Abstract
Cerebellar granule neurons are the most abundant neurons in the brain, and a critical element of the circuitry that controls motor coordination and learning. In addition, granule neuron precursors (GNPs) are thought to represent cells of origin for medulloblastoma, the most common malignant brain tumor in children. Thus, understanding the signals that control the growth and differentiation of these cells has important implications for neurobiology and neuro-oncology. Our previous studies have shown that proliferation of GNPs is regulated by Sonic hedgehog (Shh), and that aberrant activation of the Shh pathway can lead to medulloblastoma. Moreover, we have demonstrated that Shh-dependent proliferation of GNPs and medulloblastoma cells can be blocked by basic fibroblast growth factor (bFGF). But while the mitogenic effects of Shh signaling have been confirmed in vivo, the inhibitory effects of bFGF have primarily been studied in culture. Here we demonstrate that mice lacking FGF signaling in GNPs exhibit no discernable changes in GNP proliferation or differentiation. In contrast, activation of FGF signaling has a potent effect on tumor growth: treatment of medulloblastoma cells with bFGF prevents them from forming tumors following transplantation, and inoculation of tumor-bearing mice with bFGF markedly inhibits tumor growth in vivo. These results suggest that activators of FGF signaling may be useful for targeting medulloblastoma and other Shh-dependent tumors.
INTRODUCTION
Normal development requires a delicate balance between proliferation and differentiation. Too little proliferation can result in impaired tissue structure and function, while excessive proliferation can lead to cancer. One salient example of this is the development of granule neurons in the cerebellum. Mutant mice that fail to generate enough granule cells develop severe ataxia (1-3), and patients with congenital granule cell degeneration have deficits in motor coordination, language use, and cognitive function (4). Conversely, when GNP differentiation fails, cells that would normally become post-mitotic continue to proliferate and give rise to medulloblastoma (5, 6). Thus, elucidating the molecular mechanisms that control growth and differentiation of granule neurons is critical for understanding both normal cerebellar development and tumorigenesis.
GNPs are generated on the surface of the cerebellum in a structure called the external germinal layer (EGL) (7, 8). At birth, the EGL contains a single layer of undifferentiated cells, but over the next few days, these cells undergo extensive proliferation to generate a large pool of GNPs (9, 10). As new GNPs are generated, older cells exit the cell cycle and differentiate, and their cell bodies migrate inward to the internal granule layer (IGL) (11, 12). The waves of proliferation and differentiation continue until about 3 weeks of age (in mice), when all GNPs mature into granule neurons and the EGL disappears (13).
Studies from our lab and others have demonstrated that the major mitogen for GNPs is Sonic hedgehog (Shh) (14-16). Produced by Purkinje neurons, Shh acts by binding to the transmembrane protein Patched (Ptc). In the absence of Shh, Ptc acts as an antagonist of the hedgehog pathway, inhibiting the function of the signal transducing protein Smoothened (Smo). When Shh binds to Ptc, inhibition is alleviated, and Smo initiates a signaling cascade that results in activation of Gli family transcription factors and expression of hedgehog target genes (17, 18). In GNPs, these include N-myc and cyclins D1 and D2 (19, 20), and thus Shh signaling results in a robust proliferative response. Notably, mice with targeted mutations that impair Shh signaling (e.g. deletion of Shh, Smo or Gli2) exhibit decreased GNP proliferation (21-23); conversely, animals with mutations that activate the pathway (e.g. deletion of ptc) develop medulloblastoma (5, 6, 24-26). The fact that ~30% of human medulloblastomas contain activating mutations in the Shh pathway (27-29) also implies a key role for this pathway in proliferation and tumorigenesis.
But while the importance of Shh in GNP proliferation is well established, the signals that cause GNPs to exit the cell cycle and differentiate remain much less clear. One potential explanation might be decreased exposure of GNPs to Shh ligand. However, previous studies have shown that Shh continues to be expressed by Purkinje cells into adulthood (30). The fact that GNPs can exit the cell cycle despite the continued presence of Shh suggests that some other signal may override the mitogenic effects of Shh. We have previously shown that bFGF can block proliferation and promote differentiation of GNPs in the presence of Shh (14, 31), raising the possibility that FGFs might serve as differentiation factors for these cells. In addition, we have shown that bFGF can inhibit the growth of tumor cells from ptc mutant mice (31), suggesting that this pathway may hold promise for medulloblastoma therapy.
Although FGFs have been shown to inhibit proliferation of GNPs and tumor cells in vitro, their effects on these cells have not been rigorously tested in vivo. In the current study, we used conditional knockout mice to investigate the importance of FGF signaling in GNP development in vivo, and intratumoral injections to test the efficacy of bFGF as a treatment for medulloblastoma. Our results suggest that genetic ablation of FGFRs does not alter GNP differentiation in vivo. However, exposure of medulloblastoma cells to bFGF dramatically inhibits their growth. Thus, while bFGF may not be critical for GNP development, activation of the FGF pathway may be useful in the treatment of Shh-dependent tumors.
RESULTS AND DISCUSSION
FGF inhibits proliferation of GNPs during postnatal development
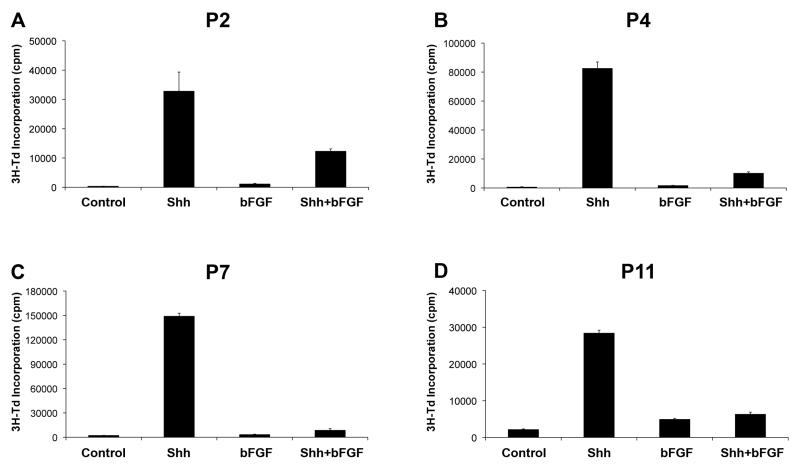
GNPs undergo proliferation and differentiation in the EGL during the first 2-3 weeks after birth. Our previous studies (31) have shown that bFGF inhibits proliferation and promotes differentiation of GNPs. To determine whether the inhibitory effects of bFGF are age-dependent, we isolated GNPs at a range of postnatal ages and measured incorporation of tritiated thymidine in response to Shh and bFGF. Consistent with our previous observations, P7 GNPs proliferated robustly in response to Shh, and this response was abolished by addition of bFGF (Figure 1C). Marked inhibition of Shh-induced proliferation was also seen at P2, P4 and P11 (Figures 1A, B and D). Thus, bFGF can inhibit proliferation of GNPs throughout postnatal development.
Figure 1. bFGF inhibits proliferation of GNPs throughout postnatal development.
GNPs were isolated from P2 (A), P4 (B), P7 (C), or P11 (D) mice and cultured in media with no added factors (Control) or 20% Shh supernatant ± bFGF (25 ng/mL) for 48 hrs. Cells were pulsed with tritiated thymidine (3H-Td) and cultured for 16hr before being assayed for thymidine incorporation. Data represent means of triplicate samples ± SEM. Differences in proliferation between Shh and Shh+bFGF were statistically significant (p < 0.0001 based on Student’s t-test) at P4, P7 and P11; at P2, the difference was 2.7-fold, but did not reach significance (p = 0.09).
bFGF promotes cell cycle exit and acts downstream of Suppressor of Fused
Our previous studies suggested that bFGF promotes cell cycle exit of GNPs and accelerates their differentiation into granule neurons (31). To better understand the mechanisms by which bFGF inhibits proliferation, we performed cell cycle analysis on cells treated with Shh ± bFGF. As shown in Supplementary Figure 1A-C, in the absence of growth factors the majority of GNPs exit the cell cycle within 48 hours (< 2% of cells in S, G2 and M phases of the cell cycle). Treatment with Shh maintains a population of GNPs in cycle (~11% of cells in S/G2/M). However, co-treatment with bFGF inhibits the effects of Shh and causes cells to accumulate in the G0/G1 phase of the cell cycle (~2% of cells in S/G2/M). We also examined whether bFGF increased apoptosis by staining cells with antibodies specific for cleaved caspase 3. As shown in Supplementary Figure 2A, bFGF did not cause a significant increase in apoptosis. Together, these data suggest that bFGF acts by promoting cell cycle exit and differentiation rather than by inducing cell death.
We previously showed that bFGF can inhibit proliferation of GNPs in response to stimulation with Shh ligand, as well as of tumor cells resulting from mutations in patched (31). These data suggest that bFGF acts on the Shh pathway at a level downstream of patched. To gain further insight into the mechanisms by which bFGF interferes with Shh signaling, we tested the effects of bFGF on cells in which the Shh pathway is activated as a consequence of a Smo mutation. NeuroD2-SmoA1 transgenic mice exhibit increased Shh pathway activation and proliferation of GNPs and develop medulloblastoma (32). As shown in Supplementary Figure 3A, we found that bFGF inhibits the basal proliferation of these cells, suggesting that it acts downstream of Smo.
Shh target genes can also be activated by mutations in Suppressor of Fused (Sufu), a negative regulator of the pathway that regulates processing, localization and transcriptional activity of Gli proteins (33-35). To determine whether bFGF could suppress activation of the pathway resulting from loss of Sufu, we tested the effects of bFGF on mouse embryo fibroblasts (MEFs) from Sufu-deficient mice (36). As shown in Supplementary Figure 3B, Sufu−/− MEFs exhibit constitutive expression of gli1 that is significantly higher than that seen in wild type MEFs. Importantly, addition of bFGF to Sufu−/− MEFS results in a 3-fold reduction in gli1 expression, suggesting that bFGF can inhibit activation of the pathway mediated by loss of Sufu. Similar results were observed when a Gli-luciferase reporter gene was used to monitor gli1 expression (data not shown). Together these data suggest that bFGF inhibits Shh signaling at a level downstream of Smo and Sufu, and proximal to the nucleus.
The inhibitory effects of bFGF are mediated by FGFR1
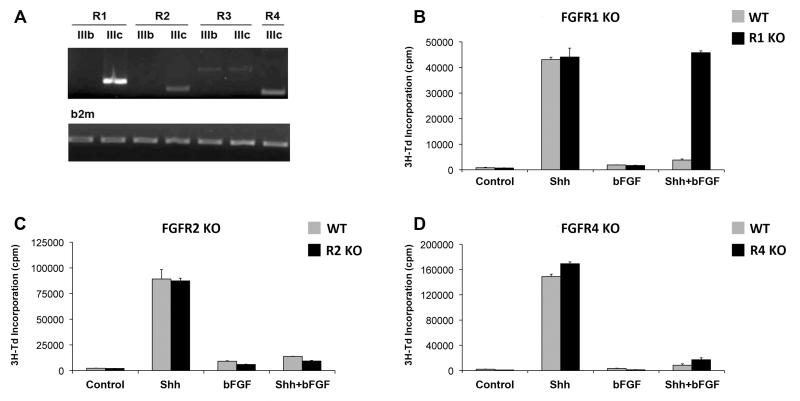
The FGF receptor family consists of four members (FGFR1-4), three of which can undergo alternative splicing to generate multiple receptor isoforms (FGFR4 exists in only one isoform) (37). To determine which receptors might mediate inhibition of Shh signaling and proliferation, we first examined the receptors expressed by GNPs. Cells were FACS-sorted from the cerebellum of Math1-GFP transgenic mice (which express green fluorescent protein (GFP) in their GNPs (38, 39)), and RNA was isolated and subjected to RT-PCR using primers for FGFR receptor isoforms. Consistent with previous studies (40), we found that GNPs express FGFR1, 2 and 4; FGFR3 was also detected in some samples, but usually at lower levels than the other receptors (Figure 2A).
Figure 2. FGF-mediated inhibition of proliferation requires FGFR1.
(A) Expression of FGFR isoforms in GNPs. RNA was isolated from FACS-sorted Math1-GFP+ cells at P7 and FGFR expression was assessed by PCR using isoform-specific primers. RT-PCR for beta 2-microglobulin was performed on each sample in parallel to control for variability among samples. (B-D) GNPs from P7 Math1-Cre;FGFR1F/F (FGFR1 KO, panel B), Math1-Cre;FGFR2F/F (FGFR2 KO, panel C) or FGFR4−/− mice (FGFR4 KO, panel D) were cultured in media containing no added factors (Control) or Shh ± bFGF for 48hrs. Cells were pulsed with 3H-Td and cultured for 16hr before being assayed for thymidine incorporation. Data represent means of triplicate samples ± SEM. Differences in proliferation between Shh and Shh+bFGF were statistically significant (p < 0.002 based on Student’s t-test) for all genotypes except FGFR1 KO mice; in those animals (panel B, black bars), proliferation in response to Shh+bFGF was not significantly different from proliferation in response to Shh alone (p = 0.66).
We have previously shown that FGF-mediated inhibition of Shh signaling can be blocked by pharmacological antagonists of FGFR kinase activity (31). Because these antagonists can act on all FGF receptors, the particular receptor or receptors required for inhibition of hedgehog signaling remained unclear. To determine which FGFRs were required for the inhibitory effects of bFGF in GNPs, we used mice lacking FGFR1, 2 and 4, the predominant FGFRs expressed in these cells. Complete loss of FGFR1 or FGFR2 results in embryonic lethality (41, 42); therefore we crossed mice carrying loxP-flanked alleles of these genes (43, 44) with Math1-Cre transgenic mice (5, 6) to generate animals lacking FGFR1 or FGFR2 in GNPs. These animals, along with germline FGFR4 knockout mice (which remain viable into adulthood (45, 46)), were used to examine the effects of loss of FGFRs on GNP responses to Shh and bFGF. GNPs from single knockout mice were cultured in the presence of Shh ± bFGF for 48 hours and then assayed for incorporation of tritiated thymidine. As shown in Figure 2B-D, GNPs from mice lacking FGFR1, FGFR2 or FGFR4 all showed robust proliferation in response to Shh. But while loss of FGFR2 or FGFR4 did not affect FGF-mediated inhibition of Shh-induced proliferation (Figure 2 C-D), loss of FGFR1 completely abrogated the inhibitory effects of bFGF (Figure 2B). Consistent with its inability to suppress Shh-induced proliferation, bFGF was also unable to inhibit Shh induction of gli1 in FGFR1-deficient GNPs (Supplementary Figure 4). FGFR1-deficient GNPs treated with Shh also showed no change in cell cycle distribution following exposure to bFGF (Supplementary Figure 1D-F). Together, these data indicate that the inhibitory effects of bFGF require signaling through FGFR1.
Our previous studies (31) demonstrated that FGF-mediated inhibition of Shh signaling is mediated, at least in part, by activation of the extracellular signal-regulated kinase (ERK). To determine whether GNPs lacking FGFR1 were still capable of activating this kinase, we stimulated cells with bFGF and examined the phosphorylation status of ERK. As shown in Supplementary Figure 5, wild type (WT) GNPs exhibited robust ERK phosphorylation in response to bFGF. In contrast, GNPs from mice lacking FGFR1 showed no increase in ERK phosphorylation. These studies suggest that loss of FGFR1 renders GNPs unresponsive to bFGF.
FGF signaling is not required for GNP differentiation
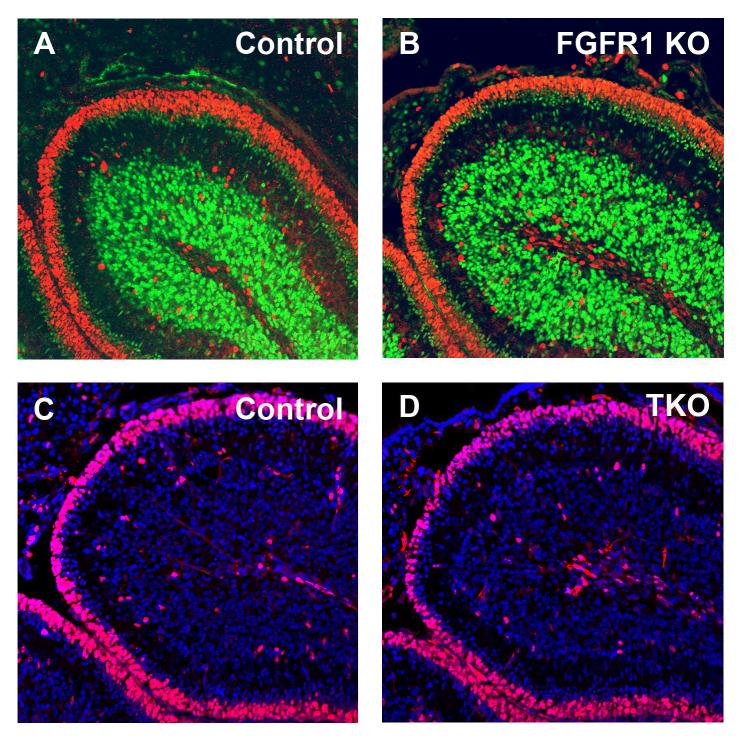
Since our studies indicated that FGFR1 was a key mediator of FGF-mediated inhibition in vitro, we next examined the effects of loss of FGFR1 on GNP differentiation in vivo. We isolated cerebella from WT and FGFR1-deficient mice at P7 and stained them with antibodies specific for markers of proliferation and differentiation. As shown in Figure 3A, WT cerebella contained a broad band of proliferating (Ki67+) cells in the outer EGL and discrete regions of differentiating (NeuN+) granule neurons in the inner EGL and IGL. Surprisingly, expression of Ki67 and NeuN in FGFR1 knockout mice was indistinguishable from that seen in WT littermates (Figure 3B). These results suggested that loss of FGFR1 does not impair GNP cell cycle exit or differentiation.
Figure 3. Loss of FGF signaling does not alter GNP growth or differentiation.
(A-B) Mid-sagittal sections from cerebella of P7 Math1-Cre;FGFR1F/F mice (FGFR1 KO, B) and littermate controls (A) were stained with anti-Ki67 (red) and anti-NeuN (green) antibodies. (C-D) Mid-sagittal sections from P7 triple knockout (Math1-Cre; FGFR1F/F; FGFR2F/F; FGFR4−/−) mice (TKO, D) and control (C) cerebella were stained with anti-Ki67 antibodies (red) and counterstained with DAPI. Note the similarity in the thickness of the EGL and in the numbers of proliferating (Ki67+) cells in mutant and WT mice.
One possible explanation for the lack of an in vivo phenotype in FGFR1-deficient mice was compensation by other FGFR receptors. To address this possibility, we generated triple knockout (TKO) mice lacking FGFRs 1, 2 and 4 in GNPs. Similar to GNPs from FGFR1 knockout mice, Shh-induced proliferation of GNPs from TKO mice was not inhibited by bFGF in culture (Supplementary Figure 6), nor did TKO GNPs treated with bFGF exhibit an increase in apoptosis (Supplementary Figure 2B). In contrast, BMP2, which inhibits GNP proliferation through a distinct mechanism (47, 48)) remained capable of inhibiting Shh-induced proliferation of TKO GNPs (Supplementary Figure 6). However, analysis of TKO cerebella showed no significant differences in proliferation or differentiation when compared to WT littermates (Figure 3C-D). Similar results were seen when cerebella were analyzed at P2, P4, P11 and P14 (data not shown). Based on these findings we conclude that FGF signaling, while capable of promoting differentiation in vitro, is not required for GNP differentiation in vivo.
FGF inhibits growth of medulloblastoma cells in vitro and in vivo
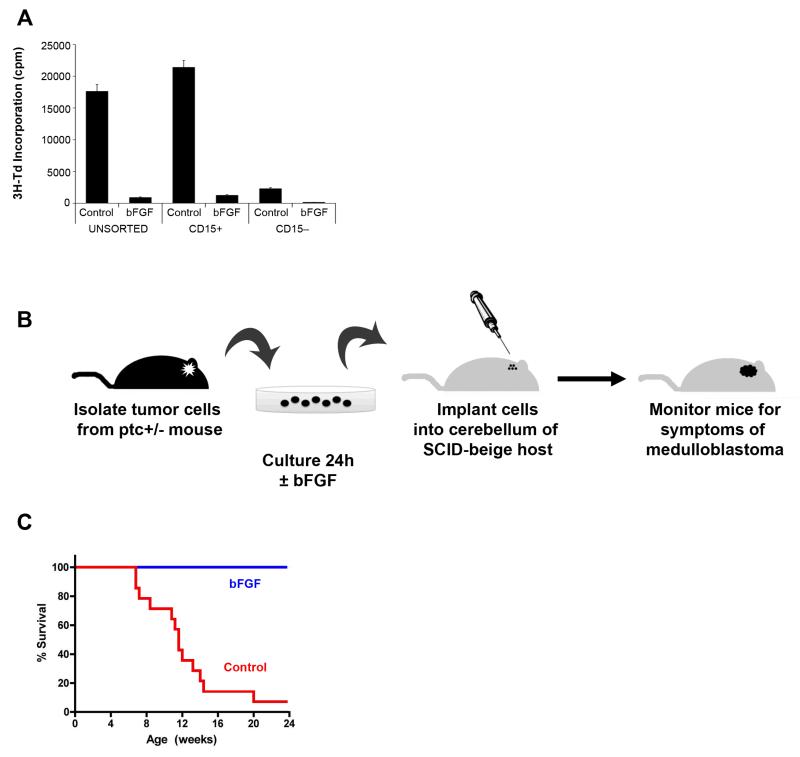
While the above experiments suggest that FGF signaling is not essential for GNP development, they leave open the possibility that it might be useful as a treatment for Shh-dependent tumors. Recent studies from our lab have indicated that tumors from ptc+/− mutant mice are propagated by a subpopulation of tumor cells that expresses the carbohydrate antigen CD15/SSEA-1 (49). To determine whether CD15+ cells are sensitive to FGF-mediated inhibition, we purified these cells by flow cytometry, cultured them in the presence or absence of bFGF, and then measured their incorporation of tritiated thymidine. As shown in Figure 4A, bFGF markedly inhibited the growth of both unfractionated and CD15+ tumor cells. The basal proliferation of CD15− cells was much lower than their CD15 counterparts, but these cells were inhibited by bFGF as well. Moreover, we did not see an increase in apoptosis when unsorted tumor cells from ptc+/− mice were cultured in the presence of bFGF, indicating that the reduced proliferation observed following bFGF treatment was not due to increased cell death (Supplementary Figure 7). These results indicated that tumor-propagating cells from ptc+/− mice are sensitive to the inhibitory effects of bFGF.
Figure 4. Exposure to bFGF prevents engraftment of medulloblastoma cells.
(A) bFGF inhibits proliferation of tumor-propagating cells. Tumor cells from ptc+/− mice were left unsorted or FACS-sorted into CD15+ and CD15− populations, and then cultured in media with no added factors (Control) or bFGF for 48 hrs. Cells were pulsed with 3H-Td and cultured for an additional 16 hrs before being assayed for thymidine incorporation. Data represent means of triplicate samples ± SEM. Differences between control and bFGF-treated cells were significant for all populations (p < 0.0001 based on unpaired Student’s t-tests). (B) ptc+/− tumor cells were cultured in the presence or absence of bFGF for 24 hrs and then transplanted into SCID-beige recipient mice. Mice were sacrificed when they displayed symptoms. (C) Survival of animals implanted with Control or bFGF-treated tumor cells. None of the mice receiving bFGF-treated tumor cells developed medulloblastoma, whereas 13/14 (93%) of mice receiving Control tumor cells developed medulloblastoma. Survival curves were significantly different (p < 0.0001) based on Log-Rank (Mantel-Cox) test.
Although bFGF inhibited the growth of tumor cells, it was possible that this inhibition was transient, or that a subpopulation of tumor cells remained capable of forming tumors even after exposure to bFGF. To test this possibility, we isolated tumor cells from ptc+/− mice and cultured them in the presence or absence of bFGF for 24hr. Cells were then transplanted into the cerebellum of SCID-beige mice and recipients were monitored for symptoms of tumor development (Figure 4B). As shown in Figure 4C, 93% of mice that received tumor cells cultured in control media went on to form tumors. In contrast, no tumors resulted from transplantation of FGF-treated cells. These studies demonstrated that the growth-inhibitory effects of bFGF are not transient, but rather, result in a long-lasting inability of cells to form tumors in vivo.
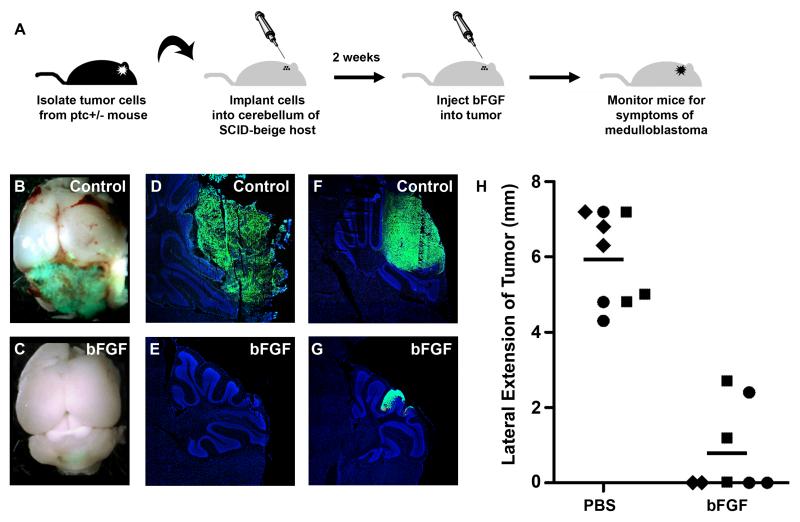
Having shown that bFGF pretreatment could abolish the tumorigenic potential of ptc+/− medulloblastoma cells, we wondered whether bFGF could also inhibit the expansion of tumor cells that were already growing in animals. To test this, we transplanted tumor cells into the cerebellum of SCID-beige mice, and after 8-12 days, injected PBS (vehicle) or bFGF into the implantation site. Animals received an additional injection 3-5 days later, and were then monitored for signs of tumor development (Figure 5A). For these experiments, we used tumors from Math1-GFP;ptc+/− mice, which allowed us to detect tumor bulk based on GFP expression (50). As shown in Figure 5B, tumors injected with PBS typically grew unchecked, encompassing a large portion of the cerebellum. In contrast, bFGF-treated tumors were barely detectable in intact cerebella (Figure 5C). Analysis of cerebellar sections revealed large numbers of GFP+ cells in PBS-treated animals, and few such cells in animals treated with bFGF (Figure 5D-G). H&E staining confirmed that the area of each tumor was comparable to the expression of GFP, both in the control and bFGF-treated animals (Supplementary Figure 8). To quantitate the reduction in tumor size following FGF treatment, we counted the number of sections in which tumor cells could be found. As shown in Figure 5H, FGF treatment resulted in a significant reduction in tumor bulk. Based on these results, we conclude that FGF is a potent inhibitor of tumor growth and may be useful for targeting ptc+/− mutant medulloblastomas and other hedgehog-dependent tumors.
Figure 5. bFGF inhibits medulloblastoma growth in vivo.
(A) Tumor cells were isolated from Math1-GFP; ptc+/− mice and immediately implanted into the cerebellum of SCID-beige mice. Eight to twelve days after transplantation, animals were injected with PBS or bFGF, and then monitored for symptoms of medulloblastoma. (B-C) Representative whole-mount pictures illustrating the presence of large GFP+ (green) tumors in Control (PBS-treated) mice (B) and the relatively small lesions detected in bFGF-treated mice (C). (D-G) Cryosections stained with DAPI (blue) showing tumors (indicated by GFP fluorescence, green) observed in representative Control (D, F) and bFGF-treated (E, G) mice. (H) Quantitative analysis of tumor size in three cohorts of mice (cohorts, consisting of animals transplanted with tumor cells from a separate Math1-GFP; ptc+/− donor, are designated by triangles, squares, or diamonds respectively). Lateral extension of each tumor was calculated as described in Materials and Methods. Lateral tumor extension was significantly different in bFGF-treated mice (mean = 0.7 mm) vs. Control mice (mean = 6.0 mm), p < 0.0001 determined by unpaired Student’s t-test.
The studies described above reinforce our previous observations that bFGF can inhibit Shh signaling and Shh-induced proliferation in GNPs and tumor cells. Moreover, they provide new insight into mechanisms of bFGF-mediated inhibition, demonstrating that (1) this requires signaling through FGFR1 and (2) that it acts on the Shh pathway at a level downstream of Smo and Sufu. Interestingly, a recent report (Lauth et al., 2010) demonstrates that oncogenic Ras can also inhibit Shh signaling, and that this involves activation of the DYRK1B kinase. Since bFGF signaling is likely to involve activation of Ras, it is tempting to speculate that bFGF-mediated inhibition involves a similar mechanism. However, it is notable that oncogenic Ras and Dyrk1B did not affect signaling in Sufu−/− cells, suggesting that the mechanisms underlying inhibition, while they might overlap with those associated with bFGF, are likely distinct.
Notwithstanding the potent inhibitory effects of bFGF in vitro, our studies demonstrate that FGF signaling is not required for normal GNP development. The fact that mice lacking FGF receptors showed no obvious phenotype is surprising, given the marked anti-mitogenic effects of FGF signaling for these cells in vitro. One possible explanation for this is that some level of FGF signaling persists even in the absence of FGF receptors. FGFs are known to activate transcription of Sprouty genes, which function as negative regulators of signaling by FGFRs and other receptor tyrosine kinases (51). In the absence of FGF signaling, this negative feedback loop could be lost, resulting in a compensatory increase in activation of the FGF pathway. Alternatively, it is possible that other extracellular signals may act independently to promote cell cycle exit and differentiation of GNPs in the postnatal cerebellum. In this regard, it is worth noting that in addition to bFGF, vitronectin, pituitary adenylate cyclase activating polypeptide (PACAP) and bone morphogenetic proteins (BMPs) 2 and 4 have all been reported to inhibit Shh-induced proliferation of GNPs in vitro (47, 48, 52, 53). To date, none of these have been shown to be required for GNP differentiation in vivo. Thus, the signals responsible for GNP cell cycle exit and differentiation remain unclear, and the possibility that multiple factors cooperate to regulate these processes remains open.
Whatever its role in GNP development, our studies clearly indicate that FGF signaling can inhibit the growth of GNP-derived tumors. This is significant for several reasons. First, it highlights the unique characteristics of the tumor-propagating cells (cancer stem cells) in these tumors. In contrast to the stem-like (CD133+, multipotent, neurosphere-forming) cells that have been shown to be important for propagation of other brain tumors (54, 55), we have found that growth of ptc-associated medulloblastoma is dependent on GNP-like cells that express the surface marker CD15 (49). Although a recent report suggested that long-term culture of ptc+/− tumor cells in FGF-containing media can expand a rare population of stem-like cells with tumorigenic potential (56), our demonstration that acute bFGF treatment – either in vitro or in vivo – dramatically inhibits subsequent tumor growth raises questions about the role of these cells in tumor maintenance and their importance as targets for therapy. Although bFGF is likely to have different effects on different classes of brain tumors (and could potentially enhance the growth of some tumor subtypes), at least for Hedgehog pathway-associated medulloblastoma, it appears to be a potent inhibitor of tumor growth.
Hedgehog antagonists have recently moved into clinical trials for a variety of types of cancer, and have begun to show some promise for treatment of medulloblastoma (57). However, early reports also suggest that resistance to these antagonists can develop quite readily (58). Therefore, it is critical to develop alternative approaches to targeting these diseases. Although bFGF does not readily cross the blood-brain barrier (59), our results suggest that intratumoral injection (perhaps aided by convection-enhanced delivery (60)) could be an effective mode of therapy. Small molecules that mimic the effects of bFGF (61-63) might also be useful for overcoming the blood brain barrier. The fact that some medulloblastoma cell lines are growth inhibited by bFGF (64, 65) suggests that our results are not confined to mouse models, and would be worth pursuing in the context of human medulloblastoma.
Materials and Methods
Animals
Math1-Cre transgenics (5, 6) were from David Rowitch at UCSF. FGFR1 conditional knockout (Fgfr1F/F) mice (43) were obtained from Chuxia Deng at the National Institutes of Health; FGFR2 conditional knockouts (Fgfr2F/F (44)) were provided by David Ornitz at Washington University. FGFR4 germline knockouts (Fgfr4−/− (66)) were maintained on a C57/BL6 background in our colony. NeuroD2-SmoA1 transgenic mice (32) were a gift from James Olson, Fred Hutchinson Cancer Center. FGFR triple knockout (TKO) mice were offspring of Fgfr1F/F; Fgfr2F/F; Fgfr4−/− x Fgfr1F/F; Fgfr2F/F; Fgfr4+/−; Math1-Cre+/− pairs. ptc+/− mice (24) were bred in our colony. Animals were bred and maintained in the Cancer Center Isolation Facility at Duke Medical Center.
Growth Factors
bFGF (Peprotech, Rocky Hill, NJ) was used at 25 ng/mL except where indicated. Shh supernatant was generated by transfecting 293T cells with Shh-N expression plasmid (David Robbins, Dartmouth Medical School, Hanover, NH) and harvesting supernatant for 3 days. Supernatant was used at 20% final concentration.
Isolation and Culture of GNPs and Tumor cells
GNPs were isolated from 2 to 11-day-old mice as described in (50). Tumor cells were isolated using a similar procedure from ptc+/− and NeuroD2-SmoA1 mice displaying signs of medulloblastoma. For isolation of CD15+ and CD15− populations, tumor cells were stained with anti-CD15 antibodies (clone MMA, BD Biosciences, San Jose, CA) and with phycoerythrin-conjugated goat-anti-mouse IgM secondary antibodies (Jackson ImmunoResearch, West Grove, PA), and then sorted using a BD FACSVantage flow cytometer. Cells were cultured on poly-D-lysine (PDL)-coated plates in Neurobasal medium containing 2% B27, 1 mM sodium pyruvate, 2 mM L-glutamine, and 1% Pen/Strep (all from Invitrogen, Carlsbad, CA).
RNA Isolation and RT-PCR
RNA was isolated by using the RNAqueous kit (Ambion, Austin, TX). Lysates were treated with DNA-free DNase treatment and removal reagents (Ambion), and RNA was quantitated on a TD-700 fluorometer (Turner BioSystems, Sunnyvale, CA) by using RiboGreen (Invitrogen). RT-PCR for fgfr1-4 was performed using primers described in (34). First-strand cDNA was synthesized using equivalent amounts of total RNA (0.1-1 μg) in a 20-μl reverse transcriptase reaction mixture (Invitrogen). PCR was performed using Platinium Taq (Invitrogen) in a 25uL reaction followed by electrophoresis on a 1.5% agarose gel. Real-time PCR for gli1 was performed as described in (50). Sufu−/− MEFs were a generous gift from Matthias Lauth and Rune Toftgard at the Karolinska Institute, Huddinge, Sweden.
Western Blotting
GNPs from P7 WT and FGFR TKO mice were cultured for 4 hours on PDL-coated plates in Neurobasal media with no additives or with 25ng/mL bFGF. Cells were washed twice with cold PBS and lysed in 200 μl RIPA buffer (0.5M Tris-HCl, pH 7.4, 1.5M NaCl, 2.5% deoxycholic acid, 10% NP-40, 10mM EDTA) with protease and phosphatase inhibitor cocktail (Thermo, Cat# 1861280) for 30 minutes on ice. Lysates were sonicated using an ultrasonicator (Misonix) at amplitude 2 for 5 seconds, and then centrifuged at 13,000 rpm for 10 min at 4°C. Protein concentrations were measured using a Bradford assay. Equal amounts of protein were separated by 8% SDS polyacylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Invitrogen, Cat# LC2006). Membranes were blocked with 5% BSA and then probed with anti-pERK and anti-total ERK (both from Santa Cruz), and with IRDye680-labeled Goat anti-Mouse IgG antibodies (Li-Cor Biosciences). All antibodies were diluted 1:3000 in Tris-buffered saline with 0.1% Tween-20. Proteins were visualized using Li-Cor Odyssey Imager.
Proliferation Assays
For measurement of thymidine incorporation, GNPs and tumor cells were cultured in PDL-coated 96-well plates at 2 × 105 cells per well. Growth factors were added at the beginning of culture, and cells were incubated for 48 h before being pulsed with [methyl-3H]-Thymidine (PerkinElmer, Fremont, CA). After 16 h, cells were harvested by using a Mach IIIM Manual Harvester 96 (Tomtec, Hamden, CT), and incorporated radioactivity was quantitated by using a Wallac MicroBeta microplate scintillation counter (PerkinElmer).
For cell cycle analysis, cells were cultured in PDL-coated plates as described above. After 48 hours, cells were fixed and permeabilized for 20 min in 100 μl of BD Cytofix/Cytoperm Solution (BD Biosciences, San Jose, CA). Cells were then washed with 1 ml of BD Perm/Wash Buffer (BD Biosciences) and resuspended in 20 μl of 7-Aminoactinomycin D (7-AAD). 300 μl of FACS buffer (5% fetal bovine serum in PBS) was added to the cells, and samples were analyzed on a FACScan Flow Cytometer (Becton Dickinson, Franklin Lakes, NJ). Cell cycle profiles were analyzed using FlowJo software (TreeStar Inc., Ashland, OR).
Immunofluorescence Staining and Analysis
For staining of frozen tissue, brains were fixed in 4% paraformaldehyde, cryoprotected in 30% sucrose, embedded in Tissue-Tek OCT (Sakura Finetek, Torrance, CA) and sectioned sagittally at a thickness of 12 μm. Sections were permeabilized with PBST (PBS + 0.1% Triton X-100), blocked with PBST + 10% normal goat serum, and incubated with primary antibodies specific for Ki67 (Abcam, Cambridge, MA) and NeuN (Millipore, Temecula, CA) overnight at 4°C followed by Alexa Fluor 594-anti-rabbit and Alexa Fluor 488-anti-mouse secondary antibodies (Molecular Probes, Eugene, OR) for 1 hr at room temperature. Sections were counterstained with DAPI (Molecular Probes) and mounted in Fluoromount-G (Southern Biotechnology Associates, Birmingham, AL). Tiled images were taken using a Zeiss LSM 510 inverted confocal microscope and related software at the Duke Light Microscopy Core Facility.
To measure apoptosis, GNPs or tumor cells were fixed in 4% PFA and stained with antibodies against cleaved caspase-3 (Cell Signaling Technology) and goat anti-rabbit IgG secondary antibodies (Jackson Immunoresearch). Following staining, immunofluorescent pictures of six randomly-selected fields were taken from each well of cultured cells using a Zeiss LSM 510 inverted confocal microscope and related software at the Duke Light Microscopy Core Facility. Cells were counted for positive cleaved caspase-3 and DAPI respectively. The ratio of cleaved caspase-3 to DAPI staining was determined for each culture condition and compared.
Stereotaxic Implantation and in vivo Treatment of Tumor Cells
Tumor cells from Math1-GFP; ptc+/− mice were transplanted into SCID-beige mice as described in (49). For bFGF pretreatment studies, animals were followed for 6 months and sacrificed when they developed symptoms of medulloblastoma (ataxia, lethargy, hydrocephaly). For in vivo bFGF treatment studies, PBS or bFGF (80ng/ml, 2.5 μl) was injected into the transplant site 8-12 days after transplantation, and again 3-5 days later. When any of the mice in a cohort exhibited symptoms, the entire cohort was sacrificed. Animals were perfused with 4% paraformaldehyde and brains were imaged by whole mount microscopy using a Leica MC 16 FA microscope, Micropublisher 5.0 RTV camera and QCapture software (QImaging, Surray, BC). Brains were then frozen in OCT, sectioned from end to end, and stained with hematoxylin and eosin or with DAPI as described above. Sections were examined for the presence of GFP+ tumor cells, and lateral tumor extension was calculated by counting the number of sections containing such cells and multiplying by the thickness of these sections.
Supplementary Material
Supplementary Figure 1. bFGF inhibits cell cycle progression. GNPs were isolated from control or FGFR1 KO mice at P7 and cultured in Neurobasal media with no supplements (Control), Shh or Shh + bFGF. Cell cycle analysis was performed after 48 hours. Shh promoted cell cycle progression in both WT and KO cells; but bFGF inhibited cell cycle progression only in WT cells.
Supplementary Figure 2. bFGF does not promote apoptosis of GNPs. Freshly isolated WT (A) and FGFR1 KO (B) GNPs were cultured for 24 hours in the presence or absence of 25 ng/mL bFGF, then fixed in 4% PFA and stained with antibodies against cleaved caspase-3 and DAPI. Confocal images of six randomly-selected fields were taken for each well. The percentage of DAPI+ cells positive for cleaved caspase-3 was determined. There was no significant change in the percentage of cleaved caspase-3 positive cells after bFGF treatment in either WT or FGFR1 KO GNPs (based on unpaired Student’s t-tests).
Supplementary Figure 3. (A) bFGF inhibits proliferation of tumor cells expressing activated Smoothened. Tumor cells from NeuroD2:SmoA1 transgenic mice were cultured on PDL-coated plates in control media or media containing 25ng/mL bFGF. After 48 hours, cells were pulsed with tritiated thymidine (3H-Td) and cultured for 16hr before being assayed for thymidine incorporation. Data represent means of triplicate samples ± SEM. Differences between control and bFGF-treated cells were statistically significant (p < 0.0001 based on unpaired Student’s t-test). (B) bFGF inhibits gli expression in WT and Sufu−/− MEFs. WT and Sufu−/− MEFs were cultured in the presence or absence of 25 ng/mL bFGF. After 24 hours, RNA was isolated and gli1 and actin were measured by RT-PCR. Data represent the ratio of gli1:actin, normalized to levels in Control WT MEFs. bFGF caused a three-fold reduction in expression of gli1 in Sufu−/− MEFs.
Supplementary Figure 4: FGFR1 is required for bFGF-mediated suppression of gli expression. GNPs were isolated from Math1-Cre; FGFR1F/F mice at P7 and cultured in control media or media containing Shh, Shh + 25 ng/ml bFGF, or Shh + 1 μM cyclopamine. After 24 hours, RNA was isolated and gli1 and actin levels were determined by real time RT-PCR. Data represent the ratio of gli1:actin, normalized to levels in Control WT GNPs.
Supplementary Figure 5. Impaired ERK phosphorylation in cells lacking FGF receptors. GNPs from FGFR TKO (Math1-Cre; Fgfr1F/F; Fgfr2F/F; Fgfr4−/−) mice and wild type littermates were treated with 25 ng/mL bFGF for 4 hours. Lysates were collected and analyzed by Western blotting for phospho-ERK (pERK) and total ERK (tERK). Increased ERK phosphorylation was observed in WT but not KO cells.
Supplementary Figure 6. Cells lacking FGFRs 1, 2 and 4 are not inhibited by bFGF. GNPs from FGFR TKO (Math1-Cre; Fgfr1F/F; Fgfr2F/F; Fgfr4−/−) mice and wild type littermates were cultured in either control (20% Mock supernatant) or Shh (20% Shh supernatant) media with or without 25ng/mL bFGF or 100ng/mL BMP2 for 48 hr. Cells were pulsed with 3H-Td and cultured for 16 hours before being assayed for thymidine incorporation. Data represent means of triplicate samples ± SEM. FGFR TKO GNPs proliferated in response to Shh; this proliferative response was not inhibited by bFGF, but was inhibited by BMP2. Differences in proliferation between Control and Shh, Shh and bFGF, and Shh and Shh+BMP were statistically significant (p < 0.0005 based unpaired Student’s t-tests) for both WT and TKO cells. Differences between Shh and Shh+bFGF were significant for WT but not for TKO cells.
Supplementary Figure 7. bFGF does not induce apoptosis of ptc+/− tumor cells. Tumor cells were cultured in the presence or absence of 25 ng/mL bFGF for 24 hr, then fixed in 4% PFA and stained for cleaved caspase-3 and DAPI. Confocal images of six randomly-selected fields were taken, and the percentage of DAPI+ cells positive for cleaved caspase-3 was determined. bFGF did not increase the ratio of cells positive for cleaved caspase-3. There was no significant change in the percentage of cleaved caspase-3 positive cells after bFGF treatment (based on unpaired Student’s t-test).
Supplementary Figure 8. H&E staining of PBS and bFGF-injected tumors. Cerebella from mice injected with ptc+/− medulloblastoma cells treated with PBS (A) or bFGF (B) were fixed, sectioned and stained with H&E. Tumor is denoted by asterisk in (A).
Acknowledgements
The authors thank Marcie Kritzik for help with the manuscript, Kerry Dorr for contributions to the early stages of this work, Albert Basson for helpful discussions, Chuxia Deng, David Ornitz and James Olson for providing mice, Matthias Lauth and Rune Toftgard for Sufu−/− MEFs, Sam Johnson for help with microscopy, Amanda Conway for help with Western blotting and Mike Cook and Beth Harvat for help with flow cytometry. This work was funded by grant number MH67916 from the National Institute of Mental Health, by funds from the Pediatric Brain Tumor Foundation of the US, and by a Leadership Award (LA1-01747) from the California Institute for Regenerative Medicine.
REFERENCES
- 1.Kofuji P, Hofer M, Millen KJ, Millonig JH, Davidson N, Lester HA, et al. Functional analysis of the weaver mutant GIRK2 K+ channel and rescue of weaver granule cells. Neuron. 1996;16(5):941–52. doi: 10.1016/s0896-6273(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 2.Hamre KM, Goldowitz D. meander tail acts intrinsic to granule cell precursors to disrupt cerebellar development: analysis of meander tail chimeric mice. Development. 1997;124(21):4201–12. doi: 10.1242/dev.124.21.4201. [DOI] [PubMed] [Google Scholar]
- 3.Mullen RJ, Hamre KM, Goldowitz D. Cerebellar mutant mice and chimeras revisited. Perspect Dev Neurobiol. 1997;5(1):43–55. [PubMed] [Google Scholar]
- 4.Pascual-Castroviejo I, Gutierrez M, Morales C, Gonzalez-Mediero I, Martinez-Bermejo A, Pascual-Pascual SI. Primary degeneration of the granular layer of the cerebellum. A study of 14 patients and review of the literature. Neuropediatrics. 1994;25(4):183–90. doi: 10.1055/s-2008-1073020. [DOI] [PubMed] [Google Scholar]
- 5.Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008 Aug 12;14(2):123–34. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008 Aug 12;14(2):135–45. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldowitz D, Hamre K. The cells and molecules that make a cerebellum. Trends Neurosci. 1998 Sep;21(9):375–82. doi: 10.1016/s0166-2236(98)01313-7. [DOI] [PubMed] [Google Scholar]
- 8.Wang VY, Zoghbi HY. Genetic regulation of cerebellar development. Nat Rev Neurosci. 2001 Jul;2(7):484–91. doi: 10.1038/35081558. [DOI] [PubMed] [Google Scholar]
- 9.Fujita S, Shimada M, Nakamura T. H3-thymidine autoradiographic studies on the cell proliferation and differentiation in the external and the internal granular layers of the mouse cerebellum. J Comp Neurol. 1966;128(2):191–208. doi: 10.1002/cne.901280206. [DOI] [PubMed] [Google Scholar]
- 10.Mares V, Lodin Z, Srajer J. The cellular kinetics of the developing mouse cerebellum. I. The generation cycle, growth fraction and rate of proliferation of the external granular layer. Brain Res. 1970;23(3):323–42. doi: 10.1016/0006-8993(70)90060-0. [DOI] [PubMed] [Google Scholar]
- 11.Fishell G, Hatten ME. Astrotactin provides a receptor system for CNS neuronal migration. Development. 1991;113(3):755–65. doi: 10.1242/dev.113.3.755. [DOI] [PubMed] [Google Scholar]
- 12.Komuro H, Rakic P. Distinct modes of neuronal migration in different domains of developing cerebellar cortex. J Neurosci. 1998;18(4):1478–90. doi: 10.1523/JNEUROSCI.18-04-01478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholson JL, Altman J. The effects of early hypo- and hyperthyroidism on the development of rat cerebellar cortex. I. Cell proliferation and differentiation. Brain Res. 1972;44(1):13–23. doi: 10.1016/0006-8993(72)90362-9. [DOI] [PubMed] [Google Scholar]
- 14.Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22(1):103–14. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 15.Wallace VA. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol. 1999;9(8):445–8. doi: 10.1016/s0960-9822(99)80195-x. [DOI] [PubMed] [Google Scholar]
- 16.Dahmane N, Ruiz i, Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999 Jun;126(14):3089–100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- 17.Jia J, Jiang J. Decoding the Hedgehog signal in animal development. Cellular and molecular life sciences: CMLS. 2006 Jun;63(11):1249–65. doi: 10.1007/s00018-005-5519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008 Sep 15;22(18):2454–72. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 19.Kenney AM, Widlund HR, Rowitch DH. Hedgehog and PI-3 kinase signaling converge on Nmyc1 to promote cell cycle progression in cerebellar neuronal precursors. Development. 2004 Jan;131(1):217–28. doi: 10.1242/dev.00891. [DOI] [PubMed] [Google Scholar]
- 20.Oliver TG, Grasfeder LL, Carroll AL, Kaiser C, Gillingham CL, Lin SM, et al. Transcriptional profiling of the Sonic hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc Natl Acad Sci U S A. 2003 Jun 10;100(12):7331–6. doi: 10.1073/pnas.0832317100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis PM, Gritli-Linde A, Smeyne R, Kottmann A, McMahon AP. Sonic hedgehog signaling is required for expansion of granule neuron precursors and patterning of the mouse cerebellum. Dev Biol. 2004 Jun 15;270(2):393–410. doi: 10.1016/j.ydbio.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Corrales JD, Blaess S, Mahoney EM, Joyner AL. The level of sonic hedgehog signaling regulates the complexity of cerebellar foliation. Development. 2006 May;133(9):1811–21. doi: 10.1242/dev.02351. [DOI] [PubMed] [Google Scholar]
- 23.Spassky N, Han YG, Aguilar A, Strehl L, Besse L, Laclef C, et al. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Dev Biol. 2008 Mar 4; doi: 10.1016/j.ydbio.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–13. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 25.Hatton BA, Villavicencio EH, Tsuchiya KD, Pritchard JI, Ditzler S, Pullar B, et al. The Smo/Smo model: hedgehog-induced medulloblastoma with 90% incidence and leptomeningeal spread. Cancer Res. 2008 Mar 15;68(6):1768–76. doi: 10.1158/0008-5472.CAN-07-5092. [DOI] [PubMed] [Google Scholar]
- 26.Lee Y, Kawagoe R, Sasai K, Li Y, Russell HR, Curran T, et al. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene. 2007 Apr 23; doi: 10.1038/sj.onc.1210467. [DOI] [PubMed] [Google Scholar]
- 27.Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006 Apr 20;24(12):1924–31. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 28.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, et al. Medulloblastoma Comprises Four Distinct Molecular Variants. J Clin Oncol. 2010 doi: 10.1200/JCO.2009.27.4324. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, et al. Integrative Genomic Analysis of Medulloblastoma Identifies a Molecular Subgroup That Drives Poor Clinical Outcome. J Clin Oncol. 2010 Dec 6; doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Traiffort E, Charytoniuk D, Watroba L, Faure H, Sales N, Ruat M. Discrete localizations of hedgehog signalling components in the developing and adult rat nervous system. Eur J Neurosci. 1999;11(9):3199–214. doi: 10.1046/j.1460-9568.1999.00777.x. [DOI] [PubMed] [Google Scholar]
- 31.Fogarty MP, Emmenegger BA, Grasfeder LL, Oliver TG, Wechsler-Reya RJ. Fibroblast growth factor blocks Sonic hedgehog signaling in neuronal precursors and tumor cells. Proc Natl Acad Sci U S A. 2007 Feb 20;104(8):2973–8. doi: 10.1073/pnas.0605770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004 Nov 1;64(21):7794–800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- 33.Cheng SY, Bishop JM. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc Natl Acad Sci U S A. 2002;99(8):5442–7. doi: 10.1073/pnas.082096999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu P, Yu F, Feron F, Pickles JO, Sneesby K, Mackay-Sim A. Basic fibroblast growth factor and fibroblast growth factor receptors in adult olfactory epithelium. Brain Res. [Research Support, Non-U.S. Gov’t] 2001 Mar 30;896(1-2):188–97. doi: 10.1016/s0006-8993(01)02173-4. [DOI] [PubMed] [Google Scholar]
- 35.Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes & development. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] 2010 Apr 1;24(7):670–82. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lauth M, Bergstrom A, Toftgard R. Phorbol esters inhibit the Hedgehog signalling pathway downstream of Suppressor of Fused, but upstream of Gli. Oncogene. [Research Support, Non-U.S. Gov’t] 2007 Aug 2;26(35):5163–8. doi: 10.1038/sj.onc.1210321. [DOI] [PubMed] [Google Scholar]
- 37.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. [Research Support, N.I.H., Extramural Research Support, U.S. Gov’t, P.H.S. Review] 2005 Apr;16(2):139–49. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Lumpkin EA, Collisson T, Parab P, Omer-Abdalla A, Haeberle H, Chen P, et al. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr Patterns. 2003 Aug;3(4):389–95. doi: 10.1016/s1567-133x(03)00089-9. [DOI] [PubMed] [Google Scholar]
- 39.Lee A, Kessler JD, Read TA, Kaiser C, Corbeil D, Huttner WB, et al. Isolation of neural stem cells from the postnatal cerebellum. Nature neuroscience. 2005 Jun;8(6):723–9. doi: 10.1038/nn1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yaguchi Y, Yu T, Ahmed MU, Berry M, Mason I, Basson MA. Fibroblast growth factor (FGF) gene expression in the developing cerebellum suggests multiple roles for FGF signaling during cerebellar morphogenesis and development. Dev Dyn. 2009 Aug;238(8):2058–72. doi: 10.1002/dvdy.22013. [DOI] [PubMed] [Google Scholar]
- 41.Yamaguchi TP, Harpal K, Henkemeyer M, Rossant J. fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes Dev. 1994 Dec 15;8(24):3032–44. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- 42.Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc Natl Acad Sci U S A. 1998 Apr 28;95(9):5082–7. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu X, Qiao W, Li C, Deng CX. Generation of Fgfr1 conditional knockout mice. Genesis. 2002 Feb;32(2):85–6. doi: 10.1002/gene.10028.abs. [DOI] [PubMed] [Google Scholar]
- 44.Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, et al. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003 Jul;130(13):3063–74. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- 45.Yu C, Wang F, Jin C, Huang X, McKeehan WL. Independent repression of bile acid synthesis and activation of c-Jun N-terminal kinase (JNK) by activated hepatocyte fibroblast growth factor receptor 4 (FGFR4) and bile acids. J Biol Chem. 2005 May 6;280(18):17707–14. doi: 10.1074/jbc.M411771200. [DOI] [PubMed] [Google Scholar]
- 46.Olson DC, Deng C, Hanahan D. Fibroblast growth factor receptor 4, implicated in progression of islet cell carcinogenesis by its expression profile, does not contribute functionally. Cell Growth Differ. 1998 Jul;9(7):557–64. [PubMed] [Google Scholar]
- 47.Rios I, Alvarez-Rodriguez R, Marti E, Pons S. Bmp2 antagonizes sonic hedgehog-mediated proliferation of cerebellar granule neurones through Smad5 signalling. Development. 2004 Jul;131(13):3159–68. doi: 10.1242/dev.01188. [DOI] [PubMed] [Google Scholar]
- 48.Zhao H, Ayrault O, Zindy F, Kim JH, Roussel MF. Post-transcriptional down-regulation of Atoh1/Math1 by bone morphogenic proteins suppresses medulloblastoma development. Genes Dev. 2008 Mar 15;22(6):722–7. doi: 10.1101/gad.1636408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Read TA, Fogarty MP, Markant SL, McLendon RE, Wei Z, Ellison DW, et al. Identification of CD15 as a Marker for Tumor-Propagating Cells in a Mouse Model of Medulloblastoma. Cancer Cell. 2009;15(1):1–13. doi: 10.1016/j.ccr.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliver TG, Read TA, Kessler JD, Mehmeti A, Wells JF, Huynh TT, et al. Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medulloblastoma. Development. 2005 May;132(10):2425–39. doi: 10.1242/dev.01793. [DOI] [PubMed] [Google Scholar]
- 51.Mason JM, Morrison DJ, Basson MA, Licht JD. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 2006 Jan;16(1):45–54. doi: 10.1016/j.tcb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 52.Pons S, Trejo JL, Martinez-Morales JR, Marti E. Vitronectin regulates Sonic hedgehog activity during cerebellum development through CREB phosphorylation. Development. 2001;128(9):1481–92. doi: 10.1242/dev.128.9.1481. [DOI] [PubMed] [Google Scholar]
- 53.Nicot A, Lelievre V, Tam J, Waschek JA, DiCicco-Bloom E. Pituitary adenylate cyclase-activating polypeptide and sonic hedgehog interact to control cerebellar granule precursor cell proliferation. J Neurosci. 2002 Nov 1;22(21):9244–54. doi: 10.1523/JNEUROSCI.22-21-09244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004 Nov 18;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 55.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006 Dec 7;444(7120):756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 56.Ward RJ, Lee L, Graham K, Satkunendran T, Yoshikawa K, Ling E, et al. Multipotent CD15+ cancer stem cells in patched-1-deficient mouse medulloblastoma. Cancer Res. 2009 Jun 1;69(11):4682–90. doi: 10.1158/0008-5472.CAN-09-0342. [DOI] [PubMed] [Google Scholar]
- 57.Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. The New England journal of medicine. 2009 Sep 17;361(12):1173–8. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yauch RL, Dijkgraaf GJ, Alicke B, Januario T, Ahn CP, Holcomb T, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009 Oct 23;326(5952):572–4. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song BW, Vinters HV, Wu D, Pardridge WM. Enhanced neuroprotective effects of basic fibroblast growth factor in regional brain ischemia after conjugation to a blood-brain barrier delivery vector. The Journal of pharmacology and experimental therapeutics. 2002 May;301(2):605–10. doi: 10.1124/jpet.301.2.605. [DOI] [PubMed] [Google Scholar]
- 60.Debinski W, Tatter SB. Convection-enhanced delivery for the treatment of brain tumors. Expert Rev Neurother. 2009 Oct;9(10):1519–27. doi: 10.1586/ern.09.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams EJ, Williams G, Howell FV, Skaper SD, Walsh FS, Doherty P. Identification of an N-cadherin motif that can interact with the fibroblast growth factor receptor and is required for axonal growth. J Biol Chem. 2001;276(47):43879–86. doi: 10.1074/jbc.M105876200. [DOI] [PubMed] [Google Scholar]
- 62.Ballinger MD, Shyamala V, Forrest LD, Deuter-Reinhard M, Doyle LV, Wang JX, et al. Semirational design of a potent, artificial agonist of fibroblast growth factor receptors. Nat Biotechnol. 1999 Dec;17(12):1199–204. doi: 10.1038/70746. [DOI] [PubMed] [Google Scholar]
- 63.Li S, Christensen C, Kohler LB, Kiselyov VV, Berezin V, Bock E. Agonists of fibroblast growth factor receptor induce neurite outgrowth and survival of cerebellar granule neurons. Dev Neurobiol. [Research Support, Non-U.S. Gov’t] 2009 Nov;69(13):837–54. doi: 10.1002/dneu.20740. [DOI] [PubMed] [Google Scholar]
- 64.Kenigsberg RL, Hong Y, Yao H, Lemieux N, Michaud J, Tautu C, et al. Effects of basic fibroblast growth factor on the differentiation, growth, and viability of a new human medulloblastoma cell line (UM-MB1) Am J Pathol. 1997 Sep;151(3):867–81. [PMC free article] [PubMed] [Google Scholar]
- 65.Vachon P, Girard C, Theoret Y. Effects of basic fibroblastic growth factor on the growth of human medulloblastoma xenografts. J Neurooncol. 2004 Mar-Apr;67(1-2):139–46. doi: 10.1023/b:neon.0000021824.41701.e5. [DOI] [PubMed] [Google Scholar]
- 66.Weinstein M, Xu X, Ohyama K, Deng CX. FGFR-3 and FGFR-4 function cooperatively to direct alveogenesis in the murine lung. Development. 1998 Sep;125(18):3615–23. doi: 10.1242/dev.125.18.3615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. bFGF inhibits cell cycle progression. GNPs were isolated from control or FGFR1 KO mice at P7 and cultured in Neurobasal media with no supplements (Control), Shh or Shh + bFGF. Cell cycle analysis was performed after 48 hours. Shh promoted cell cycle progression in both WT and KO cells; but bFGF inhibited cell cycle progression only in WT cells.
Supplementary Figure 2. bFGF does not promote apoptosis of GNPs. Freshly isolated WT (A) and FGFR1 KO (B) GNPs were cultured for 24 hours in the presence or absence of 25 ng/mL bFGF, then fixed in 4% PFA and stained with antibodies against cleaved caspase-3 and DAPI. Confocal images of six randomly-selected fields were taken for each well. The percentage of DAPI+ cells positive for cleaved caspase-3 was determined. There was no significant change in the percentage of cleaved caspase-3 positive cells after bFGF treatment in either WT or FGFR1 KO GNPs (based on unpaired Student’s t-tests).
Supplementary Figure 3. (A) bFGF inhibits proliferation of tumor cells expressing activated Smoothened. Tumor cells from NeuroD2:SmoA1 transgenic mice were cultured on PDL-coated plates in control media or media containing 25ng/mL bFGF. After 48 hours, cells were pulsed with tritiated thymidine (3H-Td) and cultured for 16hr before being assayed for thymidine incorporation. Data represent means of triplicate samples ± SEM. Differences between control and bFGF-treated cells were statistically significant (p < 0.0001 based on unpaired Student’s t-test). (B) bFGF inhibits gli expression in WT and Sufu−/− MEFs. WT and Sufu−/− MEFs were cultured in the presence or absence of 25 ng/mL bFGF. After 24 hours, RNA was isolated and gli1 and actin were measured by RT-PCR. Data represent the ratio of gli1:actin, normalized to levels in Control WT MEFs. bFGF caused a three-fold reduction in expression of gli1 in Sufu−/− MEFs.
Supplementary Figure 4: FGFR1 is required for bFGF-mediated suppression of gli expression. GNPs were isolated from Math1-Cre; FGFR1F/F mice at P7 and cultured in control media or media containing Shh, Shh + 25 ng/ml bFGF, or Shh + 1 μM cyclopamine. After 24 hours, RNA was isolated and gli1 and actin levels were determined by real time RT-PCR. Data represent the ratio of gli1:actin, normalized to levels in Control WT GNPs.
Supplementary Figure 5. Impaired ERK phosphorylation in cells lacking FGF receptors. GNPs from FGFR TKO (Math1-Cre; Fgfr1F/F; Fgfr2F/F; Fgfr4−/−) mice and wild type littermates were treated with 25 ng/mL bFGF for 4 hours. Lysates were collected and analyzed by Western blotting for phospho-ERK (pERK) and total ERK (tERK). Increased ERK phosphorylation was observed in WT but not KO cells.
Supplementary Figure 6. Cells lacking FGFRs 1, 2 and 4 are not inhibited by bFGF. GNPs from FGFR TKO (Math1-Cre; Fgfr1F/F; Fgfr2F/F; Fgfr4−/−) mice and wild type littermates were cultured in either control (20% Mock supernatant) or Shh (20% Shh supernatant) media with or without 25ng/mL bFGF or 100ng/mL BMP2 for 48 hr. Cells were pulsed with 3H-Td and cultured for 16 hours before being assayed for thymidine incorporation. Data represent means of triplicate samples ± SEM. FGFR TKO GNPs proliferated in response to Shh; this proliferative response was not inhibited by bFGF, but was inhibited by BMP2. Differences in proliferation between Control and Shh, Shh and bFGF, and Shh and Shh+BMP were statistically significant (p < 0.0005 based unpaired Student’s t-tests) for both WT and TKO cells. Differences between Shh and Shh+bFGF were significant for WT but not for TKO cells.
Supplementary Figure 7. bFGF does not induce apoptosis of ptc+/− tumor cells. Tumor cells were cultured in the presence or absence of 25 ng/mL bFGF for 24 hr, then fixed in 4% PFA and stained for cleaved caspase-3 and DAPI. Confocal images of six randomly-selected fields were taken, and the percentage of DAPI+ cells positive for cleaved caspase-3 was determined. bFGF did not increase the ratio of cells positive for cleaved caspase-3. There was no significant change in the percentage of cleaved caspase-3 positive cells after bFGF treatment (based on unpaired Student’s t-test).
Supplementary Figure 8. H&E staining of PBS and bFGF-injected tumors. Cerebella from mice injected with ptc+/− medulloblastoma cells treated with PBS (A) or bFGF (B) were fixed, sectioned and stained with H&E. Tumor is denoted by asterisk in (A).