Abstract
In vivo MRSI is an important imaging modality that has been shown in numerous research studies to give biologically relevant information for assessing the underlying mechanisms of disease and for monitoring response to therapy. The increasing availability of high field scanners and multichannel radiofrequency coils has provided the opportunity to acquire in vivo data with significant improvements in sensitivity and signal to noise ratio. These capabilities may be used to shorten acquisition time and provide increase coverage. The ability to acquire rapid, volumetric MRSI data is critical for examining heterogeneity in metabolic profiles and for relating serial changes in metabolism within the same individual during the course of the disease. In this review we discuss the implementation of strategies that use alternative k-space sampling trajectories and parallel imaging methods in order to speed up data acquisition. The impact of such methods is demonstrated using three recent examples of how these methods have been applied. These are to the acquisition of robust 3D 1H MRSI data within 5 –10 minutes at a field strength of 3T, to obtaining higher sensitivity for 1H MRSI at 7T and to using ultrafast volumetric and dynamic 13C MRSI for monitoring the changes in signals that occur following the injection of hyperpolarized 13C agents.
Keywords: spectroscopic imaging, hyperpolarized agents, spatial localization
INTRODUCTION
In vivo Magnetic Resonance Spectroscopic Imaging (MRSI) has been applied for scientific investigation and to characterize pathological states for many years. It has advantages over other metabolic imaging methods because it can provide direct correlations with anatomic imaging and physiological measurements such as diffusion, perfusion and blood oxygen level dependent (BOLD) imaging. While early studies applied 31P MRSI, the improved sensitivity of water-suppressed 1H MRSI has meant that it has been more widely used for applications in humans. Although the results of such studies have identified biologically relevant parameters for tracking spatial and temporal changes in metabolism within living systems, there are still a number of challenges in the widespread and uniform implementation of the technology. The recent availability of high field scanners, improved gradients and multi-channel radiofrequency coils has provided the possibility for dramatic improvements in the quality and flexibility of data acquisition.
A further advance in the acquisition of metabolic data is the development of instrumentation for generating hyperpolarized 13C-labeled agents that can be applied for in vivo studies of biological systems. This has opened a new field of investigation concerning the evaluation of dynamic changes in metabolite levels caused by alterations in enzyme activity. Although the majority of studies in the literature have focused on 13C1-labeled pyruvate as the agent of interest, many other agents have been polarized and have provided biologically relevant information in studies using cell bioreactors and/or animal models. One of the advantages of using such approaches in developing new therapeutic agents and in evaluating novel treatment strategies is that similar types of measurements can be made in cells, tissue slices, animal models and, ultimately, in humans.
With higher sensitivity for state of the art 1H and 13C MRSI methods has come the dual requirement of implementing pulse sequences that provide rapid spatial and frequency encoding, together with the corresponding algorithms for reconstruction of spectral data that are acquired with multi-channel and non-cartesian k-space sampling. While it is possible to use similar strategies to those developed for acquiring volumetric anatomic MRI data, there are a number of additional challenges imposed by the addition of the chemical shift dimension. In the following we review spatial encoding strategies that are being used to address those challenges and discuss three specific applications: the development of robust methods for prescribing and acquiring 1H MRSI data from the brain using clinical MR scanners, the implementation of 1H MRSI at 7T and the acquisition of in vivo hyperpolarized 13C metabolic imaging for pre-clinical and human studies.
VOLUME SELECTION AND SPATIAL ENCODING
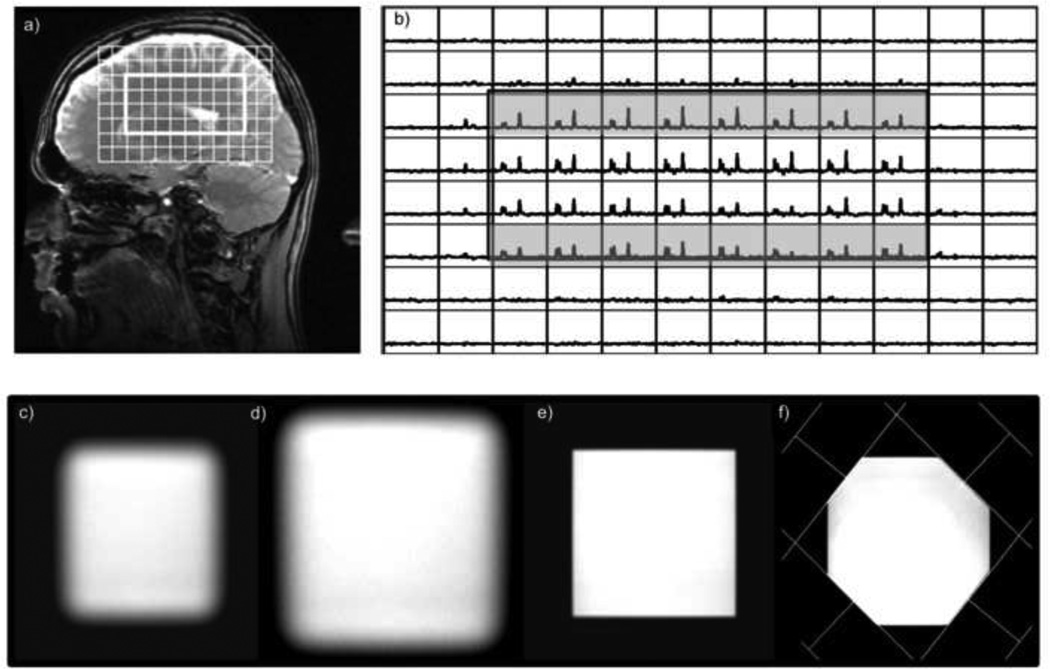
MRSI pulse sequences use a combination of frequency and spatially selective pulses to suppress unwanted signals and localize metabolites of interest to specific regions of the anatomy. A critical issue when using any form of spatially selective excitation at higher field strengths is to consider the bandwidth of the rf pulse and the frequency shift between the resonances that are being evaluated. Table 1 shows the magnitude of the chemical shift effect versus bandwidth and field strength and Figure 1 illustrates the impact of this effect upon relative levels of choline (Cho) and N-acetylaspartate (NAA) for PRESS localization and 3D spectral encoding in the brain at 3T. For this commercially available sequence, the bandwidth of the 90 degree rf pulse was 2400Hz and for the two 180 degree pulses was 936Hz. Although the middle two slices have consistent metabolite ratios, the upper slice has high NAA and low Cho, and the lower slice has low NAA with high Cho. As the relative levels of such resonances are commonly used for assessing pathology, it is clear that the effect either needs to be minimized or the relative intensities corrected when interpreting the data (1). Although it is a well-known problem, manufacturer-provide pulse sequences use pulses with relatively low bandwidth and users may not be aware of how it impacts their data. This may interfere with the performance of lipid suppression, and is especially critical for sequences that use spectral editing of coupled resonances (2,3), where achieving uniform excitation of both coupling partners is required.
Table 1.
Magnitude of chemical shift (in millimeters) associated with selection pulse with a given bandwidth at for a 100mm-wide selected region. The standard 180 degree pulses used in commercial sequences are typically 1kHz, spatial-spectral pulses are in the range of 3–5kHz and the most recent LASER pulses are in the range of 5–20kHz.
| Frequency | Water to Lipid | Cho to NAA | ||||||
|---|---|---|---|---|---|---|---|---|
| Bw (kHz) | 1.0 | 1.5 | 2.5 | 5.0 | 1.0 | 1.5 | 2.5 | 5.0 |
| Shift for 3T (mm) |
42 | 28 | 17 | 8.5 | 15 | 10 | 7 | 3.5 |
| Shift for 7T (mm) |
77 | 51 | 31 | 15.5 | 35 | 23 | 14 | 7 |
Figure 1.
Demonstration of the impact of the chemical shift artifact for 3D PRESS 1H MRSI volume selection acquired with the product sequence with a 180 degree pulse bandwidth of 936Hz: a) image showing the location of the volume and b) the corresponding spectral array (b) The chemical shift artifact is clearly seen on the top and bottom slices from the sagittal orientation. The lower row of images show c) the cross-section of a selected volume, the same volume obtained with OVERPRESS =1.2 d) without OVS, e) OVS bands that sharpen the edges and f) with additional graphically prescribed OVS bands that conform the volume to an octagon.
Three strategies have been proposed to minimize the effect of the chemical shift artifact and hence provide improved volume selection for 1H MRSI. The first is to increase the bandwidth of the selection pulses. Spatial spectral pulses with bandwidths of 3–5kHz were used for PRESS selection in early 1.5T experiments (4,5), as well as more recently in 3T and 7T applications (6,7,8). The limitation in using this approach is that the pulses are relatively long and in many cases they results in a minimum TE in the range of 70–80ms. Although they tend to have higher power requirements, recent studies using localized adiabatic selective refocusing (LASER) pulses with bandwidths of 5Khz have provided reliable volume selection with a TE of 30ms at 3T (9) and a TE of 50ms at 7T (10). The second strategy is to enlarge the size of the region selected and then cut-off the portions that are impacted using out of voxel suppression (OVS) with multiple saturation bands (11). This is illustrated in Figure 1 and not only suppresses residual lipid but can also conform the shape of the selected volume to more closely match the anatomy. The third strategy is to replace PRESS or STEAM volume selection by a spin echo sequence that excites an entire slice. In this case inversion recovery and/or OVS methods are required to remove unwanted lipid signals (12). This can provide short TE but the inversion recovery may reduce the intensities of nearby resonances (13,14).
Given accurate volume selection, the next step is to perform spatial encoding in a manner that can be reconstructed to provide an array of spectral data. Rectangular phase encoding obtains one k-space point per excitation and so improving the spatial resolution or increasing the field of view would translate into a larger matrix size and hence a longer acquisition time. As TR’s for 1H MRSI, the data acquisitions are typically in the range of 1–2 seconds, the time required to obtain 24×24 phase encodes for such data is 10–20 minutes and, when a third dimension is added to obtain volumetric coverage, the total time quickly becomes untenable. For hyperpolarized 13C MRSI the data acquisition window is typically less than 30s and the number of excitations must be kept to a minimum in order to compensate for the fact that the magnetization is non-renewable. While reducing the number of excitations by restricting k-space sampling to an octagonal or elliptical subset of the phase encode grid is often applied, this widens the true point spread function of the data and decreases the true spatial resolution.
A well-known solution that is applied routinely for MRI is use echo planar or spiral k-space trajectories. While these methods have been widely implemented in a research setting to simultaneously encode spatial and chemical information during the data acquisition window, commercially available pulse sequences and reconstructions are still very limited in the capabilities that they offer. Because both types of information are being obtained within the same time period, there is a trade-off between the spectral bandwidth and the spatial resolution of the data that can be achieved. To obtain high resolution at high field strength may therefore require the use of interleaved acquisitions. Table 2 provides representative examples of the range of spatial encoding matrices and acquisition times that have been achieved for 1H MRSI. As can be seen, spiral methods with spatial spectral pulses for PRESS volume selection were among the first to be evaluated at 1.5T and were able to provide a large 3D array of spectra within 1–2 minutes (5). Given the signal to noise ratio of data with a TE of 144ms at this field strength, however, it was necessary to obtain 8 averages for a total acquisition time of 17 minutes. For spiral acquisitions employing slice selection and LASER or GOIA (Gradient Offset Independent Adiabaticity) pulses at 3T (15) and 7T (16) it has been possible to achieve TEs of 30–45ms within less than 5 minutes. In the PEPSI sequence Posse et al (12) applied 1-D echo planar encoding in one spatial dimension with whole slice excitation and OVS to consider TE’s of the order of 10–20ms. In this case the time for acquisition time for a single slice could be as low as 64s. The choice of methodology, as well as the field strength and spatial resolution used depends upon the application being considered and the concentration of the metabolites of interest. As a general guide, a nominal spatial resolution of 1cc is readily achievable for 1H MRSI data from the brain at 3T using a multi-channel receive coil within a scan time of 5 minutes for choline, creatine, N-acetylaspartate, lactate and lipid, whereas measurements of changes in glutamate, glutamine, myo-inostitol, glutathione and γ—aminobutyric acid may require longer acquisition times or larger voxel sizes.
Table 2.
Representative examples of 2D and 3D 1H MRSI acquisition parameters with accelerated k-space encoding as reported in the literature.
| Author | Field strength | Selection | Lipid Supression | Spatial encoding | TR/TE (ms) | Acquisition time (s) |
|---|---|---|---|---|---|---|
| Posse et al 1994 (12) |
1.5T | 3D SE | 16 auto | PEPSI (RL) 32×32×8 |
2000/11 | 64s- |
| Adalsteinsson et al 1998 (5) |
1.5T | 3D PRESS spatial spectral |
OVS | SPIRAL 18×18×10 |
1100/144 | 18min (8nex) |
| Cunningham et al, 2005 (17) Zierhut et al, 2009 (18) |
3T | 3D PRESS | OVS | Flyback EPSI 16×16×16 |
1100/144 | 4.7 min |
| Ebel et al, 2005 (13) |
4T | 3D SE | IR | EPSI 50×50×18 |
1710/45 | 26min |
| Maudsley et al, 2010 (14) |
3T | 3D SE | IR | EPSI 50×50×18 |
1710/70 | 26min |
| Andronesi et al 2012 (15) |
3T | 3D LASER |
Volume selection only |
Spiral 16×16×8 22×22×12 30×30×14 |
1000/45 | 48s, 2.4min, 4.2min |
| Dydak et al, 2001 (28,29) |
3T | 2D PRESS |
OVS | 32×23 SENSE R=4 |
1500/136 | 6.5min |
| Ozturk et al, 2006a, 2006b,2009 (32,33,36) |
3T | 3D PRESS |
OVS | 16×16×8 SENSE R=4/ e-SENSE R=4 |
1100/144 | 9.5min/ 4.7min |
| Banerjee et al, 2009 (34) |
3T |
3D PRESS |
OVS | 16×16×8 GRAPPA R=4/ e-GRAPPA R=4 |
1100/144 | 9.5min/ 4.7min |
| Zhu et al, 2010 (6) | 3T | 2D SE 3 slices |
Dual band water/lipid + OVS |
27×33 ×3 SENSE R=2 AP, R=1.5 RL |
2500/144 | 10.5min |
| Lin et al, (31) | 3T | 2D SE | OVS | 16×16 PEPSI, 1-D SENSE R=2,3,4 |
2000/15 | 32–12s |
| Tsai et al 2008 (30) | 3T | 2D SE | OVS | 16×16 PEPSI, 1-D GRAPPA, R=2–4 |
2000/15 | 32–12s |
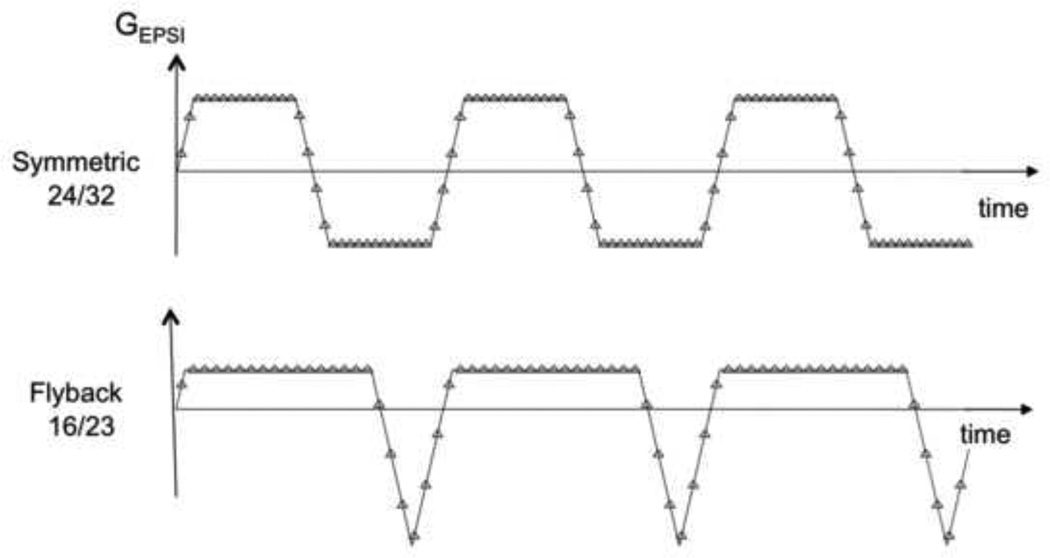
Reconstruction for spiral and generalized echo planar trajectories uses the prior knowledge of the k-space trajectory to interpolate the data onto a rectangular grid, followed by a standard fourier transform (5,12,15,16). While there is some signal loss due to imperfections in gradient performance, improvements in the current generation of hardware has made this a minor effect. The fly-back echo planar trajectories proposed by Cunningham et al (17) take advantage of rapidly switching gradients (see Figure 2). If the analysis is restricted to data on the flat part of the trapezoidal wave form, the reconstruction can be simplified to a fourier transform, followed by phase corrections that take account of differences in the temporal offset of the k-space trajectories. Using such strategies for volumetric 1H and hyperpolarized 13C MRSI data acquisitions in the prostate and brain have provided extremely robust data. For 3T 1H MRSI, the loss of efficiency due to skipping the ramp/rewind portions of the trajectory is 10–20% (17,18). For hyperpolarized 13C MRSI the effect can be as much as 40% of the waveform due to there being a factor of 4 difference in the gamma of the nucleus (19). In this case using a symmetric waveform followed by separately processing of the positive and negative lobes and combining them after corrected for phase and frequency variations is more efficient. Clearly the data points on the gradient ramps can also be included by re-gridding the data prior to reconstruction. Interleaved trajectories may also be needed for both spiral and echo planar sampling in order to achieve the spectral bandwidth and spatial resolution (20,21).
Figure 2.
Flyback and Symmetric EPSI trajectories used for 1H MRSI data acquisitions with a 1cm nominal voxel size at 3T. On the bottom row shows the calibration images and a single voxel from data obtained from each channel of an 8-element head coil from a phantom, as well as the combined spectrum.
ALTERNATIVE ENCODING METHODS
The availability of MR scanners with multiple receive channels has expanded the usage of phased array coils and raised the question of how to best combined MRSI signals from individual elements. As reviewed in the original papers (22–24), a variety of different approaches can be taken, depending on whether the goal is to correct for spatial variations in intensity of the spectra due to the non-uniform rf profiles of individual coil elements or merely obtain the best possible SNR. This can be viewed it as a special case of parallel imaging with the SENSE method that has a speed–up factor of 1 (25,26). The spectral arrays are first reconstructed separately and then added together by weighting factors that represent the reception profiles of individual coil elements. Estimates of these profiles can come either from theoretical models, empirical measurements in phantoms, filtering of in vivo images that have been acquired with minimal (usually proton density weighted) contrast, an external reference dataset (such as unsuppressed water) or from an internal metabolite that is present in all spectra at relatively high signal to noise ratio. Providing output spectra that are phase sensitive requires that either the spectral arrays from individual elements are corrected on a voxel by voxel basis before hand or that a global phase offset is estimated for each coil and included in the weighting function (27).
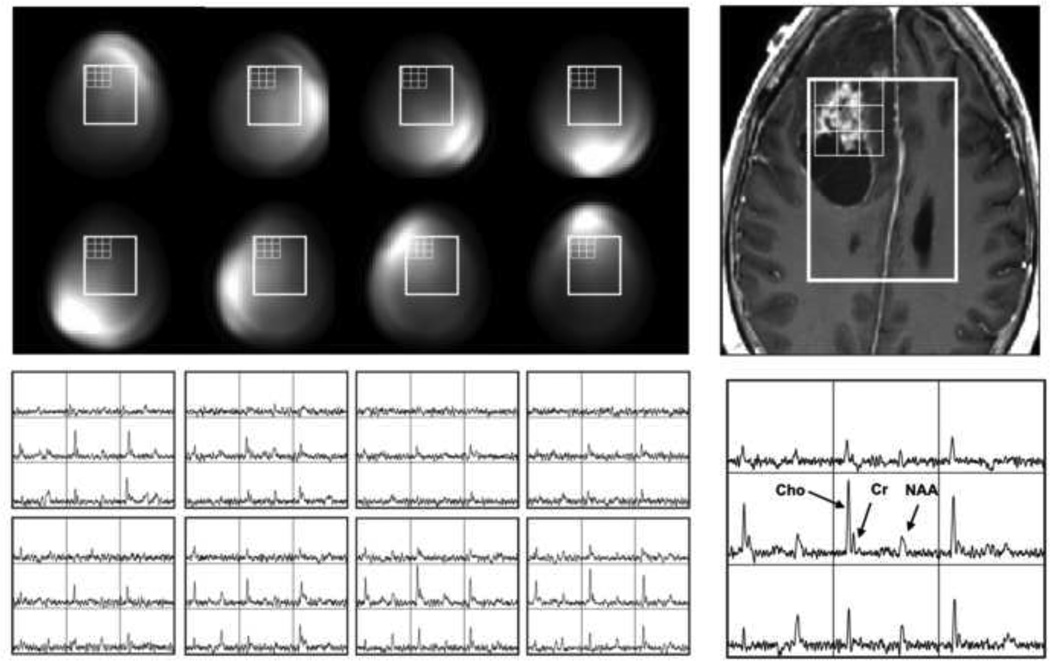
Figure 3 shows 1H MRSI data from a patient with a brain tumor that were combined using weighting functions from coil calibration images and estimates of phase offsets in each channel determined from the residual water peaks in voxels from the center of the selected volume. In both cases the SNR of the combined data is significantly improved over individual coil elements. Determining which is the best approach for combining the data from specific applications depends upon the accuracy of the phasing algorithm and the SNR of spectra from individual voxels. Using a weighting function that divides by the square root of the sum of squares of individual weights is preferred for subsequent viewing of the data because it maintains similar levels of noise in each spectrum. Direct comparison of metabolite levels between different voxels must, however, take into account that there is still a spatial weighting on individual voxels caused by the non-uniformity of the combined coil reception profiles.
Figure 3.
Demonstration of the impact of coil combination for in vivo results obtained with the same 8-element head coil as for the phantom in Figure 2. The improvement in signal to noise ratio is clearly seen for the combined spectral array.
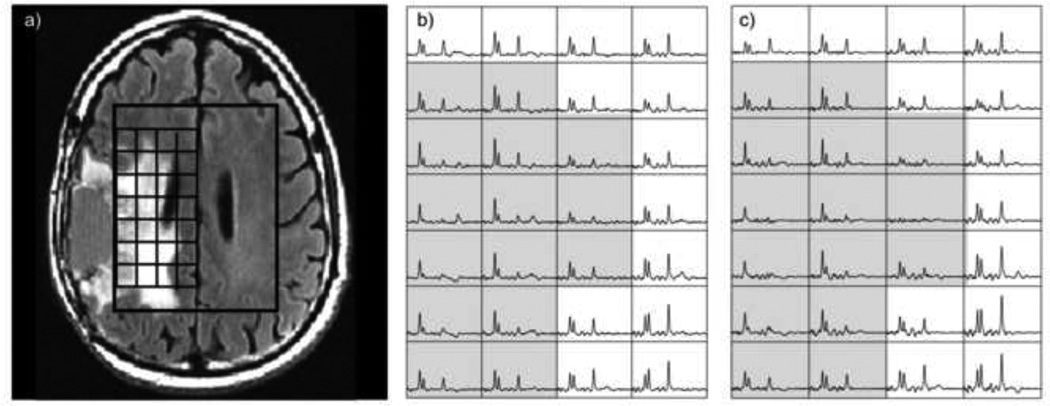
If there is enough SNR in the data to further cut back on acquisition time, the next step in reducing the number of excitations is to implement acceleration using parallel imaging. A number of studies have investigated the use of SENSE and GRAPPA for 1H MRSI were presented by Dydak et al (28,29), Tsai et al (30), Lin et al (31), Ozturk-Isik et al (32,33), Bannerjee et al (34) and Zhu et al (35). While the results demonstrated were promising, there were clearly trade-offs in terms of artifacts and reconstruction times that should be considered in using such approaches. Figure 4 gives an example of SENSE with an R factor of 4 for 1H MRSI, which reduces the acquisition time with an 8 channel head coil with elliptical phase encoding to less than 5 minutes for a 3D array of spectra without using any form of echo planar sampling (36). The coil sensitivity profiles were obtained using low resolution proton density weighted images. As can be seen, although the SENSE acquisition has a lower SNR, it gives essentially the same information as the fully sampled one.
Figure 4.
Application of elliptical SENSE for 3D 1H MRSI obtained from the brain using an *8-element head coil. The array on the left b) was fully sampled with an acquisition time of 17minutes and the one on the right c) uses SENSE with an R=4 factor (2 in RL and 2 in AP) with an acquisition time of 5 minutes. The shaded voxels highlight the region that has abnormal metabolite levels.
A new class of methods that were initially developed by Frydman and colleagues for performing ultrafast multi-dimensional NMR spectroscopy (37) utilize an approach that involves spatiotemporal encoding (SPEN) for obtaining and reconstructing signals for performing and spectroscopic imaging (38,39). These have not yet been widely applied for in vivo applications of MRSI, but preliminary data indicate that they can provide excellent results for chemical shift imaging of multiple distinct resonances. A further strategy for accelerating the acquisition of signals which are sparse in either spatial or frequency dimensions and may therefore be ideal for hyperpolarized 13C studies is compressed sensing (40–42). This applies random sampling of the acquisition space in conjunction with algorithms that perform a constrained, iterative reconstruction. The degree of undersampling that can be achieved depends upon reformulating the problem into an appropriate domain and upon the data having relatively high SNR. As is described below, initial applications to pre-clinical hyperpolarized 13C studies have demonstrated speed up factors of 4–8 for obtaining MRSI data.
AUTOMATIC PRESCRIPTION OF BRAIN 1H MRSI DATA at 3T
Despite the recent improvements in technology described above, applying volumetric 1H MRSI in a clinical setting is still challenging. Experience has shown that data quality is often compromised by lack of familiarity in prescribing the selected volume and in defining the location of graphic VSS bands that will provide adequate coverage of the region of interest. Some of this can be resolved by more extensive training, but the reliability and reproducibility of the data would benefit considerably by providing a more automated procedure for defining such parameters. This is particularly important for optimizing coverage, for making comparisons across populations and for using serial scans to evaluate response to therapy in the same subject. Automated prescription of MRI data has recently been implemented by several of the scanner manufacturers in order to ensure consistent image orientation between different scans. This is achieved by aligning a reference to earlier images from the same subject or to a brain atlas. In the following we describe recent work that has used similar strategies to simplify the acquisition of serial, volumetric 1H MRSI data.
The automated method developed by Ozhinsky et al (43) used sequence parameters that have been shown to provide robust spectral data at 3T. This included PRESS volume selection in conjunction with the BASING technique (44), which not only improves water suppression but can also be employed for J-resolved spectral editing to separate resonances such as lactate and lipid. The size of the PRESS volume was increased by a factor of 1.2 to eliminate the chemical shift artifact for data obtained with a single average and by a factor of 1.5 for acquisitions that required lactate editing (3). Five pairs of cosine modulated VSS bands were then reserved to sharpen up the selected volume and eliminate its corners so that the in-plane profile was octagonal. Nine additional oblique VSS bands were available to suppress subcutaneous lipid (11). Flyback echo planar sampling was applied in one spatial dimension with two dimensions of phase encoding to give an 18×18×16 array of spectra. The nominal voxel size is 1cc for an oblique selected volume that covered almost all of the brain in 5–8 minutes, depending on the TR being used (45). The true spatial resolution depends on whether spatial filtering is applied in the reconstruction.
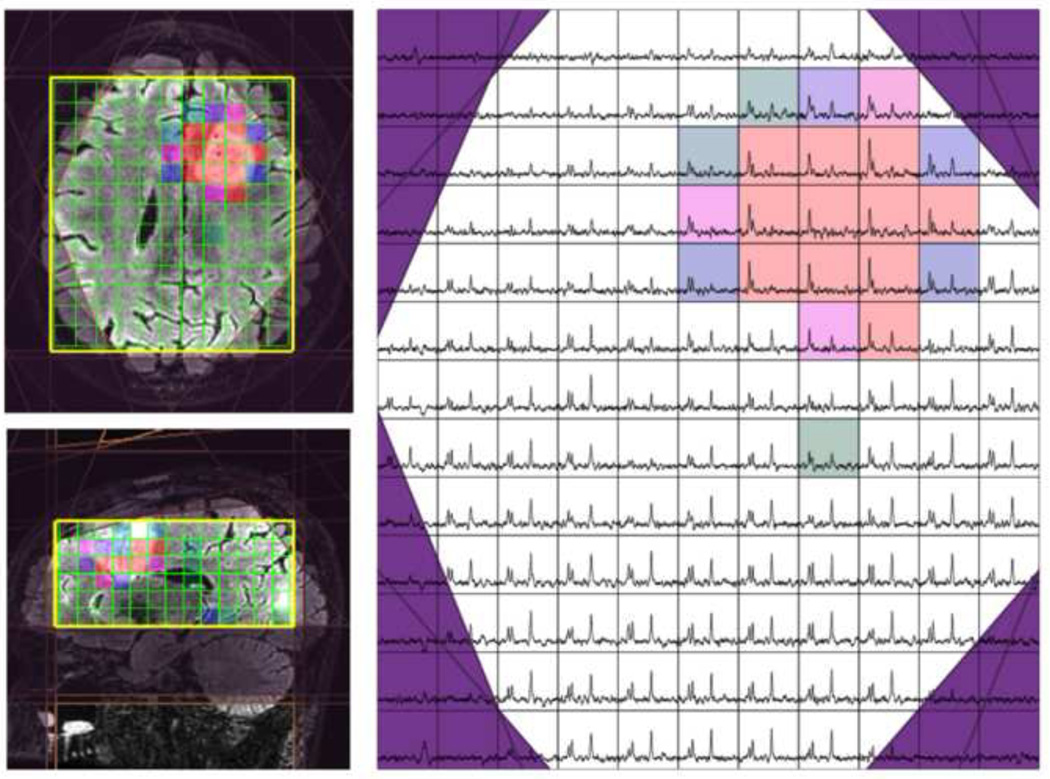
The definition of the location and orientation of the selected volume and the VSS bands was achieved automatically using the volumetric T1-weighted image that was acquired as part of the routine MRI examination. A simple k-means clustering algorithm was applied to this image in order to segment masks for brain and lipid regions. Once the spatial distribution of these entities was defined, a cost function that weights brain voxels positively and lipid voxels negatively was applied in order to simultaneously optimize the locations and size of both the selected volume and VSS bands (43). The coverage in the superior-inferior direction for the automatically selected region was 6cm and the in-plane coverage included most of the supratentorial brain. Parameters needed to define the optimal prescription were fed directly into the pulse sequence. A 3-D image highlighting the region selected was generated and presented to the operator in order to visually confirm that it was acceptable. The current implementation of the pulse sequence is flexible to allow for full automation, automatic definition of the VSS bands with manual definition of the selected volume or entirely manual prescription. An example is seen in Figure 5. The raw data were reconstructed, processed and displayed using SIVIC (46), which is an open source software package developed in our laboratory for MRSI analysis. No spatial filtering was applied in this case. Results from normal volunteers and patients with brain tumors showed that the quality was excellent and the method was particularly useful in serial studies where it was important to provide consistent coverage (43,45).
Figure 5.
3D 1H MRSI data obtained with PRESS localization, TE=144ms using the fully automated prescription strategy described by Ozhinsky et al (43) and displayed using the SIVIC package (44). The colored voxels represent areas with elevated choline and decreased NAA. The purples lines represent the location of VSS bands.
Table 3 compares the parameters used for the method just described with similar strategies that were integrated with the PEPSI sequence. Martinez et al (47) determined the OVS prescription of 16 oblique bands by segmenting the T1-weighted image from the patient and optimizing the coverage of the lipid component. Yung et al (48) used registration of the acquired image to a brain atlas and then applied an inverse transform to define the location of pre-selected OVS bands. In both cases the definition of the location of the slice used for acquisition was done manually. Comparisons of manual and automated prescription techniques have shown that it was possible to obtain much larger volumetric coverage within an acquisition time of 5 to 10 minutes while maintaining high data quality. Another benefit is that the automated methods yielded consistent prescription quality that was not dependent on operator skill, and could therefore make it possible to include 3D MRSI in clinical protocols without the need for extensive training.
Table 3.
Data acquisition parameters used for 3T studies that applied automatic prescription of saturation bands
| Author | Plane slice (mm) |
Encoding matrix |
TR/TE (ms) |
Selection | # sat bands |
Acquisition time (s) |
|---|---|---|---|---|---|---|
| Ozkinsky et al, 2011 (41) |
Axial 60mm |
Flyback EPSI (SI) 18×18×16 |
1500/144 | PRESS OP= 1.2 |
9 auto + 5×2 around PRESS box |
6.5min |
| Ozhinsky et al, 2012 (43) |
Automatic oblique 60mm |
Flyback EPSI (SI) 18×18×16 |
1100/144 | PRESS OP = 1.5 or 1.2 |
9 auto + 5×2 around PRESS box |
12min (lac- edited) 6min otherwise |
| Martinez-Ramon et al (2010) (45) |
Manual oblique 45mm |
PEPSI (RL) 32×32×8 |
2000/11 | Spin echo | 16 auto | 4.5 min |
| Yung et al (2011) (46) |
Manual oblique 52mm |
PEPSI (RL) 32×32×8 |
2000/15 | Spin echo | 16 auto from atlas |
4.4 min |
H-1 MRSI AT 7T
The introduction of ultra high field MR systems offers advantages in terms of higher SNR and improved spectral resolution compared with the clinical field strength of 3T. Studies that compared the SNR and spectral linewidth in the brain of healthy volunteers at 7T versus field strengths of 1.5T, 3T and 4T have shown an approximately linear gain in SNR with field strength, a decrease in spectral linewidth and improved quantification of metabolites (49,50). As shown in Table 4, the T1s of Cho, Cr and NAA are significantly longer and the T2’s are significantly shorter at 7T relative to 3T (51). This needs to be considered in defining protocols for acquiring MRSI data. With appropriate choice of spatial selection, k-space encoding, TR and TE, the higher field strength can be used to not only shorten acquisition time and decrease spatial resolution, but also to provide improved detection of metabolites such as glutathione that are poorly visualized at lower field strengths (52).
Table 4.
T1 and T2 relaxation times (mean±SD) for Cho, Cr and NAA at 3T and 7T that were estimated from the brain of volunteers. Statistical comparisons were performed using non-parametric Wilcoxon rank sum tests.
| Field Strength |
T1 (s) | T2 (ms) | ||||
|---|---|---|---|---|---|---|
| Cho | Cr | NAA | Cho | Cr | NAA | |
| 3T | 1.06±0.11 | 1.38±0.13 | 1.38±0.13 | 170±18 | 151±15 | 262±37 |
| 7T | 1.11±0.20 | 1.75±0.19 | 1.63±0.15 | 131±16 | 121±12 | 170±11 |
| P=0.002 | P=0.003 | P<0.001 | P<0.001 | P<0.001 | ||
At the current time, the majority of 7T MRS studies that have been reported in the literature have been in the brain and have applied single voxel localization. This is partly due to the limited number of rf coils available for studying different organ systems and partly to issues that complicate the acquisition of multi-voxel MRSI data at this field strength. One of these issues is the increased susceptibility weighting at 7T, which places a greater demand upon methods used to shim the magnetic field. Most of the shimming strategies that have been implemented apply the phase information provided by images acquired at 2 or more echo times to estimate the B0 field and then use pre-acquired shim basis sets to optimize homogeneity within the volume of interest. The acquisition time for such images is kept within 1–2 minutes by using low spatial resolution with echo planar or spiral encoding. Depending on whether the localization is volumetric or multi-slice, the shim currents are either calculated and kept constant for the entire MRSI dataset or updated dynamically on a slice-by-slice basis (53,54). In either case the coverage achieved and robustness of the data is typically less than for 3T.
A further complication is that variations in the B1 field caused by non-uniform excitation and reception are more pronounced at 7T. The use of adiabatic pulses that give similar excitation over a broad range of B1 values is important and additional effort is required to adjust the transmit power so that optimal signal is obtained from the region of interest. While the evolution of multiple transmit technologies may ultimately compensate for variations in excitation and non-uniform flip angles (55), at the current time these factors can negatively impact the quality of the data obtained. Multi-channel receiver coils are typically used in order to provide improved coverage and SNR. The variations in spatial intensity resulting from a combination of non-uniform transmit and receive need to be considered in comparing regional changes in metabolite levels.
As was shown in Table 1, the limitations on volume selection imposed by the increase in the chemical shift effect at the higher field strength can be severe. Because the motivation in going to 7T is often to improve the detection of short T2 resonances the use of whole slice, spin echo acquisitions may be preferred (56). Figure 6 illustrates an example of such 3D 1H MRSI data acquired with TE=20ms, TR=2s from a patient with a brain tumor in 10 minutes using a 32 channel receive coil with an 18×24×8 matrix, nominal voxel size 1cc, echo planar encoding in one dimension and phase encoding in the other two dimensions (21). Two interleaves were required to provide a spectral bandwidth of 1900 Hz. After applying automatically defined VSS bands to sharpen the slice profile, good quality spectra were obtained from 4 distinct slices. Water suppression used the VAPOR method and additional lipid suppression was achieved by a chemical shift selective adiabatic inversion.
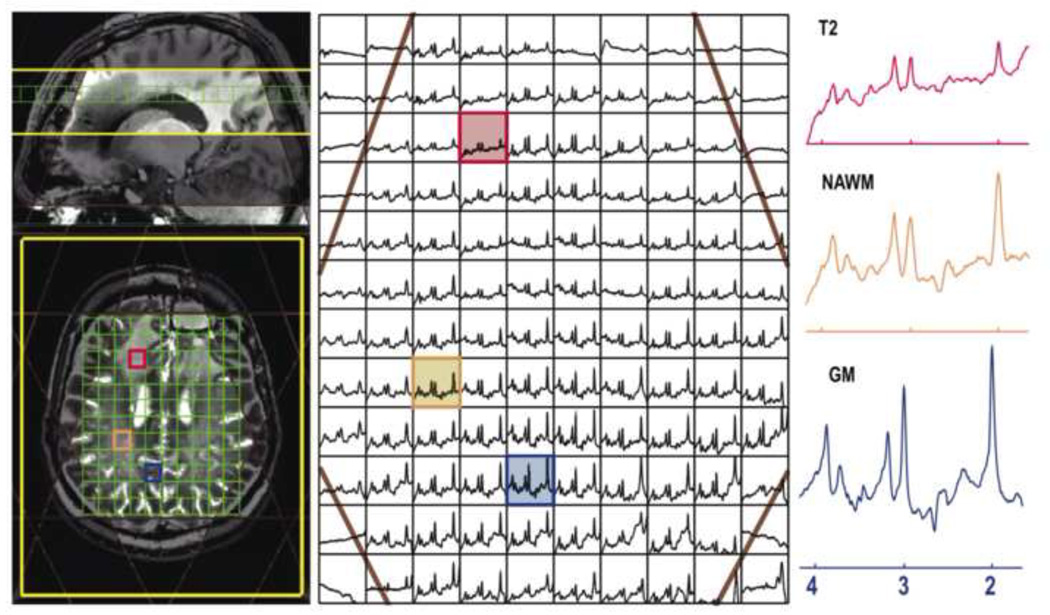
Figure 6.
One slice from a 3D MRSI dataset from a patient with anaplastic astrocytoma (TE/TR = 30/2000 ms; spectral array = 18×22×8; nominal spatial resolution = 1 cm3; total acquisition time ∼ 10 min). Note that the baseline has not been removed from the spectra that are shown. Eight VSS bands were prescribed automatically based on the position of the selected slice. The voxel labeled as T2 comes from the region of hyperintensity on the T2-weighted image, the voxel marker WM is from normal appearing white matter and the voxel labeled GM is from normal appearing grey matter.
To overcome the dual problems of minimizing the effects of short T2 and the limited B1 available at 7T, Henning et al (57) developed a sequence that combined slice selective excitation and OVS with the acquisition of free induction decays rather than echos. With this method, which they named FIDLOVS, they obtained estimates of levels of glutamate, glutamine, γ-aminobutyric acid and glutathione. Other options that applied high bandwidth pulses for volume localization included PRESS with spectral spectral pulses and TE = 80 or 35ms (58), interleaved narrow band PRESS with TE=90ms (8), slice select with semi-LASER using a TE of 50ms (10) or 19ms (59). For these longer TE acquisitions, optimizing the sub-echo time sets for point-resolved spectroscopy (PRESS) can also provide separation of glutamate, glutamine, glutathione and γ-aminobutyric acid without the need for sophisticated editing sequences (60). Increased interest in studying these metabolites for assessing abnormalities in cognitive function and for evaluating new treatment paradigms has placed a greater emphasis on developing robust tools for obtaining quantitative estimates of differences between control and patient populations.
ACQUISITION OF HYPERPOLARIZED C-13 MRSI DATA
The recent development of instrumentation for generating hyperpolarized 13C compounds and using them to evaluate in vivo metabolism (61,62) has produced new challenges for MRSI methodology. In this case the polarization is generated outside of the subject and the compound of interest is injected intravenously. T1 decay, which is typically of the order of 30–60s for the hyperpolarized agents that have been considered, is a major limitation on the acquisition protocols that can be used. Getting the agent from the polarizer to the syringe used for injection into the subject is the first hurdle to overcome. For human studies this includes not only the time required for dissolution but also for obtaining results from the quality control system that is being used to ensure the integrity of the agent. For the prototype system used in the first proof of concept human study the mean time to get from being at 1.2°K in the polarizer to being injected into the subject was 66s (63). The next issues to be considered are the rate of vascular delivery and of converting the hyperpolarized agent to its metabolic products. To make sure that the appropriate signal is captured, data must either be obtained at multiple time points or with prior knowledge of the kinetic processes involved. Another critical issue in designing the pulse sequence and data acquisition protocol is that the magnetization is not renewable and so each excitation reduces the signal available from the hyperpolarized agent. As indicated previously, using echo planar or spiral k-space trajectories in conjunction with parallel imaging and compressed sensing strategies is essential for efficiently sampling the rapidly decaying 13C signal (18,19,40–42).
Early pre-clinical studies performed in rodents used a combination of concentric phase encoding, elliptical k-space sampling and variable flip angle schemes to reduce the total sampling time and make efficient use of the available magnetization (64,65). These sequences typically took 10 to 20 sec to acquire 2D MRSI data from a single slice. Echo planar sampling using fly-back or symmetric trapezoidal trajectories was then added to obtain volumetric coverage within the same time (18). Spiral k-space sampling with multiple interleaves was applied to reduce the acquisition time by simultaneous encoding 2 spatial and 1 spectral dimensions (19).
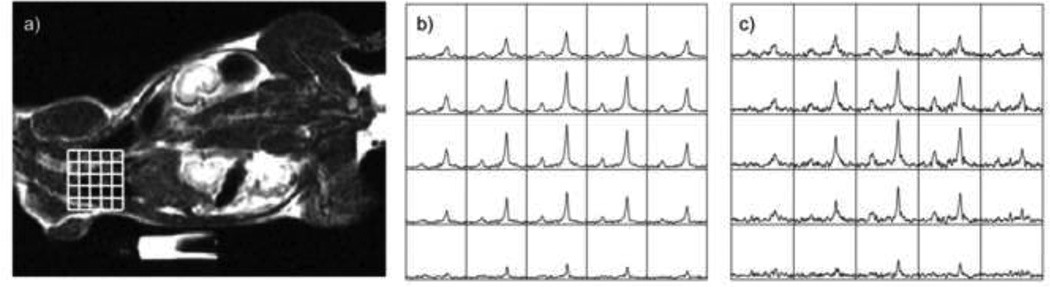
While this technology would clearly benefit from the use of parallel imaging with receiver coil arrays (35), there have been a relatively small number of such studies to date. This is partly due to the limited number of applications to larger animals or humans and partly to the limited availability of multi-nuclear, multi-channel rf coils. The example shown in Figure 7 is from a hyperpolarized 13C dataset from a rat obtained using a 3-channel coil that lay underneath the subject. An R factor of 2 was used to increase the spatial resolution by a factor of 2 without increasing the acquisition time or number of excitations (66). The fully sampled data were zero filled during reconstruction to provide a voxel by voxel comparison. In this case, the maps of coil sensitivities were obtained by replacing the rat with a large phantom containing a 13C labeled solution and acquiring another MRSI dataset. Clearly this strategy may not work for more general applications and self-calibrating parallel imaging methods such as GRAPPA are likely to be more appropriate. Issues in using such methods for hyperpolarized applications is that the encoding matrix is much smaller than for standard imaging protocols and the 13C signal comes from a relatively small region of the object of interest. Further study with simulations and comparison datasets will be needed to determine which approach is the most reliable. As the number of applications expands, this will become an essential part of the study design.
Figure 7.
Examples of 13C MRSI data obtained following injection of hyperpolarized 13C pyruvate. a) image and MRSI data obtained from the heart of a rat using b) fully sampled data and c) data reconstructed with SENSE in order to improve the spatial resolution by a factor of 2. The rf coil used had 3 element in the RL direction. The total acquisition time was 15s.
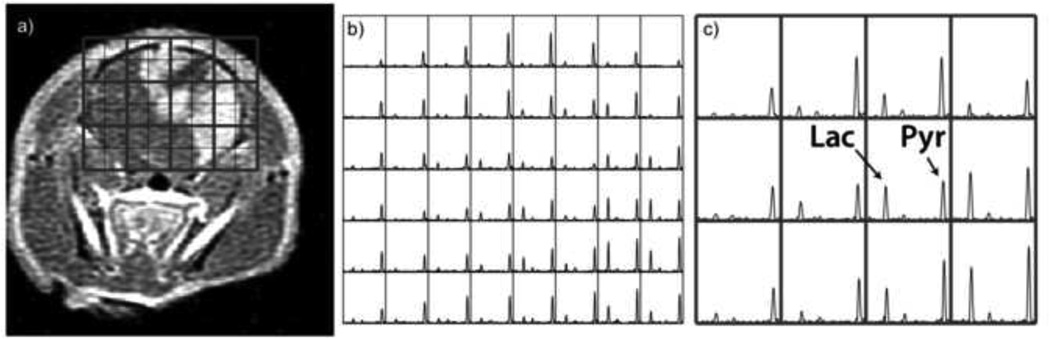
As noted previously, one approach for speeding up data acquisition that does not require multiple coils is compressed sensing (8). An example of data from the brain of a rat is shown in Figure 8. In this case, a speed up factor 4 was applied to increase the in-plane spatial resolution to better evaluate the heterogeneous tumor (42). Recent studies in other rodent models have used a 13C 3D-MRSI sequence with a factor of 7.5 acceleration by pseudorandom undersampling in kf-kx-ky space by using x and y gradient blips during a flyback readout in conjunction with non-linear reconstruction (40). These methods can be applied to acquire large arrays of spatially localized data with improved coverage or finer spatial resolution and dynamic arrays of spectral data with a time resolution of 3–5s in order to estimate rates of conversion from pyruvate to lactate (36).
Figure 8.
Application of compressed sensing with a speed-up factor of b) 4 and c) fully sampled hyperpolarized 13C MRSI data that were acquired from the brain of a rat with an implanted U87 tumor.
An alternative strategy to MRSI is to use frequency selective imaging of a single resonance using spectral-spatial RF pulses followed by spiral or echo planar imaging readouts for rapid imaging of selected metabolites (67). These methods typically have very short repetition time, allowing dynamic imaging of multiple metabolites. For most hyperpolarized 13C imaging experiments, the substrate signal is much higher than its metabolic products. In this case, multi-band spectral-spatial excitation pulses that uses a low excitation flip angle for the injected substrate to preserve its magnetization and a higher flip angle for its products can increase SNR for dynamic studies (68,69). Figure 9 is an example of 2D dynamic MRSI from tumor-bearing rat brain using multi-band RF excitation pulses with 30° and 5° flip angle for lactate and pyruvate, respectively. Similar strategies were used for evaluating changes in hyperpolarized signals for patients with prostate cancer using a volume C-13 transmit and H-1/C-13 endorectal coil for reception (see Figure 10). Whether the best solution for obtaining such data in the future is by using derivations of such MRSI sequences or by frequency selective lactate and pyruvate images is yet to be determined. In either case, it is clear that to obtain volumetric data with sufficient coverage for the fields of view relevant to studies in the human abdomen or brain it will be necessary to integrate parallel acquisition and compressed sensing strategies.
Figure 9.
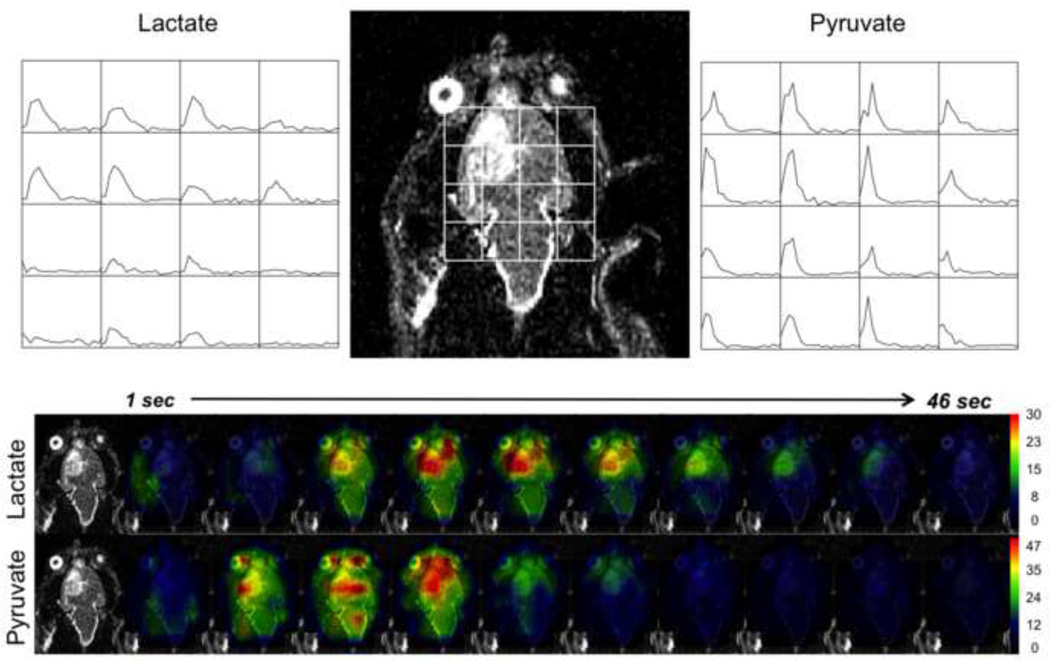
Dynamic 13C EPSI data with a time resolution of 3s from the brain of a rat with an implanted tumor obtained following injection of hyperpolarized 13C pyruvate. The arrays of dynamic curves from lactate and pyruvate cover a 45s time period starting from the end of the injection. The color overlay images represent the spatial distribution of changes in levels of lactate and pyruvate during the same period. These data were displayed using the SIVIC package (44).
Figure 10.
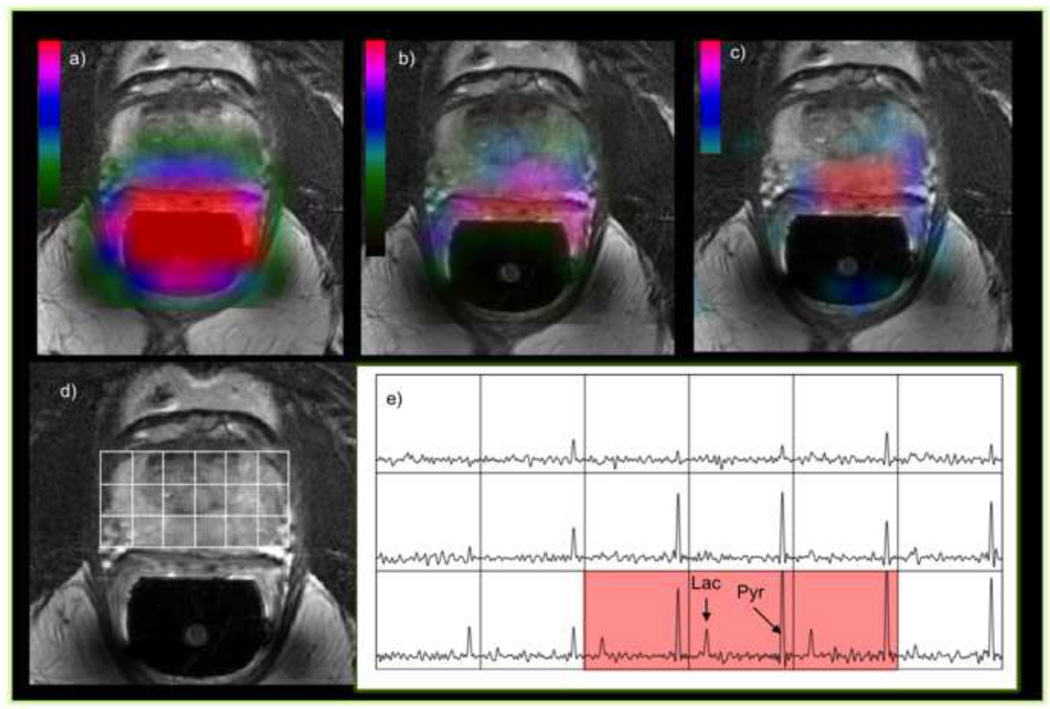
Some of the first 3-D MRSI data obtained with 1-D EPSI and 2 directions of phase encoding from a patient with prostate cancer. The left-most color overlay a) is the profile of the dual 1H/13C endorectal coil used for reception of the signals, the middle overlay b) is of pyruvate and the right most c) is of lactate. The reference image d) and spectral array e) show a portion of the 3D array that covers the tumor region. The voxels highlighted in red have lactate/pyruvate ratio that is indicative of tumor. The acquisition time for this 3D array was 12s, the matrix was 18×8×8 with a 7mm spatial resolution, a progressive flip angle, TE/TR=3/85-125ms, starting at 28s after the end of the injection of a dose of 0.43mL/Kg of hyperpolarized pyruvate.
CONCLUSIONS AND FUTURE DIRECTIONS
The field of 1H MRSI is relatively mature in that a large number of different pulses sequences and applications have been demonstrated in a research setting. These methods can be used to provide robust short and long TE volumetric data from the brain with a spatial resolution of 0.3–1cc and an acquisition time of 5–10 minutes. Similar results have been demonstrated in the prostate but the applications to other organ systems have been more limited. With the widespread availability of 3T and increasing number of 7T whole body scanners that are being installed it is time to integrate the methods that have been described here into a comprehensive package of tools that can be integrated with more routine MR imaging technologies and that is made available to the research and clinical communities. The development of hyperpolarized 13C MRSI is extremely promising for obtaining rapid metabolic data that can be used to evaluate pathology and monitor response to therapy. The growing number of applications in pre-clinical models has stimulated the development of new pulse sequences and will continue to be an active area for research into new MRSI methods.
HIGHLIGHTS.
Acquisition times for in vivo 3D 1H MRSI can be reduced to 4–5 minutes
Compressed sensing and parallel imaging are important for hyperpolarized 13C MRSI
Automated prescription of OVS gives robust 1H MRSI data with improved coverage
Spin echo 1H MRSI can provide improved quantification of short TE metabolites at 7T
Dynamic hyperpolarized 13C MRSI can be performed with 3–5 second time resolution
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Nelson SJ. Analysis of volume MRI and MR spectroscopic imaging data for the evaluation of patients with brain tumors. Magn Reson Med. 2001;46:228–239. doi: 10.1002/mrm.1183. [DOI] [PubMed] [Google Scholar]
- 2.Kaiser LG, Young K, Matson GB. Elimination of spatial interference in PRESS-localized editing spectroscopy. Magn Reson Med. 2007 Oct;58(4):813–818. doi: 10.1002/mrm.21407. [DOI] [PubMed] [Google Scholar]
- 3.Park I, Chen AP, Zierhut ML, Ozturk-Isik E, Vigneron DB, Nelson SJ. Implementation of 3 T lactate-edited 3D 1H MR spectroscopic imaging with flyback echo-planar readout for gliomas patients. Ann Biomed Eng. 2011 Jan;39(1):193–204. doi: 10.1007/s10439-010-0128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Star-Lack J, Vigneron DB, Pauly J, Kurhanewicz J, Nelson SJ. Improved solvent suppression and increased spatial excitation bandwidths for three-dimensional PRESS CSI using phase-compensating spectral/spatial spin-echo pulses. J Magn Reson Imaging. 1997 Jul-Aug;7(4):745–757. doi: 10.1002/jmri.1880070422. [DOI] [PubMed] [Google Scholar]
- 5.Adalsteinsson E, Irarrazabal P, Topp S, Meyer C, Macovski A, Spielman DM. Volumetric spectroscopic imaging with spiral-based k-space trajectories. Magn Reson Med. 1998 Jun;39(6):889–898. doi: 10.1002/mrm.1910390606. [DOI] [PubMed] [Google Scholar]
- 6.Zhu H, Ouwerkerk R, Barker PB. Dual-band water and lipid suppression for MR spectroscopic imaging at 3 Tesla. Magn Reson Med. 2010;63(6):1486–1492. doi: 10.1002/mrm.22324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu D, Chen AP, Cunningham C, Osorio JA, Nelson SJ, Vigneron DB. Spectroscopic imaging of the brain with phased-array coils at 3.0 T. Magn Reson Imaging. 2006;24(1):69–74. doi: 10.1016/j.mri.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 8.Balchandani P, Pauly J, Spielman D. Interleaved narrow-band PRESS sequence with adiabatic spatial-spectral refocusing pulses for 1H MRSI at 7T. Magnetic resonance in medicine. 2008;59:973–979. doi: 10.1002/mrm.21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheenen TW, Klomp DW, Wijnen JP, Heerschap A. Short echo time 1H-MRSI of the human brain at 3T with minimal chemical shift displacement errors using adiabatic refocusing pulses. Magn Reson Med. 2008 Jan;59(1):1–6. doi: 10.1002/mrm.21302. [DOI] [PubMed] [Google Scholar]
- 10.Scheenen TW, Heerschap A, Klomp DW. Towards 1H-MRSI of the human brain at 7T with slice-selective adiabatic refocusing pulses. Magma. 2008;21:95–101. doi: 10.1007/s10334-007-0094-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tran TK, Vigneron DB, Sailasuta N, Tropp J, Le Roux P, Kurhanewicz J, Nelson S, Hurd R. Very selective suppression pulses for clinical MRSI studies of brain and prostate cancer. Magn Reson Med. 2000;43(1):23–33. doi: 10.1002/(sici)1522-2594(200001)43:1<23::aid-mrm4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 12.Posse S, Tedeschi G, Risinger R, Ogg R, Le Bihan D. High speed 1H spectroscopic imaging in human brain by echo planar spatial-spectral encoding. Magn Reson Med. 1995 Jan;33(1):34–40. doi: 10.1002/mrm.1910330106. [DOI] [PubMed] [Google Scholar]
- 13.Ebel A, Govindaraju V, Maudsley AA. Comparison of inversion recovery preparation schemes for lipid suppression in 1H MRSI of human brain. Magn Reson Med. 2003 May;49(5):903–908. doi: 10.1002/mrm.10444. [DOI] [PubMed] [Google Scholar]
- 14.Maudsley AA, Domenig C, Sheriff S. Reproducibility of serial whole-brain MR spectroscopic imaging. NMR in biomedicine. 2010;23(3):251–256. doi: 10.1002/nbm.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andronesi OC, Gagoski BA, Sorensen AG. Neurologic 3D MR spectroscopic imaging with low-power adiabatic pulses and fast spiral acquisition. Radiology. 2012;262(2):647–661. doi: 10.1148/radiol.11110277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andronesi OC, Ramadan S, Ratai EM, Jennings D, Mountford CE, Sorensen AG. Spectroscopic imaging with improved gradient modulated constant adiabaticity pulses on high-field clinical scanners. J Magn Reson. 2010 Apr;203(2):283–293. doi: 10.1016/j.jmr.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham CH, Vigneron DB, Chen AP, Xu D, Nelson SJ, Hurd RE, Kelley DA, Pauly JM. Design of flyback echo-planar readout gradients for magnetic resonance spectroscopic imaging. Magn Reson Med. 2005;54(5):1286–1289. doi: 10.1002/mrm.20663. [DOI] [PubMed] [Google Scholar]
- 18.Zierhut ML, Ozturk-Isik E, Chen AP, Park I, Vigneron DB, Nelson SJ. (1)H spectroscopic imaging of human brain at 3 Tesla: comparison of fast three-dimensional magnetic resonance spectroscopic imaging techniques. J Magn Reson Imaging. 2009 Sep;30(3):473–480. doi: 10.1002/jmri.21834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cunningham CH, Chen AP, Albers MJ, Kurhanewicz J, Hurd RE, Yen Y-F, Pauly JM, Nelson SJ, Vigneron DB. Double spin-echo sequence for rapid spectroscopic imaging of hyperpolarized 13C. J Magn Reson. 2007;187(2):357–362. doi: 10.1016/j.jmr.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 20.Mayer D, Yen Y-F, Tropp J, Pfefferbaum A, Hurd RE, Spielman DM. Application of subsecond spiral chemical shift imaging to real-time multislice metabolic imaging of the rat in vivo after injection of hyperpolarized 13C1-pyruvate. Magn Reson Med. 2009;62(3):557–564. doi: 10.1002/mrm.22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Larson P, Chen AP, Kelley DAC, Chang SM, et al. Characterization of gliomas using MRI and short echo 1-H MRSI at 7 Tesla; Proceedings of the 20th Annual Meeting of ISMRM.2012. [Google Scholar]
- 22.Wald LL, Moyher SE, Day MR, Nelson SJ, Vigneron DB. Proton spectroscopic imaging of the human brain using phased array detectors. Magn Reson Med. 1995;34(3):440–445. doi: 10.1002/mrm.1910340322. [DOI] [PubMed] [Google Scholar]
- 23.Prock T, Collins DJ, Dzik-Jurasz AS, Leach MO. An algorithm for the optimum combination of data from arbitrary magnetic resonance phased array probes. Phys Med Biol. 2002 Jan 21;47(2):N39–N46. doi: 10.1088/0031-9155/47/2/402. [DOI] [PubMed] [Google Scholar]
- 24.Mark A. Brown, Time Domain Combination of MR spectroscopy data acquired using phased array coils. Mag. Res. Med. 2005;52:1207–1214. doi: 10.1002/mrm.20244. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Osorio JA, Ozturk-Isik E, Chen AP, Xu D, Crane JC, Cha S, Chang S, Berger MB, Vigneron DB, Nelson SJ. Considerations in applying 3-D PRESS H-1 brain MRSI with an 8-channel phased array coil at 3 T. Magn Reson Imaging. 2006 Dec;24(10):1295–1302. doi: 10.1016/j.mri.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 26.Osorio JA, Ozturk E, Xu D, Cha S, Chang S, Berger MS, Vigneron DB, Nelson SJ. 3D MRSI of Brain Tumors at 3.0 Tesla using an 8-Channel Phased Array Head Coil. J Magn Reson Imaging. 2007 Jul;26(1):23–30. doi: 10.1002/jmri.20970. [DOI] [PubMed] [Google Scholar]
- 27.Crane JC, Crawford FW, Nelson J. Grid Enabled Magnetic Resonance Scanners for Near Real-Time Medical Image Processing. Journal of Parallel and Distributed Computing. 2006;66(12):1524–1533. [Google Scholar]
- 28.Dydak U, Weiger M, Pruessmann KP, Meier D, Boesiger P. Sensitivity-encoded spectroscopic imaging. Magn Reson Med. 2001;46(4):713–722. doi: 10.1002/mrm.1250. [DOI] [PubMed] [Google Scholar]
- 29.Dydak U, Pruessmann KP, Weiger M, Tsao J, Meier D, Boesiger P. Parallel spectroscopic imaging with spin-echo trains. Magn Reson Med. 2003;50(1):196–200. doi: 10.1002/mrm.10495. [DOI] [PubMed] [Google Scholar]
- 30.Tsai SY, Otazo R, Lin YR, Chung HW, Wald LL, Wiggins GC, Lin FH. Accelerated proton echo planar spectroscopic imaging (PEPSI) using GRAPPA with a 32-channel phased-array coil. Magn Reson Med. 2008 May;59(5):989–998. doi: 10.1002/mrm.21545. [DOI] [PubMed] [Google Scholar]
- 31.Lin FH, Tsai SY, Otazo R, Caprihan A, Wald LL, Belliveau JW, Posse S. Sensitivity-encoded (SENSE) proton echo-planar spectroscopic imaging (PEPSI) in the human brain. Magn Reson Med. 2007 Feb;57(2):249–257. doi: 10.1002/mrm.21119. [DOI] [PubMed] [Google Scholar]
- 32.Ozturk-Isik E, Crane JC, Cha S, Chang SM, Berger MS, Nelson SJ. Unaliasing lipid contamination for MR spectroscopic imaging of gliomas at 3T using sensitivity encoding (SENSE) Magn Reson Med. 2006;55(5):1164–1169. doi: 10.1002/mrm.20860. [DOI] [PubMed] [Google Scholar]
- 33.Ozturk E, Banerjee S, Majumdar S, Nelson SJ. Partially parallel MR spectroscopic imaging of gliomas at 3T. Conf Proc IEEE Eng Med Biol Soc. 2006;1:493–496. doi: 10.1109/IEMBS.2006.259793. [DOI] [PubMed] [Google Scholar]
- 34.Banerjee S, Ozturk-Isik E, Nelson SJ, Majumdar S. Elliptical magnetic resonance spectroscopic imaging with GRAPPA for imaging brain tumors at 3 T. Magn Reson Imaging. 2009 Dec;27(10):1319–1325. doi: 10.1016/j.mri.2009.05.031. [DOI] [PubMed] [Google Scholar]
- 35.Zhu X, Ebel A, Ji JX, Schuff N. Spectral phase-corrected GRAPPA reconstruction of three-dimensional echo-planar spectroscopic imaging (3D-EPSI) Magn Reson Med. 2007 May;57(5):815–820. doi: 10.1002/mrm.21217. [DOI] [PubMed] [Google Scholar]
- 36.Ozturk-Isik E, Chen AP, Crane JC, Bian W, Xu D, Han ET, Chang SM, Vigneron DB, Nelson SJ. 3D sensitivity encoded ellipsoidal MR spectroscopic imaging of gliomas at 3T. Magn Reson Imaging. 2009 Nov;27(9):1249–1257. doi: 10.1016/j.mri.2009.05.028. Epub 2009 Sep 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mishkovsky M, Frydman L. Principles and progress in ultrafast multidimensional nuclear magnetic resonance. Annu Rev Phys Chem. 2009;60:429–448. doi: 10.1146/annurev.physchem.040808.090420. [DOI] [PubMed] [Google Scholar]
- 38.Ben-Eliezer N, Frydman L. Spatiotemporal encoding as a robust basis for fast three- dimensional in vivo MRI. NMR Biomed. 2011 Dec;24(10):1191–1201. doi: 10.1002/nbm.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt R, Frydman L. In vivo 3D spatial/1D spectral imaging by spatiotemporal encoding: A new single-shot experimental and processing approach. Magn Reson Med. 2012 Sep;:24. doi: 10.1002/mrm.24470. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 40.Hu S, Lustig M, Chen AP, Crane J, Kerr A, Kelley DAC, Hurd R, Kurhanewicz J, Nelson SJ, Pauly JM, Vigneron DB. Compressed sensing for resolution enhancement of hyperpolarized 13C flyback 3D-MRSI. J Magn Reson. 2008;192(2):258–264. doi: 10.1016/j.jmr.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu S, Lustig M, Balakrishnan A, Larson PEZ, Bok R, Kurhanewicz J, Nelson SJ, Goga A, Pauly JM, Vigneron DB. 3D compressed sensing for highly accelerated hyperpolarized (13)C MRSI with in vivo applications to transgenic mouse models of cancer. Magn Reson Med. 2010;63(2):312–321. doi: 10.1002/mrm.22233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park I, Hu S, Bok R, Ozawa T, Ito M, Mukherjee J, Phillips JJ, James CD, Pieper RO, Ronen SM, Vigneron DB, Nelson SJ. Evaluation of heterogeneous metabolic profile in an orthotopic human glioblastoma xenograft model using compressed sensing hyperpolarized 3D 13C magnetic resonance spectroscopic imaging. Magn Reson Med. 2012 Jul;:31. doi: 10.1002/mrm.24434. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ozhinsky E, Vigneron DB, Nelson SJ. Improved spatial coverage for brain 3D PRESS MRSI by automatic placement of outer-volume suppression saturation bands. Journal of magnetic resonance imaging: JMRI. 2011;33(4):792–802. doi: 10.1002/jmri.22507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Star-Lack J, Nelson SJ, Kurhanewicz J, Huang LR, Vigneron DB. Improved water and lipid suppression for 3D PRESS CSI using RF band selective inversion with gradient dephasing (BASING) Magn Reson Med. 1997;38(2):311–321. doi: 10.1002/mrm.1910380222. [DOI] [PubMed] [Google Scholar]
- 45.Ozhinsky E, Vigneron DB, Chang SM, Nelson SJ. Automated prescription of oblique brain 3D magnetic resonance spectroscopic imaging. Magn Reson Med. 2012 Jun;:12. doi: 10.1002/mrm.24339. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. http://sourceforge.net/apps/trac/sivic/
- 47.Martinez-Ramon M, Gallardo-Antolin A, Cid-Sueiro J, Heileman GL, Yung KT, Zheng W, Zhao C, Posse S. Automatic placement of outer volume suppression slices in MR spectroscopic imaging of the human brain. Magn Reson Med. 2010;63(3):592–600. doi: 10.1002/mrm.22275. [DOI] [PubMed] [Google Scholar]
- 48.Yung KT, Zheng W, Zhao C, Martinez-Ramon M, van der Kouwe A, Posse S. Atlas-based automated positioning of outer volume suppression slices in short-echo time 3D MR spectroscopic imaging of the human brain. Magnetic resonance in medicine. 2011;66(4):911–922. doi: 10.1002/mrm.22887. [DOI] [PubMed] [Google Scholar]
- 49.Otazo R, Mueller B, Ugurbil K, Wald L, Posse S. Signal-to-noise ratio and spectral linewidth improvements between 1.5 and 7 Tesla in proton echo-planar spectroscopic imaging. Magn Reson Med. 2006;56:1200–1210. doi: 10.1002/mrm.21067. [DOI] [PubMed] [Google Scholar]
- 50.Mekle R, Mlynarik V, Gambarota G, Hergt M, Krueger G, et al. MR spectroscopy of the human brain with enhanced signal intensity at ultrashort echo times on a clinical platform at 3T and 7T. Magn Reson Med. 2009;61:1279–1285. doi: 10.1002/mrm.21961. [DOI] [PubMed] [Google Scholar]
- 51.Li Y, Xu D, Chen AP, Vigneron DB, Nelson SJ. Proton spectroscopy of human brain at 3T and 7T: signal-to-noise ratio, spectral linewidth and relaxation times; Proceedings of the 16th Annual Meeting of ISMRM, 2008.2008. [Google Scholar]
- 52.Srinivasan R, Ratiney H, Hammond-Rosenbluth KE, Pelletier D, Nelson SJ. MR spectroscopic imaging of glutathione in the white and gray matter at 7 T with an application to multiple sclerosis. Magn Reson Imaging. 2010 Feb;28(2):163–170. doi: 10.1016/j.mri.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 53.Pan JW, Lo KM, Hetherington HP. Role of very high order and degree B(0) shimming for spectroscopic imaging of the human brain at 7 tesla. Magnetic resonance in medicine. 2012;68:1007–1017. doi: 10.1002/mrm.24122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boer VO, Klomp DW, Juchem C, Luijten PR, de Graaf RA. Multislice (1) H MRSI of the human brain at 7 T using dynamic B(0) and B(1) shimming. Magnetic resonance in medicine. 2012;68:662–670. doi: 10.1002/mrm.23288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Avdievich NI, Pan JW, Baehring JM, Spencer DD, Hetherington HP. Short echo spectroscopic imaging of the human brain at 7T using transceiver arrays. Magnetic resonance in medicine. 2009;62:17–25. doi: 10.1002/mrm.21970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu H, Soher BJ, Ouwerkerk R, Schar M, Barker PB. Spin-echo magnetic resonance spectroscopic imaging at 7 T with frequency-modulated refocusing pulses. Magn Reson Med. 2012 Jun;:12. doi: 10.1002/mrm.24357. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henning A, Fuchs A, Murdoch JB, Boesiger P. Slice-selective FID acquisition, localized by outer volume suppression (FIDLOVS) for (1)H-MRSI of the human brain at 7 T with minimal signal loss. NMR Biomed. 2009;22:683–696. doi: 10.1002/nbm.1366. [DOI] [PubMed] [Google Scholar]
- 58.Xu D, Cunningham CH, Chen AP, Li Y, Kelley DA, Mukherjee P, Pauly JM, Nelson SJ, Vigneron DB. Phased array 3D MR spectroscopic imaging of the brain at 7 T. Magn Reson Imaging. 2008 Nov;26(9):1201–1206. doi: 10.1016/j.mri.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fuchs A, Luttje M, Boesiger P, Henning A. SPECIAL semi-LASER with lipid artifact compensation for (1) H MRS at 7 T. Magn Reson Med. 2012 Apr;:19. doi: 10.1002/mrm.24292. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 60.Choi C, Dimitrov IE, Douglas D, Patel A, Kaiser LG, et al. Improvement of resolution for brain coupled metabolites by optimized (1)H MRS at 7T. NMR Biomed. 2010;23:1044–1052. doi: 10.1002/nbm.1529. [DOI] [PubMed] [Google Scholar]
- 61.Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc Natl Acad Sci U S A. 2003;100(18):10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Golman K, Zandt R, Thaning M. Real-time metabolic imaging. Proc Natl Acad Sci USA. 2006;103(30):11270–11275. doi: 10.1073/pnas.0601319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nelson S, Kurhanewicz J, Vigneron DB, Larson P, Harzstark A, Ferrone M, van Criekinge M, Chang J, Bok R, Park I, Reed G, Carvajal L, Ardenkjaer-Larson JH, Chen A, Hurd R, Odegardstuen L, Tropp J. Proof of Concept Clinical Trial of Hyperpolarized C-13 Pyruvate in Patients with Prostate Cancer; Proceedings of the 20th Annual Meeting of ISMRM.2012. [Google Scholar]
- 64.Kohler SJ, Yen Y, Wolber J, Chen AP, Albers MJ, Bok R, Zhang V, Tropp J, Nelson S, Vigneron DB, Kurhanewicz J, Hurd RE. In vivo 13 carbon metabolic imaging at 3T with hyperpolarized 13C-1-pyruvate. Magn Reson Med. 2007;58(1):65–69. doi: 10.1002/mrm.21253. [DOI] [PubMed] [Google Scholar]
- 65.Chen AP, Albers MJ, Cunningham CH, Kohler SJ, Yen Y-F, Hurd RE, Tropp J, Bok R, Pauly JM, Nelson SJ, Kurhanewicz J, Vigneron DB. Hyperpolarized C-13 spectroscopic imaging of the TRAMP mouse at 3T-initial experience. Magn Reson Med. 2007;58(6):1099–1106. doi: 10.1002/mrm.21256. [DOI] [PubMed] [Google Scholar]
- 66.Tropp J, Lupo JM, Chen A, Calderon P, McCune D, Grafendorfer T, Ozturk-Isik E, Larson PEZ, Hu S, Yen Y-F, Robb F, Bok R, Schulte R, Xu D, Hurd R, Vigneron D, Nelson S. Multichannel metabolic imaging, with SENSE reconstruction, of hyperpolarized [1-(13)C] pyruvate in a live rat at 3.0 tesla on a clinical MR scanner. JMagn Reson. 2011;208(1):171–177. doi: 10.1016/j.jmr.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lau AZ, Chen AP, Ghugre NR, Ramanan V, Lam WW, Connelly KA, Wright GA, Cunningham CH. Rapid multislice imaging of hyperpolarized 13C pyruvate and bicarbonate in the heart. Magn Reson Med. 2010;64(5):1323–1331. doi: 10.1002/mrm.22525. [DOI] [PubMed] [Google Scholar]
- 68.Larson PEZ, Kerr AB, Chen AP, Lustig MS, Zierhut ML, Hu S, Cunningham CH, Pauly JM, Kurhanewicz J, Vigneron DB. Multiband excitation pulses for hyperpolarized 13C dynamic chemical-shift imaging. J Magn Reson. 2008;194(1):121–127. doi: 10.1016/j.jmr.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Larson PEZ, Bok R, Kerr AB, Lustig M, Hu S, Chen AP, Nelson SJ, Pauly JM, Kurhanewicz J, Vigneron DB. Investigation of tumor hyperpolarized [1-13C]- pyruvate dynamics using time-resolved multiband RF excitation echo-planar MRSI. Magn. Reson. Med. 2010;63:582–5891. doi: 10.1002/mrm.22264. [DOI] [PMC free article] [PubMed] [Google Scholar]