Abstract
Objectives
A family of histone deacetylases (HDACs) mediates chromatin remodeling, and repression of gene expression. Deacetylation of histones within the HIV-1 long terminal repeat (LTR) by HDACs plays a key role in the maintenance of latency, whereas acetylation of histones about the LTR is linked to proviral expression and escape of HIV from latency. Global HDAC inhibition may adversely affect host gene expression, leading to cellular toxicities. Potent inhibitors selective for HDACs that maintain LTR repression could be ideal antilatency therapeutics.
Methods
We investigated the ability of selective HDAC inhibitors to de-repress the HIV-1 LTR in both a cell line model of latency and in resting CD4+ T cells isolated from patients who were aviremic on antiretroviral therapy (ART).
Results
We found that inhibition of class I HDACs increased acetylation of histones at the LTR, but that LTR chromatin was unaffected by class II HDAC inhibitors. In a latently infected cell line, inhibitors selective for class I HDACs were more efficient activators of the LTR than inhibitors that target class II HDACs. Class I HDAC inhibitors were strikingly efficient inducers of virus outgrowth from resting CD4+ T cells of aviremic patients, whereas HIV was rarely recovered from patient’s cells exposed to class II HDAC inhibitors.
Conclusions
Further development of selective HDAC inhibitors as part of a clinical strategy to target persistent HIV infection is warranted.
Keywords: histone deacetylase, HIV, latency, long terminal repeat, resting CD4+ T cells
Introduction
Latent infection of resting CD4+ T cells is established early during HIV-1 infection, making eradication of HIV unachievable with current ART [1–3]. Following integration of viral DNA into the cellular genome, the HIV long terminal repeat (LTR) promoter can revert to transcriptional silence in the absence of stimulation [4–6]. One of the mechanisms through which HIV latency is maintained is by the action of histone deacetylases (HDACs) at the HIV-1 LTR (reviewed in [7]).
Histone deacetylases are lysine deacetylases that modify histones and induce transcriptional repression, but can also exert influences on cellular activities that are independent of transcriptional repression (reviewed in [8]). HDACs are generally divided into three classes. The class I HDACs comprise HDAC1, 2, 3 and 8, whereas class II HDACs include 4, 5, 7 and 9 (subclass IIa) and 6 and 10 (subclass IIb). The catalytic domain of HDAC11 shares homology with both class I and II and this enzyme is sometimes classified as a class IV HDAC. The class III HDACs, the sirtuins, differ from the other classes in that they require nicotinamide adenine dinucleotide (NAD) to function and are not affected by HDAC inhibitors active against class I, II and IV.
Histone deacetylases repress transcription mainly through their ability to covalently modify the lysine tail of core histones of nucleosomes through deacetylation. Deacetylation of lysine residues on histone tails decreases the access of transcription factors to the DNA, and recruits other histone-modifying complexes that result in further transcriptional repression.
In tissue culture models of latent HIV infection, HDAC1 is recruited to the LTR by multiple DNA-binding complexes. HDAC1 recruitment by the transcription factor SV40 late specific factor (LSF) in concert with YY1 was the first mechanism reported [9]. Later studies suggested that AP-4 [10], heterodimers of the activation domain-deficient NFκB p50 subunit [11], C-myc through interaction with Sp1 [12], and CBF-1 by binding near the NFκB/NFAT enhancer element [13] could also recruit HDAC1. CTIP2 was reported to recruit HDAC1 or HDAC2 to the Sp1-binding site of the LTR [14]. Finally, it has been suggested that HDAC3 associates with the LTR [15].
Disruption of HDAC1 recruitment to the LTR by specific DNA-binding molecules, or inhibition of HDAC activity by global HDAC inhibitors leads to LTR activation and the escape of HIV from latency in both cell line models and primary cells obtained from patients [9,16,17]. Furthermore, the HDAC inhibitors (HDACis) valproic acid (VPA) and suberoylanilide hydroxamic acid (SAHA; vorinostat) induce viral outgrowth from resting CD4+ T cells of aviremic HIV-infected individuals on ART [18,19]. HDAC2 and 3 can also occupy a site at the HIV LTR, and may play a role in the repression of LTR expression [14,15]. These observations have led to the investigation of HDAC inhibition as a putative therapeutic strategy to induce HIV from latency.
As global HDAC inhibition may have adverse effect on host cells, we studied the ability of selective HDAC inhibitors to de-repress the HIV LTR in both a cell line model of latency and in CD4+ resting T cells isolated from aviremic patients on ART. We found that inhibitors that target class I HDACs 1, 2, 3 alone or in tandem with the class II HDAC6 were efficient inducers of HIV expression, yielding the outgrowth of replication-competent HIV from the resting CD4+ cells of patients. However, inhibition of HDAC6 alone, or of other class II HDACs resulted in marginal LTR activation in cell line systems, and did not result in significant recovery of virus from patient’s cells. Of note, a selective inhibitor targeting HDAC1 and 2 was not very effective at inducing LTR activation in cell lines and virus outgrowth from resting CD4+ T cells of HIV-infected individuals. These findings suggest the design of inhibitors selective for a limited array of HDACs as therapies to target persistent HIV infection.
Materials and methods
Cell culture
J89 cells, a latently infected Jurkat cell line encoding the enhanced green florescence protein (EGFP) as a marker for Tat-driven HIV LTR expression (gift of D.N. Levy) [20], were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (Sigma, St Louis, Missouri, USA), 100 U/ml penicillin and 100 ug/ml of streptomycin (Invitrogen, Carlsbad, California, USA) at 37°C under 5% CO2. Hela P4/R5 cells, a variant of HeLa Magi cells expressing both CXCR4 and CCR5 selected for low background beta-galactosidase expression via the HIV 1 minimal LTR (generous gift of N. Landau) [21], were cultured in Phenol Red-free DMEM (Invitrogen) supplemented with 10% fetal bovine serum, 0.5 mg/ml Puromycin (Sigma), 100 U/ml penicillin and 100 µg/ml streptomycin (Invitrogen).
High-throughput screening of LTR-activating compounds
Hela P4/R5 cells were seeded at 2000/well in 1536-well plates and incubated for 24 h. Test compounds were added and cells were incubated an additional 24 h and β-galactosidase activity was measured by the use of the Tropix Gal-Screen Assay Kit (Applied Biosystem, Foster City, California, USA) and read with the Perkin Elmer Envision 2101 Multilabel Luminometer.
Selective histone deacetylase inhibitors
The nonselective inhibitor valproic acid was obtained from Bedford Laboratories (Bedford, Ohio, USA). Other inhibitors were synthesized at Merck Research Laboratories. MRK1 is a selective inhibitor of the class I HDACs 1, 2, 3 and the class II HDAC6 [22]. MRK 4, apicidin and MRK 13 are selective against HDACs 1, 2 and 3 [22,23]; MRK 10 is a selective inhibitor of HDAC6 [24]. MRK 11 and MRK 14 selectivity inhibit the class II HDACs 4, 5, 6, and 7 and the class I HDAC8 [25,26]. However, siRNA knockdown of HDAC8 does not induce HIV LTR expression (A. Espeseth, unpublished observations). MRK 12 is a selective inhibitor of HDACs 1 and 2 [22]. See Table 1.
Table 1.
Selectivity of novel HDAC inhibitors.
| HDACi | Selectivity |
|---|---|
| MRK1 | HDAC1, 2, 3 and 6 |
| VPA | Nonselective |
| MRK 4 | HDAC1, 2, 3 |
| Apicidin | HDAC1, 2, 3 |
| MRK 10 | HDAC6 |
| MRK 11 | HDAC4, 5, 6, 7, 8 |
| MRK 12 | HDAC1, 2 |
| MRK 13 | HDAC1, 2, 3 |
| MRK 14 | HDAC4, 5, 6, 7, 8 |
Flow cytometry
J89 cells were washed with phosphate-buffered saline (PBS) and incubated overnight with the indicated concentration of HDACi. Cells were washed and resuspended in PBS containing 2% paraformaldehyde. GFP expression was measured by FACScan (Becton Dickinson, San Jose, California, USA), and analyzed using Cell Quest software (Macintosh, Sunnyvale, California, USA). Live cells were gated and two-parameter analysis used to differentiate GFP-associated fluorescence from background fluorescence. A total of 10 000 gated events were collected and data represent the percentage of GFP-expressing cells in total gated events.
Cytotoxicity assay
J89 cells were washed with PBS and incubated for 24 h in various concentrations of inhibitors. To measure proliferation and viability in the presence of drugs, cells were subjected to an MTT assay using a cell proliferation kit following the manufacturer’s instructions (Roche Applied Sciences, Indianapolis, Indiana, USA). The percentage of cells proliferating was calculated from cells cultured in drug-free medium.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as previously described [19] with the following modifications: J89 cells were cultured with HDACi at indicated concentrations for 4 h. Formaldehyde crossed-linked cells were lyzed for 20 min on ice using SDS Lysis buffer (Millipore, Billerica, Massachusetts, USA) and sonicated to fragment chromatin to 500–1000 base pairs. 1 × 106 sonicated cells were used to set up each immunoprecipitation reaction using 5 µg of antiacetyl histone 3 (Ac-H3, Millipore) or rabbit preimmune immunoglobulin G (Sigma). PCR on immunoprecipitated or input DNA was performed as described previously [19]. The percentage of input HIV LTR DNA was determined by comparing the cycle threshold values of each reaction to a standard curve generated from input DNA. The fold enrichment of acetylated histone proteins at the HIV-1 LTR region was calculated as a ratio of specific signal over untreated background signal.
Limiting dilution cultures of latently infected CD4+ T cells from HIV-infected donors
Lymphocytes were obtained by continuous-flow leukopheresis. Isolation of resting CD4+ T cells, recovery and quantification of replication-competent virus were performed as previously described [27]. For phytohemagluttinin (PHA) conditions, 93.6 million resting CD4+ T cells were plated in replicate dilutions of 2.5 million (36 cultures), 0.5 million (six cultures) and 0.1 million (six cultures) cells per well and stimulated with 1 µg/ml PHAL (Remel, Lenexa, Kansas, USA), a five-fold excess of allogeneic irradiated peripheral blood mononuclear cells (PBMCs) from a seronegative donor, and 20U/ml IL-2 for 24 h. For MRK HDACi, 33.5–48.6 million resting CD4+ T cells were plated in replicate dilutions of 2.5 million (12–18 cultures), 0.5 million (six cultures) and 0.1 million (six cultures) cells per well and stimulated with the indicated concentration of drugs for 24 h.
After maximum mitogen stimulation or drug exposure, cells were washed and cultured as previously described [27].Culture supernatants were collected on days 15 and 19 and assayed for virus production by p24 antigen capture ELISA (Zeptometrix, Buffalo, New York, USA). Cultures were scored as positive if p24 was detected at day 15 and was increased in concentration at day 19. A maximum likelihood method was used to calculate the infectious units per million (IUPM) of resting CD4+ T cells after exposure to PHA, or HDACi. If all cultures were negative, the IUPM was estimated to have a value lower than if 1 culture of 2.5 million cells had been positive.
Results
Histone deacetylase 1 potency corresponds with activation of the long terminal repeat
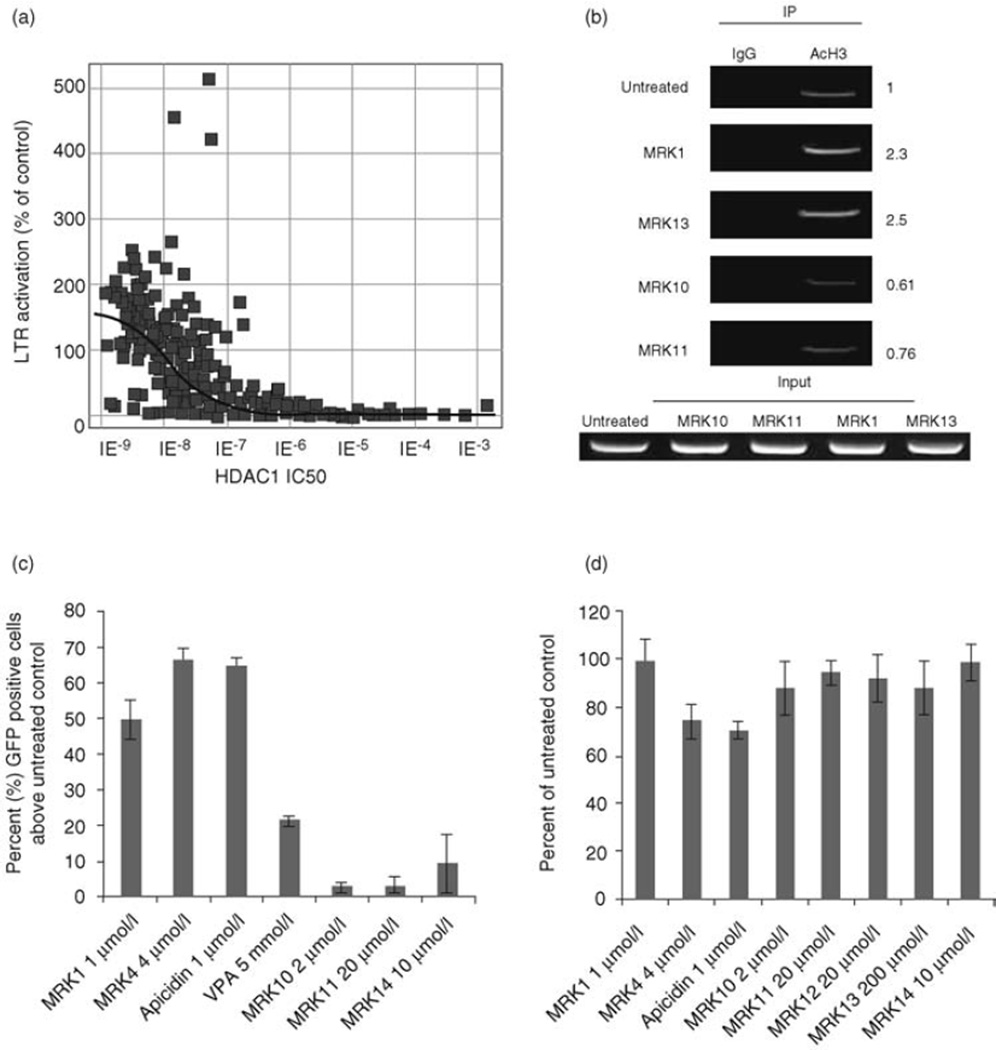
To screen for novel HDACis that induce expression of the HIV-1 promoter, candidate HDAC inhibitors from the Merck Research Laboratories chemical library were tested for the ability to up-regulate LTR expression in the P4/R5 LTR reporter cells [21]. As shown in Fig. 1a, by Spearman’s rank correlation there was a significant (P<0.0001) association between inhibitory potency against HDAC1 as measured in an in-vitro enzymatic assay, and increasing efficiency in LTR induction.
Fig. 1. Class I and nonselective HDAC inhibition induces LTR expression more effectively than class II HDAC inhibitors.
(a) HIV LTR expression is increased by HDAC inhibitors with increasing inhibitory potential for HDAC1. Hela P4/R5 cells were seeded and incubated for 24 h, followed by addition of HDAC inhibitors. β-galactosidase activity was measured after 24 h incubation with inhibitors. LTR activation is reported as the percentage β-galactosidase activity in treated cells over untreated control. (b) Class I but not class II HDAC inhibitors increase acetylation of Nuc1 at the HIV-1 LTR. J89 cells were treated with media, or HDAC inhibitors: MRK 1 or MRK 13 at class I HDAC1 IC50 (500 nmol/l and 300 nmol/l, respectively), MRK 10 or MRK 11 at class II HDAC4 IC50 (200 nmol/l and 500 nmol/l, respectively). Cells were treated for 4 h and assayed by chromatin immunoprecipitation with control rabbit IgG or anti-acetylated H3. DNA products of ChIP were quantitated in triplicate by real-time PCR. Assays are representative of three independent experiments, and real-time quantitation of the fold change relative to untreated control is shown. Only class I inhibitors demonstrate significant increases in histone H3 acetylation. (c) Class I and nonselective HDAC inhibition induces HIV expression. J89 cells were incubated overnight with the indicated concentrations of the HDACi. LTR-driven GFP production was measured by flow cytometry as described in methods. The data presented are the mean ± SE of three independent experiments. (d) Toxicity of HDAC inhibitors on J89 cells at concentrations used to measure promoter activation. J89 cells were cultured in the absence or presence of the indicated inhibitors for 24 h at the concentrations displayed. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays were performed in triplicate. The percentage of proliferating cells was calculated compared to cells cultured in standard media. ChIP, chromatin immunoprecipitation; GFP, green florescence protein; HDAC, histone deacetylase; LTR, long terminal repeat.
Class I but not class II inhibitors induce chromatin changes at the HIV long terminal repeat leading to long terminal repeat expression
Class I HDACs have been shown to maintain histones within the nucleosome-bound provirus in a hypoacetylated state, facilitating LTR repression [9–15]. Conversely, acetylation of histones by histone acetyltransferases (HATs) leads to neutralization of the net charge on lysine residues, increasing the access of transcription complexes to chromatin and recruiting other chromatin remodeling complexes, thus abrogating HDAC-mediated repression. However, the contribution of class II HDACs to this equilibrium is unexplored. We therefore compared the ability of class I and class II selective HDAC inhibitors (Table 1) to induce acetylation of nucleosome 1 (Nuc 1) of the HIV LTR by performing chromatin immunoprecipitation in J89 cells. Precipitated DNA was quantified using a set of primers spanning the Nuc 1 region of the HIV promoter. Cells were assayed after only 4 h of treatment to minimize the impact of indirect, secondary effects that may follow histone deacetylation. The percentage of input for each immunoprecipitation was calculated and the relative fold occupancy of acetylated histones reported. We found that class I but not class II inhibitors induce acetylation of histones at the HIV-LTR (Fig. 1b).
Class I selective histone deacetylase inhibitors are better than class II at inducing long terminal repeat expression
To determine the ability of selective HDAC inhibitors to induce HIV expression, selected candidate compounds were tested in the J89 Jurkat T cell model of HIV latency [20] and LTR-driven EGFP reporter gene expression measured.
Class I selective HDAC inhibitors, MRK 1, MRK 4 and Apicidin all induced LTR activation (Fig. 1c). As expected, the weak, nonselective HDAC inhibitor VPA also effectively induced LTR expression, albeit at millimolar concentrations. However, little LTR expression was induced by the class II HDAC inhibitors MRK 10, MRK 11 and MRK 14. This was not due to toxicity of VPA or MRK 10, 11 and 14, as concentrations used to induce LTR expression did not perturb cell proliferation (Fig. 1d and ref. [18]). These compounds were also evaluated in the HIV latency model cell lines AcH2, J1.1 and Hela P4/R5. A similar observation was made where only the class I and nonselective inhibitors robustly induced LTR expression (data not shown).
Class I selective histone deacetylase inhibitors induce viral outgrowth more efficiently than class II histone deacetylase inhibitors
We next examined the ability of the various HDACis to induce viral outgrowth from the resting CD4+ T cells of HIV-infected volunteers. Resting CD4+ T cells were isolated by negative selection from a total of 14 aviremic HIV-infected patients on ART (stable HIV-1 plasma RNA <50 copies/ml). Resting cells were obtained at multiple time points from six of the patients. The frequency of viral recovery after HDACi exposure in multiple limiting-dilution culture assays was compared to that after maximum mitogen activation with PHA, allogenic PBMCs, and IL-2. As observed in J89 cells, the class I HDAC inhibitor MRK 1 allowed more frequent recovery of replication-competent HIV from patient’s resting CD4+ T cells than the class II selective inhibitors, MRK 10 and MRK11 (Fig. 2). Of note, in seven of eight patients studied, there was less than a two-fold difference in the frequency of recovery of HIV in cells exposed to MRK 1 compared to PHA, a difference that is within the variance of our resting cell outgrowth assay.
Fig. 2. Recovery of replication-competent HIV from the resting CD4+ T cells of aviremic patients treated with HDAC inhibitors: MRK 1 (class I HDACs 1, 2, and 3 and class II HDAC6), VPA (nonselective), or MRK 10 or 11 (class II HDACs).
Patient cells were subjected to maximum mitogen stimulation or exposed to 2 µmol/l MRK 1, 40 µmol/l VPA, 2 µmol/l MRK 10 and 10 µmol/l MRK 11 for 24 h. Cells were washed and co-cultured with CD8-depleted PBMC as detailed in methods. Frequency of virus outgrowth from cells treated with HDACi was compared to the frequency of outgrowth from maximally stimulated cells (PHA). Each icon represents independent studies of patient cell samples; patient samples were simultaneously tested with mitogen or HDACi on the same day. Dashed lines indicate the limit of detection of the assay. HDAC, histone deacetylase; PBMC, peripheral blood mononuclear cell; VPA, valproic acid.
The nonselective inhibitor VPA was also effective at inducing virus from resting CD4+ T cells of aviremic HIV+ patients (Fig. 2), as previously demonstrated [18,19]. The surprising activity of VPA in primary cells as compared to more potent and specific HDAC inhibitors is not well understood, but could be the result of effects of VPA on other cellular enzymes, such as glycogen synthase kinase-3β.
However, whereas the class I inhibitors Apicidin and MRK4, were effective at inducing LTR expression in J89 cells (Fig. 1c), induction of virus from CD4+ resting T cells by these inhibitors was suboptimal (Fig. 3). As both MRK 4 and Apicidin appear to be slightly antiproliferative in J89 T cells (Fig. 1d), we cannot rule out the possibility that such an effect limits the recovery of virus in resting CD4+ T-cell outgrowth assays.
Fig. 3. Recovery of replication-competent HIV from resting CD4+ T cells of aviremic patients treated with Apicidin and MRK 4, inhibitors selective only for the class I HDACs 1, 2, and 3. Patient cells were subjected to maximum mitogen stimulation or exposed to 4 µmol/l MRK 4 or 1 µmol/l MRK 6 for 24 h.
Cells were washed and co-cultured with CD8-depleted PBMC as described in methods. Frequency of virus outgrowth from cells treated with HDACi was compared to outgrowth after maximal mitogen activation by PHA. Each icon represents independent studies of patient cell samples; patient samples were simultaneously tested with mitogen or HDACi on the same day. Dashed lines indicate the limit of detection of the assay. HDAC, histone deacetylase; PBMC, peripheral blood mononuclear cell.
As global HDAC inhibition may have effects on the host cell that lead to toxicities and adverse clinical outcomes, potent but selective inhibition of HDACs required to maintain LTR repression is desirable for potential therapeutics designed to disrupt HIV latency. MRK 4 and Apicidin are both selective against HDAC1–3, induce LTR expression (Fig. 1c), but may weakly induce viral outgrowth in patients’ cells due to effects on the host cell (Fig. 3).
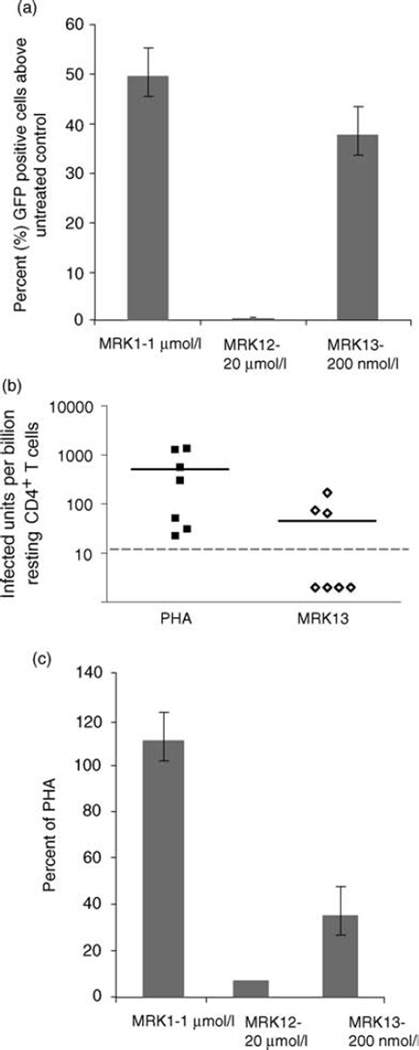
To study the effect of HDACi selective for HDACs 1–3 that are without apparent host cell toxicity, we studied two additional compounds with selectivity against HDAC1 and 2 (MRK 12) and HDAC1, 2, and 3 (MRK 13). Although activation of LTR expression in the J89 cell line by MRK 13 (which inhibits HDAC1–3) was comparable to that of MRK1 (which inhibits HDAC1–3 and 6; Fig. 4a), in six of the seven patients studied, recovery of HIV from cells exposed to MRK 13 was over three-fold less frequent than from cells exposed to PHA (Fig. 4b and c). Surprisingly, the HDAC1 and 2 inhibitors, MRK 12 performed poorly in both cell line and primary resting CD4+ T-cell assays (Fig. 4a and c).
Fig. 4. HIV expression and outgrowth is induced by inhibitors targeting HDACs 1, 2, and 3, but viral outgrowth is markedly improved by the tandem inhibition of HDAC1–2 and the class II HDAC6.
(a) J89 cells were incubated overnight with 1 µmol/l MRK 1 (selective for HDAC1, 2, 3 and 6), 2–20 µmol/l MRK 12 (HDAC1 and 2) or 200 nmol/l MRK 13 (HDAC1, 2, 3). LTR-driven GFP production was measured by flow cytometry as described in methods. The data presented are the mean ± SE of three independent experiments. (b) Resting CD4+ T cells from patients were subjected to maximum mitogen stimulation or exposed to 200–300 nmol/l MRK 13 for 24 h. Cells were washed and co-cultured with CD8-depleted PBMC as described in methods. Frequency of virus outgrowth from cells treated with MRK 13 was compared to outgrowth after maximal mitogen activation. Each icon represents independent studies of patient cell samples; patient samples were simultaneously tested with mitogen or HDACi on the same day. Dashed lines indicate the limit of detection of the assay. (c) Frequency of virus outgrowth from resting CD4+ T cells of aviremic HIV+ patients exposed to 2 µmol/l MRK 1, 2–20 µmol/l MRK 12 and 300 nmol/l MRK 13 are shown as a percentage of outgrowth obtained from corresponding maximal mitogen activation by PHA. MRK 1, n = 8; MRK 12, n = 2; MRK 13, n = 7. GFP, green florescence protein; HDAC, histone deacetylase; LTR, long terminal repeat; PBMC, peripheral blood mononuclear cell.
Discussion
Selective HDAC inhibitors induce expression of the HIV promoter and allow recovery of replication-competent HIV from the resting CD4+ T cells of ART-treated, aviremic patients. Inhibition of class I but not class II HDACs resulted in an increase of acetylated histones at the nucleosome-bound LTR. We found that inhibitors that target the class I HDACs 1, 2 and 3 were more efficient activators of the HIV LTR in a cell line model of HIV latency than inhibitors that target the class II HDACs. Class II HDAC inhibitors also performed poorly at inducing virus outgrowth from resting CD4+ T cells isolated from aviremic HIV+ patients.
MRK 12, an inhibitor selective against HDAC1 and 2 failed to activate the LTR in a cell line model of latency, and also poorly induced virus outgrowth from resting CD4+ T cells. This finding is surprising given prior studies illustrating HDAC1, and to a lesser extent HDAC2, activity at the HIV-1 LTR. However, our studies are the first to utilize selective inhibitors. HDAC1 and 2 associate with the Sin3, NuRD or CoREST corepressor complexes to repress transcription (reviewed in [28]). It seems likely that HDACs 1, 2, and 3 cooperate as part of one or more multiprotein complexes to mediate HIV LTR repression.
HDAC3 is found in complex with the nuclear hormone corepressors NCoR/SMRT. Whereas HDAC1 and 2 are reported to be global transcription repressors, HDAC3 is reported to be a more specific repressor with activity against genes involved in nuclear receptor signaling (reviewed in [28]). HDAC3 is reported to occupy a site at the HIV promoter and may play a role in suppressing transcription [15].
We investigated the ability of four inhibitors (MRK 1, MRK 4, Apicidin and MRK 13) targeting HDACs 1, 2 and 3 to induce virus outgrowth from resting CD4+ T cells. Although all four compounds induced LTR transcription in J89 cells, only MRK 1 robustly induced virus outgrowth from resting CD4+ T cells. In addition to its selectivity for HDAC1, 2, and 3, this inhibitor also targets HDAC6. However, it should be noted that HDAC6 inhibition alone has little effect on HIV LTR expression, as demonstrated (Figs 1c and 2) by an inhibitor selective for HDAC6 (MRK 10). Of note, inhibition of HDAC6 may only be relevant in the study of patient’s cells, as inhibition of HDAC1, 2, and 3 is as effective in inducing LTR expression as inhibition of HDAC1, 2, 3 and 6 in J89 cells. Interestingly, one study reported a predominantly cytoplasmic localization of HDAC6 in transformed, cancerous cells and a mostly nuclear localization in normal cells [29]. However, as HDAC6 does not appear to act directly at the HIV LTR [30], we speculate that the ability of Merck 1 to inhibit HDAC6 contributes to the outgrowth of virus from primary cells at another step in the viral lifecycle, or via other effects on the infected cell.
The mechanism by which HDAC6 might contribute to the suppression of the HIV expression requires further study. HDAC6 is a predominantly cytoplasmic enzyme, but can shuttle to the nucleus and is reported to mediate promoter repression in certain systems [29]. For example, NF-κB p50 and p65 cooperate with HDAC6 to repress transcription of the H+-K+-ATPase gene [31]. Runt-related transcription factor 2 mediates repression of the p21 promoter via its interaction with HDAC6 [32]. In yet another example of HDAC6-mediated repression, the enzyme binds to a domain of the HAT p300 leading to repression of its transcriptional activities. HDAC inhibition or siRNA knockdown of HDAC6 ablate this p300-mediated repression [33]. Regardless of the role HDAC6 may be playing in LTR repression, defining the mechanisms involved may provide additional targets for antilatency therapies.
Despite potent antiretroviral therapy, chronic HIV infection remains a formidable problem that requires novel approaches. These findings suggest that selective HDAC inhibitors may contribute to therapeutic efforts to clear persistent HIV infection.
Acknowledgements
We are grateful to D. Parker, M. Cheema, A. Duff and N. Cheng for technical assistance, L. Ngo for study coordination, R. Bosch for statistical advice, A. Kashuba, S. Fiscus, M. Kerkau, and F. Ashton of the UNC CFAR Pharmacology, Virology, Immunology, and Clinical Core facilities, and to the dedicated staff of the UNC Blood Bank. Funding for the study was provided by grants from amfAR (107168-44-RGRL) and National Institutes of Health (AI064074) to D.M.M., R00046 to the UNC GCRC and CTSA, and AI50410 to the UNC CFAR and NIH training grants T32 AI 07001-32 and T32 AI07151-30 to K.S.K. and N.M.A. Most importantly, this effort would not have been possible without the selfless contribution of study volunteers.
References
- 1.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 3.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 4.Pazin MJ, Sheridan PL, Cannon K, Cao Z, Keck JG, Kadonaga JT, et al. NF-kappa B-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes Dev. 1996;10:37–49. doi: 10.1101/gad.10.1.37. [DOI] [PubMed] [Google Scholar]
- 5.Van Lint C, Emiliani S, Ott M, Verdin E. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 1996;15:1112–1120. [PMC free article] [PubMed] [Google Scholar]
- 6.El Kharroubi A, Piras G, Zensen R, Martin MA. Transcriptional activation of the integrated chromatin-associated human immunodeficiency virus type 1 promoter. Mol Cell Biol. 1998;18:2535–2544. doi: 10.1128/mcb.18.5.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams SA, Greene WC. Regulation of HIV-1 latency by T-cell activation. Cytokine. 2007;39:63–74. doi: 10.1016/j.cyto.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 9.Coull JJ, Romerio F, Sun JM, Volker JL, Galvin KM, Davie JR, et al. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J Virol. 2000;74:6790–6799. doi: 10.1128/jvi.74.15.6790-6799.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imai K, Okamoto T. Transcriptional repression of human immunodeficiency virus type 1 by AP-4. J Biol Chem. 2006;281:12495–12505. doi: 10.1074/jbc.M511773200. [DOI] [PubMed] [Google Scholar]
- 11.Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 2006;25:139–149. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang G, Espeseth A, Hazuda DJ, Margolis DM. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase-1 to the human immunodeficiency virus type 1 promoter. J Virol. 2007;81:10914–10923. doi: 10.1128/JVI.01208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyagi M, Karn J. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 2007;26:4985–4995. doi: 10.1038/sj.emboj.7601928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marban C, Suzanne S, Dequiedt F, de Walque S, Redel L, Van Lint C, et al. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J. 2007;26:412–423. doi: 10.1038/sj.emboj.7601516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malcolm T, Chen J, Chang C, Sadowski I. Induction of chromosomally integrated HIV-1 LTR requires RBF-2 (USF/TFII-I) and Ras/MAPK signaling. Virus Genes. 2007;35:215–223. doi: 10.1007/s11262-007-0109-9. [DOI] [PubMed] [Google Scholar]
- 16.Coull JJ, He G, Melander C, Rucker VC, Dervan PB, Margolis DM. Targeted derepression of the human immunodeficiency virus type 1 long terminal repeat by pyrrole-imidazole polyamides. J Virol. 2002;76:12349–12354. doi: 10.1128/JVI.76.23.12349-12354.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ylisastigui L, Coull JJ, Rucker VC, Melander C, Bosch RJ, Brodie SJ, et al. Polyamides reveal a role for repression in latency within resting T cells of HIV-infected donors. J Infect Dis. 2004;190:1429–1437. doi: 10.1086/423822. [DOI] [PubMed] [Google Scholar]
- 18.Ylisastigui L, Archin NM, Lehrman G, Bosch RJ, Margolis DM. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS. 2004;18:1101–1108. doi: 10.1097/00002030-200405210-00003. [DOI] [PubMed] [Google Scholar]
- 19.Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, Margolis DM. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses. 2009;25:207–212. doi: 10.1089/aid.2008.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kutsch O, Benveniste EN, Shaw GM, Levy DN. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J Virol. 2002;76:8776–8786. doi: 10.1128/JVI.76.17.8776-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pescatore G, Kinzel O, Attenni B, Cecchetti O, Fiore F, Fonsi M, et al. Optimization of a series of potent and selective ketone histone deacetylase inhibitors. Bioorg Med Chem Lett. 2008;18:5528–5532. doi: 10.1016/j.bmcl.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Jones P, Altamura S, Chakravarty PK, Cecchetti O, De Francesco R, Gallinari P, et al. A series of novel, potent, and selective histone deacetylase inhibitors. Bioorg Med Chem Lett. 2006;16:5948–5952. doi: 10.1016/j.bmcl.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Hideshima T, Bradner JE, Wong J, Chauhan D, Richardson P, Schreiber SL, et al. Small-molecule inhibition of proteasome and aggresome function induces synergistic antitumor activity in multiple myeloma. Proc Natl Acad Sci U S A. 2005;102:8567–8572. doi: 10.1073/pnas.0503221102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones P, Bottomley MJ, Carfi A, Cecchetti O, Ferrigno F, Lo Surdo P, et al. 2-Trifluoroacetylthiophenes, a novel series of potent and selective class II histone deacetylase inhibitors. Bioorg Med Chem Lett. 2008;18:3456–3461. doi: 10.1016/j.bmcl.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 26.Muraglia E, Altamura S, Branca D, Cecchetti O, Ferrigno F, Orsale MV, et al. 2-Trifluoroacetylthiophene oxadiazoles as potent and selective class II human histone deacetylase inhibitors. Bioorg Med Chem Lett. 2008;18:6083–6087. doi: 10.1016/j.bmcl.2008.09.076. [DOI] [PubMed] [Google Scholar]
- 27.Archin NM, Eron JJ, Palmer S, Hartmann-Duff A, Martinson JA, Wiegand A, et al. Valproic acid without intensified antiviral therapy has limited impact on persistent HIV infection of resting CD4+ T cells. AIDS. 2008;22:1131–1135. doi: 10.1097/QAD.0b013e3282fd6df4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaquero A, Scher M, Reinberg D. Biochemistry of multiprotein HDAC complexes. In: Verdin E, editor. Histone deacetylases transcriptional regulation and other cellular functions. Totowa, New Jersey: Humana Press; 2006. p. 23. [Google Scholar]
- 29.Yoshida N, Omoto Y, Inoue A, Eguchi H, Kobayashi Y, Kurosumi M, et al. Prediction of prognosis of estrogen receptor-positive breast cancer with combination of selected estrogen-regulated genes. Cancer Sci. 2004;95:496–502. doi: 10.1111/j.1349-7006.2004.tb03239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keedy KS, Archin NM, Gates AT, Espeseth A, Hazuda DJ, Margolis DM. A limited group of class I histone deacetylases act to repress human immunodeficiency virus type-1 expression. J Virol. 2009;83:4749–4756. doi: 10.1128/JVI.02585-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W, Kone BC. NF-kappaB inhibits transcription of the H(+)-K(+)-ATPase alpha(2)-subunit gene: role of histone deacetylases. Am J Physiol Renal Physiol. 2002;283:F904–F911. doi: 10.1152/ajprenal.00156.2002. [DOI] [PubMed] [Google Scholar]
- 32.Westendorf JJ, Zaidi SK, Cascino JE, Kahler R, van Wijnen AJ, Lian JB, et al. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol Cell Biol. 2002;22:7982–7992. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Girdwood D, Bumpass D, Vaughan OA, Thain A, Anderson LA, Snowden AW, et al. P300 transcriptional repression is mediated by SUMO modification. Mol Cell. 2003;11:1043–1054. doi: 10.1016/s1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]