Abstract
Neural crest (NC) cells undergo an epithelial-to-mesenchymal transition (EMT) in order to exit from the dorsal neural tube. Similarly, ancestrally-related melanoma cells employ an EMT-like event during the initial stages of metastasis to dissociate from surrounding keratinocytes. Whether the molecular pathogenesis and cellular dynamics of melanoma metastasis resemble the embryonic NC invasion program is unclear. Here, we highlight advances in our understanding of tumor cell behaviors and plasticity, focusing on the relationship between melanoma and the neural crest invasion programs. We summarize recent discoveries of neural crest cell guidance and emerging in vivo imaging strategies that permit single cell resolution of fluorescently labeled tumor cells, with a focus on our recently developed in vivo chick embryo transplant model. Crucial to the molecular pathogenesis of metastasis, we highlight advances in gene profiling of small cell numbers, including our novel ability to gather gene expression information during distinct stages of melanoma invasion. Lastly, we present preliminary details of a comparison of specific genetic pathways associated with the early phases of melanoma invasion and known neural crest induction and migration signals. Our results suggest that malignant melanoma cells hijack portions of the neural crest program to promote plasticity and facilitate metastasis. In summary, there is considerable power in combining an in vivo model system with molecular analysis of gene expression, within the context of established developmental signaling pathways, to identify and study the molecular mechanisms of metastasis.
Keywords: Neural Crest, Melanoma, in vivo model, metastasis, chick embryo, laser capture microdissection, gene profiling, high resolution imaging
Introduction
Multipotent tumor cells and many embryonic progenitor cells share common characteristics of cell plasticity and invasiveness. The manner by which these cells invade peripheral tissues involves migration through many different microenvironments and interactions with numerous other cell types and structures. The signals between a multipotent cancer cell or embryonic progenitor cell and its microenvironment are thought to be complex and manifested in a combination of intrinsic and extrinsic cell-cell and cell-matrix interactions. Exciting studies have shown that signals potentially derived from the embryonic microenvironment can influence embryonic stem cells [Takahashi et al., 2007a; Takahashi et al., 2007b; Yu et al., 2007; Hochedlinger, 2011], multipotent tumor cells [Hendrix et al., 2007; Abbott et al., 2008] and adult cell fate and plasticity [Real et al., 2006]. One of the next logical steps is to resolve the nature of the in vivo cellular and molecular mechanisms critical to the survival and programming of cell fate and invasion. Further insights from studies at the interface of developmental and tumor biology that exploit the accessibility of the embryo may help to determine the extent of the convergence of embryonic and tumorigenic signaling pathways involved in regulating cell fate and invasive ability.
Within the developing vertebrate embryo, the neural crest (NC) represents a multipotent, highly migratory cell population that produces multiple cell types to contribute to bone and cartilage of the face, function of the heart and gut, and the entire peripheral nervous system [Le Douarin and Kalcheim, 1999; Acloque et al., 2009; Zhang et al., 2010]. NC cells undergo an epithelial-to-mesenchymal transition (EMT) to exit the neural tube, then sort into segregated migratory streams and invade the embryo along the vertebrate axis in a programmed rostral-to-caudal manner [Kulesa and Gammill, 2010]. Signals within the neural tube and the many different microenvironments encountered by the migratory NC are thought to spatially permit or inhibit NC cell movements, leading to the sculpting of NC cells into particular migratory streams that reach precise peripheral targets [Gammill and Roffers-Agarwal, 2010; Kulesa et al., 2010; Kulesa and Gammill, 2010]. Time-lapse microscopy studies have revealed a complex set of NC cell migratory behaviors that include active avoidance of some areas, but directed movement and cell-cell contact through others, supporting the hypothesis that local microenvironments shape individual NC cell trajectories [Kuriyama and Mayor, 2008; Gammill and Roffers-Agarwal, 2010; Kulesa et al., 2010; Kulesa and Gammill, 2010; Mayor and Carmona-Fontaine, 2010].
Technical advances in microscopy, cell labeling, and molecular biology have allowed scientists to extend observations made by pioneers working at the intersection of developmental biology and cancer. Early research at this interface foresaw the importance of understanding basic cellular and molecular mechanisms involving EMT and cell migration. Novel efforts pioneered the development of cell labeling and culture techniques that extended observations of cell behaviors in 2D to in situ organ culture and helped to build the knowledge base of cell behaviors in 3D [Mintz and Illmensee, 1975; Erickson et al., 1980; Davis and Trinkaus, 1981; Bilozur and Hay, 1988]. This work helped to lay the groundwork for attempts to visualize in vivo tumor cell behaviors and their response to local microenvironmental cues.
In vivo imaging strategies of the tumor microenvironment represent an intense area of study that has benefitted from new technological advances in multiphoton microscopy, fluorescent reporters of cell cycle progression, and unique windowing techniques into the living adult mouse and chick and zebrafish embryos [Condeelis and Weissleder, 2010; Fukumura et al., 2010; Giampieri et al., 2010; Friedl and Alexander, 2011; Zhou, 2011; Zhang et al., 2012]. Thus, in vivo imaging is providing unique access to tumor cell behaviors in natural microenvironments (Table 1).
Table 1.
in vivo Models of Tumor Cell Behaviors
| Animal Model | Tumor Type | Highlights/Methods | References |
|---|---|---|---|
| Chick embryo | mammalian tumor xenograft | static and dynamic imaging of cell cycle progression (CycleTrak) in migrating melanoma cells | Ridenour 2012 |
| Zebrafish embryo | mammalian tumor xenograft | dynamic imaging of tumor neovascularization and evaluation of anti-angiogenic compounds | Zhao 2011 |
| mouse | orthotopic mammalian tumor xenograft | mammary imaging window combined with photoswitchable fluorescent label provides extended intravital imaging | Kedrin 2008 |
| mouse | mammalian tumor xenograft | skin fold chamber provides orthotopic intravital imaging of early stages of tumor invasion | Alexander 2008 |
| mouse | mammalian tumor xenograft | simultaneous imaging of multiple fluorescent markers and collagen using multi-photon microscopy | Sahai 2005 |
| rat | allograft | skin flap surgery to expose tumor for timelapse imaging | Wyckoff 2000 |
Exciting investigations of tumor cell interactions within in vivo embryonic microenvironments have revealed unique insights into tumor cell behaviors [Kulesa et al., 2006; Topczewska et al., 2006; Hendrix et al., 2007; Kasemeier-Kulesa et al., 2008]. In particular, the investigation of tumor cell behaviors within the embryonic chick NC microenvironment exploits the ancestral link between the neural crest and cancer cell types, such as melanoma and neuroblastoma. The accessibility of novel cellular and molecular imaging strategies within embryos has inspired developmental and cancer biologists to move from descriptive observational studies to mechanistic analyses of the convergence of embryonic and tumorigenic signaling pathways. Here, we highlight advances in the observation of tumor cell behaviors and molecular profiling studied within the embryonic neural crest microenvironment.
The Chick Embryo Transplant Model
One of the strongest links between common mechanisms that underlie NC cell migration and cancer biology has come from the study of NC cell regulatory factors [Barrallo-Gimeno and Nieto, 2005; Betancur et al., 2010; Haldin and LaBonne, 2010; Nieto, 2011]. Work on Snail family genes expressed in early emerging NC cells, has revealed that a cell’s acquisition of migratory properties is partly due to repressing E-cadherin transcription during development [Cano et al., 2000; Bolos et al., 2003]. Studies of cell invasion in ductal carcinomas have discovered a role for SNAI1 in the metastatic cascade, thus highlighting its function and potential as an early biomarker for tumor metastatic potential [Blanco et al., 2002]. These studies suggest there is a great deal to be learned from the examination of embryonic signals guiding cell migration and their potential ability to regulate tumor cell invasion. Thus, the accessible chick embryonic NC cell microenvironment provides fertile ground to search for molecular signals common to the NC cell migratory program and cancer cell plasticity and invasion.
The development of in vivo models to study embryonic and tumor cell behaviors has a rich history. In 1975, Mintz and Illmensee investigated the concept that the mouse embryo was accessible to transplantation of tumor cells and found that signals within the embryonic microenvironment could reprogram the tumor cells to a less destructive fate [Mintz and Illmensee, 1975]. When the hypothesis of multipotent tumor cell reprogramming was investigated more recently in the zebrafish embryo, one of the results surprisingly showed that transplanted highly aggressive human melanoma cells induced zebrafish progenitor cells to form a secondary axis [Topczewska et al., 2006]. Further investigation revealed that the aggressive melanoma cells secreted Nodal, (a potent embryonic morphogen), responsible for the ectopic induction of the embryonic axis [Topczewska et al., 2006]. Thus, although the zebrafish embryo is extremely useful as a biosensor for tumor cell signals [Topczewska et al., 2006; Stewart et al., 2010; Zhang et al., 2012], one of the major limitations of this system is the lack of surgical accessibility to manipulate or transplant tissue at various developmental stages.
The avian embryo has emerged as a useful tool for analyzing both NC and tumor cell interactions, providing imaging and surgical accessibility to manipulate the NC cell migratory pathways and monitor transplanted tumor cells (Fig. 1). One of the major results of these types of studies occurred as early as the 1950’s, when tissue transplantation experiments that placed mouse sarcoma 180 cells into the chick limb bud caused NC-derived sympathetic nerve fibers to grow out and innervate the transplanted cells [Levi-Montalcini, 1952].
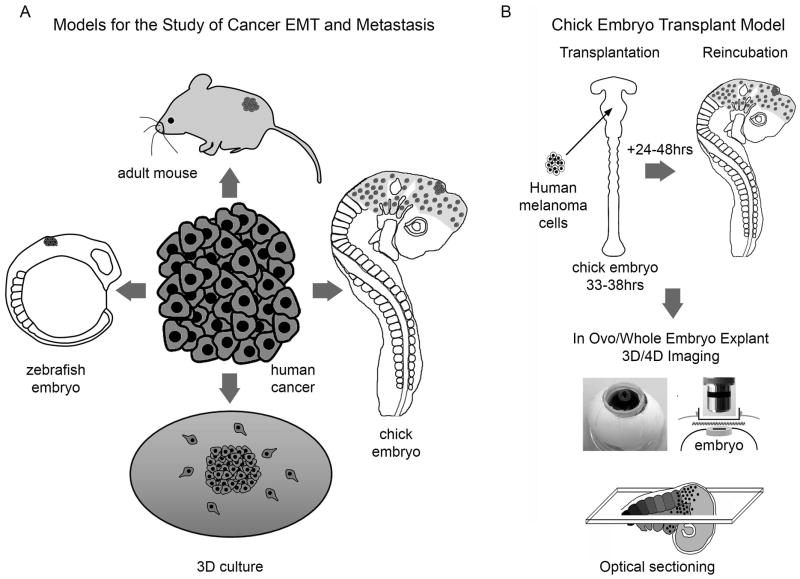
Figure 1. Models for the Study of Cancer EMT and Metastasis, including the Chick Embryo Transplant Model.
A) There are at least four model systems, to analyze human tumor cell behaviors including in vitro culture, zebrafish and chick embryos, and adult mice. B) The chick embryo transplant model permits transplantation of human tumor cells into the neural crest microenvironment and visualization of cell behaviors in vivo in 3D using a teflon window into the egg that allows oxygen transfer to the embryo.
Investigation of the tumor cell and nerve fiber interactions led to the discovery of nerve growth factor (NGF) as the secreted attractive signal from the sarcoma 180 cells [Levi-Montalcini, 1952]. If transplanted tumor cells can influence cell movements in the host embryo, the question arises as to the extent the host cell migratory pathways can influence other migratory cell types. Early studies that investigated the influence of the chick embryonic NC microenvironment employed transplantation of a variety of migratory cell types into the avian trunk NC cell migratory pathway [Erickson et al., 1980]. When transplanted sarcoma 180 cells were analyzed after embryo re-incubation, the cells were distributed along normal trunk NC pathways and usually seen as individual cells; fibroblasts, however, remained at the transplant site [Erickson et al., 1980]. More recent work supports the hypothesis that adult tumor cells can travel along host embryo NC cell migratory pathways. Injection of human SK-Mel-28 (melanoma) cells or transplantation of C8161 aggressive melanoma cells into the chick embryo neural tube revealed that the tumor cells invaded the embryo along host NC cell cranial [Kulesa et al., 2006]; summarized in Fig. 2) and trunk [Schriek et al., 2005] migratory pathways, and did not form tumors. In contrast, when B16 mouse melanoma cells were injected into non-NC cell microenvironments of the chick embryo, including the eye cup, the tumor cells formed melanomas [Oppitz et al., 2007].
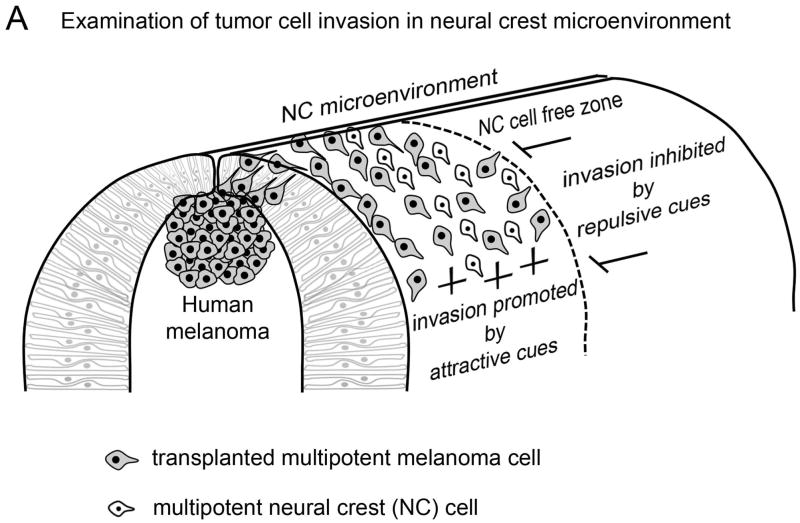
Figure 2. Examination of tumor cell invasion in neural crest microenvironment.
Human metastatic melanoma cells transplanted into the chick embryonic neural crest microenvironment follow host migratory pathways and avoid neural crest cell free zones, suggesting their invasion pattern may be influenced by neural crest microenvironmental signals.
Imaging strategies have begun to reveal an amazing complexity of melanoma and embryonic NC cell behaviors when visualized in vivo within developmental microenvironments [Kulesa et al., 2006; Kasemeier-Kulesa et al., 2008]. Interestingly, 3D confocal analysis of GFP-labeled C8161 aggressive melanoma cells within the chick embryo NC cell microenvironment revealed that tumor cells resembled host NC cell morphologies, including long filopodial extensions that contacted neighboring cells and formed follow-the-leader chain-like arrays [Kulesa et al., 2006]. When we investigated the positions of transplanted invading melanoma cells, our results showed that the tumor cells avoided typical NC cell-free zones and did not require a host NC cell scaffold to invade [Kulesa et al., 2006]. Also, the invading melanoma cells reached distant NC cell target sites in the head branchial arches and trunk sympathetic and dorsal root ganglia. This suggests that the chick embryonic NC microenvironment has a strong influence on adult tumor cell behaviors with an ancestral link to the NC. However, the extent to which the chick embryonic NC microenvironment influences the molecular invasion program of metastatic melanoma cells is still unclear, due in part to the challenge of analyzing changes in gene expression in small cell numbers.
Development of Gene Profiling for Small Cell Numbers
Whole transcriptome profiling approaches, such as RNAseq & microarray produce a wealth of information, but require large (>100pg) sample sizes [Ozsolak et al., 2010; Chen et al., 2011; Tariq et al., 2011]. In many instances, such as our transplantation of approximately 300 metastatic melanoma cells into the chick embryonic NC microenvironment, obtaining such a large amount of starting material from subpopulations of cells is not feasible. When sample size is limited, there are more directed analyses available to probe relevant genes. We describe recent advances in linear pre-amplification methods that have increased the number of genes that can be robustly analyzed from small amounts of starting material. We highlight application of this technology and its modifications to profile gene expression in metastatic melanoma cells transplanted into the chick embryo model and extracted at distinct temporal and spatial points of invasion.
In contrast to whole transcriptome profiling approaches, there is recent evidence of profiling single cells to highlight the variability within heterogeneous populations [Narsinh et al., 2011]. Profiling many single cells is commonly used to elucidate the range of expression within a population, but suffers from the increased number of cells required to produce accurate expression ranges. Another limitation is the soaring number of total reactions necessary for this broad type of analysis. We suggest that samples containing 10–50 cells strike a good balance between the sensitivity necessary to detect even the lowest expressed targets and the averaging inherent in larger populations of cells.
Several amplification protocols have been developed for expression profiling from small numbers of cells (Table 2). Many kits now eliminate the need for RNA purification, a step where template is commonly lost. Ambion’s Cells-to-Ct kit linearly amplifies cDNA using the same Taqman Gene Expression Assays employed during downstream RT-qPCR. A major advantage of ABI’s Taqman Gene Expression Assays is that each gene-specific primer set is pre-validated, a process which requires substantial time and resources. The concentration at which Taqman Assays are provided currently limits the total number of genes that can be pre-amplified per reaction for downstream RT-qPCR.
Table 2.
Methods for Profiling Small Numbers of Cells
| Method | Advantages | Disadvantages | References |
|---|---|---|---|
| Single-cell qPCR | Single cell required | Variance between cells, noise |
Taniguchi 2009 Wu 2010 |
| Single-cell RNAseq | Single cell required Transcriptome analysis | Variance between cells, noise | Tang 2009 |
| Gene-Specific PreAmp | Small sample required, pre-validated & linear amplification | Analysis of only 100 transcripts | Noutsias 2008 |
| Eberwine T7 RNA Amplification | Transcriptome analysis | Potential bias |
Van Gelder 1990 Duftner 2008 Boelens 2007 |
| RNAseq | Transcriptome analysis | Larger (>100pg) sample required |
Chen 2011 Ozsolak 2010 Tariq 2011 |
Laser capture microdissection (LCM) is a definitive method for precisely isolating single or small subpopulations of cells from complex tissue [Emmert-Buck et al., 1996; Simone et al., 1998]. This microscopy-based technique allows visualization of target tissue and harvested samples for complete confidence during sample collection. Tissue is prepared for LCM by flash freezing, essentially halting all cellular processes at the moment of tissue harvest, and cryosectioning to a single-cell thickness to avoid capture of overlapping, unwanted material. Compared with FACs purification, samples harvested by LCM better represent their in vivo biological context because they do not undergo enzymatic treatment or removal from their native microenvironment prior to isolation. Furthermore, FACs isolation is less efficient with smaller sample sizes, requiring a number of cells for calibration before isolation can begin.
We have combined tissue harvest by LCM with miniaturization of a gene-specific pre-amplification and microfluidic RT-qPCR on Fluidigm’s BioMark system to maximize the expression information gathered from small numbers of cells without sacrificing sensitivity (Fig. 3). Miniaturization of the RT-qPCR reaction on a dynamic array system requires less template and consumables per reaction, concurrently reducing the cost per RT-qPCR reaction and allowing more genes to be analyzed per sample. Proper analysis of raw expression profiling data is crucial to accurate interpretation of the data. We employ complex commercially available analysis software packages that feature data quality control measures, error propagation algorithms and endogenous control reference gene stability score calculation along with the field standard delta delta Ct method to accurately and reliably calculate relative quantities from raw expression data. Integrated downstream statistical analysis functions rapidly provide false discovery rate and confidence levels for all fold change data.
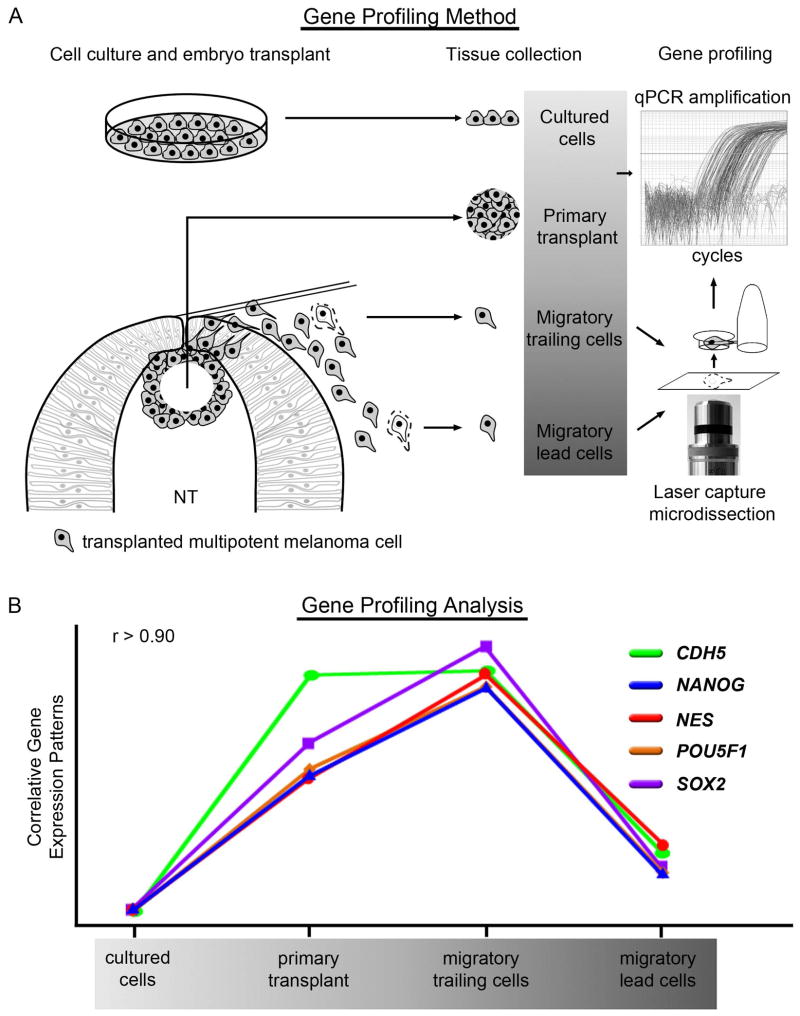
Figure 3. The combination of an in vivo model system and LCM-assisted gene profiling facilitates the identification of novel genetic interactions and putative signaling pathways.
A) Small numbers of cells were harvested from four distinct microenvironments: in vitro culture, the chick embryo dorsal neural tube (primary transplant site), and two phases of migration, trailing and lead. Cells were collected from embryonic tissue by laser capture microdissection and samples were analyzed by RT-qPCR. NT-neural tube. B) Gene expression patterns for the pluripotency markers NANOG, NES, POU5F1, and SOX2, along with the vascular endothelial cadherin CDH5, show a high degree of correlation. Gene expression patterns were derived from samples taken from the four microenvironments described above. r-values were determined using Pearson correlation.
Gene Profiling of Human Melanoma after Transplantation into the Chick Embryo
As stated above, one of the significant advantages of the chick embryo model system is access to the developing embryo, both for imaging and for molecular analysis. Using our LCM-assisted gene profiling strategy, we have interrogated gene expression profiles in human melanoma cells transplanted into the chick embryonic neural crest microenvironment. Because we hypothesize that metastatic melanoma cells exploit their ancestral relationship to the embryonic neural crest by hijacking neural crest genes associated with increased invasiveness and plasticity, we specifically examined a panel of genes postulated to play important roles in the neural crest developmental program. Small clusters (containing roughly 300 cells) of aggressive human melanoma cells (C8161) were transplanted into the dorsal neural tube at rhombomere 4 of Hamburger and Hamilton Stage 9–10 chick embryos in ovo [Hamburger and Hamilton, 1951]. This stage corresponds to the delamination and emigration of host neural crest cells in the hindbrain. Following transplantation, embryos were incubated for 24 hours, and consequently harvested and prepared for laser capture microdissection and subsequent gene expression analysis by RT-qPCR. The use of laser-assisted microdissection uniquely allowed us to capture both migrating and non-migrating melanoma cells at the same axial level with single cell resolution, and assay these cells for changes in gene expression resulting from altering microenvironments within the developing chick embryo.
To demonstrate the validity and versatility of our system, we examined the expression of CDH5 (VE-Cadherin) in transplanted melanoma cells. It was previously demonstrated that C8161 melanoma cells aberrantly express CDH5 and that VE-cadherin may play an important role in the metastatic process [Hendrix et al., 2001]. However, the mechanism through which aggressive melanoma cells aberrantly express CHD5 is unknown. In our model system, CDH5 is induced in C8161 cells following transplantation. Further induction is observed as cells delaminate from the primary transplant and exit the dorsal neural tube. However, CDH5 expression begins to regress as cells migrate away from the neural tube (Fig. 3). Importantly, a comparison of genes whose expression patterns correlate with the expression pattern observed for CHD5 revealed four genes with high correlation: NANOG, NES, POU5F1, and SOX2 (Fig. 3). These four genes are common markers of dedifferentiation and pluripotency. Like CDH5, these four genes exhibited exclusive down-regulation in lead migratory cells compared to trailing cells within close proximity to the neural tube. These changes to gene expression are likely the result of distinct microenvironmental cues, and would not be observed in vitro. Furthermore, the high correlation observed for the regulation of these genes suggests a mechanistic link between dedifferentiation and VE-cadherin expression.
Importantly, the down-regulation observed in NANOG, NES, POU5F1, and SOX2 in migrating melanoma cells succeeded the up-regulation of these markers initially observed following transplantation into the lumen of the neural tube. This suggests that the embryonic microenvironment within the neural tube promotes a dedifferentiated state. Furthermore, the down-regulation of plasticity genes was most evident when directly comparing lead migratory cells versus trailing cells that had recently exited the neural tube. This suggests that dedifferentiation may facilitate exit from the neural tube in a manner analogous to neural crest delamination, for which EMT is a requisite step [Taneyhill et al., 2007]. Then, as cells begin to migrate away from the neural tube, the loss of potent signals promoting stemness is manifest in the reduction of pluripotency markers.
Notably, mesenchymal tissue through which neural crest cells emigrate may also promote differentiation [Derby, 1982; Derby and Newgreen, 1982]. Likewise, as melanoma cells invade along neural crest routes, they may also be exposed to powerful differentiation cues, reflected by a significant decrease in the expression of pluripotency markers. This is supported by our previous findings demonstrating the re-expression of Melan A in a subset of migratory C8161 cells within our model system. Under normal culture conditions, C8161 cells are negative for Melan A gene expression. Thus, the re-expression of this marker of melanocyte differentiation suggests reprogramming toward a more differentiated state [Kulesa et al., 2006]. Indeed, the ability of melanoma cells to alter their differentiation status in concert with cell invasion has been recently documented [Giampieri et al., 2010]. Taken together, these results clearly demonstrate the dynamic regulation of gene expression with respect to changing microenvironmental signals and support the hypothesis that melanoma invasion and metastasis are facilitated by their heightened ability to sense and respond to changing microenvironments.
Perspectives
The microenvironment exerts control over the genotype and phenotype of both normal and cancer cells and plays a crucial role in the determination of cell plasticity and invasion. Clues for new therapeutic strategies may come from studies of this relationship. Yet, what is particularly challenging to the development of effective therapies is the plasticity of tumor cells. That is, tumor cell plasticity suggests high adaptivity and may represent the difference ina negative versus positive outcome.
Compelling evidence presented in the literature detailing the molecular signature of multipotent tumor cells, as well as embryonic stem and progenitor cells, demonstrate the underlying plasticity of these cell types. For example, comparative global gene analyses of aggressive and poorly aggressive human melanoma cell lines have revealed that the aggressive tumor cells express genes (and proteins) associated with multiple cellular phenotypes (including progenitor and stem cells), while simultaneously downregulating genes specific to their parental melanocytic lineage [Postovit et al., 2007]. Particularly noteworthy is evidence that molecules that play a role in embryonic NC cell guidance have common expression in highly aggressive and non-aggressive tumor cells [Kasemeier-Kulesa et al., 2008]. Aggressive melanoma cells upregulate a subset of both NC cell guidance and differentiation markers compared to their non-aggressive counterpart [Kasemeier-Kulesa et al., 2008]. Also, some aggressive melanoma cells that show a significant downregulation in pigmentation pathway-associated genes, and this dedifferentiated phenotype has been correlated with a poor clinical outcome (for review, see [Hendrix et al., 2007]). Therefore, it is tempting to speculate that a viable strategy for redifferentiating the aggressive, plastic melanoma cell phenotype back to a melanocyte-like cell, might lead to the suppression of metastasis.
New technology advances in in vivo imaging and gene profiling provided three significant advantages of analyzing human melanoma cells transplanted into an embryonic chick model. First, in vivo dynamic imaging using multiphoton microscopy of transplanted clumps of melanoma cells allowed single cell resolution of cell behaviors during the cell sorting within and separation from the transplanted cell cluster, as well as invasion into the chick embryonic NC microenvironment. Second, LCM and RT-qPCR permitted the extraction of single melanoma cells and gene expression readout of phenotypically matched sub-populations during the progressive stages of metastasis into the chick embryonic NC microenvironment. Recent advances in epigenetic profiling suggest small numbers of LCM-harvested cells may also be amenable to DNA methylation analysis by bisulfite sequencing [Paliwal et al., 2010]. Third, the direct comparison of NC and tumorigenic genes in a spatio-temporal manner allowed us to build a progressive picture of the dynamics of melanoma invasion.
Our studies provide insights into how a melanoma cell’s gene expression profile changes as a system. Highly correlative changes in gene expression observed during the course of tumor cell invasion offer the potential for identification of novel genetic interactions and signaling pathways. For example, our data demonstrated the dynamic, correlative regulation of pluripotency markers with relation to the proximity to the embryonic neural tube. Interestingly, it has been recently shown that melanoma cells taken from both primary and metastatic lesions, then grown as spheroid cultures under NC cell conditions displayed a NC cell transcriptional signature associated with high transdifferentiation capacity and similar enhanced expression of pluripotency genes examined in our study [Ramgolam et al., 2011]. Importantly, although melanoma spheroid cultures displayed higher cell plasticity, these cells did not appear to revert to a NC stem cell state [Ramgolam et al., 2011]. These data support the hypothesis that signals within the chick neural tube may prime melanoma cell plasticity and invasiveness, and that these conditions may be partially mimicked by NC culture conditions. These findings strengthen the possibility of identifying potent embryonic signals that can regulate the tumor cell phenotype.
In summary, the concept of embryonic models to work at the interface of development and cancer has ancient roots. But, when combined with new technology advances, these models provide multi-scale information on how the cancer cell behaves in a foreign microenvironment and have the potential for exciting discoveries of the boundaries of tumor cell plasticity and invasion. This increased wealth of information about tumor cell behaviors is exciting and at the same time incredibly challenging to interpret. Traditional gain- and loss-of-function studies may need to adapt from single gene to multiple gene perturbation to effectively analyze the system. Multispectral imaging approaches will need to also adapt to allow visualization of multiple gene perturbations simultaneously, highlighted by distinct fluorescent reporters. Lastly, a major challenge will be how to translate insights gained from tumor cell behaviors within the embryonic microenvironment to the human tumor microenvironment.
Acknowledgments
We gratefully acknowledge the laboratory of Dr. Mary Hendrix for providing the C8161 melanoma cell lines used in these experiments and for thoughtful and constructive discussion. CMB thanks the National Institutes of Health Ruth L. Kirschtein Postdoctoral Fellowship Program (Award Number F32CA144297) for funding and partial support from the Stowers Institute for Medical Research. PMK thanks the kind generosity of the Stowers Institute for Medical Research for funding and the American Association of Anatomists Travel Bursary for partial support to attend the TEMTIA-V meeting.
List of Abbreviations
- NC
neural crest
- EMT
epithelial-to-mesenchymal transition
- LCM
laser capture microdissection
- RT-qPCR
reverse transcription quantitative polymerase chain reaction
- NGF
nerve growth factor
- FACS
fluorescence-activated cell sort
References
- Abbott DE, Bailey CM, Postovit LM, Seftor EA, Margaryan N, Seftor RE, Hendrix MJ. The epigenetic influence of tumor and embryonic microenvironments: how different are they? Cancer Microenviron. 2008;1(1):13–21. doi: 10.1007/s12307-008-0004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119(6):1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander S, Koehl GE, Hirschberg M, Geissler EK, Friedl P. Dynamic imaging of cancer growth and invasion: a modified skin-fold chamber model. Histochem Cell Biol. 2008;130(6):1147–1154. doi: 10.1007/s00418-008-0529-1. [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132(14):3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. Annu Rev Cell Dev Biol. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilozur ME, Hay ED. Neural crest migration in 3D extracellular matrix utilizes laminin, fibronectin, or collagen. Dev Biol. 1988;125(1):19–33. doi: 10.1016/0012-1606(88)90055-3. [DOI] [PubMed] [Google Scholar]
- Blanco MJ, Moreno-Bueno G, Sarrio D, Locascio A, Cano A, Palacios J, Nieto MA. Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene. 2002;21(20):3241–3246. doi: 10.1038/sj.onc.1205416. [DOI] [PubMed] [Google Scholar]
- Boelens MC, te Meerman GJ, Gibcus JH, Blokzijl T, Boezen HM, Timens W, Postma DS, Groen HJ, van den Berg A. Microarray amplification bias: loss of 30% differentially expressed genes due to long probe - poly(A)-tail distances. BMC Genomics. 2007;8:277. doi: 10.1186/1471-2164-8-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116(Pt 3):499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2(2):76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Chen H, Liu Z, Gong S, Wu X, Taylor WL, Williams RW, Matta SG, Sharp BM. Genome-Wide Gene Expression Profiling of Nucleus Accumbens Neurons Projecting to Ventral Pallidum Using both Microarray and Transcriptome Sequencing. Front Neurosci. 2011;5:98. doi: 10.3389/fnins.2011.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis J, Weissleder R. In vivo imaging in cancer. Cold Spring Harb Perspect Biol. 2010;2(12):a003848. doi: 10.1101/cshperspect.a003848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EM, Trinkaus JP. Significance of cell-to cell contacts for the directional movement of neural crest cells within a hydrated collagen lattice. J Embryol Exp Morphol. 1981;63:29–51. [PubMed] [Google Scholar]
- Derby MA. Environmental factors affecting neural crest differentiation: melanocyte differentiation by crest cells exposed to cell-free (deoxycholate-extracted) dermal mesenchyme matrix. Cell Tissue Res. 1982;225(2):379–386. doi: 10.1007/BF00214690. [DOI] [PubMed] [Google Scholar]
- Derby MA, Newgreen DF. Differentiation of avian neural crest cells in vitro: absence of a developmental bias toward melanogenesis. Cell Tissue Res. 1982;225(2):365–378. doi: 10.1007/BF00214689. [DOI] [PubMed] [Google Scholar]
- Duftner N, Larkins-Ford J, Legendre M, Hofmann HA. Efficacy of RNA amplification is dependent on sequence characteristics: implications for gene expression profiling using a cDNA microarray. Genomics. 2008;91(1):108–117. doi: 10.1016/j.ygeno.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274(5289):998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- Erickson CA, Tosney KW, Weston JA. Analysis of migratory behavior of neural crest and fibroblastic cells in embryonic tissues. Dev Biol. 1980;77(1):142–156. doi: 10.1016/0012-1606(80)90462-5. [DOI] [PubMed] [Google Scholar]
- Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147(5):992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Fukumura D, Duda DG, Munn LL, Jain RK. Tumor microvasculature and microenvironment: novel insights through intravital imaging in pre-clinical models. Microcirculation. 2010;17(3):206–225. doi: 10.1111/j.1549-8719.2010.00029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammill LS, Roffers-Agarwal J. Division of labor during trunk neural crest development. Dev Biol. 2010;344(2):555–565. doi: 10.1016/j.ydbio.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giampieri S, Pinner S, Sahai E. Intravital imaging illuminates transforming growth factor beta signaling switches during metastasis. Cancer Res. 2010;70(9):3435–3439. doi: 10.1158/0008-5472.CAN-10-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldin CE, LaBonne C. SoxE factors as multifunctional neural crest regulatory factors. Int J Biochem Cell Biol. 2010;42(3):441–444. doi: 10.1016/j.biocel.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton H. A series of normal stages in the development of the chick embryo. Journal of Morphology. 1951;88:49–92. [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Meltzer PS, Gardner LM, Hess AR, Kirschmann DA, Schatteman GC, Seftor RE. Expression and functional significance of VE-cadherin in aggressive human melanoma cells: role in vasculogenic mimicry. Proc Natl Acad Sci U S A. 2001;98(14):8018–8023. doi: 10.1073/pnas.131209798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Seftor RE, Kasemeier-Kulesa J, Kulesa PM, Postovit LM. Reprogramming metastatic tumour cells with embryonic microenvironments. Nat Rev Cancer. 2007;7(4):246–255. doi: 10.1038/nrc2108. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K. Embryonic stem cells: testing the germ-cell theory. Curr Biol. 2011;21(20):R850–852. doi: 10.1016/j.cub.2011.09.024. [DOI] [PubMed] [Google Scholar]
- Kasemeier-Kulesa JC, Teddy JM, Postovit LM, Seftor EA, Seftor RE, Hendrix MJ, Kulesa PM. Reprogramming multipotent tumor cells with the embryonic neural crest microenvironment. Dev Dyn. 2008;237(10):2657–2666. doi: 10.1002/dvdy.21613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedrin D, Gligorijevic B, Wyckoff J, Verkhusha VV, Condeelis J, Segall JE, van Rheenen J. Intravital imaging of metastatic behavior through a mammary imaging window. Nat Methods. 2008;5(12):1019–1021. doi: 10.1038/nmeth.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesa PM, Bailey CM, Kasemeier-Kulesa JC, McLennan R. Cranial neural crest migration: new rules for an old road. Dev Biol. 2010;344(2):543–554. doi: 10.1016/j.ydbio.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesa PM, Gammill LS. Neural crest migration: patterns, phases and signals. Dev Biol. 2010;344(2):566–568. doi: 10.1016/j.ydbio.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesa PM, Kasemeier-Kulesa JC, Teddy JM, Margaryan NV, Seftor EA, Seftor RE, Hendrix MJ. Reprogramming metastatic melanoma cells to assume a neural crest cell-like phenotype in an embryonic microenvironment. Proc Natl Acad Sci U S A. 2006;103(10):3752–3757. doi: 10.1073/pnas.0506977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama S, Mayor R. Molecular analysis of neural crest migration. Philos Trans R Soc Lond B Biol Sci. 2008;363(1495):1349–1362. doi: 10.1098/rstb.2007.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin NM, Kalcheim C. The Neural Crest. Cambridge: Cambridge University Press; 1999. [Google Scholar]
- Levi-Montalcini R. Effects of mouse tumor transplantation on the nervous system. Ann N Y Acad Sci. 1952;55(2):330–344. doi: 10.1111/j.1749-6632.1952.tb26548.x. [DOI] [PubMed] [Google Scholar]
- Mayor R, Carmona-Fontaine C. Keeping in touch with contact inhibition of locomotion. Trends Cell Biol. 2010;20(6):319–328. doi: 10.1016/j.tcb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz B, Illmensee K. Normal genetically mosaic mice produced from malignant teratocarcinoma cells. Proc Natl Acad Sci U S A. 1975;72(9):3585–3589. doi: 10.1073/pnas.72.9.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsinh KH, Sun N, Sanchez-Freire V, Lee AS, Almeida P, Hu S, Jan T, Wilson KD, Leong D, Rosenberg J, Yao M, Robbins RC, Wu JC. Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells. J Clin Invest. 2011;121(3):1217–1221. doi: 10.1172/JCI44635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol. 2011;27:347–376. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- Noutsias M, Rohde M, Block A, Klippert K, Lettau O, Blunert K, Hummel M, Kuhl U, Lehmkuhl H, Hetzer R, Rauch U, Poller W, Pauschinger M, Schultheiss HP, Volk HD, Kotsch K. Preamplification techniques for real-time RT-PCR analyses of endomyocardial biopsies. BMC Mol Biol. 2008;9:3. doi: 10.1186/1471-2199-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppitz M, Busch C, Schriek G, Metzger M, Just L, Drews U. Non-malignant migration of B16 mouse melanoma cells in the neural crest and invasive growth in the eye cup of the chick embryo. Melanoma Res. 2007;17(1):17–30. doi: 10.1097/CMR.0b013e3280114f49. [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Goren A, Gymrek M, Guttman M, Regev A, Bernstein BE, Milos PM. Digital transcriptome profiling from attomole-level RNA samples. Genome Res. 2010;20(4):519–525. doi: 10.1101/gr.102129.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliwal A, Vaissiere T, Herceg Z. Quantitative detection of DNA methylation states in minute amounts of DNA from body fluids. Methods. 2010;52(3):242–247. doi: 10.1016/j.ymeth.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Postovit LM, Costa FF, Bischof JM, Seftor EA, Wen B, Seftor RE, Feinberg AP, Soares MB, Hendrix MJ. The commonality of plasticity underlying multipotent tumor cells and embryonic stem cells. J Cell Biochem. 2007;101(4):908–917. doi: 10.1002/jcb.21227. [DOI] [PubMed] [Google Scholar]
- Ramgolam K, Lauriol J, Lalou C, Lauden L, Michel L, de la Grange P, Khatib AM, Aoudjit F, Charron D, Alcaide-Loridan C, Al-Daccak R. Melanoma spheroids grown under neural crest cell conditions are highly plastic migratory/invasive tumor cells endowed with immunomodulator function. PLoS One. 2011;6(4):e18784. doi: 10.1371/journal.pone.0018784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real C, Glavieux-Pardanaud C, Le Douarin NM, Dupin E. Clonally cultured differentiated pigment cells can dedifferentiate and generate multipotent progenitors with self-renewing potential. Dev Biol. 2006;300(2):656–669. doi: 10.1016/j.ydbio.2006.09.032. [DOI] [PubMed] [Google Scholar]
- Ridenour DA, McKinney MC, Bailey CM, Kulesa PM. CycleTrak: A novel system for the semi-automated analysis of cell cycle dynamics. Dev Biol. 2012 doi: 10.1016/j.ydbio.2012.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E, Wyckoff J, Philippar U, Segall JE, Gertler F, Condeelis J. Simultaneous imaging of GFP, CFP and collagen in tumors in vivo using multiphoton microscopy. BMC Biotechnol. 2005;5:14. doi: 10.1186/1472-6750-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schriek G, Oppitz M, Busch C, Just L, Drews U. Human SK-Mel 28 melanoma cells resume neural crest cell migration after transplantation into the chick embryo. Melanoma Res. 2005;15(4):225–234. doi: 10.1097/00008390-200508000-00001. [DOI] [PubMed] [Google Scholar]
- Simone NL, Bonner RF, Gillespie JW, Emmert-Buck MR, Liotta LA. Laser-capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet. 1998;14(7):272–276. doi: 10.1016/s0168-9525(98)01489-9. [DOI] [PubMed] [Google Scholar]
- Stewart RA, Lee JS, Lachnit M, Look AT, Kanki JP, Henion PD. Studying peripheral sympathetic nervous system development and neuroblastoma in zebrafish. Methods Cell Biol. 2010;100:127–152. doi: 10.1016/B978-0-12-384892-5.00005-0. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Okita K, Nakagawa M, Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc. 2007a;2(12):3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007b;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Taneyhill LA, Coles EG, Bronner-Fraser M. Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development. 2007;134(8):1481–1490. doi: 10.1242/dev.02834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- Taniguchi K, Kajiyama T, Kambara H. Quantitative analysis of gene expression in a single cell by qPCR. Nat Methods. 2009;6(7):503–506. doi: 10.1038/nmeth.1338. [DOI] [PubMed] [Google Scholar]
- Tariq MA, Kim HJ, Jejelowo O, Pourmand N. Whole-transcriptome RNAseq analysis from minute amount of total RNA. Nucleic Acids Res. 2011;39(18):e120. doi: 10.1093/nar/gkr547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topczewska JM, Postovit LM, Margaryan NV, Sam A, Hess AR, Wheaton WW, Nickoloff BJ, Topczewski J, Hendrix MJ. Embryonic and tumorigenic pathways converge via Nodal signaling: role in melanoma aggressiveness. Nat Med. 2006;12(8):925–932. doi: 10.1038/nm1448. [DOI] [PubMed] [Google Scholar]
- Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci U S A. 1990;87(5):1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff JB, Jones JG, Condeelis JS, Segall JE. A critical step in metastasis: in vivo analysis of intravasation at the primary tumor. Cancer Res. 2000;60(9):2504–2511. [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart Slukvin R, II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang D, I, Brinas M, Binder BJ, Landman KA, Newgreen DF. Neural crest regionalisation for enteric nervous system formation: implications for Hirschsprung’s disease and stem cell therapy. Dev Biol. 2010;339(2):280–294. doi: 10.1016/j.ydbio.2009.12.014. [DOI] [PubMed] [Google Scholar]
- Zhang L, Alt C, Li P, White RM, Zon LI, Wei X, Lin CP. An optical platform for cell tracking in adult zebrafish. Cytometry A. 2012;81(2):176–182. doi: 10.1002/cyto.a.21167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Wang X, Zhao Y, Li Z, Lin S, Wei Y, Yang H. A novel xenograft model in zebrafish for high-resolution investigating dynamics of neovascularization in tumors. PLoS One. 2011;6(7):e21768. doi: 10.1371/journal.pone.0021768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH. Atomic resolution cryo electron microscopy of macromolecular complexes. Adv Protein Chem Struct Biol. 2011;82:1–35. doi: 10.1016/B978-0-12-386507-6.00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]