Abstract
Characterizing protein-protein association quantitatively has been a longstanding challenge for computer simulations. Here, a theoretical framework is put forth that addresses this challenge on the basis of detailed all-atom molecular dynamics simulations with explicit solvent. The proposed methodology relies upon independent potential of mean force (PMF) free-energy calculations carried out sequentially, wherein the biological objects are restrained in the conformation, position and orientation of the bound state, using adequately chosen biasing potentials. These restraints systematically narrow down the configurational entropy available to the system and effectively guarantee that the relevant network of interactions is properly sampled as the two proteins reversibly associate. Decomposition of the binding process into consecutive, well-delineated stages, for both the protein complex and the individual, unbound partners, offers a rigorous definition of the standard state, from which the absolute binding free energy can be determined. The method is applied to the difficult case of the extracellular ribonuclease barnase binding to its intracellular inhibitor barstar. The calculated binding free energy is −21.0 ± 1.4 kcal/mol, which compares well with the experimental value of −19.0 ± 0.2 kcal/mol. The relatively small statistical error reflects the precision and convergence afforded by the PMF-based simulation methodology. In addition to providing an accurate reproduction of the standard binding free energy, the proposed strategy offers a detailed picture of the protein-protein interface, illuminating the thermodynamic forces that underlie reversible association. The application of the present formal framework to barnase:barstar binding provides a foundation for tackling nearly any protein-protein complex.
Introduction
A host of critically important functions for cellular machinery rely on protein-protein recognition and association.1–3 Perturbation and disruption of the network of interactions underlying the formation of protein-protein complexes lead to a number of pathologies.3 Binding affinities, which reflect the natural inclination of molecular entities to associate, are key thermodynamic quantities for understanding recognition and association phenomena, and possible dysfunctions thereof. Such is the case, among many others, for G-protein-coupled receptors, the dimerization of which has been shown in a number of instances to be a prerequisite for fulfilling their cellular function,4 for β–amyloid proteins, the aggregation of which is an early event in the development of neurodegenerative diseases,5 and for proteins of the signal transduction cascade, e.g., the human GTP–hydrolyzing Ras protein interacting with the kinase Raf.6 From a purely physical standpoint, the equilibrium binding constant Keq= [A:B]/[A][B], where A and B are two molecular entities that bind in a 1:1 ratio, pertains to the relative probability of two end-states, one in which they form a stable complex, the other where they are spatially apart and non-interacting. The standard binding free energy,
| (1) |
where C∘ is the standard concentration of 1 M, is a well-defined thermodynamic quantity that is amenable, in principle, to computational analysis. The formal simplicity is, however, deceptive and the problem remains daunting. The complexity of protein–protein recognition and association,1 in particular the combinatorial explosion of possible side-chain interactions at the interface, partly explains the paucity of theoretical investigations compared to the considerable effort aimed at addressing ligand–protein binding. Several approximate treatments aimed at calculating the binding free energy of protein complexes have been proposed, including empirical knowledge-based approaches,7,8 coarse-grained potentials,9,10 and continuum dielectric solvent treatments.11 Unfortunately, none of these approximate treatments is sufficiently general to yield reliable binding free energies for protein–protein complexes that depart from the original training set, nor do they provide an atomic-level account of recognition and association phenomena.
Molecular dynamics (MD) simulations based on all-atom models and explicit solvent are, arguably, the only approach that might be able to yield a sufficiently accurate representation of the microscopic interactions in these complex systems. Computations of based on MD simulations have a long history that can be traced back to the eighties. 12,13 The most widely used approach is free energy perturbation (FEP), wherein the ligand is progressively decoupled from its environment by means of unphysical intermediate Hamiltonians. This “alchemical route” has been the object of extensive formal analysis to clarify the relationship between the decoupled ligand and the standard state. In its most sophisticated embodiment, various restraining potentials are introduced to control the fluctuations of the ligand and, thus, improve convergence. 14–17 Careful applications of the alchemical route have demonstrated that it can yield well converged results in excellent agreement with experiment, though estimating the stability of a protein-protein complex represents a significantly more arduous endeavor due to the size and the complexity of the two binding partners.18 The alchemical route, which is essentially limited to the treatment of relatively small ligands, is inapplicable to this situation: simulations during which an entire macromolecule would be decoupled from its environment are simply unmanageable due to its very large solvation free energy and a corresponding difficulty in reaching convergence.
A different strategy, which considers the potential of mean force (PMF) as a function of the physical separation of the two binding partners, offers a promising alternative. In its simplest form, the equilibrium association constant can be expressed in terms of a one-dimensional radial PMF, W(r),19,20
| (2) |
where r is the distance between the ligand and its receptor, and rmax is the cutoff distance defining the bound state. In principle, W(r) can be calculated efficiently from a series of biased simulations (it is necessary to bridge the two end-points by sampling over a series of intermediates states with overlapping distributions). Given the obvious limitations of the alchemical route, a PMF-based approach appears as the only viable option for determining protein—protein standard binding free energies. To be successful, it must, nonetheless, necessarily go beyond the rudimentary expression of Eq. 2. For example, sampling over all possible relative orientations of the two proteins–an implicit requirement of Eq. 2,19 may be difficult, if not impossible with finite-length biased simulations. Binding free energy calculations of two proteins relying on the simple one-dimensional PMF appearing in Eq. 2 require, in principle, umbrella sampling window simulations that are sufficiently long to fully sample the random tumbling of the two proteins at all separation distances. PMF calculations that do not fulfill this condition are fundamentally invalid. Such difficulties can be circumvented by introducing suitable restraining potentials during the PMF calculation,21–23 the contribution to the net binding affinity of which is evaluated in separate free-energy calculations. This approach is formally correct under any applied restraints, although a judicious choice of such restraints is necessary to ensure utmost efficiency. Recently, an extensive comparison of the alchemical and PMF routes accounting rigorously for all biasing restraints showed that equivalent results can be obtained with both methods.23
The PMF-based method with restraints was originally designed to tackle the binding of moderate-size flexible ligands and peptides,21–23 although an application of the original formulation to the case of KID and KIX domains, which are 28 and 81 residues, respectively, was successful.24 The PMF-based method was also adapted to account for the large conformational changes induced by the association of various ligands to the iGluR glutamate receptor.25 In the present contribution, the method is further extended and generalized to treat more ambitious protein–protein recognition and association problems. The extended method comprises multiple steps involving a collection of geometrical and conformational restraints enforced not only on the two protein scaffolds, but also, specifically, on the constituent amino acids that form their interfacial region. As a first application of this strategy, we determine the absolute binding affinity of the stoichiometric complex formed by the larger extracellular ribonuclease of Bacillus amyloliquefaciens (110 residues) and its intracellular inhibitor (89 residues), commonly referred to as barnase:barstar complex.26,27 The complex serves as a prototypical model system to investigate equilibrium18 and kinetic28 aspects of protein–protein recognition and association (see Figure 1). The calculated binding free energy is −21.0 ± 1.4 kcal/mol, which compares well with the experimental value of −19.0 ± 0.2 kcal/mol kcal/mol. Tracking the complete separation process from the bound to the unbound state of barnase:barstar sheds additional light into the mechanism whereby these two proteins recognize and associate and how the aqueous environment mediates these processes. While the good agreement with experiment depends on the force field and may be partly fortuitous, the noticeably small statistical error for a system of such complexity is indicative of the high precision and convergence afforded by the present PMF-based simulation methodology. In contrast, our own alternate simulations of comparable lengths (410 ns) carried out with no restraints on the interfacial side chains failed to converge. This comparison, in which even the removal of just one set of restraints during separation increases the convergence time significantly, highlights the considerable gain in computational efficiency provided by the proposed PMF-based method with restraining potentials. To the best of our knowledge, a methodology such as described here appears as one of the very few possible avenues to obtain converged binding free energies using typical molecular dynamics simulations.
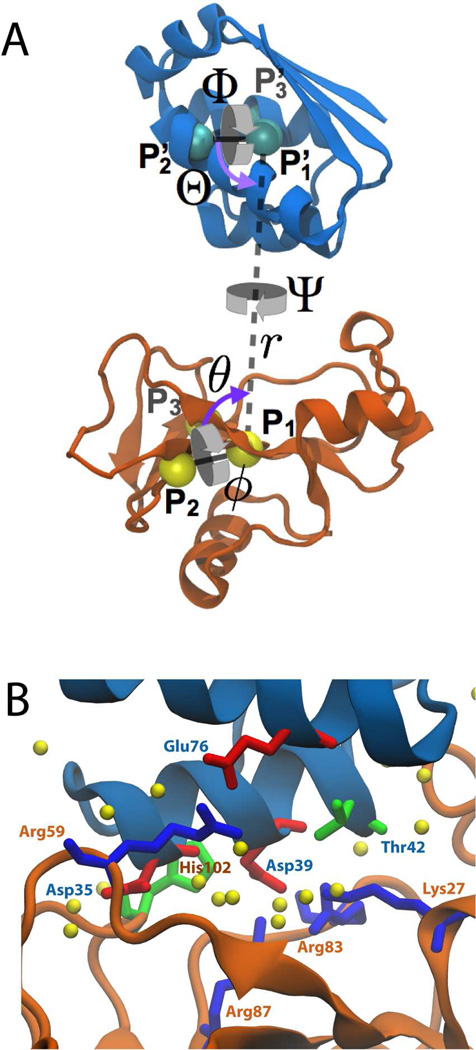
Figure 1.
Structures of barstar (blue) and barnase (orange). (A) Reference coordinates used to define the orientational and positional restraints. The spherical coordinates r (P1–P1’ distance), θ (P1’-P1-P2 angle), and ϕ (P1’-P1-P2-P3 angle), relate the position of barstar with respect to barnase. The Euler angles, Θ (P1-P1’-P2’), Φ (P1-P1’-P2’-P3’), and Ψ (P2-P1-P1’-P2’), determine the relative orientation of barstar. (B) Interface in the crystal structure. Highlighted as sticks are residues noted in the main text, colored by residue type (red for acidic, blue for basic, green for polar, and white for hydrophobic). Water molecules that hydrate the interface are shown as yellow spheres.
Methods
Calculation of
Determination of the absolute binding free energy between barstar and barnase was carried out using a generalization of the method of Woo and Roux,21 in which a series of collective geometrical and conformational restraints are applied to accelerate convergence of the PMF for separating the two proteins.
The contribution of adding each of these restraints in the bound state and then removing them in the unbound state is also computed and subtracted from the free energy resulting from the separation PMF. Many, if not all, of the necessary restraints are common to almost all applications of the method and include those on the root mean-square deviation (RMSD) of the backbone of each protein along with ones on the orientational and positional angles relating barstar to barnase.23 The former RMSD restraints serve to limit conformational changes during separation that are not germane to the free-energy calculation, namely those that involve significant deviations from the bound-state structure. The angular restraints limit the configurational entropy that results from tumbling of the two proteins when separated and also obviate the need of barstar to sample the full 8π2 of solid angle surrounding barnase in the unbound state.
To restrict fluctuations of the side chains while the proteins are separated, additional RMSD restraints were introduced on those residues known to be critical for binding or ones near them. Specifically, one restraint on the RMSD of residues 27, 59, 60, 83, 85, 87, and 102 of barnase and one on the RMSD of residues 29, 31, 33, 35, 38, 39, 42, and 76 of barstar were added. In total, four conformational restraints were applied, two on the backbones (denoted by subscripts Bn,c and Bs,c) and two on the specific set of residues at the interface (Bn,res and Bs,res); additionally, five angular restraints were applied, three on the orientation of barstar relative to barnase (denoted by subscript o, encompassing the Θ, Φ, and Ψ Euler angles as in Figure 1) and two on its relative position (denoted by a, which includes ϕ and θ).
Thus, the binding constant can be expressed as
| (3) |
and the associated binding free energy is given by Eq. 1 with C∘ = 1/1661 Å3.29 The term S* addresses the removal of the positional restraints on barstar, which is extracted from the binding site along a unidirectional axis, rather than being permitted to move freely on the sphere surrounding barnase; it is given by
| (4) |
where r* is a point far from the binding site and ua = uθ + uϕ. The term I* includes the separation PMF, W(r), which is calculated in the presence of all restraints, and is given by
| (5) |
The range of integration for I* is strictly defined over the binding site only; however, there is effectively no difference in I* between integration over a range of ±1.5 Å from the minimum compared to that over the entire range.23
While the full expression for Keq is comprised of more than ten independent free energy terms, only a subset of them need to be calculated explicitly from simulations. Some of the terms involving removing restraints in the bulk, namely and S*, can be calculated through numerical integration owing to the isotropy of space in the unbound state. Still, eight free energies relating to the conformational restraints on the backbone and interfacial residues, as well as five relating to the orientational and positional restraints remain. All of these free energies were calculated from PMFs determined sequentially, with only those restraints present that had been added prior. For example, the PMF due to the RMSD restraint on the barstar backbone in the bound state was determined bereft of other restraints, while those PMFs for the orientational restraints were determined in the presence of all RMSD restraints (see SI for a precise definition of the order). The free energy values resulting from each PMF and the simulation times needed to reach convergence are given in Table 1.
Table 1.
Free energies and simulation lengths associated to the components of
| Contribution | PMF (kcal/mol) | PMF (ns) | |
|---|---|---|---|
| −1.98±0.31 | 6 | ||
| −3.13±0.06 | 12 | ||
| −1.87±0.75 | 12 | ||
| −3.45±0.63 | 24 | ||
| −0.09±0.42 | 8 | ||
| −0.35±0.08 | 4 | ||
| −0.24±0.09 | 8 | ||
| −0.13±0.33 | 4 | ||
| −0.05±0.12 | 4 | ||
| −(1/β) ln(S*I*C∘) | −37.10±0.29 | 212 | |
| +6.61 | – | ||
| +8.05±0.31 | 21 | ||
| +5.15±0.42 | 15 | ||
| +4.15±0.53 | 18 | ||
| +3.39±0.17 | 24 | ||
| −21.04±1.43 | 372 | ||
System construction and simulation
All simulation systems, i.e., one each of barstar and barnase alone and one of the complex, were constructed starting from the 2.0-Å resolution crystallographic structure of the bound complex (PDB code 1BRS).27 Chains C and F were taken from the structure, which of the three unit-cell complexes are expected to be most representative of the native state.27 The N-terminal alanine and glutamine residues were added to barnase, and residues 40 and 82 in barstar were mutated back to cysteine in order to produce a wild-type complex. Water sufficient to encompass proteins and the complex at any separation was added along with 2–6 neutralizing sodium or chloride ions. The final system sizes were approximately 25,000 atoms for the independent proteins and 50,000 atoms for the complex. The dimensions of the molecular assembly were 70 × 70 × 108 Å3, which ensures that even at full separation, the two proteins do not interact artificially across the periodic boundaries.
Simulations were run using NAMD 2.930 and the CHARMM36 force field.31 The temperature and the pressure were maintained at 300K and 1 atm, respectively. The protocols and algorithms utilized herein can be found in reference.23
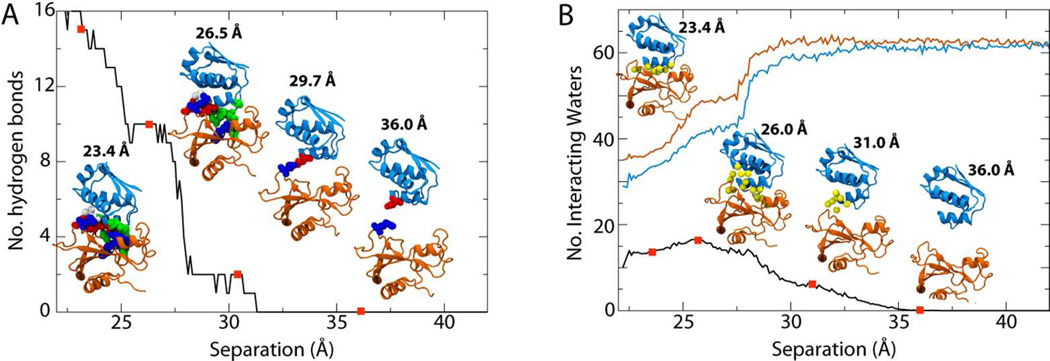
Calculation of all of the PMFs resulting from the applied restraints was accomplished using extended Adaptive Biasing Forces (eABF)23,32,33 as implemented in the collective variables module of NAMD.34 Simulations were run beyond the stated times to confirm convergence of the PMFs in individual windows. For the separation PMF, replica-exchange MD combined with umbrella sampling (REMD-US) was used.23,35 The histograms for all windows were checked for sufficient overlap required by the weighted histogram analysis method (WHAM).36 A gulf between windows was discovered at r=27.5Å, a region in which the two proteins first become truly separated, with many of the interactions simultaneously disrupted, illustrated by, e.g., a sudden drop in hydrogen bonds (see Figure 4A). Therefore, an additional window covering this region was run separately and added to the 52 used for REMD-US.
Figure 4.
Protein-protein (A) and protein-water (B) interactions as a function of barstar-barnase separation distance. The red dots along each curve correspond to the inset figures. (A) Hydrogen bonds between the two proteins. (B) The number of water molecules that interact with both proteins simultaneously are given by the black curve, those that interact with barstar in blue, and those with barnase in orange.
Results
As a prelude to the full binding free-energy calculation, an initial attempt at separation was made using steered MD (SMD).37 Conformational restraints on the backbones and angular restraints were applied on the relative orientation and position of the protein. After nearly 7 ns the interface between the two proteins, mediated by numerous salt bridges and other residue-residue interactions, was abruptly ruptured. It was apparent from the trajectory that these interactions, once broken, could not easily reform, as the side chains involved shifted away from the binding interface. Therefore, straightforward free-energy calculations based on forced separation are unable to capture all relevant states on typical simulation time scales, necessitating additional RMSD restraints on the side chains near the interface of each protein (see Methods).
In principle, it should be possible to observe spontaneous association of the unbound proteins in a sufficiently long simulation.38 Given that kon = 6.0 × 108s−1 M−118 and the volume of the box is ~ 5 × 105Å3, the average association time is expected to be 0.5 µs. To explore this idea, a 2-µs trajectory was generated, in which barstar and barnase were initially separated. The proteins first made contact within less than 100 ns, reaching a stable association within 350 ns, albeit in a non-canonical orientation in which barstar primarily interacts with the RNA-binding loop of barnase, a predicted early stage of binding.39 This state persisted for the remainder of the 2 µs. Although the complex is expected to be recovered within a few µs in this simulation, a successful effort at simulating spontaneous complex formation would probably require an ensemble of independent trajectories of a few microseconds each. However, it is worth emphasizing that estimating the actual binding free energy from unbiased simulations would still require the observation of multiple association and dissociation events, the latter occurring on a time scale that is far too long for present-day computational resources (~ 1.25 × 105s18).
Determination of the binding free energy
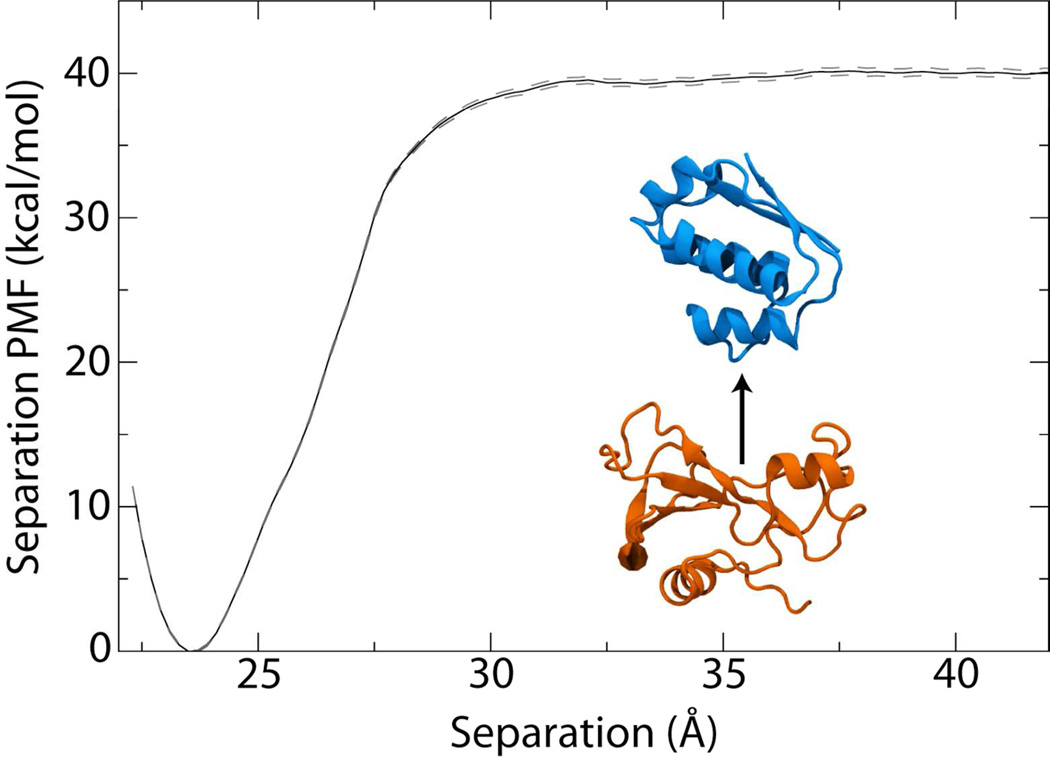
The full list of contributions to the binding free energy are given in Table 1. The separation PMF W(r), calculated using a stratification strategy, i.e., decomposition of the reaction pathway into windows, and umbrella sampling with replica-exchange MD (see Methods), accounts by far for the largest contribution. Shown in Figure 2, the well depth for the bound state is 40 kcal/mol and its related contribution to the free energy is −37.1 kcal/mol. This association free energy is nearly twice the experimental value due to the numerous restraints that minimize unfavorable entropic factors in the binding process and, thus, make sampling appreciably more efficient. Despite the enhanced sampling efficiency, the majority of the computational effort was still invested in determining W(r), which at 212 ns represents over 50% of the total simulation time. Comparison of W(r) at intermediate stages illustrates, however, that convergence of the PMF was practically achieved at much shorter times. While after 1ns/window (53 ns total), the well depth is 38 kcal/mol, after 2 ns/window (106 ns) it already reaches 39.8 kcal/mol, i.e., within 0.2 of the final well depth (see Fig. S1 in SI).
Figure 2.
Potential of mean force (PMF) for the separation of barstar and barnase (inset). The dashed lines represent one standard deviation above and below the PMF.
The remaining PMFs for conformational, orientational and positional restraints were calculated using extended adaptive biasing forces (eABF).23,32,33 Introduction of positional and orientational restraints, in particular, is justified by the virtual impossibility to sample the full range of solid angle accessible to barstar as it dissociates from barnase. As evidence of this sampling limitation, a separate, unbiased simulation of barstar in an aqueous environment reveals that its orientational auto-correlation function measured from the principal axes of the protein does not decay to zero over 150 ns (see Fig. S2). In total, eight PMFs for RMSD restraints (four in the bound state and four in the free state) and five on the angles relating the orientation (Θ, Φ, Ψ) and position (θ, ϕ) were required (see Figure 1 for a definition of the angles). The PMFs for the orientational and positional restraints in the bound state converged rapidly, requiring only 28 ns; their contribution in the unbound state was determined analytically (see Methods). The net contribution from the orientational restraints is 5.8 kcal/mol, with almost all of it originating from the loss of the orientational entropy in the isotropic unbound state . That from the positional restraints in the bound state is negligible (−0.18 kcal/mol), while in the free state is manifest in the term S*, representing the area available to barstar on the sphere surrounding barnase, and is ~18 Å2 (see SI). The loss of translational entropy, i.e., the difference between −kBT ln(S*I*C∘) and the depth of the separation PMF (40 kcal/mol), is 2.9kcal/mol, corresponding to an effective accessible volume of 12.8 Å3.
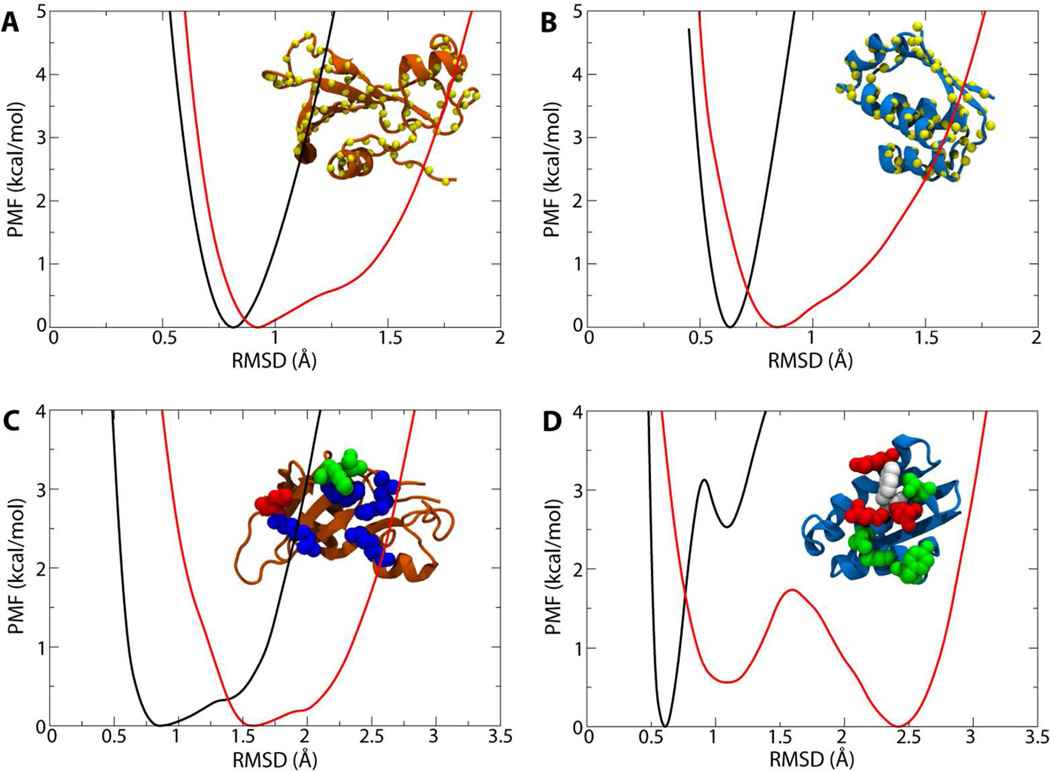
The conformational restraints (RMSD) proved more challenging, due to their rough free-energy landscapes (see Figure 3). In all cases, the well width is broader for the free state compared to the bound state, demonstrating the confining effect of binding (see the red and black curves, respectively). The four PMFs for the conformation of the backbones required a total of 60 ns and give a net contribution to the binding free energy of +2.5 kcal/mol . It has been noted based on their crystal structures that while the conformation of barnase in the bound state resembles that of the free state, the same cannot be said for barstar.27 This difference is reflected in the PMFs for restraining each protein in its bound and free states. For barnase, the minimum of the PMF in the free state is only 0.1 Å greater than that for the bound state, while for barstar, the minima are separated by 0.2 Å (all comparisons being made with reference to the bound state of each; see Figure 3A, B). As for the restraints on the interfacial residues, 72 ns were needed to resolve their contribution, which is among the largest at nearly +8 kcal/mol . This large contribution is suggestive of the significant role of side-chain orientation in optimizing the binding interface. In particular, the relative shift of the two well depths in the bimodal curves in Figure 3D for barstar provide further evidence that recovery of the bound-state orientation of the interfacial side chains will be unlikely to occur on typical simulation time scales.
Figure 3.
PMFs for the eight conformational restraints on the proteins’ RMSD. In each panel, the black curve is the PMF for the bound state, the red curve is the PMF for the bulk state of the protein, and the inset figure displays the affected atoms as spheres. (A,B) PMFs for restraints on the conformation of (A) barnase and (B) barstar. (C,D) PMFs for restraints on selected interfacial residues in (C) barnase and (D) barstar. Restrained residues are colored as in Figure 1B.
The final calculation of Keq from Eq. 3 gives an equilibrium binding constant of 2.13× 1015 M−1, i.e., a free energy of −21.0±1.4kcal/mol at a standard concentration of 1M (see Table 1 and SI for full details of the calculation), compared to an experimental binding free energy of −19.0±0.2 kcal/mol.40 The computational result is confirmed to be independent of the choice of r*, which is only required to be large enough that the two proteins no longer interact,23 with r* = 37.5 Å to 41.5 Å giving the same net free energy (see SI).
Due to the number of PMFs involved, the calculation of Keq is susceptible to an accumulation of errors. Therefore, assessing the convergence of the PMFs is paramount to assuring the accuracy of the result. The statistical error was estimated based on fluctuations of the force in each bin (when using eABF)33,41 or of the position in each window (when using REMD-US);42 the systematic error for eABF-determined PMFs was estimated by examining discontinuities in the gradient of the PMF across adjacent windows.23 Given the conservative assumptions in the error formulae, the resulting values also serve as conservative estimates.33 Nonetheless, in almost all cases, the individual errors are less than 0.5 kcal/mol, and the net error on the final binding free energy is only 1.4kcal/mol, less than 7% of the total.
Key interactions that give rise to the large binding free energy
Interactions between barstar and barnase in the crystal structure of the complex include, in part, 14 hydrogen bonds.27 For short separation distances we observe the same number of hydrogen bonds on average, shown in Figure 4A, the only difference being that the bond between Bs-Gly43 and Bn-Arg83 was not consistently maintained in simulation. These bonds involve a number of residues identified as critical to binding (ΔΔGmut_wt = 5–6 kcal/mol), including Lys27, Arg59, Arg87, and His102 in barnase and Asp35, Asp39, and Glu76 in barstar, among others.18,40,43,44 The number of hydrogen bonds quickly drops to 10 upon a separation of 2 Å from the bound state. A detailed analysis indicates those hydrogen bonds lost are two between Bn-Arg83 and Bs-Asp39, one between Bn-Arg87 and Bs-Asp39, and one between Bn-Lys27 and Bs-Thr42. This number again drops precipitously between 3.5 and 4.5 Å separation, with new ones formed between Bn-Arg59 and Bs-Asp35 persisting the longest (see far right inset of Figure 4A). Bn-Arg59 is part of the RNA-binding loop and has been previously suggested to be the point of initial contact between barstar and barnase, after which reorientation of barstar recovers the bound state.39 We note that the separation enforced here represents only one potential unbinding pathway and, thus, the specific order and timing of disrupted interactions may not be identical to the native pathway. Nonetheless, the agreement of the observed unbinding process with unrestrained simulations, namely the initial interaction of barstar with Bn-Arg59 and the subsequent reorientation, indicates that it is not qualitatively different from the true process despite the restraints on interfacial side chains.
In addition to direct interactions between barstar and barnase, binding is also mediated by a number of interfacial water molecules. From the crystal structure, nine were observed to form bridging hydrogen bonds between the two proteins.27 The hydrogen-bond networks stabilized by these water molecules have been shown to be important in the binding process, with each water-mediated interaction previously estimated to contribute 1 kcal/mol to the free energy.45 To quantify the hydration of the protein-protein interface, we determined the average number of water molecules within 3.0 Å of interfacial residues in both barnase and barstar, shown in Figure 4B. In the crystal structure nine waters were observed using this 3-Å criterion, although in simulation this increases to 13.5 on average for the bound state. As the proteins separate, the number of waters near both proteins rises to a maximum of 16.5 at 26Å, i.e., 2.5Å away from the bound state, before dropping steadily to 0 at 36 Å. However, long before this point is reached, the individual hydration of each protein reaches its bulk value at 30 Å (see the orange and blue curves in Figure 4B). The crucial role of water molecules in mediating barstar-barnase association27,45 is in contrast to many protein-protein interactions in which binding is driven by the hydrophobic effect, i.e., the expulsion of water from the interface.46–49
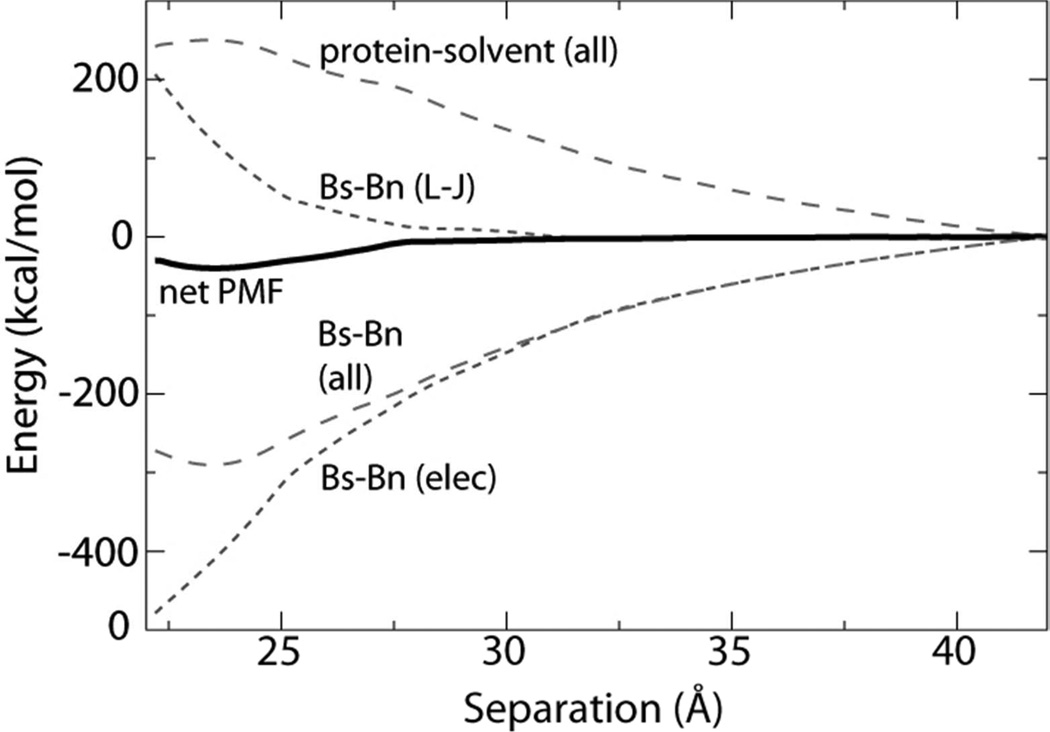
In order to determine the contribution of water to the net free energy during separation, the PMF in Figure 2 was decomposed into barstar-barnase Lennard-Jones (L–J) and electrostatic (elec.) interaction terms, as well as protein-solvent interaction. Immediately apparent from this decomposition, shown in Figure 5, is that association is governed primarily by electrostatics, in agreement with previous studies.27,28 Barstar and barnase possess both non-zero net charges (−6q and +2q, respectively) and macrodipoles. Thus, the electrostatic interaction is long range, with the behavior at large separation dominated by charge-charge attraction. Solvent reorientation screens this interaction after a separation of 10Å or more, with the sum of the two contributions, barstar-barnase electrostatics and net solvent-protein interactions, effectively canceling each other. The minimum in the combined PMF at 23.5Å arises at the point where the short-range L–J repulsion overwhelms the electrostatic attraction of the two proteins. Counterbalancing electrostatic attraction, the protein-solvent free energy increases as the two proteins are brought together, reaching a maximum at the bound-state separation. This increase is due to an entropic penalty introduced as bulk water reorganizes in the presence of the electrostatic fields of the two proteins. Assuming that the electrostatic potentials surrounding the proteins are largely insensitive to minor changes in side-chain positions, it is expected, but remains to be demonstrated, that other unbinding pathways exhibit a similar balance of energetic contributions to the free energy.
Figure 5.
Decomposition of the separation PMF (solid black line) from Figure 2 into protein-protein and protein-solvent contributions (long dashed lines). The protein-protein interaction is further partitioned into van der Waals and electrostatic contributions (short dashed lines).
In contrast to the Bs-Bn (L–J) PMF in Figure 5, the time average of the instantaneous L–J interaction energy between barstar and barnase, which reflects primarily the dispersive term and not the repulsive term, decreases upon approach (see Fig. S3). When combined with the average L–J interactions between each protein and solvent, a minimum in the net L–J interaction energy appears at the optimal separation distance. This minimum indicates that the bound state is not only a balance between electrostatics and L–J repulsion, but also between protein-protein and protein-solvent dispersion energy.
Discussion
In this paper, a theoretical framework has been proposed to determine the standard binding free energy of barnase and barstar from first principles, a complex problem at the frontiers of free-energy calculations not only due to the size of the objects, but also the intricacy of the interface formed by the bound proteins.50,51 In particular, the double-mutant experiments of Schreiber and Fersht40 underline the highly cooperative nature of the interactions that govern barnase:barstar affinity, mirrored in the non-additive contributions of the interfacial amino acids to the standard binding free energy. The size and, hence, the inertia of the two proteins, on the other hand, is manifested in an appreciably slow reorientational relaxation as they diffuse reversibly apart from each other — a process difficult to capture on the time scales ordinarily amenable to molecular dynamics. Indeed, the barstar:barnase complex was not recovered during a 2-µs simulation. These roadblocks, both conceptual and computational, explain why attempts to address the elusive problem of protein-protein recognition and association at the atomic level by means of free-energy calculations have remained scarce. They further rationalize the practical benefit of generalizing to protein-protein assemblies the formal framework developed in the context of protein-ligand binding,21–23 as well as the introduction of a set of appropriately chosen geometrical restraints to reduce the configurational space explored by the biological objects as they are separated.
From a methodological standpoint, the aforementioned generalization imposes itself almost naturally considering the complexity of the interface formed by barnase and barstar ,50,51 which consists primarily of polar and charged amino acids. The acute electrostatic complementarity of the two partner proteins,51 proposed to stabilize the dimeric complex, demands that the relevant interactions — a network of intertwined salt bridges and hydrogen bonds, be preserved in the course of the reversible association. Once disrupted, the interactions at play have a marginal probability to reform, as the participating amino acids isomerize and modify their coordination, notably with water molecules trapped between the two proteins. Though geometrical restraints acting on side chains do not preempt hydration of the polar and charged moieties when barstar moves away from barnase, they guarantee that as the proteins approach each other, the relevant interactions can be recovered, while spurious, undesirable ones are prevented from forming. The inertia of the biological objects at hand raises other concerns, excluding the possibility of estimating protein-protein standard binding free energies in the sole light of a one-dimensional separation PMF. Aside from the gain in conformational entropy induced by the separation of barstar from barnase, not easily captured in unrestrained simulations, diffusional tumbling of the two objects as they diffuse away from each other occurs over time scales likely to span that of the complete free-energy calculation reported herein (see Fig. S2). Such concerns cast doubts on the relevance in protein-protein recognition and association phenomena of orientation-averaged separation PMFs,52 which ignore variations in translational and rotational entropies.53
A noteworthy feature of the proposed staging methodology is the access it offers to physically well-defined contributions of the free energy and, hence, the glimpse it provides into the nature of the interactions that drive binding.24 While the various free energy components depend on the enforced unbinding pathway for separating the binding partners, they can provide useful insight into the various contributions to the binding free energy–as long as the enforced pathway is physically meaningful. The latter essentially corresponds to a one-dimensional curve within the subspace associated with the set of collective variables that are restrained during the protein separation (protein orientation, RMSD on a subset of backbone and side-chain atoms). The true optimal pathway within this subspace could be obtained using the string method in collective variable-space54 or the max-flux algorithm.55 Fundamentally, the optimal pathway should encounter the lowest possible free energy barrier during the separation process. From this point of view it is noteworthy that, while the separation pathway was not explicitly optimized here, there is no large barrier as the two proteins are separated (see Figure 2), suggesting that the enforced pathway is physically meaningful, albeit not unique. These considerations increase our confidence in the analysis of the various free energy contributions during the separation process. In the case of the present system, many aspects of the unbinding process were found to be in agreement with previous experiments and simulations.27,28,39,45 It has been suggested that the free-energy cost incurred in the dehydration of their interfacial residues as the two proteins associate could destabilize the dimer, on account of the loss of favorable electrostatic interactions with the surroundings.51,56,57 The electrostatic component of binding arises from direct, intermolecular and indirect, intramolecular interactions, competing with hydration. The free-energy decomposition of Figure 5 confirms that barnase:barstar recognition primarily stems from screened, long-range electrostatic interactions, which bring the two proteins together. In the short-range regime, strong, attractive electrostatic interactions are partially compensated by repulsive van der Waals forces and unfavorable interactions with water. Complementarity of electrostatic potentials,58 manifest in the oppositely charged amino acids lying at the interface, appears to be pivotal for the binding of barstar to barnase–and probably for many other protein-protein complexes. Intramolecular interactions in the unbound states, notably of residues neighboring the binding interface, are attenuated by the aqueous environment and are thus expected to be stronger in the bound states as a result of dehydration51,59 — such is the case of peripheral titratable residues in barnase, e.g., Asp54 and Glu60. Interestingly enough, while at large separation, barnase and barstar interact with a roughly equal number of water molecules, the latter protein appears to be less hydrated in the contact region of the PMF than the former. The lesser hydration of barstar can be related to its hypothesized optimal charge distribution and, thus, higher specificity, which, combined with a robust conformation, may be a prerequisite to bind barnase. Consistent with this observation, the reduced dehydration of Asp35 and Asp39 of barstar has been put forth as a mechanism to enhance binding.51 Conversely, following a similar reasoning, the somewhat lower specificity of barnase is consonant with other known functions of this enzyme, notably catalyzing RNA hydrolysis.60
The PMF-based method described here requires knowledge of the structure of a protein-protein complex of interest. This information is crucial to “guide” the separation, orientation, and conformation of the two proteins through a series of geometrically guided simulations. In these simulations, the conformational sampling of the two proteins is controlled via restraining potentials applied to the backbone and the side chains. The restraints were defined using the RMSD relative to the backbone and side-chain conformations representative of the complex. While the choice of RMSD was appropriate in the case of the barstar:barnase complex, it can be more questionable if the molecules undergo a very large conformational change upon binding. In that case different order parameters can be used within the framework of the PMF-based method to address this issue. For example, residue-residue distances were previously used to control the considerable conformational changes induced by the association of various molecules to the ligand-binding domain of the iGluR glutamate receptor.25
In the present study, we relied on the high-resolution X-ray structure of the barstar:barnase complex. Nevertheless, the method could be applied in the case of a predicted structural model as well. In this sense, the method is not aimed at discovering or predicting the structure of the complex, but at producing a reliable estimate of the binding free energy once it is known, two different tasks commonly referred to as “docking” and “scoring”, respectively. Characterizing protein-protein association quantitatively is likely to remain an extremely challenging computational endeavor for years to come. In the long-term, it is reasonable to expect that progress on this front will be achieved with the best strategies possible to perform docking and scoring. Unbiased brute-force MD simulations may play a useful role, but it is extremely unlikely that they will ever provide a single-step solution to the issue of docking and scoring of protein complexes. While it may be possible to observe the rapid and spontaneous association of two proteins in solution and discover the structure of the complex from unbiased MD, estimating the binding free energy requires statistical information about the dissociation process as well. The latter, in the case of strong binders, may be extremely slow (e.g., several hours for the barstar:barnase complex) and, therefore, out of reach of simulations. For these reasons, it is our belief that the PMF-based framework described here arguably represents one of the very few viable routes towards accurate standard binding free energies for protein-protein complexes with explicit solvent simulations.
In closing this discussion, it is important to summarize some of the limitations of the method. Enforcement of geometrical and conformational restraints to reduce the configurational entropy21–23 explored in the course of the separation of the biological objects at hand results in an artificial reaction pathway,61 from whence inferring prevalent trends about protein-protein recognition and association mechanisms is not straightforward. Furthermore, though convergence of the present set of free-energy calculations was carefully monitored, it remains a potential source of error. Additional sources other than sampling error15 could also explain the slightly discrepant experimental and theoretical free-energy estimates, chief amongst which is the use of a pairwise-additive potential energy function to describe a biological process likely to be modulated by induction phenomena. While the vast experimental data on barnase:barstar association make this study retrospective, it provides a cogent illustration of the general applicability of the methodology to virtually any protein-protein complex for which data may be more fragmentary. Additionally, even when the binding free energy is known experimentally, the developed methods permit exploration of modifications to a binding interface, including mutations designed for inhibition or optimization.
Supplementary Material
Acknowledgements
The authors are grateful to the France and Chicago collaborating in the science (FACCTS) Center for their support. The research is funded by grant MCB-0920261 from the National Science Foundation (NSF) and by grants R01-CA093577 (B.R.) and K22-AI100927 (J.C.G.) from the National Institute of Health. Simulations were carried out using the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by NSF grant number OCI-1053575. The Centre Informatique National de l’Enseignement Supérieur, Montpellier, is also gratefully acknowledged for generous provision of computer time. The 2-µs simulation of the separated proteins was carried out using on the Anton computer donated by DE Shaw Research,62 with time provided by the National Resource for Biomedical Supercomputing and the Pittsburgh Supercomputing Center through Grant RC2GM093307 from the NIH.
Footnotes
Supporting Information
Three figures and the full derivation of the binding free energy. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Chothia C, Janin J. Principles of protein-protein recognition. Nature. 1975;256:705–708. doi: 10.1038/256705a0. [DOI] [PubMed] [Google Scholar]
- 2.Jones S, Thornton JM. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stites WE. Protein-protein interactions: Interface structure, binding thermodynamics, and mutational analysis. Chem. Rev. 1997;97:1233–1250. doi: 10.1021/cr960387h. [DOI] [PubMed] [Google Scholar]
- 4.Angers S, Salahpour A, Bouvier M. Dimerization: An emerging concept for G protein–coupled receptor ontogeny and function. Annu. Rev. Pharmacol. Toxicol. 2002;42:409–435. doi: 10.1146/annurev.pharmtox.42.091701.082314. [DOI] [PubMed] [Google Scholar]
- 5.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 6.Vojtek AB, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 7.Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl. Acad. Sci. USA. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang T, Tomic S, Gabdoulline RR, Wade RC. How optimal are the binding energetics of barnase and barstar? Biophys. J. 2004;87:1618–1630. doi: 10.1529/biophysj.104.040964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YC, Tang C, Clore GM, Hummer G. Replica exchange simulations of transient encounter complexes in protein-protein association. Proc. Natl. Acad. Sci. USA. 2008;105:12855–12860. doi: 10.1073/pnas.0802460105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Periole X, Knepp AM, Sakmar TP, Marrink SJ, Huber T. Structural determinants of the supramolecular organization of G protein-coupled receptors in bilayers. J. Am. Chem. Soc. 2012;134:10959–10965. doi: 10.1021/ja303286e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gohlke H, Kiel C, Case DA. Insights into protein-protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RalGDS complexes. J. Mol. Biol. 2003;330:891–913. doi: 10.1016/s0022-2836(03)00610-7. [DOI] [PubMed] [Google Scholar]
- 12.Tembe BL, McCammon JA. Ligand-receptor interactions. Comp. Chem. 1984;8:281–283. [Google Scholar]
- 13.Hermans J, Shankar S. The free energy of xenon binding to myoglobin from molecular dynamics simulation. Isr. J. Chem. 1986;27:225–227. [Google Scholar]
- 14.Mobley DL, Chodera JD. Dill, K. A. On the use of orientational restraints and symmetry corrections in alchemical free energy calculations. J. Chem. Phys. 2006;125:084902. doi: 10.1063/1.2221683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chipot C, Pohorille A, editors. Free energy calculations. Theory and applications in chemistry and biology. Springer Verlag; 2007. [Google Scholar]
- 16.Deng Y, Roux B. Computations of standard binding free energies with molecular dynamics simulations. J. Phys. Chem. B. 2009;113:2234–2246. doi: 10.1021/jp807701h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallicchio E, Levy RM. Advances in all atom sampling methods for modeling protein-ligand binding affinities. Curr. Opin. Struct. Biol. 2011;21:161–166. doi: 10.1016/j.sbi.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schreiber G, Fersht AR. The interaction of barnase with its polypeptide inhibitor barstar studied by protein engineering. Biochemistry. 1993;32:5145–5150. doi: 10.1021/bi00070a025. [DOI] [PubMed] [Google Scholar]
- 19.Shoup D, Szabo A. Role of diffusion in ligand binding to macromolecules and cell–bound receptors. Biophys. J. 1982;40:33–39. doi: 10.1016/S0006-3495(82)84455-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jorgensen WL. Interactions between amides in solution and the thermodynamics of weak binding. J. Am. Chem. Soc. 1989;111:3770–3771. [Google Scholar]
- 21.Woo HJ, Roux B. Calculation of absolute protein–ligand binding free energy from computer simulations. Proc. Natl. Acad. Sci. USA. 2005;102:6825–6830. doi: 10.1073/pnas.0409005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gan W, Roux B. Binding specificity of SH2 domains: insight from free energy simulations. Proteins. 2009;74:996–1007. doi: 10.1002/prot.22209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gumbart JC, Roux B, Chipot C. Standard binding free energies from computer simulations: What is the best strategy? J. Chem. Theory. Comput. 2013;9:794–802. doi: 10.1021/ct3008099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dadarlat VM, Skeel RD. Dual role of protein phosphorylation in DNA activator/coactivator binding. Biophys. J. 2011;100:469–477. doi: 10.1016/j.bpj.2010.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau AY, Roux B. The hidden energetics of ligand binding and activation in a glutamate receptor. Nat. Struct. Mol. Biol. 2011;18:283–287. doi: 10.1038/nsmb.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartley RW. Barnase and barstar: two small proteins to fold and fit together. Trends Biochem. Sci. 1989;14:450–454. doi: 10.1016/0968-0004(89)90104-7. [DOI] [PubMed] [Google Scholar]
- 27.Buckle AM, Schreiber G, Fersht AR. Protein-protein recognition: Crystal structural analysis of a barnase-barstar complex at 2.0-Å resolution. Biochemistry. 1994;33:8878–8889. doi: 10.1021/bi00196a004. [DOI] [PubMed] [Google Scholar]
- 28.Schreiber G, Fersht AR. Rapid, electrostatically assisted association of proteins. Nat. Struct. Biol. 1996;3:427–431. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- 29.Deng Y, Roux B. Calculation of standard binding free energies: Aromatic molecules in the T4 lysozyme L99A mutant. J. Chem. Theory Comput. 2006;2:1255–1273. doi: 10.1021/ct060037v. [DOI] [PubMed] [Google Scholar]
- 30.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comp. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, MacKerell AD., Jr Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darve E, Pohorille A. Calculating free energies using average force. J. Chem. Phys. 2001;115:9169–9183. [Google Scholar]
- 33.Hénin J, Chipot C. Overcoming free energy barriers using unconstrained molecular dynamics simulations. J. Chem. Phys. 2004;121:2904–2914. doi: 10.1063/1.1773132. [DOI] [PubMed] [Google Scholar]
- 34.Hénin J, Forin G, Chipot C, Klein ML. Exploring multidimensional free energy landscapes using time-dependent biases on collective variables. J. Chem. Theory Comput. 2010;6:35–47. doi: 10.1021/ct9004432. [DOI] [PubMed] [Google Scholar]
- 35.Sugita Y, Kitao A, Okamoto Y. Multidimensional replica-exchange method for free-energy calculations. J. Chem. Phys. 2000;113:6042–6051. [Google Scholar]
- 36.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules. I. The Method. J. Comp. Chem. 1992;13:1011–1021. [Google Scholar]
- 37.Sotomayor M, Schulten K. Single-molecule experiments in vitro and in silico. Science. 2007;316:1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- 38.Dror RO, Pan AC, Arlow DH, Borhani DW, Maragakis P, Shan Y, Xu H, Shaw DE. Pathway and mechanism of drug binding to G-protein-coupled receptors. Proc. Natl. Acad. Sci. USA. 2011;108:13118–13123. doi: 10.1073/pnas.1104614108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spaar A, Dammer C, Gabdoulline RR, Wade RC, Helms V. Diffusional encounter of barnase and barstar. Biophys. J. 2006;90:1913–1924. doi: 10.1529/biophysj.105.075507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schreiber G, Fersht AR. Energetics of protein-protein interactions: Analysis of the barnase-barstar interface by single mutations and double mutant cycles. J. Mol. Biol. 1995;248:478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez-Gomez D, Darve E, Pohorille A. Assessing the efficiency of free energy calculation methods. J. Chem. Phys. 2004;120:3563–3578. doi: 10.1063/1.1642607. [DOI] [PubMed] [Google Scholar]
- 42.Zhu F, Hummer G. Convergence and error estimation in free energy calculations using the weighted histogram analysis method. J. Comp. Chem. 2012;33:453–465. doi: 10.1002/jcc.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartley RW. Directed mutagenesis and barnase-barstar recognition. Biochemistry. 1993;32:5978–5984. doi: 10.1021/bi00074a008. [DOI] [PubMed] [Google Scholar]
- 44.Schreiber G, Buckle AM, Fersht AR. Stability versus function: Two competing forces in the evolution of barstar. Structure. 1994;2:945–951. doi: 10.1016/s0969-2126(94)00096-4. [DOI] [PubMed] [Google Scholar]
- 45.Ikura T, Urakubo Y, Ito N. Water-mediated interaction at a protein-protein interface. Chem. Phys. 2004;307:111–119. [Google Scholar]
- 46.Cheng YK, Rossky PJ. Surface topography dependence of biomolecular hydrophobic hydration. Nature. 1998;392:696–699. doi: 10.1038/33653. [DOI] [PubMed] [Google Scholar]
- 47.Lum K, Chandler D, Weeks JD. Hydrophobicity at small and large length scales. J. Phys. Chem. B. 1999;103:4570–4577. [Google Scholar]
- 48.ten Wolde PR, Chandler D. Drying-induced hydrophobic polymer collapse. Proc. Natl. Acad. Sci. USA. 2002;99:6539–6543. doi: 10.1073/pnas.052153299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu P, Huang X, Zhou R, Berne BJ. Observation of a dewetting transition in the collapse of the melittin tetramer. Nature. 2005;437:159–162. doi: 10.1038/nature03926. [DOI] [PubMed] [Google Scholar]
- 50.Frisch C, Schreiber G, Johnson CM, Fersht AR. Thermodynamics of the interaction of barnase and barstar: changes in free energy versus changes in enthalpy on mutation. J. Mol. Biol. 1997;267:696–706. doi: 10.1006/jmbi.1997.0892. [DOI] [PubMed] [Google Scholar]
- 51.Lee LP, Tidor B. Barstar is electrostatically optimized for tight binding to barnase. Nat. Struct. Biol. 2001;8:73–76. doi: 10.1038/83082. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, Siu SWI, Gu W, Helms V. Downhill binding energy surface of the barnase-barstar complex. Biopolymers. 2010;93:977–985. doi: 10.1002/bip.21507. [DOI] [PubMed] [Google Scholar]
- 53.Tidor B, Karplus M. The contribution of vibrational entropy to molecular association The dimerization of insulin. J. Mol. Biol. 1994;238:405–414. doi: 10.1006/jmbi.1994.1300. [DOI] [PubMed] [Google Scholar]
- 54.Maragliano L, Fischer A, Vanden-Eijnden E, Ciccotti G. String method in collective variables: minimum free energy paths and isocommittor surfaces. J. Chem. Phys. 2006;125:24106. doi: 10.1063/1.2212942. [DOI] [PubMed] [Google Scholar]
- 55.Huo S, Straub JE. The MaxFlux algorithm for calculating variationally optimized reaction paths for conformational transitions in many body systems at finite temperature. J. Chem. Phys. 1997;107:5000. [Google Scholar]
- 56.Hendsch ZS, Tidor B. Do salt bridges stabilize proteins? A continuum electrostatic analysis. Protein Sci. 1994;3:211–226. doi: 10.1002/pro.5560030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nature Struct. Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 58.Lee LP, Tidor B. Optimization of binding electrostatics: Charge complementarity in the barnase-barstar protein complex. Protein Sci. 2001;10:362–377. doi: 10.1110/ps.40001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caravella JA, Carbeck JD, Duffy DC, Whitesides GM, Tidor B. Long-range electrostatic contributions to protein-ligand binding estimated using protein charge ladders, affinity capillary electrophoresis, and continuum electrostatics. J. Am. Chem. Soc. 1999;121:4340–4347. [Google Scholar]
- 60.Mossakowska DE, Nyberg K, Fersht AR. Kinetic characterization of the recombinant ribonuclease from Bacillus amyloliquefaciens (barnase) and investigation of key residues in catalysis by site-directed mutagenesis. Biochemistry. 1989;28:3843–3850. doi: 10.1021/bi00435a033. [DOI] [PubMed] [Google Scholar]
- 61.Kuttner YY, Kozer N, Segal E, Schreiber G, Haran G. Separating the contribution of translational and rotational diffusion to protein association. J. Am. Chem. Soc. 2005;127:15138–15144. doi: 10.1021/ja053681c. [DOI] [PubMed] [Google Scholar]
- 62.Shaw DE, Deneroff MM, Dror RO, Kuskin JS, Larson RH, Salmon JK, Young C, Batson B, Bowers KJ, Chao JC, Eastwood MP, Gagliardo J, Grossman JP, Ho CR, Ierardi DJ, Kolossváry I, Klepeis JL, Layman T, McLeavey C, Moraes MA, Mueller R, Priest EC, Shan Y, Spengler J, Theobald M, Towles B, Wang SC. Anton, a special-purpose machine for molecular dynamics simulation. Comm. ACM. 2008;51:91–97. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.