Abstract
Fetal alcohol spectrum disorders (FASD) results from ethanol exposure to the developing fetus and is the leading cause of mental retardation. FASD is associated with a broad range of neurobehavioral deficits which may be mediated by ethanol-induced neurodegeneration in the developing brain. An immature brain is more susceptible to ethanol neurotoxicity. We hypothesize that the enhanced sensitivity of the immature brain to ethanol is due to a limited capacity to alleviate cellular stress. Using a third trimester equivalent mouse model of ethanol exposure, we demonstrated that subcutaneous injection of ethanol induced a wide-spread neuroapoptosis in postnatal day 4 (PD4) C57BL/6 mice, but had little effect on the brain of PD12 mice. We analyzed the expression profile of genes regulating apoptosis, and the pathways of ER stress response (also known as unfolded protein response, UPR) and autophagy during these ethanol-sensitive and resistant periods (PD4 versus PD12) using PCR microarray. The expression of pro-apoptotic genes, such as caspase-3, was much higher on PD4 than PD12; in contrast, the expression of genes that regulate UPR and autophagy, such as atf6, atg4, atg9, atg10, beclin1, bnip3, cebpb, ctsb, ctsd, ctss, grp78, ire1α, lamp, lc3 perk, pik3c3, and sqstm1 was significantly higher on PD12 than PD4. These results suggest that the vulnerability of the immature brain to ethanol could result from high expression of pro-apoptotic proteins and a deficiency in the stress responsive system, such as UPR and autophagy.
Keywords: alcohol, brain development, fetal alcohol syndrome, gene expression, neurodegeneration
Introduction
Fetal alcohol spectrum disorders (FASD) are a range of disabilities that result from prenatal alcohol exposure. Children with FASD exhibit cognitive, neuropsychological and behavioral problems, and numerous secondary disabilities including depression and anxiety disorders (Hellemans et al. 2009; Riley et al. 2011). Fetal alcohol exposure is a leading non-genetic cause of mental retardation, and affects brain structure and function. The most devastating consequence of ethanol exposure is the neuronal loss in the developing central nervous system (CNS) which accounts for many symptoms shown in FASD children. Mechanisms underlying ethanol-induced neuronal loss are complex. Animal studies have demonstrated temporal windows of vulnerability to ethanol-induced neuronal loss in the developing CNS (Maier et al. 1999; Siler-Marsiglio et al. 2006). Ethanol exposure during different periods of brain development results in differences in cell loss as a function of the timing of ethanol exposure during brain development.
Ethanol is a cellular stress inducer and causes oxidative stress and endoplasmic reticulum (ER) stress in the developing brain (Chen and Luo 2009; Ke et al. 2011a). The sustained cellular stresses may result in neuronal death if they are not adequately alleviated by internal defense systems, such as the antioxidant system, ER stress response (also known as unfolded protein response, UPR) or autophagy. We have recently demonstrated that ethanol exposure can activate a neuroprotective autophagy (Chen et al. 2012). Autophagy is one of the two major pathways that accomplish regulated protein catabolism. The other one is the ubiquitin-proteasome system (UPS). Autophagy is a lysosomal degradation pathway involved in the turnover of cellular macromolecules and organelles and is essential for survival, differentiation, development and homeostasis (Levine and Kroemer 2008). Although some of these functions overlap with those of the UPS, autophagy is primarily responsible for degrading long-lived proteins and maintaining amino acid pools in the setting of chronic starvation. The autophagy pathway is uniquely capable of degrading entire organelles such as mitochondria, peroxisomes and ER, as well as intact intracellular microorganisms (Levine and Kroemer 2008). Autophagic degradation of cellular constituents can efficiently recycle essential nutrients to sustain basic biological processes. Autophagy is also used as a defense mechanism to clear intracellular microbes, misfolded proteins and damaged organelles. Constitutive clearance of cytosolic proteins by low-level basal autophagy is an additional important cytoprotective function, particularly to neurons.
The ER regulates posttranslational protein processing and transport. The ER is also the site for the biosynthesis of steroids, cholesterol and other lipids. Under some cellular stress conditions, unfolded or misfolded proteins accumulate in the ER lumen and activate a compensatory response which has been referred to as ER stress response or UPR (Ron 2002; Xu et al. 2005). UPR initiates protective responses, resulting in an overall decrease in protein synthesis, enhanced protein degradation and increased protein folding capacity of the ER (Ron 2002). However, sustained ER stress ultimately leads to apoptotic death of the cell (Xu et al. 2005; Rasheva and Domingos 2009).
We hypothesize that the brain develops more effective defense systems as it matures to alleviate environmental stresses, becoming more resistant to ethanol neurotoxicity. In this study, we demonstrated that the brain of postnatal day 4 (PD4) C57BL/6 mice was sensitive to ethanol-induced neuroapoptosis, but the brain of PD12 was quite resistant. Using pathway-specific PCR microarrays, we showed that the expression of pro-apoptotic genes, such as caspase-3, was much higher at the ethanol sensitive period (PD4). However, the expression of genes that regulate UPR and autophagy significantly increased at the ethanol-resistant period (PD12).
Materials and Methods
Materials
Animals and treatment
C57BL/6 mice were obtained from Harlan Laboratories (Indianapolis, IN) and maintained in the Animal Facility at the University of Kentucky Medical Center. All procedures were performed in accordance with the guidelines set by the NIH and the Animal Care and Use Committee of the University of Kentucky. The Institutional Animal Care & Use Committee (IACUC) has specifically approved this study. Each 4-day-old (PD4) or 12-day-old (PD12) mouse pup in a litter was assigned to either control (saline) or ethanol group. An ethanol exposure paradigm, which had been shown to induce robust neurodegeneration in mice (Olney et al. 2002; Liu et al. 2009), was employed. The mice were injected subcutaneously with saline or ethanol (2.5 g/kg, 20% solution in saline) twice at 0 h and 2 h. Eight hours after the first ethanol injection, the brains were removed. Three pups from each treatment group (saline or ethanol) on PD4 and PD12 were processed for PCR microarray and immunohistochemical (IHC) study.
Sample preparation
Protein extraction
Mice were anesthetized by intraperitoneal injection of ketamine (100 mg/kg)/xylazine (10 mg/kg), and cerebral cortices were immediately dissected. The tissues were frozen in liquid nitrogen and stored at −80°C. Proteins were extracted as previously described (Ke et al. 2011a). Briefly, tissues were homogenized in an icecold lysis buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EGTA, 1 mM PMSF, 0.5% NP-40, 0.25% SDS, 5 µg/ml leupeptin and 5 µg/ml aprotinin. Homogenates were centrifuged at 20,000 g for 30 min at 4°C and the supernatant fraction was collected.
RNA isolation
Cerebral cortices from PD4 or PD12 C57BL/6 mice were harvested and frozen. Samples were homogenized with 2 ml of TRIzol reagent (Invitrogen) using a MISONIX sonicator XL-2000 (Qsonica, LLC, Newtown, CT, USA) for 30 sec at Power 2 on ice. Total RNA was extracted from 1 ml of each homogenate according to manufacturer’s protocol. RNA was quantified using NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA). A260/A280 ratio was in the range from 1.97 to 2.05. A260/A230 ratio was between 2.14 and 2.35. One microgram of total RNA was used for first strand cDNA synthesis (RT2 First Strand Kit, SABiosciences, Frederick, MD, USA, Cat.# 330401) according to manufacturers’ protocols.
Immunohistochemistry (IHC)
After treatments, the mice were deeply anesthetized with ketamine (100 mg/kg)/xylazine (10 mg/kg), and then perfused with saline followed by 4% paraformaldehyde in 0.1 M potassium phosphate buffer (pH 7.2). The brains were removed and post-fixed in 4% paraformaldehyde for an additional 24 hours, and then transferred to a 30% sucrose solution. The brain was sectioned at 40 µm with a sliding microtome (Leica Microsystems, Wetzlar, Germany). The procedure for IHC staining has been described (Ke et al. 2011a). Briefly, free-floating sections were incubated in 0.3% H2O2 in methanol for 30 min at room temperature and then treated with 0.1% TritonX-100 for 10 min in PBS. The sections were washed with PBS three times, and then blocked with 1% BSA and 0.01% TritonX-100 for 1 hour at room temperature. The sections were incubated with an anti-active caspase-3 antibody (at dilution of 1:1000) overnight at 4°C. Negative controls were performed by omitting the primary antibody. After rinsing in PBS, sections were incubated with a biotinylated goat anti-rabbit IgG (Vector Laboratories Inc., Burlingame, CA; 1:200) for 1 hour at room temperature. The sections were washed 3 times with PBS, then incubated in avidin–biotin–peroxidase complex (Vector Laboratories Inc. 1:100 in PBS) for 1 h and developed in 0.05% 3,3’-diaminobenzidine (DAB) (Sigma-Aldrich) containing 0.003% H2O2 in PBS. The images were recorded using an Olympus microscope (BX61) equipped with a DP70 digital camera.
Analysis of genes associated with autophagy and unfolded protein response (UPR) by PCR microarray
To analyze differential expression of genes involved in autophagy and UPR during brain development, we used the RT2 Profiler PCR Array System (SABiosciences, Frederick, MD, USA, 96 well format). Mouse autophagy PCR array contained 84 key genes in the autophagic pathway (Cat.# PAMM-084Z) and UPR PCR array contained 84 key genes for the UPR pathway (Cat.# PAMM-089F). Three samples from each group and six plates for each array were used. Briefly, 102 µl of cDNA from each sample was mixed with 1350 µl of 2× RT2 SYBR Green Master mix (SABiosciences) and RNase-free H2O to the final volume 2,700 µl. 25 µl of sample was used for each well of the RT2 Profiler PCR Array plate. Quantitative Real-Time PCR was performed in a Roche Light Cycler 480 using software 1.5.OS04. After 10 min of activation at 95°C, 45 cycles were performed with the following cycle parameters: 15 sec at 95°C, 1 min at 60°C (acquisition). After finishing the last cycle a melting curve analysis was performed. Standard −ΔΔCt method was used for determining changes in gene expression during the development. SABiosciences web-based software for Standard RT2 PCR Array analysis (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php) was used to calculate experimental results including statistical analysis. The data were normalized with beta-actin.
Immunoblotting
The immunoblotting procedure has been previously described (Chen at al. 2012). Briefly, aliquots of the protein samples (30 µg) were separated on a SDS-polyacrylamide gel by electrophoresis. The separated proteins were transferred to nitrocellulose membranes. The membranes were blocked with either 5% BSA or 5% nonfat milk in 0.01 M PBS (pH 7.4) and 0.05% Tween-20 (TPBS) at room temperature for one hour. Subsequently, the membranes were probed with primary antibodies directed against target proteins overnight at 4°C. After three quick washes in TPBS, the membranes were incubated with a secondary antibody conjugated to horseradish peroxidase (Amersham, Arlington Hts. IL). The immune complexes were detected by the enhanced chemiluminescence method (Amersham). In some cases, the blots were stripped and re-probed with either an anti-tubulin or an anti-actin antibody. The density of immunoblotting was quantified with the software of Quantity One (Bio-Rad Laboratories, Hercules, CA).
Statistical analysis
Statistical analysis of obtained data was performed with SABiosciences web-based software for Standard RT2 PCR Array analysis. Student t-test was used to verify the analysis. Changes in gene expression levels with p < 0.05 were considered statistically significant.
Results
Differential effect of ethanol on caspase-3 activation in the brain of PD4 and PD12 mice
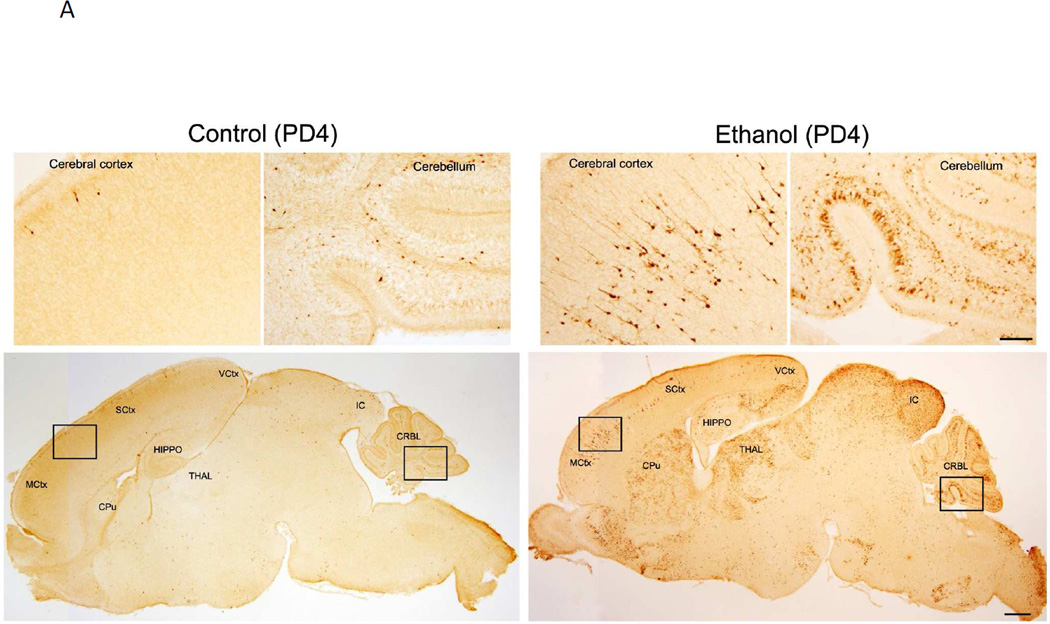
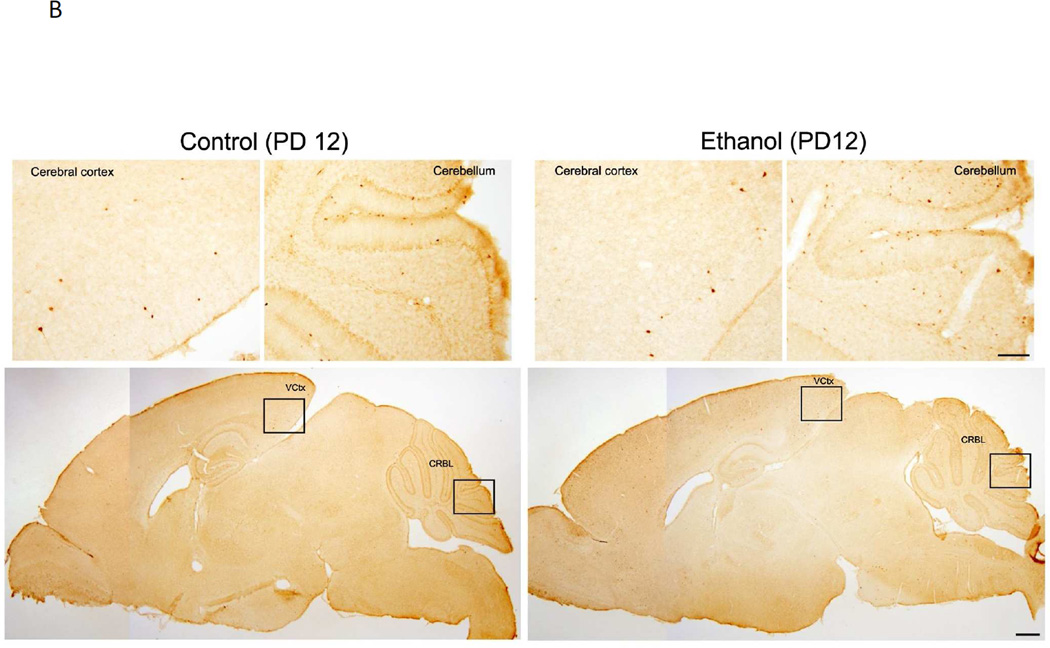
We have previously demonstrated ethanol exposure on PD7 induced a wide spread neuroapoptosis in the brain, which was indicated by the increase in the expression of active caspase-3 and Fluoro-J-C positive cells (Liu et al. 2009; Ke et al. 2011b). Consistent with the previous findings, we showed a drastic increase of immunohistochemical staining of active caspase-3 in the brain of PD4 mice (Fig. 1A). Ethanol-induced activation of caspase-3 was particularly strong in some brain regions, such as cerebral cortex, caudate putamen, thalamus, Inferior colliculus and cerebellum. On PD12, however, ethanol caused little caspase-3 activation (Fig. 1B). These results indicated a temporal window of vulnerability; i.e., brain of PD4 mice was sensitive but that of PD12 was resistant to ethanol-induced neuroapoptosis.
Figure 1. Effect of ethanol on the expression of active caspase-3 in the brain of postnatal day 4 (PD4) and (PD12) mice.
PD4 (A) and PD12 mice (B) were injected subcutaneously with ethanol (2.5 g/kg, or saline) at 0 and 2 hours as described under the Material and Methods. At 8 hours after the first injection, the brain was removed and fixed. The sagittal brain sections were processed for immunohistochemical analysis of active caspase-3 as described under the Materials and Methods. Inset regions in the lower panels (indicated by boxes) are shown in upper panel with higher magnification. Scale bar = 100 µM for lower panels. Bar = 500 µM for upper panels. MCtx: Motor cortex; SCtx: Somatosensory cortex; VCtx: Visual cortex: HIPPO: Hippocampus; CPu: Caudate putamen; THAL: Thalamus; IC: Inferior colliculus; CRBL: Cerebellum.
The expression of genes related to autophagy and UPR in the brain of PD4 and PD12 mice
We examined and compared the expression profiles of genes associated with autophagy and UPR pathways in cortex samples obtained from PD4 and PD12 mice. We have screened a total of 168 genes and presented the results showing changes greater than 1.9-fold or with p < 0.05 (Table 1). We observed changes in expression profile of genes involved in all aspects of UPR (unfolded protein binding, ubiquitination, ER associated degradation, protein folding, apoptosis, transcriptional regulation) and autophagy (vacuole formation, protein targeting to membrane/vacuole, protein transport, protease activity, chaperone mediated autophagy and co-regulation of autophagy, cell cycle and apoptosis). As the brain matured, the expression of genes involved in autophagy and UPR pathways generally increased with the exception of Casp3 and Cxcr4. The expression of Casp3 and Cxcr4 was higher on PD4 than PD12.
Table1.
Developmental expression genes associated with autophagy and UPR in the brain (PD12 versus PD4)
| Accession № | Gene symbol | Gene name | Fold change | p-value |
|---|---|---|---|---|
| NM_172669 | Ambra1 | Autophagy/beclin 1 regulator 1 | 1.64 | 0.008091 |
| NM_007471 | App | Amyloid beta (A4) precursor protein | 2.49 | 0.000004 |
| NM_001081304 | Atf6 | Activating transcription factor 6 | 2.18 | 0.000968 |
| NM_017406 | Atf6b | Activating transcription factor 6 beta | 1.79 | 0.005351 |
| NM_025770 | Atg10 | Autophagy-related 10 (yeast) | 2.66 | 0.004272 |
| NM_026217 | Atg12 | Autophagy-related 12 (yeast) | 1.57 | 0.044179 |
| NM_029846 | Atg16l1 | Autophagy-related 16-like 1 (yeast) | 1.78 | 0.008052 |
| NM_001111111 | Atg16l2 | Autophagy related 16 like 2 (S. cerevisiae) | 1.67 | 0.004659 |
| NM_026402 | Atg3 | Autophagy-related 3 (yeast) | 1.7 | 0.010828 |
| NM_174875 | Atg4a | Autophagy-related 4A (yeast) | 1.55 | 0.003280 |
| NM_174874 | Atg4b | Autophagy-related 4B (yeast) | 1.55 | 0.009190 |
| NM_175029 | Atg4c | Autophagy-related 4C (yeast) | 1.91 | 0.007793 |
| NM_153583 | Atg4d | Autophagy-related 4D (yeast) | 1.33 | 0.001714 |
| NM_053069 | Atg5 | Autophagy-related 5 (yeast) | 1.63 | 0.005571 |
| NM_001003917 | Atg9a | Autophagy-related 9A (yeast) | 3.23 | 0.001477 |
| NM_001002897 | Atg9b | ATG9 autophagy related 9 homolog B (S. cerevisiae) | 1.17 | 0.032070 |
| NM_029705 | Atxn3 | Ataxin 3 | 1.61 | 0.013019 |
| NM_007522 | Bad | BCL2-associated agonist of cell death | 1.27 | 0.013720 |
| NM_007523 | Bak1 | BCL2-antagonist/killer 1 | 1.74 | 0.013974 |
| NM_009743 | Bcl2l1 | Bcl2-like 1 | 1.44 | 0.017157 |
| NM_019584 | Becn1 | Beclin 1, autophagy related | 1.89 | 0.019423 |
| NM_007544 | Bid | BH3 interacting domain death agonist | 1.29 | 0.022037 |
| NM_009760 | Bnip3 | BCL2/adenovirus E1B interacting protein 3 | 3.71 | 0.003229 |
| NM_009810 | Casp3 | Caspase 3 | −2.26 | 0.000101 |
| NM_009812 | Casp8 | Caspase 8 | 2.01 | 0.004562 |
| NM_009877 | Cdkn2a | Cyclin-dependent kinase inhibitor 2A | 2.05 | 0.048173 |
| NM_009883 | Cebpb | CCAAT/enhancer binding protein (C/EBP), beta | 2.64 | 0.107221 |
| NM_007798 | Ctsb | Cathepsin B | 2.5 | 0.002256 |
| NM_009983 | Ctsd | Cathepsin D | 2.49 | 0.000713 |
| NM_021281 | Ctss | Cathepsin S | 3.44 | 0.009356 |
| NM_009911 | Cxcr4 | Chemokine (C-X-C motif) receptor 4 | −2.12 | 0.001083 |
| NM_178055 | Dnajb2 | DnaJ (Hsp40) homolog, subfamily B, member 2 | 2.27 | 0.087867 |
| NM_013760 | Dnajb9 | DnaJ (Hsp40) homolog, subfamily B, member 9 | 2.19 | 0.102058 |
| NM_020566 | Dnajc4 | DnaJ (Hsp40) homolog, subfamily C, member 4 | 2.2 | 0.106666 |
| NM_026013 | Dram2 | VDNA-damage regulated autophagy modulator 2 | 1.43 | 0.043783 |
| NM_010121 | Eif2ak3 | Eukaryotic translation initiation factor 2 alpha kinase 3 | 1.46 | 0.031865 |
| NM_001005331 | Eif4g1 | Eukaryotic translation initiation factor 4, gamma 1 | 1.54 | 0.002284 |
| NM_026184 | Ero1lb | ERO1-like beta (S. cerevisiae) | 2.2 | 0.143324 |
| NM_007956 | Esr1 | Estrogen receptor 1 (alpha) | 1.48 | 0.034200 |
| NM_008064 | Gaa | Glucosidase, alpha, acid | 1.87 | 0.000951 |
| NM_019749 | Gabarap | Gamma-aminobutyric acid receptor associated protein | 1.29 | 0.049942 |
| NM_020590 | Gabarapl1 | Gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 | 3.75 | 0.001987 |
| NM_026693 | Gabarapl2 | Gamma-aminobutyric acid (GABA) A receptor-associated protein-like 2 | 1.79 | 0.006892 |
| NM_024439 | H47 | Histocompatibility 47 | 2.16 | 0.123213 |
| NM_008228 | Hdac1 | Histone deacetylase 1 | 1.46 | 0.005613 |
| NM_010413 | Hdac6 | Histone deacetylase 6 | 1.35 | 0.031663 |
| NM_022331 | Herpud1 | Homocysteine-inducible, endoplasmic reticulum stress-inducible, | ||
| ubiquitin-like domain member 1 | 1.97 | 0.109860 | ||
| NM_008244 | Hgs | HGF-regulated tyrosine kinase substrate | 1.33 | 0.007340 |
| NM_010480 | Hsp90aa1 | Heat shock protein 90, alpha (cytosolic), class A member 1 | 2.12 | 0.003698 |
| NM_008301 | Hspa2 | Heat shock protein 2 | 5.48 | 0.050877 |
| NM_008300 | Hspa4 | Heat shock protein 4 | 1.49 | 0.012886 |
| NM_011020 | Hspa4l | Heat shock protein 4 like | 2.71 | 0.085738 |
| NM_031165 | Hspa8 | Heat shock protein 8 | 1.96 | 0.014241 |
| NM_013559 | Hsph1 | Heat shock 105kDa/110kDa protein 1 | 2.81 | 0.096569 |
| NM_010414 | Htt | Huntingtin | 2.15 | 0.000375 |
| NM_010512 | Igf1 | Insulin-like growth factor 1 | 1.82 | 0.000937 |
| NM_008387 | Ins2 | Insulin II | 2.06 | 0.051993 |
| NM_008326 | Irgm1 | Immunity-related GTPase family M member 1 | 2.51 | 0.002094 |
| NM_010684 | Lamp1 | Lysosomal-associated membrane protein 1 | 1.95 | 0.002805 |
| NM_025735 | Map1lc3a | Microtubule-associated protein 1 light chain 3 alpha | 2.32 | 0.003042 |
| NM_026160 | Map1lc3b | Microtubule-associated protein 1 light chain 3 beta | 1.39 | 0.007376 |
| NM_011951 | Mapk14 | Mitogen-activated protein kinase 14 | 2.19 | 0.001644 |
| NM_016700 | Mapk8 | Mitogen-activated protein kinase 8 | 1.35 | 0.027866 |
| NM_016961 | Mapk9 | Mitogen-activated protein kinase 9 | 2.86 | 0.051805 |
| NM_020009 | Mtor | Mechanistic target of rapamycin (serine/threonine kinase) | 1.46 | 0.000169 |
| NM_008689 | Nfkb1 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1, p105 | 1.63 | 0.013371 |
| NM_008720 | Npc1 | Niemann Pick type C1 | 1.14 | 0.014318 |
| NM_199469 | Nploc4 | Nuclear protein localization 4 homolog (S. cerevisiae) | 1.96 | 0.002431 |
| NM_008749 | Nucb1 | Nucleobindin 1 | 1.82 | 0.015377 |
| NM_177614 | Os9 | Amplified in osteosarcoma | 2.35 | 0.035175 |
| NM_007952 | Pdia3 | Protein disulfide isomerase associated 3 | 2.36 | 0.110736 |
| NM_181414 | Pik3c3 | Phosphoinositide-3-kinase, class 3 | 1.87 | 0.025524 |
| NM_020272 | Pik3cg | Phosphoinositide-3-kinase, catalytic, gamma polypeptide | 2.2 | 0.001131 |
| NM_001081309 | Pik3r4 | Phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 4, p150 | 1.77 | 0.003089 |
| NM_133819 | Ppp1r15b | Protein phosphatase 1, regulatory (inhibitor) subunit 15b | 1.43 | 0.018064 |
| NM_008960 | Pten | Phosphatase and tensin homolog | 1.87 | 0.000641 |
| NM_009000 | Rab24 | RAB24, member RAS oncogene family | 1.67 | 0.018972 |
| NM_009029 | Rb1 | Retinoblastoma 1 | 1.36 | 0.010937 |
| NM_026446 | Rgs19 | Regulator of G-protein signaling 19 | 1.42 | 0.002777 |
| NM_028259 | Rps6kb1 | Ribosomal protein S6 kinase, polypeptide 1 | 1.52 | 0.033948 |
| NM_027016 | Sec62 | SEC62 homolog (S. cerevisiae) | 2.47 | 0.064532 |
| NM_030685 | Serp1 | Stress-associated endoplasmic reticulum protein 1 | 2.02 | 0.079150 |
| NM_030749 | Sil1 | Endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) | 2.69 | 0.046544 |
| NM_009221 | Snca | Synuclein, alpha | 2.98 | 0.006131 |
| NM_011018 | Sqstm1 | Sequestosome 1 | 2.25 | 0.002485 |
| NM_011577 | Tgfb1 | Transforming growth factor, beta 1 | 2.2 | 0.011470 |
| NM_009425 | Tnfsf10 | Tumor necrosis factor (ligand) superfamily, member 10 | 1.66 | 0.007439 |
| NM_013881 | Ulk2 | Unc-51 like kinase 2 (C. elegans) | 1.37 | 0.012061 |
| NM_021522 | Usp14 | Ubiquitin specific peptidase 14 | 2.35 | 0.092052 |
| NM_178635 | Uvrag | UV radiation resistance associated gene | 1.8 | 0.007103 |
| NM_009503 | Vcp | Valosin containing protein | 1.97 | 0.115360 |
| NM_145940 | Wipi1 | WD repeat domain, phosphoinositide interacting 1 | 2.22 | 0.000642 |
Cerebral cortices of mice on PD4 and PD12 were removed and RNA was extracted.
The expression of genes associated with autophagy and UPR pathways was analyzed using pathway-specific PCR microarray as described under the Materials and Methods.
A total of 168 genes were screened and normalized with the expression of β-actin. The changes were expressed as a ratio of PD12 to PD4 (PD12/PD4). For example, the expression of Atf6 on PD12 was 2.18 fold greater than PD4. The changes in expression greater than 1.9-fold or with a p-value smaller than 0.05 were presented. The experiment was replicated three times.
The expression of a number of key genes that are involved in the activation of autophagy was significantly higher on PD12 than on PD4; these included several Atg genes, Becn1, Bnip3, Cdkn2a, Ctsb, Ctsd, Ctss, Gabarapl1 an 2, Irgm1, lamp1, MAPK14, Map1lc3a (Lc3), Mtor, Pik3c3, Pik3cg, Pten, Snca, Sqstm1 and Wipi1 (Table 1). These genes encode proteins involved in critical autophagic events, such as vacuole formation, protein targeting to membrane/vacuole, protein transport and protease activity. A group of genes that regulate key events of UPR were also upregulated on PD12; these included Atf6, Atxn3, Cebpb, Eif2ak3 (also known as Perk), Eif4g1, some heat shock proteins, Nploc4, Mapk9, Os9 and Sil1 (Table 1).
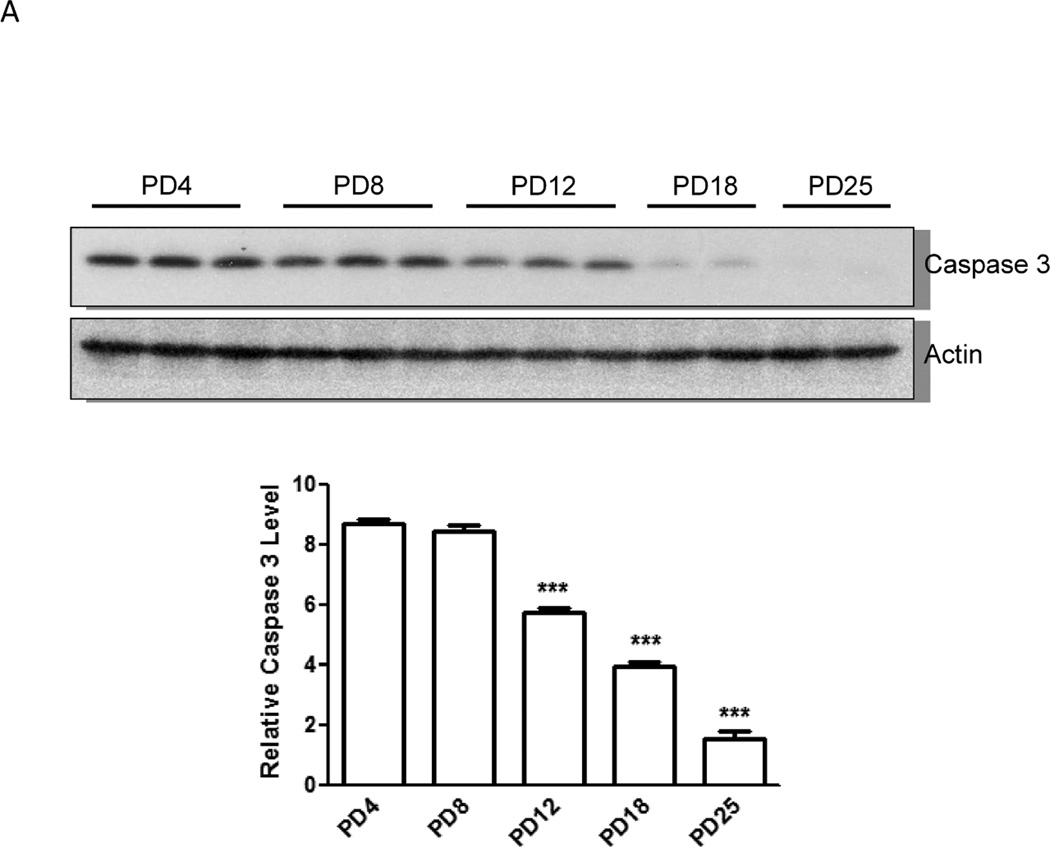
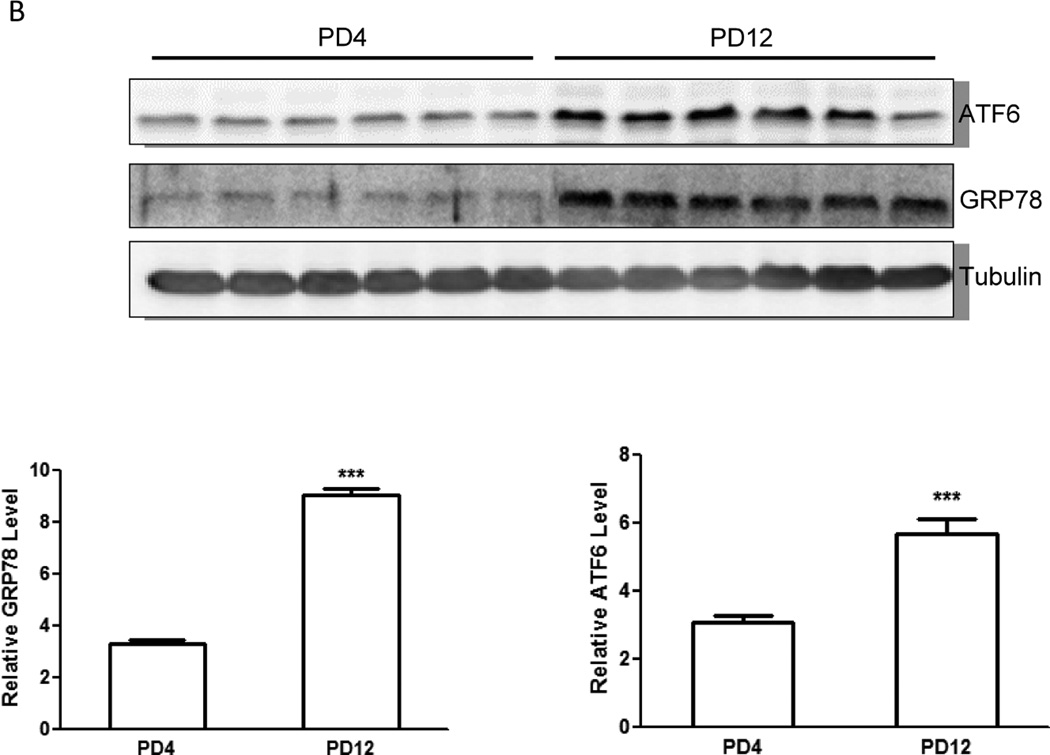
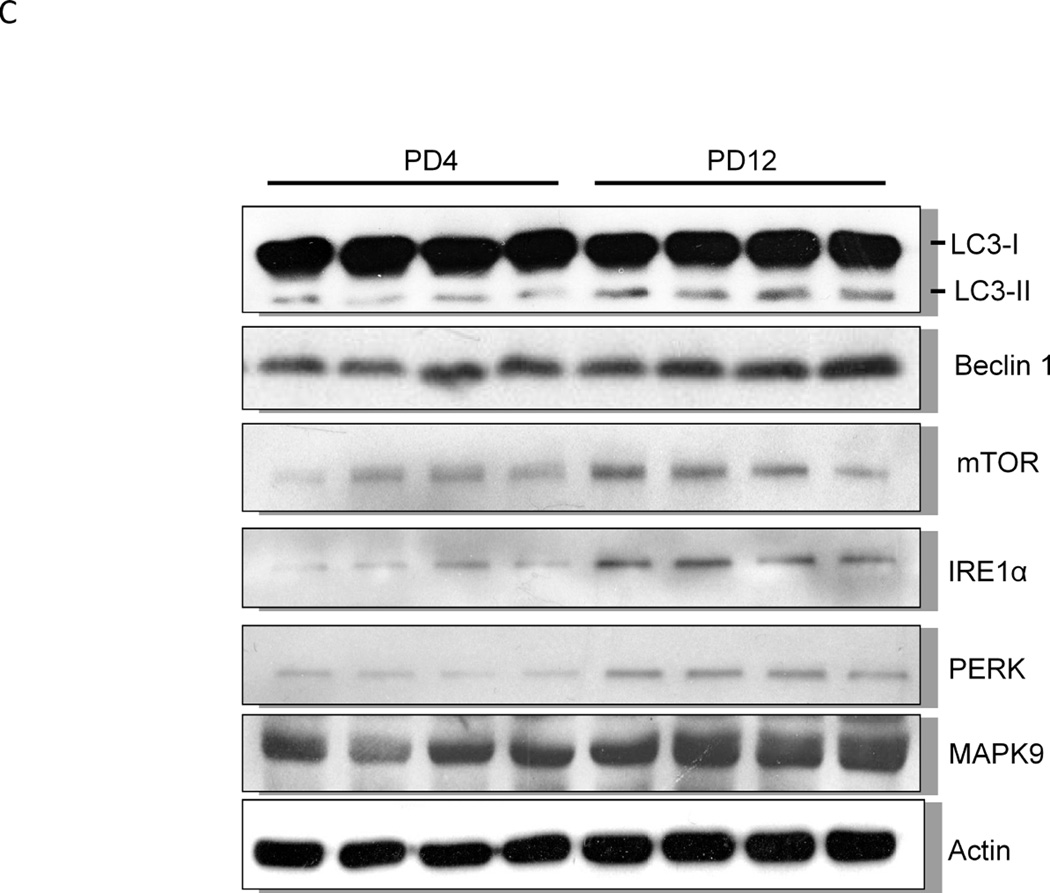
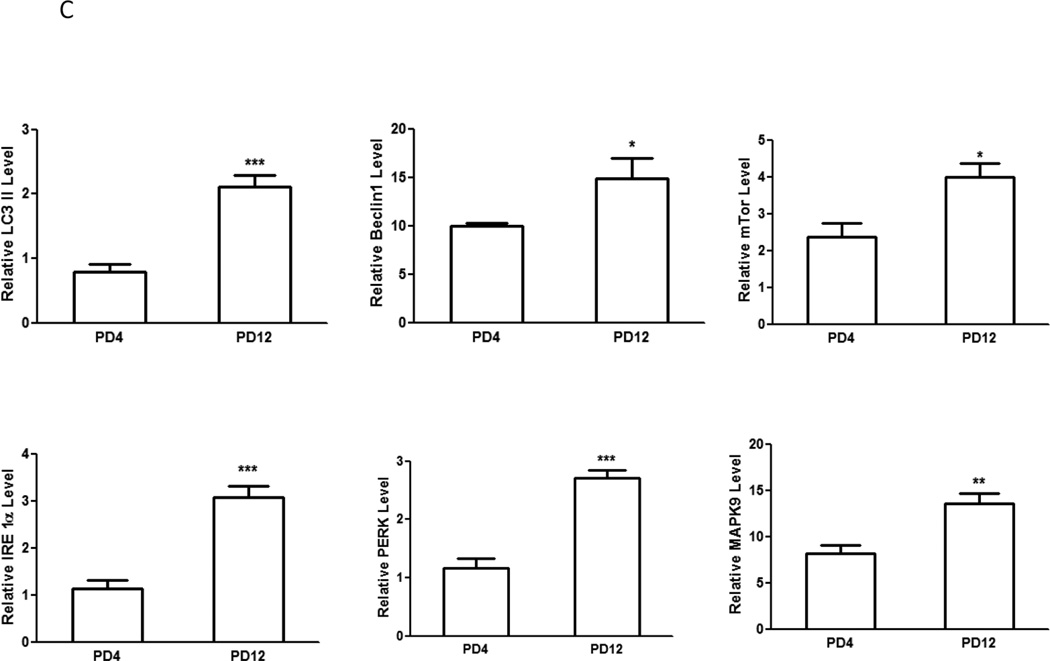
We examined the protein levels of several important genes regulating apoptosis, autophagy and UPR with immunoblotting analysis. Consistent with the pattern of gene expression, immunoblotting results confirmed a higher expression of caspase-3 protein in the brain of PD4 mice than that of PD12 (Fig. 2A). The expression of proteins associated with autophagy, such as Beclin 1, LC3-II and mTOR was significantly higher in PD12 brain (Fig. 2C). The expression of some UPR proteins, such as ATF6, Eif2ak3 (PERK), and MAPK9 was up-regulated in PD12 brain (Figs. 2B and 2C). Interestingly, the protein levels of GRP78 and IRE1α, two important UPR genes, were significantly increased as the brain matured (Figs. 2B and C), however mRNA levels exhibited little change between PD4 and PD12.
Figure 2. Developmental expression of proteins associated with UPR and autophagy pathways.
Cerebral cortices of mice at specified ages were removed and protein was extracted. A: The expression of caspase-3 was evaluated by immunoblotting as described under the Materials and Methods (top panel). Relative levels of caspase-3 were determined by densitometry and normalized to actin (bottom panel). *** denotes a significant difference from PD4 (p < 0.001). B: The expression of ATF6 and GRP78 was evaluated by immunoblotting (top panel). Relative levels of caspase-3 were determined by densitometry and normalized to tubulin (bottom panel). *** denotes a significant difference from PD4 (p < 0.001). C: The expression of LC3, beclin 1, mTOR, IRE1α, PERK and MAPK9 was evaluated by immunoblotting (top panel). Relative levels of caspase-3 were determined by densitometry and normalized to actin (bottom panel). *** denotes a significant difference from PD4 (p < 0.001); ** p < 0.01; * p < 0.05.
Discussion
In this study, we demonstrate a temporal window of vulnerability to ethanol neurotoxicity. In PD4 mice, subcutaneous (SC) injection of ethanol causes dramatic activation of caspase-3, indicative of apoptosis; whereas in PD12 mice ethanol has little effect on caspase-3 activation. Therefore, the brain of PD4 mice is sensitive to ethanol-induced neuroapoptosis while that of PD12 is resistant. The selection of SC administration of ethanol as opposed to intravenous (IV), intraperitoneal (IP) administration or gastric intubation (GI) was based on the following considerations: 1) this is a widely used model and well established to study the mechanisms of ethanol-induced apoptosis; 2) ethanol via SC administration is absorbed rapidly enough but more gradually than by IV or IP. Infant mice tolerate this more gradual sustained elevation compared to an abrupt peaking to higher levels; 3) there is less leakage of ethanol using SC administration. Although SC administration may produce some stress to animals, it is not more stressful than IV, IP or GI. Ethanol-induced neuroapoptosis is unlikely caused by the injection because the mice receiving SC injection of saline and no injection display similar intensity of active caspas-3 IHC staining (data not shown). Therefore, this model provides a useful tool to uncover the mechanisms underlying ethanol-induced neurodegeneration in the developing brain.
Caspase-3 is an executive caspase in the central position within apoptotic machinery. Caspase-3 is most frequently involved in the apoptosis of post-mitotic neurons in the developing brain (Madalosso et al. 2005). Our results indicate a high expression of caspase-3 in early postnatal days (PD4) and as the brain matures its expression levels decline (PD12). This finding is consistent with a previous study of developmental expression of caspase-3 in rats (Yakovlev et al. 2010), which showed a much higher expression of caspase-3 in the brain on PD2 than PD60. The expression profile of caspase-3 in the early postnatal days is generally consistent with the naturally occurring neuroapoptosis (programmed cell death) at this stage, indicating its important role in regulating neuroapoptosis (Spreafico et al. 1995; Madalosso et al. 2005). Therefore, high expression of caspase-3 in the early postnatal days may be responsible for enhanced sensitivity to ethanol-induced neuroapoptosis. In contrast to caspase-3, the expression of caspse-8 is higher on PD12 than PD4. Although caspase-8 may also be involved in an extrinsic apoptotic pathway, it is caspase-3 that plays an essential executive role in the regulation of apoptosis. Caspase-3 may have a non-apoptotic role in the developing brain; for example, it can regulate neural differentiation (Oomman et al. 2004; Fernando et al. 2005). High expression of caspase-3 in early postnatal days supports the notion that it may regulate neural differentiation as well.
Another gene whose expression is significantly higher on PD4 than in PD12 is chemokine (C-X-C motif) receptor 4 (CXCR4). CXCR4 is a receptor for chemokine stromal cell-derived factor 1 (SDF1 or CXCL12). SDF1/CXCR4, which were originally identified for their role in leukocyte trafficking, play an important role in neural progenitor cell proliferation and neuron migration (Shimoji et al. 2009; Zhu et al. 2009; Tiveron et al. 2010). CXCR4 mediates human immunodeficiency virus type 1 (HIV-1)-induced neurodegeneration (Mocchetti et al. 2008). CCCR4 also plays a pro-apoptotic role during neurodegeneration in the nigro-striatal system (Shimoji et al. 2009). High expression of CXCR4 on PD4 is consistent with its role in regulating proliferation of neural progenitor cells and neuron migration. It is unclear whether high levels of CXCR4 also contribute to enhanced sensitivity to ethanol.
We have previously demonstrated that ethanol induced ER stress in the developing brain (Ke et al. 2011a). Autophagy is a process of self-degradation of cellular components in which double-membrane autophagosomes sequester organelles or portions of the cytosol and fuse them with lysosomes or vacuoles for breakdown by resident hydrolases. Autophagy is up-regulated in response to extra- or intracellular stress and signals such as starvation, growth factor deprivation, ER stress, and pathogen infection (He and Klionsky 2009). The involvement of autophagy in cell death has been controversial. Since the formation of autophagosomes is frequently associated with cellular stress and cell death, some believe that autophagy may promote cell death (Galluzzi et al. 2008). However, many view autophagy as a cellular self-defense response (Shen and Codogno 2011). Particularly, in neuropathies (Huntington's, Alzheimer's and Parkinson's diseases) and ischemic heart disease, autophagy is more widely accepted as beneficial due to its role in eliminating ‘toxic assets’ and promoting cell viability (Glick et al. 2010). We have recently demonstrated that ethanol can activate autophagy in SH-SY5Y cells and the developing brain, and autophagy is protective against ethanol neurotoxicity (Chen et al. 2012).
Using pathway-specific PCR microarrays, we examined the expression of 168 genes associated with autophagy and UPR in the brain of PD4 and PD12 mice. As the brain matures, the expression of autophagy and UPR genes significantly increases (Table 1); these genes are critically involved in all aspects of autophagy and UPR. It is generally believed that UPR can trigger autophagy (He and Klionsky 2009). However, these two pathways are intermingled and many genes may regulate both UPR and autophagy, such as Atf6 and Eif2ak3 (Perk) (Vidal and Hetz 2012).
In addition to the regulation at transcriptional levels, the expression of these genes can be regulated at translational or post-translational levels. For example, although the mRNA of GRP78 displays little change between PD4 and PD12, its protein levels significantly increase on PD12. GRP78 has been a key pro-survival factor for neurons under ER stress (Wang et al. 2009). Together, these results indicate that the brain develops a more effective defense system to cope with cellular stress during the first two postnatal weeks in rodents. In other words, the enhanced sensitivity to ethanol neurotoxicity during early postnatal days likely results from a high expression of proapoptotic caspase-3 and an incomplete anti-stress system.
Ethanol also causes oxidative stress which is considered an important mechanism for ethanol neurotoxicity in the developing brain (Ikonomidou, 2009; Luo, 2012). The expression of brain antioxidant enzymes is also developmentally regulated (Khan and Black 2003). To gain an insight into the role of the antioxidant system, we will evaluate the expression of antioxidant enzymes during these ethanol-sensitive and resistant periods in our future studies.
Acknowledgements
This work was supported by grants from the National Institutes of Health (AA015407 and AA019693).
References
- Chen G, Luo J. Anthocyanins: are they beneficial in treating ethanol neurotoxicity? Neurotox Res. 2010;17(1):91–101. doi: 10.1007/s12640-009-9083-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Ke Z, Xu M, Liao M, Wang X, Qi Y, Zhang T, Frank JA, Bower KA, Shi X, Luo J. Autophagy is a protective response to ethanol neurotoxicity. Autophagy. 2012;8(11):1577–1589. doi: 10.4161/auto.21376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Vicencio JM, Kepp O, Tasdemir E, Maiuri MC, Kroemer G. To die or not to die: that is the autophagic question. Curr Mol Med. 2008;8(2):78–91. doi: 10.2174/156652408783769616. [DOI] [PubMed] [Google Scholar]
- Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221(1):3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando P, Brunette S, Megeney LA. Neural stem cell differentiation is dependent upon endogenous caspase 3 activity. FASEB J. 2005;19(12):1671–1673. doi: 10.1096/fj.04-2981fje. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans KG, Sliwowska JH, Verma P, Weinberg J. Prenatal alcohol exposure: fetal programming and later life vulnerability to stress, depression and anxiety disorders. Neurosci Biobehav Rev. 2010;34(6):791–807. doi: 10.1016/j.neubiorev.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomidou C. Triggers of apoptosis in the immature brain. Brain Dev. 2009;31(7):488–492. doi: 10.1016/j.braindev.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Ke Z, Wang X, Liu Y, Fan Z, Chen G, Xu M, Bower KA, Frank JA, Li M, Fang S, Shi X, Luo J. Ethanol induces endoplasmic reticulum stress in the developing brain. Alcohol Clin Exp Res. 2011a;35(9):1574–1583. doi: 10.1111/j.1530-0277.2011.01503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z, Liu Y, Wang X, Fan Z, Chen G, Xu M, Bower KA, Frank JA, Ou X, Shi X, Luo J. Cyanidin-3-glucoside ameliorates ethanol neurotoxicity in the developing brain. J Neurosci Res. 2011b;89(10):1676–1684. doi: 10.1002/jnr.22689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan JY, Black SM. Developmental changes in murine brain antioxidant enzymes. Pediatr Res. 2003;54(1):77–82. doi: 10.1203/01.PDR.0000065736.69214.20. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J. Mechanisms of ethanol-induced death of cerebellar granule cells. Cerebellum. 2012;11(1):145–154. doi: 10.1007/s12311-010-0219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier SE, Miller JA, Blackwell JM, West JR. Fetal alcohol exposure and temporal vulnerability: regional differences in cell loss as a function of the timing of binge-like alcohol exposure during brain development. Alcohol Clin Exp Res. 1999;23(4):726–734. doi: 10.1111/j.1530-0277.1999.tb04176.x. [DOI] [PubMed] [Google Scholar]
- Madalosso SH, Pérez-Villegas EM, Armengol JA. Naturally occurring neuronal death during the postnatal development of Purkinje cells and their precerebellar afferent projections. Brain Res Brain Res Rev. 2005;49(2):267–279. doi: 10.1016/j.brainresrev.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Mocchetti I, Bachis A, Masliah E. Chemokine receptors and neurotrophic factors: potential therapy against aids dementia? J Neurosci Res. 2008;86(2):243–255. doi: 10.1002/jnr.21492. [DOI] [PubMed] [Google Scholar]
- Olney JW, Tenkova T, Dikranian K, Muglia LJ, Jermakowicz WJ, D'Sa C, Roth KA. Ethanol-induced caspase-3 activation in the in vivo developing mouse brain. Neurobiol Dis. 2002;9(2):205–219. doi: 10.1006/nbdi.2001.0475. [DOI] [PubMed] [Google Scholar]
- Oomman S, Finckbone V, Dertien J, Attridge J, Henne W, Medina M, Mansouri B, Singh H, Strahlendorf H, Strahlendorf J. Active caspase-3 expression during postnatal development of rat cerebellum is not systematically or consistently associated with apoptosis. J Comp Neurol. 2004;476(2):154–173. doi: 10.1002/cne.20223. [DOI] [PubMed] [Google Scholar]
- Rasheva VI, Domingos PM. Cellular responses to endoplasmic reticulum stress and apoptosis. Apoptosis. 2009;14:996–1007. doi: 10.1007/s10495-009-0341-y. [DOI] [PubMed] [Google Scholar]
- Riley EP, Infante MA, Warren KR. Fetal alcohol spectrum disorders: an overview. Neuropsychol Rev. 2011;21(2):73–80. doi: 10.1007/s11065-011-9166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D. Translational control in the endoplasmic reticulum stress response. J Clin Invest. 2002;110:1383–1388. doi: 10.1172/JCI16784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HM, Codogno P. Autophagic cell death: Loch Ness monster or endangered species? Autophagy. 2011;7(5):457–465. doi: 10.4161/auto.7.5.14226. [DOI] [PubMed] [Google Scholar]
- Shimoji M, Pagan F, Healton EB, Mocchetti I. CXCR4 and CXCL12 expression is increased in the nigro-striatal system of Parkinson's disease. Neurotox Res. 2009;16(3):318–328. doi: 10.1007/s12640-009-9076-3. [DOI] [PubMed] [Google Scholar]
- Siler-Marsiglio KI, Madorsky I, Pan Q, Paiva M, Neeley AW, Shaw G, Heaton MB. Effects of acute ethanol exposure on regulatory mechanisms of Bcl-2-associated apoptosis promoter bad, in neonatal rat cerebellum: differential effects during vulnerable and resistant developmental periods. Alcohol Clin Exp Res. 2006;30(6):1031–1038. doi: 10.1111/j.1530-0277.2006.000126.x. [DOI] [PubMed] [Google Scholar]
- Spreafico R, Frassoni C, Arcelli P, Selvaggio M, De Biasi S. In situ labeling of apoptotic cell death in the cerebral cortex and thalamus of rats during development. J Comp Neurol. 1995;363(2):281–295. doi: 10.1002/cne.903630209. [DOI] [PubMed] [Google Scholar]
- Tiveron MC, Boutin C, Daou P, Moepps B, Cremer H. Expression and function of CXCR7 in the mouse forebrain. J Neuroimmunol. 2010;224(1–2):72–79. [PubMed] [Google Scholar]
- Vidal RL, Hetz C. Crosstalk between the UPR and autophagy pathway contributes to handling cellular stress in neurodegenerative disease. Autophagy. 2012;8(6):970–972. doi: 10.4161/auto.20139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Bower KA, Frank JA, Xu M, Luo J. Hypoxic Preconditioning Alleviates Ethanol Neurotoxicity: The Involvement of Autophagy. Neurotox Res. 2013 doi: 10.1007/s12640-013-9390-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Ye R, Barron E, Baumeister P, Mao C, Luo S, Fu Y, Luo B, Dubeau L, Hinton DR, Lee AS. Essential role of the unfolded protein response regulator GRP78/BiP in protection from neuronal apoptosis. Cell Death Differ. 2010;17(3):488–498. doi: 10.1038/cdd.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Peng H, Cui M, Whitney NP, Huang Y, Zheng JC. CXCL12 increases human neural progenitor cell proliferation through Akt-1/FOXO3a signaling pathway. J Neurochem. 2009;109(4):1157–1167. doi: 10.1111/j.1471-4159.2009.06043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115:2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlev A, Khafizova M, Abdullaev Z, Loukinov D, Kondratyev A. Epigenetic regulation of caspase-3 gene expression in rat brain development. Gene. 2010;450(1–2):103–108. doi: 10.1016/j.gene.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Matsumoto T, Mikami S, Nagasawa T, Murakami F. SDF1/CXCR4 signalling regulates two distinct processes of precerebellar neuronal migration and its depletion leads to abnormal pontine nuclei formation. Development. 2009;136(11):1919–1928. doi: 10.1242/dev.032276. [DOI] [PubMed] [Google Scholar]