart field contributes myocardium and smooth muscle to the arterial pole of the developing heart. Dev Biol 281: 78–90 [DOI] [PubMed] [Google Scholar]
- Wasteson P, Johansson BR, Jukkola T, Breuer S, Akyurek LM, Partanen J, Lindahl P 2008. Developmental origin of smooth muscle cells in the descending aorta in mice. Development 135: 1823–1832 [DOI] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. 2011. Targeted genome editing across species using ZFNs and TALENs. Science 333: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, et al. 2008. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 453: 524–528 [DOI] [PubMed] [Google Scholar]
- Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, Dolmetsch RE 2011. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature 471: 230–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920 [DOI] [PubMed] [Google Scholar]
- Zhang J, Lian Q, Zhu G, Zhou F, Sui L, Tan C, Mutalif RA, Navasankari R, Zhang Y, Tse HF, et al. 2011a. A human iPSC model of Hutchinson Gilford Progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell 8: 31–45 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Jiang J, Han P, Yuan Q, Zhang J, Zhang X, Xu Y, Cao H, Meng Q, Chen L, et al. 2011b. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res 21: 579–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu WZ, Xie Y, Moyes KW, Gold JD, Askari B, Laflamme MA 2010. Neuregulin/ErbB signaling regulates cardiac subtype specification in differentiating human embryonic stem cells. Circ Res 107: 776–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, et al. 2009. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell 5: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]