Abstract
Aquaporin-4 (AQP4) is the predominant water channel in the central nervous system, where it has been reported to be involved in many pathophysiological roles including water transport. In this paper, the AQP4 tetramer was modeled from its PDB structure file, embedded in a palmitoyl-oleoyl-phosphatidyl-choline (POPC) lipid bilayer, solvated in water, then minimized and equilibrated by means of molecular dynamics simulations. Analysis of the equilibrated structure showed that the central pore along the fourfold axis of the tetramers is formed with hydrophobic amino acid residues. In particular, Phe-195, Leu-191 and Leu-75, form the narrowest part of the pore. Therefore water molecules are not expected to transport through the central pore, which was confirmed by MD simulations. Each monomer of the AQP4 tetramers forms a channel whose walls consist mostly of hydrophilic residues. There are eight water molecules in single file observed in each of the four channels, transporting through the selectivity filter containing Arg-216, His-201, Phe-77, Ala-210, and the two conserved Asn-Pro-Ala (NPA) motifs containing Asn-213 and Asn-97. By using Brownian dynamics fluctuation–dissipation-theorem (BD-FDT), the overall free-energy profile was obtained for water transporting through AQP4 for the first time, which gives a complete map of the entire channel of water permeation.
Keywords: Aquaporin-4, Molecular dynamics simulations, Asn-Pro-Ala motifs, Central fourfold axis, Brownian dynamics fluctuation–dissipation-theorem (BD-FDT), Free energy computation
1. Introduction
As a family of integral membrane proteins, aquaporins (AQPs) facilitate the fast and yet highly selective flux of water and other small solutes across biological membranes. There are 13 homologous aquaporins identified in mammals, of which 6 subtypes (AQP1, AQP3, AQP4, AQP5, AQP8 and AQP9) have been reported in rodent brain cells, however, only three aquaporins have been clearly identified in human brain cells in vivo: AQP1, AQP4 and AQP9 [1–4]. Moreover, AQP1 is believed to be involved in cerebrospinal fluid (CSF) formation, for its distribution in the central nervous system (CNS) is highly concentrated in the choroid plexus of the lateral, fourth and third ventricle, and is also selectively expressed in the apical membrane microvilli of the choroid epithelium [5–7]. AQP9 is hypothesized to be a neutral solute channel being implicated in brain energy metabolism, which could facilitate the diffusion of lactate from the astrocyte to the neuron and also participate in the clearance of excess lactate in the extracellular space in astrocytes in pathological conditions [8]. In fact, AQP4 is much more abundantly expressed in the brain than AQP1 or AQP9, and now demonstrated in astrocyte foot processes surrounding capillaries, astrocyte processes, comprising the glial limiting membrane, in ependymal cells, and in subependymal astrocytes [9]. Although the major role of AQP4 is to control water movements into and out of the brain, it has been suggested to play roles in the generation of brain edema, astrocyte migration, neuronal activity, cell adhesion between astrocytes and endothelial cells, and so forth [10].
High-resolution structures of AQPs have been determined by electron microscopy and X-ray diffraction experiments, which have provided new insights into the molecular mechanisms acting in aquaporins. All known AQPs form tetrameric arrangements, and each monomer contains six membrane-spanning segments (TM1–6) with five connecting loops (A–E). Each monomer is folded into a right-handed alpha-barrel architecture, with a central channel surrounded by six transmembrane helices and connecting loops. Interestingly, there is high homology between the first and second half of AQPs; each half contains three transmembrane domains, and the two halves integrate in the membrane in opposite orientations [11]. Both loops between helices 2 (H2) and H3 (loop B, or HB), and between H5 and H6 (loop E, or HE) have a highly conserved Asn-Pro-Ala (NPA) motif. The loops meet in the center of the membrane to form the channel [11].
Molecular dynamics (MD) simulations are important tools for understanding the structure and function of biological macromolecules. Based on structures of human AQP1 and the bacterial glycerol facilitator GlpF, MD simulations demonstrated the roles played by a two-stage filter. The first stage is located in the central part of the channel at the NPA region, and the second is located on the extracellular face of the channel in the aromatic/arginine (ar/R) constriction region [12,13]. The ar/R constrictions of AQP1 and GlpF are located at the narrowest point of the channels, and different side chains at the ar/R constriction regions influence the polarity and diameter of the channel bottleneck [12,13]. The ar/R region of AQP1 has been regarded as a filter that allows the permeation of small polar solutes, whereas in GlpF this filter mechanism does not apply, resulting in quite different permeation characteristics between AQP1 and GlpF [12,13]. On the other hand, NPA constriction is vital for proton exclusion by a large electrostatic barrier. However, the contribution of both the protein's electric field and desolvation effects on the barrier are still in debate [14,15]. Obviously, MD simulations have provided remarkable insights into these water channels with high selectivity and efficiency in water or glycerol passage, while at the same time managing the exclusion of ions. Recently, the atomic structure of human AQP4 was obtained [16]. Here, we performed molecular dynamics (MD) simulations on a fully hydrated model of the AQP4 tetramer in the lipid bilayer, analyzed its structure and function, and drew its free-energy profiles of water transporting through AQP4 by using BD-FDT.
2. Materials and methods
2.1. Building a structural model of AQP4
The X-ray derived crystal structure of AQP4 was obtained from the Protein Data Bank (PDB code 3GD8). The monomeric coordinates were replicated by using the VMD software [17]. The transformation matrices provided in the PDB file were used in order to form a tetrameric structure. Consecutively the protein structure file (PSF) and Protein Data Bank (PDB) file for the AQP4 tetramer, the crystallographic water molecules, and the ions were generated using the AutoPSF plugin of VMD. To solvate the protein, Helmut Grubmüller's SOLVATE program [18] was used to fill any empty space inside the pores, as well as to surround it with water.
2.2. Placing AQP4 tetramer in a membrane
The solvated AQP4 tetramer was then placed in a model of the biological membrane. This membrane consisted of a palmitoyl-oleoyl-phosphatidyl-choline (POPC) lipid bilayer generated with the X length and Y length set to 120 Ǻ using the Membrane Builder program provided within VMD. Then, the constructed membrane patch and the partially solvated protein were aligned, moved, and assembled properly. Subsequently, it was then necessary to make room for the AQP4 in the membrane bilayer so that the protein doesn't overlap any lipid molecules. Using the beta field of the PDB file, the atoms of the “bad lipids” were marked using VMD. During this step, the majority of the lipids overlapping with the protein were removed. After measuring the water layer using the minmax option, the system was placed in a water box whose X and Y dimensions are slightly smaller than the lipid patch by using VMD's Solvate plugin, since non-equilibrated membranes tend to shrink. Lastly, K+ and Cl– ions with a concentration of 0.4 mmol/L were added throughout the system by using VMD's Autoionize Plugin, which transmutes water molecules into ions. The system consisted of 94127 atoms all together.
2.3. Running simulations of AQP4
The simulations were performed in four phases. In the first phase, the lipid tails and bulk water molecules of the system were minimized and equilibrated for 0.54 ns, while the protein, crystal-lographic water molecules, ions, and lipid head groups were fixed. In the second phase, the system was also minimized and equilibrated for 0.54 ns with the protein constrained. Because we have put our system together by hand, it has many unnatural atomic positions, therefore if we tried to do a dynamics run, it would immediately fail, with many atoms moving at very a high velocity. Thus, a “minimization” run with the NAMD software [19] was performed to guide the system to the nearest local energy minimum in the configuration space, followed by an equilibration with the protein constrained so as to permit lipids, water, and ions to adapt to the protein in its crystal form. In the third phase, the system was minimized and equilibrated again for 0.54 ns. After minimization and equilibration with the protein constrained, the lipids should be well packed around the protein, while being sure that water has not entered any forbidden regions in between the protein and the lipids. In this phase, we proceeded to release the harmonic constrains we placed on the protein in the second phase and further equilibrate the entire system. In the fourth phase, simulations were run for 11 ns. Since the protein has been equilibrated in the previous three phases and the lipid tails have been packed well, the system should be able to run simulations with the area in the XY-plane being constant, which is different from the previous simulations where the area in the XY-plane fluctuated in order to permit packing of the lipids against the protein. All simulations were performed using the CHARMM22 force field [19,20], the TIP3P water model, and the MD program NAMD2 [21], with periodic boundary conditions at constant temperature of 310 K and a constant pressure of 1 atm (NpT ensemble). The Langevin dynamics and the Langevin piston methods were used to keep the temperature and pressure constant. Full electrostatics was employed using the Particle Mesh Ewald (PME) method [22].
2.4. Free energy computation
After the long equilibrium MD simulations, we performed steered molecular dynamics (SMD) to pull four water molecules through the four channels of AQP4 simultaneously, one water molecule through each of them. We pulled 40 Å length with start position near 10 Å (Periplasm) to the end near –30 Å (cytoplasm). Two sets of pulling paths were sampled: each set has five forward paths (from the periplasm to the cytoplasm side) and five reverse paths (from cytoplasm to periplasm). The order parameter is chosen as the center-of-mass Z-coordinate of one pulled water molecule, and the pulling speed is 0.05 Å/ps. Subsequently, the Brownian dynamics fluctuation–dissipation-theorem (BD-FDT) [23,24] was used to compute the free-energy profiles for each set of the five forward and five reverses paths.
3. Results
3.1. Equilibrium molecular dynamic simulations
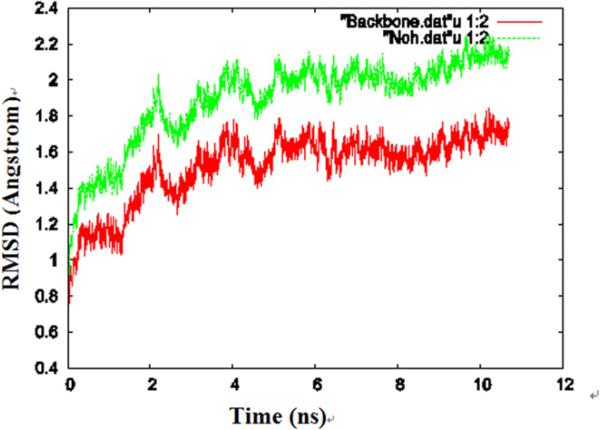
During the simulations, the protein exhibited significant fluctuation, which is demonstrated in Fig. 1, where the root mean square deviation (RMSD) of the structure of one of the AQP4 monomers relative to the crystal structure is plotted as a function of equilibration time. The change of the RMSD in the last 4 ns is relatively small (less than 0.2 Å), implying that the protein had reached a structural equilibrium with the lipids. We saved the coordinates of the last frame for analysis of its structure.
Fig. 1.
The RMSD for one of the monomers during the equilibrium molecular dynamics simulations. The RMSD of the protein heavy atoms (hydrogen atoms are excluded) is shown in green, the RMSD of the protein backbone is shown in red. (For interpretation of the references in color in this figure legend, the reader is referred to the web version of this article.)
3.2. Central pore along the fourfold axis
The tetramer was made of four monomers, which form a pore along the central fourfold axis. During the simulation, there was no water in the central pore, although water molecules were observed flowing through each of the four channels of the AQP4 tetramer. By cross-sectional analysis of the protein-membrane complex in length of 40 Ǻ along the central fourfold axis, we found that the amino acid residues involved in forming the central pore are Met-70, Asp-69, Val-71, Leu-202, Leu-75, Leu-79, Ala-198, Ser-74, Phe-195, Gly194, Leu-191, Ser-188, all of which are hydrophobic. According to our sectional analysis, Phe-195, Leu-191 and Leu-75 form the narrowest part of the pore with a size smaller than the diameter of a water molecule, which should create a hydrophobic block.
3.3. Water arrangement in the channel
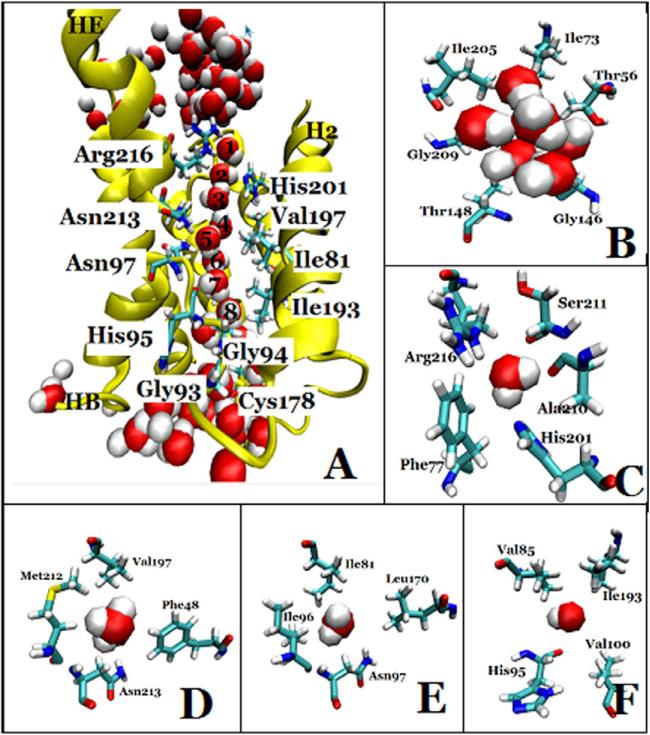
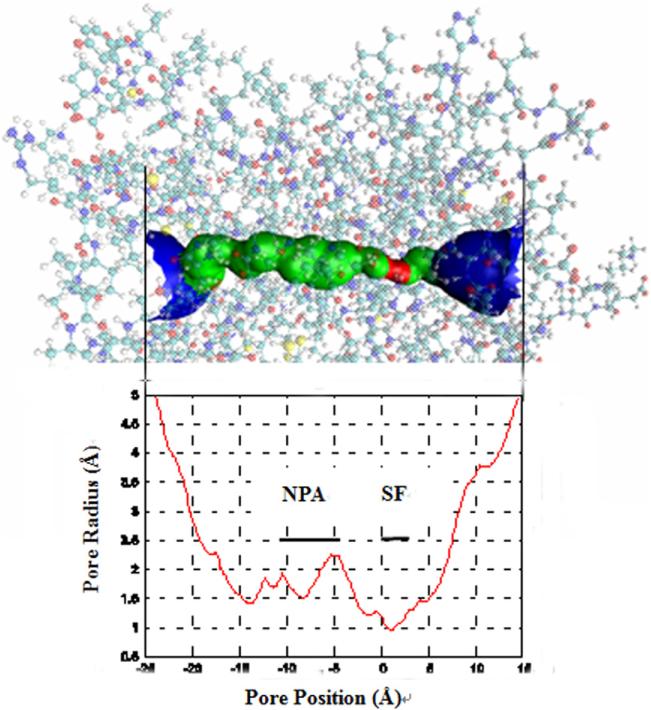
We examined the structure of the last frame, and found that there were eight water molecules forming a single file in each of the four water conducting channels of the AQP4 tetramer (Fig. 2A). Before water molecules enter into the channel, they stay in the extracellular vestibule which consists of Thr-56, Ile-73, Ile-205, Gly-209, Thr-148 and Gly-146 (Fig. 2B). Due to the selectivity filter formed by Arg-216, His-201, Phe-77, Ala-210 (Fig. 2C), the channel diameter reduces to be very small, sterically excluding the passage of molecules larger than water such as glycerol. This section has the narrowest diameter size in the channel as computed by HOLE 2.0 software (Fig. 3). In the middle section of the channel, the two conserved Asn-Pro-Ala (NPA) motifs were observed in Asn-213 and Asn-97 (Fig. 2D and Fig. 2E). Further down the channel toward the intracellular side, the water molecules interact with His-95, and the carbonyl groups of residues Val-85, Ile-193 and Val-100 in the pore (Fig. 2F). At the end of the conduction pore, the water molecules interact with residues Gly-94, Ile-174, Gly-93, and Cys-178 (Fig. 2A).
Fig. 2.
The water-conducting channel of one monomer of AQP4. (A) The overall architecture, the protein is shown in NewCartoon and important residues in Licorice drawing method, water molecules in VDW; (B) the extracellular vestibule, formed by Thr-56, Ile-73, Ile-205, Gly-209, Thr-148 and Gly-146, depicted in Licorice drawing method and water molecules in VDW; (C) the selectivity filter, formed by Arg-216, His-201, Phe-77 and Ala-210, depicted in Licorice drawing method and water molecules in VDW; (D) Asn-213, a member of the NPA motif, which interacts with water molecules in the conducting channel. Residues Asn-213, Val-197, Phe-48, and Met-212 are depicted in Licorice drawing method and water molecules in VDW; (E) Asn-97, a member of the other NPA motif, which interact with water molecules in the conducting channel. Residues Asn-97, Ile-96, Ile-81 and Leu-170 are depicted in Licorice drawing method and water in VDW; (F) His-95, which interact with water molecules in the conducting channel. Residues His-95, Val-100, Val-85 and Ile-193 are depicted in Licorice drawing method and water in VDW.
Fig. 3.
The dimension of the conducting channel's inner surface calculated using the Hole 2.0 program.
3.4. Steered molecular dynamics simulations and free energy computation
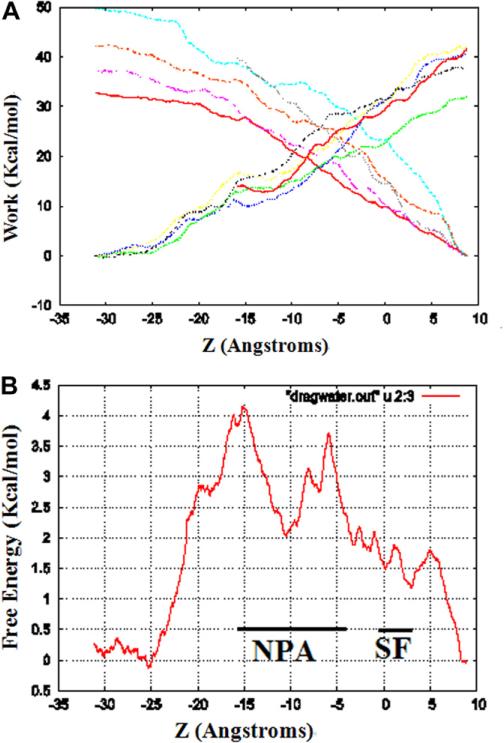
Starting from the last frame of the fully equilibrated structure of AQP4, after the equilibrium Langevin dynamics for 11 ns, we performed SMD simulations. We pulled four water molecules through the four channels of AQP4 simultaneously, one water molecule through each of them, sampling two sets of five forward and five reverse pulling paths. Subsequently, we applied the BD-FDT to each set of the five forward and five reverse paths and obtained a set of free-energy landscape. The work of the five forward and five reverse is shown in Fig. 4A, and the overall free-energy profile is shown in Fig. 4B that gives a complete map of the entire channel of water permeation.
Fig. 4.
The work curves and the free-energy profile of water permeation through the channels of AQP4. (A) Work done along the forward and reverse pulling paths. Using the last frame, four water molecules were pulled from start position at 10 Å to –30 Å through AQP4 channels, one water molecule through each channel, and five forward and five reverse paths were performed. The work is average over the four water molecules pulled through the four channels simultaneously. (B) The free-energy profile of AQP4 was computed by using the Brownian dynamics fluctuation–dissipation-theorem (BD-FDT) method according to the mechanical work done along the above pulling paths. The origin of the coordinate system is so chosen that marked for the selectivity filter (SF) NPA motifs.
4. Discussion
AQP4 has been reported to be the predominant water channel in the brain, especially in the blood–brain and brain-cerebrospinal fluid interfaces of glial cells [9,10]. In 2009, the 1.8-Å crystal structure of human AQP4 was reported [16], which revealed that each of the AQP4 monomers were formed by six and two half-length alpha-helices, tetramerizing along the crystallographic fourfold axis. The present study was aimed to develop a molecular dynamics system of an AQP4 tetramer in a lipid bilayer, and the fact that the change of the RMSD in the last few nanoseconds of the MD simulation is very small seems to demonstrate our success. Like other water channel proteins, AQP4 has high water permeability with a continuous water column inside the channel in normal physiological environments, and no empty channel exists [11–13,15,25–28]. This water permeation is referred to as single-file permeation and was demonstrated in our simulations. In our simulations, eight water molecules were presented in perfect single-file, where all the water molecules had moved in the channel with no gaps and without interchanging position at any time.
Water permeation is passively driven by osmotic gradients, so the water flow should be regulated by structural changes of the channel. To understand the pathway of water trafficking, we dissected the equilibrated AQP4-lipid system along the XY-plane, and found there are many amino acid residues on the inner surface of the channel, including the hydrophobic Phe-77, Ile-81, Val-85, Leu-170, Ile-174, Val-197, the eight backbone carbonyls of Gly-93, Gly-94, His-95, Ile-96, Gly-209, Ala-210, Ser-211, and Met-212, along with the important NPA motif members of Asn-97 and Asn-213, as determined by experimental methods. According to the cross-sectional analysis, two sites strongly interacting with water were observed, the constriction and the NPA motif, which have been demonstrated in other AQPs [11–13]. One of these is referred to as aromatic residue/arginine (ar/R) constriction, the narrowest part of the water channel, which is located close to the extracellular pore mouth and formed by four residues, i.e. Arg-216, His-201, Ala-210 and Phe-77. The histidine is able to reduce the channel diameter, and together with the highly conserved arginine, provides a hydrophilic edge in juxtaposition to an aromatic residue, which are very important for water-specific channels, sterically excluding the passage of glycerol. In Escherichia coli aquaglyceroporin GlpF, the ar/R region is formed by Trp-48, Gly-191, Phe-200 and Arg-205, while that of AQP1 is formed by Phe-56, His-180, Cys-189 and Arg-195 [13]. The reason that GlpF is more hydrophobic than AQP1 is believed to be associated with the lack of the histidine in the ar/R region of GlpF [11]. The hydrophobic Phe-77 side chain orients the water molecules such as to enforce strong hydrogen bonds to Arg-216 and His-201.
Another narrow constriction, the NPA region, is located at the center of the pore. At the end of the helices HB and HE there are two asparagines, i.e. Asn-213, Asn-97, which should act as hydrogen donors to the oxygen atoms of passing water molecular. Both of these asparagines lie on one side of the pore, and the hydrophobic side chains of Vla-197, Phe-48 and Met-212, Ile-81, Ile-96, and Leu-170 on the other of the pore. When water enters into this region, it is reoriented by the dipoles of the emanating half helices HB and HE, such that hydrogen bonds between neighboring water molecules in the chain are disrupted. Interestingly, these two asparagines, Asn-97, Asn-213, each donate its single, highly oriented hydrogen bond to a separate water molecule [16]. In contrast, in AQP0, AQP1, AQPZ and other water-selective channels, the two asparagines in the NPA motifs donate hydrogen bonds to a single water molecule [25,26,29]. Further down the channel of AQP4 toward the intracellular side, the water molecules interact with His-95, and the carbonyl groups of residues Val-85, Ile-193 and Val-100 in the channel. In the end of the conduction channel, the water molecules interact with the residues Gly-94, Ile-174, Gly-93 and Cys-178.
According to our MD simulations, before entering into the AQP4 channel, some water molecules appear in the extracellular vestibule consisting of Thr-56, Ile-73, Ile-205, Gly-209, Thr-148 and Gly-146. As we know, members of the water channel superfamily include both the aquaporins and aquaglyceroporins. In aquaglyceroporins, the vestibule is regarded to be a place for glycerol recruitment and desolvation of solutes for transport through the channel, as confirmed by the crystal structures of GlpF and PfAQP (malaria aquaglyceroporin) with glycerol molecules located in the entry vestibule [13,27,28]. In our initial structure of AQP4, three glycerol molecules were included in the extracellular vestibule, but no glycerol in whole channel from the selectivity filter to the end [16].
In the natural membranes, water channel proteins are in the form of tetramers, however, each monomer has its own channel with independent function. The tetramer is held together by extensive interactions between helices and loops of the four monomers, and has a hole in the center of the molecule. Although the tetramers were solvated in water, there are not any water molecules in this center hole throughout our simulations as long as 11 ns, although water molecules flow in single-file not only into and out of the four channels of the AQP4 tetramers. Our analysis shows the inner surface of the AQP4 channel are composed by many hydrophobic amino acids, but the hole sizes positioned at Phe-195, Leu-191, and Leu-75 are smaller than that of a water molecule, which were suggested to play important roles in the blocking of water molecules through the hole.
Accurate computation of free-energy is of essential importance in quantitative studies of biological systems because most biophysical/chemical processes are driven by the free-energy gradients. Methods to calculate free-energy differences between two states (conformations) A and B fall into two classes, equilibrium methods and, more recently developed, non-equilibrium methods. Equilibrium approaches, such as the thermodynamic integration (TI) method, require more computing resources because they are based on “full” sampling of the equilibrium ensembles involved in a given biophysical process. Non-equilibrium approaches exploit the stochastic dynamics of the system driven with some applied forces and aim to map out the free-energy landscape in terms of the potential of mean force (PMF) [30]. The free-energy difference between States A and B is extracted from the measurements of work along the transition paths connecting the two states. Recently, a non-equilibrium PMF type of approach has been developed on the basis of the Brownian dynamics fluctuation–dissipation theorems (BD-FDT), extracting the equilibrium free-energy differences from irreversible (non-equilibrium) work measurements in steered molecular dynamics (SMD) simulations [23,24]. In the present study, we computed the free energy profiles of water through AQP4 by BD-FDT, which is accurate in the limit of finitely long simulations. Using BD-FDT, a non-equilibrium PMF type of approach, we can run shorter simulations and still obtain accurate results.
In summary, we conducted MD simulations of AQP4, and computed its free-energy profile for the first time. The analysis of water molecules through the AQP4 channel, and the pore along the central fourfold axis in this paper should be helpful to the understanding of aquaporin structure and its functional implications.
Acknowledgments
The author acknowledges support from the NIH (Grant No. SC3 GM084834), and helpful discussions with Dr. Liao Chen, Guodong Hu and Yubo Zhang.
Abbreviations
- MD
molecular dynamics
- SMD
steered MD
- PDB
protein data bank
- RMSD
root mean-squared deviation
- POPC
palmitoyl-oleoyl-phosphatidylcholine
- PSF
protein structure file
- NPA
Asn-Pro-Ala
- PME
Particle Mesh Ewald
- BD-FDT
brownian dynamics fluctuation–dissipation-theorem
References
- 1.Agre P, King LS, Yasui M, et al. Aquaporin water channels-from atomic structure to clinical medicine. J. Physiol. 2002;542:3–16. doi: 10.1113/jphysiol.2002.020818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arciénega II, Brunet JF, Bloch J, et al. Cell locations for AQP1, AQP4 and AQP9 in the non-human primate brain. Neuroscience. 2010;167:1103–1114. doi: 10.1016/j.neuroscience.2010.02.059. [DOI] [PubMed] [Google Scholar]
- 3.Badaut J, Lasbennes F, Magistretti PJ, et al. Aquaporins in brain: distribution, Physiology and pathophysiology. J. Cereb. Blood Flow Metab. 2002;22:367–378. doi: 10.1097/00004647-200204000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Benga G. Water channel proteins (later called aquaporins) and relatives: past, present, and future. IUBMB Life. 2004;61:112–133. doi: 10.1002/iub.156. [DOI] [PubMed] [Google Scholar]
- 5.Masseguin C, Corcoran M, Carcenac C, et al. Altered gravity downregulates aquaporin-1 protein expression in choroid plexus. J. Appl. Physiol. 2000;88:843–850. doi: 10.1152/jappl.2000.88.3.843. [DOI] [PubMed] [Google Scholar]
- 6.Speake T, Freeman LJ, Brown PD. Expression of aquaporin 1 and aquaporin 4 water channels in rat choroid plexus. Biochim. Biophys. Acta. 2003;1609:80–86. doi: 10.1016/s0005-2736(02)00658-2. [DOI] [PubMed] [Google Scholar]
- 7.Brown PD, Davies SL, Speake T, et al. Molecular mechanisms of cerebrospinal fluid production. Neuroscience. 2004;129:957–970. doi: 10.1016/j.neuroscience.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badaut J, Regli L. Distribution, possible roles of aquaporin 9 in the brain. Neuroscience. 2004;129:971–981. doi: 10.1016/j.neuroscience.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 9.Nagelhus EA, Mathiisen TM, Ottersen OP. Aquaporin-4 in the central nervous system: cellular and subcellular distribution and coexpression with KIR4.1. Neuroscience. 2004;129:905–913. doi: 10.1016/j.neuroscience.2004.08.053. [DOI] [PubMed] [Google Scholar]
- 10.Nicchia GP, Nico B, Camassa LM, et al. The role of aquaporins-4 in the blood–brain barrier development and integrity: studies in animal and cell culture models. Neuroscience. 2004;129:935–945. doi: 10.1016/j.neuroscience.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 11.Walz T, Fujiyoshi Y, Engel A. The AQP structure and functional implications. Handb. Exp. Pharmacol. 2009;190:31–56. doi: 10.1007/978-3-540-79885-9_2. [DOI] [PubMed] [Google Scholar]
- 12.Wu B, Beitz E. Aquaporins with selectivity for unconventional permeants. Cell Mol. Life Sci. 2007;64:2413–2421. doi: 10.1007/s00018-007-7163-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Groot BL, Grubmüller H. Water permeation across biological membrances: mechanism and dynamics of aaquaporin-1 and GlpF. Science. 2001;294:2353–2357. doi: 10.1126/science.1066115. [DOI] [PubMed] [Google Scholar]
- 14.Hub JS, Grubmüller H, de Groot BL. Dynamics and energetics of permeation through aquaporins, what do we learn from molecular dynamics simulations ? Handb Exp. Pharmacol. 2009;190:57–76. doi: 10.1007/978-3-540-79885-9_3. [DOI] [PubMed] [Google Scholar]
- 15.de Groot BL, Grubmüller H. The dynamics and energetics of water permeation and proton exclusion in aquaporins. Curr. Opin. Struct. Biol. 2005;15:176–183. doi: 10.1016/j.sbi.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Ho JD, Yeh R, Sandstrom A, et al. Crystal structure of human aquaporin 4 at 1.8 Å and its mechanism of conductance. Proc. Natl. Acad. Sci. USA. 2009;106:7437–7442. doi: 10.1073/pnas.0902725106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humphrey W, Dalke A, Schulten K. VMD-visual molecular dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 18.Grubmüller H. SOLVATE v. 1.0. Theoretical Biophysics Group, Institute for Medical Optics, Ludwig-Maximilians University; Munich: 1996. [Google Scholar]
- 19.MacKerell AD, Jr., Bashford D, Bellott M, et al. Self-consistent parameterization of biomolecules for molecular modeling and condensed phase simulations. FASEB J. 1992;6:A143. [Google Scholar]
- 20.MacKerell AD, Jr., Bashford D, Bellott M, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3568–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 21.Kalé L, Skeel R, Bhandarkar M, et al. NAMD2: greater scalability for parallel molecular dynamics. J. Comp. Phys. 1999;151:283–312. [Google Scholar]
- 22.Essmann U, Perera L, Berkowitz ML, et al. A smooth Particle Mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 23.Chen LY, Bastien DA, Espejel HE. Determination of equilibrium free-energy from non-equilibrium work measurements. Phys. Chem. Chem. Phys. 2010;12:6579–6582. doi: 10.1039/b926889h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen LY. Free-energy landscape of glycerol permeation through aquaglyceroporin GlpF determined from steered molecular dynamics simulations. Biophys. Chem. 2010;151:178–180. doi: 10.1016/j.bpc.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Groot BL, Frigato T, Helms V, et al. The mechanism of proton exclusion in the aquaporin-1 water channel. J. Mol. Biol. 2003;333:279–293. doi: 10.1016/j.jmb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Harries WE, Akhavan D, Miercke LJ, et al. The channel architecture of aquaporin 0 at 2.2-Å resolution. Proc. Natl. Acad. Sci. USA. 2004;101:14045–14050. doi: 10.1073/pnas.0405274101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hub JS, de Groot BL. Mechanism of selectivity in aquaporins and aquaglyceroporins. Proc. Natl. Acad. Sci. USA. 2008;105:1198–1203. doi: 10.1073/pnas.0707662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newby ZE, O'Connell J, 3rd, Robbes-Colmenares Y, et al. Crystal structure of the aquaglyceroporin PfAQP from the malarial parasite Plasmodium falciparum. Nat. Struct. Mol. Biol. 2008;15:619–625. doi: 10.1038/nsmb.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savage DF, Egea PF, Robles-Colmenares Y, et al. Architecture and selectivity in aquaporins: 2.5 a X-ray structure of aquaporin Z. PLoS Biol. 2003;1:E72. doi: 10.1371/journal.pbio.0000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hummer G, Szabo A. Free energy reconstruction from nonequilibrium single-molecule pulling experiments. Proc. Natl. Acad. Sci. USA. 2001;98:3658–3661. doi: 10.1073/pnas.071034098. [DOI] [PMC free article] [PubMed] [Google Scholar]