Abstract
Arterial thrombosis is a major component of vascular disease, especially myocardial infarction (MI) and stroke. Current anti-thrombotic therapies such as warfarin and clopidogrel are effective in inhibiting cardiovascular events; however, there is great inter-individual variability in response to these medications. In recent years, it has been recognized that genetic factors play a significant role in drug response, and, subsequently, common variants in genes responsible for metabolism and drug action have been identified. These discoveries along with the new diagnostic targets and therapeutic strategies on the horizon hold promise for more effective individualized anti-coagulation and anti-platelet therapy.
Keywords: Pharmacogenomics, Personalized medicine, Anti-platelet therapy, Clopidogrel, Plavix, Warfarin, Coumadin, CYP2C19, CYP2C9, VKORC1, Platelet function, Cardiovascular disease, Thrombosis, Coronary artery disease, Percutaneous coronary intervention, Anti-coagulation
Introduction
Anti-coagulant and anti-platelet medications are widely prescribed drugs used for the primary and secondary treatment of a variety of pathological thrombotic processes such as thrombotic cerebrovascular and cardiovascular diseases, atrial fibrillation, pulmonary embolism, deep vein thrombosis and genetic or acquired hypercoagulability. Warfarin is the most commonly used oral anti-coagulant, whereas the most commonly used anti-platelet medications include aspirin and clopidogrel, each of which influences blood hemostasis through different mechanisms. Marked inter-individual variation in response to these commonly prescribed medications have been well-documented and represent a significant challenge to medical practice [1•]. Warfarin has a relatively narrow therapeutic index around which under-dosing may result in recurrent thrombosis and over-dosing may result in severe and life-threatening bleeding. While newer agents such as dabigatran and rivaroxaban are now available, physicians’ familiarity with warfarin, its effectiveness when dosed properly, and its low cost continue to make it the anti-coagulant treatment of choice. Likewise, clopidogrel continues to be widely prescribed due to its efficacy in the majority of patients as well as its relatively low price, while newer anti-platelet medications such as prasugrel and ticagrelor often function as alternative anti-platelet agents offered to patients for whom clopidogrel is not effective. Understanding factors that influence response to these agents offers practical opportunities for more individualized and effective therapy.
Several gene polymorphisms have been found that reproducibly contribute to inter-individual response variability to warfarin and clopidogrel. The aim of this review is to familiarize the reader with the gene polymorphisms found thus far that most significantly contribute to a given individual’s response to either of these two medications; because of the lack of genetic polymorphisms reproducibly associated with aspirin response, aspirin pharmacogenomics will not be discussed in this review, though others have covered the issue very well [2-4]. With bona fide common polymorphisms that can predict one’s response to warfarin and clopidogrel now in hand, the next challenge and active area of investigation is the development of strategies to implement these discoveries into clinical practice.
Warfarin
In 1948, warfarin was introduced as a pesticide against rodents. Recognizing its potent anti-coagulant action and potential efficacy for thrombotic disease, warfarin was approved for use in humans in 1954 and remains among the most commonly prescribed drugs today. It is derived from dicoumarol, a natural product initially isolated from sweet clover. Its synthetic form consists of both R and S enantiomers of which the S form is more active. Each form is metabolized through a different mechanism, with S-warfarin metabolized primarily by cytochrome P450 2C9 (CYP2C9) and R-warfarin metabolized predominantly by CYP3A4 [5]. Warfarin acts by inhibiting the vitamin K epoxide reductase complex by binding to the VKORC1 subunit, thus preventing reduced vitamin K-dependent gamma-carboxylation of clotting factors II, VII, IX and X, as well as proteins C and S, resulting in a potent anti-coagulant effect [6].
Dosing of warfarin typically involves a loading dose followed by daily maintenance therapy. Its therapeutic dosing is monitored by measuring activity of the extrinsic coagulation pathway using the standardized international normalized ratio (INR). There is wide inter-individual variation in the warfarin dose required to reach a therapeutic INR. Factors that markedly affect the anti-coagulant effect of warfarin include diet, particularly foods high in vitamin K, smoking, certain drugs and botanicals that affect warfarin metabolism, alcohol, body weight, and age [7]. Based upon knowledge of the mechanism of action and metabolism of warfarin, candidate gene studies have identified three genes whose common variation explains ~40%, and up to 54% of inter-individual response to warfarin dose, depending on the ancestry of the population studied. More recent genome-wide association studies (GWAS) have provided additional insights into warfarin pharmacogenomics.
CYP2C9
The CYP2C9*1 allele encodes a fully active enzyme, has the highest frequency of the 30 different alleles discovered to date, and is considered the wild-type allele. Although frequencies vary across ethnic populations, the most common decreased function alleles are CYP2C9*2 (C430T; rs1799853) and *3 (A1075C; rs1057910) alleles, which encode enzymes with 70% and 20% of wild-type enzyme activity, respectively. Multiple studies now show that patients with CYP2C9*2 and *3 alleles have greater sensitivity to warfarin, requiring lower doses to achieve a therapeutic INR [8, 9]. Among the initial important studies, Higashi and colleagues performed a retrospective study of 185 largely Caucasian patients followed in two Seattle area anticoagulation clinics, and found that compared to the *1/*1 genotype, patients with one or two copies of the *2 or *3 variant required significantly lower daily doses of warfarin [8]. Gage and colleagues examined the effect of the CYP2C9 variants in 369 patients who were taking maintenance doses of warfarin and found that the presence of the *2 or *3 variant was strongly associated with lower warfarin dose necessity; the maintenance dose was decreased by 19% per *2 allele and by 30% per *3 allele [9]. From these and other studies, it has become clear that about 13% of the variability in warfarin dose can be explained by CYP2C9 polymorphisms.
As might be expected of patients who have increased sensitivity to warfarin, several studies indicate that CYP2C9 decreased function allele carriers are at increased risk of over-anti-coagulation and bleeding events [10-12]. Higashi found that patients carrying *2 or *3 alleles experienced a bleeding rate of 10.92 per 100 patient-years, which was significantly higher than the 4.89 per 100 patient-years experienced in the *1/*1 homozygotes [8].
Some uncommon CYP2C9 decreased function alleles include *5, *6, *8, and *11. These alleles have not been studied as thoroughly but would be predicted to have similar effects as the more common CYP2C9 decreased functional alleles [13]. The allele frequencies vary noticeably among ethnic groups; for example, the frequencies of CYP2C9*2 and *3 are higher in Asian populations than Caucasian populations, which have higher frequencies of the alleles than African-American populations [12, 13]. Importantly, the allele frequency of CYP2C9*8 among African-Americans is approximately 9%, suggesting that this allele should be measured and considered in warfarin dosing algorithms [14].
VKORC1
The VKORC1 G-1639A (rs9923231) variant is located in the promoter region and results in decreased transcription as well as lower levels of VKORC1 messenger RNA (mRNA) [1•, 15]. Another VKORC1 variant, C1173T (rs9934438), is in complete linkage disequilibrium with G-1639A [16]. Decreased VKORC1 expression is associated with increased warfarin sensitivity and thus patients heterozygous (G/A) and homozygous (A/A) for the VKORC1 G-1639A variant require lower doses of warfarin compared to individuals homozygous for the wild-type VKORC1 genotype (G/G) [17]. The early important study demonstrating the potent effect of VKORC1 variants on warfarin dose was carried out by Rieder and colleagues with the same Seattle area cohort discussed above. They found that compared to those patients homozygous for the wild-type VKORC1 allele, having one of five highly correlated variants predicted an approximately 25% variance in warfarin dose. The effect of the VKORC1 variants on warfarin dose in this study was more potent than the CYP2C9 variants and accounted for 10% of the variance in warfarin dose. With respect to Rieder’s findings, it is currently estimated that the VKORC1 G-1639A variant accounts for 24% of the variation in warfarin dose [16]. The frequency of the VKORC1 -1639A allele is approximately 40% in Caucasians, 20% in African-Americans, and 85% in Asians [16, 18]. As might be expected, the VKORC1 -1639A allele has been shown to be associated with increased bleeding events and over-anti-coagulation [18, 19]. In a randomized trial of genotype-guided versus standard warfarin dosing, Anderson et al. [19] found that patients who carry variants in both CYP2C9 and VKORC1 were at a significantly increased risk of an elevated INR (INR>4) compared to all other patients.
CYP4F2
After vitamin K is reduced by the vitamin K epoxide reductase complex, the reduced form of vitamin K can used in the synthesis of coagulation factors or it can be converted into hydroxyl-vitamin K1 by the enzyme CYP4F2, encoded by the CYP4F2 gene [20]. The rs2108622 variant in CYP4F2 is a C>T nucleotide substitution that introduces a V433M missense mutation resulting in a CYP4F2 enzyme with decreased function [21]. Patients with the CYP4F2 decreased function T allele require 4-12% more warfarin per allele compared to CC homozygotes [22], accounting for approximately 1.1-7% of the inter-patient variability in warfarin dose requirement [23, 24]. The frequency of rs2108622 T alleles is approximately 25% among Caucasians and Asians, with a lower frequency of 7% observed among African-Americans [22]. Moreover, dosing models designed to include rs2108622 along with CYP2C9 and VKORC1 variants resulted in improved overall warfarin dose predictability [25]. Two GWAS identified the same three variants as being significantly associated with warfarin response [26, 27]; other genome-wide significant associations were not detected, suggesting that it is unlikely that there are other common variants that exert as large an effect.
Translation of Warfarin Pharmacogenomics into Patient Care
CYP2C9, VKORC1, and CYP4F2 genotyping is likely to have clinical utility, allowing clinicians to better individualize warfarin dosing to enhance efficacy and decrease adverse events such as bleeding. In 2007, the FDA modified the package insert of warfarin to reflect the clinical utility of VKORC1 and CYP2C9 genotype to include the statement, “lower initiation doses should be considered for patients with certain genetic variations in CYP2C9 and VKORC1 enzymes” [28]. The package insert was later modified in 2010 to include a table containing recommended daily warfarin doses for patients with various CYP2C9 and VKORC1 haplotypes [7]. There are currently several trials examining application of genetics in warfarin therapy, including the Warfarin Adverse Event Reduction For Adults Receiving Genetic Testing at Therapy Initiation (WARFARIN, 2011-2014) Trial, and the Clarification of Optimal Anticoagulation Through Genetics (COAG, 2009-2013) Trial. Both of these are NIH-funded prospective randomized clinical trials designed to determine if a dosing algorithm that includes genotypes for warfarin-response genes, such as CYP2C9 and VKORC1, can result in a greater proportion of time within the therapeutic INR range, as well as decreased warfarin-related clinical events, compared to standard dosing practices that do not include genotypes. A number of warfarin dosing algorithms and tables, such as those found at http://www.warfarindosing.org [29], have been developed that incorporate genotype to estimate warfarin dose. In addition, the Pharmacogenomics Research Network Clinical Pharmacology Implementation Committee recently published guidelines for warfarin dosing recommending that pharmacogenetic algorithm-based dosing be used when possible, and if electronic means for dosing are not available, the table-based dosing approaches are suggested [7].
Clopidogrel
Clopidogrel is an oral second-generation thienopyridine that prevents platelet activation and aggregation by irreversibly inhibiting the P2Y12 ADP receptors on the surfaces of platelets [30]. Upon ingestion, clopidogrel is absorbed by duodenal enterocytes and moves through these cells into the bloodstream. Once in circulation, approximately 85% of the clopidogrel is hydrolyzed into inactive metabolites by carboxylesterases, primarily carboxylesterase 1 (CES1), in the liver during first-pass metabolism [31]. The remainder of the drug undergoes biotransformation from an inactive pro-drug into the unstable active metabolite. This process requires two steps that involve several CYP enzymes also found in the liver. Two important CYP enzymes involved in this activation step include CYP2C19 and CYP2B6, as well as other potential enzymes including CYP1A2, CYP2C9, CYP3A4/5, and PON1 [32-34]. The active metabolite then circulates through the bloodstream, oxidizing cysteine residues and irreversibly blocking platelet P2Y12 ADP receptors. Without functional P2Y12 receptors, the Gi proteins associated with the receptors are unable to inhibit adenylyl cyclase. This causes an increase in cAMP, followed by the lack of activation of phosphoinositide 3-kinase (PI3K) and decreased expression of glycoprotein IIb/IIIa (GpIIb/IIIa). The ultimate result of this pathway is a slow-starting, long-term activation and aggregation of platelets, which clopidogrel effectively blocks [35].
Clopidogrel response variability is well established [36-41]. Patients treated with clopidogrel who demonstrate higher ex vivo platelet reactivity are at increased risk of ischemic events [42-48]. For example, Matezky and coworkers [49] found that up to 25% of subjects who received PCI with stenting for acute MI and who were placed on aspirin and clopidogrel were resistant to clopidogrel when ADP-induced platelet aggregation was assessed at day 6 of therapy. Recurrence rates for a cardiovascular event were 40% in the lowest quartile of clopidogrel response compared to only 6.7% in the upper quartile of responders. Factors that may influence variation in platelet function in response to clopidogrel include use of lipophilic statins, calcium channel blockers, proton pump inhibitors, St. John’s Wort, and smoking [50-52]. However, these factors account for only a small fraction of the variation in response.
Genetic factors have also been found to play a role in clopidogrel resistance. The Amish Pharmacogenomics of Anti-Platelet (PAPI) Study found that in healthy subjects the heritability of clopidogrel response, as measured by post-exposure ADP-stimulated platelet aggregation, was 70%. In search of specific genetic variants that influence clopidogrel response, a number of candidate gene studies and, to date, one GWAS has been performed. Following is a summary of the most salient findings.
CYP2C19
Common loss-of-function (LOF) variants in CYP2C19 are the most well-established genetic determinants of clopidogrel responsiveness. Its most common LOF variant is *2 (rs4244285), with allele frequencies of 29% in Asians, and 15% in Caucasians and Africans. Other LOF alleles include *3-*8, which are all considered rare. Those with one and two CYP2C19 LOF alleles are considered intermediate metabolizers (IM) and poor metabolizers (PM), respectively. Multiple studies have demonstrated that CYP2C19 LOF variants are associated with lower clopidogrel active metabolite concentrations [53-55], greater on-treatment residual platelet function [54-57] and poorer cardiovascular outcomes in PCI patients treated with clopidogrel [53, 58-63]; other excellent reviews have also covered much of the literature surrounding CYP2C19 [1•, 64-69]. Other studies in coronary artery disease populations with lower rates of stent placement or in patient populations with other indications for anti-platelet therapy have not shown significant effects of CYP2C19 LOF variants on clopidogrel response [70, 71]. Meta-analyses provide supporting evidence for a clinically important role of CYP2C19 LOF variants [72, 73]. For example, Jang et al. [72] estimated that carriers of one or more CYP2C19 LOF alleles had an increased risk of cardiovascular death (OR 2.18, 95% CI 1.37 to 3.47), MI (OR 1.42, 95% CI 1.12 to 1.81), and stent thrombosis (OR 2.41, 95% CI 1.76 to 3.30). These effects appear to be qualitatively consistent across ethnic populations [74-76]. By contrast, a recent meta-analysis by Holmes and coworkers [77] concluded that CYP2C19 LOF variants are not clinically significant contributors to clopidogrel response, citing issues with “treatment only” study designs and small study bias as the reasons for positive findings of other meta-analyses. We [78] and others [79, 80] suggest that a more likely explanation is that CYP2C19 genotype is an important determinant of patients’ responses to clopidogrel after receiving PCI, but possibly not in patients treated with clopidogrel for other indications [78, 81].
The burden of evidence led the FDA in March 2010 to mandate the addition of a boxed warning to clopidogrel’s label informing physicians that patients carrying CYP2C19 LOF variants may be less responsive to clopidogrel, that tests are available to assess CYP2C19*2 status, and that alternative drugs or doses are recommended in poor metabolizers [82]. The label was silent regarding CYP2C19 intermediate metabolizers and fell short of stronger language regarding use of alternative therapy. A consensus report by the American College of Cardiology Foundation Task Force on Clinical Expert Consensus Documents and the American Heart Association (ACCF/AHA) published in June 2010 recommends against routine testing for CYP2C19 LOF variants, citing that these variants only explain approximately 12% of the variation in clopidogrel response and have low positive predictive value [83]. Furthermore, at the time of its writing, prospective randomized clinical trials showing that genotype-directed therapy improves clinical outcomes had not been performed. Subsequently, in the RAPID GENE Study, 200 PCI patients were randomized to standard treatment with clopidogrel versus genotype-directed therapy in which CYP2C19*2 carriers received prasugrel. None of the 23 CYP2C19*2 carriers in the genotype-directed group had high on-treatment platelet reactivity while 7 of the 23 CYP2C19*2 carriers in the standard care group, a significantly higher number of individuals, had high on-treatment platelet reactivity [84].
At the time of this writing, no prospective randomized trials of genotype-directed therapy and clinical outcomes have been reported. The ACCF/AHA consensus statement stressed the importance of clinical judgment in choice of anti-platelet therapy, and we and others have suggested that CYP2C19 genotype may be useful in context with clinical and other factors in choosing anti-platelet therapy [85]. The Pharmacogenomics Research Network (PGRN) Clinical Pharmacogenomics Implementation Consortium (CPIC) published guidelines for CYP2C19 testing and interpretation, and a suggested algorithm for treatment (Fig. 1) [86••]. These guidelines may be useful in select patients, at least until results of properly designed and powered randomized clinical trials are available.
Fig. 1.
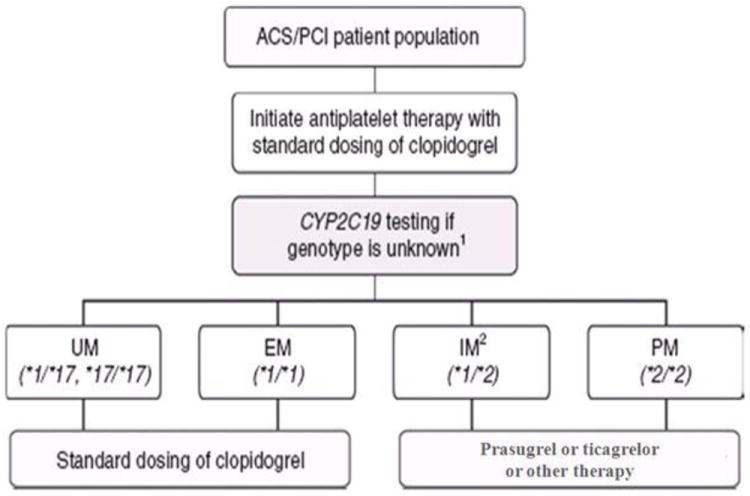
A suggested algorithm for genotype-guided anti-platelet therapy [86••]. (Adapted with permission from: Scott SA, Sangkuhl K, Gardner EE, Stein CM, Hulot JS, Johnson JA et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin Pharmacol Ther. 2011;90(2):328-32. doi:10.1038/clpt.2011.132) [86••]
In addition to LOF variants in CYP2C19, there is a gain-of-function (GOF) variant, CYP2C19*17 (rs12248560), residing in the 5’ regulatory region of the gene. This common variant, with minor allele frequencies of 21% and 16% in individuals of European and African ancestry, respectively, [87] is associated with increased transcriptional activity [63, 88]. The underlying mechanism of increased transcriptional activity likely involves hepatocyte nuclear binding to the *17 variant as evidenced by electrophoretic mobility shift assays [88].
Results of genetic association studies evaluating the effect of the CYP2C19*17 variant on clopidogrel response traits have been inconsistent. While some studies investigating patients during clopidogrel treatment have observed a decrease in cardiovascular event rates in individuals carrying the CYP2C19*17 variant [71, 89-92], others have not [53, 60, 63, 71, 93-97]. Similarly, bleeding was increased in subjects with CYP2C19*17 in some reports [91-95, 98, 99] and not associated in others [53, 71]. Although meta-analyses [91, 92] with CYP2C19*17 provide support for both a decrease in cardiovascular events and an increased risk of bleeding post treatment, CYP2C19*2 and CYP2C19*17 genotypes are in linkage disequilibrium and not independent of one another. Individuals with one or two CYP2C19*17 variant alleles are less likely to possess a copy of the CYP2C19*2 allele, and those with no copies of the CYP2C19*17 variant are more likely to have CYP2C19*2 variant alleles. Due to the linked nature of these variants, it has been suggested that enhanced response to clopidogrel in persons with one or more copies of the CYP2C19*17 variant may be due, at least in part, to the lack of CYP2C19*2 alleles in those individuals [100••]. Therefore, results reported for CYP2C19*17 should be considered cautiously unless it is clear the authors have statistically adjusted for the CYP2C19*2 variant in their association model.
ABCB1
As clopidogrel is absorbed from the intestinal lumen into the bloodstream, it must pass through intestinal enterocytes where a portion of the drug is immediately transported back into the lumen by the P-glycoprotein ATP-dependent efflux pump (ABCB1), also known as multidrug resistant 1 (MDR1). A common genetic variation in ABCB1, C3435T (rs1045642), affects gene transcription. The T allele of this polymorphism causes overexpression, which would be expected to result in greater extrusion of the drug into the intestinal lumen, less net absorption, decreased drug level in the bloodstream, and decreased response [94]. The frequency of the T allele is 57% in Caucasian, 41% in Asians (Chinese), and 11% in African descent.
Several studies have shown a modest association between the ABCB1 3435T allele and decreased clopidogrel active metabolite [101], increased on-treatment platelet reactivity [102] and cardiovascular events [103]. In addition, while determining which genes to include in a novel clopidogrel resistance risk score, which incorporates genotype and phenotype data, one group found a significant association between ABCB1 genotype and platelet reactivity as well as cardiovascular event risk [95].
Other studies have not found such an association between this ABCB1 variant and clopidogrel response, which may be due to inadequate power to discern a modest effect on clopidogrel response, specific characteristics of the patient populations, or false positive results of other studies. A recent meta-analysis examined 12 previously published studies of ABCB1 C3435T genotype. In the combined dataset, they found no association between ABCB1 genotype and on-treatment platelet reactivity, MI, ischemic stroke, all-cause mortality, stent thrombosis, or long-term major cardiovascular events. However, when stratified by loading dose, they found evidence for association between ABCB1 genotype and long-term cardiovascular events in the 300 mg loading dose group, early major adverse cardiovascular events, and bleeding; no such associations were observed in patients given the 600 mg loading dose [103]. These findings suggest that increased clopidogrel dose may be able to overcome higher efflux rates in T allele carriers.
PON1
Paraoxonase 1 (PON1) was named for its ability to metabolize paraoxon, a product of the detoxification of the insecticide parathion. PON1 is expressed in liver and is associated with HDL-cholesterol in the bloodstream. Two common variants in PON1 are A575G (rs662; Gln192Arg) and T163A (rs854560; Leu55Met), with the Gln and Met variants being associated with lower paraoxonase activity [104, 105]. Bouman and coworkers reported a significant association between PON1 Gln192Arg genotype and active clopidogrel metabolite concentration, level of platelet inhibition, and stent thrombosis [106]. These findings were remarkable since PON1 was not previously recognized to be involved in clopidogrel bioactivation. Curiously, this same study showed no effect of CYP2C19 genotype on on-treatment platelet reactivity or stent thrombosis. Subsequently, several studies failed to replicate association of Gln192Arg PON1 with a variety of endpoints including clopidogrel active metabolite levels [107], platelet function [62, 104, 107, 108], cardiovascular outcomes [104, 107, 109, 110], and stent thrombosis [107]. The reason underlying these discrepant findings are unclear. One study involving 300 patients undergoing PCI for ischemic heart disease showed a significant association between PON1 Gln192Arg genotype and on-treatment platelet reactivity at 1 and 6 months post-PCI, though with much smaller effect size than CYP2C19*2, *17, and ABCB1 genotypes [111]. These findings suggest that Bouman’s original study may have benefited from “the winner’s curse” and that PON1 genotype might have a smaller effect on clopidogrel response than initially reported, and that subsequent negative studies were not adequately powered, at least not for the stent thrombosis endpoint. Another study showed that PON1 may form a different thiol metabolite that is scarcer than clopi-H4 called Endo, which is not associated with anti-platelet response [112]. A recent analysis of PON1 Gln192Arg genotype in 424 Chinese with acute coronary syndrome found significant association with on-treatment platelet reactivity in CYP2C19*1 homozygotes but not in CYP2C19*2 carriers, suggesting interaction between clopidogrel metabolic pathways [113].
Also clouding the picture are several studies that have demonstrated that PON1 genotype may be related more to underlying cardiovascular disease risk than to clopidogrel response. A substudy of the CURE trial showed an association between PON1 genotype and cardiovascular event rates in the placebo group when the results were stratified by treatment arm [114]. These findings suggest a non-pharmacogenomic effect of PON1 genotype on cardiovascular outcome, which fits well with prior data showing PON1 to be associated with HDL particles, and that PON1 genotype is associated with enzymatic activity and the ability of HDL to prevent oxidation of LDL particles [115]. Planned large scale GWAS using an ultra-dense selection of SNPs will help resolve questions regarding associations of PON1 genetic variants with coronary events and HDL. Overall, the burden of evidence does not support a role for PON1 genotype in clopidogrel response, meaning further study will be required [116, 117].
P2RY12
The gene P2RY12 encodes the P2Y12 ADP receptor, the target for inactivation by clopidogrel on the surface of platelets. Two common linked genetic variants in this gene, G52T (rs2046934) and T744C (rs2046934), distinguish two major haplotypes, denoted H1 and H2, respectively. The H2 allele is believed to be associated with increased expression of P2RY12 [118]. A study in 225 healthy Caucasian volunteers exposed to clopidogrel showed that the H2/H2 genotype is associated with a significant decrease in inhibition of platelet aggregation in comparison to H1/H1 and H1/H2 individuals [119]. Similarly, another study in 557 clopidogrel-treated PCI patients showed H2/H2 homozygote individuals had significantly higher platelet aggregation and lower clopidogrel response [120]. In contrast, other studies, including several that examined clinical outcomes, have failed to show such associations [60, 121, 122] leading to the conclusion that if common variants in P2RY12 have an effect on clopidogrel response, this effect is small and not likely to be clinically important.
CES1
CES1 converts clopidogrel into an inactive carboxylic acid metabolite from its prodrug and thiolactone intermediate states [31]. An uncommon G/A variant (rs71647871) encodes a nonsynonymous substitution Gly143Gln resulting in marked decrease in catalytic function [123]. The frequency of the decreased function 143Gln allele is ~1%. A decreased function allele would be expected to be associated with decreased metabolism of clopidogrel into its inactive metabolite and conversely increased active metabolite levels and clopidogrel response. Indeed, in 566 healthy participants of the Pharmacogenomics of Anti-Platelet Intervention (PAPI) study, the seven 143Gln carriers had significantly higher active metabolite levels and more effective inhibition of ADP-simulated platelet aggregation. Although the variant is uncommon in the population, the effect size was found to be approximately two-fold greater than CYP2C19*2. In 330 PCI patients treated with clopidogrel, the six 143Gln carriers similarly showed more effective inhibition of platelet reactivity. In this same sample, there was a trend toward lower cardiovascular event rates in 143Gln carriers, not statistically significant perhaps due to the small sample size. Although these observations will require replication in larger studies, these data suggest that this relatively uncommon variant, present in its heterozygous form in approximately 2% of the population, may be a clinically important determinant of clopidogrel efficacy [124].
Other CYPs
As described above, other cytochrome P450 enzymes likely play roles in in vivo metabolism of clopidogrel [125-127]. Although functional variants in several of these enzymes exist and would be predicted to affect clopidogrel efficacy, to date the literature is mixed. It is likely that there are redundant mechanisms for clopidogrel metabolism rendering the effect of any single functional variant in these other CYPs small or non-existent. For example, some studies suggest a role for LOF variants in CYP2C9 in clopidogrel response [128, 129], while others do not [53, 122, 130].
Perhaps variants in other CYP genes play a role in clopidogrel response in subjects with LOF variants in CYP2C19, for whom alternate pathways may be more important. A study by Kassimis et al. showed that the *5 variant of CYP2B6 is associated with significantly higher platelet reactivity during clopidogrel treatment, but only in non-CYP2C19*2 carriers, thus showing both CYP2B6’s importance in and the profound impact of CYP2C19*2 on on-treatment platelet reactivity [130]. It is also possible that factors that affect activity of these CYPs, such as smoking or concurrent use of drugs that induce expression or inhibit action, may influence clopidogrel response in a genotype-dependent manner. A study suggests that CYP1A2 may explain in part the apparent increased clopidogrel response in smokers – the so-called smokers’ paradox – because CYP1A2 is induced by polycyclic aromatic hydrocarbons found in cigarette smoke [131]. A study by Zhou and coworkers demonstrated in Koreans that smokers carrying the CYP1A2*1F variant (rs762551) had reduced on-treatment platelet reactivity, an effect that was not apparent in non-smokers [132]. In some patients, variants in CYP3A5, a “back-up” pathway for CYP3A4, may explain interaction between clopidogrel and amlodipine, a potent CYP3A4 inhibitor. In subjects homozygous for the loss of function CYP3A5*3 variant, amlodipine causes a significant increase in on-clopidogrel platelet reactivity while no such effect was observed in carriers of at least one functional allele [133]. Another study showed that the CYP3A5*3 variant is only associated with clopidogrel response when clopidogrel is co-administered with itraconazole, a known CYP3A inhibitor [134].
Translation of Clopidogrel Pharmacogenomics into Patient Care
While approximately 12% of variation in clopidogrel response may be explained by CYP2C19 LOF variants, data suggest that a large amount of the heritability of response remains unknown. Future studies in search of genetic determinants of clopidogrel response will require much larger sample sizes of clopidogrel-treated patients and the application of genome-wide and NextGen sequencing approaches. The International Clopidogrel Pharmacogenomics Consortium (ICPC) seeks to perform a large GWAS in order to identify novel common variants for clopidogrel response [135]. The PGRN has developed a sequencing panel of 84 “pharmacogenes” (PGRN-Seq) [136]. Early data suggest the existence of much more rare variation in these genes than was previously thought [137]. As additional genetic determinants of clopidogrel response are uncovered, it is anticipated that their addition to already available CYP2C19 genetic testing will increase the clinical utility of genetic testing toward more effective individualized anti-platelet therapy.
Conclusions
In recent years, there has been much progress in our understanding of the genetic basis of variation in clopidogrel and warfarin response. Solid data now supports findings that common variants in CYP2C9 and VKORC1 alter warfarin maintenance dose requirements and prospective trials are now underway to learn whether genotype-guided therapy can increase the percent of time in the therapeutic INR range and lower adverse bleeding and thrombotic events. A large body of data now also supports a role of common LOF variants in CYP2C19 on clopidogrel response, including studies that have examined active drug levels, platelet reactivity, and clinical outcomes. There is a likely a real but smaller effect of variants in ABCB1 as determinants of clopidogrel response. At the current time, the aggregate of literature does not support a major effect by PON1, P2RY12, or other CYP genes on clopidogrel response, although variation in these genes may contribute in other ways to the complex architecture of clopidogrel response.
Acknowledgments
Adam S. Fisch has received grant support from NIH.
Footnotes
Conflict of Interest
Adam S. Fisch declares that he has no conflict of interest.
Christina G. Perry declares that she has no conflict of interest.
Sarah H. Stephens declares that she has no conflict of interest.
Richard B. Horenstein declares that he has no conflict of interest.
Alan R. Shuldiner declares that he has no conflict of interest.
References
Papers of particular interest, published recently, have been highlighted as:
-
•
Of importance
-
••
Of major importance
- 1•.Shin J. Clinical pharmacogenomics of warfarin and clopidogrel. J Pharm Pract. 2012;25(4):428–38. doi: 10.1177/0897190012448310. This paper provides a broad overview of the major alleles involved in warfarin and clopidogrel metabolism while also addressing the clinical implications of pharmacogenomic testing. [DOI] [PubMed] [Google Scholar]
- 2.Agundez JA, Martinez C, Perez-Sala D, Carballo M, Torres MJ, Garcia-Martin E. Pharmacogenomics in aspirin intolerance. Curr Drug Metab. 2009;10(9):998–1008. doi: 10.2174/138920009790711814. [DOI] [PubMed] [Google Scholar]
- 3.Goodman T, Ferro A, Sharma P. Pharmacogenetics of aspirin resistance: a comprehensive systematic review. Br J Clin Pharm. 2008;66(2):222–32. doi: 10.1111/j.1365-2125.2008.03183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faraday N, Becker DM, Becker LC. Pharmacogenomics of platelet responsiveness to aspirin. Pharmacogenomics. 2007;8(10):1413–25. doi: 10.2217/14622416.8.10.1413. [DOI] [PubMed] [Google Scholar]
- 5.Kaminsky LS, Zhang ZY. Human P450 metabolism of warfarin. Pharmacol Ther. 1997;73(1):67–74. doi: 10.1016/s0163-7258(96)00140-4. [DOI] [PubMed] [Google Scholar]
- 6.Ansell J, Hirsh J, Poller L, Bussey H, Jacobson A, Hylek E. The pharmacology and management of the vitamin K antagonists: the Seventh ACCP Conference on Antithrombotic and Thrombolytic Therapy. Chest. 2004;126(3 Suppl):204S–33S. doi: 10.1378/chest.126.3_suppl.204S. [DOI] [PubMed] [Google Scholar]
- 7.Johnson JA, Gong L, Whirl-Carrillo M, Gage BF, Scott SA, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther. 2011;90(4):625–9. doi: 10.1038/clpt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287(13):1690–8. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 9.Gage BF, Eby C, Milligan PE, Banet GA, Duncan JR, McLeod HL. Use of pharmacogenetics and clinical factors to predict the maintenance dose of warfarin. Throm Haemostasis. 2004;91(1):87–94. doi: 10.1267/THRO04010087. [DOI] [PubMed] [Google Scholar]
- 10.Zhu Y, Shennan M, Reynolds KK, Johnson NA, Herrnberger MR, Valdes R, Jr, et al. Estimation of warfarin maintenance dose based on VKORC1 (-1639 G>A) and CYP2C9 genotypes. Clin Chem. 2007;53(7):1199–205. doi: 10.1373/clinchem.2006.078139. [DOI] [PubMed] [Google Scholar]
- 11.Rettie AE, Tai G. The pharmocogenomics of warfarin: closing in on personalized medicine. Mol Interv. 2006;6(4):223–7. doi: 10.1124/mi.6.4.8. [DOI] [PubMed] [Google Scholar]
- 12.Hill CE, Duncan A. Overview of pharmacogenetics in anticoagulation therapy. Clin Lab Med. 2008;28(4):513–24. doi: 10.1016/j.cll.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Gage BF, Lesko LJ. Pharmacogenetics of warfarin: regulatory, scientific, and clinical issues. J Thromb Thrombolys. 2008;25(1):45–51. doi: 10.1007/s11239-007-0104-y. [DOI] [PubMed] [Google Scholar]
- 14.Scott SA, Jaremko M, Lubitz SA, Kornreich R, Halperin JL, Desnick RJ. CYP2C9*8 is prevalent among African-Americans: implications for pharmacogenetic dosing. Pharmacogenomics. 2009;10(8):1243–55. doi: 10.2217/pgs.09.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T, Chang CY, Jin DY, Lin PJ, Khvorova A, Stafford DW. Identification of the gene for vitamin K epoxide reductase. Nature. 2004;427(6974):541–4. doi: 10.1038/nature02254. [DOI] [PubMed] [Google Scholar]
- 16.Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352(22):2285–93. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 17.Limdi NA, Veenstra DL. Warfarin pharmacogenetics. Pharmacotherapy. 2008;28(9):1084–97. doi: 10.1592/phco.28.9.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Wang S, Barone J, Malone B. Warfarin pharmacogenomics. P & T : a peer-reviewed journal for formulary management. 2009;34(8):422–7. [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson JL, Horne BD, Stevens SM, Grove AS, Barton S, Nicholas ZP, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007;116(22):2563–70. doi: 10.1161/CIRCULATIONAHA.107.737312. [DOI] [PubMed] [Google Scholar]
- 20.Sontag TJ, Parker RS. Cytochrome P450 omega-hydroxylase pathway of tocopherol catabolism. Novel mechanism of regulation of vitamin E status. J Biol Chem. 2002;277(28):25290–6. doi: 10.1074/jbc.M201466200. [DOI] [PubMed] [Google Scholar]
- 21.Singh O, Sandanaraj E, Subramanian K, Lee LH, Chowbay B. Influence of CYP4F2 rs2108622 (V433M) on Warfarin Dose Requirement in Asian Patients. Drug Metab Pharmacok. 2011;26(2):130–6. doi: 10.2133/dmpk.DMPK-10-RG-080. [DOI] [PubMed] [Google Scholar]
- 22.Caldwell MD, Awad T, Johnson JA, Gage BF, Falkowski M, Gardina P, et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;111(8):4106–12. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borgiani P, Ciccacci C, Forte V, Sirianni E, Novelli L, Bramanti P, et al. CYP4F2 genetic variant (rs2108622) significantly contributes to warfarin dosing variability in the Italian population. Pharmacogenomics. 2009;10(2):261–6. doi: 10.2217/14622416.10.2.261. [DOI] [PubMed] [Google Scholar]
- 24.Perini JA, Struchiner CJ, Silva-Assuncao E, Suarez-Kurtz G. Impact of CYP4F2 rs2108622 on the stable warfarin dose in an admixed patient cohort. Clin Pharmacol Ther. 2010;87(4):417–20. doi: 10.1038/clpt.2009.307. [DOI] [PubMed] [Google Scholar]
- 25.Wells PS, Majeed H, Kassem S, Langlois N, Gin B, Clermont J, et al. A regression model to predict warfarin dose from clinical variables and polymorphisms in CYP2C9, CYP4F2, and VKORC1: Derivation in a sample with predominantly a history of venous thromboembolism. Thromb Res. 2010;125(6):e259–64. doi: 10.1016/j.thromres.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 26.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genetics. 2009;5(3):e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112(4):1022–7. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwarz UI, Ritchie MD, Bradford Y, Li C, Dudek SM, Frye-Anderson A, et al. Genetic determinants of response to warfarin during initial anticoagulation. N Engl J Med. 2008;358(10):999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.WARFARIN DOSING. 2012 http://www.warfarindosing.org.
- 30.Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med. 2011;364(12):1144–53. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ancrenaz V, Daali Y, Fontana P, Besson M, Samer C, Dayer P, et al. Impact of genetic polymorphisms and drug-drug interactions on clopidogrel and prasugrel response variability. Curr Drug Metab. 2010;11(8):667–77. doi: 10.2174/138920010794233521. [DOI] [PubMed] [Google Scholar]
- 32.Richter T, Murdter TE, Heinkele G, Pleiss J, Tatzel S, Schwab M, et al. Potent mechanism-based inhibition of human CYP2B6 by clopidogrel and ticlopidine. J Pharmacol Exp Therap. 2004;308(1):189–97. doi: 10.1124/jpet.103.056127. [DOI] [PubMed] [Google Scholar]
- 33.Savi P, Combalbert J, Gaich C, Rouchon MC, Maffrand JP, Berger Y, et al. The antiaggregating activity of clopidogrel is due to a metabolic activation by the hepatic cytochrome P450-1A. Throm Haemostasis. 1994;72(2):313–7. [PubMed] [Google Scholar]
- 34.Turpeinen M, Tolonen A, Uusitalo J, Jalonen J, Pelkonen O, Laine K. Effect of clopidogrel and ticlopidine on cytochrome P450 2B6 activity as measured by bupropion hydroxylation. Clin Pharmacol Ther. 2005;77(6):553–9. doi: 10.1016/j.clpt.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 35.Cattaneo M. The platelet P2Y(1)(2) receptor for adenosine diphosphate: congenital and drug-induced defects. Blood. 2011;117(7):2102–12. doi: 10.1182/blood-2010-08-263111. [DOI] [PubMed] [Google Scholar]
- 36.Gum PA, Kottke-Marchant K, Welsh PA, White J, Topol EJ. A prospective, blinded determination of the natural history of aspirin resistance among stable patients with cardiovascular disease. J Am Coll Cardiol. 2003;41(6):961–5. doi: 10.1016/s0735-1097(02)03014-0. [DOI] [PubMed] [Google Scholar]
- 37.Gurbel PA, Bliden KP, Hiatt BL, O’Connor CM. Clopidogrel for coronary stenting: response variability, drug resistance, and the effect of pretreatment platelet reactivity. Circulation. 2003;107(23):2908–13. doi: 10.1161/01.CIR.0000072771.11429.83. [DOI] [PubMed] [Google Scholar]
- 38.Gurbel PA, Tantry US. Drug insight: Clopidogrel nonresponsiveness. Nature clinical practice Cardiovascular medicine. 2006;3(7):387–95. doi: 10.1038/ncpcardio0602. [DOI] [PubMed] [Google Scholar]
- 39.Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, Alfonso F, Macaya C, Bass TA, et al. Variability in individual responsiveness to clopidogrel: clinical implications, management, and future perspectives. J Am Coll Cardiol. 2007;49(14):1505–16. doi: 10.1016/j.jacc.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 40.O’Donoghue M, Wiviott SD. Clopidogrel response variability and future therapies: clopidogrel: does one size fit all? Circulation. 2006;114(22):e600–6. doi: 10.1161/CIRCULATIONAHA.106.643171. [DOI] [PubMed] [Google Scholar]
- 41.Wang TH, Bhatt DL, Topol EJ. Aspirin and clopidogrel resistance: an emerging clinical entity. Eur Heart J. 2006;27(6):647–54. doi: 10.1093/eurheartj/ehi684. [DOI] [PubMed] [Google Scholar]
- 42.Gurbel PA, Bliden KP, Guyer K, Cho PW, Zaman KA, Kreutz RP, et al. Platelet reactivity in patients and recurrent events post-stenting: results of the PREPARE POST-STENTING Study. J Am Coll Cardiol. 2005;46(10):1820–6. doi: 10.1016/j.jacc.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 43.Gurbel PA, Becker RC, Mann KG, Steinhubl SR, Michelson AD. Platelet function monitoring in patients with coronary artery disease. J Am Coll Cardiol. 2007;50(19):1822–34. doi: 10.1016/j.jacc.2007.07.051. [DOI] [PubMed] [Google Scholar]
- 44.Angiolillo DJ, Alfonso F. Platelet function testing and cardiovascular outcomes: steps forward in identifying the best predictive measure. Throm Haemostasis. 2007;98(4):707–9. [PubMed] [Google Scholar]
- 45.Bliden KP, DiChiara J, Tantry US, Bassi AK, Chaganti SK, Gurbel PA. Increased risk in patients with high platelet aggregation receiving chronic clopidogrel therapy undergoing percutaneous coronary intervention: is the current antiplatelet therapy adequate? J Am Coll Cardiol. 2007;49(6):657–66. doi: 10.1016/j.jacc.2006.10.050. [DOI] [PubMed] [Google Scholar]
- 46.Buonamici P, Marcucci R, Migliorini A, Gensini GF, Santini A, Paniccia R, et al. Impact of platelet reactivity after clopidogrel administration on drug-eluting stent thrombosis. J Am Coll Cardiol. 2007;49(24):2312–7. doi: 10.1016/j.jacc.2007.01.094. [DOI] [PubMed] [Google Scholar]
- 47.Gurbel PA, Bliden KP, Zaman KA, Yoho JA, Hayes KM, Tantry US. Clopidogrel loading with eptifibatide to arrest the reactivity of platelets: results of the Clopidogrel Loading With Eptifibatide to Arrest the Reactivity of Platelets (CLEAR PLATELETS) study. Circulation. 2005;111(9):1153–9. doi: 10.1161/01.CIR.0000157138.02645.11. [DOI] [PubMed] [Google Scholar]
- 48.Gurbel PA, Antonino MJ, Bliden KP, Dichiara J, Suarez TA, Singla A, et al. Platelet reactivity to adenosine diphosphate and long-term ischemic event occurrence following percutaneous coronary intervention: a potential antiplatelet therapeutic target. Platelets. 2008;19(8):595–604. doi: 10.1080/09537100802351065. [DOI] [PubMed] [Google Scholar]
- 49.Matetzky S, Shenkman B, Guetta V, Shechter M, Beinart R, Goldenberg I, et al. Clopidogrel resistance is associated with increased risk of recurrent atherothrombotic events in patients with acute myocardial infarction. Circulation. 2004;109(25):3171–5. doi: 10.1161/01.CIR.0000130846.46168.03. [DOI] [PubMed] [Google Scholar]
- 50.Gilard M, Arnaud B, Le Gal G, Abgrall JF, Boschat J. Influence of omeprazol on the antiplatelet action of clopidogrel associated to aspirin. J Throm Haemostasis : JTH. 2006;4(11):2508–9. doi: 10.1111/j.1538-7836.2006.02162.x. [DOI] [PubMed] [Google Scholar]
- 51.Lau WC, Gurbel PA, Watkins PB, Neer CJ, Hopp AS, Carville DG, et al. Contribution of hepatic cytochrome P450 3A4 metabolic activity to the phenomenon of clopidogrel resistance. Circulation. 2004;109(2):166–71. doi: 10.1161/01.CIR.0000112378.09325.F9. [DOI] [PubMed] [Google Scholar]
- 52.Siller-Matula JM, Lang I, Christ G, Jilma B. Calcium-channel blockers reduce the antiplatelet effect of clopidogrel. J Am Coll Cardiol. 2008;52(19):1557–63. doi: 10.1016/j.jacc.2008.07.055. [DOI] [PubMed] [Google Scholar]
- 53.Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360(4):354–62. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 54.Kim KA, Park PW, Hong SJ, Park JY. The effect of CYP2C19 polymorphism on the pharmacokinetics and pharmacodynamics of clopidogrel: a possible mechanism for clopidogrel resistance. Clin Pharmacol Ther. 2008;84(2):236–42. doi: 10.1038/clpt.2008.20. [DOI] [PubMed] [Google Scholar]
- 55.Umemura K, Furuta T, Kondo K. The common gene variants of CYP2C19 affect pharmacokinetics and pharmacodynamics in an active metabolite of clopidogrel in healthy subjects. Journal of Throm Haemostasis : JTH. 2008;6(8):1439–41. doi: 10.1111/j.1538-7836.2008.03050.x. [DOI] [PubMed] [Google Scholar]
- 56.Fontana P, Hulot JS, De Moerloose P, Gaussem P. Influence of CYP2C19 and CYP3A4 gene polymorphisms on clopidogrel responsiveness in healthy subjects. J Throm Haemostasis : JTH. 2007;5(10):2153–5. doi: 10.1111/j.1538-7836.2007.02722.x. [DOI] [PubMed] [Google Scholar]
- 57.Brandt JT, Close SL, Iturria SJ, Payne CD, Farid NA, Ernest CS, 2nd, et al. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Throm Haemostasis : JTH. 2007;5(12):2429–36. doi: 10.1111/j.1538-7836.2007.02775.x. [DOI] [PubMed] [Google Scholar]
- 58.Trenk D, Hochholzer W, Fromm MF, Chialda LE, Pahl A, Valina CM, et al. Cytochrome P450 2C19 681G>A polymorphism and high on-clopidogrel platelet reactivity associated with adverse 1-year clinical outcome of elective percutaneous coronary intervention with drug-eluting or bare-metal stents. J Am Coll Cardiol. 2008;51(20):1925–34. doi: 10.1016/j.jacc.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 59.Collet JP, Hulot JS, Pena A, Villard E, Esteve JB, Silvain J, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet. 2009;373(9660):309–17. doi: 10.1016/S0140-6736(08)61845-0. [DOI] [PubMed] [Google Scholar]
- 60.Simon T, Verstuyft C, Mary-Krause M, Quteineh L, Drouet E, Meneveau N, et al. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360(4):363–75. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 61.Sibbing D, Stegherr J, Latz W, Koch W, Mehilli J, Dorrler K, et al. Cytochrome P450 2C19 loss-of-function polymorphism and stent thrombosis following percutaneous coronary intervention. Eur Heart J. 2009;30(8):916–22. doi: 10.1093/eurheartj/ehp041. [DOI] [PubMed] [Google Scholar]
- 62.Price MJ, Murray SS, Angiolillo DJ, Lillie E, Smith EN, Tisch RL, et al. Influence of genetic polymorphisms on the effect of high- and standard-dose clopidogrel after percutaneous coronary intervention: the GIFT (Genotype Information and Functional Testing) study. J Am Coll Cardiol. 2012;59(22):1928–37. doi: 10.1016/j.jacc.2011.11.068. [DOI] [PubMed] [Google Scholar]
- 63.Shuldiner AR, O’Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302(8):849–57. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dandona S, Roberts R. Personalized cardiovascular medicine: status in 2012. Can J Cardiol. 2012;28(6):693–9. doi: 10.1016/j.cjca.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 65.Donohue MM, Tirschwell DL. Implications of pharmacogenetic testing for patients taking warfarin or clopidogrel. Curr Neurol Neurosci. 2011;11(1):52–60. doi: 10.1007/s11910-010-0157-8. [DOI] [PubMed] [Google Scholar]
- 66.Cayla G, Silvain J, O’Connor SA, Collet JP, Montalescot G. An evidence-based review of current anti-platelet options for STEMI patients. Int J Cardiol. 2012 doi: 10.1016/j.ijcard.2012.04.160. [DOI] [PubMed] [Google Scholar]
- 67.Close SL. Pharmacogenetics and pharmacogenomics of thienopyridines: clinically relevant? FundClin Pharmacol. 2012;26(1):19–26. doi: 10.1111/j.1472-8206.2011.00983.x. [DOI] [PubMed] [Google Scholar]
- 68.Fefer P, Matetzky S. The genetic basis of platelet responsiveness to clopidogrel. A critical review of the literature. Throm Haemostasis. 2011;106(2):203–10. doi: 10.1160/TH11-04-0228. [DOI] [PubMed] [Google Scholar]
- 69.Price MJ, Tantry US, Gurbel PA. The influence of CYP2C19 polymorphisms on the pharmacokinetics, pharmacodynamics, and clinical effectiveness of P2Y(12) inhibitors. Rev Cardiovas Med. 2011;12(1):1–12. doi: 10.3909/ricm0590. [DOI] [PubMed] [Google Scholar]
- 70.Bhatt DL, Pare G, Eikelboom JW, Simonsen KL, Emison ES, Fox KA, et al. The relationship between CYP2C19 polymorphisms and ischaemic and bleeding outcomes in stable outpatients: the CHARISMA genetics study. Eur Heart J. 2012;33(17):2143–50. doi: 10.1093/eurheartj/ehs059. [DOI] [PubMed] [Google Scholar]
- 71.Pare G, Mehta SR, Yusuf S, Anand SS, Connolly SJ, Hirsh J, et al. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med. 2010;363(18):1704–14. doi: 10.1056/NEJMoa1008410. [DOI] [PubMed] [Google Scholar]
- 72.Jang JS, Cho KI, Jin HY, Seo JS, Yang TH, Kim DK, et al. Meta-analysis of cytochrome P450 2C19 polymorphism and risk of adverse clinical outcomes among coronary artery disease patients of different ethnic groups treated with clopidogrel. Am J Cardiol. 2012;110(4):502–8. doi: 10.1016/j.amjcard.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 73.Mega JL, Simon T, Collet JP, Anderson JL, Antman EM, Bliden K, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304(16):1821–30. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang F, Desta Z, Shon JH, Yeo CW, Kim HS, Liu KH, et al. Effects of clopidogrel and itraconazole on the disposition of efavirenz and its hydroxyl-metabolites: exploration of a novel CYP2B6 phenotyping index. Br J Clin Pharm. 2012 doi: 10.1111/j.1365-2125.2012.04314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zou JJ, Xie HG, Chen SL, Tan J, Lin L, Zhao YY, et al. Influence of CYP2C19 loss-of-function variants on the antiplatelet effects and cardiovascular events in clopidogrel-treated Chinese patients undergoing percutaneous coronary intervention. Eur J Clin Pharmacol. 2012 doi: 10.1007/s00228-012-1392-5. [DOI] [PubMed] [Google Scholar]
- 76.Subraja K, Dkhar SA, Priyadharsini R, Ravindra BK, Shewade DG, Satheesh S, et al. Genetic polymorphisms of CYP2C19 influences the response to clopidogrel in ischemic heart disease patients in the South Indian Tamilian population. Eur J Clin Pharmacol. 2012 doi: 10.1007/s00228-012-1381-8. [DOI] [PubMed] [Google Scholar]
- 77.Holmes MV, Perel P, Shah T, Hingorani AD, Casas JP. CYP2C19 genotype, clopidogrel metabolism, platelet function, and cardiovascular events: a systematic review and meta-analysis. JAMA. 2011;306(24):2704–14. doi: 10.1001/jama.2011.1880. [DOI] [PubMed] [Google Scholar]
- 78.Shuldiner AR, Vesely MR, Fisch A. CYP2C19 genotype and cardiovascular events. JAMA. 2012;307(14):1482. doi: 10.1001/jama.2012.443. [DOI] [PubMed] [Google Scholar]
- 79.Mega JL, Topol EJ, Sabatine MS. CYP2C19 genotype and cardiovascular events. JAMA. 2012;307(14):1482–3. doi: 10.1001/jama.2012.444. [DOI] [PubMed] [Google Scholar]
- 80.Siasos G, Tousoulis D, Stefanadis C. CYP2C19 genotype and cardiovascular events. JAMA. 2012;307(14):1483–4. doi: 10.1001/jama.2012.445. [DOI] [PubMed] [Google Scholar]
- 81.Johnson JA, Roden DM, Lesko LJ, Ashley E, Klein TE, Shuldiner AR. Clopidogrel: a case for indication-specific pharmacogenetics. Clin Pharmacol Ther. 2012;91(5):774–6. doi: 10.1038/clpt.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.FDA Drug Safety Communication: Reduced effectiveness of Plavix (clopidogrel) in patients who are poor metabolizers of the drug. 2012 http://www.fda.gov/Drugs/DrugSafety/PostmarketDrugSafetyInformationforPatientsandProviders/ucm203888.htm.
- 83.Holmes DR, Jr, Dehmer GJ, Kaul S, Leifer D, O’Gara PT, Stein CM. ACCF/AHA clopidogrel clinical alert: approaches to the FDA “boxed warning”: a report of the American College of Cardiology Foundation Task Force on clinical expert consensus documents and the American Heart Association endorsed by the Society for Cardiovascular Angiography and Interventions and the Society of Thoracic Surgeons. J Am Coll Cardiol. 2010;56(4):321–41. doi: 10.1016/j.jacc.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 84.Roberts JD, Wells GA, Le May MR, Labinaz M, Glover C, Froeschl M, et al. Point-of-care genetic testing for personalisation of antiplatelet treatment (RAPID GENE): a prospective, randomised, proof-of-concept trial. Lancet. 2012;379(9827):1705–11. doi: 10.1016/S0140-6736(12)60161-5. [DOI] [PubMed] [Google Scholar]
- 85.Roden DM, Shuldiner AR. Responding to the clopidogrel warning by the US food and drug administration: real life is complicated. Circulation. 2010;122(5):445–8. doi: 10.1161/CIRCULATIONAHA.110.973362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86••.Scott SA, Sangkuhl K, Gardner EE, Stein CM, Hulot JS, Johnson JA, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin Pharmacol Ther. 2011;90(2):328–32. doi: 10.1038/clpt.2011.132. This reference provides an implementable algorithm for approaching the patient indicated for clopidogrel using evidence established in the literature already, and in the absence of a completed, prospective, controlled, randomized, double-blinded clinical trial. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barrett JC. Haploview: Visualization and analysis of SNP genotype data. Cold Spring Harb Protoc. 2009;2009(10) doi: 10.1101/pdb.ip71. pdb ip71. [DOI] [PubMed] [Google Scholar]
- 88.Sim SC, Risinger C, Dahl ML, Aklillu E, Christensen M, Bertilsson L, et al. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther. 2006;79(1):103–13. doi: 10.1016/j.clpt.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 89.Tiroch KA, Sibbing D, Koch W, Roosen-Runge T, Mehilli J, Schomig A, et al. Protective effect of the CYP2C19 *17 polymorphism with increased activation of clopidogrel on cardiovascular events. Am Heart J. 2010;160(3):506–12. doi: 10.1016/j.ahj.2010.06.039. [DOI] [PubMed] [Google Scholar]
- 90.Bouman HJ, Harmsze AM, van Werkum JW, Breet NJ, Bergmeijer TO, Ten Cate H, et al. Variability in on-treatment platelet reactivity explained by CYP2C19*2 genotype is modest in clopidogrel pretreated patients undergoing coronary stenting. Heart. 2011;97(15):1239–44. doi: 10.1136/hrt.2010.220509. [DOI] [PubMed] [Google Scholar]
- 91.Zabalza M, Subirana I, Sala J, Lluis-Ganella C, Lucas G, Tomas M, et al. Meta-analyses of the association between cytochrome CYP2C19 loss- and gain-of-function polymorphisms and cardiovascular outcomes in patients with coronary artery disease treated with clopidogrel. Heart. 2012;98(2):100–8. doi: 10.1136/hrt.2011.227652. [DOI] [PubMed] [Google Scholar]
- 92.Li Y, Tang HL, Hu YF, Xie HG. The gain-of-function variant allele CYP2C19*17: a double-edged sword between thrombosis and bleeding in clopidogrel-treated patients. J Thromb Haemost. 2012;10(2):199–206. doi: 10.1111/j.1538-7836.2011.04570.x. [DOI] [PubMed] [Google Scholar]
- 93.Sibbing D, Koch W, Gebhard D, Schuster T, Braun S, Stegherr J, et al. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation. 2010;121(4):512–8. doi: 10.1161/CIRCULATIONAHA.109.885194. [DOI] [PubMed] [Google Scholar]
- 94.Wallentin L, James S, Storey RF, Armstrong M, Barratt BJ, Horrow J, et al. Effect of CYP2C19 and ABCB1 single nucleotide polymorphisms on outcomes of treatment with ticagrelor versus clopidogrel for acute coronary syndromes: a genetic substudy of the PLATO trial. Lancet. 2010;376(9749):1320–8. doi: 10.1016/S0140-6736(10)61274-3. [DOI] [PubMed] [Google Scholar]
- 95.Campo G, Parrinello G, Ferraresi P, Lunghi B, Tebaldi M, Miccoli M, et al. Prospective evaluation of on-clopidogrel platelet reactivity over time in patients treated with percutaneous coronary intervention relationship with gene polymorphisms and clinical outcome. J Am Coll Cardiol. 2011;57(25):2474–83. doi: 10.1016/j.jacc.2010.12.047. [DOI] [PubMed] [Google Scholar]
- 96.Bauer T, Bouman HJ, van Werkum JW, Ford NF, ten Berg JM, Taubert D. Impact of CYP2C19 variant genotypes on clinical efficacy of antiplatelet treatment with clopidogrel: systematic review and meta-analysis. BMJ. 2011;343:d4588. doi: 10.1136/bmj.d4588. 10.1136/bmj.d4588 bmj.d4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tello-Montoliu A, Jover E, Marin F, Bernal A, Lozano ML, Sanchez-Vega B, et al. Influence of CYP2C19 polymorphisms in platelet reactivity and prognosis in an unselected population of non ST elevation acute coronary syndrome. Rev Esp Cardiol (Engl) 2012;65(3):219–26. doi: 10.1016/j.recesp.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 98.Dai ZL, Chen H, Wu XY. Relationship between cytochrome P450 2C19*17 genotype distribution, platelet aggregation and bleeding risk in patients with blood stasis syndrome of coronary artery disease treated with clopidogrel. Zhong Xi Yi Jie He Xue Bao. 2012;10(6):647–54. doi: 10.3736/jcim20120608. [DOI] [PubMed] [Google Scholar]
- 99.Harmsze AM, van Werkum JW, Hackeng CM, Ruven HJ, Kelder JC, Bouman HJ, et al. The influence of CYP2C19*2 and *17 on on-treatment platelet reactivity and bleeding events in patients undergoing elective coronary stenting. Pharmacogenet Genomics. 2012;22(3):169–75. doi: 10.1097/FPC.0b013e32834ff6e3. [DOI] [PubMed] [Google Scholar]
- 100••.Gurbel PA, Tantry US, Shuldiner AR. Letter by Gurbel et al regarding article, “Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement”. Circulation. 2010;122(14):e478. doi: 10.1161/CIRCULATIONAHA.110.943548. This reference explains the importance of taking CYP2C19*2 status into account when observing the effects of CYP2C19*17 on platelet aggregation and cardiovascular outcomes since the two loci are in linkage disequilibrium. [DOI] [PubMed] [Google Scholar]
- 101.Taubert D, von Beckerath N, Grimberg G, Lazar A, Jung N, Goeser T, et al. Impact of P-glycoprotein on clopidogrel absorption. Clin Pharmacol Ther. 2006;80(5):486–501. doi: 10.1016/j.clpt.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 102.Wang XD, Zhang DF, Liu XB, Lai Y, Qi WG, Luo Y, et al. Modified clopidogrel loading dose according to platelet reactivity monitoring in patients carrying ABCB1 variant alleles in patients with clopidogrel resistance. Eur J Intern Med. 2012;23(1):48–53. doi: 10.1016/j.ejim.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 103.Su J, Xu J, Li X, Zhang H, Hu J, Fang R, et al. ABCB1 C3435T Polymorphism and Response to Clopidogrel Treatment in Coronary Artery Disease (CAD) Patients: A Meta-Analysis. PloS one. 2012;7(10):e46366. doi: 10.1371/journal.pone.0046366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lewis JP, Fisch AS, Ryan K, O’Connell JR, Gibson Q, Mitchell BD, et al. Paraoxonase 1 (PON1) gene variants are not associated with clopidogrel response. Clin Pharmacol Ther. 2011;90(4):568–74. doi: 10.1038/clpt.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mackness B, Mackness MI, Arrol S, Turkie W, Durrington PN. Effect of the molecular polymorphisms of human paraoxonase (PON1) on the rate of hydrolysis of paraoxon. Br J Pharmacol. 1997;122(2):265–8. doi: 10.1038/sj.bjp.0701390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bouman HJ, Schomig E, van Werkum JW, Velder J, Hackeng CM, Hirschhauser C, et al. Paraoxonase-1 is a major determinant of clopidogrel efficacy. Nature Med. 2011;17(1):110–6. doi: 10.1038/nm.2281. [DOI] [PubMed] [Google Scholar]
- 107.Hulot JS, Collet JP, Cayla G, Silvain J, Allanic F, Bellemain-Appaix A, et al. CYP2C19 but not PON1 genetic variants influence clopidogrel pharmacokinetics, pharmacodynamics, and clinical efficacy in post-myocardial infarction patients. Circ Cardiovas Interv. 2011;4(5):422–8. doi: 10.1161/CIRCINTERVENTIONS.111.963025. [DOI] [PubMed] [Google Scholar]
- 108.Kreutz RP, Nystrom P, Kreutz Y, Miao J, Desta Z, Breall JA, et al. Influence of paraoxonase-1 Q192R and cytochrome P450 2C19 polymorphisms on clopidogrel response. ClinPharmacol. 2012;4:13–20. doi: 10.2147/CPAA.S27822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Simon T, Steg PG, Becquemont L, Verstuyft C, Kotti S, Schiele F, et al. Effect of paraoxonase-1 polymorphism on clinical outcomes in patients treated with clopidogrel after an acute myocardial infarction. Clin Pharmacol Ther. 2011;90(4):561–7. doi: 10.1038/clpt.2011.193. [DOI] [PubMed] [Google Scholar]
- 110.Verschuren JJ, Boden H, Wessels JA, van der Hoeven BL, Trompet S, Heijmans BT, et al. Value of platelet pharmacogenetics in common clinical practice of patients with ST-segment elevation myocardial infarction. Int J Cardiol. 2012 doi: 10.1016/j.ijcard.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 111.Campo G, Ferraresi P, Marchesini J, Bernardi F, Valgimigli M. Relationship between paraoxonase Q192R gene polymorphism and on-clopidogrel platelet reactivity over time in patients treated with percutaneous coronary intervention. Journal of Throm Haemostasis : JTH. 2011;9(10):2106–8. doi: 10.1111/j.1538-7836.2011.04457.x. [DOI] [PubMed] [Google Scholar]
- 112.Gong IY, Crown N, Suen CM, Schwarz UI, Dresser GK, Knauer MJ, et al. Clarifying the importance of CYP2C19 and PON1 in the mechanism of clopidogrel bioactivation and in vivo antiplatelet response. Eur Heart J. 2012;33(22):2856–64. doi: 10.1093/eurheartj/ehs042. [DOI] [PubMed] [Google Scholar]
- 113.Wu H, Qian J, Xu J, Sun A, Sun W, Wang Q, et al. Besides CYP2C19, PON1 genetic variant influences post-clopidogrel platelet reactivity in Chinese patients. Int J Cardiol. 2012 doi: 10.1016/j.ijcard.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 114.Pare G, Ross S, Mehta SR, Yusuf S, Anand SS, Connolly SJ, et al. Effect of PON1 Q192R genetic polymorphism on clopidogrel efficacy and cardiovascular events in the Clopidogrel in the Unstable Angina to Prevent Recurrent Events trial and the Atrial Fibrillation Clopidogrel Trial with Irbesartan for Prevention of Vascular Events. Circ Cardiovas Genet. 2012;5(2):250–6. doi: 10.1161/CIRCGENETICS.111.961417. [DOI] [PubMed] [Google Scholar]
- 115.Mackness B, Mackness MI, Arrol S, Turkie W, Durrington PN. Effect of the human serum paraoxonase 55 and 192 genetic polymorphisms on the protection by high density lipoprotein against low density lipoprotein oxidative modification. FEBS Lett. 1998;423(1):57–60. doi: 10.1016/s0014-5793(98)00064-7. [DOI] [PubMed] [Google Scholar]
- 116.Lewis JP, Shuldiner AR. Paraoxonase 1 Q192R variant and clopidogrel efficacy: fact or fiction? Circ Cardiovas Genet. 2012;5(2):153–5. doi: 10.1161/CIRCGENETICS.112.962910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sibbing D, Koch W, Massberg S, Byrne RA, Mehilli J, Schulz S, et al. No association of paraoxonase-1 Q192R genotypes with platelet response to clopidogrel and risk of stent thrombosis after coronary stenting. Eur Heart J. 2011;32(13):1605–13. doi: 10.1093/eurheartj/ehr155. [DOI] [PubMed] [Google Scholar]
- 118.Szymezak J, Moreau C, Loriot MA, Durand E, Van Viet H, Desnos M, et al. High on-clopidogrel platelet reactivity: genotyping can help to optimize antiplatelet treatment. Throm Res. 2011;128(1):92–5. doi: 10.1016/j.thromres.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 119.Bura A, Bachelot-Loza C, Ali FD, Aiach M, Gaussem P. Role of the P2Y12 gene polymorphism in platelet responsiveness to clopidogrel in healthy subjects. Journal of Throm Haemostasis : JTH. 2006;4(9):2096–7. doi: 10.1111/j.1538-7836.2006.02113.x. [DOI] [PubMed] [Google Scholar]
- 120.Staritz P, Kurz K, Stoll M, Giannitsis E, Katus HA, Ivandic BT. Platelet reactivity and clopidogrel resistance are associated with the H2 haplotype of the P2Y12-ADP receptor gene. Int J Cardiol. 2009;133(3):341–5. doi: 10.1016/j.ijcard.2007.12.118. [DOI] [PubMed] [Google Scholar]
- 121.Namazi S, Kojuri J, Khalili A, Azarpira N. The impact of genetic polymorphisms of P2Y12, CYP3A5 and CYP2C19 on clopidogrel response variability in Iranian patients. Biochem Pharmacol. 2012;83(7):903–8. doi: 10.1016/j.bcp.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 122.Simon T, Bhatt DL, Bergougnan L, Farenc C, Pearson K, Perrin L, et al. Genetic polymorphisms and the impact of a higher clopidogrel dose regimen on active metabolite exposure and antiplatelet response in healthy subjects. Clin Pharmacol Ther. 2011;90(2):287–95. doi: 10.1038/clpt.2011.127. [DOI] [PubMed] [Google Scholar]
- 123.Zhu HJ, Patrick KS, Yuan HJ, Wang JS, Donovan JL, DeVane CL, et al. Two CES1 gene mutations lead to dysfunctional carboxylesterase 1 activity in man: clinical significance and molecular basis. Am J Hum Genet. 2008;82(6):1241–8. doi: 10.1016/j.ajhg.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lewis JP, Horenstein RB, Ryan K, O’Connell JR, Gibson Q, Mitchell BD, et al. The functional G143E variant of carboxylesterase 1 is associated with increased clopidogrel active metabolite levels and greater clopidogrel response. Pharmacogenet Genom. 2012 doi: 10.1097/FPC.0b013e32835aa8a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Abell LM, Liu EC. Dissecting the activation of thienopyridines by cytochromes P450 using a pharmacodynamic assay in vitro. J Pharmacol Exp Therap. 2011;339(2):589–96. doi: 10.1124/jpet.111.184895. [DOI] [PubMed] [Google Scholar]
- 126.Kazui M, Nishiya Y, Ishizuka T, Hagihara K, Farid NA, Okazaki O, et al. Identification of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metab Dispos. 2010;38(1):92–9. doi: 10.1124/dmd.109.029132. [DOI] [PubMed] [Google Scholar]
- 127.Sangkuhl K, Klein TE, Altman RB. Clopidogrel pathway. Pharmacogenet Genom. 2010;20(7):463–5. doi: 10.1097/FPC.0b013e3283385420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gremmel T, Kopp CW, Seidinger D, Koppensteiner R, Panzer S, Sunder-Plassmann R, et al. Differential impact of cytochrome 2C9 allelic variants on clopidogrel-mediated platelet inhibition determined by five different platelet function tests. Int J Cardiol. 2011 doi: 10.1016/j.ijcard.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 129.Harmsze A, van Werkum JW, Bouman HJ, Ruven HJ, Breet NJ, Ten Berg JM, et al. Besides CYP2C19*2, the variant allele CYP2C9*3 is associated with higher on-clopidogrel platelet reactivity in patients on dual antiplatelet therapy undergoing elective coronary stent implantation. Pharmacogenet Genom. 2010;20(1):18–25. doi: 10.1097/FPC.0b013e328333dafe. [DOI] [PubMed] [Google Scholar]
- 130.Kassimis G, Davlouros P, Xanthopoulou I, Stavrou EF, Athanassiadou A, Alexopoulos D. CYP2C19*2 and other genetic variants affecting platelet response to clopidogrel in patients undergoing percutaneous coronary intervention. Throm Res. 2012;129(4):441–6. doi: 10.1016/j.thromres.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 131.Zevin S, Benowitz NL. Drug interactions with tobacco smoking. An update. Clin Pharmacokinet. 1999;36(6):425–38. doi: 10.2165/00003088-199936060-00004. [DOI] [PubMed] [Google Scholar]
- 132.Zhou SF, Yang LP, Zhou ZW, Liu YH, Chan E. Insights into the substrate specificity, inhibitors, regulation, and polymorphisms and the clinical impact of human cytochrome P450 1A2. AAPS J. 2009;11(3):481–94. doi: 10.1208/s12248-009-9127-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Park KW, Kang J, Park JJ, Yang HM, Lee HY, Kang HJ, et al. Amlodipine, clopidogrel and CYP3A5 genetic variability: effects on platelet reactivity and clinical outcomes after percutaneous coronary intervention. Heart. 2012;98(18):1366–72. doi: 10.1136/heartjnl-2012-301892. [DOI] [PubMed] [Google Scholar]
- 134.Momary KM, Dorsch MP, Bates ER. Genetic causes of clopidogrel nonresponsiveness: which ones really count? Pharmacotherapy. 2010;30(3):265–74. doi: 10.1592/phco.30.3.265. [DOI] [PubMed] [Google Scholar]
- 135.ICPC - International Clopidogrel Pharmacogenomics Consortium. 2012 http://www.pharmgkb.org/page/icpc.
- 136.Gordon AS, Smith JD, Xiang Q, Metzker ML, Gibbs RA, Mardis ER, Nickerson DA, Fulton RS, Scherer SE. PGRNseq: a new sequencing-based platform for high-throughput pharmacogenomic implementation and discovery; (Program #244). Presented at the 62nd Annual Meeting of The American Society of Human Genetics; November 8, 2012; San Francisco, CA. [Google Scholar]
- 137.Fisch AS, L J, Lewis JP, Yerges-Armstrong LM, O’Connell JR, Mitchell BD, Horenstein RB, Ambulos N, Ryan K, Gibson Q, Shelton J, Shuldiner AR. Novel association of dual anti-platelet drug response with a functional variant in PPARG. Presented at the 62nd Annual Meeting of The American Society of Human Genetics; November 8, 2012; San Francisco, CA. [Google Scholar]


