Abstract
The human CCCTC-binding factor, CTCF, regulates transcription of the double-stranded DNA genomes of herpesviruses. The architectural complex cohesin and RNA Polymerase II also contribute to this organization. We profiled the occupancy of CTCF, cohesin, and RNA Polymerase on the episomal genome of the Epstein-Barr virus in a cell culture model of latent infection. CTCF colocalizes with cohesin but not RNA Polymerase. CTCF and cohesin bind specific sequences throughout the genome that are found not just proximal to the regulatory elements of latent genes, but also near lytic genes. In addition to tracking with known transcripts, RNA Polymerase appears at two unannotated positions, one of which lies within the latent origin of replication. The widespread occupancy profile of each protein reveals binding near or at a myriad of regulatory elements and suggests context-dependent functions.
Keywords: CTCF, chromatin, transcription, Epstein-Barr virus, cohesin, RNA Polymerase II
Introduction
The human CCCTC-binding factor, CTCF, regulates gene expression by organizing DNA in the three-dimensional space of the nucleus. CTCF binds tens of thousands of sites throughout the human genome (Barski et al., 2007; Kim et al., 2007) and is necessary for the assembly of chromatin loops that bring together distal DNA to regulate transcription (Hou et al., 2010; Majumder and Boss, 2010; Majumder et al., 2008; Mishiro et al., 2009). Globally, the human genome is organized by long-range interactions (Lieberman-Aiden et al., 2009) and binding sites are significantly enriched at these nodes (Botta et al., 2010). CTCF also organizes the double-stranded DNA genomes of herpesviruses. Binding sites have been identified on the Epstein-Barr virus (EBV) (Chau et al., 2006; Day et al., 2007; Tempera et al., 2010), herpes simplex virus 1 (Chen et al., 2007), and Kaposi’s sarcoma-associated herpesvirus (KSHV) (Stedman et al., 2008). Functional studies with EBV have focused on binding sites near the major promoters of latency and suggested possible roles for CTCF in the repression, activation, and insulation of latent transcripts (Chau et al., 2006; Day et al., 2007; Tempera et al., 2010). CTCF occupancy upstream of the C promoter arguably correlates with repression of EBNA2 transcription (Chau et al., 2006; Salamon et al., 2009). Mutation of a CTCF-binding site upstream of the Q promoter resulted in loss of EBNA1 transcription accompanied by the spread of repressive histone marks and CpG methylation (Tempera et al., 2010). Based on a myriad of functions, CTCF has emerged as a central component of transcriptional regulation in human and viral genomes.
CTCF may form complexes with diverse binding partners that include transcription factors, histone modifiers, and a variety of chromatin regulators (Wallace and Felsenfeld, 2007; Zlatanova and Caiafa, 2009). The CTCF protein consists of eleven zinc fingers (Filippova et al., 1996) and two flanking unstructured termini (Martinez and Miranda, 2010); both types of protein segments are capable of recruiting cofactors directly by molecular recognition (Dunker et al., 2002; Gamsjaeger et al., 2007). Specifically, the architectural complex cohesin and RNA Polymerase II (RNAP II) may play functional roles in CTCF-dependent transcriptional control. CTCF is necessary for cohesin positioning and colocalization throughout the human, mouse, and KSHV genomes (Parelho et al., 2008; Stedman et al., 2008; Wendt et al., 2008). Depletion of cohesin perturbs gene expression by the disruption of DNA looping and long-range interactions between transcriptional control elements (Hadjur et al., 2009; Mishiro et al., 2009; Nativio et al., 2009). RNAP II is thought to interact with CTCF directly through protein-protein interactions, resulting in colocalization at a subset of CTCF-binding sites (Chernukhin et al., 2007). Stalling of RNAP II tends to occur at sites of CTCF and cohesin colocalization (Wada et al., 2009), and a large fraction of CTCF sites are found within actively transcribed genes (Barski et al., 2007). The diversity and genome-wide distribution of the myriad of possible CTCF assemblies, given significant functional implications, remains actively investigated.
We initially hoped to use profiling of protein occupancy to determine the composition of CTCF complexes by identifying which proteins bind the same DNA at high resolution. Colocalization would strongly suggest molecular assembly and cooperation, perhaps characterizing the structural and functional heterogeneity or uniformity of complexes genome-wide. Indeed, we were able to do so and determined that CTCF colocalizes with cohesin but not RNAP II. In the midst of identifying binding sites, however, we also uncovered protein occupancy in unexpected regions of the EBV genome. We will discuss the functional implications of CTCF and cohesin colocalization as well as RNAP II binding outside regions of known transcription.
Results
CTCF and cohesin colocalization
CTCF binds specific sites throughout the latent EBV genome. We identified fifteen sites of CTCF occupancy at high resolution across the entire unique sequence of the EBV genome using chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) (Fig. 1, Table 1). An overrepresented sequence motif, similar to that found in the human genome (Kim et al., 2007), is detected in every region of occupancy (data not shown). During the life cycle of EBV, only a handful of regulatory elements control expression of a few latent genes, but many more genes are transcribed upon lytic activation (Kieff and Rickinson, 2007) We find CTCF-binding sites proximal to many latent regulatory elements. Occupancy is observed near the C, W, and Q promoters that drive messages encoding for the EBNA proteins. Enrichment is also detected close to start sites of the EBER-1 ncRNA and RPSM1 BART transcript, as well as the LMP-2A and LMP-1 messages. We also find, however, CTCF-binding sites proximal to many lytic genes. For example, occupancy is detected near the promoter of BZLF1, an immediate early transactivator critical for the switch to the productive cycle (Biggin et al., 1987). Both transcripts expressed during latency as well as genes activated during the lytic cycle can be found proximal to CTCF-binding sites.
Fig. 1.
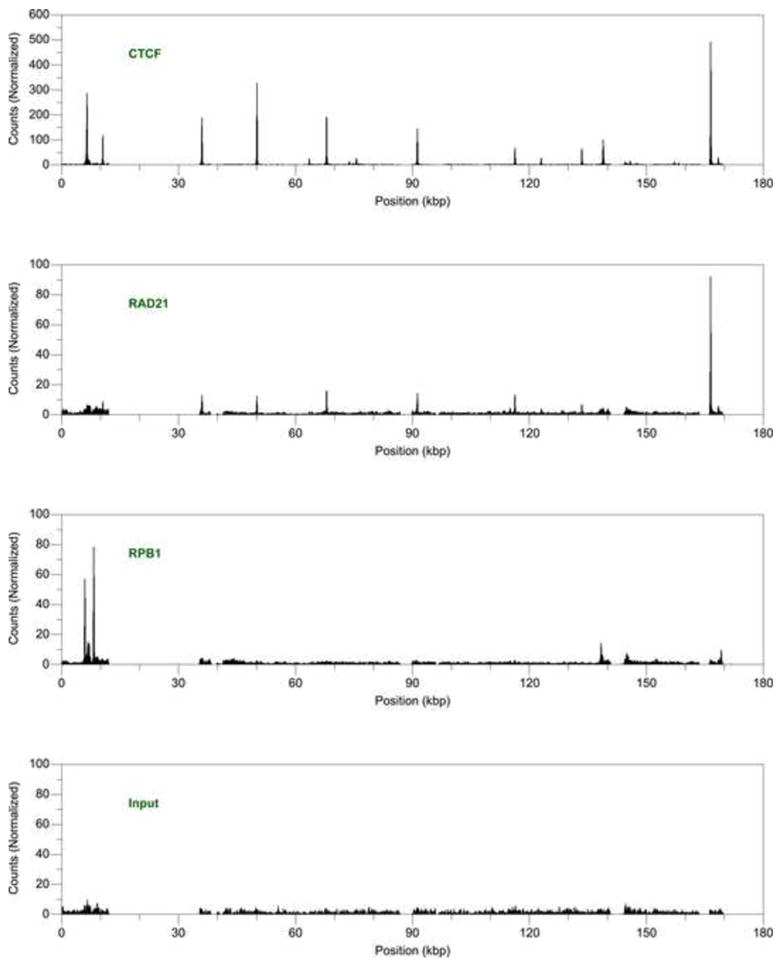
Occupancy of CTCF, cohesin, and RNA Polymerase II in the Epstein-Barr virus genome. Chromatin immunoprecipitation profiles of (A) CTCF, (B) the RAD21 subunit of cohesin, and (C) the RPB1 subunit of RNA Polymerase II in the latent Epstein-Barr virus genome of Raji cells. The profile of (D) input DNA serves as a control reference. Occupancy is calculated as enrichment over the background baseline.
Table 1.
CTCF and cohesin occupancy of the Raji EBV genome.
| CTCF-binding sites | RAD21-binding sites | Nearby transcriptional regulatory elementa |
|---|---|---|
| 6411–6580 | EBER-1 (pol III transcript) [misc RNA] | |
| 10483–10633 | C promoter (Cp) used in latency III [TATA signal] | |
| 35899–36054 | 35902–36081 | EBNA-LP, W promoter (Wp) used in earlier stages of latent infection [TATA signal] |
| 50019–50166 | 50010–50164 | EBNA-1, QUK transcript, starts from the Q promoter (Qp) in latency I [mRNA] |
| 63435–63584 | BOLF1 [TATA signal] | |
| 67872–68022 | 67892–68041 | BMRF1 [TATA signal] |
| 75554–75705 | BLRF1 [TATA signal] | |
| 91180–91331 | 91189–91339 | BZLF1 [TATA signal] |
| 116241–116386 | 116213–116378 | BGLF1 [TATA signal] |
| 122920–123090 | BDLF1 [TATA signal] | |
| 133362–133528 | BVRF1 [TATA signal] | |
| 138827–139018 | RPMS1, BART mRNAs [mRNA] | |
| 145778–145959 | LF3 [TATA signal] | |
| 166435–166588 | 166416–166570 | LMP-2A [TATA signal] |
| 168369–168541 | LMP-1 [TATA signal] |
From the human herpesvirus 4 complete genome sequence (Genbank ID: NC_007605.1).
Cohesin colocalizes with CTCF throughout the latent EBV genome. We measured the occupancy of the kleisin subunit RAD21 and identified six binding sites for cohesin dispersed throughout the EBV genome (Fig. 1, Table 1). All of these cohesin-binding sites overlap with CTCF-binding sites at high resolution; no CTCF-independent sites of cohesin occupancy are found. We did not detect RAD21 enrichment at all sites of CTCF occupancy, presumbly due to weaker efficiency of immunoprecipitation with the RAD21 antibody compared to the CTCF antibody. Similar to the distribution of CTCF-binding sites, the overlapping cohesin-binding sites are found proximal to regulatory elements of both latent and lytic genes.
RNA Polymerase II occupancy
RNAP II does not colocalize with CTCF, instead tracking with regions of known transcription as well as appearing highly enriched at unannotated positions. We measured RNAP II occupancy and identified four significant regions of binding (Fig. 1, Table 2). The particular antibody used in our experiments, 4H8, was raised against an epitope containing phosphorylated serine 5 and therefore detects occupancy both at promoters and throughout a transcribed gene (Brodsky et al., 2005; Stock et al., 2007). Binding is observed at ~ 6.5–6.9 and ~ 6.9–7.1 kbp, regions corresponding to the two EBER transcripts that span ~ 6.6–6.8 and ~ 7.0–7.1 kbp (Rosa et al., 1981). The observed peaks are distributed across the entire gene, a pattern consistent with actively elongating transcription. Although the signals did not rise above our significance threshold, we also detected some RNAP II enrichment near other known transcription units. Occupancy at ~ 138.3–138.5 kbp corresponds to the initiation site of the RPSM1 BART transcript, and the binding at ~ 169.1–169.3 kbp occurs near the promoters of the LMP genes (Fig. 1). In addition to sites of active transcription, we observed the strongest levels of enrichment at two novel positions (Fig. 1, Table 2). Recruitment occurs ~ 1 kbp upstream of the EBER-1 transcript at ~ 5.9–6.0 kbp. Occupancy is also strong at ~ 8.1–8.3 kbp, inside oriP, the proposed latent cycle origin of replication (Yates et al., 1984), at a unique sequence adjacent to FR, repeats that function in replication (Reisman et al., 1985) and transcriptional activation (Gahn and Sugden, 1995; Reisman and Sugden, 1986). Although we detected multiple RNAP II-binding sites, both at known transcription units and unannotated positions, none of these sites colocalized with CTCF.
Table 2.
RNA Polymerase II occupancy of the Raji EBV genome.
| RPB1-binding sites | Nearby transcriptional regulatory elementa |
|---|---|
| 5878–6042 | EBER-1 (pol III transcript) [misc RNA] |
| 6549–6865 | EBER-1 (pol III transcript) [misc RNA] |
| 6895–7142 | EBER-2 (pol III transcript) [misc RNA] |
| 8146–8295 | region containing oriP, the latent cycle origin of DNA replication [rep origin] |
From the human herpesvirus 4 complete genome sequence (Genbank ID: NC_007605.1).
Discussion
High-resolution mapping of protein positions with ChIP-seq on small viral episomes yields advantages over similar experiments in larger genomes: technical gains allow confident data analysis and verified annotations help generate functional hypotheses of protein function. The high copy number of episomes, ~ 45 in the case of EBV-infected Raji cells (Sternas et al., 1990), increases the signal-to-noise ratio of ChIP experiments because of deeper sequencing compared to the simultaneously examined human genome. Multifold coverage of the background consequently allowed us to statistically define an experimental baseline for experiments. This simplified peak calling to choosing a suitable enrichment over background; our conservative threshold of a 10-fold enrichment yielded an ostensible false discovery rate of 0 and strong confidence in our identification of binding sites. Viral genomes have also been well annotated and subjected to functional mutagenesis experiments. Once protein-binding sites have been determined at high resolution, we can correlate occupancy with known and tested sequence elements. These advantages yield reliable distributions of occupancy that allow us to make functional inferences based on protein positions.
Widespread distribution of CTCF and cohesin on the EBV genome reveals binding at or near both latent and lytic genes, suggesting distinct regulatory functions at different positions. Focus on CTCF function in EBV has been limited to binding sites within the latency control region, and consequently potential roles for CTCF have only involved the determination of latent promoter choice and establishment of boundaries between latent and lytic genes (Chau et al., 2006; Day et al., 2007; Tempera et al., 2010). Our experiments identified CTCF-binding sites widely dispersed on the genome, including in and around many lytic genes. We also observed colocalization with cohesin, similar to what is observed in the human and KSHV genomes (Parelho et al., 2008; Stedman et al., 2008; Wendt et al., 2008), that is likewise not limited to areas of latent transcription initiation. Because of the widespread occupancy distributions, we surmise that CTCF and cohesin may be performing functions near lytic genes separate from those previously suggested in latency control regions.
RNAP II does not colocalize with CTCF, and RNAP II positioning suggests functions separate from the transcription of latent genes. We find RNAP II at the EBER transcripts, short non-coding RNA with expression predominantly driven by RNA Polymerase III but also partially transcribed by RNAP II (Kirchner et al., 1991). We also find RNAP II at two novel positions, sites that yield the strongest enrichment of occupancy in our experiments yet do not map to any annotated transcripts. The strongest signal maps to the unique sequence adjacent to FR within OriP, the proposed origin of replication for latency. The second strongest RNAP II signal maps upstream of the EBER-1 transcript and does not appear to correspond to an annotated element. We were unable, however, to find CTCF occupancy at any of these RNAP II-binding sites. We also could not detect RNAP II and cohesin colocalization as observed in the human genome (Kagey et al., 2010). RNAP II, like CTCF and cohesin, binds near different regulatory elements throughout the genome, thus suggesting functions ranging from transcription to replication.
RNAP II occupancy detects a subset of known transcription units in the EBV-infected Raji cells, but CTCF proximity correlates with both active and repressed promoters. The two EBER RNAs are actively transcribed (Lerner et al., 1981) and strong RNAP II enrichment is observed. A complex set of alternatively spliced mRNAs, referred to as BARTs, are generated from the rightward transcription of the BamHI A fragment (Chen et al., 1992; Sadler and Raab-Traub, 1995). Different initiation sites have been proposed, but we detect a weak signal in only one promoter (Smith et al., 1993). Similarly, conflicting evidence exists regarding the identity of the transcription start site (Tao et al., 1998; Woisetschlaeger et al., 1990) for the set of alternatively spliced mRNAs that yield the EBNA genes (Allday et al., 1988; Petti et al., 1988; Wang et al., 1987). Although the W promoter is obscured from our analysis by repeat regions, we find RNAP II at neither the C nor Q promoters. This cell line also expresses the LMP1 gene (Contreras-Brodin et al., 1991), as well as LMP2A but not LMP2B (Sample et al., 1989). Although weak RNAP II enrichment is observed in the general promoter region for these cells, the peak does not correlate to an annotated element. Combining our RNAP II localization with literature describing known transcription in Raji cells gives us the opportunity to compare CTCF occupancy with polymerase activity. Again, however, neither activity nor putative repression of promoters correlates with CTCF proximity. We interpret CTCF occupancy near both active and repressed promoters as another suggestion of context-dependent gene regulation.
Combining confident, high resolution mapping of protein positions by ChIP-seq and the deep annotation of viral genomes allows us to make specific and testable inferences. What functions can we hypothesize for CTCF, cohesion, and RNAP II? In addition to operating within the latency control region, CTCF and cohesin colocalization near lytic genes may control those regulatory elements, for example by repressing lytic transcription. In addition to generating latent transcripts, occupancy at the origin may couple RNAP II recruitment with replication. CTCF, cohesin, and RNAP II all share the common potential of context-dependent functions throughout the EBV genome.
Materials and methods
Cell culture and antibodies
EBV genome-positive Raji cells (Pulvertaft, 1964) (ATCC CCL-86) were maintained in RPMI-1640 media containing 25 mM HEPES and supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) β-mercaptoethanol. Antibodies used were anti-CTCF (Upstate, 07–729), anti-Rad21 (Bethyl Labs, A300-080A), and 4H8 (Abcam, ab5408).
Chromatin immunoprecipitation followed by deep sequencing
1 × 107 Raji cells were used per ChIP-seq experiment. Crosslinking was performed by adding formaldehyde to 1% (v/v) at room temperature for 3, 10, and 5 min for CTCF, Rad21, and RNAP II immunoprecipitations, respectively. The reaction was quenched by incubation with 125 mM glycine for 10 min at 4 °C. Cells were pelleted at 1,000g for 5 min at room temperature and resuspended in 50 mM HEPES-KOH, 1 mM EDTA, 150 mM NaCl, 10% (v/v) glycerol, 0.5% (v/v) Triton X-100, Complete Protease Inhibitor Cocktail Tablets (Roche), pH 7.4, for 30 min on ice. Crude nuclei were collected by centrifugation at 600g for 5 min and lysed in 2 ml of 10 mM Tris-HCl, 1 mM EDTA, 150 mM NaCl, 5% (v/v) glycerol, 0.1% (w/v) sodium deoxycholate, 0.1%(w/v) SDS, 1% (v/v) Triton X-100, Complete Protease Inhibitor Cocktail Tablets (Roche), pH 8.0. DNA was sheared with a Bioruptor water bath sonicator (Diagenode) to produce fragments ~ 100–200 bp in size. A fraction of the sample was set aside as a genomic DNA input control and crosslinking was reversed in 10 mM Tris, 1 mM EDTA, 0.7% (w/v) SDS, 200 mg/ml proteinase K, pH 8.0, at 55 °C for 3 hrs followed by incubation at 65 °C overnight. DNA-protein complexes were purified using 10 μg epitope-specific antibodies and 100 μl protein G Dynabeads (Invitrogen) for 4 hrs at 4 °C and followed by consecutive washes, first in 10 mM Tris-HCl, 1 mM EDTA, 150 mM NaCl, 5% (v/v) glycerol, 0.1% (w/v) sodium deoxycholate, 0.1%(w/v) SDS, 1% (v/v) Triton X-100, Complete Protease Inhibitor Cocktail Tablets (Roche), pH 8.0, second in the same buffer with 500 mM NaCl, and last in 20 mM Tris-HCl, 1 mM EDTA, 250 mM LiCl, 0.5% (v/v) NP-40, 0.5% (w/v) sodium deoxycholate, Complete Protease Inhibitor Cocktail Tablets (Roche), pH 8.0. Complexes were eluted from the beads with 10 mM Tris, 1 mM EDTA, 0.7% (w/v) SDS, pH 8.0, and crosslinking was reversed as with the input control DNA.
ChIP-seq libraries were prepared by adaptor-mediated amplification (Robertson et al., 2007) with a few modifications. Both ChIP DNA and 10 ng of input DNA were repaired and phosphorylated with T4 DNA polymerase, Klenow polymerase, and T4 polynucleotide kinase. Double-stranded oligonucleotides based on genomic adapters (Illumina) were synthesized such that the product corresponded to the two adapters joined together at blunt ends and separated by deoxyuridine. After the addition of a 3′ adenine to the repaired DNA, samples were ligated overnight to the modified adapters in a 2:1 adapter:sample ratio. Ligation reactions were digested with uracil-DNA glycosylase for 30 min at 37 °C. Amplification of the DNA libraries was performed using PCR primers 1.1 and 1.2 (Illumina) for 18 cycles. The resulting PCR products were purified using Ampure beads (Agencourt). Size fractionation and purity of the finished ChIP-seq DNA libraries was monitored with a DNA 1000 Assay (Agilent). DNA concentrations were quantified using a combination of Quant-iT PicoGreen (Invitrogen) and UV absorbance to obtain a diluted final concentration of 10 nM. Clusters were generated on a Cluster Station (Illumina) and 36 cycles of sequencing performed with a Genome Analyzer II (Illumina). All ChIP-seq experiments were performed with independent replicates.
Sequence mapping and analysis
After ignoring the first basecall, 32 bp sequence fragments were obtained and mapped to the EBV reference genome (Genbank ID: NC_007605.1) using the Pipeline software (Illumina) allowing for up to 2 mismatches. Unique sequence tags were extracted and merged for each duplicate experiment. For the input, CTCF, cohesin, and RNAP II experiments, the combined data sets consisted of 3,164 plus 1,674, 11,937 plus 41,829, 4,474 plus 13,965, and 11,336 plus 7,292 sequences, respectively. Data was further processed with scripts written in AWK. Corresponding to the original DNA fragment size loaded onto the sequencer, the 32 bp tags were extended directionally by 100 bp and the number of hits per genome position counted. Each data set was then normalized to the internal baseline. Hits corresponding to deleted and repeat regions of the genome were first masked. The unmasked values were then sorted by count number and the mean and median values calculated. This set was culled in 1% increments, removing the highest values, until the calculated mean equaled less than the median. This process simulates removal of peaks from a data set until only a background remains. The last calculated mean is thus equivalent to the average number of hits within the background. All sequence hit counts were then divided by this value. This normalization results in expression of the reported peak heights relative to background. The selection criteria used for identifying a binding site were that each peak contain an approximately equal proportion of forward and reverse reads and that the maximum of each peak have a minimum ten-fold enrichment over background in both replicate experiments. Peaks coordinates are expressed as the full width at half maximum height.
Acknowledgments
We thank Clement S. Chu, the UCSF Center for Advanced Technology, and Hiten D. Madhani for access to instruments needed in deep sequencing experiments. Our research is supported by an institutional grant to JJ L. Miranda from the UCSF Fellows Program, which is funded in part by the UCSF Program for Breakthrough Biomedical Research and the Sandler Foundation. Samantha B. Cooper and Keith R. Yamamoto are supported by National Institutes of Health grant CA020535 to Keith R. Yamamoto.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allday MJ, Crawford DH, Griffin BE. Prediction and demonstration of a novel Epstein-Barr virus nuclear antigen. Nucleic Acids Res. 1988;16:4353–4367. doi: 10.1093/nar/16.10.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Biggin M, Bodescot M, Perricaudet M, Farrell P. Epstein-Barr virus gene expression in P3HR1-superinfected Raji cells. J Virol. 1987;61:3120–3132. doi: 10.1128/jvi.61.10.3120-3132.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta M, Haider S, Leung IX, Lio P, Mozziconacci J. Intra- and inter-chromosomal interactions correlate with CTCF binding genome wide. Mol Syst Biol. 2010;6:426. doi: 10.1038/msb.2010.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky AS, Meyer CA, Swinburne IA, Hall G, Keenan BJ, Liu XS, Fox EA, Silver PA. Genomic mapping of RNA polymerase II reveals sites of co-transcriptional regulation in human cells. Genome Biol. 2005;6:R64. doi: 10.1186/gb-2005-6-8-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau CM, Zhang XY, McMahon SB, Lieberman PM. Regulation of Epstein-Barr virus latency type by the chromatin boundary factor CTCF. J Virol. 2006;80:5723–5732. doi: 10.1128/JVI.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HL, Lung MM, Sham JS, Choy DT, Griffin BE, Ng MH. Transcription of BamHI-A region of the EBV genome in NPC tissues and B cells. Virology. 1992;191:193–201. doi: 10.1016/0042-6822(92)90181-n. [DOI] [PubMed] [Google Scholar]
- Chen Q, Lin L, Smith S, Huang J, Berger SL, Zhou J. CTCF-dependent chromatin boundary element between the latency-associated transcript and ICP0 promoters in the herpes simplex virus type 1 genome. J Virol. 2007;81:5192–5201. doi: 10.1128/JVI.02447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernukhin I, Shamsuddin S, Kang SY, Bergstrom R, Kwon YW, Yu W, Whitehead J, Mukhopadhyay R, Docquier F, Farrar D, Morrison I, Vigneron M, Wu SY, Chiang CM, Loukinov D, Lobanenkov V, Ohlsson R, Klenova E. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol. 2007;27:1631–1648. doi: 10.1128/MCB.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Brodin BA, Anvret M, Imreh S, Altiok E, Klein G, Masucci MG. B cell phenotype-dependent expression of the Epstein-Barr virus nuclear antigens EBNA-2 to EBNA-6: studies with somatic cell hybrids. J Gen Virol. 1991;72 ( Pt 12):3025–3033. doi: 10.1099/0022-1317-72-12-3025. [DOI] [PubMed] [Google Scholar]
- Day L, Chau CM, Nebozhyn M, Rennekamp AJ, Showe M, Lieberman PM. Chromatin profiling of Epstein-Barr virus latency control region. J Virol. 2007;81:6389–6401. doi: 10.1128/JVI.02172-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahn TA, Sugden B. An EBNA-1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J Virol. 1995;69:2633–2636. doi: 10.1128/jvi.69.4.2633-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci. 2007;32:63–70. doi: 10.1016/j.tibs.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci U S A. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff ED, Rickinson AB. Epstein-Barr Virus and Its Replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 2603–2654. [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner EA, Bornkamm GW, Polack A. Transcriptional activity across the Epstein-Barr virus genome in Raji cells during latency and after induction of an abortive lytic cycle. J Gen Virol. 1991;72:2391–2398. doi: 10.1099/0022-1317-72-10-2391. [DOI] [PubMed] [Google Scholar]
- Lerner MR, Andrews NC, Miller G, Steitz JA. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A. 1981;78:805–809. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder P, Boss JM. CTCF controls expression and chromatin architecture of the human major histocompatibility complex class II locus. Mol Cell Biol. 2010;30:4211–4223. doi: 10.1128/MCB.00327-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder P, Gomez JA, Chadwick BP, Boss JM. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J Exp Med. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez SR, Miranda JL. CTCF terminal segments are unstructured. Protein Sci. 2010;19:1110–1116. doi: 10.1002/pro.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28:1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2–H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Petti L, Sample J, Wang F, Kieff E. A fifth Epstein-Barr virus nuclear protein (EBNA3C) is expressed in latently infected growth-transformed lymphocytes. J Virol. 1988;62:1330–1338. doi: 10.1128/jvi.62.4.1330-1338.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvertaft JV. Cytology of Burkitt’s Tumour (African Lymphoma) Lancet. 1964;1:238–240. doi: 10.1016/s0140-6736(64)92345-1. [DOI] [PubMed] [Google Scholar]
- Reisman D, Sugden B. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol Cell Biol. 1986;6:3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith OL, He A, Marra M, Snyder M, Jones S. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Rosa MD, Gottlieb E, Lerner MR, Steitz JA. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol Cell Biol. 1981;1:785–796. doi: 10.1128/mcb.1.9.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler RH, Raab-Traub N. Structural analyses of the Epstein-Barr virus BamHI A transcripts. J Virol. 1995;69:1132–1141. doi: 10.1128/jvi.69.2.1132-1141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamon D, Banati F, Koroknai A, Ravasz M, Szenthe K, Bathori Z, Bakos A, Niller HH, Wolf H, Minarovits J. Binding of CCCTC-binding factor in vivo to the region located between Rep* and the C promoter of Epstein-Barr virus is unaffected by CpG methylation and does not correlate with Cp activity. J Gen Virol. 2009;90:1183–1189. doi: 10.1099/vir.0.007344-0. [DOI] [PubMed] [Google Scholar]
- Sample J, Liebowitz D, Kieff E. Two related Epstein-Barr virus membrane proteins are encoded by separate genes. J Virol. 1989;63:933–937. doi: 10.1128/jvi.63.2.933-937.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PR, Gao Y, Karran L, Jones MD, Snudden D, Griffin BE. Complex nature of the major viral polyadenylated transcripts in Epstein-Barr virus-associated tumors. J Virol. 1993;67:3217–3225. doi: 10.1128/jvi.67.6.3217-3225.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternas L, Middleton T, Sugden B. The average number of molecules of Epstein-Barr nuclear antigen 1 per cell does not correlate with the average number of Epstein-Barr virus (EBV) DNA molecules per cell among different clones of EBV-immortalized cells. J Virol. 1990;64:2407–2410. doi: 10.1128/jvi.64.5.2407-2410.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Tao Q, Robertson KD, Manns A, Hildesheim A, Ambinder RF. The Epstein-Barr virus major latent promoter Qp is constitutively active, hypomethylated, and methylation sensitive. J Virol. 1998;72:7075–7083. doi: 10.1128/jvi.72.9.7075-7083.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempera I, Wiedmer A, Dheekollu J, Lieberman PM. CTCF prevents the epigenetic drift of EBV latency promoter Qp. PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Ohta Y, Xu M, Tsutsumi S, Minami T, Inoue K, Komura D, Kitakami J, Oshida N, Papantonis A, Izumi A, Kobayashi M, Meguro H, Kanki Y, Mimura I, Yamamoto K, Mataki C, Hamakubo T, Shirahige K, Aburatani H, Kimura H, Kodama T, Cook PR, Ihara S. A wave of nascent transcription on activated human genes. Proc Natl Acad Sci U S A. 2009;106:18357–18361. doi: 10.1073/pnas.0902573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Petti L, Braun D, Seung S, Kieff E. A bicistronic Epstein-Barr virus mRNA encodes two nuclear proteins in latently infected, growth-transformed lymphocytes. J Virol. 1987;61:945–954. doi: 10.1128/jvi.61.4.945-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- Woisetschlaeger M, Yandava CN, Furmanski LA, Strominger JL, Speck SH. Promoter switching in Epstein-Barr virus during the initial stages of infection of B lymphocytes. Proc Natl Acad Sci U S A. 1990;87:1725–1729. doi: 10.1073/pnas.87.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci U S A. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J, Caiafa P. CTCF and its protein partners: divide and rule? J Cell Sci. 2009;122:1275–1284. doi: 10.1242/jcs.039990. [DOI] [PubMed] [Google Scholar]


