Abstract
The lack of bile flow from the liver into the intestine can have devastating complications including hepatic failure, sepsis, and even death. This pathologic condition known as cholestasis can result from etiologies as diverse as total parenteral nutrition (TPN), hepatitis, and pancreatic cancer. The intestinal injury associated with cholestasis has been shown to result in decreased intestinal resistance, increased bacterial translocation, and increased endotoxemia. Anecdotal clinical evidence suggests a genetic predisposition to exaggerated injury. Recent animal research on two different strains of inbred mice demonstrating different rates of bacterial translocation with different mortality rates supports this premise. In this study, a microarray analysis of intestinal tissue following common bile duct ligation (CBDL) performed under general anesthesia on these same two strains of inbred mice was done with the goal of identifying the potential molecular mechanistic pathways responsible. Over 500 genes were increased more than 2.0-fold following CBDL. The most promising candidate genes included major urinary proteins (MUPs), serine protease-1-inhibitor (Serpina1a), and lipocalin-2 (LCN-2). Quantitative polymerase chain reaction (qPCR) validated the microarray results for these candidate genes. In an in vitro experiment using differentiated intestinal epithelial cells, inhibition of MUP-1 by siRNA resulted in increased intestinal epithelial cell permeability. Diverse novel mechanisms involving the growth hormone pathway, the acute phase response, and the innate immune response are thus potential avenues for limiting cholestatic intestinal injury. Changes in gene expression were at times found to be not only due to the CBDL but also due to the murine strain. Should further studies in cholestatic patients demonstrate interindividual variability similar to what we have shown in mice, then a “personalized medicine” approach to cholestatic patients may become possible.
Keywords: Cholestasis, growth hormone, intestine, lipocalin, microarray
Introduction
Cholestasis, defined as little or no bile flow from the liver into the intestine, is a complex pathologic condition that can develop from either functional etiologies, such as hepatic parenchymal disease secondary to hepatitis, or mechanical etiologies, such as an obstructing pancreatic cancer or biliary stricture. In the pediatric population, cholestasis resulting from prolonged parenteral nutrition is by far the most common etiology. Cholestatic injury has not only a hepatic component but also an intestinal one. Failure of the intestinal barrier with decreased intestinal resistance, increased bacterial translocation, and increased episodes of sepsis has been well described (Campillo et al. 1999; Pascual et al. 2003; Frances et al. 2004); however, the exact mechanisms remain poorly understood.
Common bile duct ligation (CBDL) is a standard model of cholestasis in the literature (Georgiev et al. 2008). CBDL in mice leads to both hepatic and intestinal injuries which are precisely interrelated. We have previously found differences in the systemic inflammatory responses and outcome following CBDL between two inbred mouse strains, C57BL/6J (B6) and A/J, suggesting a genetic contribution (Alaish et al. 2005). In particular, B6 mice were significantly more likely to develop ascites following 1 week of CBDL (Alaish et al. 2005). In concordance with this observation, the frequency of mortality after CBDL was significantly higher in B6 mice compared to A/J mice on days following CBDL (Alaish et al. 2005). Interestingly, although both strains demonstrated markedly elevated plasma liver function tests following CBDL, no difference was noted in liver histology between the two ligated strains. More recently, our laboratory has shown decreased intestinal resistance and increased bacterial translocation following CBDL in these same two strains of inbred mice. Furthermore, we found genetic variation in the intestinal resistance and bacterial translocation rates, which correlated with mortality following CBDL in different strains of inbred mice (Alaish et al. 2013). Further analysis implicated an IFN-γ-mediated apoptotic-independent mechanism of tight junction disruption, which has been well described in vitro (Madara and Stafford 1989; Marano et al. 1998; Youakim and Ahdieh 1999; Bruewer et al. 2003; Clayburgh et al. 2004), as a mechanism possibly responsible for the genetic variation. Nevertheless, the 2.5-fold changes in IFN-γ gene expression following CBDL, albeit significant, did not seem monumental enough to fully explain the striking genetic influence on mortality following CBDL in the mice. In order to uncover other potential mechanisms including novel pathways, we embarked on a whole-genome microarray analysis of jejunal tissue in these two different strains of inbred mice following either a sham operation or CBDL. The differentially expressed genes reported here constitute a resource of candidate genes for roles in cholestatic intestinal injury.
Material and Methods
Animals
Male A/J and C57BL/6J (B6) mice (8 weeks old) were obtained from the Jackson Laboratory (Bar Harbor, ME) and maintained in identical environmental conditions in a pathogen-free animal facility with 12-h light–dark cycles. All mice weighed 18–25 g at the time of operation. Matriptase (St14) hypomorphic C57BL/6J mice (List et al. 2007) were bred in the Antalis laboratory. Animal studies were conducted according to protocols reviewed and approved by the University of Maryland School of Medicine Institutional Animal Care and Use Committee and adhered to guidelines promulgated by the National Institutes of Health. In accordance with these guidelines, we used the minimum number of animals to meet the rigor necessary for this series of experiments.
Experimental design
CBDL operative procedure
Mice were anesthetized by inhaled isoflurane anesthesia. The abdomen was clipped and then prepared in sterile fashion with 70% ethyl-ethanol followed by betadine. A transverse upper abdominal incision was performed. The CBD was dissected away from the portal vein and was ligated near its junction with the duodenum using aneurysm clips engineered with a precisely standardized opening/closing mechanism. The abdominal wall was then closed in a two-layer fashion using absorbable sutures. Sham-operated mice were treated identically but without dissection or ligation of the CBD. Postoperatively, animals were resuscitated with warmed subcutaneous injections of saline (1 mL) to replace losses. Mice were returned to clean cages where food and water were provided ad libitum. Buprenorphine, 0.05–0.1 mg/kg was given subcutaneously at the time of surgery and then every 8–12 h to treat postoperative pain for 48–72 h.
RNA extraction
Seven days following the surgery, the mice underwent deep general anesthesia and euthanasia by thoracotomy and cardiac exsanguination. Postoperative day 7 was chosen because this time point corresponded to our earlier finding of decreased intestinal transepithelial electrical resistance (TEER) after CBDL (Alaish et al. 2013). In addition, this time point exhibited differences in TEER based on the genetic background of the mouse (Alaish et al. 2013). These TEER findings were found in both the jejunum and ileum and correlated with differences in bacterial translocation and mortality. Further studies on jejunal tissue demonstrated differences in tight junction protein expression between CBDL and sham animals and between the strains (Alaish et al. 2013). Therefore, in this study, we chose jejunum once again; the intestinal tissue was harvested under sterile conditions. RNA extraction and purification were performed as we have previously described (Dorsey et al. 2009).
Microarray data analysis
Microarray expression profiling was performed according to the manufacturer protocols (Affymetrix, Santa Clara, CA). Briefly, total RNA was used to prepare biotinylated cRNA, followed by fragmentation and hybridization to Affymetrix arrays (Genechip Mouse 430 2.0; Affymetrix, Santa Clara, CA). The arrays were incubated for approximately 16 h, washed, stained, and scanned per Affymetrix. Differential gene expression through microarray was then performed. We utilized.cel files generated from Affymetrix profiling process for analysis. Arrays were normalized by GCRMA method implemented in gcrma R package (Bioconductor, an open source collection of software packages). Differential expression analysis was performed using limma R package (Bioconductor). First, a linear model was fitted to expression data for each gene. Empirical Bayes method was then used to assess differential expression between two conditions. A cutoff of FDR less than 0.05 was used to select significant probes. A complete data set has been submitted to the NCBI Gene Expression Omnibus (NCBI GEO #GSE47099 and NCBI Tracking System #16793295).
qPCR verification of promising candidate genes
The identification of significant changes in expression of promising candidate genes (major urinary proteins [MUPs], serine protease-1-inhibitor [Serpina1a] and lipocalin-2 [LCN2]) through microarray analysis was validated using a quantitative polymerase chain reaction (qPCR) technique. Total RNA was isolated from homogenized jejunal samples that were stored in TRIzol (Invitrogen, Grand Island, NY). The total RNA was isolated from TRIzol samples as per the manufacturer's instructions. The pellet was allowed to air dry, and the total RNA was resuspended in an appropriate volume of RNAse-free water. RNA concentrations were calculated using a NanoDrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA). Single-stranded cDNA was synthesized from 2 μg of total RNA using random hexamer primer and the First-Strand cDNA Synthesis Kit (MBI Fermentas, Hanover, MD). The specific primer sequences were designed using Beacon Designer 7.0 (Premier Biosoft International, Palo Alto, CA) and synthesized by the University of Maryland School of Medicine Biopolymer/Genomics Core. qPCR reactions were set up using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) in a total volume of 25 μL. Amplification conditions were as follows: 95°C for 3 min, 50 cycles of 95°C for 15 sec, 60°C for 15 sec, and 72°C for 20 sec. All reactions were performed using Bio-Rad iCycler instrumentation and software. All samples were normalized with 18s rRNA housekeeping gene levels with subsequent calculation of fold change in mRNA expression. Analysis was carried out in GraphPad Prism5 (San Diego, CA, USA).
Mortality following CBDL in matriptase hypomorphic B6 mice
We conducted a mortality study following CBDL in wild-type C57BL/6J mice (n = 8) and matriptase (St14) hypomorphic C57BL/6J mice (n = 11). Sham-operated mice of each strain served as controls.
Cells
Cdx2-intestinal epitheial cells (Cdx2-IEC), a transformed rat crypt IEC-6 cell line which maintains a stable differentiated phenotype upon future passages, were obtained from Dr. J.-Y. Wang (University of Maryland, Baltimore, MD). Cdx2-IEC cells were maintained at 37°C in a humidified incubator with 10% CO2 in DMEM containing 5% (v:v) fetal bovine serum (FBS), 0.5% (v:v) ITS + liquid media supplement, 0.1 million units/L penicillin, 100 mg/L streptomycin, and 4 mmol/L sopropylthio-β-d-galactoside, which served as an inducer.
Transfection of Cdx2-IEC cells with MUP-1 siRNA and FITC-dextran permeability assay
Following their sixth passage, Cdx2-IEC cells were transfected with either MUP-1 siRNA (Thermo Scientific, Inc.) or Acell Control Non-Targeting siRNA (Thermo Scientific, Inc.) as described previously (Rao et al. 2006). The siRNAs used were as follows: 80 nmol/L Control siRNA; and 20 nmol/L, 40 nmol/L, and 60 nmol/L MUP-1 siRNA. Silencing of MUP-1 in the cells was confirmed by Western blot analysis using MUP (F-3) mouse monoclonal antibody (Santa Cruz Biotechnology, Inc., Paso Robles, CA, USA). Six-well transwell plates with 12-mm-diameter inserts (Costar 3407; Corning, Inc., Kennebunk, ME, USA) were used to perform the permeability studies following the transfection. The cells were incubated on the inserts with control media for 24 h to allow proper attachment to the membrane prior to dextran administration. After 24 h, TEER was measured in both control and transfected Cdx2-IEC cells for the formation of the monolayer as described previously (El Asmar et al. 2002). The media were removed. 4-kDa FITC-dextran in control media was placed onto the apical side (top chamber); control media alone was placed on the basolateral side. The TEER was monitored for 2 h and 100-μL aliquots of the basolateral medium were collected after each 30-min time period. A sample from the top compartment at the time of the last sampling of the bottom compartment was used to normalize the samples to account for possible differences in the total fluorescence added at the beginning of the experiment. Fluorescence of the samples was quantified in a multiplate fluorescence reader in black 96-well plates; the excitation wavelength was 485 nm and emission wavelength was 538 nm.
Statistical analysis
The microarray data (reported as percent change) were analyzed using repeated measures analyses of variance (ANOVA) with false discovery rate correction to control multiple testing errors. Post hoc testing was done using Tukey's honestly significant difference (HSD). qPCR data were analyzed using ANOVA with the Bonferroni posttest. Graph Pad Prism 5 software was used. For the matriptase hypomorphic B6 mouse CBDL experiment, a Kaplan–Meier survival curve was generated with P < 0.05 considered as significant.
Results
Microarray data
As shown in Figure 1A, more than 500 genes were significantly differentially regulated in the CBDL mice compared with sham. When examining changes across strains, although there are shared genes, there are a significant number of differentially regulated genes that are unique to each strain (Fig. 1B). Table 1 shows the number of genes in each category. The first two lines of data in Table 1 demonstrate that many more genes are differentially expressed in A/J mice as compared to B6 mice following CBDL (582 vs. 137). The last two lines of data in Table 1 demonstrate that there are more genes undergoing expression changes following CBDL compared to sham (882 vs. 766). In Figure 1C, the heat map shows clustering of all differentially expressed genes by genotype. A list of all differentially expressed genes can be found in Table A1. In Figure 1D, the top 53 significantly regulated genes across the two strains are shown in a separate heat map (For both heat maps, red = upregulated genes; green = downregulated genes). Note the disparate gene expressions in the two strains.
Figure 1.
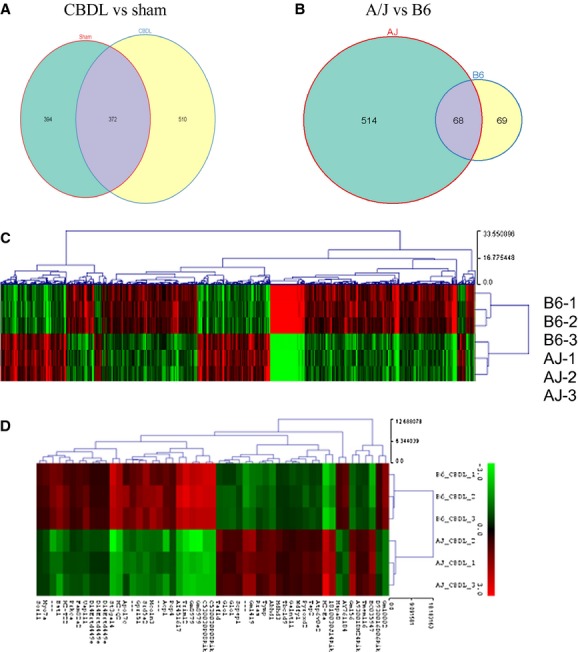
Venn diagram and heat map depicting the number of statistically significant differentially expressed genes by experimental condition and across murine strains. The venn diagram in (A) depicts the number of genes that are differentially expressed in the surgical group versus sham. In (B) we show the number of differentially expressed genes across two strains, A/J and B6. (C) The heat map shows all differentially expressed genes between strains. (D) A heat map which shows the top 53 significantly regulated genes across two strains. For both heat maps, red = upregulated genes; green = downregulated genes.
Table 1.
Differentially expressed genes with FDR < 0.05
| Total | Up | Down | |
|---|---|---|---|
| A/J CBDL versus sham | 582 | 404 | 178 |
| B6 CBDL versus sham | 137 | 76 | 61 |
| Sham A/J versus B6 | 766 | 276 | 490 |
| CBDL A/J versus B6 | 882 | 291 | 591 |
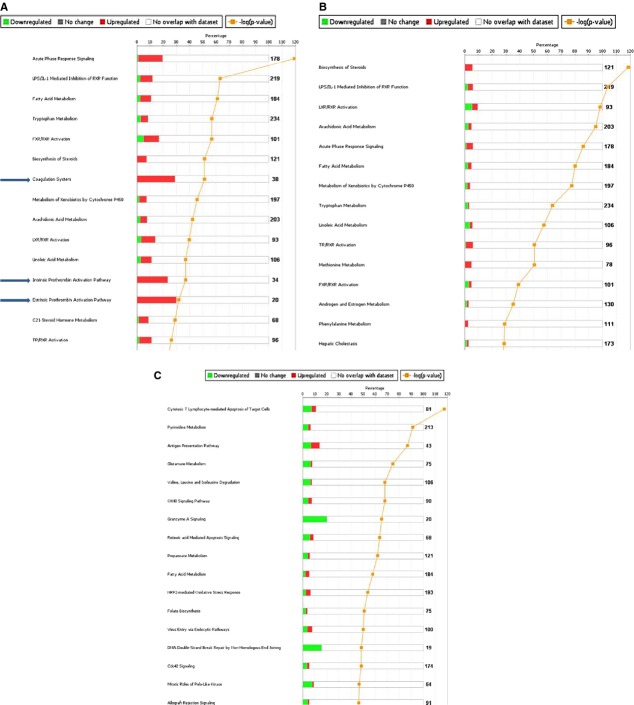
We next examined significantly enriched canonical signaling pathways. First, we evaluated those that were unique within a strain in the CBDL condition compared with sham. As shown in Figure 2A, coagulation pathways were highly upregulated in A/J CBDL mice compared with sham. In contrast, coagulation pathways were not significantly regulated in B6 CBDL versus sham (Fig. 2B). In Figure 2C, we show that there are a number of signaling pathways that are differentially regulated in A/J CBDL compared with B6 CBDL, demonstrating differential pathway activation in CBDL across these two inbred strains of mice.
Figure 2.
Significantly enriched canonical signaling pathways from differentially expressed gene sets. As denoted by the blue arrows in (A), coagulation pathways were highly upregulated in CBDL AJ mice compared with sham. In contrast, coagulation pathways were not significantly regulated in B6 CBDL versus sham (B). In (C), we show that there are a number of signaling pathways that are differentially regulated in AJ CBDL compared with B6 CBDL, demonstrating differential pathway activation in CBDL across these two inbred strains of mice.
qPCR verifies microarray results of candidate genes
The following genes struck us as particularly promising as candidate genes to limit cholestatic injury: MUPs, Serpina1a, and LCN2. They all had significant changes in gene expression following CBDL. MUPs and Serpina1a also had disparate strain expressions which could account for the phenotypic differences we see following ligation. LCN2 was chosen because it is known to play a protective role in the intestinal barrier (Berger et al. 2006). Although we did not see strain differences in LCN2, we did find striking differences between the sham and CBDL mice, which we believe is an important finding. This study not only illustrates strain differences following CBDL but also serves to illustrate intestinal gene expression changes following CBDL independent of strain. qPCR did, indeed, verify our results for MUPs, Serpina1a, and LCN2. (Fig. 3A–C).
Figure 3.

Serpina 1a, MUPs, and LCN2 gene expression changes determined by qPCR are consistent with microarray findings. (A) demonstrates a marked increase in Serpina 1a mRNA following CBDL in both strains. Also, there is a strain difference with increased Serpina 1a mRNA expression in B6 as compared to A/J mice. (B) demonstrates marked strain differences in MUP mRNA expression in the sham animals with disparate responses to CBDL. A/J sham MUP mRNA expression is low and expression increases following CBDL; whereas, in the B6 strain, MUP mRNA expression is very high in the sham group but falls in the CBDL group. (C) shows low expression of LCN2 mRNA in sham animals with marked increases following CBDL in both strains.
Matriptase (St14) hypomorphic B6 mice do not have significantly increased mortality following CBDL compared to wild-type B6 mice
Serpina1a is a serine protease inhibitor, suggesting that decreased intestinal serine protease activity could contribute to the decreased intestinal resistance associated with CBDL. Matriptase is a serine protease whose loss results in decreased intestinal resistance as measured by decreased intestinal TEER (Buzza et al. 2010). We hypothesized that when coupled with the loss of matriptase, the increase in Serpina1a which follows CBDL would result in such a large drop in intestinal resistance that mortality would increase. Although there appeared to be a trend toward increased mortality in the matriptase (St14) hypomorphic B6 mice early on following CBDL, this difference never became significant and did not persist, as can be seen in the Kaplan–Meier Survival Curve (Fig. 4).
Figure 4.

Kaplan–Meier survival curve for matriptase hypomorphic B6 and wild-type B6 mice following CBDL. The early trend toward increased mortality in the matriptase hypomorphic B6 mice is not statistically significant.
MUP-1 siRNA transfection increases permeability in Cdx2-IEC cells
Growth hormone treatment has beneficial effects on the intestine by normalizing intestinal permeability (Liu et al. 2003). We hypothesize that increased expression of MUPs by growth hormone activation (Kuhn et al. 1984) could contribute to this beneficial effect. We inhibited MUP-1 protein expression using siRNA technology. Following confirmation of inhibition of MUP-1 protein expression (Fig. 5), we performed a FITC-dextran permeability assay on Cdx2-IEC cells alone, Cdx2-IEC cells exposed to calcium-free media, Cdx2-IEC cells exposed to control siRNA, and Cdx2-IEC cells exposed to two different concentrations of MUP-1 siRNA (Fig. 6). As expected, cells exposed to calcium-free media resulted in a marked increase in cell permeability compared to Cdx2-IEC cells in control media (*P < 0.04). Cells treated with Control siRNA at 80 nmol/L and MUP-1 siRNA at 40 nmol/L concentrations were similar to untreated cells; whereas, cells treated with MUP-1 siRNA at 60 nmol/L concentration had significantly increased cell permeability compared to Cdx2-IEC cells in control media (**P < 0.0002). Indeed, permeability of cells treated with MUP-1 siRNA at 60 nmol/L concentration was similar to cells in calcium-free media.
Figure 5.
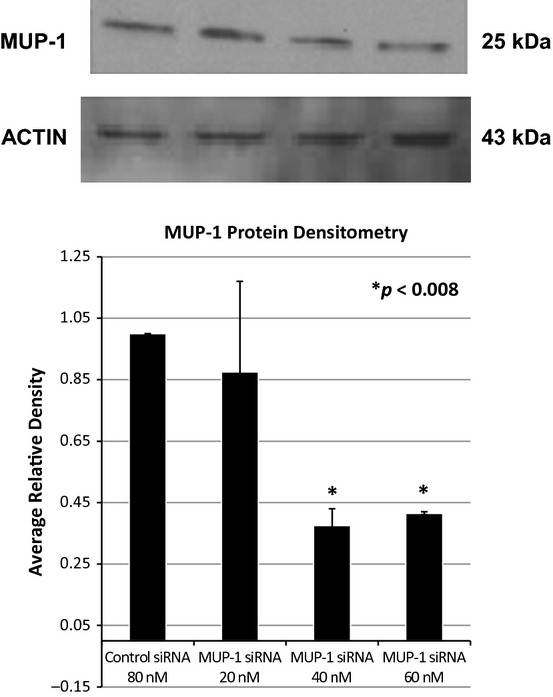
Western blot demonstrating silencing of MUP-1. Following their sixth passage, Cdx2-IEC cells were transfected with either MUP-1 siRNA (Thermo Scientific, Inc.) or Acell Control Non-Targeting siRNA (Thermo Scientific, Inc.). Western blot analysis was performed using MUP (F-3) mouse monoclonal antibody (Santa Cruz Biotechnology, Inc.).
Figure 6.
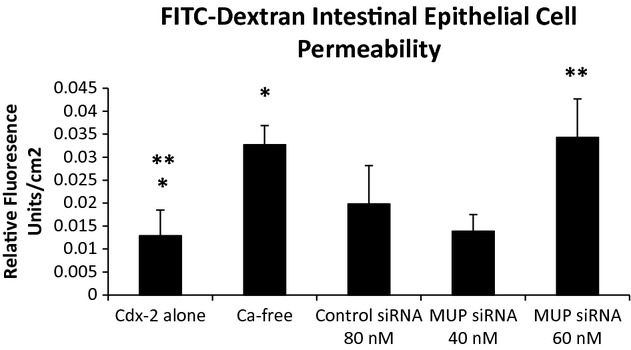
FITC-dextran permeability assay in Cdx2-IEC cells. Cells exposed to calcium-free (Ca-free) media resulted in a marked increase in cell permeability compared to Cdx2-IEC cells in control media (*P < 0.04). Cells treated with Control siRNA at 80 nmol/L and MUP-1 siRNA at 40 nmol/L concentrations were similar to untreated cells; whereas, cells treated with MUP-1 siRNA at 60 nmol/L concentration had significantly increased cell permeability compared to Cdx2-IEC cells in control media (**P < 0.0002). Indeed, permeability of cells treated with MUP-1 siRNA at 60 nmol/L concentration was similar to cells in calcium-free media.
Discussion
Cholestasis can arise from a multitude of conditions in both the adult and pediatric population. In broad terms, cholestasis may result from biliary tract obstruction or hepatic parenchymal disease. Causes of biliary tract obstruction in the adult include pancreatic carcinoma, duodenal carcinoma and cholangiocarcinoma, benign biliary strictures, and choledochal cyst; whereas, biliary atresia and choledochal cyst are seen in infants and children. Causes of hepatic parenchymal disease in both age groups include hepatitis, total parenteral nutrition, sepsis, and other more rare cholestatic syndromes. In the neonatal population, cholestasis is commonly associated with a variety of intestinal pathologies, including necrotizing enterocolitis, intestinal atresias, midgut volvulus, gastroschisis, and Hirschsprung disease. In these conditions, a baby may not be able to tolerate much or any nutrition enterally. Total parenteral (intravenous) nutrition is required. As mentioned earlier, this type of nutrition causes cholestasis and the lack of enteral nutrition worsens the cholestasis; whereas, initiation of enteral nutrition and discontinuation of parenteral nutrition result in improvement of cholestatic liver dysfunction (Javid et al. 2005).
The goal of this study was to use microarray technology to generate candidate genes from both existing and novel pathways which play a role in the development of intestinal injury following cholestasis as modeled by CBDL. Using two different inbred strains of mice, A/J and C57Bl/6J, the design of this study had the benefit of not only discovering strain-independent but also strain-dependent changes in intestinal gene expression following cholestasis. More than 500 genes were increased by more than 2.0-fold following CBDL, and the following were identified as promising candidate genes for further study: MUPs, Serpina1a, and LCN-2.
Major urinary proteins (MUPs) comprise the lipocalin superfamily of lipophilic ligand carriers which, until recently, were thought to participate exclusively in pheromone function. In a seminal article from 2009, MUP1 was shown to be a regulator for glucose and lipid metabolism, in addition to its action as a pheromone ligand to mediate chemical signaling in mice (Zhou et al. 2009). MUP production is known to be increased by testosterone, thyroid hormone, and growth hormone (GH) (Kuhn et al. 1984). The latter hormone is well known to play a significant role in intestinal healing following injury. Increased growth of the small bowel mucosa has been demonstrated in mice overexpressing bovine growth hormone (Ulshen et al. 1993). Moreover, recombinant growth hormone significantly attenuated intestinal mucosal injuries and bacterial translocation in septic rats (Yi et al. 2007), possibly through a mechanism involving the rhGH inhibition of apoptosis of intestinal mucosa cells. Endotoxemia was also reduced after GH administration in jaundiced rats (Scopa et al. 2000). Some insight into the mechanism(s) by which GH exerts it effects have been elucidated in animal models. Prophylactic treatment with growth hormone promoted IgA secretion by B lymphocytes in the plasma and in the intestine in stressed postoperative rats (Ding et al. 2004). Similar effects have been seen in patients. GH was found to attenuate the depression in cellular immunity following surgical stress in a randomized, double-blind, controlled trial of 20 patients undergoing abdominal surgery (Liu et al. 2003). In addition, GH contributes to intestinal adaptation and has been documented to enable both adult and pediatric short gut syndrome patients to graduate from total parenteral nutrition (TPN) supplementation (Byrne et al. 1995; Velasco et al. 1999; Weiming et al. 2004). Growth hormone was also shown to reduce the increase in intestinal permeability seen following abdominal surgery (Liu et al. 2003). Consequently, when our microarray analysis demonstrated MUPs 1, 2, 3, 4, 7, 11, and 20 to be significantly increased following CBDL, we sought to explore this possible mechanism further. qPCR validated the microarray results and demonstrated a >200-fold increase in MUP expression in B6 sham mice compared to A/J sham mice. Most interestingly, the two strains of mice had different responses to CBDL. MUP expression increased >5-fold in A/J mice following CBDL; however, MUP expression in B6 mice decreased greatly following CBDL, such that there was no significant difference between two strains following CBDL. We speculate that GH rises in A/J mice following CBDL to increase MUP expression which, in turn, leads to increased intestinal resistance, strengthens the intestinal barrier, and helps limit injury; whereas, in B6 mice, GH reserves and MUP expression have been depleted and simply cannot keep up with the injury. Further work is needed to clarify this.
We documented baseline expression of MUP-1 in intestinal epithelial cells. Following transfection of these cells with MUP-1 siRNA at 60 nmol/L concentration, we noticed a marked increase in cell permeability. This finding implicates increased MUP expression leading to increased intestinal resistance as a mechanism potentially responsible for the beneficial effects of growth hormone therapy.
Serpina1a is a gene on Chromosome 14 that encodes alpha-1 antitrypsin, which is a type of serine protease inhibitor, serpin. Our microarray analysis demonstrated a markedly elevated level of Serpina1a gene expression in the B6 sham mice, as well as dramatic increases in Serpina1a gene expression following CBDL in both strains. qPCR confirmed these findings. B6 sham mice had approximately 50-fold increased levels of expression compared to A/J sham mice. Furthermore, both strains exhibited approximately 100-fold increased levels of expression following CBDL when compared to the A/J sham mice.
A novel mechanism was suggested by the work of Bacher et al. (1992). They found that in gastrointestinal cell lines, the formation of tight junctions involves cellular proteases which are susceptible to protease inhibitors. Moreover, the recent work by Buzza and colleagues (2010) showing that Matriptase, a membrane-type serine protease-1, regulated epithelial barrier formation and permeability in the intestine intrigued us. Using matriptase (St14) hypomorphic mice which express less than 1% of the gene product, Buzza and colleagues (2010) demonstrated these mice to have a leaky intestinal barrier with decreased TEER and increased paracellular permeability. This raised the hypothesis that matriptase (St14) hypomorphic mice would have a higher mortality following CBDL than wild-type B6 mice. The increased expression of Serpina1a, coupled with the decreased intestinal resistance that we had found following CBDL, suggested to us that matriptase (St14) hypomorphic mice with decreased intestinal resistance and leaky tight junctions would not tolerate CBDL and the resultant further inhibition of serine protease-1 as well as the wild-type B6 mice. To our surprise, we did not find a striking effect on mortality. This might be explained in part by the work of Beliveau et al. (2009) which showed that under in vitro conditions, Serpina1a was an impotent inhibitor of matriptase compared to other serpins, such as antithrombin III. Antithrombin III may very well inhibit matriptase in vivo; however, further experiments would be necessary to confirm this. Whether antithrombin III increases mortality in matriptase hypomorphic mice following CBDL might be confounded by the known salutary effects of antithrombin III in sepsis, including the attenuation of both hepatocyte apoptosis (Huang et al. 2010) and endotoxemia-induced healing impairment in the colon (Diller et al. 2009), as well as preserved mucosal thickness and villus height following CBDL (Caglikulekci et al. 2004). Nevertheless, our results suggest that matriptase does not play a significant role in the mortality following CBDL.
A potential mechanism behind the role of Serpina1a in the cholestatic intestine might be akin to that seen in studies of lung inflammation. Alpha-1 antitrypsin inhibits the enzyme, neutrophil elastase, which is released from neutrophils and macrophages during inflammation; neutrophil elastase destroys both bacteria and host tissue. Neutrophil elastase has also been shown to play a significant role in the pathogenesis of lung injury in pulmonary fibrosis (Yamanouchi et al. 1998). Moreover, alpha-1 antitrypsin was shown to induce hepatocyte growth factor (HGF) production by human lung fibroblasts and function as an anti-inflammatory and regenerative factor in addition to its role in protease inhibition (Kikuchi et al. 2000). HGF has previously been shown to increase intestinal epithelial cell mass and function in vivo (Kato et al. 1997) and also stimulate DNA content and protein content beyond the normal adaptive response following massive small intestinal resection (Kato et al. 1998). Our data suggest that increased expression of Serpina1a would increase HGF and provide a therapeutic approach to limit the intestinal injury following cholestasis, analogous to a recent study, in which alpha-1 antitrypsin therapy was shown to decrease intestinal permeability and ameliorate acute colitis and chronic ileitis in a murine model (Collins et al. 2013).
Lipocalin-2 is an iron-sequestering protein in the antibacterial innate immune response. Upon encountering invading bacteria, the Toll-like receptor 4 (TLR4) on immune cells stimulates the transcription, translation, and secretion of lipocalin-2 (Flo et al. 2004). It binds to siderophores secreted by pathogenic bacteria, including enterochelin secreted by E. coli, and prevents bacterial iron uptake (Flo et al. 2004). LCN2−/− mice have decreased survival following E. coli infection compared to wild-type mice (Berger et al. 2006). Neutrophils isolated from LCN2−/− mice showed significantly less bacteriostatic activity compared with wild-type controls. The bacteriostatic property of the wild-type neutrophils was abolished by the addition of exogenous iron, indicating that the main function of lipocalin-2 is to limit this essential element (Berger et al. 2006). Similarly, lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine (Rafatellu et al. 2009). Our microarray analysis demonstrated significant increases in LCN2 following CBDL. qPCR confirmed this finding and is consistent with a strain-independent response to CBDL. LCN2 expression increased more than 20-fold and 40-fold, respectively, in A/J and B6 mice following CBDL. Increased LCN2 gene expression in the intestine following CBDL appears to be a mechanism whereby the host limits injury and mortality. This appears analogous to the increase in LCN2 gene expression coincident with an increase in epithelial cell apoptosis associated with intestinal adaptation following massive small bowel resection (Wildhaber et al. 2003). This is in agreement with earlier work by Devireddy et al. (2001) demonstrating lipocalin-2 to induce apoptosis in hematopoietic cells by an autocrine pathway. Thus, lipocalin-2 is a regulatory factor of intestinal growth.
In summary, our study has generated a resource of candidate genes for intestinal injury secondary to cholestasis. For Serpina1a, MUPs, and LCN2, some of the genes whose expression was most dramatically affected by CBDL, we confirmed the microarray results with qPCR. Novel mechanisms implicated by our results involve the growth hormone pathway, the acute phase response, and the innate immune response. More research is needed to further define these mechanisms; however, future therapeutic strategies might include: overexpression of Serpina1a and increased levels of HGF, upregulation of the GH pathway and increased MUP expression, and increased LCN2 expression to limit intestinal injury following cholestasis.
Appendix
Table A1.
List of all differentially expressed genes
| Probe set ID | Gene accession | Gene symbol | Gene description | Cytoband | AJ CBDL vs sham FC | AJ CBDL vs sham FDR | B6 CBDL vs sham FC | B6 CBDL vs sham FDR |
|---|---|---|---|---|---|---|---|---|
| 10566477 | NM_017371 | Hpx | hemopexin | 7 F1 | 37.4012 | 1.00E−04 | 4.7408 | 0.0435 |
| 10581605 | NM_017370 | Hp | haptoglobin | 8 D3|8 55.0 cM | 34.152 | 3.00E−04 | 6.662 | 0.0445 |
| 10505438 | NM_008768 | Orm1 | orosomucoid 1 | 4 B3|4 31.4 cM | 18.2062 | 2.00E−04 | 4.3826 | 0.0404 |
| 10414192 | NM_133653 | Mat1a | methionine adenosyltransferase I, alpha | 14 C1 | 17.981 | 0.001 | 7.268 | 0.0306 |
| 10511886 | NM_016668 // NM_016668 | Bhmt // Bhmt | betaine-homocysteine methyltransferase // betaine-homocysteine methyltransferase | 13 D1 // 13 D1 | 17.9485 | 0.0014 | 10.4211 | 0.0197 |
| 10457114 | NM_016668 | Bhmt | betaine-homocysteine methyltransferase | 13 D1 | 15.6549 | 5.00E−04 | 8.3998 | 0.0109 |
| 10578352 | NM_145594 | Fgl1 | fibrinogen-like protein 1 | 8 A4 | 13.8891 | 1.00E−04 | 4.1148 | 0.0178 |
| 10575349 | NM_146214 | Tat | tyrosine aminotransferase | 8 D3 | 12.4752 | 3.00E−04 | 4.323 | 0.0294 |
| 10449452 | NM_010220 | Fkbp5 | FK506 binding protein 5 | 17 A3.3|17 13.0 cM | 11.8521 | 0.0015 | 6.359 | 0.0294 |
| 10490989 | NM_001042611 | Cp | ceruloplasmin | 3 D | 11.1162 | 0 | 3.3852 | 0.0109 |
| 10481627 | NM_008491 | Lcn2 | lipocalin 2 | 2 A3|2 27.0 cM | 9.7186 | 8.00E−04 | 8.0914 | 0.0098 |
| 10360328 | NM_011318 | Apcs | serum amyloid P-component | 1 H3|1 94.2 cM | 9.2166 | 0 | 3.1253 | 0.0123 |
| 10451953 | NM_029796 | Lrg1 | leucine-rich alpha-2-glycoprotein 1 | 17 D|17 10.0 cM | 6.9919 | 6.00E−04 | 5.0051 | 0.0109 |
| 10574023 | NM_008630 | Mt2 | metallothionein 2 | 8 C5|8 45.0 cM | 6.9697 | 0.0206 | 8.4549 | 0.0307 |
| 10515187 | NM_007822 | Cyp4a14 | cytochrome P450, family 4, subfamily a, polypeptide 14 | 4 D1|4 49.5 cM | 4.6609 | 0.0378 | 9.8282 | 0.015 |
| 10418434 | NM_008407 | Itih3 | inter-alpha trypsin inhibitor, heavy chain 3 | 14 A2-C1 | 4.2071 | 9.00E−04 | 4.0594 | 0.0078 |
| 10543017 | NM_013743 | Pdk4 | pyruvate dehydrogenase kinase, isoenzyme 4 | 6 A1|6 0.63 cM | 3.5193 | 0.0145 | 3.492 | 0.0368 |
| 10503520 | NM_015767 | Ttpa | tocopherol (alpha) transfer protein | 4 A3|4 22.7 cM | 3.3338 | 0.003 | 2.5756 | 0.0354 |
| 10492748 | NM_010196 | Fga | fibrinogen alpha chain | 3 F1|3 44.8 cM | 3.1077 | 0.0013 | 2.2512 | 0.0307 |
| 10496727 | NM_026993 | Ddah1 | dimethylarginine dimethylaminohydrolase 1 | 3 H3 | 3.0694 | 0.001 | 2.967 | 0.0098 |
| 10583732 | NM_010700 | Ldlr | low density lipoprotein receptor | 9 A3|9 5.0 cM | 3.0527 | 0 | 2.0755 | 0.0024 |
| 10507177 | NM_001100181 | Cyp4a32 | cytochrome P450, family 4, subfamily a, polypeptide 32 | 4 D1 | 2.7853 | 0.0347 | 3.3932 | 0.0344 |
| 10527920 | NM_020010 | Cyp51 | cytochrome P450, family 51 | 5 A2|5 1.2 cM | 2.7708 | 0.001 | 2.3088 | 0.015 |
| 10362073 | NM_001161845 | Sgk1 | serum/glucocorticoid regulated kinase 1 | 10 A3 | 2.7412 | 0.0073 | 3.4689 | 0.0109 |
| 10379153 | NM_009657 | Aldoc | aldolase C, fructose-bisphosphate | 11 B5|11 44.98 cM | 2.699 | 0.0096 | 2.6246 | 0.0301 |
| 10420730 | NM_010191 | Fdft1 | farnesyl diphosphate farnesyl transferase 1 | 14 D1|14 3.0 cM | 2.6151 | 0.001 | 2.0228 | 0.0228 |
| 10515201 | NM_007823 | Cyp4b1 | cytochrome P450, family 4, subfamily b, polypeptide 1 | 4 D1|4 49.5 cM | 2.5614 | 0.001 | 1.7862 | 0.0445 |
| 10568001 | NM_133670 | Sult1a1 | sulfotransferase family 1A, phenol-preferring, member 1 | 7 F3|7 4.0 cM | 2.5199 | 1.00E−04 | 1.7899 | 0.0109 |
| 10520362 | NM_153526 | Insig1 | insulin induced gene 1 | 5 B1 | 2.4631 | 0.0014 | 1.836 | 0.0451 |
| 10412909 | NM_010191 | Fdft1 | farnesyl diphosphate farnesyl transferase 1 | 14 D1|14 3.0 cM | 2.4306 | 6.00E−04 | 2.072 | 0.0109 |
| 10424349 | NM_009270 | Sqle | squalene epoxidase | 15 D1 | 2.3683 | 0.0023 | 1.9609 | 0.0307 |
| 10478048 | NM_008489 | Lbp | lipopolysaccharide binding protein | 2 H1|2 83.0 cM | 2.3404 | 6.00E−04 | 1.8161 | 0.0194 |
| 10499483 | NM_134469 | Fdps | farnesyl diphosphate synthetase | 3 F1|3 42.6 cM | 2.2316 | 0.0044 | 2.2171 | 0.017 |
| 10412466 | NM_145942 | Hmgcs1 | 3-hydroxy-3-methylglutaryl -Coenzyme A synthase 1 | — | 2.213 | 0.0012 | 1.7749 | 0.0294 |
| 10364194 | NM_146006 | Lss | lanosterol synthase | 10 C1|10 41.1 cM | 2.1256 | 0.0051 | 1.9021 | 0.0346 |
| 10378793 | NM_025655 | Tmigd1 | transmembrane and immunoglobulin domain containing 1 | 11 B5 | 2.0643 | 0.0316 | 3.0221 | 0.0109 |
| 10365830 | — | — | — | — | 2.0533 | 0.0032 | 1.9576 | 0.0178 |
| 10542470 | NM_019946 | Mgst1 | microsomal glutathione S-transferase 1 | 6 G1 | 1.9951 | 0.0124 | 1.9203 | 0.0416 |
| 10506571 | NM_053272 | Dhcr24 | 24-dehydrocholesterol reductase | 4 C7 | 1.8836 | 0.0014 | 1.8709 | 0.0109 |
| 10450242 | NM_009780 | C4b | complement component 4B (Childo blood group) | 17 B1|17 18.8 cM | 1.8607 | 0.0124 | 1.849 | 0.0333 |
| 10585942 | NM_001033498 | Gramd2 | GRAM domain containing 2 | 9 B | 1.8264 | 0.0037 | 1.622 | 0.0375 |
| 10493548 | NM_026784 | Pmvk | phosphomevalonate kinase | 3 F1 | 1.7852 | 0.0105 | 1.808 | 0.0273 |
| 10492964 | NM_009690 | Cd5l | CD5 antigen-like | 3 F1 | 1.6284 | 0.0195 | 1.8539 | 0.0169 |
| 10524555 | NM_023556 | Mvk | mevalonate kinase | 5 F|5 64.0 cM | 1.6043 | 0.0144 | 1.63 | 0.0307 |
| 10600082 | NM_010941 | Nsdhl | NAD(P) dependent steroid dehydrogenase-like | X A7.3|X 28.87 cM | 1.5727 | 0.009 | 1.7157 | 0.0116 |
| 10573626 | NM_173866 | Gpt2 | glutamic pyruvate transaminase (alanine aminotransferase) 2 | 8 C3 | 1.5293 | 0.0438 | 1.6633 | 0.0425 |
| 10526630 | NM_015799 | Trfr2 | transferrin receptor 2 | 5 G2 | 1.5175 | 0.0197 | 1.5238 | 0.0435 |
| 10456699 | NM_177470 | Acaa2 | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacy l-Coenzyme A thiolase) | 18 E2|18 45.0 cM | 1.5095 | 0.029 | 1.6537 | 0.0273 |
| 10393970 | NM_007988 | Fasn | fatty acid synthase | 11 E2|11 72.0 cM | 1.4769 | 0.004 | 1.4846 | 0.015 |
| 10525893 | NM_030210 | Aacs | acetoacetyl-CoA synthetase | 5 F | 1.4673 | 0.0242 | 1.4896 | 0.0456 |
| 10425695 | NM_033218 | Srebf2 | sterol regulatory element binding factor 2 | 15 E1 | 1.4633 | 0.0095 | 1.4826 | 0.0233 |
| 10569972 | NM_026058 | Lass4 | LAG1 homolog, ceramide synthase 4 | 8 A1.2 | 1.4318 | 0.0145 | 1.5 | 0.0228 |
| 10570437 | NM_025785 | Fbxo25 | F-box protein 25 | 8 A1.1 | −1.3809 | 0.0356 | −1.4953 | 0.0294 |
| 10447239 | NM_026180 | Abcg8 | ATP-binding cassette, sub-family G (WHITE), member 8 | 17 E4|17 54.5 cM | −1.5584 | 0.013 | −1.8003 | 0.0109 |
| 10425945 | NM_010180 | Fbln1 | fibulin 1 | 15 E-F | −1.5672 | 0.013 | −1.601 | 0.0287 |
| 10453318 | NM_031884 | Abcg5 | ATP-binding cassette, sub-family G (WHITE), member 5 | 17 E4|17 54.5 cM | −1.6829 | 0.0069 | −1.6878 | 0.0224 |
| 10390032 | NM_153807 | Acsf2 | acyl-CoA synthetase family member 2 | 11 D | −1.8821 | 0.0041 | −2.228 | 0.0078 |
| 10492426 | NM_153807 | Acsf2 | acyl-CoA synthetase family member 2 | 11 D | −1.9567 | 0.0045 | −2.1236 | 0.0109 |
| 10518674 | NM_022020 | Rbp7 | retinol binding protein 7, cellular | 4 E2 | −2.0739 | 0.0021 | −1.8152 | 0.0258 |
| 10462442 | NM_001164724 | Il33 | interleukin 33 | 19 C2 | −2.0935 | 0.0078 | −1.9454 | 0.0354 |
| 10575833 | NM_008290 | Hsd17b2 | hydroxysteroid (17-beta) dehydrogenase 2 | 8 E1 | −2.2055 | 0.0025 | −1.9459 | 0.026 |
| 10577655 | NM_008324 | Ido1 | indoleamine 2,3-dioxygenase 1 | 8 A2 | −2.4459 | 0.0461 | −3.4593 | 0.0249 |
| 10463005 | NM_028089 | Cyp2c55 | cytochrome P450, family 2, subfamily c, polypeptide 55 | 19 C3 | −2.5559 | 0.0458 | −14.6416 | 4.00E−04 |
| 10373334 | NM_013786 | Hsd17b6 | hydroxysteroid (17-beta) dehydrogenase 6 | 10 D3 | −2.6186 | 2.00E−04 | −1.7122 | 0.0241 |
| 10502214 | NM_027816 | Cyp2u1 | cytochrome P450, family 2, subfamily u, polypeptide 1 | 3 H1 | −3.4307 | 1.00E−04 | -1.6652 | 0.0425 |
| 10514912 | NM_007860 | Dio1 | deiodinase, iodothyronine, type I | 4 C7|4 48.7 cM | −4.5064 | 0.0012 | −3.8938 | 0.0109 |
| 10480155 | NM_001081084 | Cubn | cubilin (intrinsic factor -cobalamin receptor) | 2 A1|2 9.0 cM | −4.8123 | 3.00E−04 | −2.7793 | 0.016 |
| 10585749 | NM_009992 | Cyp1a1 | cytochrome P450, family 1, subfamily a, polypeptide 1 | 9 B|9 31.0 cM | −5.6507 | 0.001 | −3.3135 | 0.0301 |
| 10513420 | NM_001134675 | Mup7 | major urinary protein 7 | 4 B3|4 | 127.8618 | 0.001 | ||
| 10513455 | NM_008647 | Mup2 | major urinary protein 2 | 4 B3 | 126.9789 | 0.001 | ||
| 10513437 | NM_001164526 | Mup11 | major urinary protein 11 | 4 B3 | 117.1801 | 0.001 | ||
| 10513428 | NM_001045550 | Mup2 | major urinary protein 2 | 4 B3 | 113.5703 | 0.001 | ||
| 10513472 | NM_008647 | Mup2 | major urinary protein 2 | 4 B3 | 104.5563 | 0.001 | ||
| 10513504 | NM_001045550 | Mup2 | major urinary protein 2 | 4 B3 | 100.7876 | 0.001 | ||
| 10513497 | NM_001045550 | Mup2 | major urinary protein 2 | 4 B3 | 94.2911 | 0.001 | ||
| 10513521 | NM_001012323 | Mup20 | major urinary protein 20 | 4 B3 | 93.1978 | 0.0021 | ||
| 10513467 | NM_001045550 | Mup2 | major urinary protein 2 | 4 B3 | 90.6275 | 0.001 | ||
| 10513512 | NM_001163011 | Mup1 | major urinary protein 1 | 4 B3|4 27.8 cM | 73.9991 | 0.001 | ||
| 10523062 | NM_009654 | Alb | albumin | 5 E1|5 50.0 cM | 61.2531 | 4.00E−04 | ||
| 10434689 | NM_013465 | Ahsg | alpha-2-HS-glycoprotein | 16 B1|16 15.0 cM | 58.1197 | 3.00E−04 | ||
| 10531149 | NM_008096 | Gc | group specific component | 5 E1|5 44.0 cM | 52.3423 | 1.00E−04 | ||
| 10492735 | NM_133862 | Fgg | fibrinogen gamma chain | 3 E3|3 41.3 cM | 38.7526 | 1.00E−04 | ||
| 10498981 | NM_181849 | Fgb | fibrinogen beta chain | 3 E3|3 48.2 cM | 38.2336 | 1.00E−04 | ||
| 10402406 | NM_009245 | Serpina1c | serine (or cysteine) peptidase inhibitor, clade A, member 1C | 12 E|12 51.0 cM | 30.3389 | 3.00E−04 | ||
| 10402399 | NM_009243 | Serpina1a | serine (or cysteine) peptidase inhibitor, clade A, member 1A | 12 E|12 51.0 cM | 23.9693 | 3.00E−04 | ||
| 10558673 | NM_021282 | Cyp2e1 | cytochrome P450, family 2, subfamily e, polypeptide 1 | 7 F5|7 68.4 cM | 21.9342 | 0.0012 | ||
| 10562169 | NM_032541 | Hamp | hepcidin antimicrobial peptide | 7 B1|7 11.0 cM | 21.1349 | 0.004 | ||
| 10434719 | NM_001102411 | Kng1 | kininogen 1 | 16 B1 | 19.6656 | 7.00E−04 | ||
| 10402409 | NM_009247 | Serpina1e | serine (or cysteine) peptidase inhibitor, clade A, member 1E | 12 E | 18.9172 | 3.00E−04 | ||
| 10548207 | NM_007376 | Pzp | pregnancy zone protein | 6 F1-G3|6 62.0 cM | 18.3515 | 3.00E−04 | ||
| 10402390 | NM_009244 | Serpina1b | serine (or cysteine) preptidase inhibitor, clade A, member 1B | 12 E|12 51.0 cM | 15.5285 | 5.00E−04 | ||
| 10498921 | NM_019911 | Tdo2 | tryptophan 2,3-dioxygenase | 3 E3 | 13.9226 | 0.0023 | ||
| 10596148 | NM_133977 | Trf | transferrin | 9 F1-F3|9 56.0 cM | 13.3568 | 4.00E−04 | ||
| 10382189 | NM_013475 | Apoh | apolipoprotein H | 11 D|11 63.0 cM | 13.1193 | 0.001 | ||
| 10553274 | NM_011314 | Saa2 | serum amyloid A 2 | 7 B4|7 23.5 cM | 10.9174 | 0.015 | ||
| 10541480 | NR_027619 | Mug-ps1 | murinoglobulin, pseudogene 1 | 6 F1 | 10.2599 | 0.0013 | ||
| 10496825 | NM_009474 | Uox | urate oxidase | 3 H2|3 75.0 cM | 10.1685 | 0.0046 | ||
| 10497337 | NM_009799 | Car1 | carbonic anhydrase 1 | 3 A1|3 10.5 cM | 9.9909 | 0.0175 | ||
| 10454192 | NM_013697 | Ttr | transthyretin | 18 A2|18 7.0 cM | 9.9844 | 0.0025 | ||
| 10463037 | NM_010003 | Cyp2c39 | cytochrome P450, family 2, subfamily c, polypeptide 39 | 19 C3 | 9.7038 | 1.00E−04 | ||
| 10398060 | NM_011458 | Serpina3k | serine (or cysteine) peptidase inhibitor, clade A, member 3K | 12 E|12 51.5 cM | 9.5497 | 0.0042 | ||
| 10541448 | NR_027619 | Mug-ps1 | murinoglobulin, pseudogene 1 | 6 F1 | 9.4334 | 0.0027 | ||
| 10541410 | NM_008645 | Mug1 | murinoglobulin 1 | 6 F1 | 9.3764 | 0.001 | ||
| 10379190 | NM_011707 | Vtn | vitronectin | 11 B5|11 45.09 cM | 8.9998 | 0.0014 | ||
| 10431915 | NM_027052 | Slc38a4 | solute carrier family 38, member 4 | 15 F1 | 8.9956 | 0.0027 | ||
| 10351546 | NM_013474 | Apoa2 | apolipoprotein A-II | 1 H3|1 92.6 cM | 8.8013 | 8.00E−04 | ||
| 10467410 | NM_145499 | Cyp2c70 | cytochrome P450, family 2, subfamily c, polypeptide 70 | 19 C3 | 8.4844 | 0.0014 | ||
| 10467319 | NM_001159487 | Rbp4 | retinol binding protein 4, plasma | 19 D1|19 38.0 cM | 7.858 | 0.0021 | ||
| 10542983 | NM_011134 | Pon1 | paraoxonase 1 | 6 A2|6 0.5 cM | 7.6708 | 0.021 | ||
| 10560618 | NM_007469 | Apoc1 | apolipoprotein C-I | 7 A3|7 4.0 cM | 7.6693 | 0.0065 | ||
| 10398075 | NM_009252 | Serpina3n | serine (or cysteine) peptidase inhibitor, clade A, member 3N | 12 F1 | 7.4314 | 0.0115 | ||
| 10368343 | NM_007482 | Arg1 | arginase, liver | 10 A4 | 7.3671 | 9.00E−04 | ||
| 10563611 | NM_009117 | Saa1 | serum amyloid A 1 | 7 B4|7 23.5 cM | 7.1769 | 0.0095 | ||
| 10434709 | NM_053176 | Hrg | histidine-rich glycoprotein | 16 B1|16 14.1 cM | 7.0319 | 0.0028 | ||
| 10513529 | NM_001039544 | Mup3 | major urinary protein 3 | 4 B3 | 6.9857 | 0.0286 | ||
| 10549154 | BC151094 | Gm766 | predicted gene 766 | 6 G3 | 6.9004 | 0.0267 | ||
| 10413615 | NM_018746 | Itih4 | inter alpha-trypsin inhibitor, heavy chain 4 | 14 B|14 11.75 cM | 6.7951 | 0.0012 | ||
| 10441753 | NM_008877 | Plg | plasminogen | 17 A1|17 7.3 cM | 6.266 | 0.0022 | ||
| 10401289 | NM_001177561 | Slc10a1 | solute carrier family 10 (sodium/bile acid cotransporter family), member 1 | 12 D1|12 37.0 cM | 6.0785 | 0.0028 | ||
| 10485027 | NM_010168 | F2 | coagulation factor II | 2 E1|2 47.5 cM | 6.048 | 0.0096 | ||
| 10402394 | NM_009246 | Serpina1d | serine (or cysteine) peptidase inhibitor, clade A, member 1D | 12 E|12 51.0 cM | 6.0378 | 0.0135 | ||
| 10363860 | NM_025807 | Slc16a9 | solute carrier family 16 (monocarboxylic acid transporters), member 9 | 10 B5.3 | 5.8753 | 0.0065 | ||
| 10434698 | NM_021564 | Fetub | fetuin beta | 16 B|16 14.1 cM | 5.7467 | 0.0098 | ||
| 10580635 | NM_053200 | Ces3 | carboxylesterase 3 | 8 C5 | 5.6566 | 0.0021 | ||
| 10526712 | NM_013478 | Azgp1 | alpha-2-glycoprotein 1, zinc | 5 G2|5 78.0 cM | 5.4824 | 0.0232 | ||
| 10506125 | NM_013913 | Angptl3 | angiopoietin-like 3 | 4 C6|4 48.0 cM | 5.2732 | 0.0124 | ||
| 10580624 | NM_007954 | Es1 | esterase 1 | 8 C5|8 43.0 cM | 5.1268 | 0.0355 | ||
| 10513630 | NM_007443 | Ambp | alpha 1 microglobulin/bikunin | 4 C1-C3|4 30.6 cM | 5.1009 | 0.0025 | ||
| 10351852 | NM_007768 | Crp | C-reactive protein, pentraxin-related | 1 H3|1 94.2 cM | 4.9882 | 0.0025 | ||
| 10363541 | NM_007494 | Ass1 | argininosuccinate synthetase 1 | 2 B|2 20.0 cM | 4.9534 | 0.0057 | ||
| 10480003 | NM_010582 | Itih2 | inter-alpha trypsin inhibitor, heavy chain 2 | 2 A1|2 1.0 cM | 4.9513 | 0.0059 | ||
| 10542615 | NM_020495 | Slco1b2 | solute carrier organic anion transporter family, member 1b2 | 6 G1|6 67.0 cM | 4.89 | 0.0212 | ||
| 10367221 | NM_133997 | Apof | apolipoprotein F | 10 D3|10 73.0 cM | 4.8785 | 0.0025 | ||
| 10551287 | NM_133657 | Cyp2a12 | cytochrome P450, family 2, subfamily a, polypeptide 12 | 7 A3 | 4.8046 | 0.0096 | ||
| 10494643 | NM_008256 | Hmgcs2 | 3-hydroxy-3-methylglutaryl -Coenzyme A synthase 2 | 3 F2.2|3 48.0 cM | 4.5815 | 0.0144 | ||
| 10563602 | NM_011316 | Saa4 | serum amyloid A 4 | 7 B4|7 23.5 cM | 4.5245 | 0.001 | ||
| 10471154 | NM_007494 | Ass1 | argininosuccinate synthetase 1 | 2 B|2 20.0 cM | 4.4979 | 0.0069 | ||
| 10357516 | NM_007576 | C4bp | complement component 4 binding protein | 1 E4|1 67.6 cM | 4.4217 | 0.001 | ||
| 10416451 | NM_019775 | Cpb2 | carboxypeptidase B2 (plasma) | 14 D2 | 4.3546 | 0.0036 | ||
| 10551293 | NM_007817 | Cyp2f2 | cytochrome P450, family 2, subfamily f, polypeptide 2 | 7 A3 | 4.2794 | 0.0394 | ||
| 10398069 | NM_009253 | Serpina3m | serine (or cysteine) peptidase inhibitor, clade A, member 3M | 12 E | 4.2775 | 0.0231 | ||
| 10438681 | NM_201375 | Kng2 | kininogen 2 | 16 B1 | 4.2464 | 0.0051 | ||
| 10367215 | NM_133996 | Apon | apolipoprotein N | 10 D3|10 76.0 cM | 4.2277 | 0.0021 | ||
| 10351015 | NM_080844 | Serpinc1 | serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 | 1 H2.1|1 84.6 cM | 4.1985 | 0.0034 | ||
| 10403322 | NM_030611 | Akr1c6 | aldo-keto reductase family 1, member C6 | 13 A2|13 8.0 cM | 4.1493 | 0.0498 | ||
| 10452316 | NM_009778 | C3 | complement component 3 | 17 E1-E3|17 34.3 cM | 4.1151 | 0.0339 | ||
| 10596718 | NM_023805 | Slc38a3 | solute carrier family 38, member 3 | 9 F1|9 63.0 cM | 4.1024 | 0.0239 | ||
| 10501555 | NM_007446 | Amy1 | amylase 1, salivary | 3 F3|3 50.0 cM | 4.1021 | 0.0069 | ||
| 10519497 | NM_054098 | Steap4 | STEAP family member 4 | 5 A1 | 3.9917 | 0.0063 | ||
| 10496001 | NM_007686 | Cfi | complement component factor i | 3 G3|3 66.6 cM | 3.9698 | 0.0014 | ||
| 10475532 | NM_021507 | Sqrdl | sulfide quinone reductase-like (yeast) | 2 F2 | 3.9595 | 0 | ||
| 10593225 | NM_001033324 | Zbtb16 | zinc finger and BTB domain containing 16 | 9 A5.3|9 23.0 cM | 3.8909 | 0.018 | ||
| 10458828 | NM_033037 | Cdo1 | cysteine dioxygenase 1, cytosolic | 18 C|18 23.0 cM | 3.7452 | 0.0025 | ||
| 10514763 | NM_146148 | C8a | complement component 8, alpha polypeptide | 4 C6 | 3.7375 | 0.0096 | ||
| 10358339 | NM_009888 | Cfh | complement component factor h | 1 F|1 74.1 cM | 3.6868 | 0.0032 | ||
| 10514532 | NM_010007 | Cyp2j5 | cytochrome P450, family 2, subfamily j, polypeptide 5 | 4 C5|4 46.5 cM | 3.6645 | 0.0256 | ||
| 10450038 | NM_020581 | Angptl4 | angiopoietin-like 4 | 17 B1 | 3.5851 | 0.0062 | ||
| 10511375 | NM_007824 | Cyp7a1 | cytochrome P450, family 7, subfamily a, polypeptide 1 | 4 A1 | 3.5797 | 0.0076 | ||
| 10423002 | NM_207216 | Ugt3a1 | UDP glycosyltransferases 3 family, polypeptide A1 | 15 A1 | 3.5454 | 0.0345 | ||
| 10483410 | NM_021022 | Abcb11 | ATP-binding cassette, sub-family B (MDR/TAP), member 11 | 2 C2|2 38.4 cM | 3.4306 | 0.0445 | ||
| 10447885 | NM_153151 | Acat3 | acetyl-Coenzyme A acetyltransferase 3 | 17 A1|17 7.55 cM | 3.3695 | 0.0023 | ||
| 10537169 | NM_009731 | Akr1b7 | aldo-keto reductase family 1, member B7 | 6 B1|6 14.0 cM | 3.3685 | 0.0032 | ||
| 10395273 | BC052902 | Gdap10 | ganglioside-induced differentiation-associated-protein 10 | 12 A3 | 3.3561 | 0.0043 | ||
| 10503502 | NM_015767 | Ttpa | tocopherol (alpha) transfer protein | 4 A3|4 22.7 cM | 3.2169 | 0.0089 | ||
| 10585015 | NM_080434 | Apoa5 | apolipoprotein A-V | 9 B | 3.2153 | 0.0026 | ||
| 10581388 | NM_008490 | Lcat | lecithin cholesterol acyltransferase | 8 D3|8 53.0 cM | 3.2103 | 0.0014 | ||
| 10482004 | BC094504 | AI182371 | expressed sequence AI182371 | 2 B | 3.1837 | 0.0281 | ||
| 10574027 | NM_013602 | Mt1 | metallothionein 1 | 8 C5|8 45.0 cM | 3.1654 | 0.0374 | ||
| 10379736 | NM_183249 | 1100001G20Rik | RIKEN cDNA 1100001G20 gene | 11 C | 3.1049 | 0.0151 | ||
| 10420899 | NM_178747 | Gulo | gulonolactone (L-) oxidase | 14 D1 | 3.0186 | 0.0345 | ||
| 10435626 | NM_013547 | Hgd | homogentisate 1, 2-dioxygenase | 16 B3|16 27.3 cM | 2.9656 | 0.0296 | ||
| 10345065 | NM_001077353 | Gsta3 | glutathione S-transferase, alpha 3 | 1 A4|1 15.0 cM | 2.9309 | 0.0145 | ||
| 10356145 | NM_030556 | Slc19a3 | solute carrier family 19 (sodium/hydrogen exchanger), member 3 | 1 C5|1 51.0 cM | 2.9123 | 0.0301 | ||
| 10533612 | NM_008277 | Hpd | 4-hydroxyphenylpyruvic acid dioxygenase | 5 F|5 67.0 cM | 2.8602 | 0.0373 | ||
| 10451451 | NM_010321 | Gnmt | glycine N-methyltransferase | 17 C|17 10.0 cM | 2.8319 | 0.0461 | ||
| 10543333 | NM_013930 | Aass | aminoadipate-semialdehyde synthase | 6 A3.1|6 4.5 cM | 2.8069 | 0.0115 | ||
| 10596166 | NM_027918 | 1300017J02Rik | RIKEN cDNA 1300017J02 gene | 9 F1 | 2.7756 | 0.0308 | ||
| 10406646 | NM_028772 | Dmgdh | dimethylglycine dehydrogenase precursor | 13 C3 | 2.7336 | 0.0244 | ||
| 10500547 | NM_153193 | Hsd3b2 | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 | 3 F2.2|3 49.1 cM | 2.6399 | 0.0032 | ||
| 10570717 | NM_001177522 | Gm14850 | predicted gene 14850 | 8 A2 | 2.5973 | 0.0148 | ||
| 10361234 | NM_008288 | Hsd11b1 | hydroxysteroid 11-beta dehydrogenase 1 | 1 H6 | 2.5957 | 0.0096 | ||
| 10507171 | NM_201640 | Cyp4a31 | cytochrome P450, family 4, subfamily a, polypeptide 31 | 4 D1 | 2.575 | 0.0484 | ||
| 10570732 | NM_001177528 | Gm15315 | predicted gene 15315 | 8 A2|8 | 2.5592 | 0.0098 | ||
| 10599826 | NM_007979 | F9 | coagulation factor IX | X A6-A7|X 22.0 cM | 2.5474 | 0.0143 | ||
| 10578916 | NM_025436 | Sc4mol | sterol-C4-methyl oxidase-like | 8 B3.1 | 2.5379 | 0.0023 | ||
| 10399365 | NM_177802 | Slc7a15 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 | 12 A1.1 | 2.5357 | 0.0361 | ||
| 10570693 | NM_007851 | Defa5 | defensin, alpha, 5 | 8 A2 | 2.5181 | 0.0129 | ||
| 10497590 | NM_007963 | Mecom | MDS1 and EVI1 complex locus | 3 A3|3 14.4 cM | 2.5047 | 0.0091 | ||
| 10545881 | NM_053096 | Cml2 | camello-like 2 | 6 C3 | 2.5013 | 0.027 | ||
| 10478523 | — | — | — | — | 2.4962 | 0.0021 | ||
| 10454944 | NM_201256 | Eif4ebp3 | eukaryotic translation initiation factor 4E binding protein 3 | 18 B2 | 2.4951 | 0.0117 | ||
| 10386683 | NM_026183 | Slc47a1 | solute carrier family 47, member 1 | 11 B2 | 2.4534 | 0.0067 | ||
| 10365559 | NM_010512 | Igf1 | insulin-like growth factor 1 | 10 C1|10 48.0 cM | 2.4375 | 0.0041 | ||
| 10500555 | NM_001161742 | Hsd3b3 | hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3 | 3 F2.2|3 49.1 cM | 2.4177 | 0.0094 | ||
| 10513412 | NM_008648 | Mup4 | major urinary protein 4 | 4 B3|4 27.8 cM | 2.3986 | 5.00E−04 | ||
| 10491846 | — | — | — | — | 2.3936 | 0.0128 | ||
| 10537306 | NM_145364 | Akr1d1 | aldo-keto reductase family 1, member D1 | 6 B1 | 2.3924 | 0.0045 | ||
| 10589099 | NM_029634 | Ip6k2 | inositol hexaphosphate kinase 2 | 9 F2 | 2.3776 | 0.0021 | ||
| 10594825 | NM_022026 | Aqp9 | aquaporin 9 | 9 D | 2.3723 | 0.0412 | ||
| 10519578 | NM_008830 | Abcb4 | ATP-binding cassette, sub-family B (MDR/TAP), member 4 | 5 A1|5 1.0 cM | 2.3672 | 0.0297 | ||
| 10590957 | NM_010242 | Fut4 | fucosyltransferase 4 | 9 A2|9 3.0 cM | 2.3611 | 0.0019 | ||
| 10447317 | NM_010137 | Epas1 | endothelial PAS domain protein 1 | 17 E4 | 2.2998 | 0.0142 | ||
| 10352000 | NM_133809 | Kmo | kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) | 1 H4 | 2.2966 | 0.0301 | ||
| 10360840 | NM_001081361 | Mosc1 | MOCO sulphurase C-terminal domain containing 1 | — | 2.2607 | 0.0105 | ||
| 10570660 | NM_001177481 | Gm10104 | predicted gene 10104 | 8 A2|8 | 2.2377 | 0.0163 | ||
| 10386460 | NM_008819 | Pemt | phosphatidylethanolamine N-methyltransferase | 11 B1.3|11 31.0 cM | 2.2274 | 0.0265 | ||
| 10576901 | NM_011388 | Slc10a2 | solute carrier family 10, member 2 | 8 A1.1|8 2.0 cM | 2.2212 | 0.0253 | ||
| 10530319 | NM_001038999 | Atp8a1 | ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 | 5 C3.1 | 2.1827 | 0.0152 | ||
| 10502050 | NM_027808 | Alpk1 | alpha-kinase 1 | 3 H1 | 2.1604 | 0.04 | ||
| 10381096 | NM_010517 | Igfbp4 | insulin-like growth factor binding protein 4 | 11 D | 2.1452 | 0.0025 | ||
| 10408689 | NM_153529 | Nrn1 | neuritin 1 | 13 A3.3 | 2.1125 | 0.0301 | ||
| 10587266 | NM_010295 | Gclc | glutamate-cysteine ligase, catalytic subunit | 9 D-E|9 42.0 cM | 2.1067 | 0.0473 | ||
| 10429588 | NM_001039720 | 9030619P08Rik | RIKEN cDNA 9030619P08 gene | 15 D3 | 2.1009 | 0.0025 | ||
| 10537146 | NM_008012 | Akr1b8 | aldo-keto reductase family 1, member B8 | 6 B1|6 13.0 cM | 2.0926 | 0.0397 | ||
| 10576774 | NM_029465 | Clec4g | C-type lectin domain family 4, member g | 8 A1.1 | 2.0888 | 0.0367 | ||
| 10542592 | NR_033555 | Gm10400 | predicted gene 10400 | — | 2.0729 | 0.0104 | ||
| 10496605 | NM_173763 | Ccbl2 | cysteine conjugate-beta lyase 2 | 3 H1 | 2.0685 | 0.0222 | ||
| 10358299 | NM_001029977 | EG214403 | predicted gene, EG214403 | 1 F | 2.0607 | 0.0386 | ||
| 10494023 | NM_011281 | Rorc | RAR-related orphan receptor gamma | 3 F2 | 2.0501 | 0.0013 | ||
| 10357363 | NM_001081756 | Nckap5 | NCK-associated protein 5 | 1 E3 | 2.0191 | 0.023 | ||
| 10506452 | AY512949 | AY512949 | cDNA sequence AY512949 | — | 2.0161 | 0.0416 | ||
| 10512895 | NM_007519 | Baat | bile acid-Coenzyme A: amino acid N-acyltransferase | 4 B1|4 22.7 cM | 2.0089 | 0.0204 | ||
| 10388430 | NM_011340 | Serpinf1 | serine (or cysteine) peptidase inhibitor, clade F, member 1 | 11 B5 | 2.0036 | 0.0181 | ||
| 10454198 | NM_026301 | Rnf125 | ring finger protein 125 | 18 A2 | 2.003 | 0.0053 | ||
| 10606989 | NM_001077364 | Tsc22d3 | TSC22 domain family, member 3 | X F1 | 1.9909 | 0.0197 | ||
| 10418455 | NM_008406 | Itih1 | inter-alpha trypsin inhibitor, heavy chain 1 | 14 A2-C1 | 1.9848 | 0.0374 | ||
| 10365482 | NM_011595 | Timp3 | tissue inhibitor of metalloproteinase 3 | 10 C1-D1|10 47.0 cM | 1.9692 | 0.0355 | ||
| 10388440 | NM_008878 | Serpinf2 | serine (or cysteine) peptidase inhibitor, clade F, member 2 | 11 B5 | 1.9646 | 0.043 | ||
| 10372682 | NM_029875 | Slc35e3 | solute carrier family 35, member E3 | 10 D2 | 1.9641 | 0.0062 | ||
| 10571657 | NM_007981 | Acsl1 | acyl-CoA synthetase long-chain family member 1 | 8 B2 | 1.9589 | 0.0041 | ||
| 10581650 | NM_011998 | Chst4 | carbohydrate (chondroitin 6/keratan) sulfotransferase 4 | — | 1.9469 | 0.039 | ||
| 10394990 | NM_026037 | Mboat2 | membrane bound O-acyltransferase domain containing 2 | 12 A1.3 | 1.9401 | 0.0244 | ||
| 10596072 | NM_001161362 | Ppp2r3a | protein phosphatase 2, regulatory subunit B'', alpha | 9 E4 | 1.9318 | 0.0021 | ||
| 10411332 | NM_008255 | Hmgcr | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | 13 D1|13 49.0 cM | 1.9292 | 0.0013 | ||
| 10571840 | NM_008278 | Hpgd | hydroxyprostaglandin dehydrogenase 15 (NAD) | 8 B3.2 | 1.9235 | 0.0025 | ||
| 10435787 | — | — | — | — | 1.9234 | 0.0109 | ||
| 10555438 | NM_199012 | Fchsd2 | FCH and double SH3 domains 2 | 7 E3 | 1.9147 | 0.0156 | ||
| 10436100 | NM_181596 | Retnlg | resistin like gamma | 16 B5|16 32.5 cM | 1.9126 | 0.0028 | ||
| 10542594 | AY512955 | Gm10210 | predicted gene 10210 | — | 1.9107 | 0.047 | ||
| 10554693 | NM_023377 | Stard5 | StAR-related lipid transfer (START) domain containing 5 | 7 D3 | 1.903 | 0.0204 | ||
| 10391744 | AK135410 | Gpatch8 | G patch domain containing 8 | 11 E1 | 1.9025 | 0.0148 | ||
| 10560624 | NM_009696 | Apoe | apolipoprotein E | 7 A3|7 4.0 cM | 1.8965 | 0.0301 | ||
| 10542880 | BC048711 | 4833442J19Rik | RIKEN cDNA 4833442J19 gene | 6 G3 | 1.8917 | 0.0144 | ||
| 10461150 | — | — | — | — | 1.8816 | 0.0397 | ||
| 10585982 | NM_173018 | Myo9a | myosin IXa | 9 B | 1.8764 | 0.019 | ||
| 10547469 | NM_001037155 | Hsn2 | hereditary sensory neuropathy, type II | 6 F1 | 1.8743 | 0.0152 | ||
| 10482762 | NM_145360 | Idi1 | isopentenyl-diphosphate delta isomerase | 13 A1 | 1.8722 | 0.0143 | ||
| 10411147 | NM_022884 | Bhmt2 | betaine-homocysteine methyltransferase 2 | 13 C3|13 49.0 cM | 1.8637 | 0.0317 | ||
| 10364038 | NM_133995 | Upb1 | ureidopropionase, beta | 10 C1 | 1.8607 | 0.0099 | ||
| 10382888 | AK171830 | 2810008D09Rik | RIKEN cDNA 2810008D09 gene | — | 1.8559 | 0.0272 | ||
| 10485170 | NM_009963 | Cry2 | cryptochrome 2 (photolyase-like) | 2 E | 1.8553 | 0.0324 | ||
| 10594988 | NM_015806 | Mapk6 | mitogen-activated protein kinase 6 | 9 D|9 38.0 cM | 1.8546 | 0.0327 | ||
| 10535938 | NM_133898 | N4bp2l1 | NEDD4 binding protein 2-like 1 | 5 G3 | 1.8517 | 0.0189 | ||
| 10556769 | NM_016870 | Acsm3 | acyl-CoA synthetase medium-chain family member 3 | 7 F3 | 1.8511 | 0.0207 | ||
| 10401149 | NM_013738 | Plek2 | pleckstrin 2 | 12 D2 | 1.8429 | 0.0098 | ||
| 10599192 | NM_028894 | Lonrf3 | LON peptidase N-terminal domain and ring finger 3 | X A2 | 1.8374 | 0.035 | ||
| 10391746 | NM_001159492 | Gpatch8 | G patch domain containing 8 | 11 E1 | 1.8331 | 0.0248 | ||
| 10403413 | NM_145360 | Idi1 | isopentenyl-diphosphate delta isomerase | 13 A1 | 1.8228 | 0.0272 | ||
| 10580955 | — | — | — | — | 1.8221 | 0.0277 | ||
| 10396730 | — | — | — | — | 1.8218 | 0.0374 | ||
| 10508800 | AY140896 | Gm3579 | predicted gene 3579 | 4 D2.3|4 | 1.8214 | 0.0197 | ||
| 10573865 | AY140896 | Gm3579 | predicted gene 3579 | 4 D2.3|4 | 1.8214 | 0.0197 | ||
| 10558961 | NM_053082 | Tspan4 | tetraspanin 4 | 7 F5 | 1.8183 | 0.0132 | ||
| 10350753 | NM_008131 | Glul | glutamate-ammonia ligase (glutamine synthetase) | — | 1.817 | 0.0285 | ||
| 10351043 | NR_028543 | Snord47 | small nucleolar RNA, C/D box 47 | 1|1 | 1.8156 | 0.0327 | ||
| 10377439 | NM_001159367 | Per1 | period homolog 1 (Drosophila) | 11 B | 1.8092 | 0.0359 | ||
| 10520388 | NM_028234 | Rbm33 | RNA binding motif protein 33 | 5 B1 | 1.8056 | 0.0379 | ||
| 10370931 | NM_021462 | Mknk2 | MAP kinase-interacting serine/threonine kinase 2 | 10 C1 | 1.7989 | 0.0023 | ||
| 10582658 | NM_007428 | Agt | angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | 8 E2|8 68.0 cM | 1.7834 | 0.0217 | ||
| 10472860 | NM_019688 | Rapgef4 | Rap guanine nucleotide exchange factor (GEF) 4 | 2 C3 | 1.7824 | 0.0118 | ||
| 10505489 | NM_021362 | Pappa | pregnancy-associated plasma protein A | 4 C1|4 32.2 cM | 1.7809 | 0.0012 | ||
| 10377751 | NM_009714 | Asgr1 | asialoglycoprotein receptor 1 | 11 B3|11 37.0 cM | 1.7793 | 0.0146 | ||
| 10351224 | NM_007976 | F5 | coagulation factor V | 1 H2.2|1 86.6 cM | 1.7735 | 0.0277 | ||
| 10372988 | NM_011391 | Slc16a7 | solute carrier family 16 (monocarboxylic acid transporters), member 7 | 10 D3 | 1.7729 | 0.0311 | ||
| 10351259 | NM_054087 | Slc19a2 | solute carrier family 19 (thiamine transporter), member 2 | 1 H2.2|1 87.0 cM | 1.7698 | 0.0242 | ||
| 10409876 | NM_007796 | Ctla2a | cytotoxic T lymphocyte-associated protein 2 alpha | 13 B2|13 36.0 cM | 1.7598 | 0.0424 | ||
| 10410877 | NM_001081176 | Polr3g | polymerase (RNA) III (DNA directed) polypeptide G | 13 C3 | 1.7388 | 0.0345 | ||
| 10424686 | BC025446 | BC025446 | cDNA sequence BC025446 | 15 D3 | 1.7378 | 0.0152 | ||
| 10351533 | NM_009803 | Nr1i3 | nuclear receptor subfamily 1, group I, member 3 | 1 H3|1 92.6 cM | 1.7285 | 0.0341 | ||
| 10533050 | NM_030704 | Hspb8 | heat shock protein 8 | 5 F|5 59.0 cM | 1.716 | 0.0394 | ||
| 10496872 | NM_133222 | Eltd1 | EGF, latrophilin seven transmembrane domain containing 1 | 3 H3-H4 | 1.7157 | 0.0233 | ||
| 10518743 | NM_173371 | H6pd | hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) | 4 E2|4 78.4 cM | 1.7094 | 0.0165 | ||
| 10487021 | NM_011774 | Slc30a4 | solute carrier family 30 (zinc transporter), member 4 | 2 E5|2 69.0 cM | 1.7066 | 0.0161 | ||
| 10595622 | — | — | — | — | 1.7056 | 0.0342 | ||
| 10595636 | — | — | — | — | 1.7056 | 0.0342 | ||
| 10595626 | — | — | — | — | 1.7034 | 0.029 | ||
| 10446425 | — | — | — | — | 1.7003 | 0.0214 | ||
| 10473160 | NM_080558 | Ssfa2 | sperm specific antigen 2 | 2 D | 1.6908 | 0.0159 | ||
| 10375229 | — | — | — | — | 1.6902 | 0.0112 | ||
| 10592938 | NM_138951 | Ttc36 | tetratricopeptide repeat domain 36 | 9 A5.2 | 1.6901 | 0.04 | ||
| 10394699 | NM_009072 | Rock2 | Rho-associated coiled-coil containing protein kinase 2 | 12 A3 | 1.6898 | 0.0376 | ||
| 10354085 | NM_019570 | Rev1 | REV1 homolog (S. cerevisiae) | 1 B | 1.6849 | 0.0097 | ||
| 10462922 | NM_019588 | Plce1 | phospholipase C, epsilon 1 | 19 D1 | 1.6829 | 0.032 | ||
| 10395259 | NM_021524 | Nampt | nicotinamide phosphoribosyltransferase | 12 B1 | 1.676 | 0.0284 | ||
| 10597875 | NM_010012 | Cyp8b1 | cytochrome P450, family 8, subfamily b, polypeptide 1 | 9 F4|9 71.0 cM | 1.6755 | 0.013 | ||
| 10344809 | NM_026493 | Cspp1 | centrosome and spindle pole associated protein 1 | 1 A2 | 1.6735 | 0.0484 | ||
| 10347748 | NM_028817 | Acsl3 | acyl-CoA synthetase long-chain family member 3 | 1 C4|1 24.1 cM | 1.6733 | 0.019 | ||
| 10375216 | NM_145962 | Pank3 | pantothenate kinase 3 | 11 A4 | 1.6733 | 0.013 | ||
| 10545877 | NM_023455 | Nat8 | N-acetyltransferase 8 (GCN5-related, putative) | 6 C3 | 1.6673 | 0.0378 | ||
| 10374453 | NM_008131 | Glul | glutamate-ammonia ligase (glutamine synthetase) | — | 1.666 | 0.0345 | ||
| 10515690 | NM_198170 | Szt2 | seizure threshold 2 | 4 D2.1 | 1.6601 | 0.0207 | ||
| 10591612 | NM_177030 | Dock6 | dedicator of cytokinesis 6 | 9 A3 | 1.6594 | 0.0225 | ||
| 10389214 | NM_011338 | Ccl9 | chemokine (C-C motif) ligand 9 | 11 C|11 47.4 cM | 1.6564 | 0.0355 | ||
| 10587688 | — | — | — | — | 1.6554 | 0.0277 | ||
| 10595620 | — | — | — | — | 1.6554 | 0.0277 | ||
| 10366275 | — | — | — | — | 1.6481 | 0.0095 | ||
| 10607089 | NM_207625 | Acsl4 | acyl-CoA synthetase long-chain family member 4 | X F2 | 1.647 | 0.0484 | ||
| 10379820 | NM_133360 | Acaca | acetyl-Coenzyme A carboxylase alpha | 11 C | 1.6452 | 0.0014 | ||
| 10414537 | NM_001161731 | Ang | angiogenin, ribonuclease, RNase A family, 5 | 14 B-C1|14 18.0 cM | 1.6406 | 0.0094 | ||
| 10505779 | NM_139306 | Acer2 | alkaline ceramidase 2 | 4 C4 | 1.6403 | 0.0141 | ||
| 10564183 | AF241256 | Snord116 | small nucleolar RNA, C/D box 116 cluster | 7 C|7 29.0 cM | 1.6393 | 0.0126 | ||
| 10544573 | NM_027852 | Rarres2 | retinoic acid receptor responder (tazarotene induced) 2 | 6 B2.3 | 1.6174 | 0.0361 | ||
| 10472436 | NM_020283 | B3galt1 | UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 | 2 C3 | 1.6168 | 0.0376 | ||
| 10515399 | NM_013807 | Plk3 | polo-like kinase 3 (Drosophila) | 4 D1 | 1.616 | 0.039 | ||
| 10407876 | NM_138654 | 5033411D12Rik | RIKEN cDNA 5033411D12 gene | 13 A2 | 1.6108 | 0.0317 | ||
| 10478847 | NM_001081005 | 1500012F01Rik | RIKEN cDNA 1500012F01 gene | 2 H3 | 1.6077 | 0.0445 | ||
| 10362896 | NM_009846 | Cd24a | CD24a antigen | 10 B2|10 26.0 cM | 1.6059 | 0.0272 | ||
| 10535956 | NM_001163493 | Stard13 | StAR-related lipid transfer (START) domain containing 13 | 5 G3 | 1.6025 | 0.0225 | ||
| 10371400 | NM_007771 | Cry1 | cryptochrome 1 (photolyase-like) | 10 C|10 46.0 cM | 1.5989 | 0.0109 | ||
| 10540122 | NM_009320 | Slc6a6 | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | 6 D1|6 38.2 cM | 1.5937 | 0.0136 | ||
| 10580953 | — | — | — | — | 1.5913 | 0.0232 | ||
| 10356082 | NM_011636 | Plscr1 | phospholipid scramblase 1 | 9 E3.3 | 1.5899 | 0.036 | ||
| 10453272 | NM_001001806 | Zfp36l2 | zinc finger protein 36, C3H type-like 2 | 17 E4 | 1.5897 | 0.013 | ||
| 10488322 | NM_001033348 | Ralgapa2 | Ral GTPase activating protein, alpha subunit 2 (catalytic) | 2 G1 | 1.5885 | 0.0019 | ||
| 10462406 | NM_001081319 | C030046E11Rik | RIKEN cDNA C030046E11 gene | 19 C1 | 1.5858 | 0.0471 | ||
| 10382797 | NM_176902 | Fam100b | family with sequence similarity 100, member B | 11 E2 | 1.583 | 0.023 | ||
| 10346328 | XR_031547 | Gm8292 | predicted gene 8292 | 1 C1.2 | 1.5828 | 0.0066 | ||
| 10459766 | NR_028560 | Scarna17 | small Cajal body-specific RNA 17 | 18|18 | 1.5813 | 0.0232 | ||
| 10461642 | NR_028560 | Scarna17 | small Cajal body-specific RNA 17 | 18|18 | 1.5813 | 0.0232 | ||
| 10474545 | NM_133649 | Slc12a6 | solute carrier family 12, member 6 | 2 E3 | 1.5812 | 0.0361 | ||
| 10471503 | BC110660 | Taf1d | TATA box binding protein (Tbp)-associated factor, RNA polymerase I, D | 9 A3 | 1.5797 | 0.0352 | ||
| 10572865 | NM_027837 | Isx | intestine specific homeobox | 8 C2 | 1.5776 | 0.0224 | ||
| 10357345 | NM_172484 | Nckap5 | NCK-associated protein 5 | 1 E3 | 1.5726 | 0.0374 | ||
| 10501494 | NM_001042711 | Amy2a5 | amylase 2a5 | 3 F3|3 50.0 cM | 1.5715 | 0.015 | ||
| 10558295 | ENSMUST00000106157 | Zranb1 | zinc finger, RAN-binding domain containing 1 | 7 F3 | 1.5699 | 0.0362 | ||
| 10553829 | NR_003376 | Gm7367 | 1110014K08Rik pseudogene | 7 C|7 | 1.5663 | 0.0376 | ||
| 10553831 | NR_003376 | Gm7367 | 1110014K08Rik pseudogene | 7 C|7 | 1.5663 | 0.0376 | ||
| 10557233 | NM_144925 | Tnrc6a | trinucleotide repeat containing 6a | 7 F3 | 1.5644 | 0.0447 | ||
| 10439667 | NM_145389 | BC016579 | cDNA sequence, BC016579 | 16 B5 | 1.5633 | 0.0355 | ||
| 10354168 | NM_018775 | Tbc1d8 | TBC1 domain family, member 8 | 1 B | 1.5603 | 0.0205 | ||
| 10418169 | ENSMUST00000079800 // ENSMUST00000079800 | 1700054O19Rik // 1700054O19Rik | RIKEN cDNA 1700054O19 gene // RIKEN cDNA 1700054O19 gene | 14 A3 // 14 A3 | 1.5539 | 0.0023 | ||
| 10509063 | NM_178257 | Il22ra1 | interleukin 22 receptor, alpha 1 | 4 D3|4 62.0 cM | 1.5466 | 0.0232 | ||
| 10391732 | NM_001159492 | Gpatch8 | G patch domain containing 8 | 11 E1 | 1.5421 | 0.0168 | ||
| 10399198 | NM_019744 | Ncoa4 | nuclear receptor coactivator 4 | 14 B | 1.5412 | 0.0444 | ||
| 10512774 | NM_178893 | Coro2a | coronin, actin binding protein 2A | 4 B1 | 1.5393 | 0.0307 | ||
| 10485633 | ENSMUST00000099651 | Gm10796 | predicted gene 10796 | — | 1.5366 | 0.0286 | ||
| 10350758 | AK080751 | A930039A15Rik | RIKEN cDNA A930039A15 gene | — | 1.5361 | 0.044 | ||
| 10427461 | NM_001136079 | Ptger4 | prostaglandin E receptor 4 (subtype EP4) | 15 A1|15 6.4 cM | 1.5341 | 0.0233 | ||
| 10417887 | NM_029104 | Zmynd17 | zinc finger, MYND domain containing 17 | 14 B | 1.531 | 0.0494 | ||
| 10436209 | NM_001033238 | Cblb | Casitas B-lineage lymphoma b | 16 B5 | 1.5287 | 0.0345 | ||
| 10587792 | NM_011636 | Plscr1 | phospholipid scramblase 1 | 9 E3.3 | 1.5275 | 0.0242 | ||
| 10469300 | NM_177268 | Ankrd16 | ankyrin repeat domain 16 | 2 A1 | 1.5231 | 0.0313 | ||
| 10497487 | — | — | — | — | 1.5217 | 0.0217 | ||
| 10510286 | NM_027985 | Mad2l2 | MAD2 mitotic arrest deficient-like 2 (yeast) | 4 E1 | 1.52 | 0.0152 | ||
| 10475866 | NM_207680 | Bcl2l11 | BCL2-like 11 (apoptosis facilitator) | 2 F3-G1 | 1.5187 | 0.0194 | ||
| 10497421 | NM_080634 | Hps3 | Hermansky-Pudlak syndrome 3 homolog (human) | 3 A2|3 12.5 cM | 1.5153 | 0.0272 | ||
| 10538356 | NM_001163640 | Chn2 | chimerin (chimaerin) 2 | 6 B3 | 1.5142 | 0.0215 | ||
| 10475946 | NM_178404 | Zc3h6 | zinc finger CCCH type containing 6 | 2 F1 | 1.5139 | 0.0465 | ||
| 10584350 | NM_009429 | Tpt1 | tumor protein, translationally-controlled 1 | 14 D3 | 1.5134 | 0.0362 | ||
| 10523856 | — | — | — | — | 1.5131 | 0.0416 | ||
| 10564159 | AF241256 | Snord116 | small nucleolar RNA, C/D box 116 cluster | 7 C|7 29.0 cM | 1.5126 | 0.0148 | ||
| 10559916 | — | — | — | — | 1.5073 | 0.0309 | ||
| 10410513 | NM_001040692 | Slc6a18 | solute carrier family 6 (neurotransmitter transporter), member 18 | 13 C1|13 42.0 cM | 1.506 | 0.0469 | ||
| 10413839 | NM_001033988 | Ncoa4 | nuclear receptor coactivator 4 | 14 B | 1.5054 | 0.048 | ||
| 10459768 | — | — | — | — | 1.5005 | 0.0316 | ||
| 10562234 | NM_001110252 | Hpn | hepsin | 7 B1 | 1.4972 | 0.0454 | ||
| 10407370 | NM_001113550 | 4833420G17Rik | RIKEN cDNA 4833420G17 gene | 13 D2.3 | 1.4954 | 0.0245 | ||
| 10517703 | — | — | — | — | 1.4948 | 0.0146 | ||
| 10497441 | NM_013755 | Gyg | glycogenin | 3 A2|3 12.5 cM | 1.4893 | 0.0056 | ||
| 10470959 | NM_172267 | Phyhd1 | phytanoyl-CoA dioxygenase domain containing 1 | 2 B | 1.4891 | 0.0376 | ||
| 10601755 | NM_019865 | Rpl36a | ribosomal protein L36A | X E3 | 1.4875 | 0.0384 | ||
| 10355050 | NM_001045513 | Raph1 | Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 | 1 C2 | 1.4843 | 0.0308 | ||
| 10514500 | NM_001004141 | Cyp2j11 | cytochrome P450, family 2, subfamily j, polypeptide 11 | 4 C5 | 1.483 | 0.045 | ||
| 10497399 | NM_001122759 | Pde7a | phosphodiesterase 7A | 3 A2|3 7.0 cM | 1.4697 | 0.0106 | ||
| 10532040 | NM_026856 | Zfp644 | zinc finger protein 644 | 5 E5|5 56.0 cM | 1.4674 | 0.0484 | ||
| 10421972 | NM_019865 | Rpl36a | ribosomal protein L36A | X E3 | 1.467 | 0.0489 | ||
| 10399478 | NM_015763 | Lpin1 | lipin 1 | 12 A1.1|12 9.0 cM | 1.4665 | 0.0214 | ||
| 10438358 | NM_213614 | 5-Sep | septin 5 | 16 A3|16 11.42 cM | 1.4654 | 0.0468 | ||
| 10544219 | NM_139294 | Braf | Braf transforming gene | 6 B1|6 15.5 cM | 1.4639 | 0.0078 | ||
| 10358894 | NM_146126 | Sord | sorbitol dehydrogenase | 2 E5|2 66.0 cM | 1.4618 | 0.0277 | ||
| 10401997 | NM_011877 | Ptpn21 | protein tyrosine phosphatase, non-receptor type 21 | 12 F1 | 1.4616 | 0.0405 | ||
| 10475437 | NM_146126 | Sord | sorbitol dehydrogenase | 2 E5|2 66.0 cM | 1.4599 | 0.0325 | ||
| 10379034 | NM_026708 | Tlcd1 | TLC domain containing 1 | 11 B5 | 1.4516 | 0.0339 | ||
| 10497935 | — | — | — | — | 1.4454 | 0.0361 | ||
| 10389025 | NM_177390 | Myo1d | myosin ID | 11 B5|11 46.0 cM | 1.443 | 0.0454 | ||
| 10389022 | NM_177390 | Myo1d | myosin ID | 11 B5|11 46.0 cM | 1.4428 | 0.0447 | ||
| 10396956 | NM_018814 | Pcnx | pecanex homolog (Drosophila) | 12 D1 | 1.4386 | 0.0122 | ||
| 10462346 | NM_021525 | Rcl1 | RNA terminal phosphate cyclase-like 1 | 19 C1 | 1.4359 | 0.0151 | ||
| 10352416 | NM_130890 | Capn8 | calpain 8 | 1 H4 | 1.4317 | 0.0233 | ||
| 10592001 | NM_011176 | St14 | suppression of tumorigenicity 14 (colon carcinoma) | 9 A4|9 17.0 cM | 1.4289 | 0.0123 | ||
| 10507110 | — | — | — | — | 1.4264 | 0.0448 | ||
| 10552030 | NM_008085 | Gapdhs | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | 7 B1 | 1.4162 | 0.0437 | ||
| 10447429 | ENSMUST00000072072 | Gm4832 | predicted gene 4832 | 17 E4 | 1.4137 | 0.035 | ||
| 10520371 | NM_028234 | Rbm33 | RNA binding motif protein 33 | 5 B1 | 1.4117 | 0.045 | ||
| 10402117 | NM_153587 | Rps6ka5 | ribosomal protein S6 kinase, polypeptide 5 | 12 E | 1.4072 | 0.048 | ||
| 10481827 | NM_001085507 | Zbtb34 | zinc finger and BTB domain containing 34 | 2 B | 1.4071 | 0.0194 | ||
| 10526656 | NM_146164 | Lrch4 | leucine-rich repeats and calponin homology (CH) domain containing 4 | 5 G2 | 1.404 | 0.0447 | ||
| 10582811 | NM_001164598 | Irf2bp2 | interferon regulatory factor 2 binding protein 2 | 8 E2 | 1.3943 | 0.0286 | ||
| 10493343 | — | — | — | — | 1.3926 | 0.0441 | ||
| 10474725 | NM_013719 | Eif2ak4 | eukaryotic translation initiation factor 2 alpha kinase 4 | — | 1.3874 | 0.0301 | ||
| 10489377 | NM_012032 | Serinc3 | serine incorporator 3 | 2 H3 | 1.3784 | 0.0274 | ||
| 10480570 | NM_001162485 | Arrdc1 | arrestin domain containing 1 | 2 A3 | 1.3734 | 0.0478 | ||
| 10366446 | NM_146010 | Tspan8 | tetraspanin 8 | 10 D2 | 1.3733 | 0.0324 | ||
| 10346970 | NM_011086 | Pikfyve | phosphoinositide kinase, FYVE finger containing | 1 C2 | 1.3711 | 0.0175 | ||
| 10384423 | NM_172496 | Cobl | cordon-bleu | 11 A1|11 6.5 cM | 1.3708 | 0.048 | ||
| 10476443 | NM_013829 | Plcb4 | phospholipase C, beta 4 | 2 F3|2 77.0 cM | 1.3705 | 0.0333 | ||
| 10534889 | NM_178162 | Agfg2 | ArfGAP with FG repeats 2 | 5 G2 | 1.369 | 0.0181 | ||
| 10518428 | NM_011929 | Clcn6 | chloride channel 6 | 4 76.4 cM | 1.3681 | 0.0262 | ||
| 10542264 | NM_001163445 | 2700089E24Rik | RIKEN cDNA 2700089E24 gene | 6 G1 | 1.3677 | 0.045 | ||
| 10497485 | — | — | — | — | 1.3662 | 0.0333 | ||
| 10349065 | NM_001122676 | Zcchc2 | zinc finger, CCHC domain containing 2 | 1 E2.1 | 1.3603 | 0.0286 | ||
| 10477311 | NM_001039939 | Asxl1 | additional sex combs like 1 (Drosophila) | 2 H1 | 1.3592 | 0.0395 | ||
| 10432122 | NM_001170711 | Asb8 | ankyrin repeat and SOCS box-containing 8 | 15 F2 | 1.3504 | 0.0362 | ||
| 10567229 | NM_001031814 | Smg1 | SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) | 7 F2 | 1.3481 | 0.0485 | ||
| 10376312 | NM_028451 | Larp1 | La ribonucleoprotein domain family, member 1 | 11 B2 | 1.3375 | 0.0311 | ||
| 10503161 | NM_001081417 | Chd7 | chromodomain helicase DNA binding protein 7 | 4 A1|4 1.0 cM | 1.3312 | 0.0407 | ||
| 10587150 | NM_001081322 | Myo5c | myosin VC | 9 D | 1.3133 | 0.0489 | ||
| 10476033 | NM_183262 | Stk35 | serine/threonine kinase 35 | 2 F1 | 1.2947 | 0.0412 | ||
| 10466530 | NM_001163144 | Pcsk5 | proprotein convertase subtilisin/kexin type 5 | 19 B|19 18.0 cM | 1.2894 | 0.0362 | ||
| 10478219 | NM_021280 | Plcg1 | phospholipase C, gamma 1 | 2 H2|2 92.0 cM | 1.2708 | 0.0381 | ||
| 10367919 | NM_029075 | Stx11 | syntaxin 11 | 10 A1 | −1.2794 | 0.048 | ||
| 10605319 | NM_145405 | Ubl4 | ubiquitin-like 4 | X A7.3|X 29.9 cM | −1.2895 | 0.048 | ||
| 10508089 | NM_025544 | Mrps15 | mitochondrial ribosomal protein S15 | 4 D2.1 | −1.2959 | 0.0479 | ||
| 10591482 | NM_016679 | Keap1 | kelch-like ECH-associated protein 1 | 9 A3 | −1.298 | 0.0397 | ||
| 10591739 | NM_001102404 | Acp5 | acid phosphatase 5, tartrate resistant | 9 A3|9 6.0 cM | −1.3007 | 0.0272 | ||
| 10551159 | XR_034166 | Gm4607 | predicted gene 4607 | 7 A3|7 | −1.3016 | 0.0479 | ||
| 10413222 | NM_134084 | Ppif | peptidylprolyl isomerase F (cyclophilin F) | 14 A3 | −1.3037 | 0.048 | ||
| 10356657 | NM_024197 | Ndufa10 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 10 | 1 D | −1.3055 | 0.0479 | ||
| 10347672 | NM_027886 | Stk11ip | serine/threonine kinase 11 interacting protein | 1 C3 | −1.3059 | 0.0438 | ||
| 10439695 | NM_019754 | Tagln3 | transgelin 3 | 16 B5 | −1.3087 | 0.0387 | ||
| 10593591 | NM_144784 | Acat1 | acetyl-Coenzyme A acetyltransferase 1 | 9 30.0 cM | −1.3115 | 0.0418 | ||
| 10432866 | NM_001003667 | Krt77 | keratin 77 | 15 F3 | −1.3141 | 0.0484 | ||
| 10378443 | NM_001011851 | Olfr412 | olfactory receptor 412 | 11 B5 | −1.3165 | 0.048 | ||
| 10487629 | NM_130884 | Idh3b | isocitrate dehydrogenase 3 (NAD+) beta | 2 F3 | −1.3214 | 0.0265 | ||
| 10432636 | NM_174992 | Smagp | small cell adhesion glycoprotein | 15 F1 | −1.3221 | 0.0376 | ||
| 10543684 | NR_030713 | Mir183 | microRNA 183 | — | −1.3247 | 0.0355 | ||
| 10411156 | NM_029153 | Scamp1 | secretory carrier membrane protein 1 | 13 D1 | −1.3284 | 0.036 | ||
| 10585417 | NM_029573 | Idh3a | isocitrate dehydrogenase 3 (NAD+) alpha | 9 A5.3 | −1.3357 | 0.0367 | ||
| 10415742 | NM_027436 | Mipep | mitochondrial intermediate peptidase | 14 D1 | −1.3368 | 0.0307 | ||
| 10406205 | NM_030711 | Erap1 | endoplasmic reticulum aminopeptidase 1 | 13 C1 | −1.3442 | 0.0454 | ||
| 10449940 | NM_022434 | Cyp4f14 | cytochrome P450, family 4, subfamily f, polypeptide 14 | 17 B1 | −1.3479 | 0.0363 | ||
| 10420225 | NM_010780 | Cma1 | chymase 1, mast cell | 14 C3|14 20.0 cM | −1.3516 | 0.0307 | ||
| 10390685 | NM_025661 | Ormdl3 | ORM1-like 3 (S. cerevisiae) | 11 D | −1.3522 | 0.0445 | ||
| 10484431 | NM_025868 | Tmx2 | thioredoxin-related transmembrane protein 2 | 2 D | −1.3536 | 0.0486 | ||
| 10515930 | BC076608 | AA415398 | expressed sequence AA415398 | 4 D2.1 | −1.3575 | 0.0362 | ||
| 10607643 | — | — | — | — | −1.3579 | 0.0374 | ||
| 10386416 | NM_001011789 | Olfr222 | olfactory receptor 222 | 11 B1.3 | −1.3596 | 0.0447 | ||
| 10488291 | NM_015754 | Rbbp9 | retinoblastoma binding protein 9 | 2 G1-H1 | −1.3617 | 0.032 | ||
| 10447551 | NM_027457 | 5730437N04Rik | RIKEN cDNA 5730437N04 gene | 17 A1 | −1.3687 | 0.0396 | ||
| 10464128 | NM_007611 | Casp7 | caspase 7 | 19 D2|19 50.0 cM | −1.3699 | 0.0383 | ||
| 10377286 | NM_001081566 | Pik3r6 | phosphoinositide-3-kinase, regulatory subunit 6 | 11 B3 | −1.3731 | 0.0233 | ||
| 10539472 | NM_019542 | Nagk | N-acetylglucosamine kinase | 6 D1 | −1.3736 | 0.0143 | ||
| 10434191 | NM_013711 | Txnrd2 | thioredoxin reductase 2 | 16 A3|16 11.2 cM | −1.3797 | 0.0324 | ||
| 10369525 | BC024943 | 2010107G23Rik | RIKEN cDNA 2010107G23 gene | 10 B4 | −1.3815 | 0.0461 | ||
| 10465826 | NM_001160356 | AI462493 | expressed sequence AI462493 | 19 A | −1.3869 | 0.0399 | ||
| 10435617 | NM_001042499 | Rabl3 | RAB, member of RAS oncogene family-like 3 | 16 B3 | −1.3924 | 0.0387 | ||
| 10495285 | NM_019972 | Sort1 | sortilin 1 | 3 F3 | −1.3932 | 0.035 | ||
| 10447354 | NM_025868 | Tmx2 | thioredoxin-related transmembrane protein 2 | 2 D | −1.3953 | 0.0338 | ||
| 10575894 | NM_026648 | Lrrc50 | leucine rich repeat containing 50 | 8 E1 | −1.3988 | 0.0418 | ||
| 10368893 | NM_145743 | Lace1 | lactation elevated 1 | 10 B2|10 25.5 cM | −1.399 | 0.0265 | ||
| 10470050 | NM_007379 | Abca2 | ATP-binding cassette, sub-family A (ABC1), member 2 | 2 A2-B|2 12.6 cM | −1.4023 | 0.0316 | ||
| 10566618 | NM_206897 | Olfr6 | olfactory receptor 6 | 7 E3 | −1.4057 | 0.0416 | ||
| 10526514 | NM_021719 | Cldn15 | claudin 15 | 5 G2 | −1.4077 | 0.0418 | ||
| 10483679 | NM_001080707 | Gpr155 | G protein-coupled receptor 155 | 2 C3 | −1.4079 | 0.035 | ||
| 10476301 | NM_001177833 | Smox | spermine oxidase | 2 F1 | −1.408 | 0.0478 | ||
| 10365116 | NM_133964 | Dohh | deoxyhypusine hydroxylase/monooxygenase | 10 C1 | −1.4203 | 0.0304 | ||
| 10386086 | NM_153079 | Nmur2 | neuromedin U receptor 2 | 11 B1.3 | −1.4289 | 0.043 | ||
| 10442370 | NM_019910 | Dcpp1 | demilune cell and parotid protein 1 | 17 A3.3|17 10.0 cM | −1.4357 | 0.043 | ||
| 10593169 | NM_023114 | Apoc3 | apolipoprotein C-III | 9 A5.2|9 27.0 cM | −1.4359 | 0.0317 | ||
| 10579508 | NM_001164679 | Ano8 | anoctamin 8 | 8 B3.3 | −1.4375 | 0.015 | ||
| 10431300 | NM_001142357 | Alg12 | asparagine-linked glycosylation 12 homolog (yeast, alpha-1,6-mannosyltransferase) | 15 E3 | −1.4389 | 0.0334 | ||
| 10585874 | NM_010421 | Hexa | hexosaminidase A | 9 B|9 29.0 cM | −1.4394 | 0.0245 | ||
| 10571302 | NM_026432 | Tmem66 | transmembrane protein 66 | 8 A4 | −1.4412 | 0.0307 | ||
| 10519652 | NR_003596 | Gm6455 | predicted gene 6455 | 5 A1 | −1.4455 | 0.0286 | ||
| 10397975 | NM_026790 | Ifi27l1 | interferon, alpha-inducible protein 27 like 1 | 12 E|12 51.0 cM | −1.4507 | 0.0331 | ||
| 10362904 | NM_130892 | Rtn4ip1 | reticulon 4 interacting protein 1 | 10 B2|10 29.0 cM | −1.4553 | 0.047 | ||
| 10352767 | NM_010892 | Nek2 | NIMA (never in mitosis gene a)-related expressed kinase 2 | 1 H6|1 103.0 cM | −1.4555 | 0.0145 | ||
| 10480699 | NM_031843 | Dpp7 | dipeptidylpeptidase 7 | 2 A3 | −1.4566 | 0.0363 | ||
| 10529454 | ENSMUST00000069741 | E130018O15Rik | RIKEN cDNA E130018O15 gene | 5 B3 | −1.4683 | 0.0429 | ||
| 10600531 | — | — | — | — | −1.4695 | 0.0218 | ||
| 10426812 | NM_010271 | Gpd1 | glycerol-3-phosphate dehydrogenase 1 (soluble) | 15 56.8 cM | −1.4746 | 0.0375 | ||
| 10519688 | NR_003596 | Gm6455 | predicted gene 6455 | 5 A1 | −1.4804 | 0.0063 | ||
| 10518679 | NM_133435 | Nmnat1 | nicotinamide nucleotide adenylyltransferase 1 | 4 E2 | −1.4814 | 0.048 | ||
| 10357418 | NM_001081078 | Lct | lactase | 1 E4 | −1.4902 | 0.0333 | ||
| 10559590 | NM_207270 | Ptprh | protein tyrosine phosphatase, receptor type, H | 7 A1 | −1.494 | 0.0286 | ||
| 10525365 | NM_001042489 | Hvcn1 | hydrogen voltage-gated channel 1 | 5 F | −1.4944 | 0.0376 | ||
| 10424929 | NM_172960 | Adck5 | aarF domain containing kinase 5 | 15 D3 | −1.4964 | 0.043 | ||
| 10351473 | NM_001033499 | Sh2d1b2 | SH2 domain protein 1B2 | 1 H3 | −1.4988 | 0.0061 | ||
| 10547976 | NM_145391 | Tapbpl | TAP binding protein-like | 6 F3 | −1.5013 | 0.041 | ||
| 10521391 | NM_030721 | Acox3 | acyl-Coenzyme A oxidase 3, pristanoyl | — | −1.5034 | 0.0098 | ||
| 10490104 | NM_011497 | Aurka | aurora kinase A | 2 H3|2 100.0 cM | −1.5077 | 0.035 | ||
| 10411633 | NM_008670 | Naip1 | NLR family, apoptosis inhibitory protein 1 | 13 D1-D3|13 54.0 cM | −1.5078 | 0.0232 | ||
| 10450145 | NM_013585 | Psmb9 | proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) | 17 B1|17 18.59 cM | −1.5167 | 0.045 | ||
| 10524681 | NM_026933 | Triap1 | TP53 regulated inhibitor of apoptosis 1 | 5 F | −1.517 | 0.0058 | ||
| 10490491 | NM_008093 | Gata5 | GATA binding protein 5 | 2 H4|2 106.0 cM | −1.5174 | 0.0361 | ||
| 10437655 | NM_011955 | Nubp1 | nucleotide binding protein 1 | 16 A1|16 3.4 cM | −1.5217 | 0.0461 | ||
| 10441530 | NM_031395 | Sytl3 | synaptotagmin-like 3 | 17 A1 | −1.5249 | 0.0443 | ||
| 10562130 | NM_194057 | Ffar1 | free fatty acid receptor 1 | 7 B1 | −1.526 | 0.0014 | ||
| 10463704 | NM_020577 | As3mt | arsenic (+3 oxidation state) methyltransferase | 19 D1 | −1.5357 | 0.0162 | ||
| 10524878 | NM_001033311 | Vsig10 | V-set and immunoglobulin domain containing 10 | 5 F | −1.5404 | 0.0046 | ||
| 10393573 | NM_011150 | Lgals3bp | lectin, galactoside-binding, soluble, 3 binding protein | 11 E | −1.5406 | 0.0119 | ||
| 10606714 | NM_013463 | Gla | galactosidase, alpha | X E-F1|X 53.0 cM | −1.5428 | 0.0277 | ||
| 10458547 | NR_028061 | Gm8615 | glucosamine-6-phosphate deaminase 1 pseudogene | 5 G3 | −1.5451 | 0.0246 | ||
| 10384378 | NM_016672 | Ddc | dopa decarboxylase | 11 A1-A4|11 7.0 cM | −1.5472 | 0.043 | ||
| 10429638 | XR_032493 | Gm9568 | glyceraldehyde-3-phosphate dehydrogenase pseudogene | 15 D3|15 | −1.5502 | 0.0482 | ||
| 10510516 | NM_019741 | Slc2a5 | solute carrier family 2 (facilitated glucose transporter), member 5 | 4 E2 | −1.5555 | 0.0065 | ||
| 10565634 | NM_008663 | Myo7a | myosin VIIA | 7 E2|7 48.1 cM | −1.5718 | 0.0053 | ||
| 10524422 | NM_010018 | Dao | D-amino acid oxidase | 5 F|5 65.0 cM | −1.575 | 0.0418 | ||
| 10566583 | AK172683 | Gm8995 | predicted gene 8995 | 7 E3 | −1.5789 | 0.0371 | ||
| 10391207 | NM_030150 | Dhx58 | DEXH (Asp-Glu-X-His) box polypeptide 58 | 11 D|11 61.5 cM | −1.5797 | 0.0135 | ||
| 10408879 | NM_001033399 | Gfod1 | glucose-fructose oxidoreductase domain containing 1 | 13 A4 | −1.5847 | 0.013 | ||
| 10505954 | NM_013690 | Tek | endothelial-specific receptor tyrosine kinase | 4 C5|4 43.6 cM | −1.5877 | 0.0422 | ||
| 10538082 | NM_133764 | Atp6v0e2 | ATPase, H+ transporting, lysosomal V0 subunit E2 | 6 B3 | −1.5918 | 0.0041 | ||
| 10545958 | NM_013471 | Anxa4 | annexin A4 | 6 D1|6 38.0 cM | −1.596 | 0.0411 | ||
| 10560474 | NM_177691 | Ppm1n | protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) | 7 A3 | −1.6066 | 0.0158 | ||
| 10577792 | NM_031257 | Plekha2 | pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 | 8 A2 | −1.623 | 0.0343 | ||
| 10455784 | NM_026240 | Gramd3 | GRAM domain containing 3 | 18 D2 | −1.6238 | 0.0361 | ||
| 10467907 | NM_145502 | Erlin1 | ER lipid raft associated 1 | 19 C3 | −1.6274 | 0.0115 | ||
| 10492102 | NM_144895 | Spg20 | spastic paraplegia 20, spartin (Troyer syndrome) homolog (human) | 3 C | −1.6317 | 0.0301 | ||
| 10377927 | NM_027445 | Rnf167 | ring finger protein 167 | 11 B4 | −1.633 | 0.0209 | ||
| 10404439 | NM_011452 | Serpinb9b | serine (or cysteine) peptidase inhibitor, clade B, member 9b | 13 A3.3|13 12.5 cM | −1.6346 | 0.0217 | ||
| 10566585 | ENSMUST00000098144 | Gm1966 | predicted gene 1966 | 7 E3 | −1.6387 | 0.0248 | ||
| 10587873 | — | — | — | — | −1.6446 | 0.037 | ||
| 10439068 | NM_145932 | Osta | organic solute transporter alpha | 16 B3 | −1.6448 | 0.035 | ||
| 10376726 | NM_001172112 | Dhrs7b | dehydrogenase/reductase (SDR family) member 7B | 11 B2 | −1.6476 | 0.0062 | ||
| 10469951 | NM_176834 | Rnf208 | ring finger protein 208 | 2 A3 | −1.6491 | 0.0225 | ||
| 10548817 | NM_025806 | Plbd1 | phospholipase B domain containing 1 | 6 G1 | −1.654 | 0.0163 | ||
| 10473356 | NM_019949 | Ube2l6 | ubiquitin-conjugating enzyme E2L 6 | 2 E1 | −1.6544 | 0.0048 | ||
| 10501235 | NM_026764 | Gstm4 | glutathione S-transferase, mu 4 | 3 F2.3 | −1.6622 | 0.0309 | ||
| 10474825 | NM_026412 | D2Ertd750e | DNA segment, Chr 2, ERATO Doi 750, expressed | 2 E5 | −1.6757 | 0.0095 | ||
| 10463716 | NM_033569 | Cnnm2 | cyclin M2 | 19 C3|19 47.0 cM | −1.6797 | 0.0023 | ||
| 10604337 | — | — | — | — | −1.6798 | 0.0307 | ||
| 10595081 | NM_012033 | Tinag | tubulointerstitial nephritis antigen | 9 D | −1.6828 | 0.0065 | ||
| 10524621 | NM_011854 | Oasl2 | 2'-5' oligoadenylate synthetase-like 2 | 5 F | −1.687 | 0.0489 | ||
| 10444810 | ENSMUST00000105041 | H2-Q1 | histocompatibility 2, Q region locus 1 | 17 B1|17 19.14 cM | −1.6883 | 0.0032 | ||
| 10584334 | NM_011734 | Siae | sialic acid acetylesterase | 9 A4|9 19.0 cM | −1.699 | 0.0032 | ||
| 10582985 | NM_009807 | Casp1 | caspase 1 | 9 A1|9 1.0 cM | −1.6991 | 0.0091 | ||
| 10488185 | NM_009751 | Bfsp1 | beaded filament structural protein 1, in lens-CP94 | 2 G | −1.7014 | 0.0096 | ||
| 10491091 | NM_009425 | Tnfsf10 | tumor necrosis factor (ligand) superfamily, member 10 | 3 A3 | −1.7131 | 0.0053 | ||
| 10583920 | NM_026189 | Eepd1 | endonuclease/exonuclease/phosphatase family domain containing 1 | 9 A4 | −1.7418 | 0.0179 | ||
| 10378572 | NM_027249 | Tlcd2 | TLC domain containing 2 | 11 B5 | −1.7499 | 0.0309 | ||
| 10383192 | — | — | — | — | −1.7677 | 0.0205 | ||
| 10559606 | NM_023440 | Tmem86b | transmembrane protein 86B | 7 A1 | −1.776 | 0.008 | ||
| 10550740 | NM_027839 | Ceacam20 | carcinoembryonic antigen-related cell adhesion molecule 20 | 7 A3 | −1.7837 | 0.0219 | ||
| 10486172 | NM_001033136 | Fam82a2 | family with sequence similarity 82, member A2 | 2 E5 | −1.7911 | 0.0045 | ||
| 10482802 | NM_139200 | Cytip | cytohesin 1 interacting protein | 2 C1.1 | −1.798 | 0.0078 | ||
| 10402347 | NM_029803 | Ifi27l2a | interferon, alpha-inducible protein 27 like 2A | 12 E | −1.807 | 0.0173 | ||
| 10519196 | NM_147776 | Vwa1 | von Willebrand factor A domain containing 1 | — | −1.8075 | 0.0079 | ||
| 10488636 | NM_001039120 | Defb26 | defensin beta 26 | 2 H1 | −1.8103 | 0.041 | ||
| 10542857 | NM_178797 | Far2 | fatty acyl CoA reductase 2 | 6 G3 | −1.8273 | 0.0308 | ||
| 10376832 | NM_007413 | Adora2b | adenosine A2b receptor | 11 B2 | −1.8282 | 0.0194 | ||
| 10558265 | NM_029609 | Lhpp | phospholysine phosphohistidine inorganic pyrophosphate phosphatase | 7 F4 | −1.8296 | 0.001 | ||
| 10569203 | NM_001142681 | Chid1 | chitinase domain containing 1 | 7 F5 | −1.8468 | 0.0112 | ||
| 10571984 | NM_001081215 | Ddx60 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 | 8 B3.1 | −1.8805 | 0.05 | ||
| 10424221 | NM_001081396 | Wdr67 | WD repeat domain 67 | 15 D1 | −1.8879 | 0.0054 | ||
| 10364134 | NM_133184 | Slc5a4a | solute carrier family 5, member 4a | 10 C1 | −1.9264 | 0.0251 | ||
| 10447904 | NM_199252 | Unc93a | unc-93 homolog A (C. elegans) | 17 A1|17 7.8 cM | −1.9295 | 0.025 | ||
| 10539186 | NM_011259 | Reg3a | regenerating islet-derived 3 alpha | 6 C3|6 33.5 cM | −1.9355 | 0.0286 | ||
| 10451287 | NM_015783 | Isg15 | ISG15 ubiquitin-like modifier | 4 E2|4 | −1.936 | 0.013 | ||
| 10447634 | NM_001142539 | Gm9992 | predicted gene 9992 | 17 A1 | −1.9524 | 0.0175 | ||
| 10389261 | NM_001037932 | Gm11437 | predicted gene 11437 | 11 C | −1.9832 | 0.0076 | ||
| 10538590 | NM_025992 | Herc5 | hect domain and RLD 5 | 6 C1|6 | −1.9883 | 0.0089 | ||
| 10510532 | NM_001085529 | Slc2a7 | solute carrier family 2 (facilitated glucose transporter), member 7 | 4 E2|4 | −2.0053 | 0.0136 | ||
| 10385500 | NM_008326 | Irgm1 | immunity-related GTPase family M member 1 | 11 B1.2 | −2.0184 | 0.0234 | ||
| 10587871 | NM_198414 | Paqr9 | progestin and adipoQ receptor family member IX | 9 E3.3 | −2.0207 | 0.0376 | ||
| 10568568 | NM_016978 | Oat | ornithine aminotransferase | 7 F3|7 63.0 cM | −2.0471 | 3.00E−04 | ||
| 10467470 | NM_019698 | Aldh18a1 | aldehyde dehydrogenase 18 family, member A1 | 19 C3 | −2.3775 | 0.0079 | ||
| 10425287 | NM_134090 | Kdelr3 | KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 | 15 E1 | −2.4335 | 0.0115 | ||
| 10376324 | NM_001135115 | Gm12250 | predicted gene 12250 | 11 B1.3 | −2.4477 | 0.0311 | ||
| 10443869 | NM_024442 | Cyp4f16 | cytochrome P450, family 4, subfamily f, polypeptide 16 | 17 B1 | −2.5218 | 0.0265 | ||
| 10566571 | NR_030719 | Gm8979 | very large inducible GTPase 1 pseudogene | 7 E3 | −2.6198 | 0.0145 | ||
| 10390748 | NM_172564 | Tns4 | tensin 4 | 11 D | −2.6277 | 0.0044 | ||
| 10462618 | NM_010501 | Ifit3 | interferon-induced protein with tetratricopeptide repeats 3 | 19 C3 | −2.6278 | 0.0357 | ||
| 10545200 | ENSMUST00000101325 | LOC100046894 | similar to Igk-C protein | — | −2.6477 | 0.0473 | ||
| 10499899 | NM_009264 | Sprr1a | small proline-rich protein 1A | 3 F1|3 45.2 cM | −2.7972 | 0.0286 | ||
| 10483074 | NM_008100 | Gcg | glucagon | 2 C1.3|2 36.0 cM | −2.7977 | 0.0019 | ||
| 10557300 | NM_007474 | Aqp8 | aquaporin 8 | 7 F3|7 61.0 cM | −2.7985 | 8.00E−04 | ||
| 10566578 | NR_030719 | Gm8979 | very large inducible GTPase 1 pseudogene | 7 E3 | −2.8066 | 0.0095 | ||
| 10430006 | NM_028064 | Slc39a4 | solute carrier family 39 (zinc transporter), member 4 | 15 D3 | −2.8188 | 0.0145 | ||
| 10390691 | NM_145434 | Nr1d1 | nuclear receptor subfamily 1, group D, member 1 | 11 D | −3.298 | 3.00E−04 | ||
| 10480633 | NM_080854 | Slc34a3 | solute carrier family 34 (sodium phosphate), member 3 | 2 A3 | −3.3614 | 0.013 | ||
| 10566366 | NM_199146 | AI451617 | expressed sequence AI451617 | 7 E3 | −3.9279 | 0.0019 | ||
| 10505451 | NM_011016 | Orm2 | orosomucoid 2 | 4 C1|4 31.4 cM | 3.9755 | 0.0307 | ||
| 10416057 | NM_013492 | Clu | clusterin | 14 D1|14 28.0 cM | 3.6483 | 0.0393 | ||
| 10452854 | NM_053188 | Srd5a2 | steroid 5 alpha-reductase 2 | 17 E2 | 3.4167 | 7.00E−04 | ||
| 10367045 | NM_009040 | Rdh16 | retinol dehydrogenase 16 | 10 D3 | 2.3984 | 0.0273 | ||
| 10523717 | NM_009263 | Spp1 | secreted phosphoprotein 1 | 5 E5|5 56.0 cM | 2.2985 | 0.0406 | ||
| 10464594 | BC034269 | BC021614 | cDNA sequence BC021614 | 19 A | 2.2581 | 0.011 | ||
| 10429140 | NM_008681 | Ndrg1 | N-myc downstream regulated gene 1 | 15 D2 | 2.087 | 0.0273 | ||
| 10469609 | NR_033225 | Gm13375 | predicted gene 13375 | 2 A3 | 2.0048 | 0.0294 | ||
| 10507671 | NM_008190 | Guca2a | guanylate cyclase activator 2a (guanylin) | 4 D2.1|4 57.0 cM | 1.8922 | 0.0482 | ||
| 10556113 | NM_016809 | Rbm3 | RNA binding motif protein 3 | X A1.1|X 2.0 cM | 1.8115 | 0.0271 | ||
| 10359917 | NM_010476 | Hsd17b7 | hydroxysteroid (17-beta) dehydrogenase 7 | 1 H3 | 1.7626 | 0.0467 | ||
| 10358454 | NM_001166409 | Rbm3 | RNA binding motif protein 3 | X A1.1|X 2.0 cM | 1.7444 | 0.0294 | ||
| 10540034 | NM_027406 | Aldh1l1 | aldehyde dehydrogenase 1 family, member L1 | 6 D1 | 1.7157 | 0.015 | ||
| 10371784 | NM_001163700 | Nr1h4 | nuclear receptor subfamily 1, group H, member 4 | 10 C2|10 50.0 cM | 1.7001 | 0.0435 | ||
| 10603469 | NM_001166409 | Rbm3 | RNA binding motif protein 3 | X A1.1|X 2.0 cM | 1.6579 | 0.0273 | ||
| 10582310 | NM_138656 | Mvd | mevalonate (diphospho) decarboxylase | 8 E1 | 1.6179 | 0.0287 | ||
| 10518568 | — | — | — | — | 1.5432 | 0.0336 | ||
| 10443898 | NM_134127 | Cyp4f15 | cytochrome P450, family 4, subfamily f, polypeptide 15 | 17 B1 | 1.5311 | 0.0468 | ||
| 10344713 | NM_016661 | Ahcy | S-adenosylhomocysteine hydrolase | 2 H1|2 89.0 cM | 1.5215 | 0.0425 | ||
| 10488816 | NM_016661 | Ahcy | S-adenosylhomocysteine hydrolase | 2 H1|2 89.0 cM | 1.5127 | 0.0377 | ||
| 10439762 | NM_016661 | Ahcy | S-adenosylhomocysteine hydrolase | 2 H1|2 89.0 cM | 1.5089 | 0.0468 | ||
| 10605055 | NM_028633 | Haus7 | HAUS augmin-like complex, subunit 7 | X A7.3 | 1.5083 | 0.015 | ||
| 10556169 | NM_025344 | Eif3f | eukaryotic translation initiation factor 3, subunit F | 7 E3 | 1.418 | 0.0317 | ||
| 10413542 | NM_009388 | Tkt | transketolase | 14 B1 | 1.4043 | 0.0425 | ||
| 10408850 | NM_001111324 | Nedd9 | neural precursor cell expressed, developmentally down-regulated gene 9 | 13 A3.3-A4 | −1.3853 | 0.0463 | ||
| 10489107 | NM_018851 | Samhd1 | SAM domain and HD domain, 1 | 2 H2 | −1.4139 | 0.0456 | ||
| 10516620 | NM_010693 | Lck | lymphocyte protein tyrosine kinase | 4 D2.2|4 59.0 cM | −1.4259 | 0.0348 | ||
| 10513256 | NM_010336 | Lpar1 | lysophosphatidic acid receptor 1 | 4 B3|4 16.0 cM | −1.434 | 0.0294 | ||
| 10604057 | NR_033443 | 6-Sep | septin 6 | X A2 | −1.4532 | 0.0468 | ||
| 10588464 | NR_029526 | Mirlet7g | microRNA let7g | — | −1.464 | 0.0301 | ||
| 10432362 | NM_026967 | Rhebl1 | Ras homolog enriched in brain like 1 | 15 F2 | −1.49 | 0.017 | ||
| 10350173 | NM_001130174 | Tnnt2 | troponin T2, cardiac | 1 E4|1 60.0 cM | −1.4944 | 0.0425 | ||
| 10547906 | NM_008479 | Lag3 | lymphocyte-activation gene 3 | 6 F2 | −1.5108 | 0.0346 | ||
| 10420659 | NM_025697 | 6330409N04Rik | RIKEN cDNA 6330409N04 gene | 14 D1 | −1.5451 | 0.0354 | ||
| 10352097 | NM_027077 | 1700016C15Rik | RIKEN cDNA 1700016C15 gene | 1 H3 | −1.5503 | 0.0468 | ||
| 10409579 | NM_019568 | Cxcl14 | chemokine (C-X-C motif) ligand 14 | 13 B1 | −1.5799 | 0.0377 | ||
| 10355960 | NM_009129 | Scg2 | secretogranin II | 1 C4|1 43.6 cM | −1.6019 | 0.0178 | ||
| 10478633 | NM_013599 | Mmp9 | matrix metallopeptidase 9 | 2 H1-H2|2 96.0 cM | −1.6069 | 0.0228 | ||
| 10501629 | NM_001080818 | Cdc14a | CDC14 cell division cycle 14 homolog A (S. cerevisiae) | 3 G1 | −1.6217 | 0.0425 | ||
| 10447056 | NM_027455 | Qpct | glutaminyl-peptide cyclotransferase (glutaminyl cyclase) | 17 E3 | −1.6326 | 0.0228 | ||
| 10407940 | — | — | — | — | −1.6681 | 0.0294 | ||
| 10605355 | — | — | — | — | −1.6826 | 0.0494 | ||
| 10593015 | NM_009850 | Cd3g | CD3 antigen, gamma polypeptide | 9 A5.2|9 26.0 cM | −1.6893 | 0.0161 | ||
| 10464999 | NM_028623 | Cst6 | cystatin E/M | 19 A|19 4.0 cM | −1.7219 | 0.0456 | ||
| 10584821 | NM_013487 | Cd3d | CD3 antigen, delta polypeptide | 9 A5.2|9 26.0 cM | −1.7359 | 0.0435 | ||
| 10561927 | NM_007467 | Aplp1 | amyloid beta (A4) precursor-like protein 1 | 7 B1|7 8.0 cM | −1.7367 | 0.0109 | ||
| 10552406 | NM_024253 | Nkg7 | natural killer cell group 7 sequence | 7 B2 | −1.7501 | 0.0178 | ||
| 10366881 | NM_007837 | Ddit3 | DNA-damage inducible transcript 3 | 10 D3 | −1.7621 | 0.0344 | ||
| 10490923 | NM_009801 | Car2 | carbonic anhydrase 2 | 3 A1|3 10.5 cM | −1.7726 | 0.0189 | ||
| 10428534 | NM_032000 | Trps1 | trichorhinophalangeal syndrome I (human) | 15 C|15 30.1 cM | −1.793 | 0.0494 | ||
| 10403821 | ENSMUST00000103558 | Tcrg-V3 | T-cell receptor gamma, variable 3 | — | −1.8213 | 0.0468 | ||
| 10516093 | NM_007558 | Bmp8a | bone morphogenetic protein 8a | 4 D2.2|4 57.4 cM | −1.8392 | 0.0307 | ||
| 10513739 | NM_011607 | Tnc | tenascin C | 4 C1|4 32.2 cM | −1.853 | 0.0348 | ||
| 10574166 | NM_153507 | Cpne2 | copine II | 8 C5 | −1.857 | 0.0346 | ||
| 10542632 | NM_010491 | Iapp | islet amyloid polypeptide | 6 G2|6 62.0 cM | −1.8827 | 0.0301 | ||
| 10541605 | NM_020001 | Clec4n | C-type lectin domain family 4, member n | 6 F3|6 55.0 cM | −1.889 | 0.017 | ||
| 10402325 | NM_023049 | Asb2 | ankyrin repeat and SOCS box-containing 2 | 12 E|12 50.0 cM | −1.9087 | 0.0492 | ||
| 10466200 | NM_027836 | Ms4a7 | membrane-spanning 4-domains, subfamily A, member 7 | 19 A | −1.9359 | 0.0172 | ||
| 10404913 | NM_026056 | Cap2 | CAP, adenylate cyclase-associated protein, 2 (yeast) | 13 A5 | −1.9619 | 0.0346 | ||
| 10420308 | NM_013542 | Gzmb | granzyme B | 14 D3|14 20.5 cM | −1.9705 | 0.0294 | ||
| 10412211 | NM_010370 | Gzma | granzyme A | 13 D|13 64.0 cM | −2.0182 | 0.0229 | ||
| 10463016 | NM_028191 | Cyp2c65 | cytochrome P450, family 2, subfamily c, polypeptide 65 | 19 C3 | −2.1252 | 0.0456 | ||
| 10551197 | NM_009999 | Cyp2b10 | cytochrome P450, family 2, subfamily b, polypeptide 10 | 7 A3|7 6.5 cM | −2.5753 | 0.0138 | ||
| 10512999 | NR_033139 | AI427809 | expressed sequence AI427809 | 4 B2 | −2.5806 | 0.0109 | ||
| 10405063 | NM_008760 | Ogn | osteoglycin | 13 A5 | −2.6878 | 0.0346 | ||
| 10512949 | NM_013454 | Abca1 | ATP-binding cassette, sub-family A (ABC1), member 1 | 4 A5-B3|4 23.1 cM | −2.753 | 0.0273 | ||
| 10366043 | NM_026268 | Dusp6 | dual specificity phosphatase 6 | 10 C3 | −3.0369 | 0.0224 | ||
| 10534493 | NM_019577 | Ccl24 | chemokine (C-C motif) ligand 24 | 5 G1 | −4.9474 | 0.0072 | ||
| 10463023 | NM_001011707 | Cyp2c66 | cytochrome P450, family 2, subfamily c, polypeptide 66 | 19 C3 | −5.082 | 0.015 |
Conflict of Interest
None declared.
References
- Alaish SM, Torres M, Ferlito M, Sun C, De Maio A. The severity of cholestatic injury is modulated by the genetic background. Shock. 2005;24:412–416. doi: 10.1097/01.shk.0000183392.83272.97. [DOI] [PubMed] [Google Scholar]
- Alaish SM, Smith AD, Timmons J, Greenspon J, Eyvazzadeh D, Murphy E, et al. Gut microbiota, tight junction protein expression, intestinal resistance, bacterial translocation and mortality following cholestasis depend on the genetic background of the host. Gut Microbes. 2013;4:1–14. doi: 10.4161/gmic.24706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher A, Griebl K, Mackamul S, Mitreiter R, Muckter H, Ben-Shaul Y. Protease inhibitors suppress the formation of tight junctions in gastrointestinal cell lines. Exp. Cell Res. 1992;200:97–104. doi: 10.1016/s0014-4827(05)80076-x. [DOI] [PubMed] [Google Scholar]
- Beliveau F, Desilets A, Leduc R. Probing the substrate specificities of matriptase, matriptase-2, hepsin, and DESC1 with internally quenched fluorescent peptides. FEBS J. 2009;276:2213–2226. doi: 10.1111/j.1742-4658.2009.06950.x. [DOI] [PubMed] [Google Scholar]
- Berger T, Togawa A, Duncan GS, Elia AJ, You-Ten A, Wakeham A, et al. Lipocalin-2-deficient mice exhibit increased sensitivity to Escherichia coli infection but not ischemia-reperfusion injury. PNAS. 2006;103:1834–1839. doi: 10.1073/pnas.0510847103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruewer M, Luegering A, Kucharzik T, Parkos CA, Madara JL, Hopkins AM, et al. Proinflammatory cytokines disrupt epithelial barrier function by apoptosis-independent mechanisms. J. Immunol. 2003;171:6164–6172. doi: 10.4049/jimmunol.171.11.6164. [DOI] [PubMed] [Google Scholar]
- Buzza MS, Netzel-Arnett S, Shea-Donahue T, Zhao A, Lin C-Y, List K, et al. Membrane-anchored serine protease matriptase regulates epithelial barrier formation and permeability in the intestine. PNAS. 2010;107:4200–4205. doi: 10.1073/pnas.0903923107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne TA, Persinger RL, Young LS, Ziegler TR, Wilmore DW. A new treatment for patients with short-bowel syndrome: growth hormone, glutamine and a modified diet. Ann. Surg. 1995;222:243–254. doi: 10.1097/00000658-199509000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caglikulekci M, Dirlik M, Scatton O, Cinel I, Ozer I, Cinel L, et al. Effect of antithrombin-III (AT-III) on intestinal epithelium changes related to obstructive icterus: experimental study in rats. Ann. Chir. 2004;129:273–277. doi: 10.1016/j.anchir.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Campillo B, Pernet P, Bories PN, Richardet JP, Devanlay M, Aussel C. Intestinal permeability in liver cirrhosis: relationship with severe septic complications. Eur. J. Gastroenterol. Hepatol. 1999;11:755–759. doi: 10.1097/00042737-199907000-00013. [DOI] [PubMed] [Google Scholar]
- Clayburgh DR, Shen L, Turner JR. A porous defense: the leaky epithelial barrier in intestinal disease. Lab. Invest. 2004;84:282–291. doi: 10.1038/labinvest.3700050. [DOI] [PubMed] [Google Scholar]
- Collins CB, Aherne CM, Ehrentraut SF, Gerich ME, McNamee EN, McManus MC, et al. Alpha-1-antitrypsin therapy ameliorates acute colitis and chronic murine ileitis. Inflamm. Bowel Dis. 2013;19:1964–1973. doi: 10.1097/MIB.0b013e31829292aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devireddy LR, Teodoro JG, Richard FA, Green MR. Induction of apoptosis by a secreted lipocalin that is transcriptionally regulated by IL-3 deprivation. Science. 2001;293:829–834. doi: 10.1126/science.1061075. [DOI] [PubMed] [Google Scholar]
- Diller R, Stratmann U, Minin E, Baumer C, von Eiff G, Huismans H, et al. ATIII attenuates endotoxemia induced healing impairment in the colon. J. Surg. Res. 2009;157:4–13. doi: 10.1016/j.jss.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Ding L, Li J-S, Li Y-S, Liu FN, Tan L. Prophylactic treatment with growth hormone improves intestinal barrier function and alleviates bacterial translocation in stressed rats. Chin. Med. J. 2004;117:264–269. [PubMed] [Google Scholar]
- Dorsey SG, Leitch CC, Renn CL, Lessans S, Smith BA, Wang XM, et al. Genome-wide screen identifies drug-induced regulation of the gene giant axonal neuropathy (Gan) in a mouse model of antiretroviral-induced painful peripheral neuropathy. Biol. Res. Nurs. 2009;11:7–16. doi: 10.1177/1099800409332726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Asmar R, Panigrahi P, Bamford P, Berti I, Not T, Coppa GV, et al. Host-dependent zonulin secretion causes the impairment of the small intestine barrier function after bacterial exposure. Gastroenterology. 2002;123:1607–1615. doi: 10.1053/gast.2002.36578. [DOI] [PubMed] [Google Scholar]
- Flo TH, Smith KD, Sato S, Rodriquez DJ, Holmes MA, Strong RK, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestering iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- Frances R, Benlloch S, Zapater P, Gonzalez JM, Lozano B, Munoz C, et al. A sequential study of serum bacterial DNA in patients with advanced cirrhosis and ascites. Hepatology. 2004;39:484–491. doi: 10.1002/hep.20055. [DOI] [PubMed] [Google Scholar]
- Georgiev P, Jochum W, Heinrich S, Jang JH, Nocito A, Dahm F, et al. Characterization of time-related changes after experimental bile duct ligation. Br. J. Surg. 2008;95:646–656. doi: 10.1002/bjs.6050. [DOI] [PubMed] [Google Scholar]
- Huang C-Y, Sheen-Chen S-M, Ho H-T, Tang RP, Eng HL. Antithrombin-III attenuates hepatocyte apoptosis in bile duct ligated rat: a striking cellular change. Surg. Innov. 2010;17:132–135. doi: 10.1177/1553350610366716. [DOI] [PubMed] [Google Scholar]
- Javid PJ, Collier S, Richardson D, Iglesias J, Gura K, Lo C, et al. The role of enteral nutrition in the reversal of parenteral-nutrition-associated liver dysfunction in infants. J. Pediatr. Surg. 2005;40:1015–1018. doi: 10.1016/j.jpedsurg.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Kato Y, Yu D, Lukish JR, Schwartz MZ. Hepatocyte growth factor enhances intestinal mucosal cell function and mass in vivo. J. Pediatr. Surg. 1997;32:991–994. doi: 10.1016/s0022-3468(97)90384-5. [DOI] [PubMed] [Google Scholar]
- Kato Y, Yu D, Schwartz MZ. Enhancement of intestinal adaptation by hepatocyte growth factor. J. Pediatr. Surg. 1998;33:235–239. doi: 10.1016/s0022-3468(98)90438-9. [DOI] [PubMed] [Google Scholar]
- Kikuchi T, Abe T, Yaekashiwa M, Tominaga Y, Mitsuhashi H, Satoh K, et al. Secretory leukoprotease inhibitor augments hepatocyte growth factor production in human lung fibroblasts. Am. J. Respir. Cell Mol. Biol. 2000;23:364–370. doi: 10.1165/ajrcmb.23.3.3942. [DOI] [PubMed] [Google Scholar]
- Kuhn NJ, Woodworth-Gutai M, Gross KW, Held WA. Subfamilies of the mouse major urinary protein (MUP) multi-gene family: sequence analysis of cDNA clones and differential regulation in the liver. Nucleic Acids Res. 1984;12:6073–6090. doi: 10.1093/nar/12.15.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- List K, Currie B, Scharschmidt TC, Szabo R, Shireman J, Molinolo A, et al. Autosomal ichthyosis with hypotrichosis syndrome displays low matriptase proteolytic activity and is phenocopied in ST14 hypomorphic mice. J. Biol. Chem. 2007;282:36714–36723. doi: 10.1074/jbc.M705521200. [DOI] [PubMed] [Google Scholar]
- Liu W, Jiang Z, Wang X, Shu H, Cui W, Wilmore DW. Impact of perioperative treatment of recombinant human growth hormone on cell immune function and intestinal barrier function: randomized, double-blind, controlled trial. World J. Surg. 2003;27:412–415. doi: 10.1007/s00268-002-6758-x. [DOI] [PubMed] [Google Scholar]
- Madara JL, Stafford J. Interferon-gamma directly affects barrier function of cultured intestinal epithelial monolayers. J. Clin. Invest. 1989;83:724–727. doi: 10.1172/JCI113938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marano CW, Lewis SA, Garulacan LA, Soler AP, Mullin JM. Tumor necrosis factor-alpha increases sodium and chloride conductance across the tight junction of CACO-2 BBE, a human intestina epithelial cell line. J. Membr. Biol. 1998;161:263–274. doi: 10.1007/s002329900333. [DOI] [PubMed] [Google Scholar]
- Pascual S, Such J, Esteban A, Zapater P, Casellas JA, Aparicio JR, et al. Intestinal permeability is increased in patients with advanced cirrhosis. Hepatogastroenterology. 2003;50:1482–1486. [PubMed] [Google Scholar]
- Rafatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao JN, Platoshyn O, Golovina VA, Liu L, Zou T, Marasa BS, et al. TRPC1 functions as a store-operated Ca2 + channel in intestinal epithelial cells and regulates early mucosal restitution after wounding. Am. J. Physiol. Gastrointest. Liver Physiol. 2006;290:G782–G792. doi: 10.1152/ajpgi.00441.2005. [DOI] [PubMed] [Google Scholar]
- Scopa CD, Koureleas S, Tsamandas AC, Spiliopoulou I, Alexandrides T, Filos KS, et al. Beneficial effects of growth hormone and insulin-like growth factor I on intestinal bacterial translocation, endotoxemia, and apoptosis in experimentally jaundiced rats. J. Am. Coll. Surg. 2000;190:423–431. doi: 10.1016/s1072-7515(99)00285-9. [DOI] [PubMed] [Google Scholar]
- Ulshen MH, Dowling RH, Fuller CR, Zimmermann EM, Lund PK. Enhanced growth of small bowel in transgenic mice overexpressing bovine growth hormone. Gastroenterology. 1993;104:973–980. doi: 10.1016/0016-5085(93)90263-c. [DOI] [PubMed] [Google Scholar]
- Velasco B, Lassaletta L, Gracia R, Tovar JA. Intestinal lengthening and growth hormone in extreme short bowel syndrome: a case report. J. Pediatr. Surg. 1999;34:1423–1424. doi: 10.1016/s0022-3468(99)90027-1. [DOI] [PubMed] [Google Scholar]
- Weiming Z, Ning L, Jieshou L. Effect of recombinant human growth hormone and enteral nutrition on short bowel syndrome. JPEN J. Parenter. Enteral Nutr. 2004;28:377–381. doi: 10.1177/0148607104028006377. [DOI] [PubMed] [Google Scholar]
- Wildhaber BE, Yang H, Coran AG, Teitelbaum DH. Gene alteration of intestinal intraepithelial lymphocytes in response to massive small bowel resection. Pediatr. Surg. Int. 2003;19:310–315. doi: 10.1007/s00383-003-1001-x. [DOI] [PubMed] [Google Scholar]
- Yamanouchi H, Fujita J, Hojo S, Yoshinouchi T, Kamei T, Yamadori I, et al. Neutrophil elastase: alpha-1-proteinase inhibitor complex in serum and bronchoalveolar lavage fluid in patients with pulmonary fibrosis. Eur. Respir. J. 1998;11:120–125. doi: 10.1183/09031936.98.11010120. [DOI] [PubMed] [Google Scholar]
- Yi C, Cao Y, Wang SR, Xu YZ, Huang H, Cui YX, et al. Beneficial effect of recombinant human growth hormone on the intestinal mucosa barrier of septic rats. Braz. J. Med. Biol. Res. 2007;40:41–48. doi: 10.1590/s0100-879x2007000100006. [DOI] [PubMed] [Google Scholar]
- Youakim A, Ahdieh M. Interferon-gamma decreases barrier function in T84 cells by reducing ZO-1 levels and disrupting apical actin. Am. J. Physiol. 1999;276:G1279–G1288. doi: 10.1152/ajpgi.1999.276.5.G1279. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Jiang L, Rui L. Identification of MUP1 as a regulator for glucose and lipid metabolism in mice. J. Biol. Chem. 2009;284:11152–11159. doi: 10.1074/jbc.M900754200. [DOI] [PMC free article] [PubMed] [Google Scholar]