Abstract
Plexiform neurofibromas commonly found in patients with Neurofibromatosis type I (NF1) have a 5% risk of being transformed into malignant peripheral nerve sheath tumors (MPNST). Germline mutations in the NF1 gene coding for neurofibromin, which is a Ras GTPase activating protein (RasGAP) and a negative regulator of Ras, result in an upregulation of the Ras pathway. We established a direct connection between neurofibromin deficiency and downstream effectors of Ras in cell lines from MPNST patients by demonstrating that knockdown of NF1 expression using siRNA in a NF1 wild type MPNST cell line, STS-26T, activates the Ras/ERK1,2 pathway and increases AP-1 binding and activity. We believe this is the first time the transactivation of AP-1 has been linked directly to neurofibromin deficiency in a disease relevant MPNST cell line. Previously, we have shown that N-Ras is constitutively activated in cell lines derived from independent MPNSTs from NF1 patients. We therefore sought to analyze the role of the N-Ras pathway in deregulating AP-1 transcriptional activity. We show that STS-26T clones conditionally expressing oncogenic N-Ras show increased phosphorylated ERK1,2 and phosphorylated JNK expression concomitant with increased AP-1 activity. MAP kinase pathways (ERK1,2 and JNK) were further examined in ST88-14, a neurofibromin-deficient MPNST cell line. The basal activity of ERK1,2 but not JNK was found to increase AP-1 activity. These experiments further confirmed the link between the loss of neurofibromin and increased activity of Ras/MAP kinase pathways and the activation of downstream transcriptional mechanisms in MPNSTs from NF1 patients.
Keywords: Neurofibromin (NF1), Activating protien 1 (AP-1), N-Ras oncogene, Extracellular signal-regulated kinase (ERK), c-Jun N-terminal kinase (JNK)
Introduction
A common clinical feature of Neurofibromatosis type I (NF1) is the plexiform neurofibroma that grows along the length of a segment of nerve and infiltrates the surrounding tissue. In about 5% of NF1 patients, plexiform neurofibromas transform into MPNSTs [1]. Critical to cell transformation is Ras activation [2] which is increased by tenfold in NF1-deficient compared to NF1-wild type cell lines [3]. Wild type neurofibromin negatively regulates Ras signaling via its functioning as a Ras GTPase activating protein (RasGAP) [4]. When this regulation is lost through mutation, the result is an overactive Ras/Raf/MEK1,2/ERK1,2 pathway. Ras, a component of the multistep process of carcinogenesis, together with its effector molecules exhibits multiple transforming properties [5]. As an example, uncontrolled proliferation, which precedes full tumorigenesis, is promoted by the deregulated Ras/Raf signaling pathway [6]. During normal growth, early in the G1 phase of cell cycle, Ras/Raf/MEK1,2/ERK1,2 signaling results in TCF/ELK-1 phosphorylation, followed by activation of the transcription factor AP-1 and increased expression of its target gene cyclin D1. Transcriptional induction of cyclin D1 enables expression of genes whose proteins are needed for S phase (reviewed in [7]). Oncogenic Ras, however, increases cyclin D1 expression through transactivation of its promoter by AP-1, shortens the G1 phase of the cell cycle, and accelerates cell growth [8]. Other Ras transforming properties include the suppression of cell cycle inhibitors, rearrangement of the cytoskeleton, and induction of angiogenesis [7].
Oncogenic Ras proteins do not transform cells in the absence of c-Jun, a component of the heterodimeric transcription factor complex AP-1 [9–11]. The absence of c-Jun or the presence of a dominant negative c-Jun results in transformation resistance in murine cells. Similarly, the in vivo expression of a dominant negative c-Jun [9, 12] in mouse skin blocks chemically induced carcinogenesis in transgenic mice [13]. Rather than a single protein, AP-1 is a hetero- or homodimeric complex made up of various combinations of members of the JUN, FOS, ATF and MAF protein families. In addition to c-Jun, other proteins that make up the AP-1 dimer such as c-Fos and FosB can transform cells in culture and induce tumors in transgenic mice (reviewed in [14]). AP-1 target genes include cell cycle regulators such as D cyclins, cyclin A, cyclin E [15], migration and invasion related genes such as matrix metalloproteinases [16–18], and angiogenesis related genes such as angioprotein-2 [19]. The key role of AP-1 in tumorigenesis and its contribution to cancer progression make it a prime target for drug development in cancer therapy [20].
The AP-1 regulatory system includes both translational and post-translational activation of AP-1 proteins in response to mitogen stimulation as well as AP-1 protein interaction with other transcription factors and co-factors. Other regulatory mechanisms include stability of AP-1 component mRNAs and protein turnover [21]. The mitogen-activated protein kinase (MAP kinase) super-family is the main mediator of mitogen stimulation of AP-1 (for review [22]). The MAP kinase pathways are responsive to Ras activation and singly or in combination may increase AP-1 protein expression or transactivation [23–26]. Previous work by the authors has shown that N-Ras as well as ERK1,2 is constitutively activated in two NF1-deficient MPNST cell lines [27]. One of these cell lines, ST88-14, has been used as a model to study the effectiveness of drugs with the potential of inhibiting tumor growth in NF1 patients [27–31].
In this study, we employed three complimentary systems to examine the regulation of AP-1 by neurofibromin deficiency through the Ras/MAP kinase pathways. These systems included: (1) the NF1-deficient MPNST ST88-14 cell line; (2) the NF1-wild type MPNST cell line STS-26T; and (3) a Tet-off cell line derived from STS-26T that conditionally expresses oncogenic N-Ras (G12V mutant). We demonstrate here that ERK1,2 pathways positively regulate AP-1 activity in ST88-14 cells. We also show that siRNA knockdown of neurofibromin expression in STS-26T cells upregulates the activity of Ras, which results in the transcriptional activation of AP-1. This is the first example of AP-1 activation in a disease-relevant MPNST cell line being directly linked to neurofibromin deficiency.
Materials and methods
Cell lines and cell culture
Human MPNST ST88-14 (a generous gift from T. Glover, University of Michigan, Ann Arbor, MI, USA) and STS-26T (a generous gift from D. Scoles, Cedars-Sinai Medical Center, Los Angeles, CA, USA) were maintained in RPMI medium 1640 (Invitrogen, Carlsbad, CA, USA) with 5% fetal bovine serum (Hyclone Laboratories, Logan, UT, USA). Cell lines were checked periodically for mycoplasma with Venor GeM Mycoplasma Detection Kit (Sigma, St. Louis, MO, USA). Cultures were propagated for no more than 3 months.
Cytogenetic characterization of ST88-14 has shown that the NF1 locus has been deleted from one allele on chromosome 17 while the gene on the remaining allele resulted in greatly reduced transcription or message instability [32]. Neurofibromin is undetectable when analyzed by western blot using a C-terminal probe for the protein [27], confirming the neurofibromin-deficient phenotype.
Construction of Tet-off gene expression cell lines
Tet-off (Clontech Laboratories Inc., Madison, WI, USA), cell lines conditionally expressing oncogenic N-Ras G12V, were established using the cell line STS-26T (NF1wt/wt). Cells were transfected with the Tet-off regulatory plasmid pRevTet-off (BD Biosciences Clontech) using Lipofectamine and Plus Reagent (Invitrogen) according to the manufacturer’s instructions. Transfected cells were selected in 400 µg/ml G418 and resistant clones were screened for expression of the reporter gene by a transient transfection with the firefly and Renilla luciferase reporter plasmids, pLP-RevTRE-luc and pRL-TK (Clontech). Expression of the luciferase reporter gene was evaluated by the Dual-Luciferase Reporter Assay System (Promega Corp., Madison, WI, USA), and two single stable parent cell lines were chosen on the basis of moderate to high activation of the reporter. These two cell lines were then transfected with pLP-RevTRE (Clontech) into which the cDNA for N-terminal 3x-hemagglutinin tagged human N-Ras G12V mutant was inserted. Double transfected clones were selected with 400 µg/ml G418 and 200 µg/ml hygromycin B in the presence of doxycycline (Invitrogen). Transgene expression was confirmed by western blotting for the hemagglutinin tag.
To assess AP-1 activity in clones that express N-Ras G12V, cells were co-transfected with pGL3 and pRL-TK (see next section), harvested over a 3 day period and quantified for luciferase activity. Results are shown for 15N clonal cell line (Fig. 3), with similar results demonstrated in an experiment with a second independent clonal cell line, 10N (data not shown).
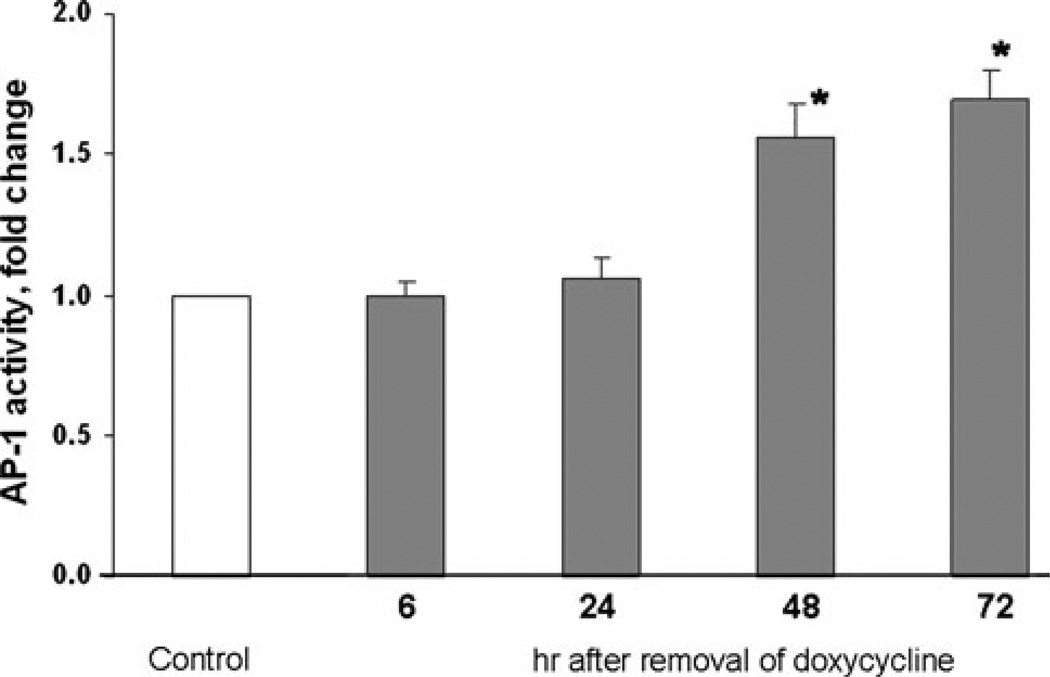
Fig. 3.
AP-1 activity progressively increases in response to N(G12V)-Ras. Tet-regulated cell line 15N was transfected with the AP-1 luciferase reporter plasmids followed by withdrawal of doxycycline in some cultures. Cells were harvested at the indicated times and lysates were subjected to the dual luciferase assay to determine AP-1 activity. Data are shown as means ± SEM for three independent experiments. Results at 48 and 72 h were significantly different (* P value < 0.05) than control samples incubated in doxycycline
Plasmids and luciferase gene reporter assay
Luciferase reporter gene assays were used to determine AP-1 transcriptional activity. Cells were seeded into 6-well plates 24 or 48 h prior to transfection. Cells were co-transfected with 0.5 µg pGL3 promoter vector (Promega, Madison, WI, USA) into which had been cloned the DNA consensus sequence AP-1 3XTRE (CGCTTGATGACTCAGCCGGAA) and 0.05 µg pRL-TK (Promega). 32 h after transfection, kinase inhibitors were added if appropriate. Cells were harvested 48 h post-transfection. Firefly and Renilla luciferase were quantified using the Dual-Luciferase Reporter Assay System (Promega) and a single sample luminometer.
Activated Ras protein pull down assay
Ras pull down assays were performed using glutathione–Sepharose beads prebound to glutathione-S-transferase (GST) fused to the Raf-RBD (Ras binding domain) [33]. Briefly, experimental and control cells were lysed in magnesium containing lysis buffer (25 mM HEPES, pH 7.5, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 10% glycerol, 10 mM MgCl2, 1 mM EDTA, 1 mM Na3VO4, 25 mM NaF). One-third of the samples were reserved for determining total Ras protein by western blotting. The remaining two-thirds of the lysates were incubated with GST–Raf-RBD beads at 4°C on a rocking platform for 90 min. Beads were centrifuged, washed 3 times with PBS and supernatant discarded. Laemmli sample buffer was added to the beads followed by boiling for 5 min. Beads were then pelleted by centrifugation and supernatants for each sample were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose and probed for active Ras. The remaining lysates were probed for total Ras. Total Ras was used to normalize active Ras data.
Electromobility shift assay (EMSA)
Nuclear extract preparation
Extracts were prepared according to a method adapted from that described by Dignam et al. [34]. Cells (107) were grown from 60 to 90% confluence in 100-mm plates. All the remaining procedures were carried out on ice. Cells were washed with ice-cold PBS followed by a wash with ice-cold Buffer A [10 mM Hepes (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol (DTT), 0.1% Igepal CA-630, 1% protease inhibitor cocktail (Sigma, St. Louis, MO, USA), 1 mM Na3VO4, 1 mM Na4P2O7.10H2O, 1 mM NaF]. After 10 min incubation in 1.5 ml Buffer A, cells were scraped into glass homogenizer (Kontes Glass Co., Vineland, NJ, USA; 15 ml) and gently homogenized for 10 strokes, pestle B. Pellet was collected by centrifugation at 5800 RCF, 4°C, 10 min and resuspended after the removal of supernatant in 150 µl Buffer C [10 mM Hepes (pH 7), 25% gylcerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1% protease inhibitor cocktail, 1 mM Na3VO4, 1 mM Na4P2O7·10H2O, 1 mM NaF, 0.5 mM phenylmethanesulfonyl fluoride (PMSF, 0.1 M stock in methanol) and 1 mM DTT]. The suspension was transferred to a microfuge tube embedded horizontally on ice and rocked for 2 h on a rocking platform at 200 rpm. The suspension was centrifuged at maximum speed for 5 min at 4°C. The supernatant was collected, aliquoted and stored at −80°C.
AP-1 binding assay
An EMSA gel shift kit (Panomics, Inc., Fremont, CA, USA) was used containing a biotin-labeled probe with the AP-1 binding sequence (5′–3′): CGCTTGATGACTCAGCCGGAA. 1–5 µg of nuclear extract was combined with labeled or unlabeled (competition reactions) probes according to the manufacturer’s instructions. Transcription factor–DNA complexes were separated on a 6% non-denaturing polyacrylamide gel and transferred to a nylon membrane. For signal detection, a chemiluminescent substrate was added to the biotin–streptavidin–horseradish peroxidase complex and the membrane was exposed to X-ray film (Pierce Biotechnology, Thermo Fisher Scientific Inc., Rockford, IL, USA).
siRNA knockdown of gene expression
Neurofibromin siRNA, control siRNA and siRNA Transfection Reagent (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) were used according to the manufacturer’s instructions. STS-26T (wt-NF1) cells were plated at 1.5 × 105 cells/10 cm plate 24 h prior to transfection. 12 µl of siRNA (50 µM) and 12 µl of siRNA Transfection Reagent, each diluted in 400 µl of Optimem (Invitrogen), were combined and after a 30 min incubation added to the 3.2 ml of Optimem in each plate. Following a 6 h incubation of cells with transfection solution, medium was replaced by growth medium. Cells were harvested 48–72 h later.
Western blot analysis
Cultures at 30 to 80% confluence were washed 3 times with ice-cold PBS, scraped and pelleted at 1,000 rpm for 5 min. Cell pellets were lysed with RIPA buffer (150 mM NaCl, 1%Triton X-100, 0.5% deoxycholic acid, 0.1% SDS, 50 mM Tris–Cl, pH 8.0) supplemented with 2% protease inhibitor cocktail, 1% PMSF (stock 10 mg/ml methanol), 1 mM Na3VO4, 1 mM Na4P2O7·10H2O, and 1 mM NaF. Secondary antibodies were conjugated to IRdye infrared dyes (Rockland Inc., Gilbersville, PA, USA). Signal was detected and the bands were quantified using the Odyssey infrared imaging system and software (Licor Biosciences, Lincoln, NE, USA).
Antibodies used in these experiments were rabbit polyclonal anti-neurofibromin [35] (#sc-67, Santa Cruz Biotechnology Inc.), mouse monoclonal anti-ras [36] (#610001, BD Biosciences), mouse monoclonal anti-phospho-ERK1,2 [37] (#9106, Cell Signaling), rabbit polyclonal anti-ERK1,2 [38] (#06-182, Upstate Cell Signaling Solutions), rabbit polyclonal anti-phospho-JNK [39] (#9251, Cell Signaling), and mouse monoclonal anti-α-tubulin [40] (T5168, Sigma–Aldrich).
Statistical analysis
A paired t test was used to determine significance when analyzing two means. One-way analysis of variance was used for multiple comparisons with post-hoc Tukey HSD tests to determine significance within the group. P < 0.05 was considered statistically significant. Results are expressed as means ± SEM.
Results
siRNA reduction of neurofibromin expression upregulates active Ras and AP-1 transcriptional activity in MPNST cells
Neurofibromin RasGAP activity accelerates the hydrolysis of GTP bound to Ras resulting in decreased Ras activity. Cell lines developed from neurofibrosarcomas surgically removed from NF1 patients express reduced neurofibromin and abnormally high Ras-GTP. To establish a system to test the link between reduced neurofibromin expression and Ras pathway activation, we used an siRNA knockdown approach in the MPNST NF1wt/wt cell line STS-26T. Three siRNA-NF1 duplexes were tested for efficacy in increasing p-ERK1,2 expression (Fig. 1a). STS-26T cells were transfected with siB and analyzed for neurofibromin expression, as well as active and total Ras expression by western blotting (Fig. 1b). Neurofibromin was reduced by 89 ± 1% while active ERK1,2 and active Ras were increased by 2.2 ± 0.4-fold and 1.6 ± 0.1-fold, respectively.
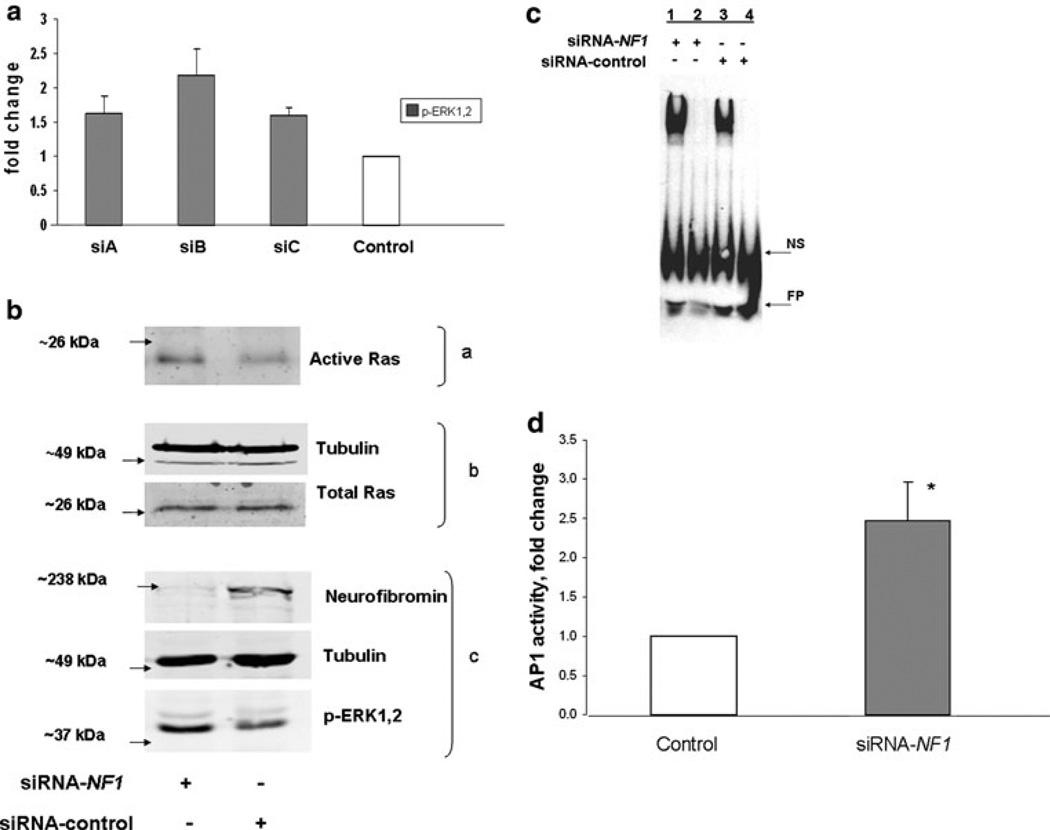
Fig. 1.
Neurofibromin knockdown increases active Ras, active ERK1,2, AP-1 binding and activity. a Three siRNA-NF1 duplexes were tested for effectiveness in upregulating the ERK1,2 pathway in STS26T. Although not significant, upregulation of ERK1,2 activity was highest with siB and this siRNA was used in subsequent experiments. Data are representative of three independent experiments and expressed as means ± SEM. b STS26T cultures were transfected with siRNA-NF1 or siRNA-control followed by harvest for western blot analysis 48 h later. Two-thirds of the cell lysate was subjected to affinity purification for active Ras (a) with the Ras-binding domain of Raf coupled to glutathione S-transferase; one-third of the lysate was reserved for determining total Ras and tubulin (b) and neurofibromin, tubulin and active ERK1,2 (c). Data are representative of three independent experiments. c STS-26T cells were transfected with siRNA, NF1 or control, and harvested 48 or 72 h post-transfection. EMSAs were performed with nuclear extracts of cells transfected with siRNA-NF1 or control siRNA. Competitive binding reactions with excess unlabeled probe were added to NF1 or control samples in lanes 2 and 4, respectively. Data are representative of three independent experiments. NS non-specific binding, FP free probe. d STS-26T cells were transfected with siRNA-NF1 or scrambled control duplexes followed 24 h later by transfection with the AP-1 luciferase reporter plasmids. Cell lysates for dual luciferase assay were prepared 48 h after second transfection. Data are presented as the fold activation relative to the activity obtained with the scrambled siRNA control. Values represent the mean ± SEM for three independent experiments. AP-1 activity in NF1 knockdown cells was found to be significantly different (* P value < 0.05) than control cells
To investigate the effect of neurofibromin knockdown on the binding of the AP-1 transcription factor with DNA, EMSAs were performed. Cells were harvested 48 h post-transfection, nuclear extracts were prepared and protein binding to the AP-1 consensus site was determined. As shown in Fig. 1c (representative of 3 independent experiments), when expression of neurofibromin was inhibited by siRNA knockdown, nuclear extracts of the cells exhibited a 1.7 ± 0.2-fold increase in AP-1 binding compared to those from control transfected cells. In addition, NF1 knockdown resulted in greater than twofold increase in AP-1 activity (Fig. 1d).
N-Ras(G12V) increases phosphorylation of ERK1,2 and JNK MAP kinases as well as AP-1 activity
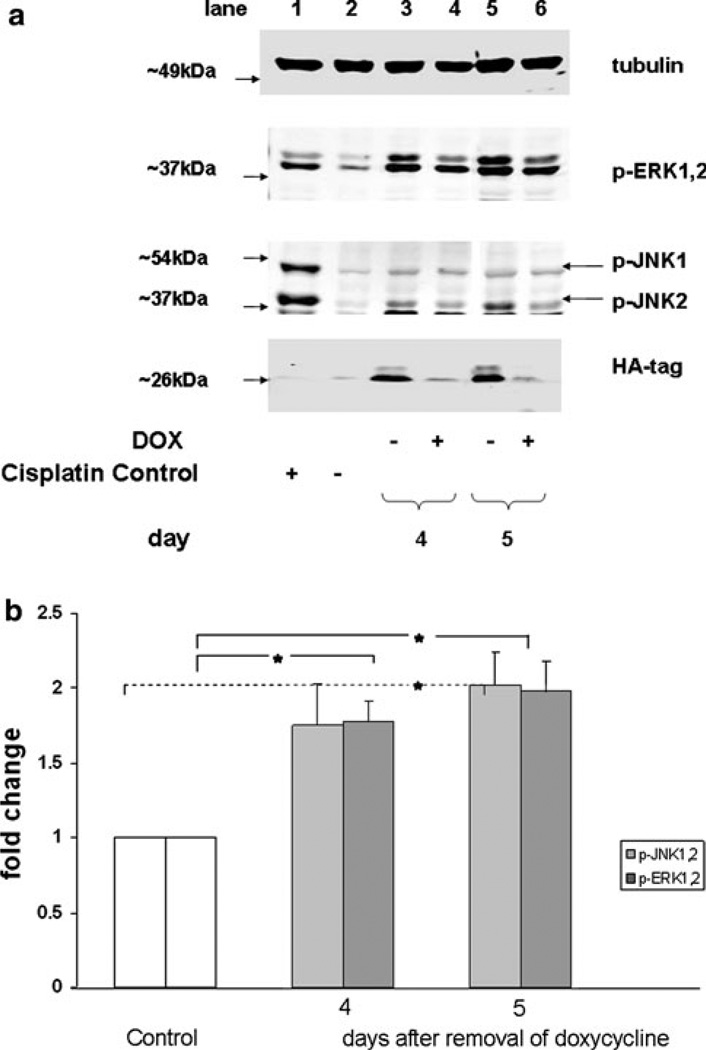
We previously demonstrated that N-Ras was the predominant activated isoform of Ras in two independent MPNST cell lines from NF1 patients [27]. However, N-Ras was not activated in the non NF1 MPNST cell line STS26T [27]. Therefore, in order to assess the role of N-Ras in AP-1 activation we constructed two independent clonal cell lines of STS26T cells (15N and 10N) that conditionally expressed the activated, mutant N-Ras (G12V) oncogene. Removal of doxycycline from the growth media turns on the expression of N-Ras (G12V). To examine changes in downstream N-Ras signaling, cell line 15N was grown in media with or without doxycycline supplementation (Fig. 2a, b) for 5 days. Induction of expression of the activated N-Ras oncogene resulted in a 1.7-fold increase in active ERK and active JNK on day 4 followed by twofold increase in activity for both proteins on day 5, Fig. 2b. To determine AP-1 activity under conditions of an upregulated Ras pathway, 15N cells were transfected with pGL3/AP-1 3XTRE and pRL-TK, dual luciferase plasmids, followed by a media change with or without supplementation of doxycycline. Cells were harvested at the times indicated to evaluate AP-1 activity by the reporter gene assay (Fig. 3). A maximum increase of 1.7-fold in AP-1 activity was observed in 15N cells 72 h after the removal of doxycycline. A similar experiment performed with the clonal cell line 10N resulted in an increase of 1.5-fold AP-1 activity at 96 h (data not shown). These data demonstrate that the induction of an activated N-Ras oncogene can increase AP-1 activity in the MPNST cell type similar to knockdown of NF1 by siRNA.
Fig. 2.
N(G12V)-Ras activation increases phosphorylation of JNK and ERK1,2 MAP kinases. a Conditional expression cell line STS26T-N-Ras (G12V) 15N was treated with and without doxycycline (DOX). Cells were harvested 4 and 5 days after the removal of doxycycline. Samples were analyzed by western blot and probed for the indicated protein. After each probe, the blot was stripped and reprobed for the subsequent protein. Detection of the hemagglutinin (HA) tag indicates the induction of N-Ras expression. To demonstrate activation (positive control) or inactivation (negative control) of MAP kinases, cell line TOV112D was treated (or not) with cis-Diammine-platinum (II) dichloride and samples were run in lanes 1 and 2. Data are representative of three independent experiments. b The bar graph quantitatively illustrates the means ± SEM for the three experiments described in a. Data are presented as the fold increase relative to the samples treated with doxycycline. Significant (* P value < 0.05) increases in p-ERK1,2 expression were observed on days 4 and 5 and in p-JNK1,2 expression on day 5 compared to control cells (doxycycline treated)
ERK1,2 and JNK1,2 regulate AP-1 activity in ST88-14 MPNST cells
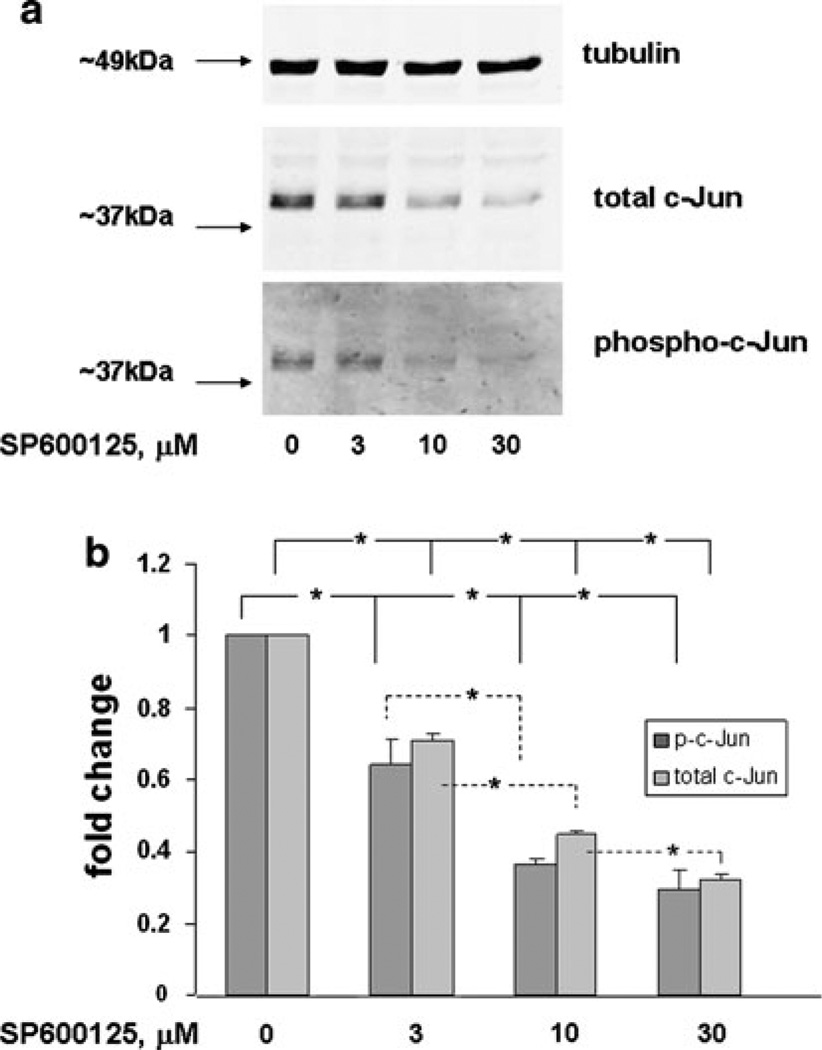
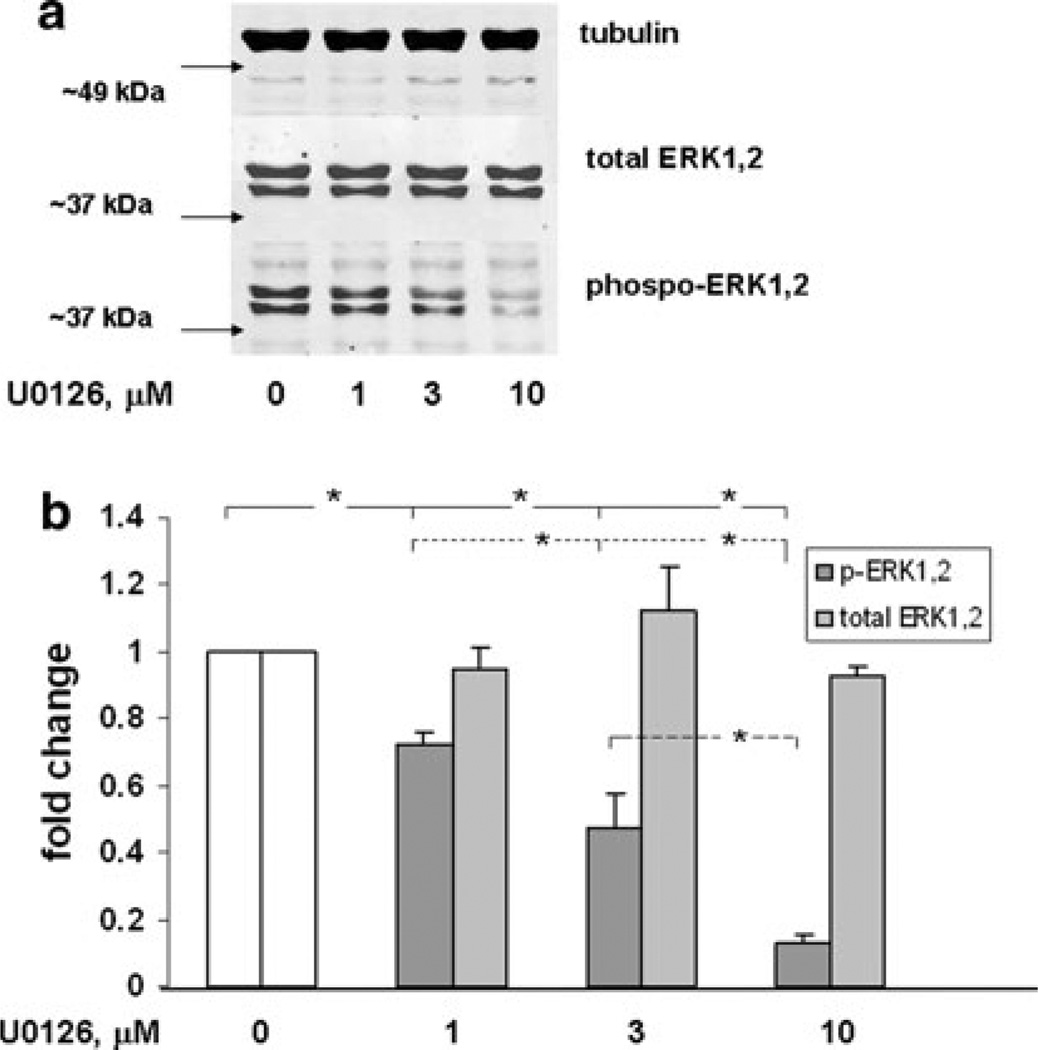
In response to external stimuli, MAP kinase families respond singly or in combination to activate AP-1 in a cell-specific manner. To determine the role of JNK and ERK MAP kinase pathways in the regulation of AP-1 in the NF1-deficient ST88-14 MPNST cell line, selective kinase inhibitors, SP600125 [41] and U0126 [40], were used. To test the effectiveness of the compounds in inhibiting substrate phosphorylation, cells were exposed to increasing concentrations of SP600125 and U0126 and then harvested to determine levels of phospho-c-Jun and p-ERK1,2, respectively. A dose-dependent response was observed for SP600125 at 3, 10 and 30 µM (Fig. 4a, b), which resulted in a reduction of the levels of both phospho-c-Jun and c-Jun. A dose-dependent response was also observed with U0126 at 1, 3, and 10 µM (Fig. 5a, b) for inhibition of the level of p-ERK1,2. Expression of total ERK1,2 protein levels, however, remained constant.
Fig. 4.
Dose response of c-Jun and protein expression and phosphorylation in ST88-14 cells treated with SP600125. a ST88-14 cells were treated with SP600125 at the concentrations indicated for 16 h. Samples were analyzed by western blot and probed for the indicated protein. After each probe the blot was stripped and reprobed for the subsequent protein. Data shown are representative of three independent experiments. b The bar graph quantitatively illustrates the means ± SEM for the three experiments described in a. For phosphoc-Jun a significant inhibition (* P value < 0.05) was observed with increased concentration of SP600125 through 10 µM. 10 µM was not different from 30 µM although there was a downward trend. For total c-Jun, significant inhibition (* P value < 0.05) was observed with all increases in concentration
Fig. 5.
Dose response of ERK1,2 protein expression and phosphorylation in ST88-14 cells triated with U0126. a ST88-14 cells were treated with U0126 at the concentrations indicated for 16 h. Samples were analyzed by western blot and probed for the indicated protein. After each probe, the blot was stripped and reprobed for the subsequent protein. Data shown are representative of three independent experiments. b The bar graph quantitatively illustrates the means ± SEM for the three experiments described in a. Phoshorylated(p)-ERK1,2 expression was significantly less (* P value < 0.05) with each increase in U0126 concentration. Total ERK1,2 does not change throughout treatments
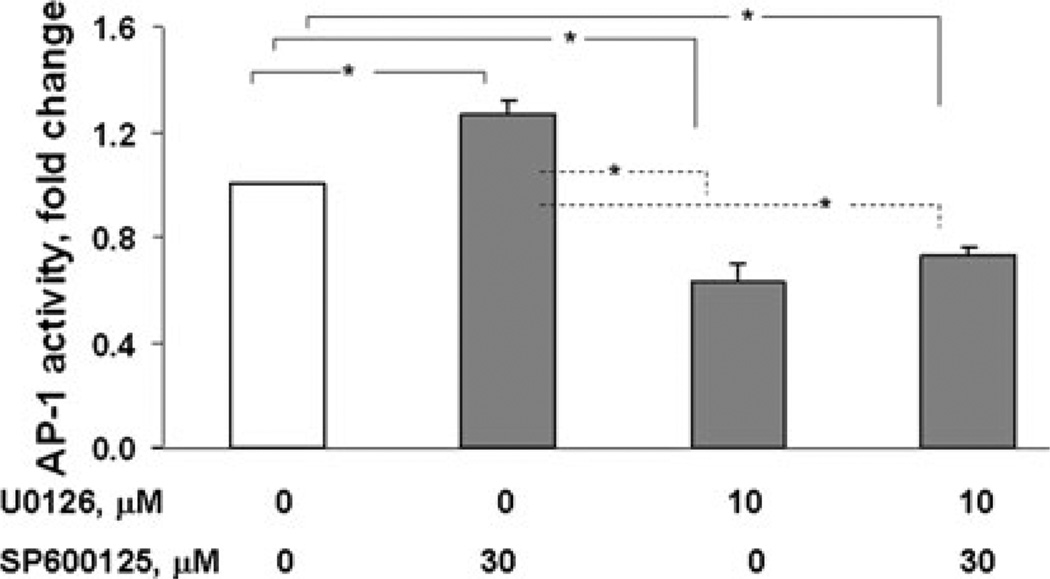
AP-1 activity in ST88-14 (NF1−/− cells) was then determined using AP-1 reporter gene assay (Fig. 6). U0126 reduced AP-1 transcriptional activity to 60% of solvent-treated controls. Conversely, inhibiting JNK signaling with SP600125 increased AP-1 activity by 25%. These data suggest that under normal growth conditions constitutive ERK1,2 signaling elevates AP-1 activity while JNK represses it.
Fig. 6.
Inhibition of MAP kinases differentially affects AP-1 activity. ST88-14 cells were transfected with the AP-1 luciferase reporter plasmids. 24 h later, the medium was changed to include the indicated inhibitors followed by 16 h of incubation, at which time cells were harvested for the dual luciferase assay. Data are expressed as percent of control comparing relative luciferase units of inhibitor-treated cells to vehicle-treated cells. The figure represents the means ± SEM of three independent experiments. Treatment with U0126 significantly (* P < 0.05) inhibited AP-1 activity whereas treatment with SP600125 significantly (* P < 0.05) increased activity
Discussion
The AP-1 transcription factor controls multiple cell processes that include proliferation, apoptosis, migration, invasion, angiogenesis, and differentiation under both basal and stimulated conditions. AP-1 can be activated by varied external stimuli (cytokines, growth factors, stress, and ultraviolet light) that signal through the MAP kinase cascades [14]. AP-1 can also respond to activated oncogenes such as Ras [42]. Therefore, it is not surprising that deregulation of AP-1 transcriptional activity is linked to tumorigenesis. In this study, we demonstrated for the first time, in a disease-relevant human MPNST cell line, that neurofibromin deficiency is directly linked to the deregulation of AP-1 activity.
We utilized MPNST cell line STS-26T (wt-NF1) to determine the effect of a neurofibromin knockdown on Ras, ERK1,2 and AP-1 activity. Human and animal studies have demonstrated increased Ras/ERK1,2 activity under conditions of neurofibromin deficiency. Murine studies have shown that fetal cells lacking neurofibromin have a high constitutive MAP kinase activity [43, 44]. Analyses of isolated human tumor cells and tissue taken from sarcomas and benign neurofibromas from NF1 patients have revealed elevated Ras-GTP levels [45, 46]. In the current study, suppression of neurofibromin expression induced Ras and ERK1,2 activation. Furthermore, neurofibromin knockdown enhanced both AP-1 consensus sequence binding and transcriptional activity, thus demonstrating a direct effect of neurofibromin deficiency in upregulating the Ras pathway and increasing AP-1 transcriptional activity.
To focus on N-Ras signaling exclusive of any other effects that neurofibromin deficiency may impose, we used STS26T cells conditionally expressing N-Ras (G12V). N-Ras (G12V) activation in 15N cells was found to increase phosphorylation of ERK1,2 and JNK. This supports previous reports that MPNST cell lines with high Ras-GTP had increases in ERK and JNK signaling pathways compared to cells with low Ras-GTP [47]. We also observed that when N-Ras (G12V) was induced in 15N and 10N cells AP-1 transcriptional activity was increased up to 1.7-fold. These experiments illustrate that activation of Ras can result in quantitatively similar levels of AP-1 activation due to neurofibromin deficiency.
Upregulation of the ERK and JNK pathways by Ras activation prompted further examination of these pathways in the NF1-deficient MPNST cell line ST88-14. Cells were treated with U0126 and SP600125, selective inhibitors of MEK1,2 and JNK1,2,3, respectively. Dose response studies demonstrated that both phospho-c-Jun and c-Jun were reduced by approximately the same amount when cells were exposed to SP600125, which suggests that there may be direct involvement of phospho-c-Jun in the reduction of c-Jun expression. The AP-1 consensus binding sequence or TPA response element (TRE) is located in the promotor of c-Jun. This sequence is constitutively occupied by c-Jun and ATF2 in many cell types [48]. Following activation by JNK, both members of the AP-1 dimer are phosphorylated inducing transcriptional activation of c-Jun. Thus, a decrease in phospho-c-Jun could reduce the induction of c-Jun expression. In contrast, the expression of ERK1,2 was maintained following U0126 treatment while p-ERK1,2 levels were reduced by close to 90%.
We found that under normal growth conditions for ST88-14, 40% of AP-1 activity can be attributed to constitutive signaling of ERK1,2. In a previous study, we determined that 10 µM U0126 inhibited cell proliferation by 30% and induced an accumulation of G1-phase cells and the loss of S-phase cells [27]. These data are consistent with the reduction in AP-1 activity observed with U0126 treatment in the current experiments. The ERK signaling pathway is a major pathway mediating cell growth by induction of the expression of D-type cyclins through the activation of AP-1. In this well documented pathway [49–51], ERK-activated ELK-1 stimulates the expression of c-Fos which binds to pre-existing c-Jun proteins leading to the activation of AP-1 and AP-1 target genes [52]. It is likely that this represents the mechanism by which ERK maintains basal AP-1 activity levels. Our data in human MPSNT cells are consistent with earlier work in which increased expression of neurofibromin was able to suppress Ras-induced AP-1 reporter gene activity in rat REF52 cells simultaneously injected with plasmids encoding full-length neurofibromin and H-Ras [53]. In contrast to U0126, where inhibition of p-ERK1,2 decreased AP-1 activation, inhibition by SP600125 results in a 20% increase in AP-1 activity. This implies that an increase of JNK activation from basal levels may be repressing AP-1 activity in these MPNST cells. This seems paradoxical because the link between activated JNK, increased phospho-c-Jun and increased activation of AP-1 is well documented (for review [48]). Our data also shows no significant difference between combined inhibition of U0126 and SP600125 compared to U0126 alone indicating that in the absence of active ERK, SP600125 appears to have no effect on AP-1 activity. This apparent paradox of JNK repression of AP-1 activation needs to be more fully studied.
In summary, in the MPNST line STS-26T containing wild type NF1 gene, the activities of Ras, ERK1,2 and AP-1 were upregulated by siRNA knockdown of neurofibromin, demonstrating the ability of the Ras-GAP protein to regulate the Ras/ERK1,2 pathway as well as downstream transcription factors. We believe this is the first time the transactivation of AP-1 has been linked directly to neurofibromin deficiency in a disease relevant MPNST cell line. STS26T cell lines engineered to constitutively express oncogenic Ras (mutant G12V) also upregulated ERK1,2, and JNK. ERK1,2 and JNK were found to have opposing roles in regulating AP-1, with ERK1,2 being dominant in increasing activity in NF1-deficient MPNST line ST88-14.
Acknowledgments
This study was supported by the Barbara and Fred Erb Endowed Chair in Cancer Genetics to MAT, as well as to the Cancer Center Support Grant of the Karmanos Cancer Institute, Wayne State University (P30CA022453). The authors are grateful for suggestions on statistical analyses provided by Dr. Judith Abrams.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Janice M. Kraniak, Programs in Molecular Biology and Genetics, Barbara Ann Karmanos Cancer Institute, 110 East Warren Avenue, Detroit, MI 48201, USA
Daochun Sun, Center for Molecular Medicine and Genetics, Wayne State University School of Medicine, Detroit, MI 48201, USA.
Raymond R. Mattingly, Programs in Molecular Biology and Genetics, Barbara Ann Karmanos Cancer Institute, 110 East Warren Avenue, Detroit, MI 48201, USA Department of Pharmacology, Wayne State University School of Medicine, Detroit, MI 48201, USA; Environmental Health Sciences Center for Molecular and Cellular Toxicology with Human Applications, Wayne State University School of Medicine, Detroit, MI 48201, USA.
John J. Reiners, Jr., Programs in Proteases, Barbara Ann Karmanos Cancer Institute, Detroit, MI 48201, USA Department of Pharmacology, Wayne State University School of Medicine, Detroit, MI 48201, USA; Institute of Environmental Health Sciences, Wayne State University School of Medicine, Detroit, MI 48201, USA; Environmental Health Sciences Center for Molecular and Cellular Toxicology with Human Applications, Wayne State University School of Medicine, Detroit, MI 48201, USA.
Michael A. Tainsky, Email: tainskym@karmanos.org, Programs in Molecular Biology and Genetics, Barbara Ann Karmanos Cancer Institute, 110 East Warren Avenue, Detroit, MI 48201, USA; Center for Molecular Medicine and Genetics, Wayne State University School of Medicine, Detroit, MI 48201, USA; Environmental Health Sciences Center for Molecular and Cellular Toxicology with Human Applications, Wayne State University School of Medicine, Detroit, MI 48201, USA; Department of Pathology, Wayne State University School of Medicine, Detroit, MI 48201, USA.
References
- 1.Levy P, Vidaud D, Leroy K, Laurendeau I, Wechsler J, Bolasco G, Parfait B, Wolkenstein P, Vidaud M, Bieche I. Molecular profiling of malignant peripheral nerve sheath tumors associated with neurofibromatosis type 1, based on large-scale real-time RT-PCR. Mol Cancer. 2004;3:20. doi: 10.1186/1476-4598-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeClue JE, Papageorge AG, Fletcher JA, Diehl SR, Ratner N, Vass WC, Lowy DR. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Reckling-hausen (type 1) neurofibromatosis. Cell. 1992;69:265–273. doi: 10.1016/0092-8674(92)90407-4. [DOI] [PubMed] [Google Scholar]
- 3.Basu TN, Gutmann DH, Fletcher JA, Glover TW, Collins FS, Downward J. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature. 1992;356:713–715. doi: 10.1038/356713a0. [DOI] [PubMed] [Google Scholar]
- 4.Xu GF, O’Connell P, Viskochil D, Cawthon R, Robertson M, Culver M, Dunn D, Stevens J, Gesteland R, White R, et al. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990;62:599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- 5.Gille H, Downward J. Multiple ras effector pathways contribute to G(1) cell cycle progression. J Biol Chem. 1999;274:22033–22040. doi: 10.1074/jbc.274.31.22033. [DOI] [PubMed] [Google Scholar]
- 6.Pruitt K, Der CJ. Ras and Rho regulation of the cell cycle and oncogenesis. Cancer Lett. 2001;171:1–10. doi: 10.1016/s0304-3835(01)00528-6. [DOI] [PubMed] [Google Scholar]
- 7.Saxena N, Lahiri SS, Hambarde S, Tripathi RP. RAS: target for cancer therapy. Cancer Invest. 2008;26:948–955. doi: 10.1080/07357900802087275. [DOI] [PubMed] [Google Scholar]
- 8.Liu JJ, Chao JR, Jiang MC, Ng SY, Yen JJ, Yang-Yen HF. Ras transformation results in an elevated level of cyclin D1 and acceleration of G1 progression in NIH 3T3 cells. Mol Cell Biol. 1995;15:3654–3663. doi: 10.1128/mcb.15.7.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson R, Spiegelman B, Hanahan D, Wisdom R. Cellular transformation and malignancy induced by ras require c-jun. Mol Cell Biol. 1996;16:4504–4511. doi: 10.1128/mcb.16.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smeal T, Binetruy B, Mercola DA, Birrer M, Karin M. Oncogenic and transcriptional cooperation with Ha-Ras requires phosphorylation of c-Jun on serines 63 and 73. Nature. 1991;354:494–496. doi: 10.1038/354494a0. [DOI] [PubMed] [Google Scholar]
- 11.Behrens A, Jochum W, Sibilia M, Wagner EF. Oncogenic transformation by ras and fos is mediated by c-Jun N-terminal phosphorylation. Oncogene. 2000;19:2657–2663. doi: 10.1038/sj.onc.1203603. [DOI] [PubMed] [Google Scholar]
- 12.Brown PH, Alani R, Preis LH, Szabo E, Birrer MJ. Suppression of oncogene-induced transformation by a deletion mutant of c-jun. Oncogene. 1993;8:877–886. [PubMed] [Google Scholar]
- 13.Young MR, Li JJ, Rincon M, Flavell RA, Sathyanarayana BK, Hunziker R, Colburn N. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc Natl Acad Sci USA. 1999;96:9827–9832. doi: 10.1073/pnas.96.17.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 15.Shen Q, Uray IP, Li Y, Krisko TI, Strecker TE, Kim HT, Brown PH. The AP-1 transcription factor regulates breast cancer cell growth via cyclins and E2F factors. Oncogene. 2008;27:366–377. doi: 10.1038/sj.onc.1210643. [DOI] [PubMed] [Google Scholar]
- 16.Bahassiel M, Karyala S, Tomlinson CR, Sartor MA, Medvedovic M, Hennigan RF. Critical regulation of genes for tumor cell migration by AP-1. Clin Exp Metastasis. 2004;21:293–304. doi: 10.1023/b:clin.0000046132.46946.dd. [DOI] [PubMed] [Google Scholar]
- 17.Kim S, Choi JH, Kim JB, Nam SJ, Yang JH, Kim JH, Lee JE. Berberine suppresses TNF-alpha-induced MMP-9 and cell invasion through inhibition of AP-1 activity in MDA-MB-231 human breast cancer cells. Molecules. 2008;13:2975–2985. doi: 10.3390/molecules13122975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan TW, Yang WH, Lin YT, Hsu SF, Li TM, Kao ST, Chen WC, Fong YC, Tang CH. Cyr61 increases migration and MMP-13 expression via alphavbeta3 integrin, FAK, ERK and AP-1-dependent pathway in human chondrosarcoma cells. Carcinogenesis. 2009;30:258–268. doi: 10.1093/carcin/bgn284. [DOI] [PubMed] [Google Scholar]
- 19.Ye FC, Blackbourn DJ, Mengel M, Xie JP, Qian LW, Greene W, Yeh IT, Graham D, Gao SJ. Kaposi’s sarcoma-associated herpesvirus promotes angiogenesis by inducing angiopoietin-2 expression via AP-1 and Ets1. J Virol. 2007;81:3980–3991. doi: 10.1128/JVI.02089-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthews CP, Colburn NH, Young MR. AP-1 a target for cancer prevention. Curr Cancer Drug Targets. 2007;7:317–324. doi: 10.2174/156800907780809723. [DOI] [PubMed] [Google Scholar]
- 21.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 22.Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Y, Liao H, Wang N, Ma KS, Verna LK, Shyy JY, Chien S, Stemerman MB. LDL-activated p38 in endothelial cells is mediated by Ras. Arterioscler Thromb Vasc Biol. 2001;21:1159–1164. doi: 10.1161/hq0701.092473. [DOI] [PubMed] [Google Scholar]
- 24.Gille H, Sharrocks AD, Shaw PE. Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature. 1992;358:414–417. doi: 10.1038/358414a0. [DOI] [PubMed] [Google Scholar]
- 25.Clarke N, Arenzana N, Hai T, Minden A, Prywes R. Epidermal growth factor induction of the c-jun promoter by a Rac pathway. Mol Cell Biol. 1998;18:1065–1073. doi: 10.1128/mcb.18.2.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M, Davis RJ. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 27.Mattingly RR, Kraniak JM, Dilworth JT, Mathieu P, Bealmear B, Nowak JE, Benjamins JA, Tainsky MA, Reiners JJ., Jr The mitogen-activated protein kinase/extracellular signal-regulated kinase kinase inhibitor PD184352 (CI-1040) selectively induces apoptosis in malignant schwannoma cell lines. J Pharmacol Exp Ther. 2006;316:456–465. doi: 10.1124/jpet.105.091454. [DOI] [PubMed] [Google Scholar]
- 28.Wojtkowiak JW, Fouad F, LaLonde DT, Kleinman MD, Gibbs RA, Reiners JJ, Jr, Borch RF, Mattingly RR. Induction of apoptosis in neurofibromatosis type 1 malignant peripheral nerve sheath tumor cell lines by a combination of novel farnesyl transferase inhibitors and lovastatin. J Pharmacol Exp Ther. 2008;326:1–11. doi: 10.1124/jpet.107.135830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barkan B, Starinsky S, Friedman E, Stein R, Kloog Y. The Ras inhibitor farnesylthiosalicylic acid as a potential therapy for neurofibromatosis type 1. Clin Cancer Res. 2006;12:5533–5542. doi: 10.1158/1078-0432.CCR-06-0792. [DOI] [PubMed] [Google Scholar]
- 30.Yan N, Ricca C, Fletcher J, Glover T, Seizinger BR, Manne V. Farnesyltransferase inhibitors block the neurofibromatosis type I (NF1) malignant phenotype. Cancer Res. 1995;55:3569–3575. [PubMed] [Google Scholar]
- 31.Dilworth JT, Wojtkowiak JW, Mathieu P, Tainsky MA, Reiners JJ, Jr, Mattingly RR, Hancock CN. Suppression of proliferation of two independent NF1 malignant peripheral nerve sheath tumor cell lines by the pan-ErbB inhibitor CI-1033. Cancer Biol Ther. 2008;7:1938–1946. doi: 10.4161/cbt.7.12.6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynolds JE, Fletcher JA, Lytle CH, Nie L, Morton CC, Diehl SR. Molecular characterization of a 17q11.2 translocation in a malignant schwannoma cell line. Hum Genet. 1992;90:450–456. doi: 10.1007/BF00220476. [DOI] [PubMed] [Google Scholar]
- 33.Mattingly RR, Felczak A, Chen CC, McCabe MJ, Jr, Rosenspire AJ. Low concentrations of inorganic mercury inhibit Ras activation during T cell receptor-mediated signal transduction. Toxicol Appl Pharmacol. 2001;176:162–168. doi: 10.1006/taap.2001.9272. [DOI] [PubMed] [Google Scholar]
- 34.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cichowski K, Santiago S, Jardim M, Johnson BW, Jacks T. Dynamic regulation of the Ras pathway via proteolysis of the NF1 tumor suppressor. Genes Dev. 2003;17:449–454. doi: 10.1101/gad.1054703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dhillon AS, Meikle S, Yazici Z, Eulitz M, Kolch W. Regulation of Raf-1 activation and signalling by dephosphorylation. EMBO J. 2002;21:64–71. doi: 10.1093/emboj/21.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zubiaur M, Fernandez O, Ferrero E, Salmeron J, Malissen B, Malavasi F, Sancho J. CD38 is associated with lipid rafts and upon receptor stimulation leads to Akt/protein kinase B and Erk activation in the absence of the CD3-zeta immune receptor tyrosine-based activation motifs. J Biol Chem. 2002;277:13–22. doi: 10.1074/jbc.M107474200. [DOI] [PubMed] [Google Scholar]
- 38.He HJ, Kole S, Kwon YK, Crow MT, Bernier M. Interaction of filamin A with the insulin receptor alters insulin-dependent activation of the mitogen-activated protein kinase pathway. J Biol Chem. 2003;278:27096–27104. doi: 10.1074/jbc.M301003200. [DOI] [PubMed] [Google Scholar]
- 39.Kujime K, Hashimoto S, Gon Y, Shimizu K, Horie T. p38 mitogen-activated protein kinase and c-jun-NH2-terminal kinase regulate RANTES production by influenza virus-infected human bronchial epithelial cells. J Immunol. 2000;164:3222–3228. doi: 10.4049/jimmunol.164.6.3222. [DOI] [PubMed] [Google Scholar]
- 40.Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, Copeland RA, Magolda RL, Scherle PA, Trzaskos JM. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 41.Han Z, Boyle DL, Chang L, Bennett B, Karin M, Yang L, Manning AM, Firestein GS. c-Jun N-terminal kinase is required for metalloproteinase expression and joint destruction in inflammatory arthritis. J Clin Invest. 2001;108:73–81. doi: 10.1172/JCI12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vandel L, Montreau N, Vial E, Pfarr CM, Binetruy B, Castellazzi M. Stepwise transformation of rat embryo fibroblasts: c-Jun, JunB, or JunD can cooperate with Ras for focus formation, but a c-Jun-containing heterodimer is required for immortalization. Mol Cell Biol. 1996;16:1881–1888. doi: 10.1128/mcb.16.5.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bollag G, Clapp DW, Shih S, Adler F, Zhang YY, Thompson P, Lange BJ, Freedman MH, McCormick F, Jacks T, Shannon K. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nat Genet. 1996;12:144–148. doi: 10.1038/ng0296-144. [DOI] [PubMed] [Google Scholar]
- 44.Zhang YY, Vik TA, Ryder JW, Srour EF, Jacks T, Shannon K, Clapp DW. Nf1 regulates hematopoietic progenitor cell growth and ras signaling in response to multiple cytokines. J Exp Med. 1998;187:1893–1902. doi: 10.1084/jem.187.11.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guha A, Lau N, Huvar I, Gutmann D, Provias J, Pawson T, Boss G. Ras-GTP levels are elevated in human NF1 peripheral nerve tumors. Oncogene. 1996;12:507–513. [PubMed] [Google Scholar]
- 46.Sherman LS, Atit R, Rosenbaum T, Cox AD, Ratner N. Single cell Ras-GTP analysis reveals altered Ras activity in a subpopulation of neurofibroma Schwann cells but not fibroblasts. J Biol Chem. 2000;275:30740–30745. doi: 10.1074/jbc.M001702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farassati F, Pan W, Yamoutpour F, Henke S, Piedra M, Frahm S, Al-Tawil S, Mangrum WI, Parada LF, Rabkin SD, Martuza RL, Kurtz A. Ras signaling influences permissiveness of malignant peripheral nerve sheath tumor cells to oncolytic herpes. Am J Pathol. 2008;173:1861–1872. doi: 10.2353/ajpath.2008.080376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 49.Dhandapani KM, Khan MM, Wade FM, Wakade C, Mahesh VB, Brann DW. Induction of transforming growth factor-beta1 by basic fibroblast growth factor in rat C6 glioma cells and astrocytes is mediated by MEK/ERK signaling and AP-1 activation. J Neurosci Res. 2007;85:1033–1045. doi: 10.1002/jnr.21182. [DOI] [PubMed] [Google Scholar]
- 50.Hirota T, Irie K, Okamoto R, Ikeda W, Takai Y. Transcriptional activation of the mouse Necl-5/Tage4/PVR/CD155 gene by fibroblast growth factor or oncogenic Ras through the Raf-MEK-ERK-AP-1 pathway. Oncogene. 2005;24:2229–2235. doi: 10.1038/sj.onc.1208409. [DOI] [PubMed] [Google Scholar]
- 51.Todisco A, Takeuchi Y, Urumov A, Yamada J, Stepan VM, Yamada T. Molecular mechanisms for the growth factor action of gastrin. Am J Physiol. 1997;273:G891–G898. doi: 10.1152/ajpgi.1997.273.4.G891. [DOI] [PubMed] [Google Scholar]
- 52.Buchwalter G, Gross C, Wasylyk B. Ets ternary complex transcription factors. Gene. 2004;324:1–14. doi: 10.1016/j.gene.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 53.al-Alawi N, Xu G, White R, Clark R, McCormick F, Feramisco JR. Differential regulation of cellular activities by GTPase-activating protein and NF1. Mol Cell Biol. 1993;13:2497–2503. doi: 10.1128/mcb.13.4.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]