Abstract
Chemical genetics has evolved into a powerful tool for studying gene function in normal- and patho-biology. PKR and PERK, two eukaryotic translation initiation factor 2 alpha (eIF2α) kinases, play critical roles in maintenance of cellular hemostasis, metabolic stability, and anti-viral defenses. Both kinases interact with and phosphorylate additional substrates including tumor suppressor p53 and nuclear protein 90. Loss of function of both kinases has been studied by reverse genetics and recently identified inhibitors. In contrast, activating probes for studying the role of catalytic activity of these kinases are not available. We identified a 3-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-5,7-dihydroxy-4H-chromen-4-one (DHBDC) as specific dual activator of PKR and PERK by screening a chemical library of 20,000 small molecules in a dual luciferase surrogate eIF2α phosphorylation assay. We present here extensive biological characterization and preliminary structure-activity relationship of DHBDC, which phosphorylate eIF2α by activating PKR and PERK but no other eIF2α kinases. These agents also activate downstream effectors of eIF2α phosphorylation; inducing CHOP and suppressing cyclin D1 expression and inhibiting cancer cell proliferation, all in a manner dependent on PKR and PERK. Consistent with the role of eIF2α phosphorylation in viral infection, DHBDC inhibits proliferation of human hepatitis C virus. Finally, DHBDC induces phosphorylation of Ikβα, and activates NF-κB pathway. Surprisingly, activation of NF-κB pathway is dependent on PERK but independent of PKR activity. These data indicate that DHBDC is an invaluable probe for elucidating the role of PKR and PERK in normal- and patho-biology.
Keywords: PKR, PERK, NF-κB, endoplasmic reticulum stress, eIF2α
Introduction
Protein Kinase R (PKR) and protein kinase R like kinase (PERK) are two ubiquitously expressed eukaryotic translation initiation factor 2 alpha (eIF2α) kinases that play critical roles in normal- and patho-biology.[1] PKR is an integral component of interferon regulated anti-viral defenses.[2] In addition to phosphorylating eIF2α to shut down protein synthesis and thereby inhibiting production of viral particles, PKR also activates NF-κB signaling with wide ranging consequences,[1d, 3] including induction of cytokines to further inhibit viral growth and spread.[4] Whether or not NF-κB activation requires catalytic activity of PKR is however, a matter of dispute. PKR has also been implicated in the RNA splicing, long-term potentiation, neuro-degeneration, and the development of type II diabetes.[5] PERK is highly expressed in endocrine pancreas and is activated by endoplasmic reticulum (ER) stress or glucose deprivation, and plays essential role in restoring ER-homeostasis and cell survival.[6] In addition to eIF2α, PERK phosphorylates nuclear factor 2 (nrf2)[7] and activates NF-κB signaling.[8] Ablation of PERK in mouse causes demise of β-cells in the pancreas and results in type-I diabetes like syndrome[9] while its inactivating mutations in humans cause Wolcott-Rallison syndrome,[10] a rare autosomal recessive disease characterized by neonatal/early-onset non-autoimmune insulin-requiring diabetes associated with skeletal dysplasia and growth retardation.[11] In addition to their roles in anti-viral defense and regulation of cell survival, both PKR and PERK have also been implicated in malignant transformation and tumorigenesis[12] with both anti- and pro-tumorigenic roles being suggested for both kinases.[13]
Forward and reverse molecular genetics approaches are important for studying gene function. Forward genetics which, requires over-expression of a gene may cause appearance of spurious phenotypes while reverse genetics will usually require total gene silencing. In addition to requiring time and expense for establishing cell and animal models, genetic approaches are also confounded by cellular adaptation as the gradual nature of overexpressing or silencing a gene may allow sufficient time for cellular/organismal adaptation. Chemical genetics, on the other hand, allows for instantaneous inhibition or activation of a gene at its physiological concentration and may even allow studying just one function of a multi-functional gene. For example, an enzyme inhibitor will allow for studying only the role of enzymatic activity of gene that may have additional cellular functions. The limitation of genetic approach for studying gene function is best demonstrated by role of PKR and NF-κB activation. Indeed, despite the conduction of multiple studies employing forward and/or reverse genetic approaches, how PKR activates NF-κB signaling remains a matter of controversy.[3c, 8, 14] This controversy may be resolved by using chemical modifiers of PKR.
Chemical inhibitors of both PKR and PERK have been reported in recent years.[15] Discovery of the chemical activators of these two kinases will provide important additional tools needed for studying their role in normal- and patho-biology. To discover PKR and/or PERK activators, we re-constituted our previously reported dual luciferase surrogate eIF2α phosphorylation (ternary complex) assay[16] in CRL-2813 human melanoma cells and screened small chemical entities in various bioactive libraries at Harvard Institute of Chemistry and Cell Biology (ICCB). Here, we report identification and extensive biological characterization of 3-(2,3-dihydroxybenzo[b][1,4]dioxin-6-yl)-5,7-dihydroxy-4H-chromen-4-one (DHBDC) and related compounds as dual specificity PKR and PERK activators. These agents phosphorylate eIF2α, induce downstream effectors of eIF2α phosphorylation including CEBP homolog protein (CHOP/GADD 153), and inhibit proliferation of cancer cells, all in a manner dependent on PKR and PERK. Importantly, DHBDC also inhibits proliferation of human hepatitis C virus (HCV) in vitro and cause phosphorylation of IκBα, and activation of NF-κB signaling. Surprisingly activation of NF-κB pathway by DHBDC is independent of PKR and is fully mediated by PERK.
RESULTS and DISCUSSION
Two eIF2α kinases, PKR and PERK, play critical roles in normal- and patho-biology but we cannot specifically and conveniently activate these two kinases to study their functions in vitro and in vivo. Small chemical activators of PKR and PERK are therefore, highly valuable for studying the functions of these two kinases.
High throughput campaign and identification of eIF2α kinase activators
We have previously reported generation of cell based dual luciferase reporter surrogate assay for identifying agents that induce eIF2α phosphorylation.[16-17] Briefly, this assay takes advantage of the fact that phosphorylation of eIF2α reduces the overall rate of translation initiation while paradoxically increasing the translation of a subset of mRNAs containing multiple upstream open reading frames (ORF) in their 5’untranslated regions (5’UTR). These include mRNA coding for activating transcription factor 4 (ATF-4). We generated an expression plasmid that drives transcription of firefly (F) and renilla (R) luciferase open reading frames (ORF) under the control of a bi-directional promoter/enhancer complex.[16-17] In this assay, F luciferase ORF is fused to 5’UTR of ATF-4 mRNA that has multiple uORFs while R luciferase mRNA is fused to a 5’UTR lacking any uORFs. Compounds that cause phosphorylation of eIF2α would increase F luciferase while decreasing the R luciferase expression, resulting in an increased F/R luciferase ratio (F/R).
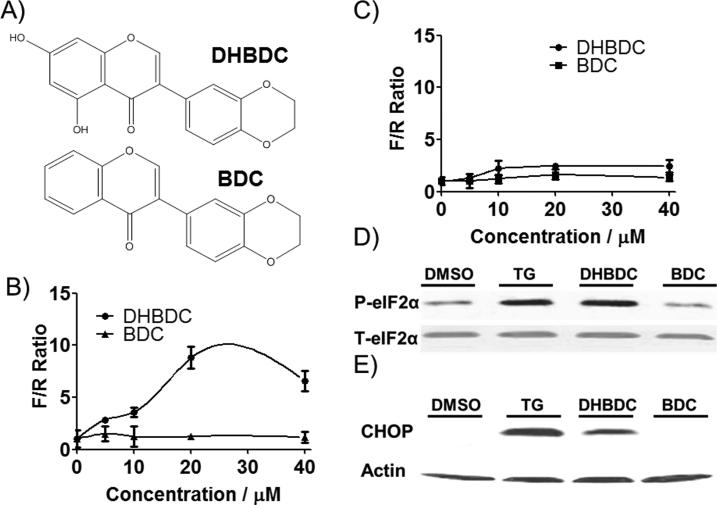
We stably transfected CRL-2813 cells with this bi-directional dual luciferase expression vector (Figure S1) and selected stable cell lines. We chose CRL-2813 cells because they were very sensitive to modifiers of eIF2α phosphorylation.[18] To calculate the activity scores for test compounds, the F/R ratios in compound treated wells were normalized to vehicle (DMSO) treated wells (F/R = 1). After screening various ICCB libraries we selected 38 compounds (Table S1) for confirmation in the same assay. Hit compounds were also counter-screened for activation of inositol requiring 1 (Ire1) catalyzed X-box binding protein 1 (Xbp-1) mRNA splicing; this counter-screen eliminates agents that induce eIF2α phosphorylation by causing ER-stress. These studies identified 3-(2,3-dihydroxybenzo[b][1,4]dioxin-6-yl)-5,7-dihydroxy-4H-chromen-4-one (DHBDC) that was active in the surrogate eIF2α phosphorylation assay without any effect on a similar dual luciferase Ire1 activity assay. We obtained this compound as well as several close analogs for in-depth characterization and molecular target identification. Figure 1A shows structure of DHBDC and its inactive analog, 3-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-4H-chromen-4-one (BDC). Figure 1B shows that DHBDC but not BDC increase F/R in the surrogate reporter assay for eIF2α phosphorylation, while Figure 1C shows that both compounds are inactive in dual luciferase surrogate Ire1 activity assay. To determine whether DHBDC increases F/R ratio by causing eIF2α phosphorylation, we treated cells with DHBDC or BDC and blotted extracts with antibodies specific to phosphorylated eIF2α, total eIF2α or the downstream target and effector of phosphorylated eIF2α, CHOP/GADD153. As shown in Figure 1D, DHBDC but not BDC induces eIF2α phosphorylation and expression of CHOP. Taken together, these data indicate that DHBDC induces eIF2α phosphorylation and thereby activates its downstream effectors independent of general endoplasmic reticulum stress response.
Figure 1.
Characterization of hit compounds. A) Structure of active compound DHBDC and its inactive analog BDC. B) Dose dependent activity of DHBDC and BDC in the dual luciferase surrogate eIF2α phosphorylation assay. C) Activity of the DHBDC and BDC in dual luciferase Xbp-1 mRNA splicing counter-assay. D) Induction of eIF2α phosphorylation and E) CHOP by DHBDC and BDC, Thapsigargin (TG) is used as a positive control. F/R = F to R luciferase ratio. The experiment was conducted in triplicate and each experiment was independently performed three times. Data are shown as Mean±S.E.M.
Identifying eIF2α kinase(s) activated by DHBDC
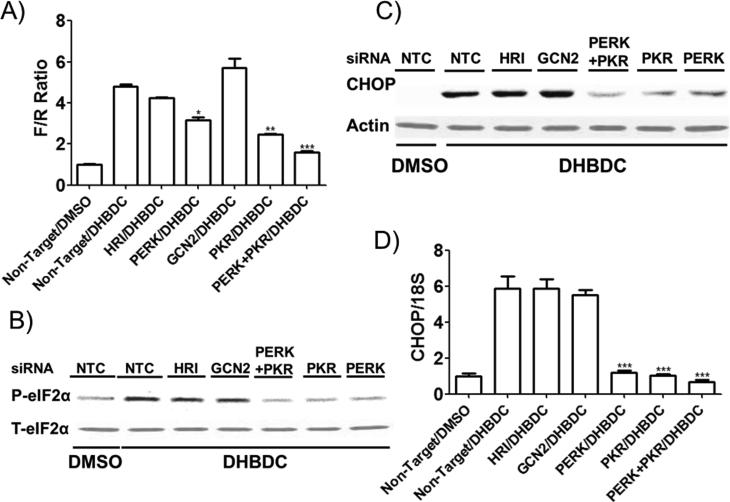
To determine whether induction of eIF2α phosphorylation is caused by kinase activation and if so to identify the responsible kinase(s), we transfected reporter CRL-2813 cells with siRNAs directed against each of the four eIF2α kinases; PERK, PKR, heme regulated inhibitor kinase (HRI), or general response depressible 2 (GCN2) individually or in all combinations using non-targeted siRNA transfected cells as control. Treatment of transfected cells with DMSO or DHBDC indicated that knocking down either PERK or PKR significantly reduce while simultaneously knocking down both PERK and PKR almost totally abrogate DHBDC's activity (Figure 2A) in this assay. To confirm these results, we investigated the dependence of DHBDC induced eIF2α phosphorylation as well as expression of its downstream effector, CHOP, on PKR and PERK. Figures 2B, 2C, and 2D demonstrate that induction of eIF2α phosphorylation (2B) and CHOP protein (2C) and mRNA (2D) expression is mediated by PKR and PERK. These data indicate that DHBDC is a dual activator of PKR and PERK.
Figure 2.
Identifying PKR and PERK as the molecular targets of DHBDC. A) Expression of all four eF2α kinases was knocked-down individually or in all combination by transfecting cell with siRNAs specific to all four eIF2α kinases or non-targeted siRNA and the effect of DHBDC on the ternary complex assay was determined. Only PKR and PERK dual knock-down is shown. B) Expression of all four eF2α kinases was knocked-down individually or PKR and PERK expression was knocked down simultaneously by transfecting cells with specific siRNAs. Effect of DHBDC treatment on eIF2α phosphorylation in the transfected cells was determined by western blot analysis with antibodies specific to phosphorylated and total eIF2α. C) and D) Expression of all four eF2α kinases was knocked-down as in B and the effect of DHBC treatment on CHOP mRNA expression was determined by real-time PCR (C) and CHOP protein by western blot analysis (D). NTC = Non-Target siRNA Control. F/R = F to R luciferase ratio. The experiment was conducted in triplicate and each experiment was independently performed three times. Data are shown as Mean±S.E.M.
Specificity of DHBDC
To determine if DHBDC specifically activates PKR and PERK or is a promiscuous kinase activator, we evaluated effects of DHBDC on the activity of three unrelated protein kinase pathways, PI3K-Akt, Ras-Raf-MAPK, and mTOR pathways. We treated serum starved mouse embryonic fibroblasts with DHBDC or BDC in the presence or absence of serum and determined the levels of phosphorylated Akt, MAPK, and eIF4E binding protein 4E-BP1 by Western blot analysis of cell extracts. As shown in Figure S2, neither DHBDC nor its inactive analog BDC had any effect on the phosphorylation of Akt, MAPK, or 4E-BP1, indicating that DHBDC is a specific activator of PERK and PKR.
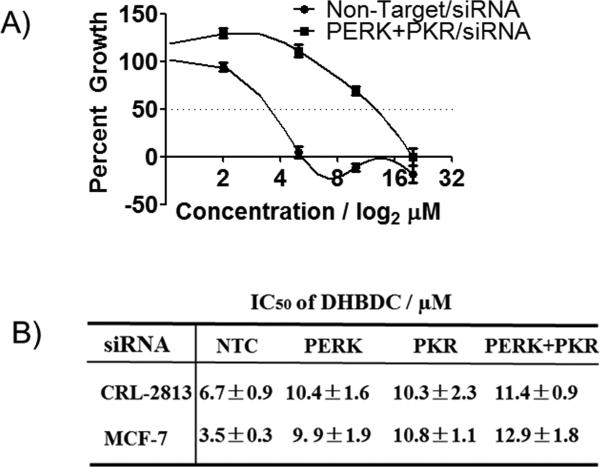
To determine if DHBDC can be utilized for probing the role of PKR and/or PERK in normal- and patho-biology, we determined effect of this aspect, cell proliferation, as a biological response parameter because induction of eIF2α phosphorylation results in inhibition of cell proliferation.[19] Furthermore, cell proliferation is a critical biologic response parameter for measuring the specificity of a compound because it can be influenced by many pathways; a compound with many off-target effects could therefore inhibit cell proliferation independent of its presumed target. We reasoned that if DHBDC is specific activator of PKR and PERK, then it should inhibit cell proliferation in a PKR and PERK dependent manner. To determine whether this is the case, we knocked down the expression of PKR, PERK or both kinases in CRL-2813 melanoma and MCF-7 human breast cancer cells and determined effect of DHBDC on cell proliferation using non-targeted siRNA transfected cells as controls. MCF-7 cells were added to these studies because knock-down efficiency is higher in these cells than in CRL-2813 cells (~90% in MCF-7 cells v.s. ~70% in CRL-2813 cells). As shown in Figure 3A and 3B knocking down expression of PERK, PKR, or both significantly reduced the sensitivity of cells to DHBDC. These data further confirm that DHBDC is a specific dual PKR and PERK activator.
Figure 3.
Dependence of anti-proliferative activity of DHBDC on PKR and PERK. CRL-2813 human melanoma or MCF-7 cells were transfected with siRNAs targeting PKR, PERK, or both and treated with various doses of DHBDC for three days and cell proliferation was quantified by SRB assay. A) Growth inhibition curve for Non-Target Control and both PERK and PKR siRNAs in MCF-7 cells. B) Calculated IC50 for Non-Target Control, PERK, PKR, and both PERK and PKR siRNAs in CRL-2813 and MCF-7 cells. NTC = Non-Target Control. The experiment was conducted in triplicate and each experiment was independently performed three times. Data are shown as Mean±S.E.M.
DHBDC inhibits expression of pro-proliferative genes
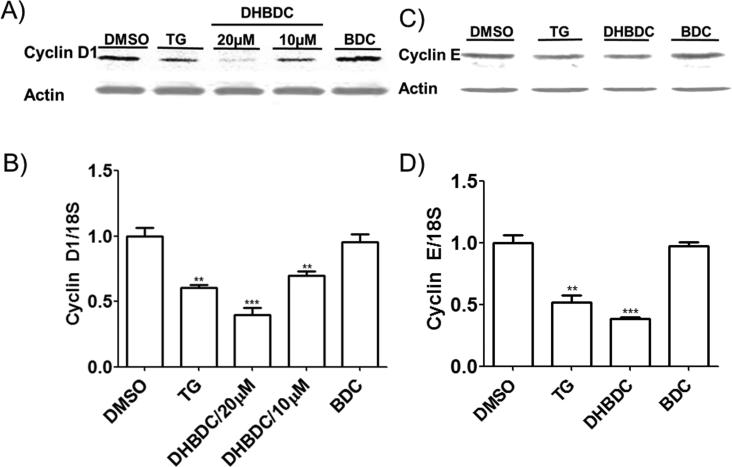
Phosphorylation of eIF2α inhibits translation initiation with profound effects on the expression of pro-proliferative proteins.[16, 19] Furthermore, PKR and PERK activation has been tied to repression of cyclin D1 expression.[1e] To determine if DHBDC preferentially inhibit expression of pro-proliferative proteins, we determined its effect on the expression of cyclin D1, cyclin E, and β-actin by Western blot. As shown in Figure 4A and 4C, DHBDC inhibited expression of cyclin D1 and to a lesser extent cyclin E proteins with minimal effect on the expression of β-actin. To determine if these effects were translational we quantified the mRNA levels of the same panel by real-time quantitative PCR. As shown in Figure 4B, while cyclin D1 mRNA levels were dose-dependently decreased by DHBDC, this was not sufficient to account for the dramatic reduction in the expression of Cyclin D1 protein. The effect of DHBDC on cyclin E mRNA accumulation was negligible (Figure 4D). These data indicate that DHBDC preferentially inhibits expression of oncogenic proteins both transcriptionally and post-transcriptionally.
Figure 4.
DHBDC inhibits expression of cyclin D1 and cyclin E. A) and B) CRL-2813 cells were treated with 10 or 20 μM DHBDC, 20 μM BDC or 50 nM TG and expression of Cyclin D1 protein was determined by Western blot (A) and cyclin D1 mRNA was quantified by real-time PCR (B). C) and D) CRL-2813 cells were treated with 20 μM DHBDC, 20 μM BDC or 50 nM TG and expression of Cyclin E protein was determined by Western blot (C) and cyclin E mRNA was quantified by real-time PCR (D). β-actin was utilized as a loading control for Western blot analysis. The experiment was conducted in triplicate and each experiment was independently performed three times. Data are shown as Mean±S.E.M.
Preliminary structure activity relationship studies
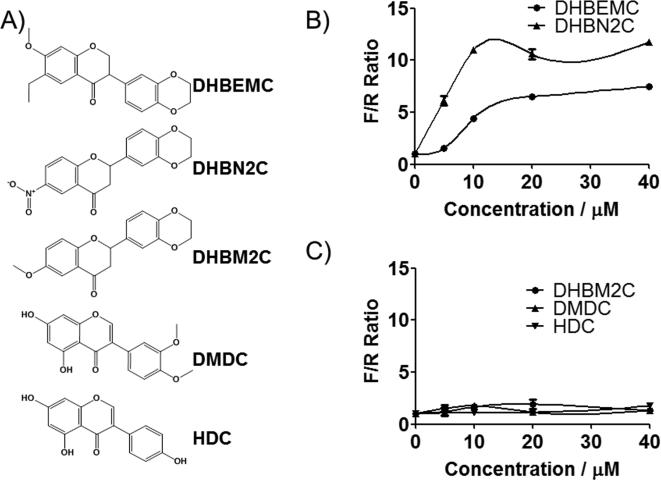
The dual PKR and PERK activator used in these studies, DHBDC, contains an activated alkene in the form of α,β-unsaturated ketone conjugate. This Michael-type acceptor functionality can be a source of cellular toxicity or a target for enhanced metabolic clearance. The inactive compound, BDC, also contains the same Michael-type acceptor suggesting that activation of PKR or PERK does not involve this moiety. To further investigate this matter and to obtain preliminary data on the role of the 5,7-dihydroxyls and the chromen moiety in the activation of PKR or PERK, we obtained additional related derivatives for testing (Figure 5A). These were 3-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-6-ethyl-7-methoxychroman-4-one (DHBEMC), 2-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-6-nitrochroman-4-one (DHBN2C), 2-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-6-methoxychroman-4-one (DHBM2C), 3-(3,4-dimethoxyphenyl)-5,7-dihydroxy-4H-chromen-4-one (DMDC), and 5,7-dihydroxy-3-(4-hydroxyphenyl)-4H-chromen-4-one (HDC). Two of the DHBDC-related analogs that did not contain Michael-type acceptor, namely DHBEMC and DHBN2C, were still active, demonstrating conclusively that Michael-type acceptor is dispensable for the activation of PKR and/or PERK (Figure 5B). Furthermore, in these two compounds, the hydroxyl groups are replaced by other substituents on the ring, indicating that these groups are not strictly required for activity. The [1,4]dioxin ring, on the other hand, is essential for the compounds’ activity as simple opening of this ring (DMDC) or its replacement with hydroxyl group (HDC) resulted in the complete loss of activity (Figure 5C). These data demonstrate that the PKR and PERK kinase activation by DHBDC requires intact 2,3-dihydrobenzo[b][1,4]dioxine ring system as well as the substituted chroman-4-one system and is independent of the presence of a Micheal-type acceptor present in the primary hit compound.
Figure 5.
Choromone ring and electron withdrawing substitution of phenyl ring is necessary but Michael-type acceptor is dispensable for the activity of DHBDC. A) Structure of various DHBDC analogs obtained for the preliminary SAR. B) and C). Activity of these analogs in the surrogate eIF2α phosphorylation assay. Reporter cell lines described in Figure S1 were treated with the indicated concentration of each compound and F/R ratio was determined. F/R = ratio of F to R luciferase. The experiment was conducted in triplicate and each experiment was independently performed three times. Data are shown as Mean±S.E.M.
Divergence of catalytic targets for PKR and PERK
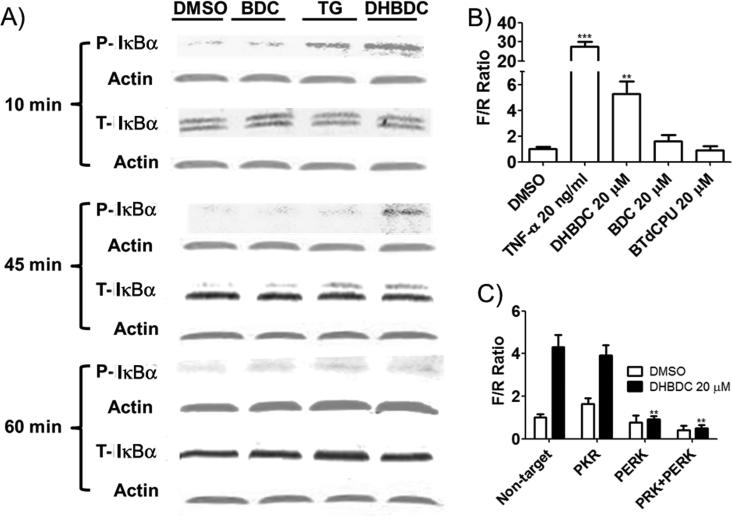
To further probe cellular functions of PKR we took note of the reported effects of these two kinases on NF-κB activity. PKR interacts with and activates NF-κB but whether this activation is dependent on the kinase activity has long been debated.[3c, 20] Similarly, PERK activates NF-κB presumably by depleting IκBαthrough eIF2α phosphorylation dependent suppression of its synthesis.[8] If the activation of NF-κB is dependent on the catalytic activity of PKR and/or PERK, DHBDC should activate this pathway; otherwise the compound should have no effect. To determine which the case is, we treated CRL-2813 cells with DHBDC or its inactive analog BDC and measured phosphorylation of IκBα. As shown in Figure 6A, DHBDC but not inactive analog BDC induced phosphorylation of IκBα, which preceded the time dependent disappearance of this protein. DHBDC had no effect on the phosphorylation or the expression of the p65 subunit of NF-κB (S3). To determine the functional significance of IκBα phosphorylation, we investigated the effect of DHBDC or BDC on activation of NF-κB transcriptional activity in a reporter gene assay using tumor necrosis factor alpha (TNFα) as positive control. We transfected CRL-2813 cells with F luciferase expression plasmid driven by NF-κB dependent promoter and R luciferase expression plasmid driven by thymidine kinase (TK) promoter and treated transfected cells with DHBDC, BDC, TNFα or DMSO. As expected from its effect on the IκBα phosphorylation, DHBDC but not BDC induced expression of NF-κB responsive promoter driven F luciferase with no effect on the expression of TK-promoter driven R luciferase, the internal control (Figure 6B). To determine if the activation of NF-κB transcriptional activity was dependent on eIF2α phosphorylation, we took advantage of BTdCPU, a well-characterized activator of another eIF2α kinase, HRI.[16] If the activation of NF-κB by DHBCD was mediated by eIF2α phosphorylation, then BTdCPU should display similar activity as DHBDC in the reporter gene assay. Figure 6B shows that the HRI activator BTdCPU does not have any effect on the NF-κB reporter gene assay. This data indicate that DHBDC activates NF-κB signaling independently of eIF2α phosphorylation. We then sought to determine whether activation of NF-κB dependent promoter by DHBDC is mediated by PKR and/or PERK. We co-transfected CRL-2813 cells with siRNA targeting PKR and/or PERK and F luciferase and R luciferase expression plasmids; 48 hours after transfection treated cells with DMSO or DHBDC for an additional 10 hours and determined the reporter activity by dual luciferase assay. To our surprise, knocking down PKR expression had no effect on the DHBDC activation of NF-κB reporter while knocking down PERK expression totally abrogated DHBDC induction of NF-κB responsive reporter driven F-luciferase (Figure 6C). Taken together, these data indicate clearly that at least in these cells, PERK but not PKR catalytically activates NF-κB signaling and that this activity is independent of eIF2α phosphorylation.
Figure 6.
DHBDC induces phosphorylation of IκBα and activates NF-κB promoters. A) CRL-2813 cells were treated for the indicated times with vehicle, DHBDC, TG or BDC, cell lysates were separated by SDS-PAGE and probed with antibodies specific to phosphorylated and total IκBα. B) Cells were transfected with a NF-κB responsive promoter driven F luciferase reporter plasmid and TK-promoter driven R luciferase reporter, treated with vehicle, TNF, DHBDC, BDC or BTdCPU and F/R ratio was determined. Almost all the observed change in F/R was accounted for by increased expression of F luciferase. C) Cells were co-transfected with non-targeted siRNA or siRNAs targeting PKR, PERK, or both and NF-κB promoter driven F luciferase and TK-promoter driven R luciferase plasmids used in A and B above. Cells were treated with DHBDC and F/R ratio was determined by dual luciferase assay. F/R = ratio of F to R luciferase. The experiment was conducted in triplicate and each experiment was independently performed three times. Data are shown as Mean±S.E.M.
Another often cited target of PKR and/or PERK is tumor suppressor p53[21] whose phosphorylation is thought to cause its proteasome dependent degradation. To determine if this is a universal effect mediated by the catalytic activity of PKR and/or PERK we determined p53 levels in cells treated with DMSO, DHBDC, or its inactive analog, BDC. Thapsigargin (TG) was used as non-specific activator of PERK. Figure S4 shows that these agents have no effect on p53 expression, indicating that the effects of PKR and/or PERK on p53 expression may be cell-type specific rather than universal.
DHBDC inhibits Hepatitis C Virus (HCV)
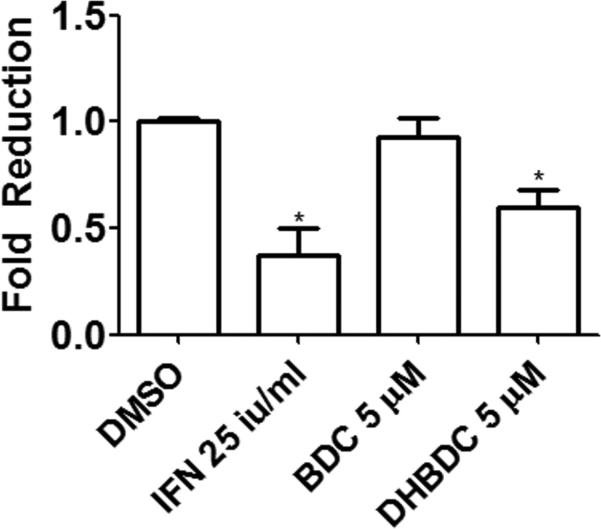
Based on the role of eIF2α phosphorylation on cellular defense against viral infections, we tested the effect of DHBDC and its inactive analog BDC on HCV infection. Human Huh7.5.1 hepatoma cells were treated with 5 μM DHBDC or BDC, interferon alpha (PBL) 25IU/ml, or DMSO. 24 hours later, fully infectious JFH1 HCV was added to cells at the MOI of 1 and after 48 hour further incubation the percent of infected cells was determined. Figure 7 shows that DHBDC significantly reduced JFH1 infection compared to cells treated with DMSO (40% reduction). IFN alone reduced infection by roughly 60%. While preliminary, these results are consistent with a possible antiviral effect expected of a PKR and PERK activating agents.
Figure 7.
DHBDC inhibits HCV infection. Huh7.5.1 human hepatoma cells were treated with 5 μM DHBDC or BDC, interferon alpha (PBL) 25IU/ml, or DMSO for 24 hours fallowed by addition of fully infectious JFH1 HCV. After 48h additional culture cells were fixed in 4% PFA, permeabilized and stained with anti-HCV primary and Alexa-488 tagged secondary antibody then Hoechst DNA stain. Cells were then imaged on an automated microscope at 4X for percent infected cells. Percent infection for all groups was normalized to that of DMSO treated cells. The experiment was conducted in triplicate and each experiment was independently performed three times. Data are shown as Mean±S.E.M.
Mechanism of kinase activation by DHBDC
To determine the direct mechanism of PKR and PERK activation by DHBDC we hypothesized that the compound must act on a common element involved in the activation of these two kinases. Both PKR and PERK bind an inhibitory protein, p58IPK. Disruption of p58IPK's interaction with PKR or PERK results in the activation of these kinases. We therefore sought to determine if DHBDC activates PKR and PERK by inhibiting their interaction with p58IPK. We treated cells with DHBDC or vehicle, immunoprecipitated lysates with PKR or p58IPK specific antibodies, separated the immuncomplexes by SDS-PAGE and immonoblotted with antibodies against PKR and p58IPK. As shown in Figure S5, treatment with DHBDC had no effect on the interaction of PKR with p58IPK. Similarly DHBDC did not have any discernible effect on the interaction of PKR with its interaction partner and activator, the protein activator of PKR (PACT). Our attempts at identifying interaction of DHBDC with PKR by NMR chemical shift assay were not successful due to limiting solubility of DHBDC in the NMR buffer. The precise molecular mechanism of PKR and/or PERK activation by DHBDC must therefore await additional studies that could be facilitated by development of a more soluble analog.
Numbers of kinase inhibitors have long been utilized to probe the role of their targets in normal- and patho-biology and as drugs for treatment of human disorders. The repertoire of kinase activators on the other hand is very limited. We report here that a dual PKR and PERK activator, DHBDC, induces eIF2α phosphorylation as well as expression of its downstream effector, CHOP, suppresses expression of cyclin D1 and cyclin E, and inhibits cell proliferation; all mediated by PKR and PERK. We further demonstrate that DHBDC but not its inactive analog BDC induces IκBα phosphorylation and activates NF-κB signaling. Activation of the NF-κB signaling is independent of eIF2α phosphorylation because BTdCPU, which activates another eIF2α kinase, HRI, has no effect on NF-κB reporter assay. Perhaps the biggest surprise of our studies is that activation of NF-κB pathway by DHBDC is fully mediated by PERK activity with no contribution from PKR (Figure 6C). Finally DHBDC appear to reduce human HCV infectivity (Figure 7).
In summary, the dual PKR and PERK activator described here significantly expands the repertoire of tools needed for probing the role of these kinases in normal- and patho-biology and may form basis for a new drug development program.
MATERIALS and METHODS
Plasmids and Ternary complex assay
The dual luciferase expression vector and other plasmids used for these studies are described in.[17] The ternary complex assay, surrogate for eIF2α phosphorylation, has been described elsewhere.[16] Briefly, the pBISA vector, which contains seven copies of the tetracycline-regulated transactivator response element (TRE), is flanked on both sides by minimal human cytomegalovirus (CMV) minimal promoters allowing bi-directional transcription of ORFs cloned into two multiple cloning sites (MCSs) . F and R luciferases were subcloned into MCS-I and MCS-II, respectively. This plasmid, designated pBISA-DL, transcribes two mRNAs that contain the 90 nucleotide plasmid derived 5'UTR (same sequence in both mRNAs), and the ORF encoding either F or R luciferase ORF followed by a polyadenylation sequence. This plasmid was further modified by inserting the 5'UTR of ATF-4 mRNA into MCS-I in front of the F luciferase mRNA. Transcription from this unit generates an mRNA that contains the F luciferase ORF preceded by a 5'UTR composed of 90 nucleotides derived from the plasmid and 267 nucleotides derived from the 5'UTR of ATF-4 mRNA. Transcription from the other unit generates an mRNA that contains the R luciferase ORF proceeded only by the 90-nucleotide plasmid-derived sequence in the 5'UTR (pBISA-DL(ATF-4)).[16]
Stable and transient transfection
Stable cell lines utilized in this study are generated as described elsewhere.[16] Briefly cells were seeded at the density of 105 in 60-mm dish (stable transfection) or 104 cells per well of 96-well plate (transient transfection) and transfected one day later using the Lipofectamine 2000 (Invitrogen). For selection of stable cell lines, transfected cells were transferred to 100-mm plates and selected with appropriate antibiotics.
Compound screening
Cells were seeded at the density of 3,000 cells per well in 384-well plates, and allowed to attach overnight at 37°C, 5% CO2. Compounds were added in 100 nl of a 10 mM DMSO stock solution for a final screening concentration of 33 μM, using pin transfer (Seiko Compound Transfer Robot). Cells were then incubated in the presence of compound for an additional ten hours at 37°C, 5% CO2. Plates were allowed to equilibrate to room temperature for thirty minutes. F luciferase reporter activity was read by the addition of 10 μl of Dual Glo Luciferase reagent (Promega Inc., Madison, WI), followed by 15 minutes incubation at room temperature to allow for adequate signal buildup. Luminescence counting was conducted on a Microbeta Trilux using a 1 second read time. R luciferase reporter activity was measured following addition of 10 μl Stop and Glo Luciferase reagent (Promega) and incubation identical to the one carried out for the F luciferase. The data calculations were carried out as the ratio of firefly to renilla luciferase signal.
Western blotting
Cells cultured under recommended media conditions were plated and maintained in serum-containing media without antibiotics in 140 mm plates (Nunc) until reaching 70% confluence. Cells were then treated with compounds for 6 hours, unless otherwise indicated by figure legends, washed with cold PBS once, and lysed with M-PER Mammalian Protein Extraction Reagent (Pierce) for 30 minutes on ice. The cell lysates were centrifuged at 12,000 RPM for 15 min and the supernatants were transferred to fresh tubes and the protein concentrations were determined by BCA (Pierce). Equal amount of proteins were mixed with Laemmli Sample Buffer, heated at 100°C for 5 min and separated by SDS-PAGE and probed with anti-phosphoserine-51-eIF2α (P-eIF2α), anti-total eIF2α-specific antibodies (T-eIF2α) (Biosource International, Hopkinton, MA), anti-CHOP, anti-Cyclin D1 or anti Actin (Santa Cruz Biotechnology, CA) essentially as described[22].
Cell growth assay
Cells were seeded in 96-well plates and maintained for 5 days in the presence of 0.5 to 20 μM of individual compound, and cell proliferation was measured by the sulforhodamine B (SRB) assay as described.[23] Briefly, at the end of a 5-day treatment, cells were fixed in 10% cold trichloroacetic acid, cell number was estimated by measuring the remaining bound dye of sulforhodamine B after washing. The percentage of growth was calculated by using the equation: 100× [(T-T0)/(C-T0)], where T and C represent the absorbance in treated and control cultures at Day 5, and T0 represents absorbance of cells fixed at the time of compound addition (time zero), respectively. If T is less than T0, cell death has occurred and can be calculated from 100× [(T-T0)/T0].
Real time PCR
For real time PCR, total RNA was extracted with FastLane Cell SYBR Green Kit (Qiagen, Gaithersburg, MD) according to manufacturer's protocol. 1-Step Real-time PCR was performed on a Bio-Rad iCycler IQ5 system by using B-R 1-Step SYBR Green qRT-PCR Kit (Quanta BioSciences, Gaithersburg, MD) according to manufacturer's specifications. The thermal cycler conditions were as follows: 10 minutes at 50°C, hold for 5 minutes at 95°C, followed by 2-step PCR for 45 cycles of 95°C for 15 seconds followed by 60°C for 30 seconds. All PCR reactions were performed in triplicate in at least 2 independent PCR runs. Mean values of these repeated measurements were used for calculation. To calibrate the results, all the transcripts quantities were normalized to 18S rRNA. The following primers were used in real-time PCR reactions: Human CHOP (5’ AGAACCAGGAAACGGAAACAGA 3’ and 5’ TCTCCTTCATGCGCTGCTTT 3’); Human 18s rRNA (5’ CGGCGACGACCCATTCGAAC 3’ and 5’ GAATCGAACCCTGATTCCCCGTC 3’) Primers for Human Cyclin D1 and Cyclin E were purchased from Qiagen (QuantiTect Primer Assay), Human Cyclin D1 (QT00495285), Human Cyclin E (QT00041986).
Evaluation of HCV infection
Huh7.5.1 human hepatoma cells were cultured in 96 well black clear bottom plates overnight in DMEM supplemented with 15% Fetal Bovine Serum (Gibco). Cells were treated with compounds, interferon alpha (PBL) 25IU/ml, or DMSO for 24hours. Fully infectious JFH1 HCV was added to cultures for 48h followed by fixation in 4% Paraformaldehyde, permeabilization using 0.2% triton, and staining with anti-HCV primary antibody followed by Alexa-488 tagged secondary antibody then Hoechst DNA stain. Cells were then imaged on an automated microscope (Molecular Devices ImageXpress Micro) at 4X followed by analysis for percent infected cells and total cells per well. Data was analyzed by first calculating the percent infection in each well then comparing percent infection to DMSO treated wells to normalize the data in each well to DMSO treated wells.
Statistical analysis
The data were expressed as the mean ± S.E.M. and were analyzed by one-way ANOVA using Graphpad 5.0 (GraphPad Software Inc.; San Diego, CA, USA). A P value of less than 0.05 was considered to be statistically significant.
Supplementary Material
AKNOWLEDGEMENTS
This work was supported by NIH grants #R21AG032546 and #1RO1CA152312 to BHA. We thank J.A. Halperin (Brigham and Women's Hospital/Harvard Medical School) for critical comments on the manuscript; and C. Shamu, J. Smith, S. Rudnicki, S.M. Johnston, D. Flood, J. Nale and D. Wrobel from ICCB-Longwood for technical assistant and advice.
REFERENCES
- 1.a Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Molecular cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]; b Raven JF, Koromilas AE. Cell Cycle. 2008;7:1146–1150. doi: 10.4161/cc.7.9.5811. [DOI] [PubMed] [Google Scholar]; c Baltzis D, Qu LK, Papadopoulou S, Blais JD, Bell JC, Sonenberg N, Koromilas AE. Journal of virology. 2004;78:12747–12761. doi: 10.1128/JVI.78.23.12747-12761.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Takada Y, Ichikawa H, Pataer A, Swisher S, Aggarwal BB. Oncogene. 2007;26:1201–1212. doi: 10.1038/sj.onc.1209906. [DOI] [PubMed] [Google Scholar]; e Raven JF, Baltzis D, Wang S, Mounir Z, Papadakis AI, Gao HQ, Koromilas AE. J Biol Chem. 2008;283:3097–3108. doi: 10.1074/jbc.M709677200. [DOI] [PubMed] [Google Scholar]
- 2.a Sen A, Pruijssers AJ, Dermody TS, Garcia-Sastre A, Greenberg HB. Journal of virology. 2011;85:3717–3732. doi: 10.1128/JVI.02634-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Rothenburg S, Seo EJ, Gibbs JS, Dever TE, Dittmar K. Nature structural & molecular biology. 2009;16:63–70. doi: 10.1038/nsmb.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Okonski KM, Samuel CE. Journal of virology. 2013;87:756–766. doi: 10.1128/JVI.02270-12. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Kang JI, Kwon SN, Park SH, Kim YK, Choi SY, Kim JP, Ahn BY. Virus research. 2009;142:51–56. doi: 10.1016/j.virusres.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 3.a von Holzen U, Pataer A, Raju U, Bocangel D, Vorburger SA, Liu Y, Lu X, Roth JA, Aggarwal BB, Barber GN, Keyomarsi K, Hunt KK, Swisher SG. Clin Cancer Res. 2007;13:6032–6039. doi: 10.1158/1078-0432.CCR-06-2932. [DOI] [PubMed] [Google Scholar]; b Zamanian-Daryoush M, Mogensen TH, DiDonato JA, Williams BR. Mol Cell Biol. 2000;20:1278–1290. doi: 10.1128/mcb.20.4.1278-1290.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Bonnet MC, Weil R, Dam E, Hovanessian AG, Meurs EF. Mol Cell Biol. 2000;20:4532–4542. doi: 10.1128/mcb.20.13.4532-4542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Gil J, Garcia MA, Gomez-Puertas P, Guerra S, Rullas J, Nakano H, Alcami J, Esteban M. Mol Cell Biol. 2004;24:4502–4512. doi: 10.1128/MCB.24.10.4502-4512.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Nallagatla SR, Toroney R, Bevilacqua PC. Current opinion in structural biology. 2011;21:119–127. doi: 10.1016/j.sbi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Clerzius G, Gelinas JF, Gatignol A. Reviews in medical virology. 2011;21:42–53. doi: 10.1002/rmv.674. [DOI] [PubMed] [Google Scholar]
- 5.a Nakamura T, Furuhashi M, Li P, Cao H, Tuncman G, Sonenberg N, Gorgun CZ, Hotamisligil GS. Cell. 2010;140:338–348. doi: 10.1016/j.cell.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Couturier J, Morel M, Pontcharraud R, Gontier V, Fauconneau B, Paccalin M, Page G. J Biol Chem. 2010;285:1272–1282. doi: 10.1074/jbc.M109.041954. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Bullido MJ, Martinez-Garcia A, Tenorio R, Sastre I, Munoz DG, Frank A, Valdivieso F. Neurobiology of aging. 2008;29:1160–1166. doi: 10.1016/j.neurobiolaging.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 6.Feng D, Wei J, Gupta S, McGrath BC, Cavener DR. BMC cell biology. 2009;10:61. doi: 10.1186/1471-2121-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullinan SB, Diehl JA. J Biol Chem. 2004;279:20108–20117. doi: 10.1074/jbc.M314219200. [DOI] [PubMed] [Google Scholar]
- 8.Deng J, Lu PD, Zhang Y, Scheuner D, Kaufman RJ, Sonenberg N, Harding HP, Ron D. Mol Cell Biol. 2004;24:10161–10168. doi: 10.1128/MCB.24.23.10161-10168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Molecular cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]; b Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, Diehl JA. Mol Cell Biol. 2003;23:7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Julier C, Nicolino M. Orphanet journal of rare diseases. 2010;5:29. doi: 10.1186/1750-1172-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y, Taylor SI, Tan SL, Sonenberg N. Endocrine reviews. 2003;24:91–101. doi: 10.1210/er.2002-0018. [DOI] [PubMed] [Google Scholar]
- 12.a Yoon CH, Lee ES, Lim DS, Bae YS. Proc Natl Acad Sci U S A. 2009;106:7852–7857. doi: 10.1073/pnas.0812148106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Mounir Z, Krishnamoorthy JL, Robertson GP, Scheuner D, Kaufman RJ, Georgescu MM, Koromilas AE. Science signaling. 2009;2:ra85. doi: 10.1126/scisignal.2000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a Follo MY, Finelli C, Mongiorgi S, Clissa C, Bosi C, Martinelli G, Blalock WL, Cocco L, Martelli AM. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, U.K. 2008;22:2267–2269. doi: 10.1038/leu.2008.122. [DOI] [PubMed] [Google Scholar]; b Pataer A, Swisher SG, Roth JA, Logothetis CJ, Corn PG. Cancer biology & therapy. 2009;8:245–252. doi: 10.4161/cbt.8.3.7386. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Vorburger SA, Pataer A, Swisher SG, Hunt KK. American journal of pharmacogenomics : genomics-related research in drug development and clinical practice. 2004;4:189–198. doi: 10.2165/00129785-200404030-00006. [DOI] [PubMed] [Google Scholar]; d Nawrocki ST, Carew JS, Dunner K, Jr., Boise LH, Chiao PJ, Huang P, Abbruzzese JL, McConkey DJ. Cancer Res. 2005;65:11510–11519. doi: 10.1158/0008-5472.CAN-05-2394. [DOI] [PubMed] [Google Scholar]; e Bobrovnikova-Marjon E, Grigoriadou C, Pytel D, Zhang F, Ye J, Koumenis C, Cavener D, Diehl JA. Oncogene. 2010;29:3881–3895. doi: 10.1038/onc.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Sequeira SJ, Ranganathan AC, Adam AP, Iglesias BV, Farias EF, Aguirre-Ghiso JA. PloS one. 2007;2:e615. doi: 10.1371/journal.pone.0000615. [DOI] [PMC free article] [PubMed] [Google Scholar]; g Blais JD, Addison CL, Edge R, Falls T, Zhao H, Wary K, Koumenis C, Harding HP, Ron D, Holcik M, Bell JC. Mol Cell Biol. 2006;26:9517–9532. doi: 10.1128/MCB.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; h Delgado Andre N, De Lucca FL. Cancer letters. 2007;258:118–125. doi: 10.1016/j.canlet.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 14.Gil J, Rullas J, Garcia MA, Alcami J, Esteban M. Oncogene. 2001;20:385–394. doi: 10.1038/sj.onc.1204109. [DOI] [PubMed] [Google Scholar]
- 15.a Eley HL, McDonald PS, Russell ST, Tisdale MJ. Cancer chemotherapy and pharmacology. 2009;63:651–659. doi: 10.1007/s00280-008-0782-y. [DOI] [PubMed] [Google Scholar]; b Wang H, Blais J, Ron D, Cardozo T. Chemical biology & drug design. 2010;76:480–495. doi: 10.1111/j.1747-0285.2010.01048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Axten JM, Medina JR, Feng Y, Shu A, Romeril SP, Grant SW, Li WH, Heerding DA, Minthorn E, Mencken T, Atkins C, Liu Q, Rabindran S, Kumar R, Hong X, Goetz A, Stanley T, Taylor JD, Sigethy SD, Tomberlin GH, Hassell AM, Kahler KM, Shewchuk LM, Gampe RT. Journal of medicinal chemistry. 2012;55:7193–7207. doi: 10.1021/jm300713s. [DOI] [PubMed] [Google Scholar]; d Harding HP, Zyryanova AF, Ron D. J Biol Chem. 2012;287:44338–44344. doi: 10.1074/jbc.M112.428987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen T, Ozel D, Qiao Y, Harbinski F, Chen L, Denoyelle S, He X, Zvereva N, Supko JG, Chorev M, Halperin JA, Aktas BH. Nat Chem Biol. 2011;7:610–616. doi: 10.1038/nchembio.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziegeler G, Ming J, Koseki JC, Sevinc S, Chen T, Ergun S, Qin X, Aktas BH. J Biol Chem. 2010;285:15408–15419. doi: 10.1074/jbc.M110.113365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denoyelle S, Chen T, Chen L, Wang Y, Klosi E, Halperin JA, Aktas BH, Chorev M. Bioorg Med Chem Lett. 2012;22:402–409. doi: 10.1016/j.bmcl.2011.10.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a Aktas H, Fluckiger R, Acosta JA, Savage JM, Palakurthi SS, Halperin JA. Proc Natl Acad Sci U S A. 1998;95:8280–8285. doi: 10.1073/pnas.95.14.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Chen L, Aktas BH, Wang Y, He X, Sahoo R, Zhang N, Denoyelle S, Kabha E, Yang H, Freedman RY, Supko JG, Chorev M, Wagner G, Halperin JA. Oncotarget. 2012;3:869–881. doi: 10.18632/oncotarget.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonnet MC, Daurat C, Ottone C, Meurs EF. Cellular signalling. 2006;18:1865–1875. doi: 10.1016/j.cellsig.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Baltzis D, Pluquet O, Papadakis AI, Kazemi S, Qu LK, Koromilas AE. J Biol Chem. 2007;282:31675–31687. doi: 10.1074/jbc.M704491200. [DOI] [PubMed] [Google Scholar]
- 22.Aktas H, Cai H, Cooper GM. Mol Cell Biol. 1997;17:3850–3857. doi: 10.1128/mcb.17.7.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palakurthi SS, Fluckiger R, Aktas H, Changolkar AK, Shahsafaei A, Harneit S, Kilic E, Halperin JA. Cancer Res. 2000;60:2919–2925. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.