Abstract
While organized screening programs in industrialized countries have significantly reduced cervical cancer incidence, cytology-based screening has several limitations. Equivocal or mildly abnormal Pap tests require costly retesting or diagnostic work-up by colposcopy and biopsy. In low-resource countries, it has been difficult to establish and sustain cytology-based programs. Advances in understanding human papillomavirus biology and the natural history of human papillomavirus-related precancers and cancers have led to the discovery of a range of novel biomarkers in the past decade. In this article, we will discuss the potential role of new biomarkers for primary screening, triage and diagnosis in high-resource countries and their promise for prevention efforts in resource constrained settings.
Keywords: accuracy, biomarkers, cervical cancer, HPV, prevention, risk prediction, screening
Cervical cancer: incidence & burden
Invasive cervical cancer (ICC) is a significant cause of cancer-related morbidity and mortality among women worldwide, with substantial geographic variation [1]. Many industrialized countries have achieved significant successes in reducing ICC burden over the past six decades, and with annual incidence rates between 4 and 14 per 100,000, ICC no longer ranks even among the top ten cancers in these settings. The low incidence is achieved through substantial healthcare investments for screening programs and diagnostic workup in these countries. On the other hand, cervical cancer is the leading cancer among women in many resource-constrained settings of the developing world, where incidence and mortality rates are about five- to six-times higher [1]. Rates are highest in sub-Saharan Africa, South-Central Asia and parts of South America, where ICC represents from a sixth up to a fifth of all cancers among women [1].
Persistent infections with carcinogenic human papillomavirus (HPV) genotypes have long been established as the necessary, but not sufficient, cause of ICC [2,3]. Organized prevention programs in industrialized settings have relied on early detection of HPV-associated dysplastic changes in exfoliated cervical cells (‘Pap smear’) that reflect underlying precancerous lesions [4]. Cervical cancer screening has been a success owing to the long period, typically extending over many years, carcinogenic HPV infections take to progress to precancerous lesions (cervical intraepithelial neoplasia, grade 3 or CIN3) and ICC, and the availability of relatively safe and effective methods of treatment of cervical precancer. Yet concerns about the substantial cost burden associated with screening, limited accuracy of cytology and complications of unnecessary treatment have prompted research and development of more efficient approaches for cervical cancer prevention. Over the past two decades, substantial improvements in understanding the natural history of HPV-associated cervical carcinogenesis as well as advancements in molecular technologies have led to the availability of novel screening tests that provide alternatives or adjunctive methods to cytology. Prominent among these are the HPV-DNA based screening assays, already widely used as adjunctive methods for primary screening and for triage of equivocal cytology. At the same time, HPV vaccines have been developed and introduced that have high efficacy in preventing HPV infections when administered in HPV-naive populations. There is unanimous agreement that screening efforts have to continue, and screening algorithms have to be made more efficient, since it will take years to decades to see an effect of HPV vaccination on reduction in cancer incidence. In addition, newer screening approaches are needed to anticipate continuous changes in disease prevalence in populations with increasing vaccination coverage.
Biomarker principles
In this article, we discuss the current evidence and opportunities for improvement of cervical cancer screening through the use of novel biomarkers. In many countries, Pap cytology is still the primary screening test, either alone or in conjunction with HPV testing (predominantly in the USA) [5–8]. In some European countries, a switch to primary HPV screening followed by cytology triage has now been recommended [9].
New biomarkers may have potential use in primary screening, as triage tests for primary cytology screening, and as triage tests for primary HPV screening. For any biomarker to be useful, the test result has to influence clinical management. Management options include direct referral for treatment, referral to colposcopy to confirm precancer histologically, increased surveillance through more intensive screening or release to routine screening. The management options should be chosen based on an individual’s risk of precancer and cancer, indicated by screening test results and other risk indicators such as age [10,11].
When assessing screening options, it is important to consider physical and financial harm associated with unnecessary tests and procedures. False-positive test results may cause anxiety, lead to overtreatment of women, increase risks of obstetric complications, and thus increase the downstream costs of a screening program. The goal of cervical cancer screening programs is to prevent cancer, not to treat cervical intraepithelial neoplasia. Currently, a treatment threshold of CIN2 or worse lesions is widely used, despite the fact that a large percentage of CIN2 lesions spontaneously regress [12]. Furthermore, there is increasing evidence that even CIN3 is a heterogeneous group; only about 30–50% of large CIN3s are estimated to invade to cancer over a long time period [13,14]. An important area of cervical cancer biomarker research focuses on the identification of markers for cervical lesions that likely progress to cancer. It is important to note that risk thresholds and available resources can vary substantially between populations and may lead to different screening recommendations based on the optimal trade-offs between benefits and harms.
In the first part of this article, we briefly summarize the evidence on biomarkers that have been widely evaluated and are already in limited use in clinical practice. In the second part, we discuss biomarkers in discovery and validation phases that have the promise to improve screening in the future.
HPV life cycle & natural history & the basis for biomarker selection
The HPV genome consists of a circular double-stranded, 8000 bp long DNA with three regions:
The upper regulatory region which functions as a transcription and replication control region;
An ‘early’ region encoding proteins (E1, E2, E4, E5, E6, E7) for replication, regulation and modification of the host cytoplasm and nucleus;
A ‘late’ region encoding the viral capsid proteins (L1, L2).
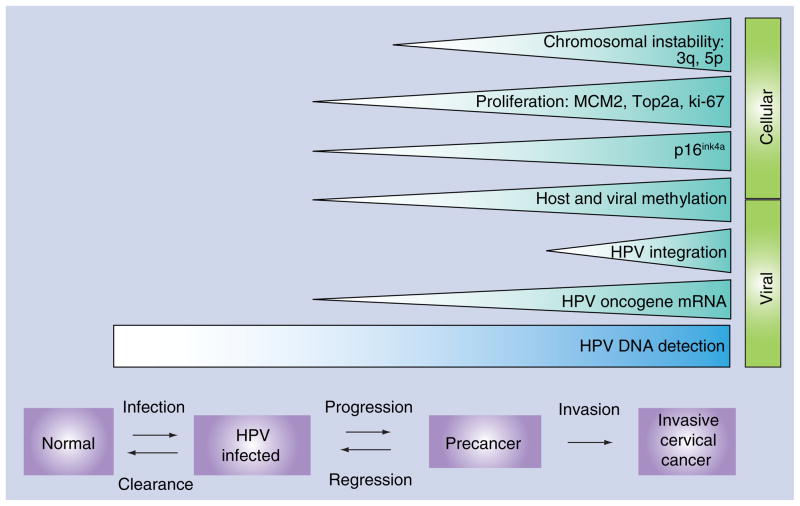
The prominent areas of research focused on biomarker discovery and validation are conceptually based on events in the HPV life cycle and natural history of HPV-dependent cervical carcinogenesis. While the phylogenetic taxonomy and classification of papillomaviruses continues to be refined, 13 HPV genotypes (HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68) are considered carcinogenic while some others (HPV types 26, 53, 66, 67, 70, 73, 82) are considered possibly carcinogenic in humans [15]. The molecular mechanisms of how HPV causes cancer have been extensively studied. Two viral oncoproteins, E6 and E7, interfere with key cellular pathways that control cell proliferation and apoptosis. Specifically, E7 disrupts pRb from its binding to E2F and triggers uncontrolled cell cycling. E6 interferes with p53 and abrogates apoptosis, which would normally occur in cells with uncontrolled cell proliferation. E6 and E7 induce substantial chromosomal instability in transformed cells, even at precancerous stages [15,16]. While biomarker discovery continues in multiple directions, current biomarker candidates can be broadly categorized into two groups, viral or cellular markers (Figure 1). The biomarker research pipeline extends from discovery (in vitro/preclinical studies) to early stage validation, and then to validation in randomized clinical trials [17]. A tabular representation of the state of availability and status of regulatory approval of commercially marketed biomarkers is presented in Table 1.
Figure 1. Human papillomavirus natural history and cellular and viral biomarkers used in cervical cancer screening.
HPV infection happens shortly after sexual initiation. Most infections clear spontaneously, but a few carcinogenic HPV infections may persist and initiate oncogenic changes in epithelial cells at the cervical transformation zone. In a small fraction of cases, these persistent abnormalities may progress to invasive cervical cancer in the absence of early detection and treatment. Viral and cellular biomarkers indicating key steps of the functional progression model (HPV infection, precancer and invasive cancer) have been discovered, with some currently in early discovery stages, while others have already been commercialized.
HPV: Human papillomavirus.
Table 1.
Major classes of biomarkers being developed and validated for use in cervical cancer prevention research.
| Type of biomarker | Test format | Application | Quality of evidence/regulatory approval | Manufacturers and test names |
|---|---|---|---|---|
| Viral markers | ||||
| Detection of carcinogenic HPV DNA (and HPV genotyping) | Signal amplification (e.g., Digene Hybrid Capture-2) Target genome amplification by PCR (e.g., Amplicor®, Linear Array®) |
Primary screening Triage of equivocal cytology | Large population-based studies and randomized trials Many tests licensed for use in the USA and Europe, many in final regulatory stages |
Qiagen: Digene hc2, careHPV™, QIAensemble™† Roche: Amplicor®, Cobas® 4800†, Linear Array®‡ Cervista® HPV HR† CLART® HPV2‡ Autogenomics: Infiniti® HR-HPV QUAD‡ BioRad: HR-HPV Dx PCR Innogenetics: InnoLiPA™‡ Multimetrix: Multiplex HPV Genotyping Kit‡ Greiner: Papillocheck® HPV-Screening‡ Abbott: RealTime HR HPV®† Not commercialized: GP 5+/6+ EIA‡ |
| Detection of E6/E7 mRNA | Nucleic acid sequence-based amplification Transcription-mediated amplification In situ hybridization |
Adjunct to primary HPV-based screening Triage of equivocal or mildly abnormal cytology |
Multiple clinical studies published Large population-based studies underway |
GenProbe: Aptima® Norchip: PreTect® Proofer‡ BioMerieux: NucliSENS EasyQ® HPV‡ IncellDx: HPV OncoTect®‡ |
| Detection of HPV protein | Immunostaining of histology and cytology slides (L1) ELISA (E6) |
Adjunct to primary HPV-based screening Triage of equivocal or mildly abnormal cytology |
Some clinical studies published | Cytoimmun: Cytoactiv® ArborVita: AVantage™ HPV E6 |
| Cellular markers | ||||
| p16ink4a (also with addition of Ki-67) | Immunostaining of histology and cytology slides ELISA |
Primary screening Triage of equivocal or mildly abnormal cytology |
Multiple clinical studies published Large population-based studies underway |
mtm Laboratories: CINtec® and CINtec® PLUS |
| MCM2 and TOP2A | Immunostaining of histology and cytology slides | Primary screening Triage of equivocal or mildly abnormal cytology |
Some clinical studies published | Becton Dickinson: ProEx™ C |
Results include partial HPV genotyping.
Genotyping assay.
HPV: Human papillomavirus; MCM2: Minichromosome maintenance protein 2; TOP2A: Topoisomerase IIA.
The limitations of cervical cytology
Cytology was introduced in the early 1950s as a primary screening method as part of annual preventive examinations, even though it was never subject to evaluation for effectiveness in randomized trials. The declining incidence rates in settings in Northern America, Europe, and Australia have provided widely accepted proof of the effectiveness of cytology-based screening [1,18]. Screening with cytology has become a well-established component of standard preventative care in most industrialized settings [19,20]. For example, more than 80% of women surveyed in a US study reported receiving Pap smears in the past 3 years [18]. In fact, over half of incident cases of ICC in the US continue to occur in women who have never or rarely been screened [21]. Although the clinical sensitivity of a single Pap smear is quite modest (60–70%) [22,23], the success of cytology-based screening is achieved by frequently repeated Pap testing, causing substantial cost burdens on the healthcare system [24,25]. Multiple efforts have focused on improving the accuracy and cost effectiveness of cytology-based screening protocols [26]. The introduction of liquid-based cytology has decreased the proportion of inadequate slides, and has permitted reflex testing for other molecular markers [27,28]. Yet false-negative rates associated with cytology continue to be substantial, primarily since cytological detection still relies on visual identification and subjective interpretation of morphologic changes induced by carcinogenic HPV [27]. About 10–15% of women with equivocal (atypical squamous cell of undetermined significance [ASC-US]) [29] and mildly abnormal (low grade squamous intraepithelial lesions [LSIL]) [30] results have an underlying CIN3. Since ASC-US and LSIL represent significant proportions of cytological results, further workup is required to make management decisions [29]. It is expected that cytological screening will be especially challenged in HPV-vaccinated populations, as the reduction of CIN2+ prevalence will be much higher than the reduction of low-grade abnormalities, further decreasing the signal-to-noise ratio of cytology testing [31,32]. Finally, most promising efforts to curb rising healthcare costs rely on prolonging screening intervals through improvements in negative predictive value of the screening test, a weakness of low-sensitivity cytology screening [32].
The role of HPV DNA testing in cervical cancer screening
Testing for HPV DNA, the necessary cause of virtually all ICC, provides a biologically salient approach for screening [33,34]. The detection of HPV in cervical scrapings was one of the first, and to date is the most widely evaluated, alternative to cytology [26,35]. Current HPV detection technologies are focused on hybridization with signal amplification of HPV DNA (e.g., Digene Hybrid Capture® 2 (hc2) (by Qiagen) [36]; Cervista® HPV HR (by Hologic) [37]) or genomic amplification using PCR (e.g., Amplicor® HPV Test and Cobas® HPV Test (by Roche) [38,39]), with most results reported as aggregate presence or absence of carcinogenic HPV types.
The primary benefit of using HPV testing is the high sensitivity and high negative predictive value, since the absence of carcinogenic HPV indicates an extremely low risk of CIN3/ICC for 5–10 years, thereby allowing for safe prolonging of screening intervals [32]. The role of HPV DNA testing as a solitary primary screening test (to replace cytology) or as an adjunct to cytological screening has been evaluated in large randomized trials over the past decade [40–46]. Results show overwhelming evidence that HPV DNA testing has a higher sensitivity in comparison with cytology for detection of CIN3 [26,47,48]. Yet its utility is constrained by its limitation of lower specificity than cytology, since the majority of HPV infections are transient and would not progress to cervical dysplasia [49,50]. The high prevalence of benign and self-limiting HPV infections, and the low prevalence of cervical cancer precursors (let alone ICC), in the second and third decades of life further limit the use of HPV DNA testing for these age groups. Hence, the use HPV DNA testing in primary screening is currently primarily focused on women 30 years or older [9]. At any age, however, a single negative HPV DNA test indicates a very low risk of precancer over the next 5–10 years and allows clinicians to extend screening intervals safely [51].
Since the FDA approval of Digene hc2 as a test for triage of ASC-US cytology in 2000, its use has increased steadily in the USA [29,52]. In the ALTS trial, it was found that while HPV testing was deemed to have utility in distinguishing women with ASC-US who were at risk for pre-cancer, it was limited in its discriminating capacity for mildly abnormal (LSIL) cytology given the high background prevalence of carcinogenic HPV in this population [53]. The availability of genotype-specific information for HPV could potentially provide additional risk stratification in HPV-positive women. This may be of particular relevance in the detection of HPV types 16 and 18, since HPV 16-associated lesions are more likely to be persistent and have higher carcinogenicity than other HPV types [54,55], and since HPV 18 is more associated with lesions within the endocervical canal that are frequently missed by cytology [56]. Indeed, some newer HPV DNA detection assays are able to provide type-specific information for HPV 16/18 [39,57–61] (Table 1). A typical application is HPV16/18 genotyping in HPV-positive, cytology-negative women. Positivity for HPV16/18 may warrant earlier referral to colposcopy because of the higher risk associated with these types. However, it remains to be determined in clinical studies and cost-effectiveness analyses whether HPV genotyping provides sufficient risk stratification in a screening population.
In the USA, cytology and HPV DNA co-testing are being widely used. In Canada and many European settings, a strategy with primary screening by HPV testing followed by cytology triage of HPV DNA positives (‘sequential’ or ‘two-stage’ testing) was proposed [32] and has been evaluated in multiple randomized clinical trials [62]. This strategy takes advantage of the high negative predictive value of HPV DNA testing and maximizes sensitivity, while reserving cytology for those who have higher likelihood of dysplastic lesions. The reliance on cytology, with subjective interpretation and substantial inter-observer variability, along with potential for sampling/collection errors, however, remains a challenge.
Novel biomarkers in cervical cancer prevention
Given limitations in use of both cytology and HPV DNA based approaches as standalone tests for screening, the focus of cervical cancer prevention research has been on development and validation of new disease-specific biomarkers of HPV-associated transformation [63–66]. The underlying biological basis and utility of some prominent biomarkers is discussed below and their functional relevance in relation with various stages of cervical carcinogenesis is schematically presented in Figure 1.
E6/E7 mRNA detection
The progression from a transient to a transforming HPV infection is characterized by a strong increase of HPV E6/E7 mRNA and protein expression [67]. Multiple studies have evaluated the role of detection of mRNA transcripts in cervical scrapings to identify cervical precancers [68–76]. At least two commercial platforms are currently available: PreTect® Proofer (Norchip [marketed as NucliSENS EasyQ® by BioMerieux in some European markets]) and APTIMA® (GenProbe) (Table 1). In a recent meta-analysis by Burger and colleagues [77], 11 studies that evaluated HPV E6/E7-based mRNA detection against HPV DNA testing for detection of CIN2+ reference standard were summarized. Given the considerable heterogeneity, pooling of data was not possible. A ‘best evidence synthesis’ for E6/E7 mRNA HPV testing accuracy was provided, that reflected a sensitivity ranging between 0.41 to 0.86 for the PreTect Proofer/NucliSENS Easy Q assays while a higher range – from 0.90 to 0.95 – for the APTIMA assay. The specificity ranged from 0.63 to 0.97 and from 0.42 to 0.61 for the PreTect Proofer/NucliSENS EasyQ and APTIMA assays, respectively. The considerable difference in sensitivity (and specificity) between PreTect Proofer/EasyQ and APTIMA may in part be explained by the difference in type coverage: The former tests detect only five types (HPV16, 18, 31, 33, 45), while the latter covers 14 types (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68).
p16ink4a
The biomarker most widely evaluated is p16ink4a, a cyclin-dependent kinase inhibitor that is markedly overexpressed in cancerous and precancerous cervical tissue. p16ink4a is a cellular correlate of the increased expression of the viral oncoprotein E7 that disrupts a key cell cycle regulator, pRb, in transforming HPV infections. The disturbance of the Rb pathway leads to a compensatory overexpression of p16ink4a through a negative feedback loop [64]. The resultant overexpression and cellular accumulation of p16ink4a is a specific marker of cervical precancerous lesions and can be measured through immunocytochemical staining of histology and cytology slides and using ELISA assays [78].
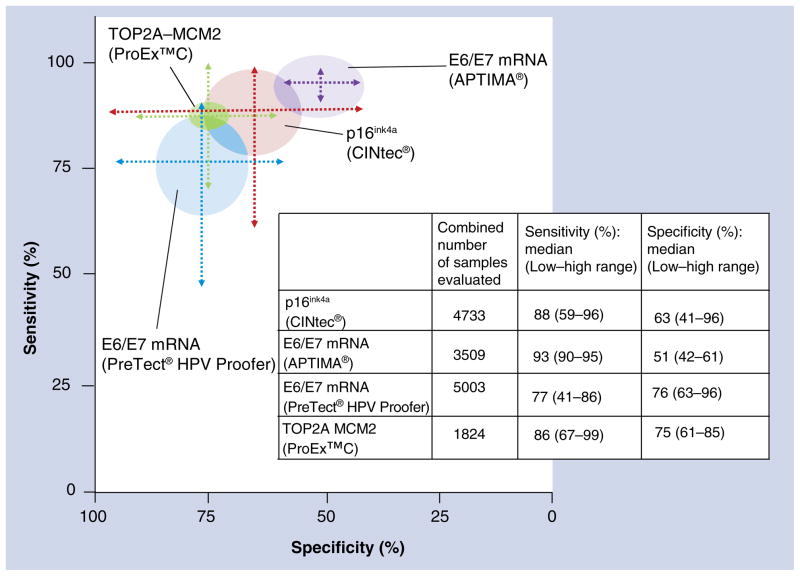
A commercially available CE-marked assay (CINtec®, mtm Laboratories) has been widely validated. Liquid-based cytology systems such as ThinPrep®, SurePath™, CYTO-screen system® and others have been used in these studies. p16ink4a has been evaluated as a standalone test and as an adjunct to cytology [79–84] or HPV testing [80,85,86]. The role of p16ink4a based detection in screening and triage has been reviewed in previous articles [63,87]. These reviews noted substantial heterogeneity in methods used for defining p16ink4a positivity in the cytology application, including quantitative and morphologic approaches. The sensitivity has ranged between 0.59 and 0.96 and the specificity has ranged between 0.41 and 0.96 for the detection of CIN2+ lesions in clinical studies, reflecting the heterogeneity in test interpretation and analyzed populations (Figure 2). Recently, a dual immunostain of p16ink4a with Ki-67 (CINtec® PLUS) has been introduced that is supposed to substantially simplify and standardize the evaluation of stained slides [80,84].
Figure 2. Graphical representation of the range of estimates of sensitivity and specificity of studies evaluating commercially available biomarkers at the cervical intraepithelial neoplasia grade 2+ threshold.
The sensitivity ranges are plotted along the y-axis and specificity along the x-axis. The median values of the range of estimates of sensitivity and specificity are used as the points of convergence of the sensitivity and specificity range lines. The circles around each graph reflect the combined total number of samples used in the studies that were summarized. This graphical representation does not weigh the studies by sample size, and the median value does not reflect a computed summary/pooled measure. The purpose of this graph is to visualize the wide range of performance estimates reported in studies evaluating these biomarkers. The heterogeneity is related to various factors, including but not limited to differences in targeted populations, differences in clinical end points and heterogeneity of biomarker performance.
Markers of aberrant S-phase induction
The cell cycle activation mediated by HPV oncogenes in transforming infections is characterized by aberrant S-phase induction. An assay detecting two proteins indicating aberrant S-phase induction, topoisomerase IIA (TOP2A) and minichromosome maintenance protein 2 (MCM2) is commercially available (ProEx™ C by Becton Dickinson) [88]. Few clinical studies with limited sample size have shown that it has a sensitivity ranging between 0.67 and 0.99 and specificity ranging between 0.61 and 0.85 (Figure 2) [89–94].
Other biomarkers undergoing clinical validation
Other cellular makers such as CK13 and CK14 [95], MCM5 and CDC6 [96], Survivin [97] and CEA [98] have also been evaluated in various stages of development. Most are marked by nonuniformity in determination of end points and limited sample sizes. Other viral markers such as HPV L1 capsid protein [99–101] and E6 oncoprotein detection [102,103] have been evaluated in a limited number of small studies, but more evidence is needed to determine their utility.
Biomarkers for low-resource settings
In the context of resource-constrained settings, the failure to establish and sustain cytology-based screening has necessitated research on operationally simple and less resource-intensive approaches for cancer prevention and control [104]. Visual methods such as visual inspection with acetic acid and visual inspection with Lugol’s Iodine provide immediate in vivo detection of visually apparent precancerous cervical lesions and the potential to link screening results and same-visit treatment by cryotherapy (or appropriate referral for cryotherapy-ineligible lesions). While visual inspection with acetic acid/visual inspection with Lugol’s Iodine have been extensively evaluated [105,106] and have high operational feasibility in the hands of nonphysician health providers, they miss anywhere between 20 and 50% of true disease due to variations in definitions of disease positivity, inherent subjectivity in test results, and challenges in quality assurance and control [105,107]. There is a huge need for utilizing novel biologically-based approaches in resource-constrained settings of the developing world for improving access and accuracy of screening [108]. careHPV™ is a new assay developed by Qiagen that is a low-cost adaptation of the Digene hc2 assay and can be performed rapidly (<2 h) without access to running water or electricity, an ideal solution for operation in field settings [109]. This assay has been shown to have performance characteristics approaching those of hc2 [109], and in conjunction with simpler alternatives like visual inspection it may permit effective single visit strategies (‘screen-and-treat’) by same-day results and linkage to cryotherapy [35,104]. Yet, further research on adaptation of these strategies is needed to avoid overtreatment, given the different age distributions of HPV prevalence worldwide [110]. Novel biomarkers that reflect measurement of an advanced disease process end point, such as overexpression of p16ink4a or HPV E6 protein detection [102,103], are also being evaluated in these settings, with the goal of achieving an optimal balance of sensitivity and specificity for very infrequent testing. Additional efforts are also being undertaken to evaluate biomarker assays using noninvasive and user-operated screening methods (e.g., self-sampling or urine-based sampling) that can address challenges in improving access to cervical cancer prevention services in these settings.
Biomarkers in discovery & early validation phases
Many discovery approaches for cervical cancer screening biomarkers are currently underway. Improved understanding of cancer epigenetics has led to a strong focus on methylation markers for biomarker development in many cancer sites, including the cervix. In addition, several recurring chromosomal imbalances have been observed for cervical precancer and are currently being explored as biomarkers. Finally, we briefly discuss potential markers earlier in the development stage, such as miRNA and proteomic markers.
Epigenetic markers: DNA methylation
Methylation of CpG sites within the genome occurs at varying levels during carcinogenesis. While tumors are often hypomethylated in repetitive regions of DNA such as LINE elements, promoter regions of tumor suppressor genes may become hypermethylated, frequently leading to decreased expression of important regulatory proteins (reviewed in [111]). Since DNA methylation is a stable analyte that can be detected in many biospecimens, and changes in methylation patterns that occur early in carcinogenesis are often retained in invasive tumors, they represent potentially clinically useful biomarkers.
Most work in the cervical cancer field has focused on candidate genes that were identified by gene expression profiling in cervical cancer cell lines and tissue or have been suggested to play a role in tumor development in sites other than the cervix. Very few studies have taken advantage of microarray technologies or other profiling approaches to identify differentially methylated genes [112–114]. Broadening the scope of research to include previously unknown genes offers an opportunity to identify novel markers that could be useful clinically. In addition, most methylation markers that have been studied extensively come from studying the host genome. However, there is growing evidence that methylation of HPV DNA may also be important in cervical carcinogenesis and could provide additional biomarkers for screening and prognosis.
Host methylation
To date, methylation of many genes has been studied in cervical cancer. Table 2 lists individual genes where the methylation status in clinical samples has been published in at least three studies and summarizes the methylation frequency for each gene in normal, high-grade and cancerous samples. There is a great diversity in the roles these genes play in normal cellular processes, ranging from apoptosis to cell–cell interactions. In general, these genes are negative regulators of cell growth and motility, therefore it is conceivable that they are more frequently methylated, and presumably silenced, in cervical cancer and its precursor lesions.
Table 2.
Methylation markers studied in cervical specimens.
| Gene | Number of studies | Methylation frequency (number positive) | Full name | Biological function | ||
|---|---|---|---|---|---|---|
| NL | HGCIN† | Ca | ||||
| DAPK | 22 | 0.068 (33) | 0.296 (158) | 0.582 (659) | Death-associated protein kinase-1 | Serine-threonine kinase; positive mediator of IFN-γ-induced apoptosis |
| RASSF1 | 17 | 0.031 (10) | 0.102 (31) | 0.141 (175) | Ras association (RalGDS/AF-6) domain family member-1 | Ras effector protein; microtubule regulation, cell migration, proliferation and apoptosis |
| CDH1 | 15 | 0.159 (37) | 0.129 (36) | 0.521 (456) | Cadherin 1, E-cadherin | Calcium-dependent cell adhesion glycoprotein |
| CDKN2A/p16 | 15 | 0.049 (17) | 0.131 (26) | 0.220 (187) | Cyclin-dependent kinase inhibitor 2A | Inhibits CDK4 kinase; regulation of cell-cycle control in G1 |
| MGMT | 12 | 0.091 (33) | 0.124 (37) | 0.183 (124) | 0–6 methylguanine-DNA methyltransferase | DNA repair |
| RARB | 12 | 0.045 (15) | 0.130 (40) | 0.343 (169) | Retinoic acid receptor-β | Regulates gene expression in response to thyroid–steroid hormones |
| CADM1 | 10 | 0.256 (43) | 0.385 (106) | 0.657 (236) | Cell adhesion molecule 1 | Intracellular adhesion |
| FHIT | 10 | 0.072 (21) | 0.020 (2) | 0.398 (268) | Fragile histidine triad gene | Diadenosine 5′,5‴-P1, P3-triphosphate hydrolase; purine metabolism |
| TIMP3 | 9 | 0 (0) | 0.107 (6) | 0.189 (82) | TIMP metallopeptidase inhibitor 3 | Matrix metalloproteinase; degradation of the extracellular matrix |
| TERT | 7 | 0.156 (12) | 0.388 (73) | 0.628 (120) | Telomerase reverse transcriptase | Enzymatic component of telomerase; responsible for the addition of short repeats to the ends of chromosomes or telomeres |
| CDH13 | 5 | 0.177 (25) | 0.047 (7) | 0.391 (79) | Cadherin 13, H-cadherin | Calcium-dependent cell adhesion glycoprotein |
| PAX1 | 4 | 0 (0) | 0.356 (36) | 0.917 (33) | Paired box 1 | Pattern formation during embryogenesis |
| TFPI2 | 4 | 0.200 (20) | 0.342 (13) | 0.721 (88) | Tissue factor pathway inhibitor 2 | Regulation of plasmin-mediated matrix remodeling |
| CCNA | 3 | 0.108 (8) | 0.387 (24) | 0.696 (94) | Cyclin A2 | Activates CDK2 kinases; promotes G1/S and G2/M transitions |
| MAL | 3 | 0.098 (4) | 0.577 (71) | 0.942 (227) | T-lymphocyte maturation-associated protein | Candidate linker protein in T-cell signaling; implicated in myelin biogenesis and function in the nervous system; formation, stabilization and maintenance of glycosphingolipid-enriched membrane microdomains |
| TWIST | 3 | 0.0928 (4) | 0.403 (27) | 0.362 (68) | Twist homolog 1 | Transcription factor; differentiation and cell lineage determination |
Inclusion criteria: genes that have been studied in normal, high-grade and cancer samples; genes that showed a low level of methylation (<20%) in normal samples that increased in precancerous lesions and/or cancer samples; genes that have been reported in at least three studies; or genes that have been utilized in a marker panel.
Includes CIN2, CIN3 and HSIL in calculations.
Ca: Cervical cancer; HGCIN: High-grade cervical intraepithelial neoplasia; NL: No lesion.
A recent review of methylation markers in cervical cancer detection reported that a total of 68 genes had been analyzed in 51 studies using clinical samples [115]. Since this review, additional genes have been reported in the literature that show different methylation levels when comparing cervical cancers to normal samples (Table 2). However, identifying changes in methylation patterns in cervical precancers will be important for biomarkers that can be used in early detection. Some studies have shown that the frequency of DNA methylation of candidate genes increases with increasing severity of the cervical lesion, suggesting that these changes occur early in cancer development and are a potential source of biomarkers for early detection of cervical cancer (reviewed in [115] and [116–120], summarized in Table 2).
A few differentially methylated genes have been formally studied as diagnostic tools for detection of cervical precancer (summarized in Table 3), including single markers and marker panels. Table 3 summarizes the performance of panels of methylation markers for the detection of cervical precancer. Although some candidates have shown promising results, further studies are needed to confirm that host methylation markers can be useful for cervical cancer prevention.
Table 3.
Diagnostic performance of methylation markers.
| Genes analyzed | Study (year) | Sample size | End point | AUC | Sensitivity (%) | Specificity (%) | Ref. |
|---|---|---|---|---|---|---|---|
| Individual markers | |||||||
|
| |||||||
| SOX1 | Lai (2010) | 185 | CIN3+ | 0.95 | 87.7 | 81.6 | [142] |
| Lai (2008) | 257 | CIN2+ | NR | 26.0 | 97.0 | [112] | |
|
| |||||||
| LMX1A | Lai (2010) | 185 | CIN3+ | 0.90 | 76.7 | 88.4 | [142] |
| Lai (2008) | 257 | CIN2+ | NR | 22.0 | 90.0 | [112] | |
|
| |||||||
| NKX6–1 | Lai (2010) | 185 | CIN3+ | 0.97 | 93.2 | 97.3 | [142] |
| Lai (2008) | 257 | CIN2+ | NR | 58.0 | 67.0 | [112] | |
| WT1 | Lai (2008) | 257 | CIN2+ | NR | 52.0 | 84.0 | [112] |
|
| |||||||
| PAX1 | Lai (2010) | 185 | CIN3+ | 0.89 | 78.1 | 90.5 | [142] |
| Huang (2010) | 73 | CIN3+ | NR | 71.4 | 93.3 | [143] | |
| Lai (2008) | 257 | CIN2+ | NR | 54.0 | 99.0 | [112] | |
|
| |||||||
| Marker panels | |||||||
|
| |||||||
| RARβ/TWIST/MGMT | Kim (2009) | 209 | HSIL | 0.80 | 78.7 | 82.2 | [144] |
|
| |||||||
| SOX1/HOXA11/CADM1 | Apostolidou (2009) | 59 | HSIL | 0.91 | 84.4 | 76.0 | [145] |
|
| |||||||
| CCNA1/C13ORF18 | Yang (2009) | 185 | CIN2+ | NR | 47.2 | 96.4 | [146] |
|
| |||||||
| CADM1/MAL† | Overmeer (2011) | 261 | CIN3+ | NR | 70.0 | 78.0 | [147] |
| Hesselink (2011)‡ | 250 | CIN3+ | 0.72 | 86.8 | 43.4 | [148] | |
|
| |||||||
| CDH13/DAPK/RARβ/TWIST1 | Feng (2005) | 319 | CIN3+ | NR | 74.0 | 95.0 | [149] |
Inclusion criteria: genes or panels were analyzed in clinical samples and diagnostic performance was reported.
Study populations were restricted to HPV-positive women only.
Multiple cut-offs were reported in the article.
NR: Not reported.
Viral methylation
Preliminary work has suggested that understanding methylation of the HPV genome could lead to additional biomarkers for the detection of cervical cancer and its progression. The promoter regions of E6 and E7 are more frequently methylated in the later stages of tumor progression and the methylation level has been correlated to E6 mRNA expression [121,122]. In addition, methylation of CpGs within L1 have been shown to be elevated in high grade lesions [123,124]. The functional relevance of this phenomenon is currently not known [125].
The data for methylation markers, both host and viral, in cervical cancer screening has come from small, heterogeneous studies, limiting the evidence of their clinical utility. Although some small panels such as CADM1 and MAL have promise as triage tests for HPV-positive women, the best panel or marker combination has yet to be identified and validated. The addition of other genes such as DAPK, RARβ, TWIST or other viral markers to CADM1 and MAL may be necessary to increase the diagnostic performance of the panel.
Chromosomal abnormalities
Cervical carcinomas are characterized by a high degree of genomic instability with many recurrent chromosomal amplifications and deletions. Based on studies in clinical cervical cancer samples, several regions are typically lost in cervical carcinogenesis (2q, 3p, 4p, 5q, 6q, 11q, 13q and 18q) while other regions are amplified (1q, 3q, 5p and 8q; reviewed in [64]). Some of these alterations can be detected in precancerous lesions (CIN3) [126–128].
Gain of 3q is the most consistently reported chromosomal abnormality in cervical cancer. One gene within this region that is of particular interest in carcinogenesis is TERC. A large multicenter study in China recently confirmed previous small studies and showed that TERC amplification could serve as an effective triage test for HPV-positive women who have ASC-US or LSIL cytological diagnoses [129]. In addition, in a small study of women who had repeat pap smears available, TERC amplification was only seen in patients who progressed to a diagnosis of CIN3+ in follow-up tests from an initial diagnosis of CIN1/2 [130]. A separate study showed that amplification of 3q had a high negative predictive value for the development of CIN2+ in women with LSIL cytology results [131]. Together this suggests that amplification of this region could be useful in determining which women have clinically relevant HPV infections and need to be referred to coloposcopy and which women can be monitored by continued screening.
Other potentially important genes located within regions of gain or loss have not been definitively identified, however, Fitzpatrick et al. showed a correlation between expression of mRNA and either loss (6p and 4q) or gain (3q and 12q) of chromosomal DNA for genes within these regions in cervical cancer samples [132]. This could lead to the identification of other genes that are significant in cervical cancer development.
On the horizon
miRNAs
miRNAs, short noncoding RNAs, are responsible for negatively regulating the expression of genes by binding to the mRNA and preventing its translation. Abnormalities in miRNA expression patterns have been seen in a number of tumors and these changes have been suggested to have prognostic value for other cancers (reviewed in [133]). Profiles of cervical tumors and cancer cell lines have identified miRNAs that have increased (miR-21, miR-127 and miR-199a) and decreased (miR-143, miR214, miR-218 and miR-34a) expression in cancer compared with normal tissue, suggesting a role for miRNAs in cervical carcinogenesis [134–136]. Since these changes in expression are seen in early, precancerous lesions [137,138], they hold promise as biomarkers for cervical cancer screening; however, they have not been formally studied in this way.
Proteomics
The field of proteomics and the identification of differentially expressed proteins in biospecimens is a growing area of research. Most proteomic work in cervical cancer has focused on comparing cancer specimens to normal samples to identify potential markers for tumors. A serum-based study with 165 patients identified three peaks by MALDI-TOF that were different between cancer patients and healthy volunteers [139]. In a validation data set, these biomarkers showed a sensitivity of 87.5% and specificity of 90% in the detection of cervical cancer.
Some studies have successfully used alternative biospecimens for the identification of protein markers, including cervical–vaginal fluid from colposcopy exams and cervical mucus [140,141]. suggesting that proteomic research does not need to be restricted to serum- or tissue-based assays. However, most proteomic studies have been small, few have studied precancerous lesions, and any differentially expressed proteins need to be validated in larger studies.
Future perspective
Cervical cancer prevention is at a transition from cytology-based screening programs to HPV-based prevention. With primary prevention using vaccines and secondary prevention using a highly sensitive HPV DNA test with long-term negative predictive value at hand, extending screening intervals will be crucial for these programs to work. New biomarkers will be important to decide who among the HPV-positive women needs to be referred for further evaluation or treatment. Large studies are currently underway for various triage biomarker candidates. It can be expected that the first screening programs based on primary HPV testing and new biomarkers as secondary tests will be implemented in a few years. It will be important to reserve treatment for those women who are at risk of developing cancer, rather than treating any high-grade lesion. Prospective biomarkers may play an important role in these therapy decisions. The implementation of new prevention strategies will be very different in each healthcare setting; in a few years, we can expect to see a wide variety of cervical cancer prevention programs existing in parallel.
Conclusion
Large randomized trials have shown that primary HPV screening for replacing cytology-based screening is feasible and is likely to be cost-effective because it allows the safe extension of screening intervals after a negative HPV test. Disease-specific biomarkers such as p16ink4a, HPV E6/E7 mRNA, or novel methylation assays may serve as secondary markers after a positive HPV DNA test to identify women with prevalent precancers who require immediate colposcopy or treatment. At this point, these markers are not sufficiently validated for introduction into HPV-based screening programs. The identification of prognostic biomarkers that can predict progression to invasive cancers is an important, but challenging area of biomarker research. It is important to note that there is substantial heterogeneity in biomarker discovery and validation studies. Even for some commercial assays, the heterogeneity in study design and disease ascertainment limit the comparability of results and the generation of high quality meta-analyses. Due to space constraints, we were not able to cover the methodological aspects of cervical cancer biomarker validation [17]. The promising results obtained with a ruggedized HPV test in low-resource settings are very exciting. Similar to industrialized countries, low-cost versions of disease-specific biomarkers could be used to identify women who need immediate treatment. In both high- and low-resource settings, incorporating biomarker data with a risk-based approach to screening will help to identify assay combinations that achieve optimal risk stratification.
Executive summary.
Concerns about substantial cost burden associated with screening, limited accuracy of cytology, and complications of unnecessary treatment have prompted research and development of more efficient approaches for cervical cancer prevention.
New biomarkers may have potential use in primary screening, as triage tests for primary cytology screening and as triage tests for primary human papillomavirus (HPV) screening.
Detection of HPV E6/E7 mRNA and protein expression, as well as markers of cellular proliferation such as p16ink4a/Ki-67 immunostaining have been widely validated and are in limited clinical use, mainly as triage markers after primary HPV DNA screening.
Candidate biomarkers will be increasingly available for clinical validation through expansion in new technologies. Markers of viral and host methylation changes, markers demonstrating chromosomal imbalances, miRNA, and proteomics markers are some of the most likely candidates that may progress further in the developmental pipeline to clinical correlative studies.
In both high- and low-resource settings, incorporating biomarker data with a risk based approach to screening will help to identify assay combinations that achieve optimal risk stratification.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189(1):12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 3.Munoz N, Bosch FX, de Sanjose S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348(6):518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 4.Ferenczy A, Franco E. Cervical-cancer screening beyond the year 2000. Lancet Oncol. 2001;2(1):27–32. doi: 10.1016/S1470-2045(00)00192-3. [DOI] [PubMed] [Google Scholar]
- 5.Jordan J, Arbyn M, Martin-Hirsch P, et al. European guidelines for quality assurance in cervical cancer screening: recommendations for clinical management of abnormal cervical cytology, part 1. Cytopathology. 2008;19(6):342–354. doi: 10.1111/j.1365-2303.2008.00623.x. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell H. The price of guidelines: revising the national guidelines for managing Australian women with abnormal pap smears. Sex Health. 2006;3(1):53–55. doi: 10.1071/sh05027. [DOI] [PubMed] [Google Scholar]
- 7.Barzon L, Giorgi C, Buonaguro FM, Palu G. Guidelines of the Italian Society for Virology on HPV testing and vaccination for cervical cancer prevention. Infect Agent Cancer. 2008;3:14. doi: 10.1186/1750-9378-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright TC, Jr, Massad LS, Dunton CJ, Spitzer M, Wilkinson EJ, Solomon D. 2006 consensus guidelines for the management of women with abnormal cervical cancer screening tests. Am J Obstet Gynecol. 2007;197(4):346–355. doi: 10.1016/j.ajog.2007.07.047. [DOI] [PubMed] [Google Scholar]
- 9.Meijer CJ, Berkhof J, Castle PE, et al. Guidelines for human papillomavirus DNA test requirements for primary cervical cancer screening in women 30 years and older. Int J Cancer. 2009;124(3):516–520. doi: 10.1002/ijc.24010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katki HA, Wacholder S, Solomon D, Castle PE, Schiffman M. Risk estimation for the next generation of prevention programmes for cervical cancer. Lancet Oncol. 2009;10(11):1022–1023. doi: 10.1016/S1470-2045(09)70253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castle PE, Sideri M, Jeronimo J, Solomon D, Schiffman M. Risk assessment to guide the prevention of cervical cancer. Am J Obstet Gynecol. 2007;197(4):356.e1–356.e6. doi: 10.1016/j.ajog.2007.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castle PE, Schiffman M, Wheeler CM, Solomon D. Evidence for frequent regression of cervical intraepithelial neoplasia-grade 2. Obstet Gynecol. 2009;113(1):18–25. doi: 10.1097/AOG.0b013e31818f5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13▪▪.McCredie MR, Sharples KJ, Paul C, et al. Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: a retrospective cohort study. Lancet Oncol. 2008;9(5):425–434. doi: 10.1016/S1470-2045(08)70103-7. Retrospective analysis of women with cervical intraepithelial neoplasia grade 3 who were unethically left without adequate treatment in New Zealand. The study found that the cumulative 30-year risk of untreated cervical intraepithelial neoplasia grade 3 to develop invasive cancer was 31.3% (95% CI: 22.7–42.3) [DOI] [PubMed] [Google Scholar]
- 14.Schiffman M, Rodriguez AC. Heterogeneity in CIN3 diagnosis. Lancet Oncol. 2008;9(5):404–406. doi: 10.1016/S1470-2045(08)70110-4. [DOI] [PubMed] [Google Scholar]
- 15.Bouvard V, Baan R, Straif K, et al. A review of human carcinogens – Part B: biological agents. Lancet Oncol. 2009;10(4):321–322. doi: 10.1016/s1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- 16.Duensing S, Munger K. Mechanisms of genomic instability in human cancer: insights from studies with human papillomavirus oncoproteins. Int J Cancer. 2004;109(2):157–162. doi: 10.1002/ijc.11691. [DOI] [PubMed] [Google Scholar]
- 17.Arbyn M, Ronco G, Cuzick J, Wentzensen N, Castle PE. How to evaluate emerging technologies in cervical cancer screening? Int J Cancer. 2009;125(11):2489–2496. doi: 10.1002/ijc.24774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solomon D, Breen N, McNeel T. Cervical cancer screening rates in the United States and the potential impact of implementation of screening guidelines. CA Cancer J Clin. 2007;57(2):105–111. doi: 10.3322/canjclin.57.2.105. [DOI] [PubMed] [Google Scholar]
- 19.Yabroff KR, Saraiya M, Meissner HI, et al. Specialty differences in primary care physician reports of papanicolaou test screening practices: a national survey, 2006 to 2007. Ann Intern Med. 2009;151(9):602–611. doi: 10.7326/0003-4819-151-9-200911030-00005. [DOI] [PubMed] [Google Scholar]
- 20.Saraiya M, Berkowitz Z, Yabroff KR, Wideroff L, Kobrin S, Benard V. Cervical cancer screening with both human papillomavirus and Papanicolaou testing vs Papanicolaou testing alone: what screening intervals are physicians recommending? Arch Intern Med. 2010;170(11):977–985. doi: 10.1001/archinternmed.2010.134. [DOI] [PubMed] [Google Scholar]
- 21.Tangka FK, O’Hara B, Gardner JG, et al. Meeting the cervical cancer screening needs of underserved women: the National Breast and Cervical Cancer Early Detection Program, 2004–2006. Cancer Causes Control. 2010;21(7):1081–1090. doi: 10.1007/s10552-010-9536-3. [DOI] [PubMed] [Google Scholar]
- 22.Nanda K, McCrory DC, Myers ER, et al. Accuracy of the Papanicolaou test in screening for and follow-up of cervical cytologic abnormalities: a systematic review. Ann Intern Med. 2000;132(10):810–819. doi: 10.7326/0003-4819-132-10-200005160-00009. [DOI] [PubMed] [Google Scholar]
- 23.Cong X, Cox DD, Cantor SB. Bayesian meta-analysis of Papanicolaou smear accuracy. Gynecol Oncol. 2007;107(1 Suppl 1):S133–S137. doi: 10.1016/j.ygyno.2007.08.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myers ER, McCrory DC, Subramanian S, et al. Setting the target for a better cervical screening test: characteristics of a cost-effective test for cervical neoplasia screening. Obstet Gynecol. 2000;96(5 Pt 1):645–652. doi: 10.1016/s0029-7844(00)00979-0. [DOI] [PubMed] [Google Scholar]
- 25.MacDonald CF. Assessing secondary prevention methods for cervical cancer: costs and benefits in managed care. Am J Manag Care. 2008;14(6 Suppl 1):S185–S192. [PubMed] [Google Scholar]
- 26.Schiffman M, Wentzensen N, Wacholder S, Kinney W, Gage JC, Castle PE. Human papillomavirus testing in the prevention of cervical cancer. J Natl Cancer Inst. 2011;103(5):368–383. doi: 10.1093/jnci/djq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siebers AG, Klinkhamer PJ, Grefte JM, et al. Comparison of liquid-based cytology with conventional cytology for detection of cervical cancer precursors: a randomized controlled trial. JAMA. 2009;302(16):1757–1764. doi: 10.1001/jama.2009.1569. [DOI] [PubMed] [Google Scholar]
- 28.Kulasingam SL, Hughes JP, Kiviat NB, et al. Evaluation of human papillomavirus testing in primary screening for cervical abnormalities: comparison of sensitivity, specificity, and frequency of referral. JAMA. 2002;288(14):1749–1757. doi: 10.1001/jama.288.14.1749. [DOI] [PubMed] [Google Scholar]
- 29.Arbyn M, Buntinx F, Van Ranst M, Paraskevaidis E, Martin-Hirsch P, Dillner J. Virologic versus cytologic triage of women with equivocal Pap smears: a meta-analysis of the accuracy to detect high-grade intraepithelial neoplasia. J Natl Cancer Inst. 2004;96(4):280–293. doi: 10.1093/jnci/djh037. [DOI] [PubMed] [Google Scholar]
- 30.Arbyn M, Paraskevaidis E, Martin-Hirsch P, Prendiville W, Dillner J. Clinical utility of HPV-DNA detection: triage of minor cervical lesions, follow-up of women treated for high-grade CIN: an update of pooled evidence. Gynecol Oncol. 2005;99(3 Suppl 1):S7–S11. doi: 10.1016/j.ygyno.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 31.Huh W, Einstein MH, Herzog TJ, Franco EL. What is the role of HPV typing in the United States now and in the next five years in a vaccinated population? Gynecol Oncol. 2010;117(3):481–485. doi: 10.1016/j.ygyno.2010.01.037. [DOI] [PubMed] [Google Scholar]
- 32.Dillner J, Rebolj M, Birembaut P, et al. Long term predictive values of cytology and human papillomavirus testing in cervical cancer screening: joint European cohort study. BMJ. 2008;337:A1754. doi: 10.1136/bmj.a1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castellsague X, Diaz M, de Sanjose S, et al. Worldwide human papillomavirus etiology of cervical adenocarcinoma and its cofactors: implications for screening and prevention. J Natl Cancer Inst. 2006;98(5):303–315. doi: 10.1093/jnci/djj067. [DOI] [PubMed] [Google Scholar]
- 34.Arbyn M, Sasieni P, Meijer CJ, Clavel C, Koliopoulos G, Dillner J. Chapter 9: Clinical applications of HPV testing: a summary of meta-analyses. Vaccine. 2006;24(Suppl 3):S3/78–89. doi: 10.1016/j.vaccine.2006.05.117. [DOI] [PubMed] [Google Scholar]
- 35.Cuzick J, Arbyn M, Sankaranarayanan R, et al. Overview of human papillomavirus-based and other novel options for cervical cancer screening in developed and developing countries. Vaccine. 2008;26(Suppl 10):K29–K41. doi: 10.1016/j.vaccine.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 36.Eder PS, Lou J, Huff J, Macioszek J. The next-generation Hybrid Capture High-Risk HPV DNA assay on a fully automated platform. J Clin Virol. 2009;45(Suppl 1):S85–S92. doi: 10.1016/S1386-6532(09)70013-7. [DOI] [PubMed] [Google Scholar]
- 37.Bartholomew DA, Luff RD, Quigley NB, Curtis M, Olson MC. Analytical performance of Cervista® HPV 16/18 genotyping test for cervical cytology samples. J Clin Virol. 2011;51(1):38–43. doi: 10.1016/j.jcv.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 38.De Francesco MA, Gargiulo F, Schreiber C, Ciravolo G, Salinaro F, Manca N. Comparison of the AMPLICOR human papillomavirus test and the hybrid capture 2 assay for detection of high-risk human papillomavirus in women with abnormal PAP smear. J Virol Methods. 2008;147(1):10–17. doi: 10.1016/j.jviromet.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 39.Stoler MH, Wright TC, Jr, Sharma A, Apple R, Gutekunst K, Wright TL. High-risk human papillomavirus testing in women with ASC-US cytology: results from the ATHENA HPV study. Am J Clin Pathol. 2011;135(3):468–475. doi: 10.1309/AJCPZ5JY6FCVNMOT. [DOI] [PubMed] [Google Scholar]
- 40.Mayrand MH, Duarte-Franco E, Rodrigues I, et al. Human papillomavirus DNA versus Papanicolaou screening tests for cervical cancer. N Engl J Med. 2007;357(16):1579–1588. doi: 10.1056/NEJMoa071430. [DOI] [PubMed] [Google Scholar]
- 41▪▪.Bulkmans NW, Berkhof J, Rozendaal L, et al. Human papillomavirus DNA testing for the detection of cervical intraepithelial neoplasia grade 3 and cancer: 5-year follow-up of a randomised controlled implementation trial. Lancet. 2007;370(9601):1764–1772. doi: 10.1016/S0140-6736(07)61450-0. This trial, conducted within the regular screening program in The Netherlands, was among the first to demonstrate the utility of primary human papillomavirus (HPV) screening over cytology in earlier detection of clinically relevant cervical lesions. [DOI] [PubMed] [Google Scholar]
- 42.Naucler P, Ryd W, Tornberg S, et al. Efficacy of HPV DNA testing with cytology triage and/or repeat HPV DNA testing in primary cervical cancer screening. J Natl Cancer Inst. 2009;101(2):88–99. doi: 10.1093/jnci/djn444. [DOI] [PubMed] [Google Scholar]
- 43.Leinonen M, Nieminen P, Kotaniemi-Talonen L, et al. Age-specific evaluation of primary human papillomavirus screening vs conventional cytology in a randomized setting. J Natl Cancer Inst. 2009;101(23):1612–1623. doi: 10.1093/jnci/djp367. [DOI] [PubMed] [Google Scholar]
- 44▪▪.Sankaranarayanan R, Nene BM, Shastri SS, et al. HPV screening for cervical cancer in rural India. N Engl J Med. 2009;360(14):1385–1394. doi: 10.1056/NEJMoa0808516. This study, conducted in India, was the first to prove the effectiveness of HPV DNA testing in reducing both incidence and mortality due to cervical cancer, in comparison with cytology, visual inspection, and no intervention. [DOI] [PubMed] [Google Scholar]
- 45.Kitchener HC, Almonte M, Thomson C, et al. HPV testing in combination with liquid-based cytology in primary cervical screening (ARTISTIC): a randomised controlled trial. Lancet Oncol. 2009;10(7):672–682. doi: 10.1016/S1470-2045(09)70156-1. [DOI] [PubMed] [Google Scholar]
- 46.Ronco G, Giorgi-Rossi P, Carozzi F, et al. Efficacy of human papillomavirus testing for the detection of invasive cervical cancers and cervical intraepithelial neoplasia: a randomised controlled trial. Lancet Oncol. 2010;11(3):249–257. doi: 10.1016/S1470-2045(09)70360-2. [DOI] [PubMed] [Google Scholar]
- 47.Koliopoulos G, Arbyn M, Martin-Hirsch P, Kyrgiou M, Prendiville W, Paraskevaidis E. Diagnostic accuracy of human papillomavirus testing in primary cervical screening: a systematic review and meta-analysis of non-randomized studies. Gynecol Oncol. 2007;104(1):232–246. doi: 10.1016/j.ygyno.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 48.Lynge E, Rebolj M. Primary HPV screening for cervical cancer prevention: results from European trials. Nat Rev Clin Oncol. 2009;6(12):699–706. doi: 10.1038/nrclinonc.2009.167. [DOI] [PubMed] [Google Scholar]
- 49.Wentzensen N, Gravitt PE, Solomon D, Wheeler CM, Castle PE. A study of Amplicor human papillomavirus DNA detection in the atypical squamous cells of undetermined significance-low-grade squamous intraepithelial lesion triage study. Cancer Epidemiol Biomarkers Prev. 2009;18(5):1341–1349. doi: 10.1158/1055-9965.EPI-08-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kinney W, Stoler MH, Castle PE. Special commentary: patient safety and the next generation of HPV DNA tests. Am J Clin Pathol. 2010;134(2):193–199. doi: 10.1309/AJCPRI8XPQUEAA3K. [DOI] [PubMed] [Google Scholar]
- 51▪.Katki HA, Kinney WK, Fetterman B, et al. Cervical cancer risk for women undergoing concurrent testing for human papillomavirus and cervical cytology: a population-based study in routine clinical practice. Lancet Oncol. 2011;12(7):663–672. doi: 10.1016/S1470-2045(11)70145-0. This study, conducted in a large population within a routine clinical practice setting in the USA, confirmed that HPV DNA testing without cytology co-testing may be sufficiently sensitive for primary screening of cervical cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moriarty AT, Schwartz MR, Eversole G, et al. Human papillomavirus testing and reporting rates: practices of participants in the College of American Pathologists Interlaboratory Comparison Program in Gynecologic Cytology in 2006. Arch Pathol Lab Med. 2008;132(8):1290–1294. doi: 10.5858/2008-132-1290-HPTARR. [DOI] [PubMed] [Google Scholar]
- 53▪▪.The Atypical Squamous Cells of Undetermined Significance/Low-Grade Squamous Intraepithelial Lesions Triage Study (ALTS) Group. . Human papillomavirus testing for triage of women with cytologic evidence of low-grade squamous intraepithelial lesions: baseline data from a randomized trial. J Natl Cancer Inst. 2000;92(5):397–402. doi: 10.1093/jnci/92.5.397. This multicenter clinical trial in the USA demonstated that HPV DNA testing is an effective strategy for the triage of equivocal atypical squamous cell cytology, but not for triage of mildly abnormal low grade squamous intraepithelial lesion cytology results. [DOI] [PubMed] [Google Scholar]
- 54.Xi LF, Koutsky LA, Castle PE, et al. Human papillomavirus type 16 variants in paired enrollment and follow-up cervical samples: implications for a proper understanding of type-specific persistent infections. J Infect Dis. 2010;202(11):1667–1670. doi: 10.1086/657083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castle PE, Rodriguez AC, Burk RD, et al. Short term persistence of human papillomavirus and risk of cervical precancer and cancer: population based cohort study. BMJ. 2009;339:b2569. doi: 10.1136/bmj.b2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Costa S, Negri G, Sideri M, et al. Human papillomavirus (HPV) test and PAP smear as predictors of outcome in conservatively treated adenocarcinoma in situ (AIS) of the uterine cervix. Gynecol Oncol. 2007;106(1):170–176. doi: 10.1016/j.ygyno.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 57.Einstein MH, Martens MG, Garcia FA, et al. Clinical validation of the Cervista HPV HR and 16/18 genotyping tests for use in women with ASC-US cytology. Gynecol Oncol. 2010;118(2):116–122. doi: 10.1016/j.ygyno.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 58.Geraets DT, Lenselink CH, Bekkers RL, van Doorn LJ, Quint WG, Melchers WJ. Universal human papillomavirus genotyping by the digene HPV Genotyping RH and LQ Tests. J Clin Virol. 2011;50(4):276–280. doi: 10.1016/j.jcv.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 59.Jamison J, Wilson RT, Carson J. The evaluation of human papillomavirus genotyping in cervical liquid-based cytology specimens; using the Roche Linear Array HPV genotyping assay. Cytopathology. 2009;20(4):242–248. doi: 10.1111/j.1365-2303.2009.00643.x. [DOI] [PubMed] [Google Scholar]
- 60.Brismar-Wendel S, Froberg M, Hjerpe A, Andersson S, Johansson B. Age-specific prevalence of HPV genotypes in cervical cytology samples with equivocal or low-grade lesions. Br J Cancer. 2009;101(3):511–517. doi: 10.1038/sj.bjc.6605165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wong AK, Chan RC, Nichols WS, Bose S. Human papillomavirus (HPV) in atypical squamous cervical cytology: the Invader HPV test as a new screening assay. J Clin Microbiol. 2008;46(3):869–875. doi: 10.1128/JCM.01424-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arbyn M, Ronco G, Meijer CJ, Naucler P. Trials comparing cytology with human papillomavirus screening. Lancet Oncol. 2009;10(10):935–936. doi: 10.1016/S1470-2045(09)70296-7. [DOI] [PubMed] [Google Scholar]
- 63▪.Cuschieri K, Wentzensen N. Human papillomavirus mRNA and p16 detection as biomarkers for the improved diagnosis of cervical neoplasia. Cancer Epidemiol Biomarkers Prev. 2008;17(10):2536–2545. doi: 10.1158/1055-9965.EPI-08-0306. This review is a comprehensive compilation of mRNA and p16 based biomarkers in cervical cancer screening. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64▪.Wentzensen N, von Knebel Doeberitz M. Biomarkers in cervical cancer screening. Dis Markers. 2007;23(4):315–330. doi: 10.1155/2007/678793. This descriptive review highlights the biological underpinning and clinical implications of current and future biomarkers in cervical cancer screening. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mayrand MH, Franco EL. Integrating novel primary- and secondary-prevention strategies: the next challenge for cervical cancer control. Future Oncol. 2010;6(11):1725–1733. doi: 10.2217/fon.10.141. [DOI] [PubMed] [Google Scholar]
- 66.Szarewski A, Ambroisine L, Cadman L, et al. Comparison of predictors for high-grade cervical intraepithelial neoplasia in women with abnormal smears. Cancer Epidemiol Biomarkers Prev. 2008;17(11):3033–3042. doi: 10.1158/1055-9965.EPI-08-0508. [DOI] [PubMed] [Google Scholar]
- 67▪.Doorbar J. Papillomavirus life cycle organization and biomarker selection. Dis Markers. 2007;23(4):297–313. doi: 10.1155/2007/613150. This review discusses the underlying biology and pathogenesis of HPV and its implications for selection of biomarkers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cattani P, Zannoni GF, Ricci C, et al. Clinical performance of human papillomavirus E6 and E7 mRNA testing for high-grade lesions of the cervix. J Clin Microbiol. 2009;47(12):3895–3901. doi: 10.1128/JCM.01275-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dockter J, Schroder A, Hill C, Guzenski L, Monsonego J, Giachetti C. Clinical performance of the APTIMA HPV Assay for the detection of high-risk HPV and high-grade cervical lesions. J Clin Virol. 2009;45(Suppl 1):S55–S61. doi: 10.1016/S1386-6532(09)70009-5. [DOI] [PubMed] [Google Scholar]
- 70.Coquillard G, Palao B, Patterson BK. Quantification of intracellular HPV E6/E7 mRNA expression increases the specificity and positive predictive value of cervical cancer screening compared with HPV DNA. Gynecol Oncol. 2011;120(1):89–93. doi: 10.1016/j.ygyno.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 71.Keegan H, McInerney J, Pilkington L, et al. Comparison of HPV detection technologies: Hybrid capture 2, PreTect HPV-Proofer and analysis of HPV DNA viral load in HPV16, HPV18 and HPV33 E6/E7 mRNA positive specimens. J Virol Methods. 2009;155(1):61–66. doi: 10.1016/j.jviromet.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 72.Ratnam S, Coutlee F, Fontaine D, et al. Clinical performance of the PreTect HPV-Proofer E6/E7 mRNA assay in comparison with that of the Hybrid Capture 2 test for identification of women at risk of cervical cancer. J Clin Microbiol. 2010;48(8):2779–2785. doi: 10.1128/JCM.00382-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reuschenbach M, Clad A, von Knebel Doeberitz C, et al. Performance of p16INK4a-cytology, HPV mRNA, and HPV DNA testing to identify high grade cervical dysplasia in women with abnormal screening results. Gynecol Oncol. 2010;119(1):98–105. doi: 10.1016/j.ygyno.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 74.Sorbye SW, Fismen S, Gutteberg TJ, Mortensen ES. HPV mRNA test in women with minor cervical lesions: experience of the University Hospital of North Norway. J Virol Methods. 2010;169(1):219–222. doi: 10.1016/j.jviromet.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 75.Trope A, Sjoborg K, Eskild A, et al. Performance of human papillomavirus DNA and mRNA testing strategies for women with and without cervical neoplasia. J Clin Microbiol. 2009;47(8):2458–2464. doi: 10.1128/JCM.01863-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu R, Belinson SE, Du H, et al. Human papillomavirus messenger RNA assay for cervical cancer screening: the Shenzhen Cervical Cancer Screening Trial I. Int J Gynecol Cancer. 2010;20(8):1411–1414. doi: 10.1111/IGC.0b013e3181f29547. [DOI] [PubMed] [Google Scholar]
- 77.Burger EA, Kornor H, Klemp M, Lauvrak V, Kristiansen IS. HPV mRNA tests for the detection of cervical intraepithelial neoplasia: a systematic review. Gynecol Oncol. 2011;120(3):430–438. doi: 10.1016/j.ygyno.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 78.Balasubramanian A, Hughes J, Mao C, et al. Evaluation of an ELISA for p16INK4a as a screening test for cervical cancer. Cancer Epidemiol Biomarkers Prev. 2009;18(11):3008–3017. doi: 10.1158/1055-9965.EPI-09-0328. [DOI] [PubMed] [Google Scholar]
- 79.Passamonti B, Gustinucci D, Recchia P, et al. Expression of p16 in abnormal pap-tests as an indicator of CIN2+ lesions: a possible role in the low grade ASC/US and L/SIL (Ig) cytologic lesions for screening prevention of uterine cervical tumours. Pathologica. 2010;102(1):6–11. [PubMed] [Google Scholar]
- 80.Petry KU, Schmidt D, Scherbring S, et al. Triaging Pap cytology negative, HPV positive cervical cancer screening results with p16/ Ki-67 Dual-stained cytology. Gynecol Oncol. 2011 doi: 10.1016/j.ygyno.2011.02.033. [DOI] [PubMed] [Google Scholar]
- 81.Sung CO, Kim SR, Oh YL, Song SY. The use of p16(INK4A) immunocytochemistry in “Atypical squamous cells which cannot exclude HSIL” compared with “Atypical squamous cells of undetermined significance” in liquid-based cervical smears. Diagn Cytopathol. 2010;38(3):168–171. doi: 10.1002/dc.21164. [DOI] [PubMed] [Google Scholar]
- 82.Guo M, Warriage I, Mutyala B, et al. Evaluation of p16 immunostaining to predict high-grade cervical intraepithelial neoplasia in women with Pap results of atypical squamous cells of undetermined significance. Diagn Cytopathol. 2010;39(7):482–488. doi: 10.1002/dc.21415. [DOI] [PubMed] [Google Scholar]
- 83.Denton KJ, Bergeron C, Klement P, Trunk MJ, Keller T, Ridder R. The sensitivity and specificity of p16(INK4a) cytology vs HPV testing for detecting high-grade cervical disease in the triage of ASC-US and LSIL pap cytology results. Am J Clin Pathol. 2010;134(1):12–21. doi: 10.1309/AJCP3CD9YKYFJDQL. [DOI] [PubMed] [Google Scholar]
- 84.Schmidt D, Bergeron C, Denton KJ, Ridder R. p16/ki-67 dual-Stain cytology in the triage of ASCUS and LSIL papanicolaou cytology: Results from the european equivocal or mildly abnormal papanicolaou cytology study. Cancer Cytopathology. 2011;119(3):158–166. doi: 10.1002/cncy.20140. [DOI] [PubMed] [Google Scholar]
- 85.Zeng WJ, Li Y, Fei HL, et al. The value of p16ink4a expression by fluorescence in situ hybridization in triage for high risk HPV positive in cervical cancer screening. Gynecol Oncol. 2011;120(1):84–88. doi: 10.1016/j.ygyno.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 86▪▪.Carozzi F, Confortini M, Dalla Palma P, et al. Use of p16-INK4A overexpression to increase the specificity of human papillomavirus testing: a nested substudy of the NTCC randomised controlled trial. Lancet Oncol. 2008;9(10):937–945. doi: 10.1016/S1470-2045(08)70208-0. This study, nested within a large randomized clinical trial in Italy, showed that HPV testing with p16ink4a triage leads to a significant increase in sensitivity compared with conventional cytology, but no substantial increase in referral to colposcopy. [DOI] [PubMed] [Google Scholar]
- 87.Tsoumpou I, Arbyn M, Kyrgiou M, et al. p16(INK4a) immunostaining in cytological and histological specimens from the uterine cervix: a systematic review and meta-analysis. Cancer Treat Rev. 2009;35(3):210–220. doi: 10.1016/j.ctrv.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Halloush RA, Akpolat I, Jim Zhai Q, Schwartz MR, Mody DR. Comparison of ProEx C with p16INK4a and Ki-67 immunohistochemical staining of cell blocks prepared from residual liquid-based cervicovaginal material: a pilot study. Cancer. 2008;114(6):474–480. doi: 10.1002/cncr.23951. [DOI] [PubMed] [Google Scholar]
- 89.Siddiqui MT, Cohen C, Nassar A. Detecting high-grade cervical disease on ASC-H cytology: role of BD ProEx C and Digene Hybrid Capture II HPV DNA testing. Am J Clin Pathol. 2008;130(5):765–770. doi: 10.1309/AJCPWW6V2KGXODUI. [DOI] [PubMed] [Google Scholar]; Future Microbiol. 2011;6(9) [Google Scholar]
- 90.Siddiqui MT, Hornaman K, Cohen C, Nassar A. ProEx C immunocytochemistry and high-risk human papillomavirus DNA testing in papanicolaou tests with atypical squamous cell (ASC-US) cytology: correlation study with histologic biopsy. Arch Pathol Lab Med. 2008;132(10):1648–1652. doi: 10.5858/2008-132-1648-PCIAHH. [DOI] [PubMed] [Google Scholar]
- 91.Guo M, Baruch AC, Silva EG, et al. Efficacy of p16 and ProExC immunostaining in the detection of high-grade cervical intraepithelial neoplasia and cervical carcinoma. Am J Clin Pathol. 2011;135(2):212–220. doi: 10.1309/AJCP1LLX8QMDXHHO. [DOI] [PubMed] [Google Scholar]
- 92.Kelly D, Kincaid E, Fansler Z, Rosenthal DL, Clark DP. Detection of cervical high-grade squamous intraepithelial lesions from cytologic samples using a novel immunocytochemical assay (ProEx C) Cancer. 2006;108(6):494–500. doi: 10.1002/cncr.22288. [DOI] [PubMed] [Google Scholar]
- 93.Tambouret RH. Diagnostic importance of ProEx C in gynecologic cytopathology. Ann Pathol. 2008;28(Spec No 11):S92–S93. doi: 10.1016/j.annpat.2008.09.030. [DOI] [PubMed] [Google Scholar]
- 94.Pinto AP, Schlecht NF, Woo TY, Crum CP, Cibas ES. Biomarker (ProEx C, p16(INK4A), and MiB-1) distinction of high-grade squamous intraepithelial lesion from its mimics. Mod Pathol. 2008;21(9):1067–1074. doi: 10.1038/modpathol.2008.101. [DOI] [PubMed] [Google Scholar]
- 95.Baak JP, Kruse AJ, Robboy SJ, Janssen EA, van Diermen B, Skaland I. Dynamic behavioural interpretation of cervical intraepithelial neoplasia with molecular biomarkers. J Clin Pathol. 2006;59(10):1017–1028. doi: 10.1136/jcp.2005.027839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Murphy N, Ring M, Heffron CC, et al. Quantitation of CDC6 and MCM5 mRNA in cervical intraepithelial neoplasia and invasive squamous cell carcinoma of the cervix. Mod Pathol. 2005;18(6):844–849. doi: 10.1038/modpathol.3800361. [DOI] [PubMed] [Google Scholar]
- 97.Branca M, Giorgi C, Santini D, et al. Survivin as a marker of cervical intraepithelial neoplasia and high-risk human papillomavirus and a predictor of virus clearance and prognosis in cervical cancer. Am J Clin Pathol. 2005;124(1):113–121. doi: 10.1309/L8BWF431WU9AC8FJ. [DOI] [PubMed] [Google Scholar]
- 98.Liang J, Mittal KR, Wei JJ, Yee H, Chiriboga L, Shukla P. Utility of p16INK4a, CEA, Ki67, P53 and ER/PR in the differential diagnosis of benign, premalignant, and malignant glandular lesions of the uterine cervix and their relationship with Silverberg scoring system for endocervical glandular lesions. Int J Gynecol Pathol. 2007;26(1):71–75. doi: 10.1097/01.pgp.0000225851.97739.9f. [DOI] [PubMed] [Google Scholar]
- 99.Yu L, Wang L, Zhong J, Chen S. Diagnostic value of p16INK4A, Ki-67, and human papillomavirus L1 capsid protein immunochemical staining on cell blocks from residual liquid-based gynecologic cytology specimens. Cancer Cytopathol. 2010;118(1):47–55. doi: 10.1002/cncy.20061. [DOI] [PubMed] [Google Scholar]
- 100.Huang MZ, Li HB, Nie XM, Wu XY, Jiang XM. An analysis on the combination expression of HPV L1 capsid protein and p16INK4a in cervical lesions. Diagn Cytopathol. 2010;38(8):573–578. doi: 10.1002/dc.21258. [DOI] [PubMed] [Google Scholar]
- 101.Hoshikawa S, Sano T, Yoshida T, Ito H, Oyama T, Fukuda T. Immunohistological analysis of HPV L1 capsid protein and p16 protein in low-grade dysplastic lesions of the uterine cervix. Pathol Res Pract. 2010;206(12):816–820. doi: 10.1016/j.prp.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 102.Schweizer J, Lu PS, Mahoney CW, et al. Feasibility study of a human papillomavirus E6 oncoprotein test for diagnosis of cervical precancer and cancer. J Clin Microbiol. 2010;48(12):4646–4648. doi: 10.1128/JCM.01315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang HP, Walmer DK, Merisier D, et al. A pilot analytic study of a research-level, lower-cost human papillomavirus 16, 18, and 45 test. J Virol Methods. 2011;176(1–2):112–114. doi: 10.1016/j.jviromet.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sankaranarayanan R, Boffetta P. Research on cancer prevention, detection and management in low- and medium-income countries. Ann Oncol. 2010;21(10):1935–1943. doi: 10.1093/annonc/mdq049. [DOI] [PubMed] [Google Scholar]
- 105.Sauvaget C, Fayette JM, Muwonge R, Wesley R, Sankaranarayanan R. Accuracy of visual inspection with acetic acid for cervical cancer screening. Int J Gynaecol Obstet. 2011;113(1):14–24. doi: 10.1016/j.ijgo.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 106.Sankaranarayanan R, Esmy PO, Rajkumar R, et al. Effect of visual screening on cervical cancer incidence and mortality in Tamil Nadu, India: a cluster-randomised trial. Lancet. 2007;370(9585):398–406. doi: 10.1016/S0140-6736(07)61195-7. [DOI] [PubMed] [Google Scholar]
- 107.Zhao FH, Lin MJ, Chen F, et al. Performance of high-risk human papillomavirus DNA testing as a primary screen for cervical cancer: a pooled analysis of individual patient data from 17 population-based studies from China. Lancet Oncol. 2010;11(12):1160–1171. doi: 10.1016/S1470-2045(10)70256-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Goldie SJ, Gaffikin L, Goldhaber-Fiebert JD, et al. Cost-effectiveness of cervical-cancer screening in five developing countries. N Engl J Med. 2005;353(20):2158–2168. doi: 10.1056/NEJMsa044278. [DOI] [PubMed] [Google Scholar]
- 109▪.Qiao YL, Sellors JW, Eder PS, et al. A new HPV-DNA test for cervical-cancer screening in developing regions: a cross-sectional study of clinical accuracy in rural China. Lancet Oncol. 2008;9(10):929–936. doi: 10.1016/S1470-2045(08)70210-9. This study, conducted in China, was the first to demonstrate the performance of a new lower cost HPV DNA test for use in resource-limited settings. [DOI] [PubMed] [Google Scholar]
- 110.Clifford G, Franceschi S, Diaz M, Munoz N, Villa LL. Chapter 3: HPV type-distribution in women with and without cervical neoplastic diseases. Vaccine. 2006;24(Suppl 3):S3/26–34. doi: 10.1016/j.vaccine.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 111.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 112.Lai HC, Lin YW, Huang TH, et al. Identification of novel DNA methylation markers in cervical cancer. Int J Cancer. 2008;123(1):161–167. doi: 10.1002/ijc.23519. [DOI] [PubMed] [Google Scholar]
- 113.Sova P, Feng Q, Geiss G, et al. Discovery of novel methylation biomarkers in cervical carcinoma by global demethylation and microarray analysis. Cancer Epidemiol Biomarkers Prev. 2006;15(1):114–123. doi: 10.1158/1055-9965.EPI-05-0323. [DOI] [PubMed] [Google Scholar]
- 114.Wang SS, Smiraglia DJ, Wu YZ, et al. Identification of novel methylation markers in cervical cancer using restriction landmark genomic scanning. Cancer Res. 2008;68(7):2489–2497. doi: 10.1158/0008-5472.CAN-07-3194. [DOI] [PubMed] [Google Scholar]
- 115.Wentzensen N, Sherman ME, Schiffman M, Wang SS. Utility of methylation markers in cervical cancer early detection: appraisal of the state-of-the-science. Gynecol Oncol. 2009;112(2):293–299. doi: 10.1016/j.ygyno.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.de Wilde J, Kooter JM, Overmeer RM, et al. hTERT promoter activity and CpG methylation in HPV-induced carcinogenesis. BMC Cancer. 2010;10:271. doi: 10.1186/1471-2407-10-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Flatley JE, McNeir K, Balasubramani L, et al. Folate status and aberrant DNA methylation are associated with HPV infection and cervical pathogenesis. Cancer Epidemiol Biomarkers Prev. 2009;18(10):2782–2789. doi: 10.1158/1055-9965.EPI-09-0493. [DOI] [PubMed] [Google Scholar]
- 118.Neyaz MK, Kumar RS, Hussain S, et al. Effect of aberrant promoter methylation of FHIT and RASSF1A genes on susceptibility to cervical cancer in a North Indian population. Biomarkers. 2008;13(6):597–606. doi: 10.1080/13547500802078859. [DOI] [PubMed] [Google Scholar]
- 119.Overmeer RM, Henken FE, Bierkens M, et al. Repression of MAL tumour suppressor activity by promoter methylation during cervical carcinogenesis. J Pathol. 2009;219(3):327–336. doi: 10.1002/path.2598. [DOI] [PubMed] [Google Scholar]
- 120.Yang N, Nijhuis ER, Volders HH, et al. Gene promoter methylation patterns throughout the process of cervical carcinogenesis. Cell Oncol. 2010;32(1–2):131–143. doi: 10.3233/CLO-2009-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brandsma JL, Sun Y, Lizardi PM, et al. Distinct human papillomavirus type 16 methylomes in cervical cells at different stages of premalignancy. Virology. 2009;389(1–2):100–107. doi: 10.1016/j.virol.2009.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hublarova P, Hrstka R, Rotterova P, et al. Prediction of human papillomavirus 16 e6 gene expression and cervical intraepithelial neoplasia progression by methylation status. Int J Gynecol Cancer. 2009;19(3):321–325. doi: 10.1111/IGC.0b013e31819d8a5c. [DOI] [PubMed] [Google Scholar]
- 123.Kalantari M, Calleja-Macias IE, Tewari D, et al. Conserved methylation patterns of human papillomavirus type 16 DNA in asymptomatic infection and cervical neoplasia. J Virol. 2004;78(23):12762–12772. doi: 10.1128/JVI.78.23.12762-12772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Turan T, Kalantari M, Calleja-Macias IE, et al. Methylation of the human papillomavirus-18 L1 gene: a biomarker of neoplastic progression? Virology. 2006;349(1):175–183. doi: 10.1016/j.virol.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 125.Kalantari M, Chase DM, Tewari KS, Bernard HU. Recombination of human papillomavirus-16 and host DNA in exfoliated cervical cells: a pilot study of L1 gene methylation and chromosomal integration as biomarkers of carcinogenic progression. J Med Virol. 2010;82(2):311–320. doi: 10.1002/jmv.21676. [DOI] [PubMed] [Google Scholar]
- 126.Hidalgo A, Schewe C, Petersen S, et al. Human papilloma virus status and chromosomal imbalances in primary cervical carcinomas and tumour cell lines. Eur J Cancer. 2000;36(4):542–548. doi: 10.1016/s0959-8049(99)00323-8. [DOI] [PubMed] [Google Scholar]
- 127.Umayahara K, Numa F, Suehiro Y, et al. Comparative genomic hybridization detects genetic alterations during early stages of cervical cancer progression. Genes Chromosomes Cancer. 2002;33(1):98–102. doi: 10.1002/gcc.1215. [DOI] [PubMed] [Google Scholar]
- 128.Yang YC, Shyong WY, Chang MS, et al. Frequent gain of copy number on the long arm of chromosome 3 in human cervical adenocarcinoma. Cancer Genet Cytogenet. 2001;131(1):48–53. doi: 10.1016/s0165-4608(01)00510-6. [DOI] [PubMed] [Google Scholar]
- 129.Jiang J, Wei LH, Li YL, et al. Detection of TERC amplification in cervical epithelial cells for the diagnosis of high-grade cervical lesions and invasive cancer: a multicenter study in China. J Mol Diagn. 2010;12(6):808–817. doi: 10.2353/jmoldx.2010.100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Heselmeyer-Haddad K, Sommerfeld K, White NM, et al. Genomic amplification of the human telomerase gene (TERC) in pap smears predicts the development of cervical cancer. Am J Pathol. 2005;166(4):1229–1238. doi: 10.1016/S0002-9440(10)62341-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jalali GR, Herzog TJ, Dziura B, Walat R, Kilpatrick MW. Amplification of the chromosome 3q26 region shows high negative predictive value for nonmalignant transformation of LSIL cytologic finding. Am J Obstet Gynecol. 2010;202(6):581E581–E585. doi: 10.1016/j.ajog.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 132.Fitzpatrick MA, Funk MC, Gius D, et al. Identification of chromosomal alterations important in the development of cervical intraepithelial neoplasia and invasive carcinoma using alignment of DNA microarray data. Gynecol Oncol. 2006;103(2):458–462. doi: 10.1016/j.ygyno.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 133.Cortez MA, Ivan C, Zhou P, Wu X, Ivan M, Calin GA. microRNAs in cancer: from bench to bedside. Adv Cancer Res. 2010;108:113–157. doi: 10.1016/B978-0-12-380888-2.00004-2. [DOI] [PubMed] [Google Scholar]
- 134.Lee JW, Choi CH, Choi JJ, et al. Altered MicroRNA expression in cervical carcinomas. Clin Cancer Res. 2008;14(9):2535–2542. doi: 10.1158/1078-0432.CCR-07-1231. [DOI] [PubMed] [Google Scholar]
- 135.Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67(13):6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- 136.Martinez I, Gardiner AS, Board KF, Monzon FA, Edwards RP, Khan SA. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene. 2008;27(18):2575–2582. doi: 10.1038/sj.onc.1210919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li B, Hu Y, Ye F, Li Y, Lv W, Xie X. Reduced miR-34a expression in normal cervical tissues and cervical lesions with high-risk human papillomavirus infection. Int J Gynecol Cancer. 2010;20(4):597–604. doi: 10.1111/IGC.0b013e3181d63170. [DOI] [PubMed] [Google Scholar]
- 138.Li Y, Liu J, Yuan C, Cui B, Zou X, Qiao Y. High-risk human papillomavirus reduces the expression of microRNA-218 in women with cervical intraepithelial neoplasia. J Int Med Res. 2010;38(5):1730–1736. doi: 10.1177/147323001003800518. [DOI] [PubMed] [Google Scholar]
- 139.Liu C, Pan C, Shen J, Wang H, Yong L, Zhang R. Discrimination analysis of mass spectrometry proteomics for cervical cancer detection. Med Oncol. 2010;20(4):597–604. doi: 10.1007/s12032-010-9740-8. [DOI] [PubMed] [Google Scholar]
- 140.Panicker G, Ye Y, Wang D, Unger ER. Characterization of the human cervical mucous proteome. Clin Proteomics. 2010;6(1–2):18–28. doi: 10.1007/s12014-010-9042-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zegels G, Van Raemdonck GA, Coen EP, Tjalma WA, Van Ostade XW. Comprehensive proteomic analysis of human cervical–vaginal fluid using colposcopy samples. Proteome Sci. 2009;7:17. doi: 10.1186/1477-5956-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lai HC, Lin YW, Huang RL, et al. Quantitative DNA methylation analysis detects cervical intraepithelial neoplasms type 3 and worse. Cancer. 2010;116(18):4266–4274. doi: 10.1002/cncr.25252. [DOI] [PubMed] [Google Scholar]
- 143.Huang TH, Lai HC, Liu HW, et al. Quantitative analysis of methylation status of the PAX1 gene for detection of cervical cancer. Int J Gynecol Cancer. 2010;20(4):513–519. doi: 10.1111/IGC.0b013e3181c7fe6e. [DOI] [PubMed] [Google Scholar]
- 144.Kim JH, Choi YD, Lee JS, Lee JH, Nam JH, Choi C. Assessment of DNA methylation for the detection of cervical neoplasia in liquid-based cytology specimens. Gynecol Oncol. 2010;116(1):99–104. doi: 10.1016/j.ygyno.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 145.Apostolidou S, Hadwin R, Burnell M, et al. DNA methylation analysis in liquid-based cytology for cervical cancer screening. Int J Cancer. 2009;125(12):2995–3002. doi: 10.1002/ijc.24745. [DOI] [PubMed] [Google Scholar]
- 146.Yang N, Eijsink JJ, Lendvai A, et al. Methylation markers for CCNA1 and C13ORF18 are strongly associated with high-grade cervical intraepithelial neoplasia and cervical cancer in cervical scrapings. Cancer Epidemiol Biomarkers Prev. 2009;18(11):3000–3007. doi: 10.1158/1055-9965.EPI-09-0405. [DOI] [PubMed] [Google Scholar]
- 147.Overmeer RM, Louwers JA, Meijer CJ, et al. Combined CADM1 and MAL promoter methylation analysis to detect (pre-) malignant cervical lesions in high-risk HPV-positive women. Int J Cancer. 2010 doi: 10.1002/ijc.25890. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 148.Hesselink AT, Heideman DA, Steenbergen RD, et al. Combined promoter methylation analysis of CADM1 and MAL: an objective triage tool for high-risk human papillomavirus DNA-positive women. Clin Cancer Res. 2011;17(8):2459–2465. doi: 10.1158/1078-0432.CCR-10-2548. [DOI] [PubMed] [Google Scholar]
- 149.Feng Q, Balasubramanian A, Hawes SE, et al. Detection of hypermethylated genes in women with and without cervical neoplasia. J Natl Cancer Inst. 2005;97(4):273–282. doi: 10.1093/jnci/dji041. [DOI] [PubMed] [Google Scholar]