Abstract
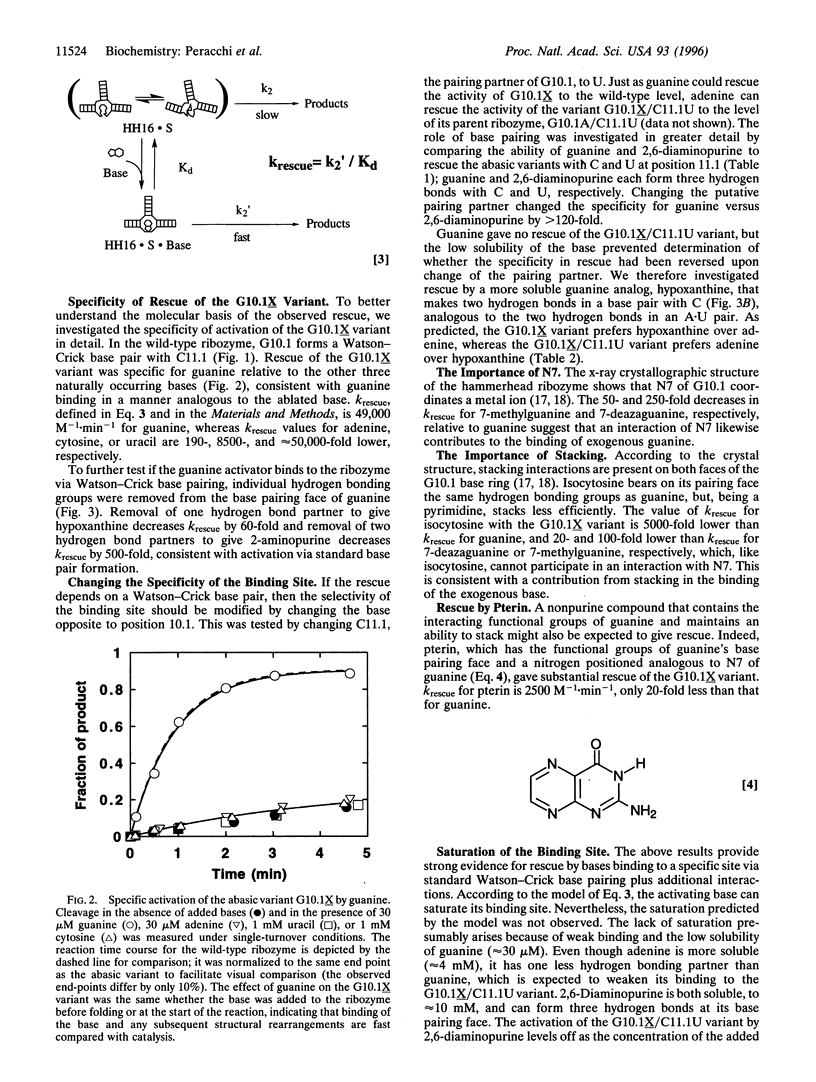
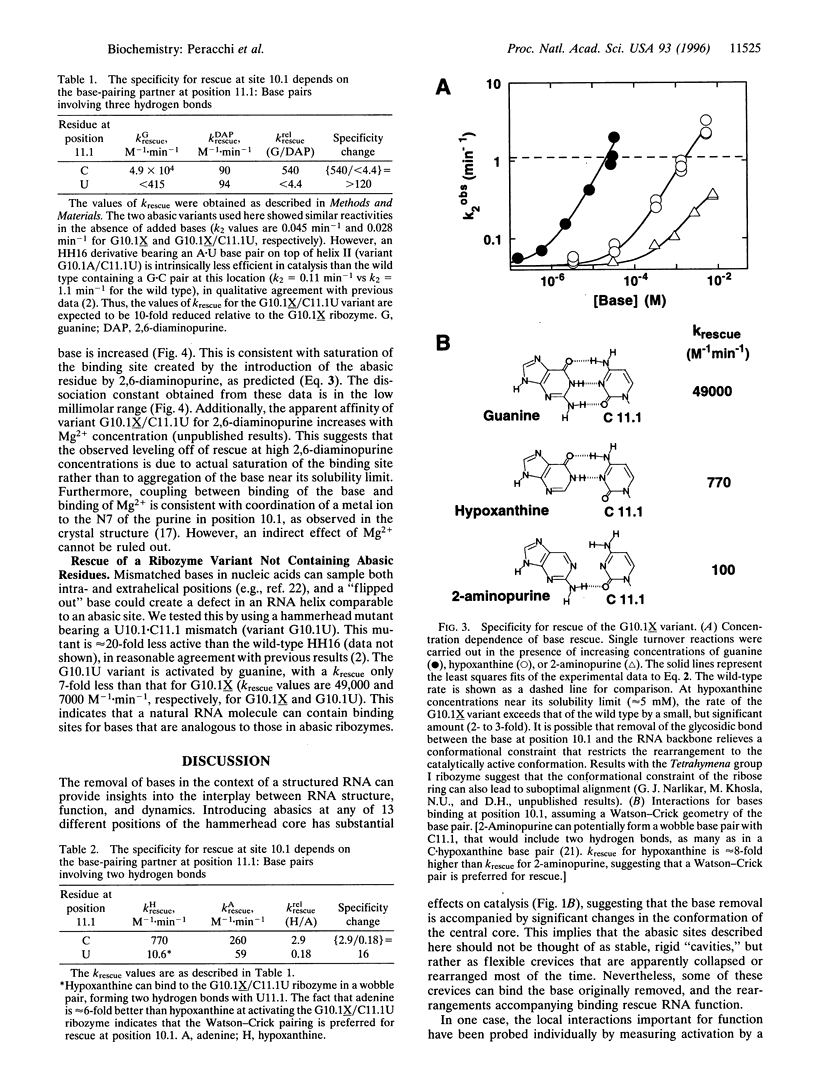
We have synthesized 13 hammerhead ribozyme variants, each containing an abasic residue at a specific position of the catalytic core. The activity of each of the variants is significantly reduced. In four cases, however, activity can be rescued by exogenous addition of the missing base. For one variant, the rescue is 300-fold; for another, the rescue is to the wild-type level. This latter abasic variant (G10.1X) has been characterized in detail. Activation is specific for guanine, the base initially removed. In addition, the specificity for guanine versus adenine is substantially altered by replacing C with U in the opposite strand of the ribozyme. These results show that a binding site for a small, noncharged ligand can be created in a preexisting ribozyme structure. This has implications for structure-function analysis of RNA, and leads to speculations about evolution in an "RNA world" and about the potential therapeutic use of ribozymes.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson P., Monforte J., Tritz R., Nesbitt S., Hearst J., Hampel A. Mutagenesis of the hairpin ribozyme. Nucleic Acids Res. 1994 Mar 25;22(6):1096–1100. doi: 10.1093/nar/22.6.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner S. A., Ellington A. D., Tauer A. Modern metabolism as a palimpsest of the RNA world. Proc Natl Acad Sci U S A. 1989 Sep;86(18):7054–7058. doi: 10.1073/pnas.86.18.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaber M., Baase W. A., Gassner N., Matthews B. W. Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent. J Mol Biol. 1995 Feb 17;246(2):317–330. doi: 10.1006/jmbi.1994.0087. [DOI] [PubMed] [Google Scholar]
- Bratty J., Chartrand P., Ferbeyre G., Cedergren R. The hammerhead RNA domain, a model ribozyme. Biochim Biophys Acta. 1993 Dec 14;1216(3):345–359. doi: 10.1016/0167-4781(93)90001-t. [DOI] [PubMed] [Google Scholar]
- Connell G. J., Yarus M. RNAs with dual specificity and dual RNAs with similar specificity. Science. 1994 May 20;264(5162):1137–1141. doi: 10.1126/science.7513905. [DOI] [PubMed] [Google Scholar]
- Cuniasse P., Fazakerley G. V., Guschlbauer W., Kaplan B. E., Sowers L. C. The abasic site as a challenge to DNA polymerase. A nuclear magnetic resonance study of G, C and T opposite a model abasic site. J Mol Biol. 1990 May 20;213(2):303–314. doi: 10.1016/S0022-2836(05)80192-5. [DOI] [PubMed] [Google Scholar]
- Eriksson A. E., Baase W. A., Wozniak J. A., Matthews B. W. A cavity-containing mutant of T4 lysozyme is stabilized by buried benzene. Nature. 1992 Jan 23;355(6358):371–373. doi: 10.1038/355371a0. [DOI] [PubMed] [Google Scholar]
- Fagan P. A., Fàbrega C., Eritja R., Goodman M. F., Wemmer D. E. NMR study of the conformation of the 2-aminopurine:cytosine mismatch in DNA. Biochemistry. 1996 Apr 2;35(13):4026–4033. doi: 10.1021/bi952657g. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Guest C. R., Hochstrasser R. A., Sowers L. C., Millar D. P. Dynamics of mismatched base pairs in DNA. Biochemistry. 1991 Apr 2;30(13):3271–3279. doi: 10.1021/bi00227a015. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Hertel K. J., Herschlag D., Uhlenbeck O. C. A kinetic and thermodynamic framework for the hammerhead ribozyme reaction. Biochemistry. 1994 Mar 22;33(11):3374–3385. doi: 10.1021/bi00177a031. [DOI] [PubMed] [Google Scholar]
- Hertel K. J., Pardi A., Uhlenbeck O. C., Koizumi M., Ohtsuka E., Uesugi S., Cedergren R., Eckstein F., Gerlach W. L., Hodgson R. Numbering system for the hammerhead. Nucleic Acids Res. 1992 Jun 25;20(12):3252–3252. doi: 10.1093/nar/20.12.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenison R. D., Gill S. C., Pardi A., Polisky B. High-resolution molecular discrimination by RNA. Science. 1994 Mar 11;263(5152):1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- Lord R. C., Thomas G. J., Jr Raman studies of nucleic acids. II. Aqueous purine and pyrimidine mixtures. Biochim Biophys Acta. 1967 Jun 20;142(1):1–11. [PubMed] [Google Scholar]
- Matulic-Adamic Jasenka, Beigelman Leonid, Portmann Stefan, Egli Martin, Usman Nassim. Synthesis and Structure of 1-Deoxy-1-phenyl-beta-D-ribofuranose and Its Incorporation into Oligonucleotides. J Org Chem. 1996 May 31;61(11):3909–3911. doi: 10.1021/jo960091b. [DOI] [PubMed] [Google Scholar]
- McConnell T. S., Cech T. R. A positive entropy change for guanosine binding and for the chemical step in the Tetrahymena ribozyme reaction. Biochemistry. 1995 Mar 28;34(12):4056–4067. doi: 10.1021/bi00012a024. [DOI] [PubMed] [Google Scholar]
- Pley H. W., Flaherty K. M., McKay D. B. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994 Nov 3;372(6501):68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- Pyle A. M., McSwiggen J. A., Cech T. R. Direct measurement of oligonucleotide substrate binding to wild-type and mutant ribozymes from Tetrahymena. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8187–8191. doi: 10.1073/pnas.87.21.8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi J. J. Ribozymes. Curr Opin Biotechnol. 1992 Feb;3(1):3–7. doi: 10.1016/0958-1669(92)90117-2. [DOI] [PubMed] [Google Scholar]
- Ruffner D. E., Stormo G. D., Uhlenbeck O. C. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990 Nov 27;29(47):10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- Sassanfar M., Szostak J. W. An RNA motif that binds ATP. Nature. 1993 Aug 5;364(6437):550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- Scott W. G., Finch J. T., Klug A. The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell. 1995 Jun 30;81(7):991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- Sigurdsson S. T., Eckstein F. Structure-function relationships of hammerhead ribozymes: from understanding to applications. Trends Biotechnol. 1995 Aug;13(8):286–289. doi: 10.1016/S0167-7799(00)88966-0. [DOI] [PubMed] [Google Scholar]
- Toney M. D., Kirsch J. F. Direct Brønsted analysis of the restoration of activity to a mutant enzyme by exogenous amines. Science. 1989 Mar 17;243(4897):1485–1488. doi: 10.1126/science.2538921. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Usman N., Cedergren R. Exploiting the chemical synthesis of RNA. Trends Biochem Sci. 1992 Sep;17(9):334–339. doi: 10.1016/0968-0004(92)90306-t. [DOI] [PubMed] [Google Scholar]
- Zhang X. J., Baase W. A., Matthews B. W. Multiple alanine replacements within alpha-helix 126-134 of T4 lysozyme have independent, additive effects on both structure and stability. Protein Sci. 1992 Jun;1(6):761–776. doi: 10.1002/pro.5560010608. [DOI] [PMC free article] [PubMed] [Google Scholar]


