Abstract
Purpose
In MS-based studies to discover urinary protein biomarkers, an important question is how to analyze the data to find the most promising potential biomarkers to be advanced to large-scale validation studies. Here, we describe a ‘systems biology-based’ approach to address this question.
Experimental design
We analyzed large-scale LC-MS/MS data of urinary exosomes from renal allograft recipients with biopsy-proven evidence of immunological rejection or tubular injury. We asked whether bioinformatic analysis of urinary exosomal proteins can identify protein groups that correlate with biopsy findings and whether the protein groups fit with general knowledge of the pathophysiological mechanisms involved.
Results
LC-MS/MS analysis of urinary exosomal proteomes identified more than 1000 proteins in each pathologic group. These protein lists were analyzed computationally to identify Biological Process and KEGG Pathway terms that are significantly associated with each pathological group. Among the most informative terms for each group were: “sodium ion transport” for tubular injury; “immune response” for all rejection; “epithelial cell differentiation” for cell-mediated rejection; and “acute inflammatory response” for antibody-mediated rejection. Based on these terms, candidate biomarkers were identified using a novel strategy to allow a dichotomous classification between different pathologic categories.
Conclusions and clinical relevance
The terms and candidate biomarkers identified make rational connections to pathophysiological mechanisms, suggesting that the described bioinformatic approach will be useful in advancing large-scale biomarker identification studies toward a validation phase.
Keywords: exosome, proteomics, rejection, transplant, urine
Introduction
The main objective of the present study is to develop a bioinformatic strategy that can reveal exploitable patterns in protein mass spectrometry data from urinary samples. The question being addressed is how to proceed from mass spectrometry-generated lists of proteins to a choice of candidate biomarkers for future testing in validation clinical trials. Here we describe and illustrate an approach derived from the field of systems biology that uses computational tools to discover biologically meaningful differences among experimentally derived protein lists [1, 2]. This approach is based on the idea that the objective of biomarker discovery studies should be to identify biological processes that are deranged rather than to look for specific proteins that may stand out. The rationale has been presented previously [3]. Briefly, it is based on the idea that protein biomarkers that “make sense” from the perspective of pathophysiology are more likely to succeed in the clinical setting than randomly discovered proteins that have uncertain connection to the relevant disease mechanisms (see Discussion). In this study, as a ‘proof of principle’, we apply such an approach to proteins detected by LC-MS/MS (liquid chromatography-tandem mass spectrometry) analysis of urinary exosome samples from three groups of renal transplant patients with different pathologic findings in the allograft biopsy: tubular injury, cell-mediated rejection, and antibody-mediated rejection. The bioinformatic approach outlined identified biological processes and candidate biomarkers associated with the different patient groups that fit with known pathophysiological mechanisms.
Materials and methods
Patients and samples
Urine samples were collected prospectively from adult renal transplant recipients followed at the Johns Hopkins Comprehensive Transplant Center. These patients underwent a kidney graft biopsy, either for cause (due to rise in baseline serum creatinine) or as protocol biopsy in patients with stable renal function. All patients received standard immunosuppressive treatment with prednisone, mycophenolate, and tacrolimus. Exclusion factors included combined transplantation of kidney and other organs, evidence of infection in the kidney (bacterial or viral such as BK/polyoma, CMV, or Herpes), primary glomerular disease (de novo or recurrent), post-transplant lymphoproliferative disorder (PTLD). Kidney graft biopsies were processed for routine studies as previously described including staining with hematoxilin-eosin (H&E), periodic acid-Schiff (PAS) methenamine silver, and Masson’s trichrome for light microscopy examination [4]. C4d staining by immunofluorescence was routinely performed on frozen sections of transplant kidney biopsies (alternatively C4d immunoperoxidase-based stain was also available on sections from paraffin embedded tissue). Biopsies were graded for cellular- and antibody-mediated rejection according to the Banff score [5, 6]. Based on the graft biopsy findings, patients were assigned to four groups: Non-Specific Findings (N), mild to moderate Tubular Injury (TI), Cell-Mediated Rejection (CMR), or Antibody-Mediated Rejection (AMR). The study protocol was approved by the Johns Hopkins Medical Institutions Review Board.
Freshly voided urine (20-200 ml) was collected in the morning, before the biopsy (but was not the first morning urine sample), and was processed immediately to isolate urinary exosomes using the differential centrifugation procedure described by Gonzales et al [7]. Processed protein samples solubilized in Laemmli reagent were pooled for each pathological group for analysis by LC-MS/MS.
Sample processing and mass spectrometry analysis
Triplicate sets of 200 μg of urinary exosomal proteins pooled from each pathological group (7 TI, 6 CMR, 3 AMR, and 2 N, supporting Information Table S1) were separated by one dimensional SDS/PAGE electrophoresis using 10% polyacrylamide gels. After staining with Coomassie blue, the gels were de-stained, cut in multiple slices, dehydrated, reduced, alkylated, and subjected to trypsin digestion at 37°C overnight to obtain peptides, which were reconstituted in 0.1% formic acid for analysis as described [8]. LC-MS/MS was performed on triplicate aliquots of digested protein samples using LTQ Orbitrap XL (two runs) and LTQ Orbitrap Velos (one run) (Thermo Scientific, Waltham, MA). Reversed-phase C18 chromatographic separation of trapped peptides was carried out on a prepacked Beta Basic C18 PicoFrit column (75 μm i.d. × 10 cm length; New Objective) at 300 nL/min using the following gradient: 2–5% solvent B for 2 min; 5–45% solvent B for 45 min; 45–50% solvent B for 5 min; 50–95% solvent B for 5 min (solvent A: 0.1% formic acid in 98% water, 2% acetonitrile; solvent B: 0.1% formic acid in 100% acetonitrile). To identify peptide sequences, the MS data was searched against National Center for Biotechnology Information (NCBI) Reference Sequence human protein database (April 8, 2010, 38767 entries) which included a list of common contaminating proteins and scored using SEQUEST algorithm on Proteome Discoverer software (ver. 1.1, Thermo Scientific). Precursor ion tolerance was 50 ppm, while fragment ion tolerance was 1.0 Da. Two missed trypsin cleavage sites were allowed. Static modifications included carbamidomethylation of cysteine (+57.021 Da) and variable modifications included oxidation of methionine (+15.995 Da). The peptide false discovery rate was limited to < 2% for individual peptides using the target-decoy approach [9]. All MS spectral files associated with this manuscript may be downloaded from ProteomeCommons.org Tranche using the following hashes:
-
LTQ Orbitrap XL Data 1st set
p7DyXnHc+EyANszEZXPsThX/EzRPhJR0YmfDYGZJoRREeKgRm00/jM47tDEH5Gh0wzO/Bk N5BcA0IQPCCN2QmJAWs4UAAAAAAAAfow==
-
LTQ Orbitrap XL Data 2nd set
RD+8p8/vHKT7QhWVYe0hMsVKKEZitqPGBgv60DDvoGuOQ1ZaGz6Bpgmy+o/2FSOe+Y4v 1BfFtzlZcKEOJ43T7HwEVkUAAAAAAAAiHg==
-
LTQ Orbitrap Velos Data
DlI2b2OfY46ui/Fln5JivUyHW03ZxU4Pv5xapK4xl9FB4wdSyW1Ffo4gQ38tjNwZo4SehiBbar RU0k8QiwrOiS5phrYAAAAAAAACyg==
For the evaluation of relative abundance of exosomal proteins among groups, the spectral counts were normalized by dividing by the number of theoretical tryptic peptides detectable by LC-MS/MS (criteria for predicting a detectable tryptic peptide are as followed: theoretical m/z = 300-2000 for charge states +1 to +3, peptide length = 6-35, possible missed cleavage sites < 3). These values were further normalized by dividing by the median value for each group to account for potential differences in sample size.
Bioinformatic analysis
Identified proteins were classified as ‘extrinsic to exosomes’ if they possessed the Gene Ontology Cellular Component term of “extracellular space”, but lacked the terms “anchor” and “cytosol”. The remaining proteins were placed in the ‘intrinsic to exosomes’ subgroup. Protein lists were analyzed using the DAVID bioinformatic tool (Database for Annotation, Visualization and Integrated Discovery, NIAID, Bethesda, MD http://david.abcc.niifcrf.gov/) [10] to determine what Gene Ontology Biological Process terms are over-represented relative to the concatenated lists from all four conditions. The DAVID tool was also applied to determine what KEGG pathways (Kyoto Encyclopedia of Genes and Genomes) [11] are over-represented in each pathological group.
Results
Proteomes were profiled by LC-MS/MS in urinary exosome samples from renal allograft recipients with acute decrease in renal function and distinct biopsy findings: tubular injury (TI), cell-mediated rejection (CMR), and antibody-mediated rejection (AMR) (supporting Information Table S1). We also examined urinary exosomes of two allograft recipients with stable renal function undergoing “protocol” kidney graft biopsy, with non-specific findings (N). We selected cases that were stereotypical of each category, recognizing that real-world cases often display less distinct histology. Exosome samples were pooled within each histology class (supporting Information Table S1) and analyzed by LC-MS/MS. 1989 urine exosomal proteins were identified in renal transplant patients (false discovery rate < 2%, target-decoy analysis). This total exceeds the 1160 proteins reported in the Urinary Exosome Protein Database (http://dir.nhlbi.nih.gov/papers/lkem/exosome/). A list of all identified proteins along with associated median-normalized spectral counts, a measure of relative abundance (Materials and methods), can be accessed online through http://helixweb.nih.gov/ESBL/Database/EXORT/ (username: clp, password: Esbl!@#$, login to be removed upon acceptance of this paper). Each group had a large subset of uniquely identified proteins, TI: 353 proteins; CMR: 322 proteins; and AMR 165 proteins, with 1073 proteins present in more than one group.
We classified the identified proteins into one of two subgroups (Materials and methods): ‘intrinsic to exosomes’ or ‘extrinsic to exosomes’ (see the online database). The ‘extrinsic’ proteins are presumably plasma proteins that have crossed the glomerular filter and are nonspecifically associated with exosomes. The ‘intrinsic’ proteins were used for bioinformatic analysis. Overall, 92% of the proteins identified were classified as intrinsic, suggesting that they were a component of exosomes (or similar membranous structures) released from kidney cells.
Interestingly, there seems to be a higher proportion of extrinsic proteins in the transplanted control group than in the non-transplanted group (chi-square, p < 0.0001) when comparing the most abundant proteins (based on spectral counts) found in the group of transplant recipients with non-specific findings versus the most abundant proteins found in a group of healthy, non-transplanted volunteers that we studied previously [7], (supporting Information Table S2). Although these results need to be confirmed on a larger number of observations in transplanted patients without kidney pathology, they suggest that transplanted kidneys excrete higher levels of plasma proteins than non-transplanted kidneys even with no obvious pathological changes suggestive of tubular injury or rejection, and are also consistent with previously noted differences in the proteomic profile of urine from patients with transplanted or native kidneys [12, 13].
Identification of biological processes over-represented in groups
We analyzed the intrinsic exosomal proteins in each group to determine what characteristics distinguish the groups based on the biological processes that they take part in. This analysis was conducted with an online bioinformatic analysis tool called “DAVID” (Materials and methods), using the Gene Ontology (GO) Database, which attaches hierarchical descriptors to all annotated proteins coded by the human genome. We applied the Biological Process GO terms to identify which Biological Process terms are represented statistically more frequently in a list of exosomal proteins from a given patient group than from the entire set of exosomal proteins.
We asked whether any GO Biological Process terms are over-represented in the urinary exosomal proteins from kidneys of each patient group with abnormal biopsy findings (compared with the list of all proteins identified in transplanted kidneys). Table 1 shows the GO Biological Processes identified in each group with p < 0.05 (Fisher Exact). A comparison of Table 1A, 1B, and 1C reveals that several terms were enriched in more than one group. For example the term “immune response” is found in both graft rejection groups. Thus, proteins populating the “immune response” list can be deemed potential discriminators between rejection and tubular injury, but not between the two forms of rejection. supporting Information Table S3 shows proteins in this “immune response” list (cell-mediated and antibody-mediated rejection combined). Each pathological group in Table 1 is associated with at least one unique term (represented in Bold). As discussed below, most of these appear to be related to the underlying pathophysiology.
Table 1.
Gene Ontology Biological Process terms over-represented in lists of exosomal proteins associated with different biopsy findings. Categories unique to a given group are shown in Bold.
|
No.
Proteins |
p-value |
FDR
(%) |
|
|---|---|---|---|
| A. Tubular Injury | |||
| GO:0007264 Small GTPase mediated signal transduction | 64 | 0.0036 | 6.2 |
| G0:0006814 Sodium ion transport | 26 | 0.0050 | 8.6 |
| GO:0010033 Response to organic substance | 73 | 0.0073 | 12.4 |
| B. Cell-Mediated Rejection | |||
| GO:0030029 Actin filament-based process | 47 | 0.0005 | 0.8 |
| GO:0030855 Epithelial cell differentiation | 21 | 0.0018 | 3.2 |
| GO:0007264 Small GTPase mediated signal transduction | 62 | 0.0021 | 3.7 |
| GO:0030036 Actin cytoskeleton organization | 41 | 0.0026 | 4.5 |
| GO:0007010 Cytoskeleton organization | 63 | 0.0031 | 5.5 |
| GO:0006955 Immune response | 49 | 0.0118 | 19.1 |
| GO:0043603 Cellular amide metabolic process | 16 | 0.0118 | 19.2 |
| C. Antibody-Mediated Rejection | |||
| GO:0007264 Small GTPase mediated signal transduction | 64 | <0.0000 | 0.0 |
| GO:0015031 Protein transport | 97 | <0.0000 | 0.1 |
| GO:0045184 Establishment of protein localization | 97 | 0.0001 | 0.1 |
| GO:0008104 Protein localization | 103 | 0.0003 | 0.5 |
| GO:0010033 Response to organic substance | 64 | 0.0018 | 3.1 |
| GO:0007242 Intracellular signaling cascade | 105 | 0.0034 | 6.0 |
| GO:0044057 Regulation of system process | 26 | 0.0039 | 6.7 |
| GO:0006986 Response to unfolded protein | 13 | 0.0063 | 10.6 |
| GO:0002526 Acute inflammatory response | 18 | 0.0066 | 11.1 |
| GO:0010035 Response to inorganic substance | 28 | 0.0083 | 13.7 |
|
GO:0051605 Protein maturation by peptide bond
cleavage |
15 | 0.0086 | 14.2 |
| GO:0006955 Immune response | 44 | 0.0094 | 15.5 |
For the Tubular Injury group, the Gene Ontology Biological Process term “sodium ion transport” was the only unique term (Table 1A). This protein list (Table 2) includes a large number of transporters, many of them are sodium-coupled solute transporters expressed in the apical brush border membrane of the proximal tubule. Based on the transcriptomic databases of proximal tubule, medullary thick ascending limb, and inner medullary collecting duct [14], [15], 25 out of all 29 proteins (86%) in Table 2 were expressed with relatively high signal values in proximal tubule. This finding is presumably indicative of proximal tubule damage and therefore makes sense from a pathophysiological perspective.
Table 2.
Protein list for Gene Ontology Biological Process “sodium ion transport” enriched in urinary exosomes from transplants with Tubular Injury compared with all transplants. Asterisks indicate proteins that were expressed with relatively high signal values in proximal tubule based on the transcriptomic databases of proximal tubule, medullary thick ascending limb, and inner medullary collecting duct [14], [15].
| Protein Name | GI Accession | Gene Symbol |
|---|---|---|
| sodium/potassium-transporting ATPase subunit alpha-1 isoform a * | 21361181 | ATP1A1 |
| sodium/potassium-transporting ATPase subunit beta-1 * | 4502277 | ATP1B1 |
| sodium/potassium-transporting ATPase subunit gamma isoform 2 * | 11125764 | FXYD2 |
| solute carrier family 12 member 3 isoform 1 | 186910315 | SLC12A3 |
| solute carrier family 12 member 3 isoform 3 | 186910319 | SLC12A3 |
| solute carrier family 12 member 1 isoform A | 134254459 | SLC12A1 |
| solute carrier family 12 member 2 * | 4506975 | SLC12A2 |
| solute carrier family 13 member 1 * | 19923838 | SLC13A1 |
| solute carrier family 13 member 2 isoform b * | 4506979 | SLC13A2 |
| solute carrier family 13 member 2 isoform c * | 225637552 | SLC13A2 |
| solute carrier family 13 member 3 isoform a * | 31377715 | SLC13A3 |
| solute carrier family 13 member 3 isoform b * | 58761541 | SLC13A3 |
| sodium-dependent phosphate transport protein 1 * | 154800443 | SLC17A1 |
| sodium-dependent phosphate transport protein 4 isoform a * | 148743791 | SLC17A3 |
| solute carrier family 22 member 5 * | 4507005 | SLC22A5 |
| solute carrier family 22 member 4 * | 24497490 | SLC22A4 |
| solute carrier family 23 member 1 isoform a * | 44680145 | SLC23A1 |
| sodium-dependent phosphate transport protein 2B isoform a | 110611906 | SLC34A2 |
| sodium-coupled neutral amino acid transporter 2 * | 21361602 | SLC38A2 |
| electrogenic sodium bicarbonate cotransporter 1 isoform 2 * | 4507025 | SLC4A4 |
| sodium-coupled monocarboxylate transporter 1 * | 167466278 | SLC5A8 |
| sodium/glucose cotransporter 1 * | 4507031 | SLC5A1 |
| sodium/glucose cotransporter 5 isoform 2 * | 109659836 | SLC5A10 |
| sodium/myo-inositol cotransporter 2 * | 17941285 | SLC5A11 |
| sodium-coupled monocarboxylate transporter 2 * | 157671931 | SLC5A12 |
| sodium/glucose cotransporter 2 * | 4507033 | SLC5A2 |
| sodium/glucose cotransporter 4 isoform 2 * | 206597483 | SLC5A9 |
| sodium- and chloride-dependent creatine transporter 1 isoform 3 * | 218563758 | SLC6A8 |
| sodium/hydrogen exchanger 3 * | 194239733 | SLC9A3 |
In the Cell-Mediated Rejection group (Table 1B), there are several unique terms, including three similar terms: “actin filament based process”, “actin cytoskeleton organization”, and “cytoskeleton organization”. The proteins in the former group are shown in supporting Information Table S4. An additional unique term in the Cell-Mediated Rejection group is “epithelial cell differentiation” (Table 3). These are predominantly proteins involved in intermediate filament organization and include four different uroplakin isoforms. The latter are expressed in the renal ureter and pelvis, and point to the possibility that the transitional epithelium of the urinary drainage system is a target tissue in cell-mediated allograft rejection.
Table 3.
Protein list for Gene Ontology Biological Process “epithelial cell differentiation” enriched in urinary exosomes from transplants with Cell-Mediated Rejection compared with all transplants.
| Protein Name | GI Accession | Gene Symbol |
|---|---|---|
| ammonium transporter Rh type C | 7706683 | RHCG |
| annexin A1 | 4502101 | ANXA1 |
| cornifelin | 26024195 | CNFN |
| desmoplakin isoform I | 58530840 | DSP |
| envoplakin | 156104874 | EVPL |
| ezrin | 161702986 | EZR |
| filaggrin | 60097902 | FLG |
| glutathione S-transferase Mu 3 | 23065552 | GSTM3 |
| hornerin | 57864582 | HRNR |
| periplakin | 45439327 | PPL |
| protein-glutamine gamma-glutamyltransferase K | 4507475 | TGM1 |
| tumor susceptibility gene 101 protein | 5454140 | TSG101 |
| uroplakin-1a | 5902148 | UPK1A |
| uroplakin-1b | 49619237 | UPK1B |
| uroplakin-2 precursor | 5803215 | UPK2 |
| uroplakin-3a isoform 1 | 5902152 | UPK3A |
In the Antibody-Mediated Rejection group (Table 1C), several unique terms appear, including three similar terms relevant to protein trafficking. The proteins in the “protein localization” group are listed in supporting Information Table S5. Additional unique terms in the Antibody-Mediated Rejection group are “response to unfolded protein“ (supporting Information Table S6) and “acute inflammatory response” (Table 4). The latter category is dominated by components of the complement pathway. These categories refer to aspects of pathophysiology that appear logical in the context of current knowledge about antibody-mediated rejection (see Discussion).
Table 4.
Protein list for Gene Ontology Biological Process “acute inflammatory response” enriched in urinary exosomes from transplanted kidneys with Antibody-Mediated Rejection compared with all transplants.
| Protein Name | GI Accession | Gene Symbol |
|---|---|---|
| 14-3-3 protein zeta/delta | 208973244 | YWHAZ |
| alpha-1-antichymotrypsin precursor | 50659080 | SERPINA3 |
| C4b-binding protein alpha chain precursor | 4502503 | C4BPA |
| complement C1q subcomponent subunit B precursor | 87298828 | C1QB |
| complement C1q subcomponent subunit C precursor | 166235903 | C1QC |
| complement component C7 precursor | 45580688 | C7 |
| complement component C8 gamma chain precursor | 166197660 | C8G |
| complement component C9 precursor | 4502511 | C9 |
| complement factor B preproprotein | 67782358 | CFB |
| complement receptor type 1 isoform F precursor | 86793036 | CR1 |
| complement receptor type 1 isoform S precursor | 86793109 | CR1 |
| inter-alpha-trypsin inhibitor heavy chain H4 isoform 1 precursor | 31542984 | ITIH4 |
| inter-alpha-trypsin inhibitor heavy chain H4 isoform 2 precursor | 262050538 | ITIH4 |
| mannan-binding lectin serine protease 2 isoform 1 precursor | 21264363 | MASP2 |
| mannan-binding lectin serine protease 2 isoform 2 precursor | 21264361 | MASP2 |
| peroxiredoxin-2 isoform a | 32189392 | PRDX2 |
| plasma protease C1 inhibitor precursor | 73858570 | SERPING1 |
| PREDICTED: complement C3-like, partial | 169218213 | LOC100133511 |
| serotransferrin precursor | 4557871 | TF |
| tuberin isoform 4 | 116256350 | TSC2 |
Identification of KEGG biological pathways over-represented in pathological groups
We next analyzed the data to determine what characteristics distinguish the groups based on the KEGG biological pathways that they take part in, asking the question: “What KEGG pathways are represented statistically more frequently in a list of urinary exosomal proteins from a given patient group than in the list of all identified intrinsic exosomal proteins in all groups?”
The findings are summarized in Table 5. There were no unique KEGG pathways for exosomal proteins in the Tubular Injury group. One pathway was over-represented in the two Rejection groups, namely “endocytosis” (supporting Information Table S7 shows the “endocytosis” proteins combined from the two Rejection groups). Two KEGG pathways were uniquely over-represented in the list of exosomal proteins found in the Cell-Mediated Rejection group, viz. “tight junction” (supporting Information Table S8) and “glutathione metabolism” (supporting Information Table S9). Interestingly the “tight junction” KEGG pathway with the Cell-Mediated Rejection group proteins include three proteins that are critically involved in epithelial polarization through PDZ interactions, viz. Crumbs3 (CRB3), PALS1 (MPP5), and PATJ (INADL) [16], and may be important in the recovery of tubular integrity damaged by “Tubulitis”, a signature pathologic lesion in this type of rejection. Also as seen in Table 5, four KEGG pathways were uniquely over-represented in the Antibody-Mediated Rejection group, viz. “antigen processing and presentation” (Table 6), “neurotrophin signaling pathway”, “pathways in cancer” and “pathogenic E. coli infection” (supporting Information Tables S10-S12). The “antigen processing and presentation” list is dominated by major histocompatibility complex proteins and heat shock proteins.
Table 5.
KEGG pathways over-represented in lists of urinary exosomal proteins associated with different biopsy findings. Pathways unique to a histological category are indicated in Bold.
| No. Proteins | p-value | FDR (%) | |
|---|---|---|---|
| A. Tubular Injury | |||
| none | |||
| B. Cell-Mediated Rejection | |||
| hsa04530:Tight junction | 28 | 0.01 | 16.62 |
| hsa04144:Endocytosis | 40 | 0.02 | 19.59 |
| hsa00480:Glutathione metabolism | 16 | 0.06 | 53.13 |
| C. Antibody-Mediated Rejection | |||
| hsa04612:Antigen processing and presentation | 15 | <0.01 | 4.20 |
| hsa04722:Neurotrophin signaling pathway | 22 | 0.01 | 6.73 |
| hsa05200:Pathways in cancer | 25 | 0.01 | 11.71 |
| hsa05130:Pathogenic Escherichia coli infection | 20 | 0.01 | 14.04 |
| hsa04144:Endocytosis | 36 | 0.01 | 16.30 |
Table 6.
List of proteins identified in urinary exosomes of patients in Antibody-Mediated Rejection group that are present in KEGG pathway “antigen processing and presentation”.
| Protein Name | GI Accession | Gene Symbol |
|---|---|---|
| calreticulin precursor | 4757900 | CALR |
| cathepsin S isoform 1 preproprotein | 23110962 | CTSS |
| HLA class I histocompatibility antigen, A-1 alpha chain precursor | 24797067 | HLA-A |
| major histocompatibility complex, class I, B precursor | 17986001 | HLA-B |
| major histocompatibility complex, class I, B precursor | 17986001 | HLA-B |
| HLA class II histocompatibility antigen, DM alpha chain precursor | 18765715 | HLA-DMA |
| HLA class II histocompatibility antigen, DM beta chain precursor | 4504399 | HLA-DMB |
| HLA class II histocompatibility antigen, DR alpha chain precursor | 52426774 | HLA-DRA |
| heat shock protein HSP 90-alpha isoform 2 | 154146191 | HSP90AA1 |
| heat shock protein HSP 90-alpha isoform 2 | 154146191 | HSP90AA1 |
| heat shock protein HSP 90-beta | 20149594 | HSP90AB1 |
| heat shock 70 kDa protein 1A/1B | 194248072 | HSPA1A |
| heat shock 70 kDa protein 1A/1B | 194248072 | HSPA1A |
| heat shock 70 kDa protein 1-like | 124256496 | HSPA1L |
| heat shock-related 70 kDa protein 2 | 13676857 | HSPA2 |
| heat shock 70 kDa protein 6 | 34419635 | HSPA6 |
| heat shock 70 kDa protein 6 | 34419635 | HSPA6 |
| heat shock cognate 71 kDa protein isoform 2 | 24234686 | HSPA8 |
| proteasome activator complex subunit 1 isoform 1 | 5453990 | PSME1 |
A strategy for identification of candidate biomarker proteins from Biological Processprotein lists
The approach described above selects groups of MS-identified proteins associated via Gene Ontology Biological Process terms or KEGG Pathways that relate to pathophysiological mechanisms in different patient groups. In full-scale biomarker studies, the next task would be to select candidate biomarkers from the protein groups that are predicted from the mass spectrometry data to have the potential to discriminate patient groups. In this study, we developed a strategy to identify candidate biomarkers from our ‘proof-of-principle’ data using the protein lists associated with the over-represented Biological Processes for each pathologic category (Tables 2-4 and supporting Information Tables S3-6). In this context, a first practical goal would be to identify pairs of proteins that will allow a dichotomous classification into Rejection vs. Tubular Injury categories (Figure 1). A second goal would be to choose protein pairs that allow samples to be categorized as Antibody-Mediated Rejection vs. Cell-Mediated Rejection (Figure 2). Selecting pairs of proteins that may have sufficient discriminating power between two different conditions, based on ratios of their abundance, would avoid the need of normalization factors, a main hurdle in optimizing assay conditions for urine biomarkers.
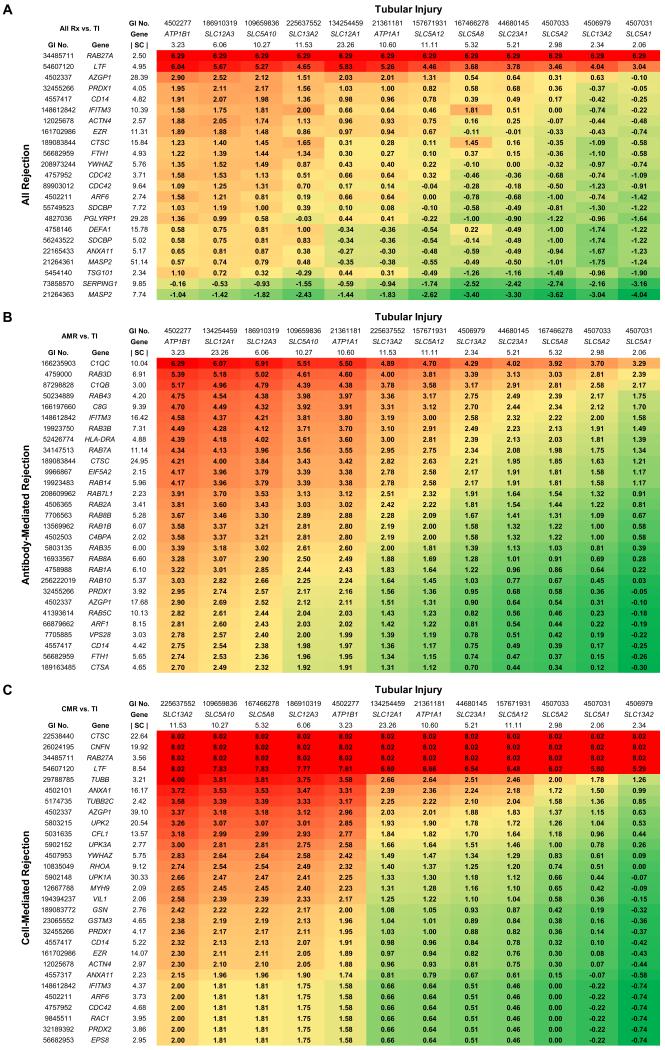
Figure 1. Heat map representation of discrimination factors for candidate protein biomarker pairs in discrimination of (A) All Rejection versus Tubular Injury; (B) Antibody-Mediated Rejection versus Tubular Injury; and (C) Cell-Mediated Rejection versus Tubular Injury.
Proteins for All Rejection were selected from the “immune response” list (supporting Information Table S3) and common proteins enriched in both Antibody-Mediated Rejection and Cell-Mediated Rejection. Proteins for Antibody-Mediated Rejection were selected from the “acute inflammatory response” list (Table 4), the “protein localization” list (supporting Information Table S5), and the “response to unfolded proteinȁ list (supporting Information Table S6). Proteins for Cell-Mediated Rejection were selected from the “epithelial cell differentiation” list (Table 3) and the “actin filament based process” list (supporting Information Table S4). Proteins for Tubular Injury were selected from the “sodium ion transport” list (Table 2). Discrimination factors (colored boxes) are log2(RX/RTI), where RX is the median-normalized spectral count ratio of the two proteins for indicated Rejection group and RTI is the ratio of the two proteins in Tubular Injury. Colors stratify discrimination factors from high (red) to low (green). The spectral counts shown for specific proteins represent the median-normalized spectral count values (|SC|) obtained in tubular injury (across top) or in appropriate rejection state (left); values for ‘all rejection’ represent average of cell-mediated and antibody-mediated values. Proteins are listed by both the GI accession number and the official gene symbol. Full annotation of the candidate biomarkers listed can be obtained using the GI accession number at the Entrez Protein online site (http://www.ncbi.nlm.nih.gov/protein).
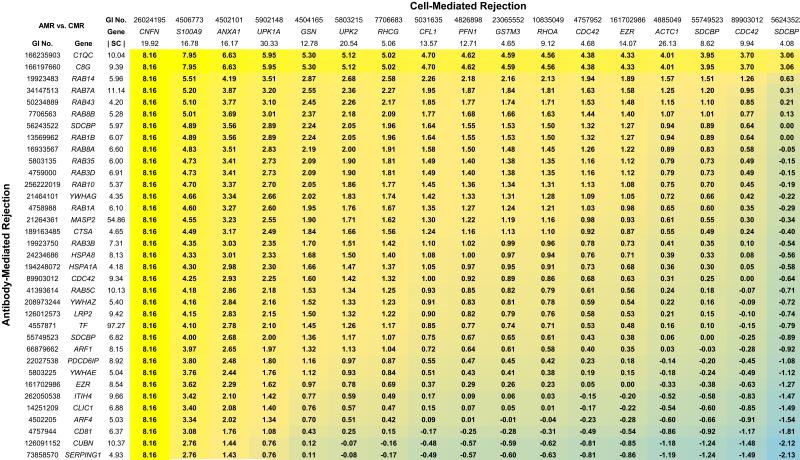
Figure 2. Heat map representation of discrimination factors for candidate protein biomarker pairs in discrimination of Antibody-Mediated Rejection versus Cell-Mediated Rejection.
Proteins for Antibody-Mediated Rejection were selected from the “acute inflammatory response” list (Table 4), the “protein localization” list (supporting Information Table S5), and the “response to unfolded protein” list (supporting Information Table S6). Proteins for Cell-Mediated Rejection were selected from the “epithelial cell differentiation” list (Table 3) and the “actin filament based process” list (supporting Information Table S4). Discrimination factors (colored boxes) are log2(RAMR/RCMR), where RAMR is the median-normalized spectral count ratio of the two proteins for the Antibody-Mediated Rejection group and RCMR is the ratio of the two proteins in the Cell-Mediated Rejection group. Colors stratify discrimination factors from high (yellow) to low (blue). The spectral counts shown for specific proteins represent the median-normalized spectral count values (|SC|) obtained in the appropriate rejection states. Proteins are listed by both the GI accession number and the official gene symbol. Full annotation of the candidate biomarkers listed can be obtained using the GI accession number at the Entrez Protein online site (http://www.ncbi.nlm.nih.gov/protein).
As rationalized in the Discussion, criteria for selection of candidate proteins from the Gene Ontology Biological Process lists obtained in the present study were: 1) maximization of the spectral count ratio between the two states being discriminated and 2) high absolute abundance based on spectral counts. Figure 1A shows a heat map representation of the dichotomous classification between All Rejection vs. Tubular Injury. The numbers for each pair of proteins are discrimination factors between the two conditions for a protein pair [Values are log2(RR/RTI), where RR is the median-normalized spectral count ratio of the two proteins for All Rejection and RTI is the ratio of the two proteins in Tubular Injury]. The best discrimination factors are the highest positive numbers. Figure 1B shows the same type of information for the comparison between Antibody-Mediated Rejection versus Tubular Injury and Figure 1C shows comparable information for Cell-Mediated Rejection versus Tubular Injury. Figure 2 shows the dichotomous classification between Antibody-Mediated Rejection and Cell-Mediated Rejection in which the median-normalized spectral count ratio between potential markers are represented in the same manner as in Figure 1. All proteins presented in Figures 1 and 2 were also selected from the Biological Process protein lists only if their absolute abundance levels in the pathologic category associated with a particular protein list were at least 2-fold above the median value for that pathologic category (median-normalized spectral count values > 2).
Discussion
Typically biomarker development studies involve at least three distinct stages: discovery, validation, and implementation [17]. Here, we describe a bioinformatic approach to the analysis of discovery data. We used state-of-the-art mass spectrometry techniques for proteomic analysis of urinary exosomes from renal transplant recipients with renal dysfunction and biopsy-proven tubular injury, cell-mediated rejection, or antibody-mediated rejection. Although this approach was developed in the context of urinary proteomics data, it theoretically could be applied to analysis of data from other body fluids.
A central question addressed in this study is: “how best to analyze large-scale mass spectrometry data from urinary biomarker discovery studies to find proteins that have a meaningful connection to the pathophysiology of the disease process under study?” Optimally, candidate protein biomarkers must be sufficiently abundant to allow measurement in whole urine and should ‘make sense’ in terms of the pathophysiological processes involved [3]. The latter requirement is based on three considerations: a) measurements of biomarkers consistent with current pathophysiological knowledge will more readily be integrated with other types of results in future clinical studies; b) such biomarkers will be more readily acceptable and useful to the clinical nephrology community than those with no known connection to pathophysiology; and c) biomarkers that fit with recognized mechanisms are less likely to be false positives.
Because of the considerations outlined in the foregoing Discussion, we have devised a ‘process-oriented’ approach to bioinformatic interpretation of the protein mass spectrometry discovery data. Thus, we are not looking for specific proteins that are enriched in individual patient groups, but rather we wish to identify classes of proteins (involved in specific biological processes) that can be shown statistically to be associated with individual patient groups. To implement this approach, we used the online computer utility, DAVID (see Materials and methods), to identify either Gene Ontology Biological Process (GO-BP) terms or KEGG Pathway terms that are selectively associated with lists of proteins identified in urinary exosomes in each renal transplant subgroup. Based on this approach, we can summarize the key observations in our ‘proof of principle’ samples as follows:
The only GO-BP term significantly associated with the Tubular Injury exosomal protein list was “sodium ion transport” (Table 1A). The list of proteins in this group (Table 2) consists chiefly of transporter proteins characteristic of the proximal tubule brush border but also includes proteins from the thick ascending limb of Henle (SLC12A1) and distal convoluted tubule (SLC12A3). These findings are consistent with the idea that tubular injury is seen mainly in proximal tubule, thick ascending limb of Henle, and distal convoluted tubule, where ATP demand is high because of the energy cost of transepithelial solute transport. This study did not single out any of the proteins previously proposed to be associated with ischemic tubule injury such as KIM-1 [18], Hsp72 [19], klotho [20], IL-6 [21], NGAL [22], L-FABP [23], netrin-1 [24], or fetuin-A [25], presumably because the pathophysiology is dependent on the context (transplanted kidney with immunosuppression) and because our bioinformatic analysis focused on functional classes of proteins.
Proteins classified with the GO-BP term “immune response” were enriched in the two rejection groups but not the Tubular Injury group (Table 1). This classification makes sense in the context of the immunological nature of rejection. The proteins in this group are dominated by proteins of the major histocompatibility complex and complement components (supporting Information Table S3).
The GO-BP term “epithelial cell differentiation” is uniquely associated with Cell-Mediated Rejection (Table 1B). Despite its name, this category is made up largely of proteins associated with stratified epithelia including the transitional epithelium of the pelvis and ureter (Table 3). For example, we identified four uroplakins, abundant proteins that form a large part of the surface area of the umbrella cells of transitional epithelia [26, 27]. This result is compatible with the hypothesis that cell-mediated rejection may affect multiple epithelial cells of the transplanted organ, including the pelvis and ureter that are not affected in the other two pathologies.
Several GO-BP terms are associated with proteins in the Antibody-Mediated Rejection group including “acute inflammatory response” (Table 1C). Several complement components were found in the “acute inflammatory response” category (Table 4), a finding that fits with the proposed roles for the complement pathway in the pathophysiology of antibody-mediated rejection, and with current efforts to devise therapies based on inhibition of terminal complement activation with targeted monoclonal antibodies [28].
In a full-blown discovery study, the ultimate objective would be the development and testing of immunoassays (or other assays) for selected biomarker proteins. A major problem in designing useful urinary assays is normalization. The need for normalization arises from the fact that water excretion is highly variable, depending on physiological factors, rendering measurement of the absolute concentrations of a given biomarker substance virtually useless for making practical comparisons. Much has been written about the use of various measures such as creatinine, osmolality, Tamm-Horsfall protein, etc., to normalize urinary measurements. Such normalizing factors add work and variability to the read-out. However, because many clinical problems (including the one described in this paper) involve the development of a dichotomous classification that distinguishes one state from another, the normalization problem may be obviated; it is theoretically sufficient to form ratios of measured biomarker abundances, thereby canceling out the normalization factor. In the example developed for this study, two dichotomies are seen: 1) between rejection and tubular injury and 2) between cell-mediated rejection and antibody-mediated rejection. Consequently, in such studies it is rational to identify pairs of biomarker proteins, which, when measured under arbitrary loading conditions, give a concentration ratio that correlates with disease state. If the assays are linear, the ratio should be independent of water excretion rate or sample size, and separate normalization should be unnecessary.
Given the above observations, we selected candidate biomarkers representative of the biological processes identified by bioinformatic analysis. Operationally, we identified pairs of candidate biomarker proteins that potentially could allow a dichotomous classification into Rejection vs. Tubular Injury (Figure 1) and, among those classified as Rejection, will allow samples to be categorized as Antibody-Mediated vs. Cell-Mediated (Figure 2). Among the proteins in Figures 1 and 2, SERPING1 [29] and S100A9 [30] have already been reported to be candidate biomarker proteins for acute rejection in renal allografts. Many membrane protein candidates were identified and have not been reported in the literatures associated with renal allograft rejection. This observation is likely to be due to the use of urinary exosomes in this study. Urinary exosomes are tiny (40–80 nm) membranous structures secreted by every renal tubule epithelial cell type, as well as podocytes and transitional epithelia from the urinary collecting system [31].
In summary, the object of this paper was to describe and illustrate an approach to bioinformatic analysis of biomarker discovery data in order to facilitate identification of candidate biomarkers for future testing in validation clinical trials. For this purpose, we prepared samples from a small number of actual renal transplant patients with different pathological changes in the allograft. The primary goal of this study was not to validate protein biomarkers for different processes that could cause an elevation of serum creatinine in renal transplant patients. Optimally, such validation studies would warrant the analysis of larger numbers of samples than used here. Nevertheless, data presented here, when combined with results from other biomarker studies [12] [13] [32] [33] [34] [35], could be useful in the design of validation studies for a limited number of protein biomarker candidates that make pathophysiological sense.
Supplementary Material
Statement of clinical relevance.
A rising serum creatinine concentration in renal transplant patients raises a diagnostic dilemma: Is the rise in creatinine due to rejection or to tubular injury? If the former is true, what is the pathophysiological basis of the rejection? Presently these questions are best answered by examination of tissue from renal biopsy, an invasive procedure that requires hours to provide a diagnosis. The ultimate long-term objective of the present line of investigation is to devise immunological assays that can predict the biopsy results, possibly allowing earlier start of appropriate therapeutic intervention pending biopsy findings. The question addressed in this paper is: “how best to analyze large-scale mass spectrometry data from urinary biomarker discovery studies to find proteins that have a meaningful connection to the pathophysiology of the disease process under study?” To address this, we used state-of-the-art mass spectrometry techniques for analysis of urinary exosomes from patients with renal transplants, including those with biopsy evidence of tubular injury, cell-mediated rejection, and antibody-mediated rejection. We have developed a ‘process-oriented’ approach to bioinformatic interpretation of the protein mass spectrometry discovery data that facilitates identification of candidate biomarkers for future testing in validation clinical trials.
Acknowledgements
Parts of this study were funded with a grant from the National Kidney Foundation of Maryland (to SMB) and the operating budget of Division of Intramural Research, National Heart, Lung, and Blood Institute (Project ZO1-HL001285 to MAK). Mass spectrometry was conducted in the National Heart, Lung, and Blood Institute Proteomics Core Facility (director, Marjan Gucek). A portion of the data described in this study was presented in Abstract form at the American Society of Nephrology Renal Week 2009.
Footnotes
Conflict of interest statement
The authors have no competing financial interests to disclose.
References
- [1].Khositseth S, Pisitkun T, Slentz DH, Wang G, et al. Quantitative protein and mRNA profiling shows selective post-transcriptional control of protein expression by vasopressin in kidney cells. Mol Cell Proteomics. 2011;10:M110–004036. doi: 10.1074/mcp.M110.004036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gunaratne R, Braucht DW, Rinschen MM, Chou CL, et al. Quantitative phosphoproteomic analysis reveals cAMP/vasopressin-dependent signaling pathways in native renal thick ascending limb cells. Proc Natl Acad Sci U S A. 2010;107:15653–15658. doi: 10.1073/pnas.1007424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Knepper MA. Common sense approaches to urinary biomarker study design. J Am Soc Nephrol. 2009;20:1175–1178. doi: 10.1681/ASN.2009030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bagnasco SM, Tsai W, Rahman MH, Kraus ES, et al. CD20-positive infiltrates in renal allograft biopsies with acute cellular rejection are not associated with worse graft survival. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2007;7:1968–1973. doi: 10.1111/j.1600-6143.2007.01885.x. [DOI] [PubMed] [Google Scholar]
- [5].Racusen LC, Colvin RB, Solez K, Mihatsch MJ, et al. Antibody-mediated rejection criteria - an addition to the Banff 97 classification of renal allograft rejection. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2003;3:708–714. doi: 10.1034/j.1600-6143.2003.00072.x. [DOI] [PubMed] [Google Scholar]
- [6].Solez K, Colvin RB, Racusen LC, Haas M, et al. Banff 07 classification of renal allograft pathology: updates and future directions. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2008;8:753–760. doi: 10.1111/j.1600-6143.2008.02159.x. [DOI] [PubMed] [Google Scholar]
- [7].Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, et al. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol. 2009;20:363–379. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- [10].Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- [11].Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Clarke W, Silverman BC, Zhang Z, Chan DW, et al. Characterization of renal allograft rejection by urinary proteomic analysis. Ann Surg. 2003;237:660–664. doi: 10.1097/01.SLA.0000064293.57770.42. discussion 664-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].O’Riordan E, Orlova TN, Podust VN, Chander PN, et al. Characterization of urinary peptide biomarkers of acute rejection in renal allografts. Am J Transplant. 2007;7:930–940. doi: 10.1111/j.1600-6143.2007.01733.x. [DOI] [PubMed] [Google Scholar]
- [14].Huling JC, Pisitkun T, Song JH, Yu MJ, et al. Gene expression databases for kidney epithelial cells. Am J Physiol Renal Physiol. 2012;302:F401–407. doi: 10.1152/ajprenal.00457.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wright J, Morales MM, Sousa-Menzes J, Ornellas D, et al. Transcriptional adaptation to Clcn5 knockout in proximal tubules of mouse kidney. Physiol Genomics. 2008;33:341–354. doi: 10.1152/physiolgenomics.00024.2008. [DOI] [PubMed] [Google Scholar]
- [16].Wang Q, Margolis B. Apical junctional complexes and cell polarity. Kidney Int. 2007;72:1448–1458. doi: 10.1038/sj.ki.5002579. [DOI] [PubMed] [Google Scholar]
- [17].Granger CB, Van Eyk JE, Mockrin SC, Anderson NL. National Heart, Lung, And Blood Institute Clinical Proteomics Working Group report. Circulation. 2004;109:1697–1703. doi: 10.1161/01.CIR.0000121563.47232.2A. [DOI] [PubMed] [Google Scholar]
- [18.Han WK, Bailly V, Abichandani R, Thadhani R, Bonventre JV. Kidney Injury Molecule-1 (KIM-1): a novel biomarker for human renal proximal tubule injury. Kidney Int. 2002;62:237–244. doi: 10.1046/j.1523-1755.2002.00433.x. [DOI] [PubMed] [Google Scholar]
- [19].Barrera-Chimal J, Perez-Villalva R, Cortes-Gonzalez C, Ojeda-Cervantes M, et al. Hsp72 is an early and sensitive biomarker to detect acute kidney injury. EMBO Mol Med. 2011;3:5–20. doi: 10.1002/emmm.201000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Aiello S, Noris M. Klotho in acute kidney injury: biomarker, therapy, or a bit of both? Kidney Int. 2010;78:1208–1210. doi: 10.1038/ki.2010.367. [DOI] [PubMed] [Google Scholar]
- [21].Dennen P, Altmann C, Kaufman J, Klein CL, et al. Urine interleukin-6 is an early biomarker of acute kidney injury in children undergoing cardiac surgery. Crit Care. 2010;14:R181. doi: 10.1186/cc9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bolignano D, Donato V, Coppolino G, Campo S, et al. Neutrophil gelatinase-associated lipocalin (NGAL) as a marker of kidney damage. Am J Kidney Dis. 2008;52:595–605. doi: 10.1053/j.ajkd.2008.01.020. [DOI] [PubMed] [Google Scholar]
- [23].Matsui K, Kamijo-Ikemori A, Hara M, Sugaya T, et al. Clinical significance of tubular and podocyte biomarkers in acute kidney injury. Clin Exp Nephrol. 2011;15:220–225. doi: 10.1007/s10157-010-0384-y. [DOI] [PubMed] [Google Scholar]
- [24].Ramesh G, Krawczeski CD, Woo JG, Wang Y, Devarajan P. Urinary netrin-1 is an early predictive biomarker of acute kidney injury after cardiac surgery. Clin J Am Soc Nephrol. 2010;5:395–401. doi: 10.2215/CJN.05140709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhou H, Pisitkun T, Aponte A, Yuen PS, et al. Exosomal Fetuin-A identified by roteomics: a novel urinary biomarker for detecting acute kidney injury. Kidney Int. 2006;70:1847–1857. doi: 10.1038/sj.ki.5001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Riedel I, Liang FX, Deng FM, Tu L, et al. Urothelial umbrella cells of human ureter are heterogeneous with respect to their uroplakin composition: different degrees of urothelial maturity in ureter and bladder? Eur J Cell Biol. 2005;84:393–405. doi: 10.1016/j.ejcb.2004.12.011. [DOI] [PubMed] [Google Scholar]
- [27].Wu XR, Kong XP, Pellicer A, Kreibich G, Sun TT. Uroplakins in urothelial biology, function, and disease. Kidney Int. 2009;75:1153–1165. doi: 10.1038/ki.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Stegall MD, Gloor JM. Deciphering antibody-mediated rejection: new insights into mechanisms and treatment. Curr Opin Organ Transplant. 2010;15:8–10. doi: 10.1097/MOT.0b013e3283342712. [DOI] [PubMed] [Google Scholar]
- [29].Sigdel TK, Kaushal A, Gritsenko M, Norbeck AD, et al. Shotgun proteomics identifies proteins specific for acute renal transplant rejection. Proteomics Clin Appl. 2010;4:32–47. doi: 10.1002/prca.200900124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Eikmans M, Roos-van Groningen MC, Sijpkens YW, Ehrchen J, et al. Expression of surfactant protein-C, S100A8, S100A9, and B cell markers in renal allografts: investigation of the prognostic value. J Am Soc Nephrol. 2005;16:3771–3786. doi: 10.1681/ASN.2005040412. [DOI] [PubMed] [Google Scholar]
- [31].Knepper MA, Pisitkun T. Exosomes in urine: who would have thought…? Kidney Int. 2007;72:1043–1045. doi: 10.1038/sj.ki.5002510. [DOI] [PubMed] [Google Scholar]
- [32].Quintana LF, Sole-Gonzalez A, Kalko SG, Banon-Maneus E, et al. Urine proteomics to detect biomarkers for chronic allograft dysfunction. J Am Soc Nephrol. 2009;20:428–435. doi: 10.1681/ASN.2007101137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ling XB, Sigdel TK, Lau K, Ying L, et al. Integrative urinary peptidomics in renal transplantation identifies biomarkers for acute rejection. J Am Soc Nephrol. 2010;21:646–653. doi: 10.1681/ASN.2009080876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Srivastava M, Eidelman O, Torosyan Y, Jozwik C, et al. Elevated expression levels of ANXA11, integrins beta3 and alpha3, and TNF-alpha contribute to a candidate proteomic signature in urine for kidney allograft rejection. Proteomics Clin Appl. 2011;5:311–321. doi: 10.1002/prca.201000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Metzger J, Chatzikyrkou C, Broecker V, Schiffer E, et al. Diagnosis of subclinical and clinical acute T-cell-mediated rejection in renal transplant patients by urinary proteome analysis. Proteomics Clin Appl. 2011;5:322–333. doi: 10.1002/prca.201000153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.