Abstract
The recent human outbreak of H7N9 avian influenza A virus has caused worldwide concerns. Receptor binding specificity is critical for viral pathogenicity, and still not thoroughly studied for this emerging virus. Here, we evaluated the receptor specificity of the haemagglutinin (HA) of two human H7N9 isolates (A/Shanghai/1/13 and A/Anhui/1/13) through a solid-phase binding assay and a flow cytometry-based assay. In addition, we compared it with those from several HAs from human and avian influenza viruses. We observed that the HAs from the novel H7 isolates strongly interacted with α2,3-linked sialic acids. Importantly, they also showed low levels of binding to α2,6-linked sialic acids, but significantly higher than other avian H7s.
As of 7 June 2013, a total of 132 laboratory-confirmed cases of human infection with avian influenza A virus (IAV) H7N9, including 32 deaths have been reported to the World Health Organization (WHO, 2013). Although the number of cases greatly increased during the month of April 2013, there is still no evidence of human-to-human transmission so far (WHO, 2013). Thus, the increased cases in humans suggested better transmission from avian sources to humans than in the case of other avian viruses.
Some receptor restrictions for avian influenza viruses in human airways account for the reduced ability of avian strains to establish infections in humans (Kuiken et al., 2006; Matrosovich et al., 2004; Neumann & Kawaoka, 2006; Nicholls et al., 2007; Peiris et al., 2007; van Riel et al., 2006). The capacity of the IAV to infect birds or humans is partially defined by the binding specificity of the haemagglutinin (HA), the major glycoprotein on the influenza virus surface. In general, HAs from human strains preferentially bind sialic acids attached through an α2,6 linkage to the terminal galactose (SAα2,6) of the oligosaccharides on the cell surface. These types of linkages are predominant in the respiratory epithelia in the upper respiratory tract of humans and other mammals such as ferrets (Shinya et al., 2006). In contrast, the HA of avian strains bind preferentially to α2,3-linked sialic acids (SAα2,3), which are abundant in the avian intestinal tract and are also present in the human lower respiratory tract (Nicholls et al., 2007; Pillai & Lee, 2010). Binding to SAα2,6 receptors is one of the requirements for efficient replication in the human upper respiratory tract which facilitates respiratory droplet-based transmission. Efficient human-to-human transmission of emerging influenza viruses by respiratory droplets is a prerequisite for rapid spread throughout the human population, and animal viruses with this ability pose a major pandemic threat potential. Analysis of the first three available HA sequences from the novel Chinese H7N9 viruses (A/Shanghai/1/13, A/Shanghai/2/13 and A/Anhui1/13) indicated the presence of amino acid residues in the receptor binding site (RBS) that are associated with enhanced binding of H5 and H7 HAs to SAα2,6. The A/Shanghai/1/13 HA possesses an A138S (H3 numbering) mutation (Table 1), which has been reported to confer SAα2,6 binding ability to an Indonesian H5N1 swine isolate (Nidom et al., 2010). Usually, alanine is conserved in this position in avian viruses and the presence of a serine could indicate adaptation to the human host in this virus (Kageyama et al., 2013; Nidom et al., 2010). However, this mutation is only present in the A/Shanghai/1/13 isolate and was not detected in any of the later H7N9 isolates [Global Initiative on Sharing All Influenza Data (GISAID) database, 16 May 2013]. HAs of A/Anhui/1/13 and A/Shanghai/2/13 possess two mutations that are also associated with increased binding to SAα2,6. G186V as well as Q226L are described to enhance SAα2,6 binding in H7 IAV (Belser et al., 2008; Srinivasan et al., 2013; Yang et al., 2010). L226 in combination with S228 plays a synergic role on SAα2,6 binding of human H3N2 isolates and introduction of these two mutations into H7 HAs led to increased SAα2,6 binding as well (Srinivasan et al., 2013). However, all novel Chinese H7N9 isolates so far possess a G228. It is of note that two human isolates have acquired an isoleucine in position 226 whereas A/Shanghai/1/13 and a chicken isolate show the avian prototypic glutamine at this position (Table 1). G186V and Q226L are found in almost all Chinese H7N9 isolates so far (Table 1), including viruses of human, chicken, pigeon and environmental origin (Table 1). The presence of abundant SAα2,6 receptors has been demonstrated in chickens and quail (Guo et al., 2007; Wan & Perez, 2006), and avian viruses with increased binding to SAα2,6 have been isolated from chickens (Matrosovich et al., 2001; Pawar et al., 2012; Watanabe et al., 2011). Therefore, the G186V and Q226L mutations could represent adaptations to chicken (or quail) rather than adaptions to the human host. Further, an H7 isolate possessing an E186 and exhibiting binding to SAα2,6 was unable to transmit via respiratory droplets in the ferret model (Belser et al., 2008). In addition to the mentioned mutations all Chinese H7N9 isolates possess the T160A substitution which leads to the loss of a glycosylation site and has been associated with increased SAα2,6 binding activity in an H5N1 isolate (Gao et al., 2013; Kageyama et al., 2013; Wang et al., 2010). However, this mutation is present in most recent H7 isolates from both the American as well as the Eurasian lineage, and many of these HAs do not exhibit binding to SAα2,6 (Figs 1 and 2). A recent transmission study with A/Shanghai/2/13 (HA identical to A/Anhui/1/13) indeed showed very limited transmission from ferret to ferret via aerosol droplets (one animal out of three) and no transmission from pigs to ferrets in the same setting (Zhu et al., 2013).
Table 1. Mutations associated with a change in receptor specificity of novel Chinese H7N9 human and avian isolates and amino acids in those corresponding positions in the two H7s (in bold) included in the receptor binding characterization.
| Strain name | aa 138 | N158 glycosylation site | aa 186 | aa 226 |
| A/Shanghai/1/2013 | S | No | G | Q |
| A/Shanghai/2/2013* | A | No | V | L |
| A/Shanghai/3/2013 | A | No | V | L |
| A/Shanghai/4/2013* | A | No | V | L |
| A/Shanghai/4664T/2013 | A | No | V | L |
| A/Anhui/1/2013 | A | No | V | L |
| A/Fujian/1/2013* | A | No | V | L |
| A/Hangzhou/1/2013 | A | No | V | I |
| A/Hangzhou/2/2013 | A | No | V | L |
| A/Hangzhou/3/2013 | A | No | V | L |
| A/Zhejiang/1/2013† | A | No | V | I |
| A/Zhejiang/2/2013* | A | No | V | L |
| A/Zhejiang/DTID-ZJU01/2013* | A | No | V | L |
| A/pigeon/Shanghai/S1069/2013 | A | No | V | L |
| A/chicken/Shanghai/S1053/2013* | A | No | V | L |
| A/chicken/Jiangsu/K27/2013‡ | A | No | V | L |
| A/chicken/Jiangsu/K89/2013* | A | No | V | L |
| A/chicken/Zhejiang/DTID-ZJU0/2013 | A | No | V | Q |
| A/environment/Shanghai/S1088/2013* | A | No | V | L |
| A/environment/Hangzhou/34/2013 | A | No | V | L |
Identical to A/Anhui/1/13.
Identical to A/Hangzhou/1/13.
Identical to A/Hangzhou/3/13.
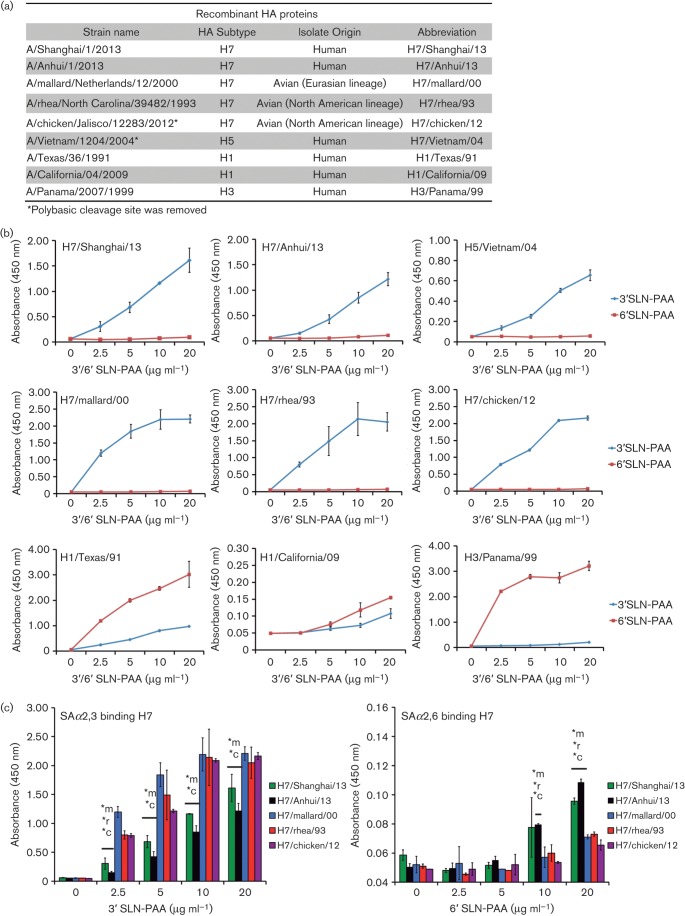
Fig. 1.
Receptor binding specificity of recombinant HAs by solid-phase binding assay. (a) Recombinant HA proteins included in the assay. (b) Binding affinity to SAα2,3 versus SAα2,6 glycans for every HA tested. (c) Binding affinity to SAα2,3 or SAα2,6 glycans of the H7 proteins. Statistical analysis: t-test; *m, *r and *c indicate P≤0.05 when compared to H7/mallard/00, H7/rhea/93 or H7/chicken/12, respectively. Results in (b) and (c) are shown as means±sd (mean of two replicates).
Fig. 2.
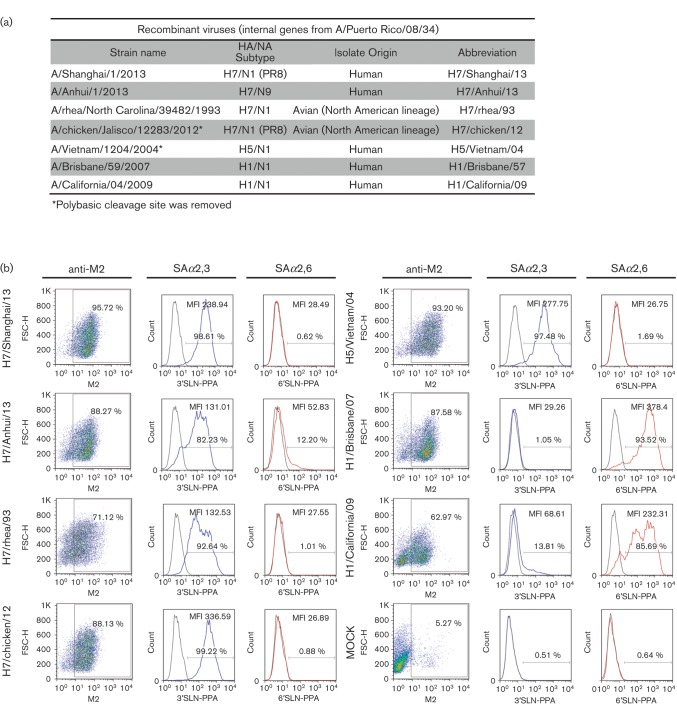
Receptor binding specificity of HAs using a flow cytometry-based binding assay. (a) Recombinant influenza A viruses included in the assay. (b) Percentages of infected cells (anti-M2 antibody-positive cells) are shown in the dot plots. Positive M2 cells were gated (except in the mock-infected samples, where we gated the total cell population) and the percentage of cells binding to 3′SLN-PAA or to 6′SLN-PAA, as well as the mean of fluorescence intensity (MFI) of the binding cells, was calculated (indicated in each histogram).
In order to shed light on the receptor binding specificity of the novel Chinese H7N9 isolates, we aimed to investigate the binding of A/Shanghai/1/13 and A/Anhui/1/13 HA to SAα2,3 and SAα2,6 substrates. The amino acid sequence of the HA of A/Shanghai/1/13 is so far unique; the sequence of the A/Anhui/1/13 HA, however, is identical to the sequence of A/Shanghai/2/13 and to that of many other isolates (Table 1). The study was performed using a solid-phase binding assay as well as a flow cytometry-based assay as previously described (Ramos et al., 2011).
For the solid-phase binding assay, we used recombinant HAs derived from H7N9 strains, and European or North American lineage H7 viruses (A/mallard/Netherlands/12/2000 H7N3, A/rhea/North Carolina/39482/1993 H7N1 and A/chicken/Jalisco/12283/2012 H7N3), as well as human H5N1, H1N1 and H3N2 strains (Fig. 1a). In order to generate the recombinant HAs, we cloned the HA sequence in frame into a modified pFastBac baculo transfer vector (Invitrogen) that features a C-terminal T4 trimerization domain and a hexahistidine tag and expressed recombinant protein in the baculovirus expression system as described previously (Goff et al., 2013; Krammer et al., 2012). For the solid-phase binding assay, 96-well ELISA plates were coated with the specific HA at 20 µg ml−1. After blocking and washing, we added the polyacrylamide (PAA)–biotin-conjugated glycans Neu5Acα2,3Galβ1,4GlcNAc-PAA (3′SLN-PAA) and Neu5Acα2,6Galβ1,4GlcNAc-PAA (6′SLN-PAA), provided by the Consortium of Functional Glycomics, at the specified concentrations. Then, we incubated the plates with streptavidin–HRP (R&D Systems), developed the assay with the substrate o-phenylenediamine (Invitrogen) and determined the absorbance at 450 nm with a microplate reader (BioTek).
As shown in Fig. 1(b), all the H7 tested, including those corresponding to either human or avian strains, showed clear preference for binding to SAα2,3, as indicated by the high levels of absorbance upon incubation with 3′SLN-PAA. In the case of H5/Vietnam/04, as previously described by us and others (Ramos et al., 2011; Stevens et al., 2006; Yamada et al., 2006), we also observed a strong binding preference for SAα2,3 residues. In contrast, the H1 and H3 HAs from human viruses, as expected, showed preferential binding to 6′SLN-PAA. When we compared the relative binding for SAα2,3 of the several H7s included in this study, we observed that the HA corresponding to those viruses isolated from human cases of avian influenza (H7/Shanghai/13, H7/Anhui/13) bound with less affinity to 3′SLN-PAA than the avian isolates tested (H7/mallard/00, H7/rhea/93 or H7/chicken/12) (Fig. 1c). Interestingly, the H7 from human H7N9 isolates showed low levels of binding to the SAα2,6 substrate, which were significantly higher than the ones detected in the case of the H7N1 and H7N3 avian isolates (Fig. 1c).
In order to confirm these results, we studied the glycan binding properties of these two H7N9 viruses using a flow cytometry binding assay as described previously (Ramos et al., 2011). Briefly, Madin–Darby canine kidney (MDCK) epithelial cells were infected at an m.o.i. of 5 with the recombinant viruses indicated in Fig. 2(a). At 20 h post-infection, the cells were harvested, washed with cold PBS, and incubated with 20 µg ml−1 of the biotinylated glycans 3′SLN-PAA or 6′SLN-PAA, and anti-M2 antibody E10 (Mount Sinai Hybridoma Shared Research Facility) (Tan et al., 2012) for 1 h at 4 °C. The cells were then washed with PBS containing 1 % BSA and incubated with streptavidin–FITC (Jackson Immunoresearch) and secondary anti-mouse rhodamine antibody (Jackson Immunoresearch). GS4071 (the free carboxylate of oseltamivir) was added during both incubations to avoid cleavage of the sialic acids of the synthetic polymers. Flow cytometry was performed using a FACScan flow cytometer (Becton Dickinson) and analysed with FlowJo software.
The recombinant viruses were generated by reverse genetics as described previously (Fodor et al., 1999). They expressed the HA and neuraminidase (NA) or only HA (as specified in Fig. 2a) from the indicated viruses and the rest of the genes from A/Puerto Rico/8/1934 (PR8). First, we analysed the percentage of infected cells by gating the population of cells positive for anti-M2 staining (Fig. 2b). We observed a high percentage of infected cells in all cases, ranging between 62.95 and 95.72 % of the total cells. Next, we evaluated the proportion of cells that bound to the SAα2,3 or SAα2,6 biotinylated glycans within the population of M2-positive cells, which express also the HA of interest on their surface. Additionally, we evaluated the mean fluorescence intensity (MFI) of the 3′/6′SLN-PAA positive population (Fig. 2b). In this case, we observed that the H7/Shanghai/13 HA bound similarly to the HA from H7N1 and H7N3 avian isolates (H7/rhea/93 and H7/chicken/12) and the H5 from H5/Vietnam/04 to the SAα2,3 substrate (values >92 % binding cells and MFI >131.01). In all these cases, virtually no binding to the SAα2,6 substrate (<2 % binding cells) was detected. However, whilst H7/Anhui/13 HA also showed strong binding to the SAα2,3 glycan (82 % binding cells, MFI 131.01), we could observe some binding to SAα2,6 (12.20 % binding cells, MFI 52.83), which was higher than for the rest of IAVs included in the assay. Importantly, although these data clearly indicated some affinity for the human-like receptor SAα2,6 for H7/Anhui/13 HA, the interacting capacity was significantly lower than the H1N1 human isolates H1/Brisbane/07 and H1/California/09 (Fig. 2b).
Overall, the data from both assays indicated that, whilst strong affinity for SAα2,3 residues is maintained in the novel H7N9 IAV, mutations A138S in the HA of A/Shanghai/1/13, and G186V and Q226L in HA of A/Anhui/1/13 might be responsible for the increased interaction for SAα2,6 as compared with other H7 avian viruses, facilitating adaptation of this novel strain to the human host. Surprisingly, while using the solid-phase binding assay, we detected some levels of binding to the SAα2,6 glycan for H7/Shanghai/13 HA, but this was not the case when we used the flow cytometry-based assay. These differences could be due to the different setting in which the HA is allowed to interact with the sialylated substrate, since in one case recombinant HA was used and in the other case HA was expressed on the surface of infected cells. Therefore, the use of both systems combined indicates that, whilst the HA from both H7N9 isolates weakly interact with SAα2,6, this interaction is slightly stronger for H7/Anhui/13. Of interest, mutations G186V and Q226L in HA of H7/Anhui/13 are also present in most of the H7N9 viruses isolated from human, avian or environmental sources (Table 1; sequences were obtained from the GISAID).
These data are in accordance with two recent reports that evaluated the receptor binding of the HA from the human H7N9 to the same sialyl-glycans using two different assays from the ones used in our study (Xiong et al., 2013; Zhou et al., 2013). Xiong et al. (2013) compared the receptor specificity of A/Anhui/1/13 and an avian H7N3 using biolayer interferometry. Similarly to our observations, they reported that the human H7N9 showed a lower avidity for SAα2,3 and a higher avidity for SAα2,6 than the avian H7N3, whilst retaining the SAα2,3 binding preference. Zhou et al. (2013) obtained results consistent with ours using a similar experimental setting to the first one used in this study. They used a solid-phase binding assay, where they coated ELISA plates with different concentrations of the sialyl-glycans and detected virus binding with anti-HA antibodies. Using this assay, they also reported binding of the H7N9 viruses to SAα2,6 (which is higher in the case of A/Anhui/1/13 than A/Shanghai/1/13), but in both cases this binding to SAα2,6 was lower than the strong binding reported for both viruses to SAα2,3 substrates.
Further analysis of the interaction of the H7N9 virus with sialyl-receptors using glycan arrays has also been recently reported (Belser et al., 2013; Watanabe et al., 2013). Whilst our studies as well as the ones reported by Xiong et al. (2013) and Zhou et al. (2013) provide quantitative information using a limited number of polymers, glycan arrays are a powerful technology to elucidate the qualitative range of glycans that interact with the influenza virus HA. Using these assays, Belser et al. (2013) showed that A/Shanghai/1/13 bound to a broader array of SAα2,3 than SAα2,6, whilst A/Anhui/1/13 showed mixed binding to SAα2,3 and SAα2,6 glycans. Watanabe et al. (2013), however, found mixed binding to SAα2,3 and SAα2,6 in the case of A/Shanghai/1/13, and a clear reduction in binding to SAα2,3 and an increased interaction with the same type of SAα2,6 using a human H1N1 virus. Interestingly, when they performed this assay in the presence of a neuraminidase inhibitor, they observed a clear increase in the number of SAα2,3 glycans, indicating that their previous observations might reflect the combined activities of the HA and NA proteins.
Our data are also consistent with a recent report that showed weak binding of A/Anhui/1/13 HA to tissue samples from the human respiratory tract (Tharakaraman et al., 2013). Additionally, transmission experiments in ferrets and pigs, two of the most established models to evaluate possible adaptation to airborne transmission in humans, demonstrated limited aerosol transmission in ferrets and no transmission in pigs (Watanabe et al., 2013; Zhu et al., 2013).
In summary, we analysed the receptor binding specificity of the novel H7N9 influenza virus from two human isolates. Whilst these H7 HAs showed weak binding for SAα2,6 as compared with H7 HAs from other non-H7N9 avian isolates that do not possess the mutations associated with a change in receptor specificity (Table 1), they bound strongly to SAα2,3. This low but significant binding affinity to SAα2,6 might have facilitated avian-to-human transmission of the virus (Han et al., 2013). However, based on data from other H7 strains (Belser et al., 2008) that exhibit a similar SAα2,6 binding activity and from transmission studies conducted with the novel H7N9 strains in ferrets (Zhu et al., 2013), we would argue that this virus is not able to transmit readily from human to human via aerosol transmission. This notion is supported by recent data from Chinese laboratories, which tested 20 739 human cases of upper respiratory tract infection in March and April 2013 for the presence of H7N9 and found only six positives, suggesting that, so far, there is no spread of this novel virus in the local population (Xu et al., 2013).
Acknowledgements
We thank Peter Palese for helpful advice and discussions and Anice Lowen and Randy A. Albrecht for sharing recombinant viruses. We would also like to thank Jennifer Debeauchamp and Richard Webby (St Jude Children’s Research Hospital, TN, USA) for sharing the plasmids for generation of the recombinant HA from A/Anhui/1/13. We thank the Flow Cytometry Shared Facility in the Icahn School of Medicine at Mount Sinai for assistance and the Consortium of Functional Glycomics for sharing reagents. Parts of the data will be published on the Consortium of Functional Glycomics website. Furthermore, we would like to thank GISAID for making H7N9 sequencing data publicly available. This study is partly funded by NIH/NIAID contract Center for Research on Influenza Pathogenesis (CRIP) HHSN266200700010C (A. G. -S. and A. F. -S.) and Influenza Pathogenesis & Immunology Research Center (IPIRC) HHSN266200700006C (J. S.) as part of the CEIRS Network, as well as NIH/NIAID 1PO1AI90935, 1R01AI073450 and DARPA HR0011-11-C-0094 (A. F. -S.). F. K. was supported by an Erwin Schrödinger fellowship by the Austrian Science Fund (FWF).
References
- Belser J. A., Blixt O., Chen L. M., Pappas C., Maines T. R., Van Hoeven N., Donis R., Busch J., McBride R. & other authors (2008). Contemporary North American influenza H7 viruses possess human receptor specificity: implications for virus transmissibility. Proc Natl Acad Sci U S A 105, 7558–7563 10.1073/pnas.0801259105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser J. A., Gustin K. M., Pearce M. B., Maines T. R., Zeng H., Pappas C., Sun X., Carney P. J., Villanueva J. M. & other authors (2013). Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature [Epub ahead of print] 10.1038/nature12391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor E., Devenish L., Engelhardt O. G., Palese P., Brownlee G. G., García-Sastre A. (1999). Rescue of influenza A virus from recombinant DNA. J Virol 73, 9679–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R., Cao B., Hu Y., Feng Z., Wang D., Hu W., Chen J., Jie Z., Qiu H. & other authors (2013). Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368, 1888–1897 10.1056/NEJMoa1304459 [DOI] [PubMed] [Google Scholar]
- Goff P. H., Krammer F., Hai R., Seibert C. W., Margine I., García-Sastre A., Palese P. (2013). Induction of cross-reactive antibodies to novel H7N9 influenza virus by recombinant Newcastle disease virus expressing a North American lineage H7 subtype hemagglutinin. J Virol 87, 8235–8240 10.1128/JVI.01085-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C. T., Takahashi N., Yagi H., Kato K., Takahashi T., Yi S. Q., Chen Y., Ito T., Otsuki K. & other authors (2007). The quail and chicken intestine have sialyl-galactose sugar chains responsible for the binding of influenza A viruses to human type receptors. Glycobiology 17, 713–724 10.1093/glycob/cwm038 [DOI] [PubMed] [Google Scholar]
- Han J., Jin M., Zhang P., Liu J., Wang L., Wen D., Wu X., Liu G., Zou Y. & other authors (2013). Epidemiological link between exposure to poultry and all influenza A(H7N9) confirmed cases in Huzhou city, China, March to May 2013. Euro Surveill 18, 20418. [PubMed] [Google Scholar]
- Kageyama T., Fujisaki S., Takashita E., Xu H., Yamada S., Uchida Y., Neumann G., Saito T., Kawaoka Y., Tashiro M. (2013). Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill 18, 20453. [PMC free article] [PubMed] [Google Scholar]
- Krammer F., Margine I., Tan G. S., Pica N., Krause J. C., Palese P. (2012). A carboxy-terminal trimerization domain stabilizes conformational epitopes on the stalk domain of soluble recombinant hemagglutinin substrates. PLoS ONE 7, e43603 10.1371/journal.pone.0043603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken T., Holmes E. C., McCauley J., Rimmelzwaan G. F., Williams C. S., Grenfell B. T. (2006). Host species barriers to influenza virus infections. Science 312, 394–397 10.1126/science.1122818 [DOI] [PubMed] [Google Scholar]
- Matrosovich M. N., Krauss S., Webster R. G. (2001). H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology 281, 156–162 10.1006/viro.2000.0799 [DOI] [PubMed] [Google Scholar]
- Matrosovich M. N., Matrosovich T. Y., Gray T., Roberts N. A., Klenk H. D. (2004). Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci U S A 101, 4620–4624 10.1073/pnas.0308001101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G., Kawaoka Y. (2006). Host range restriction and pathogenicity in the context of influenza pandemic. Emerg Infect Dis 12, 881–886 10.3201/eid1206.051336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls J. M., Chan M. C., Chan W. Y., Wong H. K., Cheung C. Y., Kwong D. L., Wong M. P., Chui W. H., Poon L. L. & other authors (2007). Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat Med 13, 147–149 10.1038/nm1529 [DOI] [PubMed] [Google Scholar]
- Nidom C. A., Takano R., Yamada S., Sakai-Tagawa Y., Daulay S., Aswadi D., Suzuki T., Suzuki Y., Shinya K. & other authors (2010). Influenza A (H5N1) viruses from pigs, Indonesia. Emerg Infect Dis 16, 1515–1523 10.3201/eid1610.100508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar S. D., Parkhi S. S., Koratkar S. S., Mishra A. C. (2012). Receptor specificity and erythrocyte binding preferences of avian influenza viruses isolated from India. Virol J 9, 251 10.1186/1743-422X-9-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J. S., de Jong M. D., Guan Y. (2007). Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev 20, 243–267 10.1128/CMR.00037-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S. P., Lee C. W. (2010). Species and age related differences in the type and distribution of influenza virus receptors in different tissues of chickens, ducks and turkeys. Virol J 9, 5 10.1186/1743-422X-7-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos I., Bernal-Rubio D., Durham N., Belicha-Villanueva A., Lowen A. C., Steel J., Fernandez-Sesma A. (2011). Effects of receptor binding specificity of avian influenza virus on the human innate immune response. J Virol 85, 4421–4431 10.1128/JVI.02356-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya K., Ebina M., Yamada S., Ono M., Kasai N., Kawaoka Y. (2006). Avian flu: influenza virus receptors in the human airway. Nature 440, 435–436 10.1038/440435a [DOI] [PubMed] [Google Scholar]
- Srinivasan K., Raman R., Jayaraman A., Viswanathan K., Sasisekharan R. (2013). Quantitative description of glycan-receptor binding of influenza a virus H7 hemagglutinin. PLoS ONE 8, e49597 10.1371/journal.pone.0049597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J., Blixt O., Tumpey T. M., Taubenberger J. K., Paulson J. C., Wilson I. A. (2006). Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312, 404–410 10.1126/science.1124513 [DOI] [PubMed] [Google Scholar]
- Tan G. S., Krammer F., Eggink D., Kongchanagul A., Moran T. M., Palese P. (2012). A pan-H1 anti-hemagglutinin monoclonal antibody with potent broad-spectrum efficacy in vivo. J Virol 86, 6179–6188 10.1128/JVI.00469-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharakaraman K., Jayaraman A., Raman R., Viswanathan K., Stebbins N. W., Johnson D., Shriver Z., Sasisekharan V., Sasisekharan R. (2013). Glycan receptor binding of the influenza A virus H7N9 hemagglutinin. Cell 153, 1486–1493 10.1016/j.cell.2013.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riel D., Munster V. J., de Wit E., Rimmelzwaan G. F., Fouchier R. A., Osterhaus A. D., Kuiken T. (2006). H5N1 virus attachment to lower respiratory tract. Science 312, 399 10.1126/science.1125548 [DOI] [PubMed] [Google Scholar]
- Wan H., Perez D. R. (2006). Quail carry sialic acid receptors compatible with binding of avian and human influenza viruses. Virology 346, 278–286 10.1016/j.virol.2005.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lu B., Zhou H., Suguitan A. L., Jr, Cheng X., Subbarao K., Kemble G., Jin H. (2010). Glycosylation at 158N of the hemagglutinin protein and receptor binding specificity synergistically affect the antigenicity and immunogenicity of a live attenuated H5N1 A/Vietnam/1203/2004 vaccine virus in ferrets. J Virol 84, 6570–6577 10.1128/JVI.00221-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Ibrahim M. S., Ellakany H. F., Kawashita N., Mizuike R., Hiramatsu H., Sriwilaijaroen N., Takagi T., Suzuki Y., Ikuta K. (2011). Acquisition of human-type receptor binding specificity by new H5N1 influenza virus sublineages during their emergence in birds in Egypt. PLoS Pathog 7, e1002068 10.1371/journal.ppat.1002068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Kiso M., Fukuyama S., Nakajima N., Imai M., Yamada S., Murakami S., Yamayoshi S., Iwatsuki-Horimoto K. & other authors (2013). Characterization of H7N9 influenza A viruses isolated from humans. Nature [Epub ahead of print] 10.1038/nature12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2013). WHO risk assessment. Human infections with avian influenza A(H7N9) virus. http://www.who.int/influenza/human_animal_interface/influenza_h7n9/RiskAssessment_H7N9_07Jun13.pdf [Google Scholar]
- Xiong X., Martin S. R., Haire L. F., Wharton S. A., Daniels R. S., Bennett M. S., McCauley J. W., Collins P. J., Walker P. A. & other authors (2013). Receptor binding by an H7N9 influenza virus from humans. Nature 499, 496–499 10.1038/nature12372 [DOI] [PubMed] [Google Scholar]
- Xu C., Havers F., Wang L., Chen T., Shi J., Wang D., Yang J., Yang L., Widdowson M.-A., Shu Y. (2013). Monitoring avian influenza A(H7N9) virus through national influenza-like illness surveillance, China Emerg Infect Dis 19, 496–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S., Suzuki Y., Suzuki T., Le M. Q., Nidom C. A., Sakai-Tagawa Y., Muramoto Y., Ito M., Kiso M. & other authors (2006). Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature 444, 378–382 10.1038/nature05264 [DOI] [PubMed] [Google Scholar]
- Yang H., Chen L. M., Carney P. J., Donis R. O., Stevens J. (2010). Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoS Pathog 6, e1001081 10.1371/journal.ppat.1001081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang D., Gao R., Zhao B., Song J., Qi X., Zhang Y., Shi Y., Yang L. & other authors (2013). Biological features of novel avian influenza A (H7N9) virus. Nature 499, [Epub ahead of print]. 10.3201/eid1908.130662 [DOI] [PubMed] [Google Scholar]
- Zhu H., Wang D., Kelvin D. J., Li L., Zheng Z., Yoon S. W., Wong S. S., Farooqui A., Wang J. & other authors (2013). Infectivity, transmission, and pathology of human-isolated H7N9 influenza virus in ferrets and pigs. Science 341, 183–186 10.1126/science.1239844 [DOI] [PubMed] [Google Scholar]