Abstract
The development of the placenta is imperative for successful pregnancy establishment, yet the earliest differentiation events of the blastocyst-derived trophectoderm that forms the placenta remains difficult to study in humans. Human embryonic stem cells (hESC) display a unique ability to form trophoblast cells when induced to differentiate either by the addition of exogenous BMP4 or by the formation of cellular aggregates called embryoid bodies (EBs). While mouse trophoblast stem cells have been isolated from blastocyst outgrowths, mouse ESC do not spontaneously differentiate into trophoblast cells
In this review, we focus on addressing the similarities and differences between mouse trophoblast stem cell differentiation and human ESC-derived trophoblast differentiation. We discuss the functional and mechanistic diversity that is found in different species models. Of central importance are the unique signaling events that trigger downstream gene expression that create specific cellular fate decisions. We support the idea that we must understand the nuances that hESC differentiation models display so that investigators can choose the appropriate model system to fit experimental needs.
Introduction
Theories of embryological development date back to Aritstotle’s time (382-322 B.C.) with the concept of epigenesis, where it was thought that the embryo developed from an amorphous mass derived from the mother. Aristotle believed that the male contribution of sperm was what gave the soul to this mass and helped guide development (Aristotle, translated by Peck 1943). Other early thinkers believed in the preformationist theory where a mini-individual (homunculus) existed within the germ cell and initiated embryonic development (Magner, 2002). While current knowledge has advanced beyond these early hypotheses, a deeper understanding of the events in early embryogenesis and the key regulators involved in the establishment of a healthy pregnancy remains a goal only incompletely realized. Early pregnancy loss is thought to occur in 10- 25% of all clinically recognized pregnancies, and preeclampsia and other hypertensive disorders that can be linked to placental biology affect 5-8% of pregnancies in the US (http://www.americanpregnancy.org/pregnancycomplications/miscarriage.html/; http://www.preeclampsia.org/health-information/faq). Thus, the basic developmental mechanisms that direct placentation are of high clinical relevance.
The first differentiation event in the preimplantation mammalian embryo is the formation of the trophectoderm that will contribute the trophoblast compartment of the placenta. The responsibilities of the trophoblasts include signaling the presence of the conceptus to the maternal reproductive and immune systems, and acquiring the vital nutrition necessary for fetal growth during pregnancy. Since placentation is the earliest morphogenetic event in pregnancy, animal models and embryos have contributed significantly to studies of placental development, with mouse trophoblast stem cells providing an important research tool while a fully equivalent cell line has not been isolated in primates. The isolation of human embryonic stem cells (hESC) from blastocyst stage embryos has provided a unique and powerful embryonic surrogate to begin understanding human development, and overcoming the obvious ethical limitations of working with human embryos (Thomson, et al. 1998). These hESC have been used to identify approaches that induce trophoblast differentiation, aimed to provide an understanding of the mechanisms, which support a commitment to the trophoblast lineage in embryonic development. Herein we will review the similarities and differences, where known, in mouse and human trophoblast differentiation and placental development. The differentiation of trophoblast cells from human embryonic stem cells will be highlighted on a functional and mechanistic level, presenting current thinking on the signaling events necessary to achieve trophoblast differentiation.
Trophoblast Development
Mouse placental development
During the initial stages of placental development, both mouse and human pregnancy presents a deep interstitial implantation, and the development of a hemochorial placenta where the trophoblasts are in direct contact with the maternal blood (Pijnenborg, et al. 1981). Although both are hemochorial, organization that allows the placental trophoblast to interface with maternal blood differs between the two. In the mouse, the fetal blood vessels within the placenta are interconnected to form complex capillary networks among which maternal blood vessels intertwine and thus form a placental labyrinthe (Cross 2005; Rossant and Cross 2001). The trophoblasts line channels through which the maternal blood circulates within the labyrinth, forming the exchange surface between fetal and maternal blood. In distinction, in the human (as well as in old world nonhuman primates), a villous placenta forms in which the trophoblasts develop villi that arborize into terminal branches that have few interconnections (Kingdom, et al. 2000). Within these villi, the fetal vasculature develops, and since the villi have a trophoblast surface and display extensive branching, a large surface area is created for gas and nutrient exchange between the mother and fetus. Thus, the organization of the maternal-fetal exchange surface is distinct between these two placentas.
Differences between the human and mouse placenta can also be seen in the morphology and phenotype of the trophoblasts that arise during development. In the mouse, at the time of implantation, the trophectoderm cells that lie away from the inner cell mass (ICM) halt division but undergo endoreduplication, thus forming the trophoblast giant cells. These cells eventually form the outer regions of the ectoplacental cone surrounding the conceptus (Cross 2005; Rossant and Cross 2001). The ectoplacental cone is also composed of diploid trophoblast cells that give rise to the spongiotrophoblast that forms the outer structural layer of the placenta (Cross 2005; Rossant and Cross 2001). The syncytiotrophoblasts within the mouse placenta are multinucleate cells that lie within the labyrinthe and are the direct interface for gas and nutrient exchange between the maternal and fetal vasculatures.
Both the trophoblast giant cells and the spongiotrophoblast secrete many factors that support the establishment and maintenance of pregnancy. These factors include hormones, angiogenic and tissue remodeling factors like placental lactogens, proliferin, vascular endothelial growth factor (VEGF), matrix metalloproteinases, and urokinase type plasminogen activator (uPA) (Achen, et al. 1997; Cross 2005; Groskopf, et al. 1997; Rossant and Cross 2001; Soares, et al. 1996; Teesalu, et al. 1998; Teesalu, et al. 1999; Vuorela, et al. 1997). We will not discuss mouse placental physiology in detail here, and the reader is referred to other excellent reviews for further detailed discussion (Cross 2005; Rossant and Cross 2001).
Human placental development
As with the mouse blastocyst, the human blastocyst upon apposition and adhesion to the uterine luminal epithelium rapidly penetrates to the endometrial stroma, where the formation of a multinuclear syncytium, and proliferating cytotrophoblasts advance embryonic remodeling of the superficial endometrium to become surrounded by maternal tissues (Carter and Pijnenborg 2011). As the human placenta continues its development, the cytotrophoblasts are the main proliferating trophoblasts that give rise to the cytotrophoblast columns. The cytotrophoblasts fuse to form the syncytiotrophoblasts that cover the branch-like protrusions (villi) that erupt from the cell columns. The syncytiotrophoblasts is the primary endocrine and transport interface, directly exposed to maternal blood in the intervillous space that forms from the erosion of maternal decidual vessels during implantation and early placental growth (Benirschke, et al. 2006). Growth factors such as vascular endothelial growth factor (VEGF), angiopoietins, angiostatins, and placental growth factor (PlGF) act within the human villi to control placental villous vasculogenesis and angiogenesis (Kingdom et al. 2000). Also arising from the cytotrophoblasts in the human placenta are the extravillous cytotrophoblasts that leave the cell column and migrate into the maternal decidua. Further discussion of these cells appears below.
Human and mouse placentation: remodeling of maternal tissues
A notable similarity between mouse and human placental development is the migration of the trophoblastic cells into the maternal decidua and their remodeling of the spiral arterioles (Pijnenborg et al. 1981). In the mouse, the trophoblast giant cells invade the maternal arterioles during the early postimplantation period, followed by the glycogen trophoblasts that carry out later interstitial migration and invasion resulting in dilated arteries that lack elastic lamina and smooth muscle cells but are lined with trophoblasts that form canals that carry maternal blood to the base of the placenta (Adamson, et al. 2002; Cross 2005). The maternal blood then percolates back through the labyrinthe space providing nutrients to fetus via the syncytiotrophoblast transport (Adamson et al. 2002; Cross 2005). In the human, the extravillous trophoblasts reside in the trophoblast cell columns and undergo an epithelial-mesenchymal transition, where they migrate, invade, and remodel the maternal spiral arterioles. More specifically, the interstitial extravillous trophoblasts will migrate into the stroma of the maternal decidua and come into close contact with uterine immune cells that aid in vessel remodeling (Kaufmann, et al. 2003). The endovascular extravillous trophoblasts will appear within the lumen of maternal vessels but there is still controversy on the mode of invasion of the extravillous trophoblasts in the human placenta (Kaufmann et al. 2003). One possibility is that trophoblasts from the decidua come into close contact with the outside of the vessels (intravasation), remove the smooth muscle, and eventually line the vessels.
Alternatively, trophoblasts presumably enter vessels proximal to the placenta, arrive in the lumen, move opposite to the maternal blood flow, and proceed to remodel and line the maternal vessels (extravasation) (Kaufmann et al. 2003; Pijnenborg, et al. 2006). Regardless of the pathway that they take, a significant outcome of the initial invasion in the human is to create vascular plugs in the maternal vessels thereby decreasing the arterial pressure towards the implantation site (Hamilton and Boyd 1966). The extended remodeling creates dilated maternal vessels resulting in a low pressure, high output environment providing ample nutrition to the developing fetus (Kingdom et al. 2000; Pijnenborg, et al. 1980; Pijnenborg, et al. 1982). Much work still remains to uncover the details regarding trophoblast differentiation and function during formation of the placental-decidual interface, therefore the development of tractable model systems is imperative for the field.
Trophoblast Lineage and Stem Cell Differentiation
Trophoblast stem cells (TSC) were first isolated from the polar trophectoderm and from the extraembryonic ectoderm of mouse blastocysts (Tanaka, et al. 1998). Maintenance of the undifferentiated state and self-renewal in mouse TSC are controlled by a combination of fibroblast growth factor 4 (FGF4), heparin, and secreted factors from fetal fibroblasts (Rielland, et al. 2008; Tanaka et al. 1998). Activin A or transforming growth factor beta1 (TGFB1) can replace conditioned medium from fetal fibroblasts to maintain mouse TSC (Erlebacher, et al. 2004; Roberts and Fisher 2011). It was subsequently demonstrated that TSC could also be directly derived from mouse embryonic stem cells through repression of the Pit1-Octamer-Unc86 (POU) transcription factor Oct4 and in the presence of FGF4 and feeder cells (Niwa, et al. 2000).
Transcriptional Control
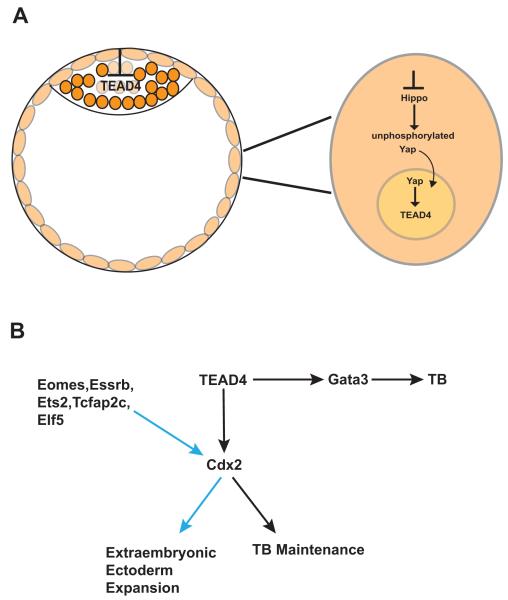
Mouse molecular genetics has provided extensive information regarding the molecular mechanisms that drive differentiation of trophoblast stem cells and placental development. Of central importance are the genes involved in trophoblast lineage development. Table 1 lists the factors discussed in this review, and gives a brief description of the significance of the genes discussed in more detail below. Although the caudal-type homeobox gene Cdx2 has been classified as a key regulator in determining trophoblast fate, it is itself regulated by the transcription factor Tead4. Tead4 mutants are capable of producing embryonic stem cells, but fail to produce the trophectoderm lineage or TSC (Yagi, et al. 2007). Moreover, disruption of the Tead4 gene after implantation results in viable offspring and conversely, over-expression of Tead4 results in TSC-like cell lines, pinpointing the necessity of Tead4 expression for trophoblast lineage commitment in the developing embryo (Nishioka, et al. 2009; Senner and Hemberger 2010; Yagi et al. 2007). Interestingly, Hippo signaling that is associated with the epithelial-to-mesenchymal transition in mammals is also linked to TEAD4 transcriptional activity. Inactive Hippo signaling in the outside cells of the developing embryo allows the Hippo co-activator Yap to remain unphosphorylated and thus translocate to the nucleus where it assists in Tead4 activation (Figure 1A) (Nishioka et al. 2009; Senner and Hemberger 2010). Recent studies have shown that in an evolutionarily conserved mechanism, the nuclear localization of Tead4 in the outer blastomeres of the embryo leads to a trophoblast-specific transcriptional program that is selectively impaired in the inner cells of the embryo and thus allows the inner cells to become the ICM (Figure 1A) (Home, et al. 2012).
Table 1.
Genes with significant relevance to trophoblast differentiation and placental development.
| Gene Symbol | Summary of significance (see text for details) |
|---|---|
| Fgf4 | Maintenance of mouse TSC proliferation |
| Tgfb1 | Maintenance of mouse TSC proliferation |
| Oct4 (Pou5f1) | ESC pluripotency transcription factor |
| Cdx2 | Directs mouse trophoblast lineage selection and proliferation |
| Tead4 | Trophoblast lineage selection upstream of Cdx2 |
| Hippo | Indirect Tead4 regulator |
| Yap | Tead4 coactivator |
| Trim (ectodermin) | Nodal expression regulation |
| Nodal | TGFB1 regulator, trophoblast differentiation |
| Bmp4 | Inner cell mass maintenance (mouse) |
| Gata3 | Promotes trophoblast differentiation |
| Eomes | Mouse TSC maintenance |
| Esrrb | Mouse TSC maintenance |
| Ets2 | Enhances Cdx2 action |
| Tcfap2c (tfap2c) | Trophoblast differentiation, enhances Cdx2 action |
| Elf5 | Trophoblast lineage determination, Cdx2, Eomes interactions |
| Hand1 | Trophoblast giant cell differentiation (mouse) |
| Mdfi | Trophoblast giant cell differentiation (mouse) |
| Mash2/Ascl2 | Maintenance of mouse spongiotrophoblast |
| HASH2 | Expressed in human cytotrophoblasts |
| Egfr | Maintenance of mouse spongiotrophoblast |
| Gcm1 | Mouse syncytiotrophoblast differentiation |
| Eset | Methylation site-related silencing of trophoblast genes |
| Sox2 | Pluripotency factor for ESC |
| Nanog | Pluripotency factor for ESC |
| Rex1 | Inner cell mass marker |
| Fgf5 | Epiblast marker |
| Klf4 | Pluripotency factor for ESC |
| Lif | Maintenance of mouse ESC self-renewal |
Figure 1.
Schematic diagram of the cell signaling that dictates trophoblast (TB) differentiation in the mouse. A. The nuclear localization of TEAD4 in the outer blastomeres of the embryo leads to a trophoblast-specific transcriptional program, which is selectively impaired in the inner cells of the embryo (ie. the ICM). Inactive Hippo signaling maintains Yap in an unphosphorylated state, resulting in translocation of Yap to the nucleus and thus induction of the TEAD4 transcriptional program in the outer cells of the embryo. B. Trophoblast differentiation is not limited to Cdx2 induction (Gata3 induction results in trophoblast differentiation) and likewise Cdx2 induction is not only limited to trophoblast differentiation but also results in extraembryonic ectoderm expansion. Black arrows indicate positive induction pathways. Blue arrows indicated enhancement of Cdx2 leading to extraembryonic ectoderm expansion.
Paracrine signaling initiated by factors including FGF and EGF from the mouse ICM is also necessary to support trophoblast and TSC self-renewal. During later developmental stages, the Smad4 inhibitor, ectodermin (Trim) determines the appropriate amount of Nodal activity derived from the epiblast, thus titrating the balance between the trophoblast undifferentiated and differentiated state (Morsut, et al. 2010; Roberts and Fisher 2011). FGF4-stimulated expression of Cdx2 in TSC, in turn lead to Cdx2 binding to an FGF4-responsive enhancer element in the promoter region of BMP4 that results in growth factors that maintain the ICM (Murohashi, et al. 2010; Roberts and Fisher 2011).
Once induction of the trophoblast fate has been initiated, Cdx2 maintains trophoblast/TSC function (Figure 1B). Tead4 also regulates the transcription factor Gata3 to induce trophoblast differentiation from ESC but Gata3 functions independently of Cdx2 (Ralston, et al. 2010). Interestingly, transcription factor binding sites for Tcfap2 found in the mouse that mediate Cdx2-independent repression of the pluripotency marker Oct4 are not found in humans and cattle, suggesting a crucial difference between the molecular cues that initiate trophoblast development in different species (Berg, et al. 2011; James, et al. 2012). The homeobox transcription factor Eomesodermin (Eomes) is expressed after blastocyst development in response to Cdx2 expression but also enhances Cdx2 and aids in extraembryonic ectoderm expansion (Rossant and Cross 2001; Russ, et al. 2000; Senner and Hemberger 2010). Similarly, the nuclear hormone receptor Esrrb, and the transcription factors Ets2, Tcfap2c, and Elf5 all enhance Cdx2 expression, and direct extraembryonic ectoderm expansion. Essrb deletion results in trophoblast defects where TSC isolation is impaired (Chawengsaksophak, et al. 1997; Donnison, et al. 2005; Luo, et al. 1997; Rossant and Cross 2001; Russ et al. 2000; Senner and Hemberger 2010; Werling and Schorle 2002; Yamamoto, et al. 1998). More specifically, trophoblasts can be induced by over-expression of Cdx2 or by Tcfap2c in ESC independently whereas Elf5 activation requires Cdx2 and Tcfap2c co-expression in order to derive trophoblasts from ESC (Kuckenberg, et al. 2010; Senner and Hemberger 2010). Mutations in the genes that directly support the differentiation of the trophoblast giant cells (Hand1 and Mdfi) and the maintenance of the spongiotrophoblast (Mash2 and Egfr) also result in impaired development of each respective trophoblast subtype thus illustrating coordinated regulation necessary for trophoblast differentiation (Guillemot, et al. 1994; Kraut, et al. 1998; Riley, et al. 1998; Rossant and Cross 2001; Sibilia and Wagner 1995; Threadgill, et al. 1995). Similar to the mouse spongiotrophoblasts that offer structural support to the placenta, the human cytotrophoblasts offer support and express the basic helix-loop-helix (bHLH) transcription factor HASH2, a human homologue of the murine Ascl2 (Mash2) (Benirschke et al. 2006; Hamilton and Boyd 1960; Janatpour, et al. 1999; Roberts, et al. 2004). The syncytiotrophoblasts express the transcription factor glial cell missing 1 (GCM1) as do their functional counterpart in the mouse labyrinth, also termed syncytioptrophoblasts (Anson-Cartwright, et al. 2000; Janatpour et al. 1999; Roberts et al. 2004).
Epigenetic regulation of trophoblast lineage-specific transcription factors
Recently, studies on the epigenetic regulation of lineage-specific transcription factors in mouse have highlighted methylation patterns associated with expressing/repressing embryo-specific genes in the trophoblast lineage. More specifically, the trivalent histone/lysine footprint H3K9me3, H3K4me2/3, and H3K27me3 or the bivalent H3K4me3 and H3K9me3 footprint have been shown to silence embryonic genes in cells developing into trophoblasts (Azuara, et al. 2006; Bernstein, et al. 2006; Boyer, et al. 2006; Senner and Hemberger 2010). Conversely, the methyltransferase Eset is recruited by Oct4 to silence trophoblast genes via histone H3K9 methylation in the ICM and ESC in a selective way that is still not well understood but may involve SUMOylation of Oct4 (Senner and Hemberger 2010; Yeap, et al. 2009). At center stage of epigenetic regulation in regards to trophoblast development is the transcription factor Elf5. Elf5 seems to be a key regulator in cell fate decisions for trophoblast development in the embryo by maintaining Cdx2 and Eomes gene expression (Donnison et al. 2005; Ng, et al. 2008). Elf5 is hypomethylated and therefore expressed in TSC, but methylated, and therefore silenced, in ESC (Ng et al. 2008). Subsequent studies have identified a “TSC-like” compartment in the villous cytotrophoblasts of the human placenta where ELF5+/CDX2+ cells reside (Hemberger, et al. 2010). In addition, these authors determined that the human ESC lines that they derived expressed negligible amounts of ELF5 compared to trophoblast cell lines and an 8-week human placental sample, and moreover, the ESC-derived trophoblasts did not express ELF5 at all leading to the conclusion that hESC-derived cells should not be considered to be authentic trophoblasts (Hemberger et al. 2010).
These results may depend on the ESC-derived trophoblast preparations used for study. The trophoblast cell preparations used were established cultures having undergone selective expansion based on hCG expression following embryoid body (EB) formation (Hemberger et al. 2010). However, using the H1 ESC line, we have recently found ELF5 mRNA differentially expressed through the initial 1-4 weeks during trophoblast outgrowth culture derived from EBs of various sizes and (Gerami-Naini, Giakoumopoulos, et al., in preparation). This underscores that trophoblast derivation methods might result in the formation of different trophoblast subtypes depending on the state of the parental hESC line, and the differentiation paradigm employed. It is clear that heterogeneity of hESC can profoundly influence trophoblast differentiation potential (Pera, et al. 2004; Xu, et al. 2002). Thus, as discussed further below, the field will benefit from complementary approaches being taken to define models for trophoblast differentiation during embryonic development.
Cell Signaling and Trophoblast Differentiation
Mouse, Human, and Non-Human Primates TSC
TSC have offered many insights into mouse placental development but recapitulating the characteristics of their mouse counterparts have yet to be isolated and established from many other relevant species. Rhesus macaque and human TSC-like lines have been established but do not express the same transcriptional repertoire found in the mouse-derived TSC (Douglas, et al. 2009; Harun, et al. 2006; Vandevoort, et al. 2007). The rhesus-derived lines lack expression of CDX2 but express many trophoblast markers such as chorionic gonadotrophin, CD9, KRT7, POU5F1, and EOMES (Kamei, et al. 2002; Roberts and Fisher 2011; Vandevoort et al. 2007). Also, of central importance is that although isolated by blastocyst outgrowth similarly to mouse TSC, the rhesus cell lines are able to proliferate in the absence of feeder layers or bFGF (Roberts and Fisher 2011; Vandevoort et al. 2007).
In the human study, EBs were generated from ESC and multiple rounds of cellular aggregation and disaggregation were used in combination with human chorionic gonadotrophin (hCG) as a marker, to isolate their TSC lines (Harun et al. 2006). Similarly to the mouse, human TSC lines were maintained in “TSC” medium as described by Tanaka et al. 1998 in the presence of FGF4 thus variations exist in line derivation and maintenance that make comparisons challenging between mice and primates (Harun et al. 2006). The human-derived lines express CDX2, HLAG, CD9 and KRT7 but do not express EOMES and after some time in culture, these lines fuse to form syncytium (Harun et al. 2006). More recently, a trophoblast progenitor cell niche has been identified from the chorion of the human placenta providing yet another renewable source of multipotent cells reported to be capable of differentiating into all three trophoblast subtypes (Genbacev, et al. 2011). That these human and other cell lines express differentiation markers such as hCG suggests that they are more highly differentiated than mouse TSC, which lack expression of endocrine markers such as mouse placental lactogens (Douglas et al. 2009; Tanaka et al. 1998).
The naïve vs. primed cellular state
Species differences during cell fate decisions may be due to the signaling differences that dictate the potency of the cells within their cellular compartments in vivo. These differences are apparent when comparing human ESC (hESC) to mouse ESC (mESC) and mouse epiblast stem cells (mEpiSC). Mouse EpiSC can be isolated from both pre- and post-implantation embryos and it has been established that hESC have similar gene expression and cell signaling profiles to mEpiSC compared to mESC, suggesting that hESC represent a more advanced embryonic stage than mESC (Najm, et al. 2011; Tesar, et al. 2007). Since a given ESC line provides a “snapshot” in time of a transient developmental state defining the expression and functional profiles associated with each line are critical. These differences may illuminate the proposed “naïve” vs. “primed” pluripotent state in which established stem cell lines exist in vitro (De Los Angeles, et al. 2012; Nichols and Smith 2009). Mouse ESC derived from female embryos exist in a naïve pluripotent state consisting of two active X chromosomes; the expression of pluripotency markers Oct4, Sox2, and Nanog; and are able to form chimeras that result in germline transmission (De Los Angeles et al. 2012). Human ESC and mEpiSC exist in a primed state in vitro, where cells display one active and one inactive X chromosome, express the above mentioned pluripotency markers, but are not able to form chimeras or incorporate into the germline (De Los Angeles et al. 2012). Although differences do exist, important similarities are also found between hESC and mESC. Briefly, hESC and mESC both 1.) express the ICM marker REX1; 2.) do not express the epiblast marker FGF5; and 3.) express KLF4, which is also used to reprogram somatic cells into an ESC-like state (Adjaye, et al. 2005; Darr, et al. 2006; Greber, et al. 2010; Pelton, et al. 2002). Interestingly, KLF4 can also be used to revert primed mEpiSC into an ESC-like naïve state (Guo, et al. 2009; Takahashi, et al. 2007; Yu, et al. 2007), similar to the reprogramming of “terminally-differentiated” somatic cells such as fibroblasts to induced pluripotent stem cells (iPSC) (Yu, et al. 2007; Takahashi, et al. 2007). Interestingly, a recent study has shown that derivation of hESC from a female embryo in 5% oxygen compared to 20% oxygen results in ESC with two active X chromosomes, therefore the potential to obtain developmental equivalents to the naïve pluripotent state found in mESC exists (De Los Angeles et al. 2012; Lengner, et al. 2010).
FGF at the top of the signaling cascade
Of central importance in providing the signaling cues that dictate mESC, mEpiSC, and hESC self-renewal and differentiation are fibroblast growth factors 2 and 4 (FGF2,-4). In the mouse, Leukemia Inhibitory Factor (LIF) is required for ESC self-renewal and FGF/ERK signaling drives differentiation of ESC (Burdon, et al. 1999). When inhibitors block FGF receptor tyrosine kinases, Map Kinase ERK Kinase (MEK), and Glycogen Synthase Kinase (GSK) signaling (termed 3i), or MEK and GSK alone (termed 2i), naïve pluripotent cells can be derived from mouse embryos (Nichols and Smith 2009; Silva and Smith 2008; Ying, et al. 2008). Conversely, hESC require FGF2/ERK to self-renew and can be induced to differentiate by the canonical WNT/beta-catenin signaling pathway (Sumi, et al. 2008). Activin/Nodal signaling also contribute to hESC self-renewal, and BMP-induced trophoblast differentiation is dependent on inhibition of Activin/Nodal signaling (Wu, et al. 2008). Transforming Growth Factor beta (TGFb) and FGF2 work together to sustain the hESC in an undifferentiated state by maintaining NANOG, OCT4, and SOX2 in addition to inhibiting BMP signaling expression (Xu, et al. 2008). More specifically, TGFb signaling through SMAD 2/3 promotes enhanced NANOG expression in hESC thus maintaining an undifferentiated state whereas TGFb signaling through SMAD 1/5/8, which is generally found to be repressed in undifferentiated hESC, results in decreased NANOG expression leading to ESC differentiation (Xu et al. 2008). In addition, it has been shown that ERK2 phosphorylates OCT4 at multiple sites outside the OCT4 DNA binding domains, possibly due to FGF2 acting directly on OCT4 thus resulting in self-renewal of hESC (Brumbaugh, et al. 2012).
Similarly to hESC, culturing mEpiSC in the presence of BMP and Activin A will also maintain an undifferentiated cellular state, but FGF signaling appears to play different roles in each cell type. In mEpiSC, FGF2 stabilizes the epiblast state by inhibiting differentiation to neuroectoderm and inhibiting the reversion to a mESC-like state but in hESC, FGF synergizes with SMAD 2/3 signaling resulting in NANOG gene expression and maintenance of self-renewal (Greber et al. 2010). Thus, each cell line and the subsequent data generated from each cell line provide unique and specific results to the individual line. Moreover, caution should be taken when directly translating results from one species to another.
BMP-induced differentiation
One of the early breakthroughs in the use of hESC to model trophoblast differentiation was the discovery that treatment of hESC with BMP4 or other similar ligands (BMP2, BMP7, GDF5) induced uniform trophoblast differentiation, in a time- and dose-dependent manner (Xu et al. 2002). Since this seminal observation, the role of BMP also as a regulator of somatic and extraembryonic lineages becomes more apparent (Greber 2011; Xu et al. 2002). On the other hand, Pera, et al., (2004) found that BMP2 resulted in cells displaying extra-embryonic endoderm characteristics and further showed that treatment of hESC with BMP-antagonist noggin resulted in neural precursors (Pera et al. 2004). In addition, along with WNT/b-catenin signaling, BMP establishes posterior primitive streak and mesoderm progenitors from hESC, but delaying BMP signals results in the differentiation of anterior primitive streak and endoderm progenitors (Sumi et al. 2008). Yu, et al., (2011) showed that in the presence of high concentrations of FGF2, BMP4-induced differentiation of hESC results in mesendoderm (an epiblast-derived progenitor of mesoderm or endoderm) rather than extraembryonic trophoblast differentiation (Yu, et al. 2011). More specifically, it was shown that FGF2, acting through MEK/ERK signaling and in the presence of BMP4, results in the prolonged expression of NANOG that results in FGF-independent, BMP4-induction of mesendoderm (Greber 2011; Yu et al. 2011). Complementary to this study, it has also been shown that the induction of mesoderm from BMP4/FGF2-treated hESC also involves the well-established extraembryonic marker CDX2 and that the mesoderm marker Brachyury (T) precedes CDX2 induction (Bernardo, et al. 2011; Greber 2011). These studies have prompted the conclusion that the cells differentiated in the presence of BMP4 are not trophoblasts but indeed cells of the extraembryonic mesoderm (Bernardo et al. 2011; Greber 2011; Yu et al. 2011).
This conclusion has been challenged by others, who suggest that the choice of using T as a marker for mesoderm may not be valid because T expression has been found in teratocarcinoma cells and in trophoblast cell lines (Ezashi, et al. 2012; Gokhale, et al. 2000). In addition, Ezashi, et al., (2012) have reported that the gene expression profiles indicating embryonic and extra-embryonic endoderm (ie. FLK1, VCAM1, and TBX4) are expressed differentially between the H9 hESC line used by Bernardo, et al., 2011 and the H1 ESC line that they use before and after BMP4-induced differentiation (Ezashi et al. 2012). Moreover, the ELF5 gene was found to be inactive by methylation status in the BMP4-treated cells by Bernardo, et al., (2011) in opposition to what is seen in mTSC, suggesting that trophoblast was not being faithfully differentiated. However, a small subset of the cells actually expressed the ELF5 protein but the methylation status of the ELF5 promoter was not determined therefore indicating a potential early differentiation state of a trophoblast subpopulation (Ezashi et al. 2012). Finally, Bernardo, et al., (2011) did not detect the non-polymorphic surface class I molecule, HLA-G on BMP-derived trophoblasts, which is most often used as a placental marker. In contrast, the H1 hESC line was reported to express HLA-G mRNA when induced to form trophoblasts with BMP4 at even low doses of 10 ng/mL (Das, et al. 2007; Ezashi et al. 2012; Xu et al. 2002). Thus, it remains controversial as to whether BMP4-treated hESC truly differentiate into trophoblast cells.
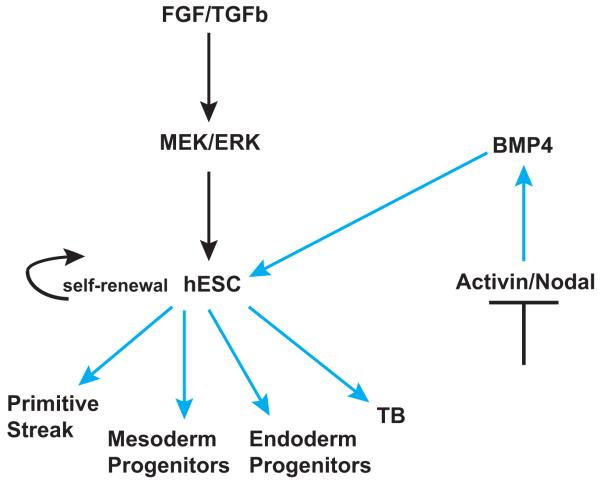
A recent study helping to clarify the differences in gene expression that may arise when ESC cells are primed for differentiation has defined putative progenitor signatures for mesoderm, vascular endothelium, and trophoblast from hESC (Drukker, et al. 2012). Interestingly, when hESC were treated with 100 ng/mL of BMP4 for three days, screened with a monoclonal antibody library using flow cytometry, and subsequently FACS sorted, three distinct progenitor populations were derived (Drukker et al. 2012). Global gene expression analysis indicated trophoblast-specific gene induction, and cell cultures of these progenitors were able to form syncytium. They did not form teratomas when engrafted into immunodeficient mice, but rather formed mesenchymal tissues and epithelial structures (Drukker et al. 2012). Therefore the point is made that BMP4 differentiation induction may be bi-directional and that potentially a small subset of cells truly undergo differentiation toward the trophoblast lineage and are not an artifact of differentiation toward the mesoderm lineage (Figure 2).
Figure 2.
Schematic of the cell signaling that dictates hESC self-renewal and differentiation induction by BMP4. Controversy exists as to whether BMP4 signaling results in trophoblast (TB) lineage differentiation or whether BMP4 along with the continuous presence of FGF result in primitive streak and mesendoderm derivitives and not trophoblast. Black arrows indicate inductive pathways for hESC self-renewal. Blue arrows indicate differentiation induction of hESC.
Moreover, comparative transcriptome analysis of hESC-derived trophoblasts by BMP4 treatment and mural trophectoderm cells isolated from human blastocysts revealed 138 genes in common between the groups with similarities in major canonical pathways and proteins secreted that are involved in the implantation process (Aghajanova, et al. 2012). Additionally, it was determined that trophoblast cells derived on days 8, 10, and 12 of BMP4 treatment were more consistent on a transcriptional level to trophectoderm cells than trophoblasts derived on days 0, 2, 4, and 6 of BMP4 treatment thus further supporting BMP4 treatment of hESC as a viable model to study trophoblast differentiation and development (Aghajanova et al. 2012).
The embryoid-body model for trophoblast differentiation
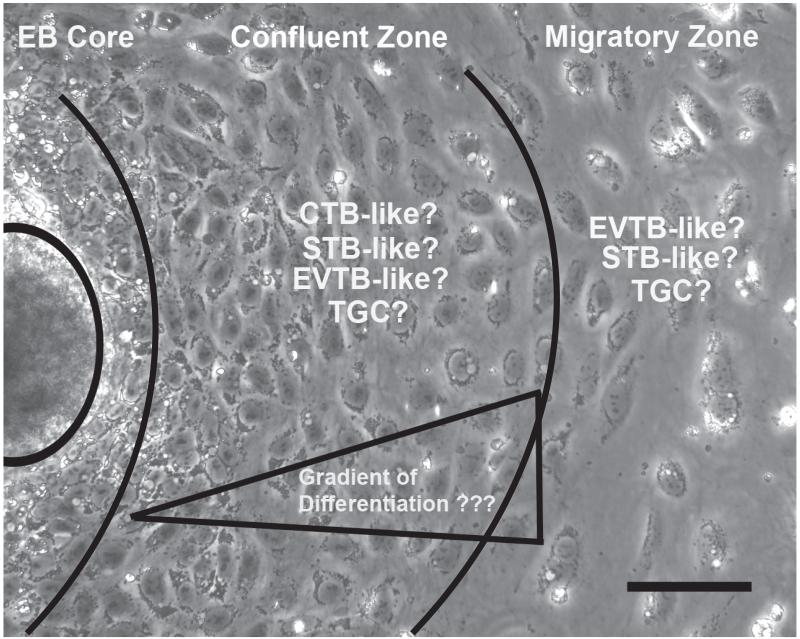
An alternative approach for the formation of trophoblasts from hESC was established which entails forming suspension cultures of aggregates or EBs from undifferentiated hESC (Gerami-Naini, et al. 2004). This system has proven amenable for studies investigating the spatial interactions between cells and the surrounding extracellular matrix (Gerami-Naini et al. 2004; Giakoumopoulos, et al. 2010). Differentiation in this paradigm is achieved by EB formation, the simultaneous withdrawal of FGF2 from the culture medium, and the addition of fetal bovine serum (FBS). When EBs were maintained in suspension culture hCG, progesterone, and estradiol-17b secretion was initiated compared to unconditioned culture medium alone (Gerami-Naini et al. 2004). Adherent cultures of these EBs led to outgrowths of cells with epithelial morphology that maintained hCG, progesterone, and steroid hormone secretion (Figure 3). Moreover, when these suspended EBs were in 3-dimensional culture with Matrigel “rafts”, cellular protrusions appeared and placental hormone secretion was significantly higher compared to EBs maintained in suspension culture or cells in 2-dimensional adherent culture (Gerami-Naini et al. 2004).
Figure 3.
Photomicrograph depicts EB-derived trophoblast outgrowths. Once the EB has adhered to the surface, outgrowths grow and move away from the EB core. The undifferentiated EB mass is circled left. With the EB core still maintaining the potential to differentiate into other tissue derivatives, interests lie in isolating cells near to the core, which make up a confluent cellular zone and isolating the cells at the farthest edge away from the core, which constitute the migratory zone of cells. Studies still remain to elucidate the potential gradient of differentiation that might exist resulting in various trophoblast subtypes. CTB=cytotrophoblasts, STB=syncytiotrophoblasts, EVTB=extravillous cytotrophoblasts, TGC=trophoblast giant cells. Scale bar=100μm.
We have attempted to elucidate the specific extracellular matrix component providing the cues to support EB-derived trophoblast hormone secretion by allowing EBs to adhere to various extracellular matrices in 2-dimensions. Regardless of the matrix utilized, hormone secretion was similar under all conditions, suggesting that adhesion per se, rather than a specific interaction, is adequate for maintaining hormone secretion (Gerami-Naini, et al., in preparation). EB-derived trophoblast outgrowths are able to display migratory characteristics, as is seen in extravillous cytotrophoblasts, when placed in a migration chamber and display enhanced migration in the presence of secreted factors from endothelial cells (Golos, et al. 2010). In addition, incorporating placental fibroblasts into EBs with an aggregation protocol followed by suspension culture provides cellular cues that enhanced placental hormone secretion (Giakoumopoulos et al. 2010). Moreover, Harun, et al., 2006 utilized the EB model system to derive their TSC lines (Harun et al. 2006). Thus, the EB system has provided a useful and adaptable model for trophoblast derivation.
Recent efforts in our lab have been to utilize a system that aggregates EBs of uniform size and shape thus alleviating some of the heterogeneity that results from traditional methods where EBs were formed from undifferentiated colonies of heterogeneous size by light enzymatic digestion. Improving the consistency among EBs will be a valuable refinement of the alternative to BMP4 treatment for trophoblast differentiation.
One of the more problematic issues in the use of hESC to derive trophoblasts remains the uncertainty as to the heterogeneity of the “trophoblast” population obtained. The outgrowths that result from adherent EB culture (Figure 3), display morphologically distinct cells as cells grow away from the EB “core”. Similar heterogeneity of differentiation was demonstrated by Ezashi, et al., (2012) and spatial heterogeneity in functional individual markers such as hCG expression was dependent on cellular location within the hESC colony when differentiation was induced by BMP4 treatment.
Thus, an important goal for refining the use of hESC models for the formation of trophoblasts is a better definition of cell heterogeneity, and the formulation of approaches to achieve more highly purified populations of cells. This quest to isolate a pure differentiated population is ongoing and Drukker, et al., (2012) clearly stated the necessity to do so by saying, “Without purifying true pluripotent cells and differentiating cells, it is difficult to determine, for example, whether low mRNA levels of differentiating genes detected in cultures of mESCs (or hESCs) reflect priming of lineage specification programs in undifferentiated cells or the presence of small populations of differentiating cells.” By using selective surface markers for subpopulation isolation, highly purified differentiated cells can be obtained and thus genomic/proteomic libraries can be created for universal reference before embarking on a new experimental protocol or manipulation.
Afterword
Extrapolating in vitro studies with hESC to the in vivo process of embryo implantation and placenta formation is a major challenge for the field. One important consideration is that the hESC lines that are used may actually give different results due to differences in isolation methods, passage number, maintenance of pluripotency, culture density, and specific culture conditions, such as extracellular matrix or soluble factors used for differentiation induction. Thus, standardized protocols and careful descriptions of cellular detail is important. In addition, emerging evidence indicates that the time course of differentiation is crucial and current studies suggest that BMP4-induced differentiation of hESC results in differential lineage induction at different time points and under different culture conditions (e.g., in the presence of FGF2). Finally, careful selection of markers used for lineage identification, and attention to the specificity of the expression profile generated by spatial and temporal expression patterns produced in vivo is a critical area for further study. It is hoped that in the future, defining the signaling nuances that exist between the pluripotent, primed, and differentiated state of ESC, will lead toward the development of robust protocols for human TSC derivation that will aid in understanding human embryo implantation and subsequent development of the placenta, the organ that, is a prerequisite of our own existence.
Acknowledgments
Many lab members contributed to the studies in the Golos lab discussed in this paper, particularly Behzad Gerami-Naini, Oksana Dovzhenko, Mark Garthwaite and Maureen Durning., and we gratefully acknowledge their assistance.
Funding.
This work was supported by NIH grants RR021876, HD38843, HD34215 and HD53925 to T.G.G., and P51OD011106/P51RR000167 to the WNPRC. M. Giakoumopoulos was supported in part by T32 HD041921. This research was conducted in part at a facility constructed with support from Research Facilities Improvement Program grant numbers RR15459-01 and RR020141-01. This publication’s contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
Footnotes
Declaration of interest:
There is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Achen MG, Gad JM, Stacker SA, Wilks AF. Placenta growth factor and vascular endothelial growth factor are co-expressed during early embryonic development. Growth Factors. 1997;15:69–80. doi: 10.3109/08977199709002113. [DOI] [PubMed] [Google Scholar]
- Adamson SL, Lu Y, Whiteley KJ, Holmyard D, Hemberger M, Pfarrer C, Cross JC. Interactions between trophoblast cells and the maternal and fetal circulation in the mouse placenta. Dev Biol. 2002;250:358–373. doi: 10.1016/s0012-1606(02)90773-6. [DOI] [PubMed] [Google Scholar]
- Adjaye J, Huntriss J, Herwig R, BenKahla A, Brink TC, Wierling C, Hultschig C, Groth D, Yaspo ML, Picton HM, et al. Primary differentiation in the human blastocyst: comparative molecular portraits of inner cell mass and trophectoderm cells. Stem Cells. 2005;23:1514–1525. doi: 10.1634/stemcells.2005-0113. [DOI] [PubMed] [Google Scholar]
- Aghajanova L, Shen S, Rojas AM, Fisher SJ, Irwin JC, Giudice LC. Comparative transcriptome analysis of human trophectoderm and embryonic stem cell derived trophoblasts reveal key participants in early implantation. Biol Reprod. 2012;86:1–21. doi: 10.1095/biolreprod.111.092775. [DOI] [PubMed] [Google Scholar]
- Anson-Cartwright L, Dawson K, Holmyard D, Fisher SJ, Lazzarini RA, Cross JC. The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat Genet. 2000;25:311–314. doi: 10.1038/77076. [DOI] [PubMed] [Google Scholar]
- Aristotle, Peck Al. Generation of animals. W. Heinemann; Harvard University Press; London Cambridge, Mass.: 1943. [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Benirschke K, Kaufmann P, Baergen RN. Pathology of the human placenta. Springer; New York: 2006. [Google Scholar]
- Berg DK, Smith CS, Pearton DJ, Wells DN, Broadhurst R, Donnison M, Pfeffer PL. Trophectoderm lineage determination in cattle. Dev Cell. 2011;20:244–255. doi: 10.1016/j.devcel.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, Callery EM, Trotter MW, Hemberger M, Smith JC, et al. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell. 2011;9:144–155. doi: 10.1016/j.stem.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brumbaugh J, Hou Z, Russell JD, Howden SE, Yu P, Ledvina AR, Coon JJ, Thomson JA. Phosphorylation regulates human OCT4. Proc Natl Acad Sci U S A. 2012;109:7162–7168. doi: 10.1073/pnas.1203874109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon T, Chambers I, Stracey C, Niwa H, Smith A. Signaling mechanisms regulating self-renewal and differentiation of pluripotent embryonic stem cells. Cells Tissues Organs. 1999;165:131–143. doi: 10.1159/000016693. [DOI] [PubMed] [Google Scholar]
- Carter AM, Pijnenborg R. Evolution of invasive placentation with special reference to non-human primates. Best Pract Res Clin Obstet Gynaecol. 2011;25:249–257. doi: 10.1016/j.bpobgyn.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K, James R, Hammond VE, Kontgen F, Beck F. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature. 1997;386:84–87. doi: 10.1038/386084a0. [DOI] [PubMed] [Google Scholar]
- Cross JC. How to make a placenta: mechanisms of trophoblast cell differentiation in mice--a review. Placenta. 2005;26(Suppl A):S3–9. doi: 10.1016/j.placenta.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Darr H, Mayshar Y, Benvenisty N. Overexpression of NANOG in human ES cells enables feeder-free growth while inducing primitive ectoderm features. Development. 2006;133:1193–1201. doi: 10.1242/dev.02286. [DOI] [PubMed] [Google Scholar]
- Das P, Ezashi T, Schulz LC, Westfall SD, Livingston KA, Roberts RM. Effects of fgf2 and oxygen in the bmp4-driven differentiation of trophoblast from human embryonic stem cells. Stem Cell Res. 2007;1:61–74. doi: 10.1016/j.scr.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Los Angeles A, Loh YH, Tesar PJ, Daley GQ. Accessing naive human pluripotency. Curr Opin Genet Dev. 2012;22:272–282. doi: 10.1016/j.gde.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnison M, Beaton A, Davey HW, Broadhurst R, L’Huillier P, Pfeffer PL. Loss of the extraembryonic ectoderm in Elf5 mutants leads to defects in embryonic patterning. Development. 2005;132:2299–2308. doi: 10.1242/dev.01819. [DOI] [PubMed] [Google Scholar]
- Douglas GC, VandeVoort CA, Kumar P, Chang TC, Golos TG. Trophoblast stem cells: models for investigating trophectoderm differentiation and placental development. Endocr Rev. 2009;30:228–240. doi: 10.1210/er.2009-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drukker M, Tang C, Ardehali R, Rinkevich Y, Seita J, Lee AS, Mosley AR, Weissman IL, Soen Y. Isolation of primitive endoderm, mesoderm, vascular endothelial and trophoblast progenitors from human pluripotent stem cells. Nat Biotechnol. 2012;30:531–542. doi: 10.1038/nbt.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlebacher A, Price KA, Glimcher LH. Maintenance of mouse trophoblast stem cell proliferation by TGF-beta/activin. Dev Biol. 2004;275:158–169. doi: 10.1016/j.ydbio.2004.07.032. [DOI] [PubMed] [Google Scholar]
- Ezashi T, Telugu BP, Roberts RM. Model systems for studying trophoblast differentiation from human pluripotent stem cells. Cell Tissue Res. 2012 doi: 10.1007/s00441-012-1371-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genbacev O, Donne M, Kapidzic M, Gormley M, Lamb J, Gilmore J, Larocque N, Goldfien G, Zdravkovic T, McMaster MT, et al. Establishment of human trophoblast progenitor cell lines from the chorion. Stem Cells. 2011;29:1427–1436. doi: 10.1002/stem.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerami-Naini B, Dovzhenko OV, Durning M, Wegner FH, Thomson JA, Golos TG. Trophoblast differentiation in embryoid bodies derived from human embryonic stem cells. Endocrinology. 2004;145:1517–1524. doi: 10.1210/en.2003-1241. [DOI] [PubMed] [Google Scholar]
- Giakoumopoulos M, Siegfried LM, Dambaeva SV, Garthwaite MA, Glennon MC, Golos TG. Placental-derived mesenchyme influences chorionic gonadotropin and progesterone secretion of human embryonic stem cell-derived trophoblasts. Reprod Sci. 2010;17:798–808. doi: 10.1177/1933719110371853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale PJ, Giesberts AM, Andrews PW. Brachyury is expressed by human teratocarcinoma cells in the absence of mesodermal differentiation. Cell Growth Differ. 2000;11:157–162. [PubMed] [Google Scholar]
- Golos TG, Giakoumopoulos M, Garthwaite MA. Embryonic stem cells as models of trophoblast differentiation: progress, opportunities, and limitations. Reproduction. 2010;140:3–9. doi: 10.1530/REP-09-0544. [DOI] [PubMed] [Google Scholar]
- Greber B. When BMP meets FGF. Cell Stem Cell. 2011;9:91–92. doi: 10.1016/j.stem.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Greber B, Wu G, Bernemann C, Joo JY, Han DW, Ko K, Tapia N, Sabour D, Sterneckert J, Tesar P, et al. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell. 2010;6:215–226. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Groskopf JC, Syu LJ, Saltiel AR, Linzer DI. Proliferin induces endothelial cell chemotaxis through a G protein-coupled, mitogen-activated protein kinase-dependent pathway. Endocrinology. 1997;138:2835–2840. doi: 10.1210/endo.138.7.5276. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner AL. Essential role of Mash-2 in extraembryonic development. Nature. 1994;371:333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton WJ, Boyd JD. Development of the human placenta in the first three months of gestation. J Anat. 1960;94:297–328. [PMC free article] [PubMed] [Google Scholar]
- Hamilton WJ, Boyd JD. Trophoblast in human utero-placental arteries. Nature. 1966;212:906–908. doi: 10.1038/212906a0. [DOI] [PubMed] [Google Scholar]
- Harun R, Ruban L, Matin M, Draper J, Jenkins NM, Liew GC, Andrews PW, Li TC, Laird SM, Moore HD. Cytotrophoblast stem cell lines derived from human embryonic stem cells and their capacity to mimic invasive implantation events. Hum Reprod. 2006;21:1349–1358. doi: 10.1093/humrep/del017. [DOI] [PubMed] [Google Scholar]
- Hemberger M, Udayashankar R, Tesar P, Moore H, Burton GJ. ELF5-enforced transcriptional networks define an epigenetically regulated trophoblast stem cell compartment in the human placenta. Hum Mol Genet. 2010;19:2456–2467. doi: 10.1093/hmg/ddq128. [DOI] [PubMed] [Google Scholar]
- Home P, Saha B, Ray S, Dutta D, Gunewardena S, Yoo B, Pal A, Vivian JL, Larson M, Petroff M, et al. Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc Natl Acad Sci U S A. 2012;109:7362–7367. doi: 10.1073/pnas.1201595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James JL, Carter AM, Chamley LW. Human placentation from nidation to 5 weeks of gestation. Part I: What do we know about formative placental development following implantation? Placenta. 2012;33:327–334. doi: 10.1016/j.placenta.2012.01.020. [DOI] [PubMed] [Google Scholar]
- Janatpour MJ, Utset MF, Cross JC, Rossant J, Dong J, Israel MA, Fisher SJ. A repertoire of differentially expressed transcription factors that offers insight into mechanisms of human cytotrophoblast differentiation. Dev Genet. 1999;25:146–157. doi: 10.1002/(SICI)1520-6408(1999)25:2<146::AID-DVG9>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Kamei T, Jones SR, Chapman BM, KL MC, Dai G, Soares MJ. The phosphatidylinositol 3-kinase/Akt signaling pathway modulates the endocrine differentiation of trophoblast cells. Mol Endocrinol. 2002;16:1469–1481. doi: 10.1210/mend.16.7.0878. [DOI] [PubMed] [Google Scholar]
- Kaufmann P, Black S, Huppertz B. Endovascular trophoblast invasion: implications for the pathogenesis of intrauterine growth retardation and preeclampsia. Biol Reprod. 2003;69:1–7. doi: 10.1095/biolreprod.102.014977. [DOI] [PubMed] [Google Scholar]
- Kingdom J, Huppertz B, Seaward G, Kaufmann P. Development of the placental villous tree and its consequences for fetal growth. Eur J Obstet Gynecol Reprod Biol. 2000;92:35–43. doi: 10.1016/s0301-2115(00)00423-1. [DOI] [PubMed] [Google Scholar]
- Kraut N, Snider L, Chen CM, Tapscott SJ, Groudine M. Requirement of the mouse I-mfa gene for placental development and skeletal patterning. EMBO J. 1998;17:6276–6288. doi: 10.1093/emboj/17.21.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuckenberg P, Buhl S, Woynecki T, van Furden B, Tolkunova E, Seiffe F, Moser M, Tomilin A, Winterhager E, Schorle H. The transcription factor TCFAP2C/AP-2gamma cooperates with CDX2 to maintain trophectoderm formation. Mol Cell Biol. 2010;30:3310–3320. doi: 10.1128/MCB.01215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Luo J, Sladek R, Bader JA, Matthyssen A, Rossant J, Giguere V. Placental abnormalities in mouse embryos lacking the orphan nuclear receptor ERR-beta. Nature. 1997;388:778–782. doi: 10.1038/42022. [DOI] [PubMed] [Google Scholar]
- Magner L. A History of the Life Sciences. Marcel Dekker, Inc; New York: 2002. [Google Scholar]
- Morsut L, Yan KP, Enzo E, Aragona M, Soligo SM, Wendling O, Mark M, Khetchoumian K, Bressan G, Chambon P, et al. Negative control of Smad activity by ectodermin/Tif1gamma patterns the mammalian embryo. Development. 2010;137:2571–2578. doi: 10.1242/dev.053801. [DOI] [PubMed] [Google Scholar]
- Murohashi M, Nakamura T, Tanaka S, Ichise T, Yoshida N, Yamamoto T, Shibuya M, Schlessinger J, Gotoh N. An FGF4-FRS2alpha-Cdx2 axis in trophoblast stem cells induces Bmp4 to regulate proper growth of early mouse embryos. Stem Cells. 2010;28:113–121. doi: 10.1002/stem.247. [DOI] [PubMed] [Google Scholar]
- Najm FJ, Chenoweth JG, Anderson PD, Nadeau JH, Redline RW, McKay RD, Tesar PJ. Isolation of epiblast stem cells from preimplantation mouse embryos. Cell Stem Cell. 2011;8:318–325. doi: 10.1016/j.stem.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng RK, Dean W, Dawson C, Lucifero D, Madeja Z, Reik W, Hemberger M. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol. 2008;10:1280–1290. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Pelton TA, Sharma S, Schulz TC, Rathjen J, Rathjen PD. Transient pluripotent cell populations during primitive ectoderm formation: correlation of in vivo and in vitro pluripotent cell development. J Cell Sci. 2002;115:329–339. doi: 10.1242/jcs.115.2.329. [DOI] [PubMed] [Google Scholar]
- Pera MF, Andrade J, Houssami S, Reubinoff B, Trounson A, Stanley EG, Ward-van Oostwaard D, Mummery C. Regulation of human embryonic stem cell differentiation by BMP-2 and its antagonist noggin. J Cell Sci. 2004;117:1269–1280. doi: 10.1242/jcs.00970. [DOI] [PubMed] [Google Scholar]
- Pijnenborg R, Dixon G, Robertson WB, Brosens I. Trophoblastic invasion of human decidua from 8 to 18 weeks of pregnancy. Placenta. 1980;1:3–19. doi: 10.1016/s0143-4004(80)80012-9. [DOI] [PubMed] [Google Scholar]
- Pijnenborg R, Robertson WB, Brosens I. Trophoblast invasion and formation of the basal plate in the human placenta. Bibl Anat. 1982:69–73. [PubMed] [Google Scholar]
- Pijnenborg R, Robertson WB, Brosens I, Dixon G. Review article: trophoblast invasion and the establishment of haemochorial placentation in man and laboratory animals. Placenta. 1981;2:71–91. doi: 10.1016/s0143-4004(81)80042-2. [DOI] [PubMed] [Google Scholar]
- Pijnenborg R, Vercruysse L, Hanssens M. The uterine spiral arteries in human pregnancy: facts and controversies. Placenta. 2006;27:939–958. doi: 10.1016/j.placenta.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, Guo G, Robson P, Draper JS, Rossant J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137:395–403. doi: 10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- Rielland M, Hue I, Renard JP, Alice J. Trophoblast stem cell derivation, cross-species comparison and use of nuclear transfer: new tools to study trophoblast growth and differentiation. Dev Biol. 2008;322:1–10. doi: 10.1016/j.ydbio.2008.07.017. [DOI] [PubMed] [Google Scholar]
- Riley P, Anson-Cartwright L, Cross JC. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat Genet. 1998;18:271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- Roberts RM, Ezashi T, Das P. Trophoblast gene expression: transcription factors in the specification of early trophoblast. Reprod Biol Endocrinol. 2004;2:47. doi: 10.1186/1477-7827-2-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RM, Fisher SJ. Trophoblast stem cells. Biol Reprod. 2011;84:412–421. doi: 10.1095/biolreprod.110.088724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- Senner CE, Hemberger M. Regulation of early trophoblast differentiation-lessons from the mouse. Placenta. 2010;31:944–950. doi: 10.1016/j.placenta.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Sibilia M, Wagner EF. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science. 1995;269:234–238. doi: 10.1126/science.7618085. [DOI] [PubMed] [Google Scholar]
- Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares MJ, Chapman BM, Rasmussen CA, Dai G, Kamei T, Orwig KE. Differentiation of trophoblast endocrine cells. Placenta. 1996;17:277–289. doi: 10.1016/s0143-4004(96)90051-x. [DOI] [PubMed] [Google Scholar]
- Sumi T, Tsuneyoshi N, Nakatsuji N, Suemori H. Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/beta-catenin, Activin/Nodal and BMP signaling. Development. 2008;135:2969–2979. doi: 10.1242/dev.021121. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Okita K, Nakagawa M, Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc. 2007;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Teesalu T, Blasi F, Talarico D. Expression and function of the urokinase type plasminogen activator during mouse hemochorial placental development. Dev Dyn. 1998;213:27–38. doi: 10.1002/(SICI)1097-0177(199809)213:1<27::AID-AJA3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Teesalu T, Masson R, Basset P, Blasi F, Talarico D. Expression of matrix metalloproteinases during murine chorioallantoic placenta maturation. Dev Dyn. 1999;214:248–258. doi: 10.1002/(SICI)1097-0177(199903)214:3<248::AID-AJA8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris RC, et al. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science. 1995;269:230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- Vandevoort CA, Thirkill TL, Douglas GC. Blastocyst-derived trophoblast stem cells from the rhesus monkey. Stem Cells Dev. 2007;16:779–788. doi: 10.1089/scd.2007.0020. [DOI] [PubMed] [Google Scholar]
- Vuorela P, Hatva E, Lymboussaki A, Kaipainen A, Joukov V, Persico MG, Alitalo K, Halmesmaki E. Expression of vascular endothelial growth factor and placenta growth factor in human placenta. Biol Reprod. 1997;56:489–494. doi: 10.1095/biolreprod56.2.489. [DOI] [PubMed] [Google Scholar]
- Werling U, Schorle H. Transcription factor gene AP-2 gamma essential for early murine development. Mol Cell Biol. 2002;22:3149–3156. doi: 10.1128/MCB.22.9.3149-3156.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Zhang W, Chen G, Cheng L, Liao J, Jia N, Gao Y, Dai H, Yuan J, Xiao L. Combinatorial signals of activin/nodal and bone morphogenic protein regulate the early lineage segregation of human embryonic stem cells. J Biol Chem. 2008;283:24991–25002. doi: 10.1074/jbc.M803893200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- Xu RH, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, Yu J, Antosiewicz-Bourget J, Tian S, Stewart R, et al. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, Buonanno A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134:3827–3836. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Flannery ML, Kupriyanov S, Pearce J, McKercher SR, Henkel GW, Maki RA, Werb Z, Oshima RG. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 1998;12:1315–1326. doi: 10.1101/gad.12.9.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeap LS, Hayashi K, Surani MA. ERG-associated protein with SET domain (ESET)-Oct4 interaction regulates pluripotency and represses the trophectoderm lineage. Epigenetics Chromatin. 2009;2:12. doi: 10.1186/1756-8935-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lins derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu P, Pan G, Yu J, Thomson JA. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell. 2011;8:326–334. doi: 10.1016/j.stem.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]