Abstract
Safe and effective vaccines offer the best intervention for disease control. One strategy to maximize vaccine immunogenicity without compromising safety is to rationally design molecular complexes that mimic the natural structure of immunogenic microbes, but without the disease-causing components. Here we use highly programmable DNA nanostructures as platforms to assemble a model antigen and CpG adjuvants together into nanoscale complexes with precise control of the valency and spatial arrangement of each element. Our results from immunized mice show that compared to a mixture of antigen and CpG molecules, the assembled antigen-adjuvant-DNA complexes induce strong and long-lasting antibody responses against the antigen without stimulating a reaction to the DNA nanostructure itself. This result demonstrates the potential of DNA nanostructures to serve as general platforms for the rational design and construction of a variety of vaccines.
Keywords: DNA nanostructure, synthetic vaccine, mouse immunization
The goal of developing safer and more effective vaccines has been a priority since human beings began fighting disease through vaccination over 1000 years ago1. Many of the vaccines that are currently administered were derived from live attenuated organisms, killed whole organisms, or subunit vaccines1,2. Although live vaccines have the advantage of inducing a strong immune response, there is a risk that the attenuated organism will revert back to a virulent form, which is detrimental to the public health. Killed or inactivated whole organisms and subunit vaccines do not pose the same serious health risk; however, they tend to induce weaker or ineffective immune responses and often require multiple doses for enhanced efficacy2. Recombinant DNA technology has facilitated the assembly of subunit proteins into virus-like particles (VLPs)3,4 that resemble the structure of natural viruses but without containing their genetic materials, representing a major breakthrough in vaccine development. Immunogenic epitopes displayed from the VLPs were shown to induce a strong immune response and thus, VLPs have been extensively explored as an effective and safe platform to assemble the epitopes of interest against many pathogens and tumor cells 3,5. However, sometimes, it is challenging to incorporate antigenic epitopes into VLPs at defined positions and configurations because of the inherent uncertainties in engineering epitope-VLP fusion proteins4.
Alternatively, nanotechnology provides researchers with a robust platform for the assembly of subunit vaccines. In particular, biodegradable polymers such as poly (D,L-lactide-co-glycolide) (PLGA)2 have been used to encapsulate vaccine antigens and adjuvants. These subunit vaccines have been shown to increase antigen delivery and antigen presenting cell (APC) targeting, thereby enhancing the immunogenicity of the antigen6,7. Today, DNA nanotechnology is recognized as a highly programmable and robust way to self-assemble heterogeneous nanostructures. A variety of different two- and three- dimensional DNA nanostructures8–14 have been constructed and used for precisely organizing biochemical molecules15–18 and targeted cellular transport and delivery19–22. Gaining control over structural features such as particle size and shape, epitope valency, and configuration is highly desirable and long sought after in vaccine development and DNA nanostructures present an opportunity to exert such control. Several research groups have assembled multiple adjuvant elements on a DNA nanostructure and found increased immunostimulation in vitro23,24 and ex vivo25. Here we provide the first evidence that antigens and adjuvants assembled by DNA nanostructures induce strong antibody responses in vivo, highlighting the potential of DNA-nanostructures to serve as new platforms for vaccine construction.
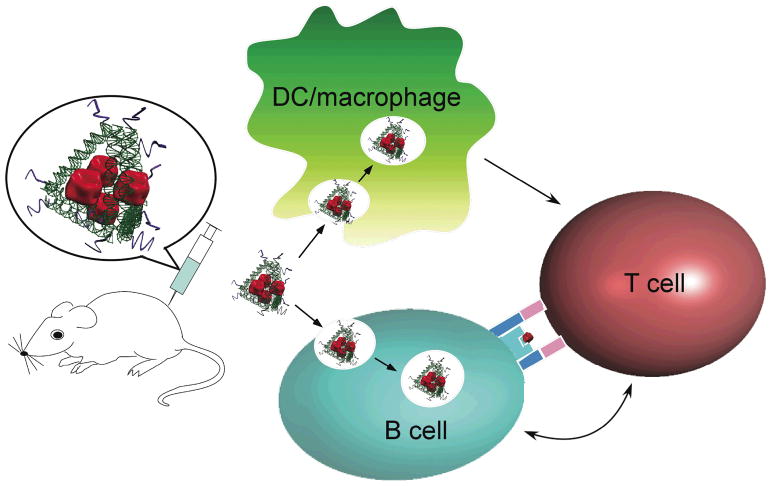
We used a tetrahedral DNA nanostructure26–28 as a scaffold to assemble a model antigen, streptavidin (STV), and a representative adjuvant, CpG oligo-deoxynucleotides (ODN)29 (mouse-specific ODN-1826), into a synthetic vaccine complex (Figure 1). This vaccine complex resembles a natural viral particle in both size and shape27,30, where the STV and CpG ODN elements are located at particular positions (Figures 1 and Figure S1). The complex was tested both in vitro and in vivo for its immunogenicity, particularly its ability to elicit an antibody response against the model antigen, STV.
Figure 1.
Schematic design of the DNA scaffolded adjuvant-antigen vaccine complex. The CpG ODN adjuvant molecules (Supplementary information and Figure S1) are depicted as curved purple ribbons in the model. The model antigen (streptavidin) is shown in red and the tetrahedral DNA scaffold is represented by green helices. The injected vaccine complexes bind specifically to B cells and non-specifically to dendritic cells and macrophages. The complexes are internalized by the three types of antigen-presenting cells, disassembled, and the individual peptide antigens are subsequently presented to T cells to activate B cell response and antibody production.
Targeted delivery of the antigen to antigen presenting cells (APC), including macrophages, dendritic cells (DCs) and B cells, is a vital first step in initiating an effective immune response. Previous studies have shown that the size, shape, surface charge, hydrophobicity, hydrophilicity, and receptor interactions of an antigen can influence its uptake by APCs30. After internalization, the targets are processed and presented to T cells for T cell activation. It has been demonstrated that co-localization of antigens and adjuvants within the same APCs can augment antigen presentation and T cell activation6. Finally, activated T cells assist in the differentiation of antigen-specific B cells and the production of the antibodies that are specific to the target antigen, as illustrated in Figure 1. Given the recent report that DNA nanostructures increase the amount of CpG adjuvant molecules that are internalized by APCs24,25, we speculated that DNA nanostructures would also increase the amount of antigen taken by APCs, thereby promoting co-delivery of the antigen and CpG to the same APC population.
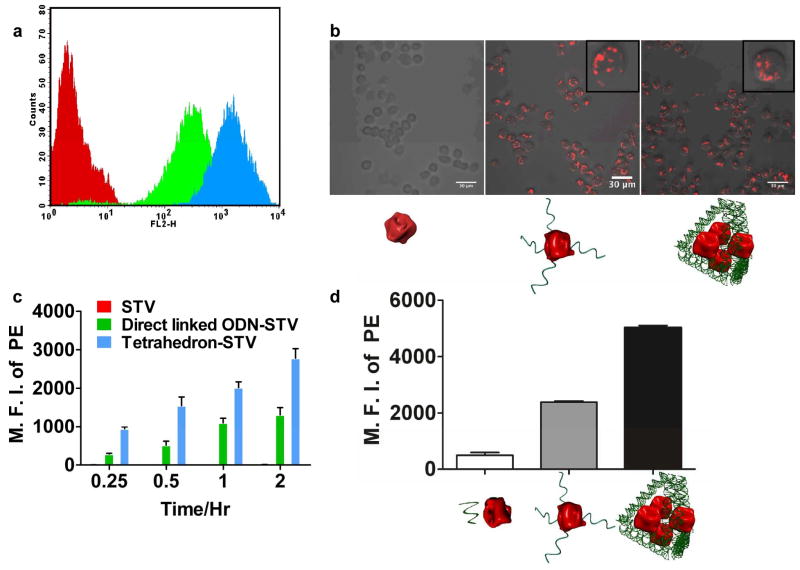
To test this hypothesis we loaded fluorescently labeled model antigen, phycoerythrin conjugated streptavidin (PE-STV), onto the DNA tetrahedron and used flow cytometry to track the internalization of the complex in a mouse macrophage-like cell line (RAW 264.7). As shown in Figures 2a and 2c, internalization of the tetrahedron-PE-STV complex occurs quickly (within 15 minutes) in the RAW 264.7 cells. Confocal microscope analysis (Supplementary Information) of the sample confirmed that the PE fluorescent signal was present inside the cells (Figure 2b) and that the antigens localized to the lysosomes after incubating for 2hrs (Figure S4). The fluorescent signal in the tetrahedron-PE-STV group continued to increase up to 6 hours, while no fluorescent increase was observed in the control group treated with only PE-STV (Figure 2c). This result indicates that the DNA scaffold enhances cellular uptake of the antigen. This finding was further substantiated in primary DCs (Figures 2d, details in SI), but not in a mouse B cell line that lacked the specific antibody required to bind STV (Figure S5). Furthermore, after an incubation time of 5 hrs with fetal bovine serum (FBS) at room temperature, the tetrahedron scaffold remained stable, as more than 88.7% of the DNA band was still retained (Figure S3), which may be sufficient for in vivo capture by APCs. Our in vitro study, together with previous reports of DNA assemblies facilitating adjuvant uptake19,24,25,31,32, suggest that DNA nanostructures can promote delivery of both assembled antigens and adjuvant to APCs, which is a prerequisite for induction of an effective immune response.
Figure 2.
Antigen internalization in RAW 264.7 cells and primary DCs. (a) Representative flow cytometry result showing the cellular PE fluorescence in RAW 264.7 cells after 30 minute incubation with PE-STV and/or DNA scaffolds. The red, green, and blue traces correspond to STV, directly linked ODN-STV and tetrahedron-STV (Supplementary Information), respectively. (b) Representative confocal microscopy images showing internalization of PE-STV in RAW 264.7 cells measured from the flow cytometry intensity. The insets show zoom-in images of representative cells. The cartoons below each panel represent STV, directly linked ODN-STV and tetrahedron-STV, respectively. (c) Histogram showing time-dependent cellular internalization of PE-STV in RAW 264.7 cells. The mean fluorescent intensity (MFI) of PE is plotted against the length of incubation time. Each column represents the average of three parallel measurements, and error bars are generated from the standard deviation. (d) Histogram showing the cellular internalization of PE-STV in primary DCs after 2 hour incubation. Each column represents the average of two parallel measurements and error bars are generated from standard error of the mean value. The cartoons shown below each panel represent free DNA + STV, directly linked ODN-STV and tetrahedron-STV, respectively.
We next compared the immunogenicity of the fully assembled tetrahedron-STV-CpG ODN vaccine complexes in inducing anti-STV antibody responses in a BALB/c mouse model to those of an unassembled mixture of STV and CpG ODN, or STV alone. Specifically, we followed the antibody response in three groups of mice injected with different combinations of CpG ODN and STV: 1) STV only; 2) free STV mixed with CpG; and 3) tetrahedron-STV-CpG ODN complex (Supplementary Information, Figure S1). The directly linked STV-CpG complex was included as a positive control (Figure S6). As outlined in Figure 3a, after two immunizations with the DNA scaffolded vaccine complex followed by a challenge of STV protein only, serum was collected from each mouse group and the level of anti-STV IgG antibodies was assessed using an enzyme-linked immunosorbent assay (ELISA). Over a period of 70 days, we found that mice immunized with the fully assembled tetrahedron-STV-CpG ODN complex developed a much higher level of anti-STV IgGs than the free CpG+STV (Figure 3b). This reflects the development of long-term immunity against the antigen, presumably due to the persistence of long-lived antibody secreting plasma cells and/or generation of STV-specific memory B cells.
Figure 3.
Antibody response in BALB/c mice. (a) Immunization protocol. The time intervals between the primary and secondary immunization and the antigen challenge are 28 days and 24 days, respectively. Details see Supporting Information. (b) Anti-STV IgG level after antigen challenge. The average antibody level was determined from the results of at least eight mice per group and is plotted here. The error bars are generated from the standard deviation. (c) Specific memory B cell response in mice assessed by ELISPOT. The average was calculated from results of at least eight mice per group and the asterisk indicates a p value of less than 0.05 as determined by an unpaired student t test.
To directly evaluate the long-term immunity induced by various immunization regimes, we applied an enzyme-linked immunosorbent spot (ELISPOT) assay that allows numeration of STV-specific memory B cells present in the spleen cells of immunized mice. Specifically, after in vitro stimulation with STV, memory B cells are converted into antibody-secreting cells (ASCs) which are detected by the ELISPOT assay. As shown in Figure 3c, significantly elevated levels of specific ASCs were found in mice immunized with the tetrahedron-STV-CpG ODN complex compared to those immunized with free CpG+STV and STV only. Thus, the tetrahedron scaffolded-STV-CpG ODN complexes induce a stronger and longer lasting anti-STV antibody response, due in part to the generation of STV-specific memory B cells.
Beyond the tetrahedral DNA nanostructure described above, we also constructed a branch-shaped structure31–33 for antigen-adjuvant co-assembly (Supplementary Information, Figure S1). The antigen assembled by this branched DNA structure (designated as Branch-CpG, Figure S1) induces an antibody response at a level intermediate between that of free CpG+STV and the tetrahedron-STV-CpG ODN complex (Figure S7b). Interestingly, in the in vitro experiment antigen internalization for the branch-STV complex is lower than that for the tetrahedron-STV complex, but higher than that for free STV (Figure S7a). Taken together, the different DNA nanostructures appear to influence both the in vitro cellular uptake of the antigen, and the in vivo induction of antigen-specific antibody responses. This is likely because of differences in the size, shape or stability of the DNA nanostructures which may affect their ability to deliver the attached antigen and adjuvant to APCs. While the actual mechanisms still remain to be elucidated, the observed correlation between an elevated level of antigen internalization and a stronger antibody response may provide us with a screening tool to predict or identify the optimal DNA nanostructures for subsequent vaccine construction and test in vivo.
In addition to efficacy, the safety of a vaccine platform is another important parameter in vaccine design. Any non-targeted immune responses, including those against the platform itself, should be minimized. We should point out that the amount of antigen and CpG ODN used in our antigen-adjuvant-DNA complex to induce a specific immune response is lower than reported elsewhere34, implying the reduced chance of this complex to cause overt non-specific activation often associated with injection of free adjuvant. Furthermore, any immune reaction mounted against the double stranded DNA scaffold could result in tissue damage and trigger autoimmunity; for example, anti-double stranded DNA (anti-dsDNA) antibodies are implicated in the pathogenesis of many autoimmune diseases including systemic lupus erythematosus35. We measured the level of anti-dsDNA antibodies in the mouse serum 18 days post secondary immunization, a time when the anti-STV antibody level was still very high and anti-dsDNA antibodies, if present, would be detected with the highest sensitivity. Using two independent methods, we observed no detectable level of anti-dsDNA antibodies in the tetrahedron-STV-CpG ODN group (Figure 4).
Figure 4.
Response against the double-stranded DNA scaffold. (a) Results analyzed by anti-dsDNA antibody ELISA kit. Relative OD indicates the ratio between the measured OD405 for each sample and that of a standard calibrator provided by the manufacture. (b) Confocal microscopy images assessing the anti-dsDNA antibody by ANA kit. i) and ii) slides incubated with positive and negative control serum provided by manufacture; iii) and iv) slides incubated with mouse serum from the Free CpG + STV group; v) and vi) slides incubated with mouse serum from Tetrahedron-CpG−STV group
In addition to evaluating the level of anti-dsDNA antibody production, we performed ELISA analysis to determine whether there are antibodies generated against the tetrahedron-shaped structure. Similarly, no antibodies were detected in the mouse serum 18 days post secondary immunization (Figure S8). Taken together, these results indicate that the antigen-adjuvant-DNA complex is relatively safe and that the response induced by the vaccine complex is specific to the antigen and not the DNA platform.
In summary, we demonstrated that a DNA scaffold can be used to construct an antigen-adjuvant complex that elicits a strong and specific antibody response in vivo, without inducing an undesirable response against the scaffold itself. The close proximity of the antigen and adjuvant is critical to enhance the immunogenicity of a vaccine; directly linking CpG ODN to an antigen has been shown to induce a strong B cell response34 (Figure S6), however, such direct linkages may be subject to certain limitations when constructing more complex vaccines. The ability to construct multivalent and multi-specific antigen-adjuvant complexes remains a challenge. Programmable DNA nanostructures provide an excellent platform for construction of vaccines with multivalency and three-dimensional configuration, mimicking VLPs. With well-established protein-DNA conjugation techniques, it is now feasible to attach a single antigen or adjuvant on a DNA strand. Thus, multivalent and multispecific vaccine complexes can be easily achieved by incorporating multiple antigen- or adjuvant-loaded single stranded DNAs into the same structure. The three dimensional arrangement of each of the immunogenic components can be readily controlled through the rational design of the scaffold sequences. With the rapid development of protein-DNA and small molecule-DNA conjugation techniques, it may be possible to construct even more complex vaccines in which many different immunogenic epitopes, “danger signals” that stimulate immune cells, or various therapeutic components such as siRNA19 can be assembled on the same DNA scaffold to enhance the efficacy of the complex vaccines. Our work demonstrates the potential of DNA nanostructures to serve as general platforms for vaccine development.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of Health (CA141021 and DA 030045 to Y. C). We thank Dr. Chengde Mao for kindly providing information on the design and assembly of the tetrahedron DNA. We also thank Dr. Jeanette Nangreave for editing the manuscript. Dr. Yang Xu is a visiting scholar from the State Key Lab of Molecular Oncology, Cancer Institute & Hospital, PUMC & CAMS of China.
Footnotes
Author contributions
Y. C., H. Y. and Y. L. conceived the project and designed the project. Y. C., H. Y., Y. L. and X. L. designed the experiment. X. L., Y. X., T. Y. and C. C. performed the experiments. X. L. analyzed the data and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Supporting Information. Details of the nanostructure design, experimental assembly, post-assembly characterization, and sequences of all DNA oligonucleotides used in this study. This material is available free of charge via the Internet at http://pubs.acs.org.
Contributor Information
Hao Yan, Email: Hao.Yan@asu.edu.
Yung Chang, Email: Yung.Chang@asu.edu.
References
- 1.Fenner F. Smallpox and its eradication. World Health Organization; Geneva: 1988. [Google Scholar]
- 2.Peek LJ, Middaugh CR, Berkland C. Adv Drug Deliv Rev. 2008;60:915–928. doi: 10.1016/j.addr.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noad R, Roy P. Trends in Microbiology. 2003;11:438–444. doi: 10.1016/s0966-842x(03)00208-7. [DOI] [PubMed] [Google Scholar]
- 4.Sytkowski AJ, Lunn ED, Davis KL, Feldman L, Siekman S. Proc Natl Acad Sci U S A. 1998;95:1184–1188. doi: 10.1073/pnas.95.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beyer T, Herrmann M, Reiser C, Bertling W, Hess J. Curr Drug Targets Infect Disord. 2001;1:287–302. doi: 10.2174/1568005014605973. [DOI] [PubMed] [Google Scholar]
- 6.Krishnamachari Y, Salem AK. Adv Drug Deliv Rev. 2009;61:205–217. doi: 10.1016/j.addr.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasturi SP, Skountzou I, Albrecht RA, Koutsonanos D, Hua T, Nakaya HI, Ravindran R, Stewart S, Alam M, Kwissa M, Villinger F, Murthy N, Steel J, Jacob J, Hogan RJ, Garcia-Sastre A, Compans R, Pulendran B. Nature. 2011;470:543–547. doi: 10.1038/nature09737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodman RP, Schaap IA, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 9.Seeman NC. J Theor Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 10.Shih WM, Quispe JD, Joyce GF. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 11.Zhang C, Su M, He Y, Zhao X, Fang PA, Ribbe AE, Jiang W, Mao C. Proc Natl Acad Sci U S A. 2008;105:10665–10669. doi: 10.1073/pnas.0803841105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shih WM. Nature. 2010;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H. Science. 2011;332:342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 14.Dietz H, Douglas SM, Shih WM. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auyeung E, Cutler JI, Macfarlane RJ, Jones MR, Wu JS, Liu G, Zhang K, Osberg KD, Mirkin CA. Nature Nanotechnology. 2012;7:24–28. doi: 10.1038/nnano.2011.222. [DOI] [PubMed] [Google Scholar]
- 16.Aldaye FA, Palmer AL, Sleiman HF. Science. 2008;321:1795–1799. doi: 10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]
- 17.Chhabra R, Sharma J, Liu Y, Rinker S, Yan H. Adv Drug Deliv Rev. 2010;62:617–625. doi: 10.1016/j.addr.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 19.Lee H, Lytton-Jean AK, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M, Peng CG, Charisse K, Borodovsky A, Manoharan M, Donahoe JS, Truelove J, Nahrendorf M, Langer R, Anderson DG. Nat Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walsh AS, Yin H, Erben CM, Wood MJ, Turberfield AJ. ACS Nano. 2011;5:5427–5432. doi: 10.1021/nn2005574. [DOI] [PubMed] [Google Scholar]
- 21.Douglas SM, Bachelet I, Church GM. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 22.Surana S, Bhat JM, Koushika SP, Krishnan Y. Nat Commun. 2011;2:340. doi: 10.1038/ncomms1340. [DOI] [PubMed] [Google Scholar]
- 23.Mohri K, Nishikawa M, Takahashi N, Shiomi T, Matsuoka N, Ogawa K, Endo M, Hidaka K, Sugiyama H, Takahashi Y, Takakura Y. ACS Nano. 2012 doi: 10.1021/nn300727j. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Pei H, Zhu B, Liang L, Wei M, He Y, Chen N, Li D, Huang Q, Fan C. ACS Nano. 2011;5:8783–8789. doi: 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- 25.Schuller VJ, Heidegger S, Sandholzer N, Nickels PC, Suhartha NA, Endres S, Bourquin C, Liedl T. ACS Nano. 2011;5:9696–9702. doi: 10.1021/nn203161y. [DOI] [PubMed] [Google Scholar]
- 26.Zhang C, Su M, He Y, Leng Y, Ribbe AE, Wang G, Jiang W, Mao C. Chem Commun (Camb) 2010;46:6792–6794. doi: 10.1039/c0cc02363a. [DOI] [PubMed] [Google Scholar]
- 27.Zhang C, Tian C, Guo F, Liu Z, Jiang W, Mao C. Angew Chem Int Ed Engl. 2012;51:3382–3385. doi: 10.1002/anie.201108710. [DOI] [PubMed] [Google Scholar]
- 28.He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 29.Klinman DM. Nat Rev Immunol. 2004;4:249–258. doi: 10.1038/nri1329. [DOI] [PubMed] [Google Scholar]
- 30.Bachmann MF, Jennings GT. Nat Rev Immunol. 2010;10:787–796. doi: 10.1038/nri2868. [DOI] [PubMed] [Google Scholar]
- 31.Nishikawa M, Matono M, Rattanakiat S, Matsuoka N, Takakura Y. Immunology. 2008;124:247–255. doi: 10.1111/j.1365-2567.2007.02762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rattanakiat S, Nishikawa M, Funabashi H, Luo D, Takakura Y. Biomaterials. 2009;30:5701–5706. doi: 10.1016/j.biomaterials.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 33.Churchill ME, Tullius TD, Kallenbach NR, Seeman NC. Proc Natl Acad Sci U S A. 1988;85:4653–4656. doi: 10.1073/pnas.85.13.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klinman DM, Barnhart KM, Conover J. Vaccine. 1999;17:19–25. doi: 10.1016/s0264-410x(98)00151-0. [DOI] [PubMed] [Google Scholar]
- 35.Ceppelini R, Polli E, Celada FA. Proceedings of the Society for Experimental Biology and Medicine. 1957;96:572–574. doi: 10.3181/00379727-96-23544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.