Abstract
Subsets of mammalian adult stem cells reside in the quiescent state for prolonged periods of time. This state, which is reversible, has long been viewed as dormant and with minimal basal activity. Recent advances in adult stem cell isolation have provided insights into the epigenetic, transcriptional and post-transcriptional control of quiescence and suggest that quiescence is an actively maintained state in which signalling pathways are involved in maintaining a poised state that allows rapid activation. Deciphering the molecular mechanisms regulating adult stem cell quiescence will increase our understanding of tissue regeneration mechanisms and how they are dysregulated in pathological conditions and in ageing.
Stem cells are undifferentiated, long-lived cells that are unique in their abilities to produce differentiated daughter cells and to retain their stem cell identity by self-renewal1. Most mammalian adult tissues contain resident stem cells, which proliferate to compensate for tissue loss throughout the life of the organism. They possess remarkable proliferative capacity, allowing them to engage in massive and repetitive regenerative activities in response to tissue damage. A subset of tissue-specific adult stem cells persists in the quiescent state for prolonged periods of time2. Whereas quiescence is not an essential characteristic that defines stem cells, dysregulation and loss of quiescence often results in an imbalance in progenitor cell populations ultimately leading to stem cell depletion3. As a result, tissue replenishment is affected during homeostasis and following damage. Thus, deciphering the regulation of quiescence will contribute much to our understanding of how tissue regeneration is accomplished in physiological and pathological settings and may lead to new therapeutic strategies for tissue maintenance or repair.
The concept of cellular quiescence has changed over time. Previously, it was thought that cells become quiescent by default, because of challenges to continued pro liferation such as nutrient deprivation or contact inhibition. Now, it is believed that cells, particularly stem cells, adopt the quiescent state to preserve key functional features. Recently, much attention has focused on the active regulation of the quiescent state as well as the properties of stem cells that persist in a quiescent state. Such properties allow them to withstand metabolic stress and to preserve genomic integrity over a lifetime.
In this Review, we summarize recent advances in the field of stem cell quiescence and discuss the characteristics and regulation of the quiescent state. Beginning with a historical summary of studies of the cell cycle and the existence of a quiescent state, we focus on the identification of stem cell populations that reside in the G0 phase of the cell cycle, the molecular signatures of this state and the regulatory mechanisms that maintain cells in the quiescence state. Finally, we examine specific properties of quiescent stem cells that assure survival over extended periods of time, and we present a model of the quiescent state as a ‘poised state’ rather than a dormant state.
The G0 phase of the cell cycle
Historically, the G0 phase of the cell cycle was referred to as an inactive, non-cycling state. It was first recognized and described as a state in which cells have irreversibly exited the cell cycle, as exemplified by terminally differentiated cells such as neurons or cardiomyocytes or, more recently, senescent cells (BOX 1). Such cells do not re-enter the cell cycle except in response to extraordinary experimental stimuli. By contrast, the discovery of another type of G0 phase, namely the quiescent state, is characterized by the ability of cells to re-enter the cell cycle in response to normal physiological stimuli.
Box 1: Reversibility of the G0 state of the cell cycle.
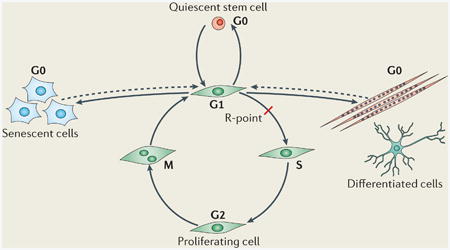
Somatic cells are able to enter reversible (quiescent) or irreversible (senescent and differentiated) G0 states from the G1 phase of the cell cycle before the restriction point (R-point). Once cells reach the R-point, they are committed to the next round of the cell cycle (see the figure). Subpopulations of stem cells reside in the quiescent state and enter the cell cycle when they become activated in response to extrinsic signals. The fate of a cell is determined during G1, and cells differentiate, become senescent or re-enter the quiescent state. Senescent cells are dysfunctional cells that have ceased proliferation and are permanently withdrawn from the cell cycle148. Increasing evidence suggests that senescence has a role in suppressing malignant tumour formation148. Moreover, the accumulation of senescent cells in aged tissues causes tissue damage due to factors that these cells secrete149, and removal of these cells may delay tissue ageing150. Unravelling the mechanisms that regulate cellular senescence may provide clues as to how the relative reversibility of different G0 states is controlled and have broad implications for tissue regeneration, ageing and cancer.
Analogous to differentiated, non-cycling cells in mammals, some types of amphibians possess mature differentiated cells that are able to dedifferentiate and proliferate to regenerate lost tissues and even entire appendages151. In these amphibians, such as newts, differentiated multinucleate myotubes are able to undergo cellularization to generate mononucleated cells152. Surprisingly, intracellular pathways that mediate the remarkable regenerative capacity of these organisms seem to be intact in mammals. For example, myonuclei in terminally differentiated mammalian myotubes have been reported to exhibit cell cycle re-entry when exposed to an extract derived from regenerating newt limbs153. Also, overexpression of the homeobox-containing transcriptional repressor MSX1 in mammalian myotubes, a protein that is specifically expressed in undifferentiated cells in developing limb buds154, has been reported to cellularize differentiated myotubes into proliferating mononucleated cells155. Furthermore, recent studies suggest that both terminally differentiated cells and senescent cells are able to re-enter the cell cycle by inhibiting tumour suppressors such as p53 and RB156,157. Together, these results indicate that intrinsic mechanisms inducing ‘irreversibly’ arrested cell types to enter the cell cycle (dotted arrows) are intact, although repressed, in mammalian cells.
Discovery of quiescence
The existence of a quiescent state was hypothesized on the basis of early cell cycle studies (BOX 1). In 1951, Howard and Pelc used radioactive labelling techniques to study the timing of DNA replication during cell division, thereby defining the four phases of the cell cycle4. Interestingly, the concept of quiescence arose from the observation that not all cells in a population proliferate at similar rates. The term ‘growth fraction’ was used to describe the cell population that is actively proliferating. In somatic tissues, some cells continuously divide, while other cells exist in a non-proliferative state during homeostasis but are able to respond to extrinsic stimuli and re-enter the cell cycle to begin proliferating5.
For years, debate continued about the nature of the state of cells that are non-cycling but able to proliferate in response to extrinsic stimuli. Some investigators considered these cells to be in a prolonged G1 phase, and others postulated that they could be in a cell cycle phase that is distinct from G1 and termed this non-proliferative state G0 (which is also referred to as the quiescent state)6. Subsequent studies demonstrated that sub-optimal conditions such as high cell density7 or serum insufficiency8 could drive cells into this quiescent state. In 1974, Pardee provided evidence for a distinct quiescent state and demonstrated the existence of a restriction point (R-point) in G1 that determines cell fates: cells in G1 can become quiescent before the R-point but commit to enter a mitotic cell cycle after the R-point9 (BOX 1). The author hypothesized that normal mammalian cells possess unique regulatory mechanisms to shift from a quiescent state to a proliferative state and that dysregulation of these mechanisms might result in malignant transformation. In 1985, Zetterberg and Larsson discovered that serum deprivation results in the inhibition of protein synthesis in all cell cycle states but that only cells in early G1 exit the cell cycle and become quiescent10. Together, these early studies suggested the existence of a quiescent state, access to which is restricted. To date, the molecular control of quiescence still remains to be fully elucidated.
The diversity of quiescent states
Many unicellular organisms reside in the quiescent state for a prolonged period of time to survive in unfavourable environments11. Quiescence is also a state of growth cessation that occurs in multicellular organisms. For example, studies of seed dormancy revealed that plants utilize this state to preserve the capacity for growth, thereby circumventing an unfavourable environment12. In mammals, the ability of tissue stem cells to reside in the quiescent state is crucial for proper homeostasis and regeneration of many tissue types. Quiescent stem cells are able to respond to stimuli that originate from their niche environment by activating and entering the cell cycle (BOX 2). Interestingly, tissue stem cells are not the only population of cells in G0 that are able to resume proliferation and contribute to tissue regeneration. For example, mature hepatocytes are capable of entering the cell cycle and contribute to liver regeneration in the case of partial hepatectomy13. Thus, both stem cells and differentiated cells can reside in a reversible G0 phase.
Box 2: Quiescent stem cells and the stem cell niche.
In addition to the intrinsic mechanisms that regulate stem cell quiescence, the stem cell niche (that is, a specific microenvironment that surrounds stem cells and has important regulatory functions) is essential for stem cell maintenance, including the maintenance of quiescence158. First described by Schofield in 1978 (REF. 159), stem cell niches have been identified for all types of adult stem cells in mammalian tissues. In malignant tissues, cancer stem cells are thought to take advantage of the niche that supports normal stem cell behaviours158,160. To understand the role of the stem cell niche, it is necessary to determine the composition of the niche (for a review, see REF 158). Local cellular stem cell niche components include other cell types such as those of the vasculature and interstitium, as well as matrix proteins and constituents. Soluble factors, either secreted from nearby cells or from distant sources, can influence stem cell function, resulting in alternative stem cell fates161. The recent development of new genetic tools has provided insights into the interaction between the niche and the stem cell. Using heterochronic parabiosis to study muscle stem cell (MuSC) ageing, it has been demonstrated that systemic niche factors are crucial regulators of quiescent stem cell function that change with age162. Another study, using a transgenic reporter of the regulatory factor SCF (stem cell factor), identified the major sources of SCF in the haematopoietic niche in bone marrow163. The interaction between the niche and quiescent stem cells is also relevant in stem cell ageing. Intriguingly, the disruption of the niche has been linked to the decline of stem cell function during the process of ageing24. Analysis of injured muscle has revealed an age–dependent decrease in the expression of the Notch ligand Delta-like 1, resulting in decreased Notch signalling and impaired MuSC proliferation164. By contrast, aged muscle fibres secrete fibroblast growth factor 2 (FGF2), which induces MuSCs to divide more frequently, resulting in the disruption of stem cell quiescence24. Thus, a better understanding of how quiescent stem cells interact with the niche will provide important insights into the components that regulate stem cell quiescence.
Identification of quiescent stem cells
Our understanding of the characteristics of quiescent stem cells has been limited by the rarity of this population in many tissue compartments. Quiescent stem cells have been identified by their low RNA content14,15 and their lack of cell proliferation markers16, as well as by label retention as an indication of low turnover. Label retention as an indication of quiescence is based on the concept that once cells have incorporated a label, rapidly dividing cells lose the label quickly, whereas quiescent or very slowly cycling cells retain the label for extended periods of time. Identification and localization of cells in the quiescent state have relied primarily on techniques that allow the analysis of the incorporation and then retention of labels such as 5′-bromo-2′-deoxyuridine (BrdU)17,18, tritiated thymidine19,20 or, more recently, the use of H2B–GFP21–24 or H2B–YFP25. For decades, label retention was considered to be an essential property of adult stem cells26. However, it has become increasingly apparent that the use of label retention alone is insufficient to identify adult stem cells. Recently, evidence has suggested the coexistence of reserve (quiescent) and active (proliferating) stem cell pools in high-turnover tissue compartments2,27. The use of a lineage tracing approach based on label retention has provided new insights into the nature and function of label-retaining cells (LRCs) in the gut25. Whereas active stem cells function during normal homeostasis, quiescent LRCs seem to serve as a reserve pool of stem cells, only called into action upon tissue injury. In addition, interconversion of reserve and active intestinal stem cell (ISC) populations has also been observed previously25,28–30.
Similar to the gut, skin is another high-turnover tissue in which both quiescent and active stem cells are present. The mammalian epidermis consists of regions that contain hair follicles interspersed with interfollicular epidermis. Hair follicle morphogenesis relies on both quiescent and active stem cells. Quiescent stem cells that are responsible for regenerating the hair follicles lie within the bulge of the hair follicles21,31,32, which can be visualized using advanced imaging techniques33. Interestingly, adult epidermal homeostasis seems to rely solely on active stem cells34, whereas quiescent stem cells in the bulge are involved in the process of wound healing but not normal homeostasis35. Lineage tracing experiments have also facilitated the identification of long-lived stem cells in the mammary epithelium36,37 and in glandular epithelia such as that found in the prostate38.
In low-turnover tissues such as liver or muscle, the use of label-retention techniques is well-suited for the identification of quiescent stem cells. However, it has been proposed that more than one type of low-turnover stem cells exist in a given tissue. In the muscle compartment, the existence of a low-turnover population of fibrogenic and adipogenic progenitor cells that is functionally distinct from muscle stem cells has recently been proposed39,40. Thus, in either high- or low-turnover tissues, techniques such as lineage tracing are needed to identify quiescent stem cells and to study their function.
Molecular signatures of quiescent stem cells
Recent advances in genetic approaches and high-throughput analyses of various stem cell subpopulations have provide d valuable information on the molecular signatures of quiescent stem cells in different tissue compartments. These findings have not only revealed unique signatures of the quiescent state but also provided potential avenues for identifying and characterizing regulatory pathways, networks and determinants of the quiescent state.
Transcript profiles
Transcript profiling was traditionally limited to bulk-differentiated tissues due to a lack of cell purification techniques and a need for large amounts of RNA to perform such analysis. To understand the transcriptomes of quiescent stem cells, much effort had been focused on various techniques to purify and characterize stem cell populations. Prospective isolation of quiescent stem cells by fluorescence-activated cell sorting (FACS) was first used to purify haematopoietic stem cells (HSCs)41 and has quickly become a standard technique for isolating stem cells. To date, FACS techniques have been devised for the isolation of muscle stem cells (MuSCs)15, ISCs42, hair follicle stem cells (HFSCs)31,43, neural stem cells (NSCs)44 and many other stem cell populations. These advances in purifying subpopulations of stem cells have allowed the use of high-throughput techniques such as microarray and RNA-sequencing to further our understanding of the transcriptomes of these stem cells.
Facilitated by advanced isolation techniques, high-throughput gene expression analyses of quiescent stem cells and their differentiated progeny have provided important information regarding the identities of genes that are important for lineage determination and differentiation. In particular, a comparison of gene expression profiles of different types of quiescent stem cells, including HSCs45, MuSCs15 and HFSCs43, reveals a gene signature that is common to these quiescent stem cells (TABLE 1).
Table 1. Quiescent stem cell gene signature*.
| Function | Downregulated genes‡ | Upregulated genes‡ |
|---|---|---|
| Cell cycle progression and checkpoint control | ANLN, BIRC5, CCNA2, CCNB1, CCNE2, SGOL1 | CCND3, PDK1 |
|
| ||
| DNA replication and chromosome segregation | MCM4, PCNA, RRM2, TOP2A | |
|
| ||
| Mitochondrial function | CYCS, MTCH2, SLC25A5 | |
|
| ||
| Chromatin and nucleosome assembly | H2AFZ, HAT1 | SMARCA2 |
|
| ||
| Regulation of transcription | FOXO3, EZH1, PRDM5, PTOV1, ZFP30, ZBTB20, PHF1, CTDSP1, THRA, TEF | |
|
| ||
| RNA processing | DDX39 | DICER1 |
|
| ||
| Other | 2810417H13Rik, CAPZA1, HADHB, IDH3A, KPNA2, PGK1 | A930001N09Rik, BCAS3, DDX3Y, GABARAPL1, GLTSCR2, ITM2A, IL18, ZYX, EPHX1, CLSTN1, GSTK1, 5730403B10Rik, DDT, IVD, FHL1, NDRG2, GRINA, PIK3R1, FYN, CHKB, PINK1, ULK2, DNAJB9, PFDN5, CTSF, CRIM1, SEPP1, GABBR1, GRB10, BBS2, RPS14, IGF2R, SELENBP1, RNF167, MAP1LC3A |
Comparison of microarray data sets revealed a gene signature that is common to quiescent haematopoietic stem cells (HSCs), muscle stem cells (MuSCs) and hair follicle stem cells (HFSCs). Selected genes (30 out of 71 genes) are shown and grouped on the basis of pathways in which they are presumed to function. Genes exhibiting expression level changes that are shared among the stem cell compartments are listed under ‘other’. Consistent with the dormant phenotype of a quiescent stem cell, genes that are involved in cell cycle progression, DNA replication or mitochondrial functions are mostly downregulated in quiescent stem cells.
For a full list of gene definitions see Supplementary information S1 (table)).
As expected from a non-proliferative phenotype, the signature reveals the downregulation of genes that are involved in DNA replication and cell cycle progression. Examples of genes that are downregulated in all three quiescent cell types (HSCs, MuSCs and HFSCs) include genes encoding cyclin A2, cyclin B1, cyclin E2 and survivin, which control various aspects of cell cycle progression15,43,45. Cyclin A2 and cyclin E2 are important regulators of cell cycle checkpoints46,47. HSCs that lack cyclin A2 are unable to proliferate in vitro, indicating the essential role of cyclin A2 in HSC proliferation48. Whereas cyclin B1 binds to cyclin-dependent kinase 1 (CDK1) and promotes entry into mitosis49, survivin has important roles in the regulation of microtubule dynamics during mitosis50. Moreover, downregulated genes correlated with the proliferation status (including genes such as proliferating cell nuclear antigen (PCNA) and mini chromosome maintenance complex component 4 (MCM4)) and with mitochondrial function (for example cytochromec (CYCS))15,43,45. As mitochondrial biogenesis is required for stem cell activation, low expression of CYCS reflects low metabolic activity of the quiescent stem cell. Conversely, genes that are upregulated in quiescent stem cells include genes encoding signalling molecules involved in transcriptional regulation and stem cell fate decisions such as forkhead box O3 (FOXO3) and enhancer of zeste homolog 1 (EZH1). It is likely that there are transcriptional signatures that are unique to specific populations of quiescent stem cells. However, it is also possible that gene products, the expression levels of which change as stem cells progress from the quiescent state to the activated state, constitute signalling pathways that are common to various different stem cell populations. This may reveal mechanisms that specifically relate to the induction or maintenance of quiescence.
Other than protein-coding genes, profiling of non-coding RNAs such as microRNAs (miRNAs) has also revealed the function of various miRNAs in regulating stem cell quiescence51,52. miRNA signatures have recently been identified in multiple quiescent stem cell populations such as the HSCs52, NSCs52, MuSCs51,52 and HFSCs53. Similar to the gene expression analysis, miRNA profiling of HSCs, NSCs and MuSCs and their differentiated progenies has led to a common miRNA signature of stem cell activation from quiescence, which suggests an important role of miRNA pathways in regulating stem cell quiescence post-transcriptionally52.
Characterizing the transcriptional landscape of quiescent stem cells is likely to provide information on common gene expression patterns that maintain quiescence, such as genes that are involved in cell cycle regulation, as well as specific patterns that relate to quiescent stem cells in particular lineages in various tissue compartments.
Epigenetic profiles
Recent epigenetic studies have shed light on how chromatin states contribute to maintaining stem cells in a poised state for lineage progression. Knowledge gained from embryonic stem (ES) cells and induced pluripotent stem (iPS) cells can be applied to quiescent adult stem cells. Studies of histone methylation have revealed the epigenetic landscape as one of the key determinants of gene expression54,55. Trimethylation of histone H3 at Lys4 (H3K4me3) and H3K27me3 are of particular interest because of their roles in the positive and negative regulation of transcription, respectively54. Chromatin regions that are marked by both H3K4me3 and H3K27me3, which are termed bivalent domains, are frequently located in close proximity to transcription start sites56. Many genes that carry such bivalent chromatin patterns are master regulators of cell lineages and are thought to maintain ES cells in a poised state to allow flexibility for lineage choices.
In view of regulators that govern chromatin modifications, conditional knockouts of the H3K27 methyl-transferases EZH1 and EZH2 in HFSCs revealed an essential role of chromatin modification in hair follicle homeostasis and wound repair57. In muscle, deletion of EZH2 impairs MuSC proliferation and derepresses gene expressions of non-muscle lineages58. By contrast, overexpression of EZH2 in HSCs prevents HSC exhaus-tion59, whereas HSCs are lost when EZH1 is ablated60. Together, these studies suggest important roles of H3K27 methyltransferases in regulating stem cell quiescence in an epigenetic manner.
In tissue compartments such as muscle and skin, where prospective isolation of large quantities of stem cells is feasible, it is now possible to use genome-wide chromatin immunoprecipitation followed by sequencing (ChIP–seq) to obtain epigenetic profiles of the resident stem cells. In contrast to observations in ES cells, few genes are marked by bivalent domains in lineage-restricted, quiescent HFSCs61. Intriguingly, in both quiescent HFSCs and MuSCs, thousands of genes are marked by the H3K4me3 mark (which is associated with active transcription)61,62, suggesting a permissive chromatin state for transcription. However, it has previously been proposed that H3K4me3 marks genes for transcriptional activation but does not necessarily predict whether these genes are being actively transcribed63. Given the low transcriptional output in quiescent stem cells, it is likely that not all genes marked by H3K4me3 are indeed being actively transcribed but may reflect the fact that quiescent stem cells are, in general, less differentiated than their proliferating progeny and that this epigenetic mark identifies genes that may be transcribed upon activation. This correlation and the significance of this epigenetic signature remain to be demonstrated experimentally.
Molecular regulation of quiescence
Although transcriptional and epigenetic profiling may be of value to provide molecular signatures of quiescent stem cells and may point to pathways that are important for the induction or maintenance of the quiescent state, each pathway needs to be tested in studies of stem cell quiescence in vivo to determine the functional relevance. In the following section, we highlight genes and pathways for which experimental evidence supports an important contribution to the regulation of the quiescent state.
p53 and RB protein
p53, a master regulator of diverse cellular processes, especially those involved in the maintenance of genomic integrity, has an important role in regulating stem cell quiescence64. p53 deficiency in HSCs promotes cell cycle entry with a reduction in the number of quiescent HSCs64. The mechanism by which p53 mediates HSC quiescence is independent of the CDK inhibitor p21, which is an important regulator of cell cycle progression at G1 that has previously been shown to regulate HSC quiescence65. In addition to p53, another crucial regulator of the cell cycle, the tumour suppressor RB, has also been implicated as a regulator of stem cell quiescence. Early studies of RB revealed that one of its major roles is to inhibit cell cycle progression66. The cell cycle progresses normally when RB is inactivated by phosphorylation facilitated by different cyclin–CDK complexes67. In NSCs, genetic ablation of RB together with p53 triggers NSC over-proliferation, resulting in a brain tumour phenotype68. In ES cells, ablation of all three RB family members (RB, p107 and p130) results in impaired differentiation and an increase in cell turnover under growth arrest conditions69. Genetic ablation of RB in quiescent MuSCs results in a vast increase of muscle stem and progenitor cells and an acceleration of cell cycle re-entry70 (FIG. 1a). Muscle progenitors that lack RB do not differentiate due to their inability to exit the cell cycle70. Similarly, the quiescent HSC pool is lost when all three RB family proteins are conditionally ablated. This HSC depletion is accompanied by an expansion of early haematopoietic progenitors and an impairment of the reconstitution potential in transplantation experiments71.
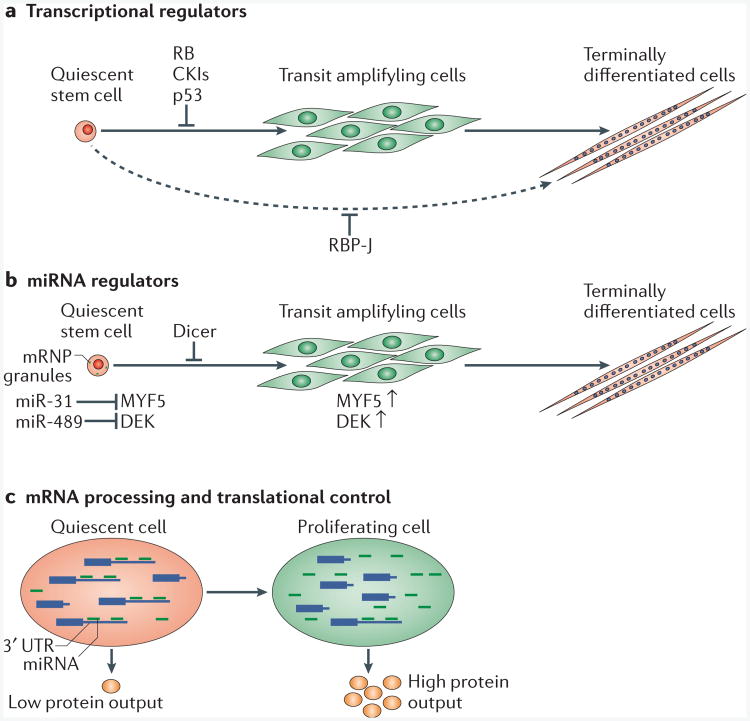
Figure 1. Molecular regulation of stem cell quiescence.
Recent data suggest that the state of quiescence is actively regulated by different molecular mechanisms. a | Quiescence regulators such as RB, cyclin-dependent kinase inhibitors (CKIs) and p53 negatively regulate the activation of quiescent stem cells. In quiescent muscle stem cells, the loss of the downstream effector of the Notch signalling pathway, RBP-J, promotes spontaneous activation and terminal differentiation, in some cases without cell division. b | Quiescent stem cells are actively regulated by post-transcriptional mechanisms. The loss of the microRNA (miRNA)-processing enzyme Dicer or specific miRNAs promotes quiescent stem cell activation. c | Differential mRNA processing alters the susceptibility of mRNAs to miRNA regulation. In quiescent cells, distal polyadenylation signals (PASs) are used to generate mRNA transcripts with long 3′ untranslated regions (3′ UTRs). In proliferating cells, proximal PASs are used, which decrease the number of miRNA target sites on the transcripts, allowing some transcripts to escape miRNA-mediated inhibition and leading to increased protein expression.
CDK inhibitors (CKIs)
Many CKIs, including p21, p27 and p57, are expressed in quiescent stem cells and promote cell cycle arrest by inhibiting CDKs (FIG. 1a). As shown in stem cell compartments such as HSCs and NSCs, inhibition of p21 results in an increase in stem cell proliferation and in a decrease in the quiescent stem cell population65,72. In NSCs, the loss of p21 does not seem to alter the lineage fate. The reduced number of quiescent stem cells correlates with impaired self-renewal capacity of p21-deficient cells, which ultimately results in an exhaustion of the stem cell pool65,72.
Interestingly, inhibition of p27 does not affect the number or self-renewal of HSCs, but increases the size of the haematopoietic progenitor pool73. The fact that p57 deficiency has no effect on HSC quiescence might be due to the functional overlap with other CKIs74,75. Previously, it was shown that p27 and p57 bind to heat shock cognate protein 70 (HSC70), a molecular chaperone involved in the nuclear import of specific proteins. p27 and p57 control nuclear transport of the HSC70–cyclin D1 complex and regulate the cell cycle entry of HSCs75. In double-knockout mice lacking both p57 and p27, the loss of CKIs promotes nuclear import of the HSC70–cyclin D1 complex and concomitant RB phosphorylation. As a result, HSC quiescence is severely impaired75. These studies suggest that CKIs are functionally important for the maintenance of stem cell quiescence.
Notch signaling
Notch signalling is involved in tissue maintenance and contributes to cell fate decisions during tissue regeneration76,77. This pathway is an important regulator of proliferation and cell fate commitment of transit amplifying progenitors in many tissue compartments76,77. Recent evidence has demonstrated that Notch signalling also has a role in regulating stem cell quiescence. In MuSCs, genetic ablation of RBP-J, the DNA binding factor that is essential for mediating canonical Notch signalling, results in a depletion of the quiescent stem cell pool78,79. The loss of quiescence is associated with spontaneous activation and premature differentiation of stem cells78,79 (FIG. 1a). In adult NSCs, cell fate is determined by the levels of Notch activity, and quiescence is promoted by high Notch activity80. In contrast to its role in muscle and brain, Notch signalling is not required for quiescent HSC maintenance81, highlighting the complex context-dependent role of this pathway in regulating stem cell quiescence. In fact, Notch signalling promotes differentiation of stem cell progeny in the interfollicular epidermis and hair follicles82,83. However, whether Notch may also have a role in the regulation of stem cell quiescence in these compartments remains to be determined as, for example in muscle, Notch signalling may both promote quiescence and be important in lineage progression of stem cell progeny78,79,84.
Post-transcriptional regulation by miRNAs
Since the discovery of the founding miRNA, lin-4, a small non-coding RNA that regulates several crucial genes during development in Caenorhabditis elegans85, hundreds of conserved miRNAs have been discovered in vertebrates86. Over the past decade, it is clear that these small non-coding RNAs have important roles in the post-transcriptional regulation of diverse cellular processes. miRNAs bind to the 3′ untranslated region (3′ UTR) of target mRNAs, resulting in their cleavage or trans-lational repression87. This mode of post-transcriptional regulation has emerged as an important aspect in the control of stem cell quiescence, as recently demonstrated in HSCs88 and MuSCs51. In HSCs, miR-126 controls stem cell quiescence by attenuating multiple components in the PI3K–AKT signalling pathway88. Interestingly, reducing miR-126 activity allows HSC proliferation without inducing exhaustion88. Conversely, overexpression of miR-126 in HSCs impairs cell cycle entry, resulting in a lower haematopoietic contribu-tion88. By contrast, conditional knockout of the miRNA processing factor Dicer triggers spontaneous activation of quiescent MuSCs51. MuSCs subsequently undergo apoptosis, which is similar to the finding in HSCs89. Furthermore, many quiescence-specific miRNAs have been identified in MuSCs, and it was demonstrated that one miRNA, miR-489, is an important regulator of the quiescent state51. miR-489 functions to prevent MuSC proliferation by suppressing the oncogene DEK51,90. In another study, the Myf5 (myogenic factor 5) mRNA and its regulatory miRNA miR-31 were found to be sequestered in mRNA ribonucleoprotein particle (mRNP) granules in quiescent MuSCs91. This report suggests that quiescent MuSCs are primed for differentiation, as the storage of mRNAs makes then readily available for the activation of differentiation programmes. These studies provide evidence for the spatial and temporal regulation of miRNAs in the quiescent state and the active regulation of stem cell quiescence by post-transcriptional mechanisms (FIG. 1b).
The finding of distinct miRNA expression patterns in quiescent stem cells suggests that miRNAs are important regulatory components of the quiescent state. Although the underlying mechanisms have yet to be determined, the importance of transcript 3′ UTRs as targets for miRNAs suggests that stem cell quiescence is controlled, at least in part, by mechanisms that alter 3′ UTR length and thus the susceptibility to regulation by miRNAs92–95. 3′ UTR length of a transcript can be modified by mechanisms such as alternative splicing or alternative cleavage and polyadenylation96,97. There is much interest in the role of alternative polyadenylation in controlling several aspects of stem cell function, many of which specifically relate to changes in 3′ UTRs98. The differential susceptibility of a myogenic factor, paired box 3 (PA×3), to miRNA regulation has been reported in quiescent MuSCs that were isolated from different muscle groups92. It has previously been shown that T cell activation from the quiescent state is associated with widespread shortening of 3′ UTRs, thereby circumventing the regulatory role of mRNA-targeting miRNAs during activation94. Shortening of 3′ UTRs seems to correlate with proliferation in many cell types94, including aberrant proliferation in the case of cancer cells95 or during somatic cell reprogramming99 (FIG. 1c). In addition, miRNA regulation can act as a fine-tuning mechanism to modify target gene expression. mRNAs can be partially repressed when both miRNA and target mRNA are co-expressed100. A change in the expression levels of miRNAs can tip the balance and result in repression or activation of many functionally important target genes101,102. In the same way, miRNAs may repress genes that are required for stem cell activation. By tipping the balance, genes that are functionally important for activation can be derepressed and participate in the process rapidly.
Survival mechanisms in quiescent cells
Long-lived, non-dividing quiescent stem cells may accumulate damage from environmental stress (for example, oxidative stress caused by the accumulation of reactive oxygen species (ROS)), similarly to any long-lived cell such as a post-mitotic neuron or cardio-myocyte103. Environmental stress may lead to damage of cellular constituents, including DNA, a process that has been proposed to underlie the ageing of cells and tissue and to limit lifespan104,105. Accordingly, quiescent stem cells seem to have adopted specific mechanisms to respond to environmental stresses and, thus, to maintain cellular integrity and assure long-term survival. These mechanisms are likely to be different from their proliferating progeny, which can be subject to selection during proliferative expansion and are capable of diluting out damaged cellular components during cell division.
Signalling to protect from environmental stress
Studies of the FOXO family of transcription factors have revealed that this pathway is functionally important in quiescent stem cells to safeguard these cells from environmental stress. In the mammalian system, FOXO family members (which are FOXO1, FOXO3, FOXO4 and FOXO6) have important roles in various cellular processes in a PI3K– AKT pathway-dependent manner106. HSCs depleted of FOXO1, FOXO3 and FOXO4 exhibit a marked increase in ROS and the propensity to exit from quiescence107 (FIG. 2a). Interestingly, administration of the antioxidant N-acetyl-L-cysteine is able to rescue this FOXO-deficient phenotype in HSCs107. In NSCs, FOXO3 regulates the size of the NSC pool108. NSCs devoid of FOXO3 are defective in self-renewal, highlighting the importance of this pathway in regulating stem cell quiescence and survival.
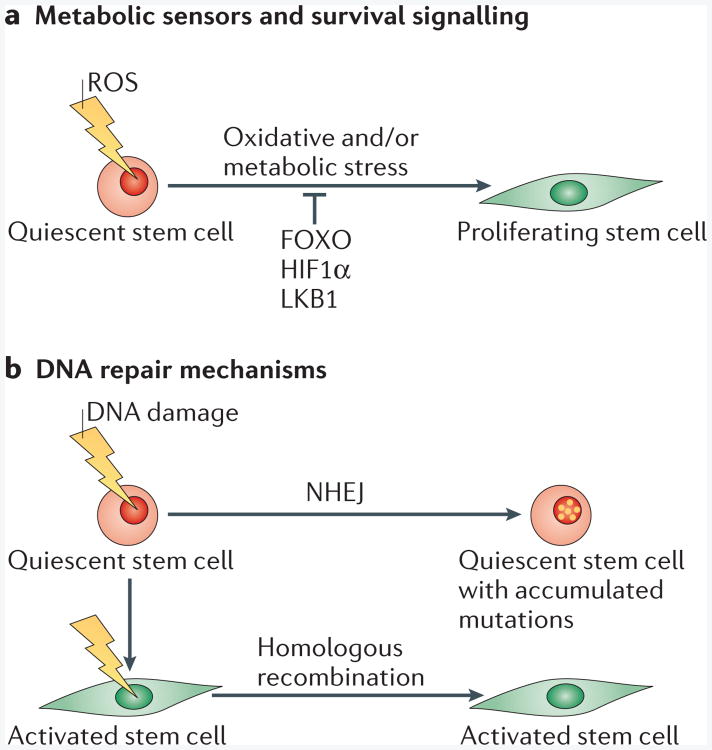
Figure 2. Survival mechanisms of quiescent and activated stem cells.
a| Metabolic sensors an d effectors such as forkhead box O (FOXO), hypoxia-inducible factor 1α (HIF1α) and liver kinase B1 (LKB1) are expressed in quiescent stem cells. Expression of these molecules is essential for metabolic functions and survival of quiescent stem cells in adverse environments. These factors protect quiescent stem cells from oxidative stress caused by the accumulation of reactive oxygen species (ROS). Quiescent stem cells devoid of these pathways have an increased propensity to become activated and fail to maintain the stem cell pool. b | Non-homologous end-joining (NHEJ) and homologous recombination are pathways that repair DNA double-strand breaks (DSBs). Homologous recombination is a high-fidelity mechanism that uses homologous templates as guides for DSB repair, whereas NHEJ directly ligates the ends of the DSBs. DSBs that have been repaired by NHEJ can be imprecise when the overhangs at the DSBs are not compatible. In quiescent stem cells, the error-prone NHEJ mechanism is used for DSB repair, which suggests that DNA mutations accumulate in these cells throughout their life. MYF5, myogenic factor 5.
Metabolic sensors and response mechanisms
The survival of quiescent cells depends on intrinsic mechanisms to sustain metabolic function during persistent environmental stresses. In an extreme case, quiescent MuSCs and HSCs were found viable in post-mortem tissue109. The remarkable ability to survive in such adverse conditions suggests that quiescent stem cells may have unique protective mechanisms, many of which are described above. Recent findings suggest that at least some stem cell populations reside in poorly oxygenated niches110–112, and this has sparked interests in understanding how stem cells regulate their metabolic demand in such hypoxic environments. Interestingly, quiescence is induced when cultured haematopoietic cells are grown under hypoxic conditions113,114. During normal homeostasis, HSCs express hypoxia inducible factor 1α (HIF1α), a basic helix–loop–helix transcription factor that is expressed in mammalian cells growing in hypoxic conditions. The level of HIF1α is important for HSC quiescence115. Inhibition of ubiquitin-mediated degradation of HIF1α results in over-stabilization of HIF1α protein and induction of HSC quiescence116 (FIG. 2a). Conversely, HSCs in HIF1α-knockout mice are not able to maintain quiescence, and these mice exhibit HSC depletion116. Liver kinase B1 (LKB1), a regulator of AMP-activated protein kinase (AMPK), mammalian target of rapamycin complex 1 (mTORC1) and FOXO pathways, links sensing and metabolism and is required for maintaining energy homeostasis. Moreover, LKB1 is thought to be the master regulator of cellular metabolism by limiting cell growth in unfavourable conditions such as hypoxia117. Interestingly, upon genetic ablation of LKB1, HSC quiescence is lost and is accompanied by an increase in progenitor cell proliferation and eventual depletion of HSCs118–120 (FIG. 2a). The effect of LKB1 on HSC quiescence is cell autonomous, as shown by transplantation experiments in which LKB1-deficient HSCs are not able to rescue lethally irradiate d recipient mice118–120.
In keeping with their ability to sense and respond to environmental cues related to the metabolic state, quiescent cells rely on autophagic processes for survival, and the induction of autophagy seems to be important in the regulation of stem cell activation. Autophagy is a lysosomal degradation pathway that is involved in cytoplasmic organelle recycling, preserving the healthy state of cells by removing damaged components121. Conditional knockout of the essential autophagy gene Atg7 in the haematopoietic system results in a reduced number of stem and progenitor cells of multiple lineages and the accumulation of aberrant mitochondria and ROS. This suggests that autophagy is essential for maintenance of HSC quiescence122. Autophagy is also induced in HSCs in which Lkb1 is conditionally ablated, which implies that autophagy may act as a compensatory mechanism to rescue the metabolic stress in these mutants118.
Preservation of genomic integrity
In addition to environmental stress, quiescent stem cells can also be subjected to DNA damage during normal homeostatic turnover, and quiescent stem cells depend on DNA repair mechanisms for survival123. Among the most detrimental DNA mutations are double-strand breaks (DSB), and cells have specialized mechanisms to repair these mutations. In mammalian systems, two major mechanisms, namely homologous recombination and non-homologous end-joining (NHEJ), mediate DSB repair124. Whereas NHEJ is an error-prone DNA repair mechanism, homologous recombination is a high-fidelity DSB repair mechanism. Homologous recombination uses a long homologous sequence to guide repair in the S phase and G2 phase of the cell cycle, in which sister chromatids are available as templates125. By contrast, NHEJ does not require a template and is predominantly used in G1 phase of the cell cycle125. Consistent with these findings, a recent study demonstrated that quiescent HSCs preferentially use NHEJ to repair DSBs126, whereas homologous recombination has been reported to occur more predominantly in proliferating progenitor cells. Intriguingly, this suggests that although DSBs are repaired in quiescent stem cells, mutations may accumulate in these cells as a consequence of using error-prone repair mechanisms (FIG. 2b).
In considering mechanisms by which quiescent stem cells preserve genomic integrity, John Cairns proposed the immortal strand hypothesis that was based on the idea that stem cells possess unique mechanism to safeguard their DNA by non-randomly segregating sister chromosomes during mitosis127. In this model, the oldest template DNA strands are preferentially segregated to the self-renewed stem cells to avoid the accumulation of replication-induced mutations in the stem cell pool. Emerging evidence has demonstrated that stem cells in different tissue compartments exhibit template strand segregation during cell divisions128. There is mounting evidence of asymmetric chromosome segregation in stem cells such as MuSCs17,18,129, ISCs20,130,131 and NSCs132. However, the mechanisms by which stem cells engage in asymmetric chromosome segregation remain to be determined133,134. Furthermore, asymmetric chromosome segregation does not occur universally in stem cell compartments, and its prevalence may differ depending on the experimental paradigm. During normal tissue homeostasis, neither template strand segregation nor label retention was observed in one study of HSCs135. Similarly, evidence of asymmetric chromosome segregation was lacking in certain studies of epidermal stem cells136,137 and ISCs138. Further studies investigating the mechanisms that regulate template strand segregation may reveal how, and the extent to which, stem cell populations use this intriguing cellular function to preserve genomic integrity in the quiescent state.
Quiescence as a poised state
Recent discoveries suggest that the quiescent state is not just a passive state but, instead, actively regulated by different intrinsic mechanisms. It seems that quiescent stem cells have the ability to sense environmental changes and respond by re-entering the cell cycle for proliferation. How does a quiescent stem cell respond to such stimuli rapidly? In one extreme scenario, a quiescent stem cell would maintain the expression of all necessary components that are required for activation and proliferation. However, given the low metabolic state of a quiescent stem cell, this seems unlikely. We thus propose that quiescent stem cells are poised for activation by specific energetically favourable mechanisms that are compatible with the low metabolic state of quiescence and that allow for rapid and global responses needed for activation (FIG. 3). One such example is the regulation of the quiescent state by miRNAs (see above). From an energetics point of view, it seems favourable for a quiescent stem cell to alter the expression of specific miRNAs, as each of these in turn affect a pool of target genes. It has previously been shown that the miR-16 family of miRNAs has a role in regulating G0 to G1 transition139. Intriguingly, silencing of transcripts that are downregulated by miR-16 also affects cell cycle progression139. As miRNAs can affect a number of target genes that are important in a shared pathway, we propose that this is the case in the regulation of quiescence and activation. Further investigations of the regulatory factors that affect alternative cleavage and polyadenylation of mRNAs or miRNA expression will provide a better understanding of how the quiescent state is actively regulated by these transcriptional and post-transcriptional mechanisms.
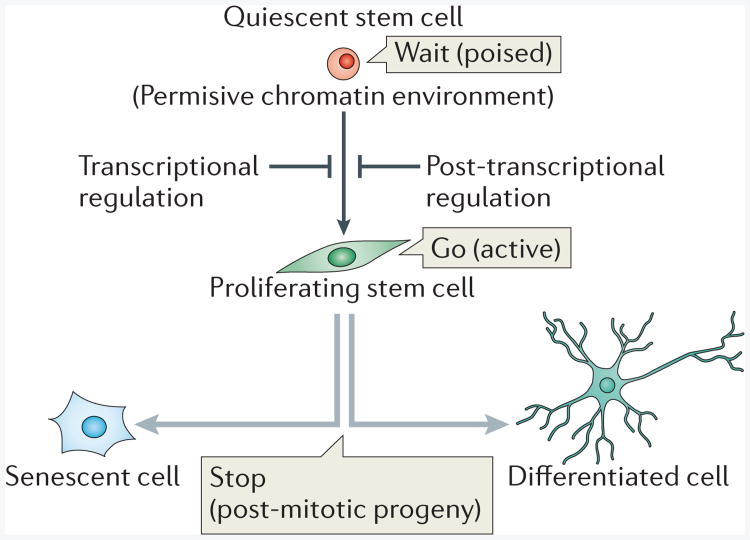
Figure 3. Quiescent stem cells are poised for activation.
A proposed model of how the quiescent state of a stem cell constitutes a poised state for activation. Quiescent stem cells are actively regulated at the epigenetic, transcriptional and post-transcriptional level. The epigenetic landscape keeps the chromatin in a permissive state, which allows rapid transcriptional activation. Additional layers of transcriptional and post-transcriptional control safeguard quiescent stem cells to enable precise stem cell activation when necessary.
In view of the epigenetic control of stem cell quiescence, loci required for stem cell activation are possibly marked by permissive histone marks. Transcriptional activation of loci that are marked by H3K4me3 at their transcription start sites may depend on additional trans-criptional and post-transcriptional mechanisms. To understand whether loci are actively transcribed, it is crucial to consider occupancy of RNA polymerase II (Pol PII) at methylated histones as well as the Pol II phosphorylation status (FIG. 4). Pol II activity is tightly regulated by phosphorylation of its carboxy-terminal domain (CTD)140. Phosphorylation of Ser5 of the Pol II CTD by transcription factor IIH (TFIIH) is required for transcription initiation, whereas phosphorylation of Ser2 of the Pol II CTD, of DSIF (DRB sensitivity inducing factor) and of NELF (negative elongation factor) by pTEFb (positive transcription elongation factor) is needed for transcription elongation141–143. In various cell types, including ES cells, Pol II often occupies promoter regions and not the gene body. This is consistent with evidence of promoter-proximal pausing which indicates some form of post-initiation regulation144–146. Interestingly, Ser2 phosphorylation of the Pol II CTD is absent in many adult quiescent stem cells, which indicates a lack of transcription elongation in these cells147. MYC controls Pol II-mediated elongation and pause release, and it can activate a large number of genes that promote rapid proliferation144. The existence of mechanisms in ES cells that allow silent genes to be activated in a precise and synchronous fashion may also be applicable to the regulation of adult stem cell activation from the quiescent state. The identification of genes that are poised for stem cell activation may provide insight into how the quiescent state can rapidly respond to changes in their environment.
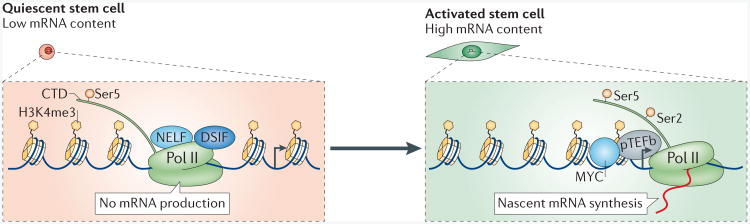
Figure 4. Transcriptional control of stem cell quiescence.
Current data suggest that RNA polymerase II (Pol II) is paused at transcription start sites where histone H3 is trimethylated at Lys 4 (H3K4me3; which is a permissive histone mark). Quiescent stem cells have low mRNA content. The carboxy-terminal domain (CTD) of PolII is phosphorylated at Ser5 but not the Ser2, which indicates transcriptional initiation but not transcriptional elongation. Many types of quiescent stem cells lack Ser2 phosphorylation at the Pol II CTD, which suggests that transcriptional elongation does not occur in these cells, and hence the mRNA levels are low. In quiescent stem cells, Pol II is associated with the negative elongation factors DSIF (DRB sensitivity inducing factor) and NELF. Upon stem cell activation, phosphorylation of DSIF, of NELF (negative elongation factor) and of Ser2 of the Pol II CTD, in combination with the expression of MYC, which is recruited to promoters together with transcription elongation factors (such as pTEFb (positive transcription elongation factor b)), transform the state of promoter proximal pausing into productive elongation and lead to mRNA synthesis.
In addition, genes that are necessary for lineage progression can be poised by transcriptional and post-transcriptional mechanisms. Epigenetic profiling of the H3K4me3 mark in quiescent HFSCs or quiescent MuSCs (T.H.C and T.A.R, unpublished observations) has revealed a number of genes that are marked by this permissive histone mark61,62. In support of the hypothesis that the quiescent state is poised for activation, many of these H3K4me3-rich genes (most of which are expressed at low levels) are functionally important for activation and proliferation. Thus, gene expression and epigenetic profiling during stem cell lineage progression will provide important insights into genes and pathways that are programmed for activation in quiescent stem cells, rendering those cells poised for activation.
Perspective and concluding remarks
Advances in genetic approaches such as lineage tracing have now allowed the design of prospective isolation techniques for the purification of rare cell populations such as quiescent stem cells. Similarly, the use of conditional gene ablation approaches allows the functional analysis of individual genes and pathways in such rare populations in physiological settings. As demonstrated by studies of different stem cell compartments, one of the consequences of inhibiting essential signalling pathways that maintain the quiescent state is the premature activation or differentiation of stem cells. This is often followed by the exhaustion of the stem cell pool and results in impaired tissue homeo-stasis and regeneration, highlighting the importance of maintaining stem cell quiescence for tissue and organismal health. In some tissues, quiescent stem cells seem to serve as a reserve pool of stem cells and are only called into action upon tissue injury. A better understanding of stem cell quiescence and the intrinsic mechanisms by which such cells sense and respond to environmental signals will undoubtedly aid the design of new therapeutic approaches based on enhancing stem cell functionality.
Supplementary Material
Acknowledgments
Research in the Rando laboratory is supported by awards from the Glenn Foundation for Medical Research, the US National Institutes of Health (P01 AG036695, R37 MERIT Award AG023806, R01 AR056849, and R01 AR062185), the Muscular Dystrophy Association and the Department of Veterans Affairs (Merit Review) to
Glossary
- Progenitor cell
Proliferating stem cell progeny that can differentiate into specific cell types
- Heterochronic parabiosis
Whereby an old animal is surgically connected to a young animal to promote the establishment of a single, shared circulatory system between the two
- Lineage tracing
The process of identifying all progeny of a single cell
- Transit amplifying progenitors
Progenitor cells that replicate rapidly with very short cell cycle times for progenitor cell expansion
Footnotes
Competing interests statement: The authors declare no competing financial interests
Further Information: Thomas Rando's homepage: http://randolab.stanford.edu
Supplementary Information: See online article: S1 (table)
References
- 1.Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- 2.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nature Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 4.Howard A, Pelc SR. Synthesis of deoxyribonucleic acid in normal and irradiated cells and its relation to chromosome breakage. Hered (Lond) [Suppl] 1953;6:261–273. [Google Scholar]
- 5.Baserga R. Biochemistry of the cell cycle: a review. Cell Prolifer. 1968;1:167–191. [Google Scholar]
- 6.Patt HM, Quastler H. Radiation effects on cell renewal and related systems. Physiol Rev. 1963;43:357–396. doi: 10.1152/physrev.1963.43.3.357. [DOI] [PubMed] [Google Scholar]
- 7.Stoker MGP. The Leeuwenhoek Lecture, 1971: tumour viruses and the sociology of fibroblasts. Proc R Soc Series B Biol Sci. 1972;181:1–17. doi: 10.1098/rspb.1972.0038. [DOI] [PubMed] [Google Scholar]
- 8.Temin HM. Stimulation by serum of multiplication of stationary chicken cells. J Cell Physiol. 1971;78:161–170. doi: 10.1002/jcp.1040780202. [DOI] [PubMed] [Google Scholar]
- 9.Pardee AB. A restriction point for control of normal animal cell proliferation. Proc Natl Acad Sci USA. 1974;71:1286–1290. doi: 10.1073/pnas.71.4.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zetterberg A, Larsson O. Kinetic analysis of regulatory events in G1 leading to proliferation or quiescence of Swiss 3T3 cells. Proc Natl Acad Sci USA. 1985;82:5365–5369. doi: 10.1073/pnas.82.16.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray JV, et al. ‘Sleeping beauty’: quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2004:68, 187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amen RD. A model of seed dormancy. Bot Rev. 1968;34:1–31. [Google Scholar]
- 13.Fausto N. Liver regeneration and repair: hepatocytes, progenitor cells, and stem cells. Hepatology. 2004;39:1477–1487. doi: 10.1002/hep.20214. [DOI] [PubMed] [Google Scholar]
- 14.Hüttmann A, Liu SL, Boyd AW, Li CL. Functional heterogeneity within rhodamine123loHoechst33342lo/spprimitive hemopoietic stem cells revealed by pyronin Y. Exp Hematol. 2001;29:1109–1116. doi: 10.1016/s0301-472x(01)00684-1. [DOI] [PubMed] [Google Scholar]
- 15.Fukada S, et al. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- 16.Gerdes J, Schwab U, Lemke H, Stein H. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer. 1983;31:13–20. doi: 10.1002/ijc.2910310104. [DOI] [PubMed] [Google Scholar]
- 17.Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 2007;5:e102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinin V, Gayraud-Morel B, Gomès D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nature Cell Biol. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 19.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 20.Potten CS, Hume WJ, Reid P, Cairns J. The segregation of DNA in epithelial stem cells. Cell. 1978;15:899–906. doi: 10.1016/0092-8674(78)90274-x. [DOI] [PubMed] [Google Scholar]
- 21.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. Reports a conditional genetic approach by expressing an H2B—GFP fusion protein to label slow-cycling cells and identifies LRCs in the skin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson A, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 23.Foudi A, et al. Analysis of histone 2B–GFP retention reveals slowly cycling hematopoietic stem cells. Nature Biotechnol. 2008;27:84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakkalakal JV, Jones KM, Basson MA, Brack AS. The aged niche disrupts muscle stem cell quiescence. Nature. 2012;490:355–360. doi: 10.1038/nature11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buczacki SJA, et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. Describes a strategy to genetically mark quiescent crypt cells in the gut and provides a new method to document the behaviour of LRCs over time. [DOI] [PubMed] [Google Scholar]
- 26.Bickenbach JR. Identification and behavior of label-retaining cells in oral mucosa and skin. J Dent Res. 1981;60:1611–1620. doi: 10.1177/002203458106000311011. [DOI] [PubMed] [Google Scholar]
- 27.Grompe M. Tissue stem cells: new tools and functional diversity. Cell Stem Cell. 2012;10:685–689. doi: 10.1016/j.stem.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeda N, et al. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. Using genetic approaches, this paper demonstrates that quiescent ISCs could be interconverted to active ISCs and displayed stem cell identity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barker N, et al. Identification of stem cells in smallintestine and colon by marker gene. Lgr5 Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. Using knock-in reporter alleles, this paper shows the expression of LGR5 in the crypt in actively cycling basal columnar cells and suggests that LGR5 is a stem cell marker in the intestine, multiple adult tissues and cancers. [DOI] [PubMed] [Google Scholar]
- 30.Barker N, et al. Lgr5+vestem cells drive self-renewal in the stomach and build long-lived gastric units. in vitro Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 31.Morris RJ, et al. Capturing and profiling adult hair follicle stem cells. Nature Biotechol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 32.Jaks V, et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nature Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 33.Rompolas P, et al. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature. 2012;487:496–499. doi: 10.1038/nature11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clayton E, et al. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- 35.Ito M, et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nature Med. 2005;11:1351–1354. doi: 10.1038/nm1328. Shows that ablation of hair follicle stem cells in the bulge leads to complete loss of hair follicles but the survival of the epidermis, suggesting that quiescent hair follicle stem cells are only called into action during would repair but not during normal homeostasis of the epidermis. [DOI] [PubMed] [Google Scholar]
- 36.Van Keymeulen A, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 37.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 38.Ousset M, et al. Multipotent and unipotent progenitors contribute to prostate postnatal development. Nature Cell Biol. 2012;14:1131–1138. doi: 10.1038/ncb2600. [DOI] [PubMed] [Google Scholar]
- 39.Joe AWB, et al. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nature Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nature Cell Biol. 2010;12:143–152. doi: 10.1038/ncb2014. References 39 and 40 reveal the existence of a fibrogenic and/or adipogenic progenitor population that resides in skeletal muscle. Together with muscle stem cells, these progenitors facilitate myogenesis in response to muscle injury. [DOI] [PubMed] [Google Scholar]
- 41.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 42.Muñoz J, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 44.Pastrana E, Cheng LC, Doetsch F. Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proc Natl Acad Sci USA. 2009;106:6387–6392. doi: 10.1073/pnas.0810407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forsberg EC, et al. Molecular signatures of quiescent, mobilized and leukemia-initiating hematopoietic stem cells. PLoS ONE. 2010;5:e8785. doi: 10.1371/journal.pone.0008785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lauper N, et al. Cyclin E2: a novel CDK2 partner in the late G1 and S phases of the mammalian cell cycle. Oncogene. 1998;17:2637–2643. doi: 10.1038/sj.onc.1202477. [DOI] [PubMed] [Google Scholar]
- 48.Kalaszczynska I, et al. Cyclin A is redundant in fibroblasts but essential in hematopoietic and embryonic stem cells. Cell. 2009;138:352–365. doi: 10.1016/j.cell.2009.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pines J, Hunter T. Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell. 1989;58:833–846. doi: 10.1016/0092-8674(89)90936-7. [DOI] [PubMed] [Google Scholar]
- 50.Li F, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 51.Cheung TH, et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482:524–528. doi: 10.1038/nature10834. Proposes that the miRNA pathway is essential for the maintenance of muscle stem cell quiescence and identifies the quiescence-specific miRNA miR-489 that functions as a regulator of muscle stem cell quiescence by inhibiting DEK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arnold CP, et al. microRNA programs in normal and aberrant stem and progenitor cells. Genome Res. 2011;21:798–810. doi: 10.1101/gr.111385.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, Stokes N, Polak L, Fuchs E. Specific microRNAs are preferentially expressed by skin stem cells to balance self-renewal and early lineage commitment. Cell Stem Cell. 2011;8:294–308. doi: 10.1016/j.stem.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernstein BE, Meissner A, Lander EL. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 55.Li G, et al. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 57.Ezhkova E, et al. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25:485–498. doi: 10.1101/gad.2019811. Suggests that the H3K27 methyltransferases EZH1 and EZH2 are essential for hair follicle and wound repair and demonstrates an important role of the Polycomb group complex in regulating the hair follicle lineage in an epigenetic manner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Juan AH, et al. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011;25:789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamminga LM, et al. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107:2170–2179. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hidalgo I, et al. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell. 2012;11:649–662. doi: 10.1016/j.stem.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 61.Lien WH, et al. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell. 2011;9:219–232. doi: 10.1016/j.stem.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woodhouse S, Pugazhendhi D, Brien P, Pell JM. Ezh2 maintains a key phase of muscle satellite cell expansion but does not regulate terminal differentiation. J Cell Sci. 2013;126:565–579. doi: 10.1242/jcs.114843. [DOI] [PubMed] [Google Scholar]
- 63.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y, et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009;4:37–48. doi: 10.1016/j.stem.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng T, et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287:1804–1808. doi: 10.1126/science.287.5459.1804. This is one of the earliest papers to suggest stem cell quiescence is actively regulated. Shows, using p21-deficent mice that HSC proliferation increases and the proportion of quiescent HSCs decreases. [DOI] [PubMed] [Google Scholar]
- 66.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 67.Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 68.Jacques TS, et al. Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes. EMBO J. 2010;29:222–235. doi: 10.1038/emboj.2009.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G1control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 2000;14:3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hosoyama T, Nishijo K, Prajapati SI, Li G, Keller C. Rb1 gene inactivation expands satellite cell and postnatal myoblast pools. J Biol Chem. 2011;286:19556–19564. doi: 10.1074/jbc.M111.229542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Viatour P, et al. Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell Stem Cell. 2008;3:416–428. doi: 10.1016/j.stem.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kippin TE, Martens DJ, van der Kooy D. p21 loss compromises the relative quiescence of forebrain stem cell proliferation leading to exhaustion of their proliferation capacity. Genes Dev. 2005;19:756–767. doi: 10.1101/gad.1272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cheng T, Rodrigues N, Dombkowski D, Stier S, Scadden DT. Stem cell repopulation efficiency but not pool size is governed by p27kip1. Nature Med. 2000;6:1235–1240. doi: 10.1038/81335. [DOI] [PubMed] [Google Scholar]
- 74.Matsumoto A, et al. p57 is required for quiescence and maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2011;9:262–271. doi: 10.1016/j.stem.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 75.Zou P, et al. p57Kip2and p27Kip1cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell Stem Cell. 2011;9:247–261. doi: 10.1016/j.stem.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 76.Luo D, Renault VM, Rando TA. The regulation of Notch signaling in muscle stem cell activation and postnatal myogenesis. Semin Cell Dev Biol. 2005;16:612–622. doi: 10.1016/j.semcdb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 77.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 78.Bjornson CRR, et al. Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells. 2012;30:232–242. doi: 10.1002/stem.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mourikis P, et al. A critical requirement for Notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells. 2012;30:243–252. doi: 10.1002/stem.775. References 78 and 79 demonstrate that Notch signalling has a role in regulating muscle stem cell quiescence, by showing that genetic ablation of RBP-J specifically in quiescent muscle stem cells results in spontaneous cell cycle entry and, ultimately, the depletion of the stem cell pool. [DOI] [PubMed] [Google Scholar]
- 80.Chapouton P, et al. Notch activity levels control the balance between quiescence and recruitment of adult neural stem cells. J Neurosci. 2010;30:7961–7974. doi: 10.1523/JNEUROSCI.6170-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maillard I, et al. Canonical Notch signaling is dispensable for the maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2008;2:356–366. doi: 10.1016/j.stem.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Estrach S, Cordes R, Hozumi K, Gossler A, Watt FM. Role of the Notch ligand Delta1 in embryonic and adult mouse epidermis. J Invest Dermatol. 2007;128:825–832. doi: 10.1038/sj.jid.5701113. [DOI] [PubMed] [Google Scholar]
- 83.Williams SE, Beronja S, Pasolli HA, Fuchs E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature. 2011;470:353–358. doi: 10.1038/nature09793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 85.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 86.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540–1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 87.Bartel DP. microRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 88.Lechman ER, et al. Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell. 2012;11:799–811. doi: 10.1016/j.stem.2012.09.001. Suggests that the HSC pool size could be fine-tuned by altering the level of miR-126 in HSCs and shows that miR-126 regulates HSC activation by targeting the PI3K—AKT— glycogen synthase kinase 3β pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guo S, et al. microRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci USA. 2010;107:14229–14234. doi: 10.1073/pnas.0913574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ageberg M, Gullberg U, Lindmark A. The involvement of cellular proliferation status in the expression of the human proto-oncogene. DEK Haematologica. 2006;91:268–269. [PubMed] [Google Scholar]
- 91.Crist CG, Montarras D, Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell. 2012;11:118–126. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 92.Boutet SC, et al. Alternative polyadenylation mediates microRNA regulation of muscle stem cell function. Cell Stem Cell. 2012;10:327–336. doi: 10.1016/j.stem.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miura P, Amirouche A, Clow C, Bélanger G, Jasmin BJ. Brain-derived neurotrophic factor expression is repressed during myogenic differentiation by miR-206. J Neurochem. 2012;120:230–238. doi: 10.1111/j.1471-4159.2011.07583.x. [DOI] [PubMed] [Google Scholar]
- 94.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nature Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mueller AA, Cheung TH, Rando TA. All's well that ends well: alternative polyadenylation and its implications for stem cell biology. Curr Opin Cell Biol. 2013;25:22–32. doi: 10.1016/j.ceb.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS ONE. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mukherji S, et al. microRNAs can generate thresholds in target gene expression. Nature Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 102.Farh KKH, et al. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 103.Rando TA. Stem cells, ageing and the quest for immortality. Nature. 2006;441:1080–1086. doi: 10.1038/nature04958. [DOI] [PubMed] [Google Scholar]
- 104.Kregel KC, Zhang HJ. An integrated view of oxidative stress in aging: basic mechanisms, functional effects, and pathological considerations. Am J Physiol Regul Integr Comp Physiol. 2007;292:R18–R36. doi: 10.1152/ajpregu.00327.2006. [DOI] [PubMed] [Google Scholar]
- 105.Rossi DJ, Jamieson CHM, Weissman IL. Stems cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 106.Tothova Z, Gilliland DG. FoxO transcription factors and stem cell homeostasis: insights from the hematopoietic system. Cell Stem Cell. 2007;1:140–152. doi: 10.1016/j.stem.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 107.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 108.Renault VM, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5:527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Latil M, et al. Skeletal muscle stem cells adopt a dormant cell state post mortem and retain regenerative capacity. Nature Commun. 2012;3:903. doi: 10.1038/ncomms1890. [DOI] [PubMed] [Google Scholar]
- 110.Mounier R, Chrétien F, Chazaud B. Blood vessels and the satellite cell niche. Curr Top Dev Biol. 2011;96:121–138. doi: 10.1016/B978-0-12-385940-2.00005-X. [DOI] [PubMed] [Google Scholar]
- 111.Simsek T, et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7:380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Parmar K, Mauch P, Vergilio JA, Sackstein R, Down JD. Distribution of hematopoietic stem cells in the bone marrow according to regional hypoxia. Proc Natl Acad Sci USA. 2007;104:5431–5436. doi: 10.1073/pnas.0701152104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shima H, et al. Acquisition of G0state by CD34-positive cord blood cells after bone marrow transplantation. Exp Hematol. 2010;38:1231–1240. doi: 10.1016/j.exphem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 114.Hermitte F, Brunet de la Grange P, Belloc F, Praloran V, Ivanovic Z. Very low O2concentration (0.1%) favors G0 return of dividing CD34+cells. Stem Cells. 2006;24:65–73. doi: 10.1634/stemcells.2004-0351. [DOI] [PubMed] [Google Scholar]
- 115.Suda T, Takubo K, Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell. 2011;9:298–310. doi: 10.1016/j.stem.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 116.Takubo K, et al. Regulation of the HIF-1α level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7:391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 117.Blagih J, Krawczyk CM, Jones RG. LKB1 and AMPK: central regulators of lymphocyte metabolism and function. Immunol Rev. 2012;249:59–71. doi: 10.1111/j.1600-065X.2012.01157.x. [DOI] [PubMed] [Google Scholar]
- 118.Gurumurthy S, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gan B, et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468:701–704. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468:653–658. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mortensen M, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lombard DB, et al. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 124.Sancar A, Lindsey-Boltz LA, Uuml;nsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 125.Shrivastav M, Haro LPD, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 126.Mohrin M, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7:174–185. doi: 10.1016/j.stem.2010.06.014. Proposes that HSCs preferentially use a more error-prone DNA repair mechanism in the quiescent state and that HSCs might accumulate mutations as a consequence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 128.Charville GW, Rando TA. Stem cell ageing and non-random chromosome segregation. Phil Trans R Soc B Biol Sci. 2011;366:85–93. doi: 10.1098/rstb.2010.0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rocheteau P, Gayraud-Morel B, Siegl-Cachedenier I, Blasco MA, Tajbakhsh S. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell. 2012;148:112–125. doi: 10.1016/j.cell.2011.11.049. [DOI] [PubMed] [Google Scholar]
- 130.Falconer E, et al. Identification of sister chromatids by DNA template strand sequences. Nature. 2009;463:93–97. doi: 10.1038/nature08644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci. 2002;115:2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- 132.Karpowicz P, et al. Support for the immortal strand hypothesis: neural stem cells partition DNA asymmetrically. in vitro J Cell Biol. 2005;170:721–732. doi: 10.1083/jcb.200502073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rando TA. The immortal strand hypothesis: segregation and reconstruction. Cell. 2007;129:1239–1243. doi: 10.1016/j.cell.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 134.Lansdorp PM. Immortal strands? Give me a break. Cell. 2007;129:1244–1247. doi: 10.1016/j.cell.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 135.Kiel MJ, et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449:238–242. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sotiropoulou PA, Candi A, Blanpain C. The majority of multipotent epidermal stem cells do not protect their genome by asymmetrical chromosome segregation. Stem Cells. 2008;26:2964–2973. doi: 10.1634/stemcells.2008-0634. [DOI] [PubMed] [Google Scholar]
- 137.Kuroki T, Murakami Y. Random segregation of DNA strands in epidermal basal cells. Jpn J Cancer Res. 1989;80:637–642. doi: 10.1111/j.1349-7006.1989.tb01690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schepers AG, Vries R, van den Born M, van de Wetering M, Clevers H. Lgr5 intestinal stem cells have high telomerase activity and randomly segregate their chromosomes. EMBO J. 2011;30:1104–1109. doi: 10.1038/emboj.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Linsley PS, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription. in vivo Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wada T, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yamaguchi Y, et al. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 143.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 144.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wade JT, Struhl K. The transition from transcriptional initiation to elongation. Curr Opin Genet Dev. 2008;18:130–136. doi: 10.1016/j.gde.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Freter R, Osawa M, Nishikawa S. Adult stem cells exhibit global suppression of RNA polymerase II serine-2 phosphorylation. Stem cells. 2010;28:1571–1580. doi: 10.1002/stem.476. [DOI] [PubMed] [Google Scholar]
- 148.Campisi J. Cellular senescence: putting the paradoxes in perspective. Curr Opin Genet Dev. 2011;21:107–112. doi: 10.1016/j.gde.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Coppé JP, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:e301. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Brockes JP, Kumar A. Plasticity and reprogramming of differentiated cells in amphibian regeneration. Nature Rev Mol Cell Biol. 2002;3:566–574. doi: 10.1038/nrm881. [DOI] [PubMed] [Google Scholar]
- 152.Kumar A, Velloso CP, Imokawa Y, Brockes JP. Plasticity of retrovirus-labelled myotubes in the newt limb regeneration blastema. Dev Biol. 2000;218:125–136. doi: 10.1006/dbio.1999.9569. [DOI] [PubMed] [Google Scholar]
- 153.McGann CJ, Odelberg SJ, Keating MT. Mammalian myotube dedifferentiation induced by newt regeneration extract. Proc Natl Acad Sci USA. 2001;98:13699–13704. doi: 10.1073/pnas.221297398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Simon HG, et al. Differential expression of myogenic regulatory genes and Msx-1 during dedifferentiation and redifferentiation of regenerating amphibian limbs. Dev Dynam. 1995;202:1–12. doi: 10.1002/aja.1002020102. [DOI] [PubMed] [Google Scholar]
- 155.Odelberg SJ, Kollhoff A, Keating MT. Dedifferentiation of mammalian myotubes induced by msx1. Cell. 2000;103:1099–1109. doi: 10.1016/s0092-8674(00)00212-9. [DOI] [PubMed] [Google Scholar]
- 156.Beauséjour CM, et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 2003;22:4212–4222. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pajcini KV, Corbel SY, Sage J, Pomerantz JH, Blau HM. Transient Inactivation of Rb and ARF yields regenerative cells from postmitotic mammalian muscle. Cell Stem Cell. 2010;7:198–213. doi: 10.1016/j.stem.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hsu YC, Fuchs E. A family business: stem cell progeny join the niche to regulate homeostasis. Nature Rev Mol Cell Biol. 2012;13:103–114. doi: 10.1038/nrm3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells. 1978;4:7–25. [PubMed] [Google Scholar]
- 160.Lander AD, et al. What does the concept of the stem cell niche really mean today? BMC Biol. 2012;10:19. doi: 10.1186/1741-7007-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Brack AS, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–810. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 162.Conboy IM, et al. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- 163.Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Conboy IM, Conboy MJ, Smythe GM, Rando TA. Notch-mediated restoration of regenerative potential to aged muscle. Science. 2003;302:1575–1577. doi: 10.1126/science.1087573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.