Abstract
Exosomes biomarkers mediating important biological process, especially in the systemic disease diagnostics and therapeutics, yet the protective exosomal vesicle structure hinders rapid, simple detection of the harbored molecules. We have established a new method, the electric field-induced release and measurement (EFIRM), which can simultaneously disrupt exosomes to release the contents and on-site monitoring the harbored exosomal RNA/proteins biomarkers. When exposed to a nonuniform electrical field, exosomal RNA and proteins are rapidly released. Bio-recognition of these biomolecules is carried out concurrently. We tested the hypothesis that the lung cancer cell line, H460 stably transfected with hCD63-GFP, would shed hCD63-GFP expressing exosomes that could be detected in serum and saliva. We confirmed in vivo that H460-CD63-GFP shed exosomes were transported to blood and saliva. This result demonstrates for the first time tumor-shed exosomes were detected in saliva, in addition to blood, presenting a new translational utility of exosome-based biomarker detection in saliva.
Keywords: Exosome, Electrochemical sensors, Tumor biomarkers, Lung cancer, Salivary diagnostics
1. Introduction
Exosome research, based on detection of encapsulated biomarkers, has shown great potential in disease therapeutics and diagnostics (personalized medicine) (Rosell et al., 2009). Exosomes are lipid-encapsulated microvesicles that are released by various cells into body fluids (Fevrier and Raposo, 2004; Smalheiser, 2007; Stoorvogel et al., 2002; Valadi et al., 2007; van Niel et al., 2006), including blood (Taylor and Gercel-Taylor, 2010), urine (Miranda et al., 2010; Pisitkun et al., 2004), milk (Johansson et al., 2006), and saliva (Lau and Wong, 2012; Palanisamy et al., 2010; Sharma et al., 2010). By remote communication via the harbored bio-molecules, exosomes regulate a variety of cellular pathways in recipient cells (Gibbings et al., 2009; Schorey and Bhatnagar, 2008; Smalheiser, 2007; van Niel et al., 2006) that relate to the progression and prognosis of disease conditions. The particular lipid composition of the vesicle and the presence of the harbored protein, nucleic acid, and other constituents confer protection to the vesicle against degradation and contribute to its stability in the extracellular environment (Houseley et al., 2006; Schmid and Jensen, 2008). Therefore, for translational and clinical applications, exosome diagnostics requires: (1) rapid, exosome-specific extraction, (2) simple, effective biomarker release from exosomes, and (3) highly sensitive, specific detection of endogenous bio-molecules. Conventional exosome diagnostic methods comprise a two-step procedure, including exosome extraction from body fluids, followed by the specific detection of harbored biomarkers that are associated with a variety of diseases (Chen et al., 2010). These technologies are labor-intensive and may be complicated by interference. To increase our understanding of exosome-associated biology and cellular communicative processes, there is an urgent need for a rapid, simple, accurate, real-time method for monitoring exosome-harbored biomolecules.
The vesiclar structure of exosomes hinders the detection of exosome-harbored bio-molecules. Therefore, for detection and quantification, it is necessary to release the harbored bio-molecules. Detection sensitivity is closely related to the efficiency of the releasing process. One of the traditional methods for releasing the vesicle encapsulated constituents is to apply lysis reagents. The lysis reaction time typically varies from several minutes to several hours. This lysis process may compromise the stability of the bio-molecules after release from the exosome into the lysis milieu. The rapid degradation of released bio-molecules implicates a need for detection technologies that can make accurate assessments within seconds to minutes. Our previous studies showed that mRNA levels were reduced to less than 40% within 1 min of release from macromolecules into saliva (Park et al., 2006). Stabilization reagents are therefore necessary for accurate exosome biomarker detection. The combination of exosome releasing agents and stabilization reagents renders the process complicated and in addition, may introduce interference.
An electric field, particularly one with a non-uniform profile, can stimulate vesicle deformation in biological samples and direct the flow of the released bio-molecules (Wei et al., 2009a, 2006). An electric field can cause redistribution or polarization of lipid vesicular structures that protect bio-molecules. The non-uniform electrical field will either rupture the membrane or disrupt the tertiary structure of the exosomal lipid bilayer, which causes temporary pore formation; in both cases, the harbored bio-molecules can be released. By applying a special cyclic square wave electrical field (csw E-field), we have shown that mRNA could be released directly from exosome in the saliva sample (Wei et al., 2009a). The total release process lasted approximately 200 s. In addition, unlike the high voltage associated with a direct current electric field, the csw E-field generates voltage on the order of several hundred mV, which is non-disruptive for most biomolecules.
In this paper, we report the development a method for directly detecting harbored exosomal bio-molecules with electrical field-induced release and measurement (EFIRM). We utilized affinity-CD63-based magnetic beads to extract the exosomes from biological samples. A csw E-field is then applied to release, collect, and incubate the exosome-harbored biomolecules (Wei et al. 2009a, b). To demonstrate the analytical performance of EFIRM, we performed detection of exosomal GAPDH mRNA, a housekeeping gene mRNA present in most exosomes and the stably transfected hCD63-GFP fusion protein expressed as an exosomal surface protein marker.
2. Materials and methods
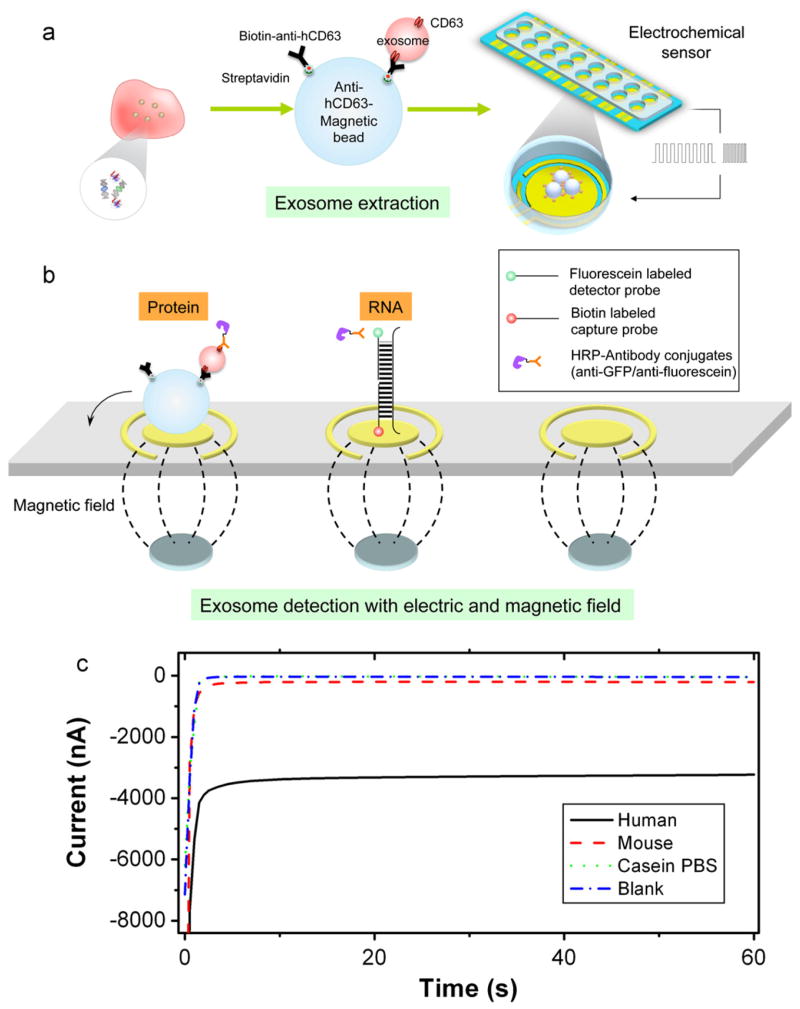
The technology described here integrated exosome extraction enhanced with electrical field and magnetic bead combined with simultaneous biomolecule release and detection. The detection was performed with amperometric electrochemical sensor technology (Wei et al., 2009b) based on an array of 16 bare gold electrode chips and workstation (GeneFluidics, USA). Each unit of the array had a working electrode, a counter electrode, and a reference electrode, which are all bare gold prior the treatment. The entire procedure is illustrated in Scheme 1 and the detailed experimental procedure is described in the supplementary documents.
Scheme 1.
Schematic of the electric field-induced release and measurement (EFIRM) system. (a) Anti-hCD63 antibodies conjugation and exosome extraction; (b) magnetic force assisted exosome extraction and electric filed induced release and (c) amperometric readout of the anti-human CD63 beads with human, mouse, casein PBS and blank samples. Signals were readout by anti-hCD63-HRP.
2.1. Magnetic bead-based exosome extraction
For rapid, effective extraction, magnetic bead-based technology was applied to isolate and accumulate exosomes. The magnetic beads (Invitrogen, USA) were approximately 1–2 μm in diameter, measured by TEM. Briefly, the streptavidin-coated magnetic beads (Invitrogen, USA) were mixed with biotinylated anti-hCD63, which is specific to human CD63 (Ancell, USA) on a mixer for 30 min at room temperature. Then, 10 μL of samples, including serum, or saliva were incubated with the anti-hCD63-conjugated magnetic beads in casein-PBS (Invitrogen, USA). Samples were mixed for 2 h at room temperature to form exosome–magnetic bead complexes. Next, exosome–magnetic bead complexes were attracted onto the electrochemical sensor by applying an array of magnets. The unattached species were removed by washing.
2.2. Exosomal GAPDH mRNA release and measurement
For GAPDH mRNA detection, the electrodes were pre-coated with a matrix of conducting polymer. The surface of the electrode was pre-coated with oligonucleotide capture probes with sequences specific for human GAPDH and a biotin label at the 5′ end. The immobilization of the capture probe was carried out in an electrochemical polymerization by applying a csw E-field for 20 cycles of 9 s at −300 mV followed by 1 s at +200 mV (200 s total) (Wei et al., 2009b). After the exosome extraction step, the exosome–magnetic bead complex was collected onto the capture probe-coated electrode with an array of magnets placed underneath the electrochemical sensor. When loading the sample onto the electrode, 10 nM of detector probe (with a fluorescein-labeled 3′ end) was mixed in with the exosome–magnetic bead complexes. The csw E-field was then applied to release the harbored GAPDH mRNA from the exosomes. The csw-E field was 20 cycles of 9 s at −300 mV followed by 1 s at +200 mV (200 s total).
Capture probe: 5′-Biotin-AGGTCCACCACTGACACGTTG
Detector probe: 5′-GCAGTGGGGACACGGAAGGCC-Fluorescein-3′
In parallel, samples from the same batches were lysed with 0.5% Triton-X 100 (Sigma, USA) for 20 min at room temperature to compare release and protection efficiencies.
The released mRNA hybridized with the oligonucleotide capture and detector probes. Then, we added 150 unit/ml of anti-fluorescein antibody conjugated to horseradish peroxidase (HRP; 1:1000 dilution; Roche, USA). Finally, the 3,3′,5,5′-tetramethyl-benzidine (TMB) substrate for horseradish peroxidase was loaded, and an amperometric signal was readout. (Scheme 1c)
2.3. EFIRM measurements of exosomal hCD63-GFP
For the hCD63-GFP protein detection, there was no need to disrupt the endosome, because the protein was membrane-bound and the GFP portion was accessible to the medium. Therefore, the rabbit anti-GFP antibody conjugated to HRP (Invitrogen, USA) was mixed with the exosome-hCD63-magnetic bead complex in solution for 1 h at room temperature. After washing, the mixture was collected on the electrodes with the applied magnetic field. Then, the TMB substrate was added, and amperometric measurements were carried out. GFP protein levels are expressed relative to the negative control. This was calculated as the ratio between the sample and the blank casein PBS buffer.
3. Results and discussion
3.1. Affinity capture of exosomes by immuno anti-CD63-magnetic beads
Effective exosome extraction requires efficiency and specificity. Magnetic beads are efficient tool for exosome extraction, because they facilitate exosome capturing and isolation via exosome surface biomarkers. We conjugate anti-human CD63 antibody (hCD63) to magnetic beads to capture exosomes that expressed exosome membrane-expressed CD63 (Schorey and Bhatnagar, 2008) (Scheme 1).
Transmission electron microscopy (TEM) was used to examine exosomes after extraction from human saliva with anti-hCD63 antibody-conjugated magnetic beads (Fig. 1a and b). CD63 is a typical exosomal surface marker (Denzer et al., 2000b; Escola et al., 1998). The TEM images showed exosomes located on the surface of the magnetic beads. The sizes of the extracted particles ranged from 70 to 100 nm, consistent with the size distribution of typical exosomes (Johansson et al., 2006; Ogawa et al., 2008; Schorey and Bhatnagar, 2008) (Fig. 1b). The exosome extraction efficiency was approximately 85%, determined by comparing the levels of hCD63 found in extracted exosomes to the levels found in exosome-depleted human saliva (Supplementary Fig. 1S). To demonstrate the specificity of the magnetic bead-based exosome extraction, anti-hCD63 coated magnetic beads were applied to mouse saliva, which lacks the hCD63 protein. The TEM results showed no mouse-saliva exosomes attached to the beads (Fig. 1c). Furthermore, when beads were coated with streptavidin only without anti-hCD63, no structures resembling exosomes were observed (Fig. 1d).
Fig. 1.
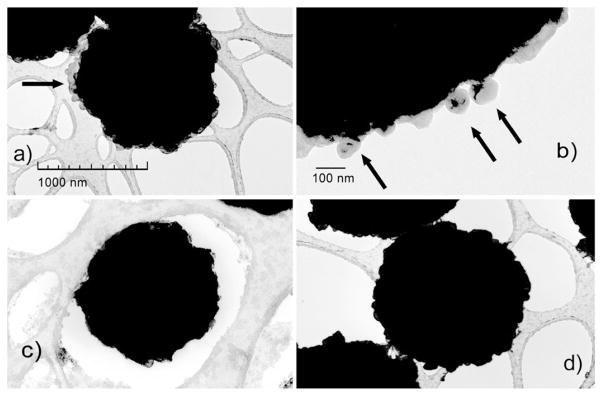
Transmission electron microscopy (TEM) images of magnetic bead based exosome extraction with (a) exosomes (arrows) from human saliva with anti-hCD63-conjugated magnetic beads; (b) magnified view of human exosomes (arrows) on a bead; (c) mouse saliva after extraction with no apparent exosome and (d) human saliva after extraction with magnetic beads coated only with streptavidin (negative control).
The traditional methods for extracting exosomes from body fluids are based on ultra-centrifugation or precipitation (Gonzalez-Begne et al., 2009; Palanisamy et al., 2010). Multiple steps of ultra-centrifugation can isolate vesicles with the size characteristics of exosomes with additional bio-imaging or molecular biology analyses. Recently, other methodologies have been developed, including precipitation combined with centrifugation (Taylor and Gercel-Taylor, 2010). Those technology are not based on specificity and are time consuming, usually takes several hours to several days.
In contrast, the magnetic bead based extraction is based on affinity capture of exosome-specific markers, like CD63, which appear on most types of exosomes (Denzer et al., 2000a). The total exosome-extraction time (from sample loading) was 30 min at room temperature. The EFIRM has major advantages over the conventional ultra-centrifugation and precipitation based methodologies. For exosomes without the surface marker CD63, other capture probes can be attached to the magnetic bead, like exosome-specific lectin (Caby et al., 2005; Kesimer et al., 2009; Wubbolts et al., 2003) (also see Supplementary Fig. 3S).
3.2. Release of harbored mRNA from human saliva exosomes via csw electrical field
Previous studies suggested that the vesicle structure of exosomes protects exosomal mRNA from degradation agents in the surrounding mileu (Park et al., 2009). For example, Triton-X 100 has been shown to disrupt the integrity of salivary exosomes, but this leads to endogenous RNA degradation by RNases in the saliva matrix (Park et al., 2006). Therefore, a rapid release process and immediate detection are necessary to effectively measure and quantify exosomal mRNA. We tested whether the csw E-field would facilitate the release and detection of RNA from human saliva exosomes (Fig. 2a). For comparison, the release efficiency was measured using Triton-X 100-based lysis carried out on the same human saliva exosomes, captured by the same magnetic beads, and performed in parallel with the csw E-field assay. We probed for GAPDH mRNA, because it is present in most exosomes. The released RNA would hybridize to a DNA probe complementary to the mRNA target, and the amount would be measured with a quick readout. The results would provide proof-of-concept that EFIRM technology could be used to detect exosome-harbored mRNA, or not.
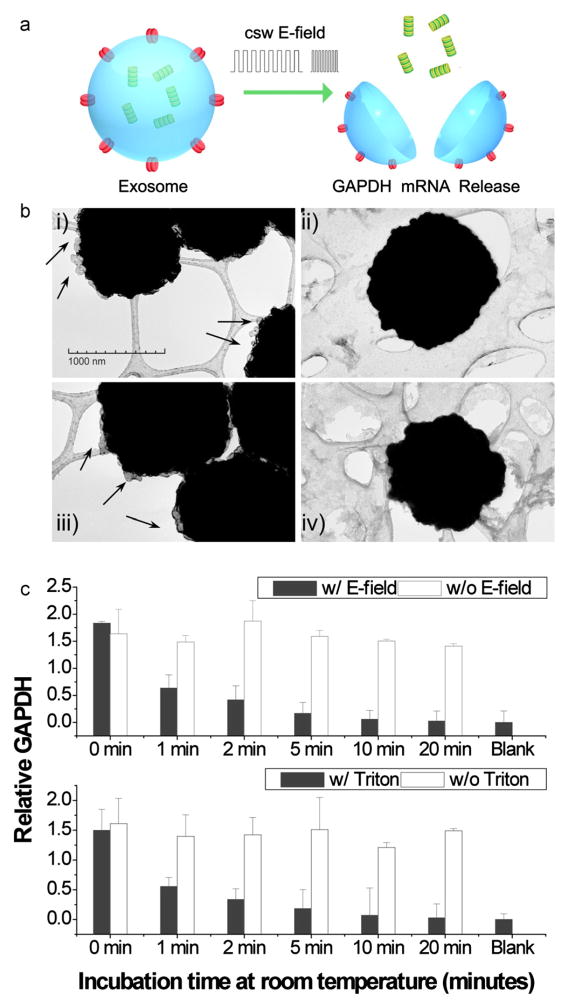
Fig. 2.
E-field and Triton X-100 lysis compared for releasing exosomal GAPDH mRNA from human saliva. (a) Schematic illustration of an exosome disrupted with E-field and RNA released. (b) TEM images before (left) and after (right) E-field (top) or Triton X-100 (bottom) treatment. (i and iii) Exosomes (arrows) attached to anti-hCD63 magnetic beads; (ii and iv) Exosomes were disrupted after treating with (ii) a csw E-field for 200 s, or (iv) with Triton-X 100 for 20 min. The background is the lacey support film for TEM. (c) GAPDH mRNA by EFIRM (top) or Triton X-100 (bottom).
The exosome–magnetic bead complexes were examined by TEM before and after treatment with the csw E-field or Triton-X 100 (Fig. 2b). After both treatments, the exosomes on the beads were no longer observed. Measurements of released RNA (filled bars) are presented as the ratio between the mRNA signal and the blank control (casein PBS buffer with no exosomes). Positive controls (without [w/o] electrical field treatment in the release step, open bars) were undegraded mRNA measured in untreated exosome samples; therefore, these mRNAs were not exposed to saliva enzymes. The levels of GAPDH mRNAs were measured at different time points by EFIRM after the application of electrical field or Triton X-100 (Fig. 2c). The results showed that the initially high GAPDH mRNA levels decreased gradually over the incubation time for both E-field and Triton X-100 disrupted samples. In contrast, the untreated exosomes (positive controls) maintained constant GAPDH mRNA levels during the experiment. These data suggested that both E-field and Triton-X detergent released the GAPDH mRNA from human saliva exosomes (see also Supplementary Fig. 2S). With exposure to the saliva, the signals dropped to less than 40% within one min. After 20 min, the readouts decreased down to the background level. The kinetics of the GAPDH mRNA signal decay supported the assumption that the exosome protected and stabilized endogenous mRNA. Without the exosome protection, the harbor GAPDH mRNA will rapidly decay.
The relative GAPDH levels were similar when released by either the E-field or by Triton X lysis. This indicated that the two methods had similar efficacies for releasing RNA from exosomes. However, the electric field is directly applied to the extracted exosomes, without introduction of extra reagents; thus, the whole process takes about 200 s at room temperature, which facilitates multiple, parallel sample analysis. In contrast, traditional assays for exosomal mRNA/protein detection are typically performed for 1–24 h, and additional reagents and methods must be applied, like Western blotting or ELISA for protein detection and q-PCR for nucleic acid detection.
3.3. Performance of EFIRM technology to simultaneously detect exosomal surface proteins and harbored mRNA
The sensitivity of EFIRM for detecting exosomal GAPDH mRNA and hCD63-GFP protein was determined by benchmarking the results against conventional methods of Western blotting for detecting the hCD63-GFP protein and nested q-PCR for detecting the GAPDH mRNA. Western blotting and EFIRM showed similar protein detection sensitivities at the same dilutions (Fig. 3a). The electrochemical level of GAPDH mRNA by EFIRM and the CT value on a log10 scale from q-PCR also showed a similar trend, but different concentrations (Fig. 3b). These results demonstrated that the sensitivity of EFIRM was comparable to that of conventional methods for detecting both mRNA and GFP protein.
Fig. 3.
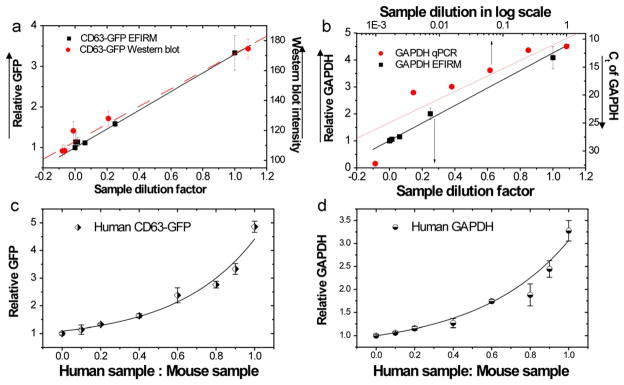
EFIRM compared to Western blot and q-PCR measurements for detection of hCD63-GFP protein and GAPDH mRNA in exosomes shed from H460 cells. (a) Titration of the GFP moiety of hCD63-GFP by EFIRM and Western blotting. (b) Titration of GAPDH mRNA by EFIRM and q-PCR. Data in (a) and (b) are fitted by a linear model. The PCR readout is presented on a log scale. The human specificity of EFIRM measurements was tested by detecting human exosomal (c) CD63-GFP protein and (d) GAPDH mRNA in mixtures of endosomes from human lung cancer H460 cells and interfering exosomes from mouse Lewis lung carcinoma LL2/LLC1 cells.
We then tested the discriminating ability of EFIRM technology for detecting human protein (hCD63-GFP) and mRNA (hGAPDH mRNA) endosomal targets in the presence of interfering mouse exosomes. The detection probes were based on human-specific sequences for the GAPDH mRNA and the human CD63 protein. EFIRM measurements were performed on exosomal hCD63-GFP protein and GAPDH mRNA on mixtures of human and mouse exosomes at different ratios. The corresponding electrochemical signals are presented in Fig. 3(c) and (d) vs the ratio between human and mouse exosome. The pure human exosomes (ratio=1) showed higher signal intensities than the pure mouse exosomes (ratio=0) (Fig. 3c and d). Even when the human exosomes were highly outnumbered by mouse exosomes (human exosome: mouse exosome=0.2), the signal remained above 2 standard deviations from the pure mouse exosome sample. This result showed that the EFIRM technology was highly specific in the detection of both human GAPDH mRNA and hCD63-GFP protein.
3.4. EFRIM technology for detecting exosomal hCD63-GFP in body fluids
We explored a translational application of EFIRM to address a central question in salivary diagnostics: can saliva be used to detect distal systemic diseases. The hypothesis to be tested is that does tumor-shed exosomes traffic through the body of the host via the vasculature can further traffic into saliva, contributing the exosomal content to saliva constituents. This hypothesis was tested using EFIRM technology with an in vivo mouse tumor model system. Nude mice were with the human lung cancer cell line H460 expressing hCD63-GFP. Nude mice were injected intrapleuraly with 1 × 106 H460 human lung cancer cells (n=9) or 100 μl saline (n=11). After 20 days, serum and saliva were collected. EFIRM was then to measure hCD63-GFP positive exosomes in mouse saliva and serum. Fig. 4 shows the correlation between exosomal hCD63-GFP levels in saliva and serum. All samples from tumor bearing group had measurable hCD63-GFP levels by EIFRM in both serum and saliva.
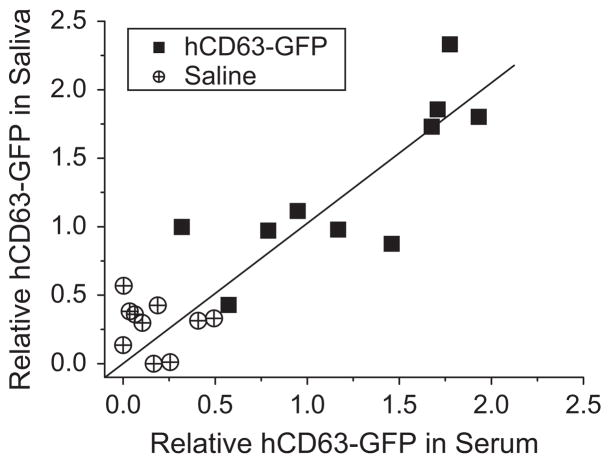
Fig. 4.
Correlation between human CD63-GFP levels in saliva and serum measured with EFIRM technology. Samples are from mice that had been injected with a human lung cancer cell line that expressed CD63-GFP (solid squares) or with saline alone (cross and circles). The linear fitting was carried out between the saliva and the serum relative readout (R=0.79).
Each point represents the ratio of electrochemical current readings from serum (X-axis) and saliva (Y-axis). Relative values of the signal were calculated and nomarlized then the blank control was substracted with signal measured from casein–PBS buffer. Data points close to the line (slope=1) indicate that the serum and saliva had similar protein concentrations; data points below the line indicate a higher signal in serum than in saliva, and vice versa. The linear fitting was carried out between the saliva and the serum relative readout (R=0.79). Therefore, the human CD63-GFP-exosome levels from serum and saliva was fairly well correlated in the 20 mouse samples (both serum and saliva were measured for each mouse). Furthermore, the mice injected with hCD63-GFP and those injected with saline (control group) showed significant differences in the hCD63-GFP positive exosomes in both serum and saliva. The low values measured in the saline group suggested that the saline injection did not generate high GFP signals in either serum or saliva. The detection required only 10 μl of raw sample in an on-site measurement. The time from the raw sample loading to detection was approximately 3 h. This data suggested that GFP in exosome shed by distal tumor can traffic into the vasculature. More importantly it is the first demonstration that distal tumor-shed GFP in exosome can traffic further into saliva and that the serum and saliva quantitation of tumor-derived exosome is fairly linear.
4. Conclusions
The molecules harbored in exosomes play important roles in biological science. A highly desirable goal for exosome research is the rapid, simple, simultaneous tracking and quantification of exosome harbored molecules. In this work, we present the electric field induced release and measurement (EFIRM) technique for measuring harbored biomolecules in exosomes. EFIRM uses an electrical field to simultaneously disrupt exosomes to release the contents and measure the harbored exosomal RNA/proteins. With EFIRM, we confirmed that in vivo H460-CD63-GFP shed exosomes were transported to blood and saliva, which demonstrates for the first time tumor-shed exosomes were detected in saliva, in addition to blood
The translational application of EFIRM its detection of tumor-shed GFP from exosomes in the serum and saliva of tumor-bearing mice in a linear manner is a significant step towards credentialing the use of saliva and its constituent biomarker for systemic disease detection. It also opens up a novel facet of molecular diagnostics in that the non-invasive detection of tumor-specific constituents of protein, microRNA, mRNA as well as tumor-specific mutation profiles maybe feasible in saliva.
Supplementary Material
Acknowledgments
This work was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1TR000124 (to FW); the Felix & Mildred Yip Endowed Professorship and the Barnes Family Fund (to DTWW).
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bios.2012.12.046.
Footnotes
Disclosures
David Wong is co-founder of RNAmeTRIX Inc., a molecular diagnostic company. PeriRx LLC sublicensed intellectual properties pertaining to molecular diagnostics from RNAmeTRIX. David Wong is a consultant to PeriRx.
Contributor Information
Fang Wei, Email: frada@ucla.edu.
Jieping Yang, Email: jiepyang@ucla.edu.
David T.W. Wong, Email: dtww@ucla.edu.
References
- Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. International Immunology. 2005;17 (7):879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- Chen C, Skog J, Hsu CH, Lessard RT, Balaj L, Wurdinger T, Carter BS, Breakefield XO, Toner M, Irimia D. Lab on a Chip. 2010;10 (4):505–511. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ. Journal of Cell Science. 2000a;113 (Pt 19):3365–3374. doi: 10.1242/jcs.113.19.3365. [DOI] [PubMed] [Google Scholar]
- Denzer K, Kleijmeer MJ, Heijnen HFG, Stoorvogel W, Geuze HJ. Journal of Cell Science. 2000b;113 (19):3365–3374. doi: 10.1242/jcs.113.19.3365. [DOI] [PubMed] [Google Scholar]
- Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. Journal of Biological Chemistry. 1998;273 (32):20121–20127. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- Fevrier B, Raposo G. Current Opinion in Cell Biology. 2004;16 (4):415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Nature Cell Biology. 2009;11 (9):1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Begne M, Lu B, Han X, Hagen FK, Hand AR, Melvin JE, Yates JR. Journal of Proteome Research. 2009;8 (3):1304–1314. doi: 10.1021/pr800658c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D. Nature Reviews Molecular Cell Biology. 2006;7 (7):529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Johansson SM, Admyre C, Rahman QK, Filen JJ, Lahesmaa R, Norman M, Neve E, Scheynius A, Gabrielsson S. Journal of Immunology. 2006;176:S184–S184. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- Kesimer M, Scull M, Brighton B, DeMaria G, Burns K, O’Neal W, Pickles RJ, Sheehan JK. FASEB Journal. 2009;23 (6):1858–1868. doi: 10.1096/fj.08-119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CS, Wong DTW. PLOS ONE. 2012;7 (3):e33037. doi: 10.1371/journal.pone.0033037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Bond DT, McKee M, Skog J, Paunescu TG, Da Silva N, Brown D, Russo LM. Kidney International. 2010;78 (2):191–199. doi: 10.1038/ki.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Kanai-Azuma M, Akimoto Y, Kawakami H, Yanoshita R. Biological & Pharmaceutical Bulletin. 2008;31 (6):1059–1062. doi: 10.1248/bpb.31.1059. [DOI] [PubMed] [Google Scholar]
- Palanisamy V, Sharma S, Deshpande A, Zhou H, Gimzewski J, Wong DT. PLOS ONE. 2010;5(1) doi: 10.1371/journal.pone.0008577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park NJ, Li Y, Yu TW, Brinkman BMN, Wong DT. Clinical Chemistry. 2006;52 (6):988–994. doi: 10.1373/clinchem.2005.063206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, Wong DT. Clinical Cancer Research. 2009;15 (17):5473–5477. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisitkun T, Shen RF, Knepper MA. Proceedings of the National Academy of Sciences USA. 2004;101 (36):13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosell R, Wei J, Taron M. Clinical Lung Cancer. 2009;10 (1):8–9. doi: 10.3816/CLC.2009.n.001. [DOI] [PubMed] [Google Scholar]
- Schmid M, Jensen TH. Trends in Biochemical Sciences. 2008;33 (10):501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Schorey JS, Bhatnagar S. Traffic. 2008;9 (6):871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Rasool HI, Palanisamy V, Mathisen C, Schmidt M, Wong DT, Gimzewski JK. ACS Nano. 2010;4 (4):1921–1926. doi: 10.1021/nn901824n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR. Biology Direct. 2007;2:35. doi: 10.1186/1745-6150-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. Traffic. 2002;3 (5):321–330. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- Taylor DD, Gercel-Taylor C. Gynecologic Oncology. 2008;110 (1):13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Nature Cell Biology. 2007;9 (6):654–U672. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Journal of Biochemistry. 2006;140 (1):13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- Wei F, Liao W, Xu Z, Yang Y, Wong DT, Ho CM. Small. 2009a;5 (15):1784–1790. doi: 10.1002/smll.200900369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Patel P, Liao W, Chaudhry K, Zhang L, Arellano-Garcia M, Hu S, Elashoff D, Zhou H, Shukla S, Shah F, Ho CM, Wong DT. Clinical Cancer Research. 2009b;15 (13):4446–4452. doi: 10.1158/1078-0432.CCR-09-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Qu P, Zhai L, Chen C, Wang H, Zhao XS. Langmuir. 2006;22 (14):6280–6285. doi: 10.1021/la060602h. [DOI] [PubMed] [Google Scholar]
- Wubbolts R, Leckie RS, Veenhuizen PT, Schwarzmann G, Mobius W, Hoernschemeyer J, Slot JW, Geuze HJ, Stoorvogel W. Journal of Biological Chemistry. 2003;278 (13):10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.