Abstract
In this issue of Structure, Nolan and colleagues present the structure of BMP antagonist, PRDC, which adopts a head-to-tail dimer with distinct structure and inhibitory mechanism compared to other dimeric antagonists of TGF-beta superfamily, such as noggin.
The transforming growth factor-beta (TGF-β) superfamily is comprised of a diversified family of secreted signaling proteins, with more than 30 members in humans and other vertebrates to regulate hundreds of genes (Hinck, 2012). The proteins of the superfamily evolved as developmental factors responsible for embryonic patterning and morphogenesis in invertebrates, but have further evolved to regulate numerous extraembryonic functions as organisms have diversified. These include, but are not limited to, the regulation of bone and muscle mass by proteins such as BMP-7, BMP-9, and GDF-8, regulation of gonadal function by the activins, inhibins, GDF-9, and BMP-15, regulation of the many different cell types of the adaptive immune system by the TGF-βs, and regulation of the differentiation of embryonic stem cells by activin A and nodal.
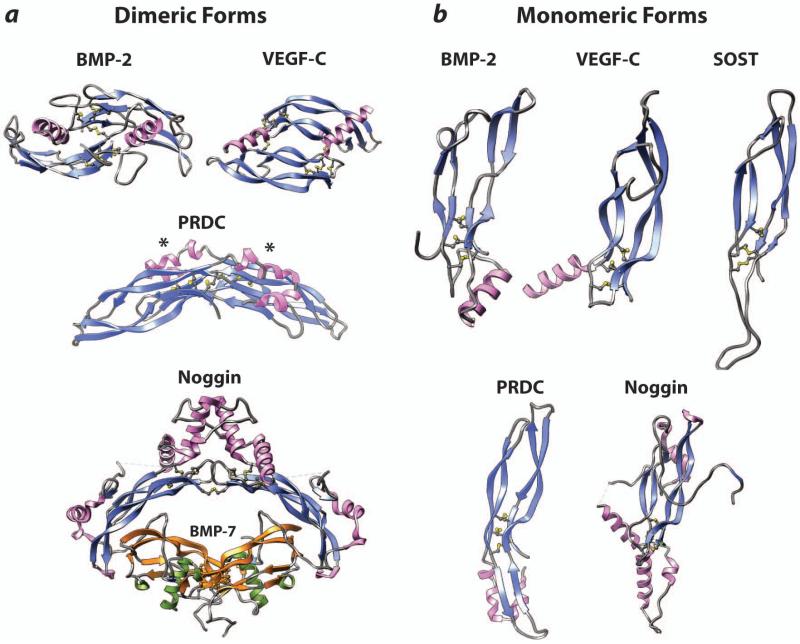
TGF-βs, BMPs, GDFs and other proteins of the superfamily are structurally similar, consisting of two extended monomers held together in most, but not all cases, by a single disulfide bond (Fig. 1a) (Hinck, 2012). The monomers of all superfamily members include a cystine knot, which is formed by three disulfides, the first and second of which form bridges a few residues apart on adjacent β-strands while a third passes through the eight residue ring formed by the first and second disulfide. The extended β-sheet structure, together with the stabilizing cystine knot, is known as a growth factor fold. This fold is present in a number of other secreted signaling proteins, including nerve growth factor (NGF), platelet derived growth factors (PDGF), vascular endothelial growth factor (VEGF), and others (Fig. 1b). These signaling proteins are also active as disulfide-linked dimers, though the arrangement of monomers differs and is responsible for their signaling through distinct receptors and disparate activities (Fig 1a).
Figure 1. The malleability of the cystine knot growth factor fold.
(a) Dimeric forms of the cystine-knotted signaling proteins, BMP-2 and VEGF-C, and the BMP antagonists, PRDC and noggin. Disulfide bonds that form the cystine knot, as well as those that form the inter-chain disulfide(s) in BMP-2, VEGF-C, and noggin, are depicted using a ball- and-stick representation. Asterisks on the PRDC structure designate the BMP binding site as identified through site-directed mutagenesis and accompanying functional studies. (b) Monomeric forms of the cystine-knotted signaling proteins and the BMP antagonists shown in panel a (shown also is the BMP antagonist SOST). Disulfide bonds that form the cystine knot are depicted using a ball-and-stick representation as in panel a.
The proteins of the TGF-β superfamily signal by binding and bringing together two transmembrane receptors, known as receptor types I and II. The assembly of these receptors into heteromeric complexes leads to the activation of the type I receptor kinase, which in turn activates cytoplasmic effectors, known as Smads (Massague et al., 2005). There are seven type I receptors and five type II receptors in most vertebrate species, and among these, the type I receptors couple to two different classes of Smads: the more recently evolved members of the superfamily, including the TGF-βs, activins, nodal, and some of the GDFs and BMPs (GDF-9, -11, and -15 and BMP-15), bind and signal through type I receptors that activate R-Smads 2, 3, while the more distantly related GDFs (GDF-1, -3, -5, -7, and -10) and BMPs (BMP-2, -3, -4, -5, -6, -7, -8, -9, and -10) bind and signal through type I receptors that couple to and activate R-Smads 1, 5, and 8 (Hinck, 2012). This restricts the functional diversity that can be attained through intrinsic differences in signaling. The diversity of signaling is instead dependent upon the unique patterns with which the superfamily ligands are targeted to different cells and tissues and the context-dependent manner by which cells respond upon activation of Smads (Massague and Wotton, 2000).
The targeting of superfamily signaling proteins is largely mediated by secreted antagonists, which bind the signaling proteins and block the receptor binding sites. The antagonists are structurally diverse, ranging from large multidomain proteins, such as follistatin and chordin, to smaller single domain proteins with a cystine knot growth factor fold, such as those of the DAN family and noggin (Bragdon et al., 2011). The structural diversity of the antagonists stands in contrast to the signaling proteins and is thought to be responsible for the specificity of most toward a limited subset of signaling proteins. Though the secreted antagonists have vital roles targeting superfamily signaling proteins to specific cells and tissues, there is at present only a limited molecular understanding of their diverse molecular structures and inhibitory mechanisms (Cash et al., 2009; Groppe et al., 2002).
The focus of current discussion is the DAN family antagonist, known as protein related to DAN and cerebus, or PRDC. Though all nine members of the DAN family include a cystine-knot motif and adopt a growth factor fold, they differ significantly in their inhibitory potencies and the subset of signaling proteins they target. The most potent DAN family antagonists, DAN, PRDC, and Gremlin, are thought to only antagonize BMPs and other superfamily signaling proteins, while the least potent of the DAN family antagonists, SOST and USAG-1, also bind the coreceptor LRP5/6 to antagonize Wnt signaling. The only structural information available for the DAN family of antagonists is SOST, which includes an even number of cysteines and is monomeric (Fig. 1b) (Veverka et al., 2009). This stands in contrast to the PRDC, which through prior studies had been shown to form a highly stable non-covalent dimer, even though it includes an odd number of cysteines, including one (C120) that is positionally conserved with the cysteine that forms the inter-chain disulfide in TGF-βs and other proteins of the TGF-β superfamily (Kattamuri et al., 2012). The reason for this did not become apparent until the structure of PRDC was determined and it was shown that PRDC forms a head-to-tail dimer with an interface one-and-a-half times larger than the signaling proteins of the TGF-β superfamily and with extensive hydrogen bonding between the exposed β-strand of finger 2 (Fig. 1a). This alternative manner of dimerization positions the monomers so that C120 is incapable of forming the inter-chain disulfide characteristic of most proteins of the TGF-β superfamily. The authors further showed that the residues of PRDC responsible for binding BMPs reside largely on the convex surface of the PRDC dimer in the cysteine-rich DAN domain. These findings suggest that PRDC achieves high affinity for BMP dimers due to multivalent binding (dimeric PRDC binding to dimeric BMPs). The more distantly related cystine-knot BMP antagonist noggin was also previously shown to bind BMPs with high affinity by forming a dimer, but this differs in two significant ways relative to PRDC. The first is that, unlike PRDC, noggin forms an unusual head-to-head dimer that is stabilized both by a single interchain disulfide, and by the addition of several helical segments that pack against one another at the dimer interface (Fig. 1a). The second is that noggin also forms an arch-like structure, but unlike PRDC, it uses its concave surface, together with an extended clamp-like structure, to nearly fully surround the signaling protein and block the type I and type II receptor binding sites.
The structure of PRDC is significant since it shows how the same cystine-knotted growth factor fold can be modified to form two entirely distinct BMP antagonists. The finding that alternate arrangements of the cystine-knotted growth factor have given rise to distinct antagonists with distinct specificities is perhaps not surprising given the diversity of the dimeric structures among the different classes of cystine-knotted growth factors (TGF-β, PDGF, NGF, VEGF, etc.) and their binding to distinct receptors. The structure of PRDC nevertheless reiterates the malleability of this important structural motif and the many ways in which it has evolved to expand and diversify cell signaling. The future studies of other DAN family antagonists, such as gremlin, cerebus, coco, and others, therefore promise to offer plenty of additional surprises. We are looking forward to seeing what else this domain can do!
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Bragdon B, Moseychuk O, Saldanha S, King D, Julian J, Nohe A. Bone morphogenetic proteins: a critical review. Cell Signal. 2011;23:609–620. doi: 10.1016/j.cellsig.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Cash JN, Rejon CA, McPherron AC, Bernard DJ, Thompson TB. The structure of myostatin:follistatin 288: insights into receptor utilization and heparin binding. The EMBO journal. 2009;28:2662–2676. doi: 10.1038/emboj.2009.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppe J, Greenwald J, Wiater E, Rodriguez-Leon J, Economides AN, Kwiatkowski W, Affolter M, Vale WW, Izpisua Belmonte JC, Choe S. Structural basis of BMP signalling inhibition by the cystine knot protein Noggin. Nature. 2002;420:636–642. doi: 10.1038/nature01245. [DOI] [PubMed] [Google Scholar]
- Hinck AP. Structural studies of the TGF-betas and their receptors - insights into evolution of the TGF-beta superfamily. FEBS Lett. 2012;586:1860–1870. doi: 10.1016/j.febslet.2012.05.028. [DOI] [PubMed] [Google Scholar]
- Kattamuri C, Luedeke DM, Nolan K, Rankin SA, Greis KD, Zorn AM, Thompson TB. Members of the DAN family are BMP antagonists that form highly stable noncovalent dimers. J Mol Biol. 2012;424:313–327. doi: 10.1016/j.jmb.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Massague J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veverka V, Henry AJ, Slocombe PM, Ventom A, Mulloy B, Muskett FW, Muzylak M, Greenslade K, Moore A, Zhang L, et al. Characterization of the structural features and interactions of sclerostin: molecular insight into a key regulator of Wnt-mediated bone formation. J Biol Chem. 2009;284:10890–10900. doi: 10.1074/jbc.M807994200. [DOI] [PMC free article] [PubMed] [Google Scholar]