Abstract
The mass spectrometry-based “omics” sub-discipline that focuses on comprehensive, often exploratory, analyses of endogenous peptides involved in cell-to-cell communication is oftentimes referred to as peptidomics. While the progress in bioanalytical technology development for peptide discovery has been tremendous, perhaps the largest advances have involved robust quantitative mass spectrometric approaches and data mining algorithms. These efforts have accelerated the discovery and validation of biomarkers, functionally important posttranslational modifications, and unexpected molecular interactions, information that aids drug development. In this article we outline the current approaches used in quantitative peptidomics and the technical challenges that stimulate new advances in the field, while also reviewing the newest literature on functional characterizations of endogenous peptides using quantitative mass spectrometry.
Introduction
In contrast to gene expression analyses, peptidomics probes the final gene products of prohormones that directly mediate the observed biological effect; the resulting data provides detailed information about system alterations and suggests the pathways involved. Due to their small size (3–50 amino acids), endogenous peptides, such as neuropeptides, as well as peptide hormones, growth factors, and intracellular products of proteolysis, are not easily accessible for assay using traditional gel-based proteomics methods and therefore, are typically studied using affinity and molecular techniques. The important roles that endogenous peptides play in physiology encourage the development of quantitative measurements for understanding peptide regulation and perturbation-induced changes in numerous pathologies. Mass spectrometry (MS) not only provides the ability to simultaneously deduce structural information without prior knowledge on the peptide complement present, but can also be used to quantify peptide amounts.
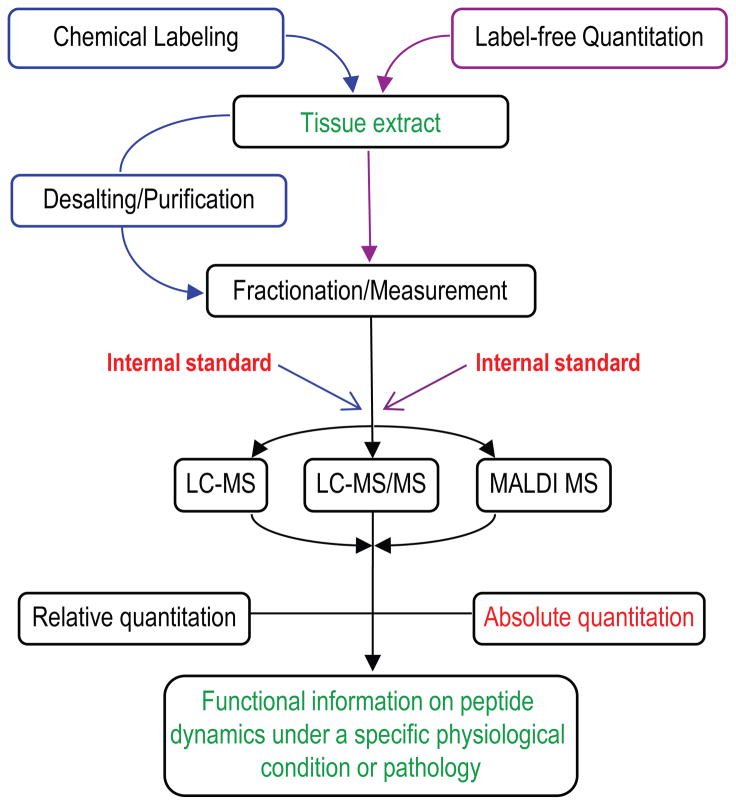
Whereas quantitative peptide-centered proteomic analyses have become indispensable for high throughput interrogation of proteins in complex biological samples, quantitative peptidomic studies are becoming more commonly used for peptide investigation. A number of options for peptide quantitation are available; a thorough understanding of how these approaches work enables the user to select the most appropriate measurement protocols for a given experiment (Figure 1).
Figure 1.
Current quantitative peptidomics approaches.
Current approaches for quantitation with mass spectrometry
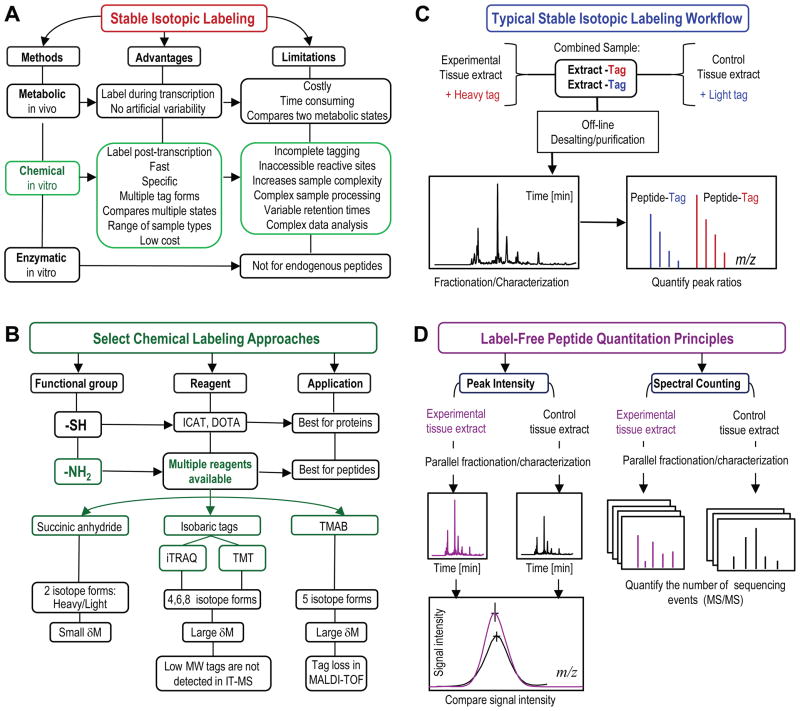
In MS, differences in peptide abundance between sample cohorts representing different physiological conditions can be deduced from a statistical comparison of the measured signal for each of the detected peptides, and can also be determined based on how many times a specific peptide is detected in a specified measurement. On a number of MS platforms, signal intensity has been shown to be proportional to the amount of detected peptide, as long as it falls within the linear dynamic range of the detector [1]. To account for inconsistencies related to the nature of the sample, sample preparation, or instrument performance, distinct approaches utilizing the specificity of MS detection exist for relative quantitative measurements of peptides in complex biological samples. Figure 2 illustrates some of the more commonly used methods for peptide quantitation using MS.
Figure 2.
Mass-spectrometry based strategies for quantitation of endogenous peptides. A) Routes of isotope introduction in the stable isotopic labeling methods. B) Overview of select popular chemical labeling methods. C) Typical stable isotopic labeling workflow. D) Label-free quantitation strategies. Key: ICAT–isotope-coded affinity tag; DOTA–1,4,7,10-tetraazacyclododecane-N,N′,N″,N‴-tetraacetic acid; iTRAQ–isobaric tags for relative or absolute quantitation; TMT–tandem mass tags; TMAB–trimethylammoniumbutyryl-based tags; IT-MS–ion trap mass spectrometer; MALDI-TOF–matrix-assisted laser desorption/ionization time-of-flight mass spectrometer.
Chemical labeling of endogenous peptides
The unique ability of MS to differentiate compounds by mass has fostered the use of isotopically labeled internal standards [2]. Isotope incorporation into the analyte structure provides an accurate measure of the relative abundance of the peptides; this is because MS detection is independent of the isotopic composition of homologous compounds [3]. Perhaps the most accurate quantitative comparison can be achieved by incorporating an isotopically enriched element (via appropriate amino acids, salts, etc.) into the growth media or the diet for the labeling of proteins during synthesis in cultured cells or in whole animals, respectively. This approach is more suitable for protein labeling in cell lines or simpler organisms, and is less easily adapted for larger animals having longer lifetimes [4].
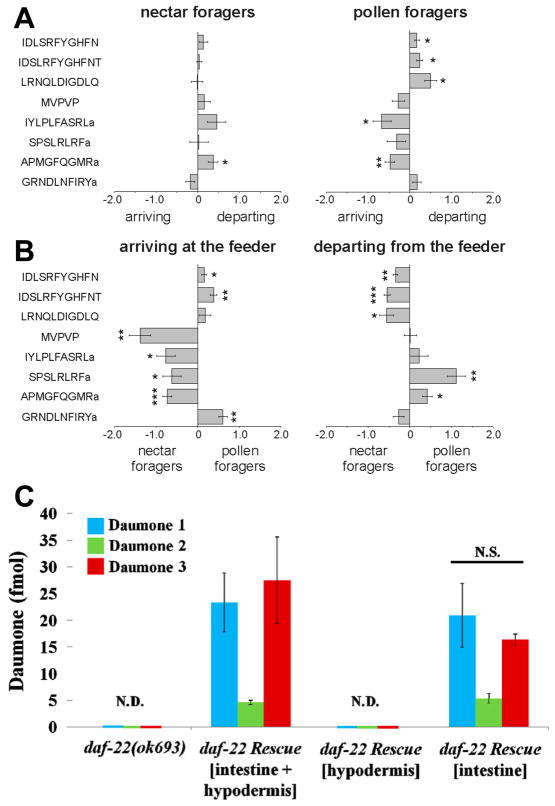
In contrast to metabolic labeling, in vitro chemical labeling of peptides performed after tissue harvest is fast and can be performed via numerous routes (Figure 2A, B). Currently, the reported ion-based quantification strategies using commercial reagents (iTRAQ, TMT) are the most common chemical labeling methods because they allow multiplexed assessments; however, the majority of published reports involve bottom-up proteomics studies. We find that incorporation of stable isotopes appears to be a preferred and less-expensive approach for quantitation of endogenous peptides in variety of animal models via well-developed protocols (Figure 2C) [5,6]. For example, trimethylammoniumbutyryl (or TMAB)-based tags have been used to evaluate the role of protein convertase 1/3 and 2 in the processing of peptides in the mammalian brain [7,8], to compare the peptidome of human cell lines [9], and to investigate the peptidome of brain slices [10]. Peptides modified via a simple reaction with deuterated succinic anhydride can be characterized and quantified using a majority of mass spectrometers including electrospray ionization and matrix-assisted laser desorption/ionization (MALDI) sources [1,11]. As one exiting example highlighted in Figure 3A, B, we determined neuropeptides that had level changes between behaviorally distinct honey bees [12]. The same labeling protocol was used to show the role of processing enzymes in the non-classical processing of spinal cord peptides and morphine responses [13], and in the production of amidated peptides under genetic and dietary manipulations in mammals [14]. Both iTRAQ and isotope incorporation have been successfully adapted for the relative quantitation of nanoliter volume samples of brain releasates from Aplysia, and to show that insulin C peptide levels are three times higher at their release sites than at their cellular locations [11]. In another study, triplex dimethyl labeling was employed to determined changes in neuropeptide concentrations in the central nervous system of Rhodnius prolixus in relation to feeding [15].
Figure 3.
Differences in brain peptide abundances between four behaviorally different honey bee forager groups. A) Relative differences in peptide abundances between arriving and departing nectar or pollen foragers. B) Relative differences in peptide abundances between nectar and pollen foragers arriving or departing from the feeder. x axis: normalized, log2 transformed and centered peptide ratios. Negative and positive values indicate the direction of change. MVPVP = MVPVPVHHMADELLRNGPDTVI. Indicated significance levels (Student’s t test): *, P < 0.05; **, P < 0.005; ***, P < 0.0005. Used with permission from ref. [12]. C)Absolute quantitation of daumone pheromone using stable isotope internal standard. The amount of pheromones (in 2 μL injection volume) in daf-22rescue worms. Quantitation was performed for three pheromones present in the wormbody of 20 young adult worms each, from the daf-22 (ok693)and the daf-22 rescue strains. Values represent means ± standard deviation from four independent sets. N.D., “not detected”; N.S., “not significant” relative to the amount of pheromones in the daf-22 rescue worms (intestinal and hypodermal). (Used with permission from ref. [27]. Copyright 2013 American Chemical Society.)
A newer development in in vitro labeling is the metal-chelate approach that tags amino- and thiol groups with a macrocyclic bifunctional chelator of rare earth metals, 1,4,7,10-tetraazacyclododecane-N,N′,N″,N‴-tetraacetic acid (DOTA). DOTA labeling is certainly suitable for quantitation of extreme peptide changes [16], such as in enzymatic reactions, rather than the typically subtle alterations found in physiological peptide dynamics.
Label-free quantitation of endogenous peptides
Label-free quantitation is based on comparisons of the MS signals or the number of tandem (MS/MS) mass spectra for unmodified peptides across multiple experiments (Figure 2D) [7,17]. A significant advantage to using label-free methods is that there is no limit to the number of samples/conditions that can be compared, and they can accommodate small-volume samples that are less suitable for derivatization with labeling reagents. In addition, these methods extend to peptides having N-terminal posttranslational modifications, the ones omitted by labeling methods that rely on modifications of free amine groups. This methodology is widely used in biomarker research, particularly in conjunction with MALDI MS, as we discuss below.
Another label-free strategy, spectral counting, is based on the linear correlation between peptide abundance and the number of sequencing events for each of the detected peptides [7]. This up and coming method is made possible by the growing numbers of high resolution mass analyzers, such as orbitrap, quadrupole time-of-flight, and Fourier transform-ion cyclotron resonance, as well as the sophisticated supporting software. Recent studies have demonstrated the utility of spectral counting for measuring endogenous peptides by exploring day–night changes in peptide expression profiles for the mammalian suprachiasmatic nucleus [18], and by examining peptides involved in the embryogenesis of Japanese quails [19] and endogenous peptides in the nucleus accumbens of morphine-dependent rats [20]. Spectral counting and peak intensity comparisons showcase the power of MS as an exploration technique because it combines structural and qualitative characterization in one experiment, without additional sample modifications.
Alternatively, when measurement of a known subset of peptides in the sample is sufficient, selected reaction monitoring (SRM, also referred to as multiple reaction monitoring, MRM) allows quantitation based on comparisons of small numbers of sequence-specific fragment ions (transition ions) over time [21,22]. The use of SRM for quantification of endogenous peptides has been less common, but we expect its use to grow. Examples of SRM applied to endogenous peptide characterization include alpha- and gamma-endorphin quantitation in rat brain [23], alpha melanocyte stimulating hormone in mouse pituitary [24], and the neuropeptide Y family [25]. Introduction of hybrid quadrupole instruments with ion trap, time-of-flight, and orbitrap mass analyzers may increase the application of SRM in quantitative peptidomics in the near future.
Absolute quantitation in peptidomics
Biological studies increasingly require information on absolute amounts or concentrations of bioactive compounds, which is possible in MS with the addition of a reference peptide/s synthesized with or without stable isotopes as internal standards [26]. The approach is not yet common and may not have widespread use due to the need for custom standards for each peptide under consideration. The cost and analyte preselection requirement make this approach best suited for targeted analysis of specific peptides in the identified pathways or those that are involved in well-characterized physiological functions, as was the case in the determination of the nematode pheromones in Caenorhabditis elegans worm bodies and in the liquid culture medium (Figure 3C) [27]. Synthetic standards that do not incorporate selected isotopic labels can be used with standard addition methods and permit absolute quantitation of physiologically active neuropeptides from samples as small as a few cells [11].
Challenges that drive new developments in quantitative peptidomics
Important considerations when performing quantitative peptide measurements are the multitude of global influences that can affect measurement accuracy, ranging from biased sampling to the quality of the acquired mass spectral data and the efficiency of the mathematical tools used for data assessment. The quantitation of endogenous peptide dynamics remains difficult and time-consuming, but it is these challenges and limitations that have stimulated recent technological and methodological advances.
Peptidomic investigations heavily rely on liquid separations prior to MS analysis (Figure 1); however, a universal peptide sampling strategy does not yet exist. Tissue extracts are the typical sample type, and extraction of all of the peptides from tissue remains elusive because of the broad range of local peptide concentrations and the diverse chemical properties of peptides. In mass-limited samples, such as individual cells or defined brain nuclei, few peptides are abundant enough to exceed the sensitivity limit of current MS technologies. The requirement for substantial amounts of sample material frequently restricts peptidomics approaches to the analysis of either larger anatomically defined structures (whole brain) or pooled samples [7,8,12–14,18]. An alternative that is suitable for quantitative profiling of minute individual samples is the single-step extraction of signal peptides using 2,5-dihydroxybenzoic acid (DHB), a commonly used matrix for MALDI MS [28]. The method is suitable for fractionation of peptides by capillary electrophoresis using DHB as a background electrolyte [29]. As peptide extraction efficacy varies with the solvent and procedure used, Zhang et al. [30] introduced a mixing on column strategy that permits separate loading of aqueous and organic peptide extracts onto the same high-performance liquid chromatography column for simultaneous analysis, which improves peptide representation in the liquid chromatography (LC-MS) data.
Sampling that truly reflects the in vivo state of the tissue is imperative for finding potential biomarkers using quantitative MS-based analysis, but as yet is not entirely possible. Inevitable post-mortem degradation of ubiquitous proteins in tissue samples, and resultant truncated peptides, may easily overwhelm the signals from endogenous signaling peptides during an MS measurement [31]. A newer automated technology introduced by Svensson et al. [32] combines heat and pressure under vacuum to increase tissue temperature rapidly and uniformly, thus making sampling more reproducible and better suited for quantitative measurements. The peptide profile for a tissue sample using this technology is comparable to tissue collected with animal sacrifice by focused microwave irradiation, which is thought to closely reflect the in vivo peptidome.
The development of mass spectrometers with increasingly high mass-resolving power provide the ability to resolve overlapping isotopic peak patterns and identify different molecular species of the same or similar nominal mass. This has led to the identification and quantification of hundreds of peptides in an individual experiment [13,18]. High resolution mass spectrometers, however, generate gigabytes of complex data, which create the need for robust software tools to achieve effective MS data processing in a quantitative manner. Instrument manufacturers and software developers have launched commercial and open source interactive tools for processing of label-free and labeling data, as well as for protein/peptide biomarker profiling applications that are often compatible with both LC-MS and direct MS workflows [17,33–35].
Quantitative peptidomics for the functional characterization of peptides
As evidenced in recent NCBI literature, quantitative peptidomics is commonly used to investigate biomarkers and study the regulation of proteolytic activity. However, a number of fundamental research studies have also employed quantitative MS-based strategies in various animal models to track changes in endogenous peptide levels associated with specific behavioral phenotypes [12,36], addiction and psychiatric disorders [6,37], circadian rhythms [18], and obesity [38].
Peptides make good biomarkers as they are numerous in body fluids, cells, and tissues; collectively they provide higher diagnostic power than single protein markers [39]. The urine peptidome is frequently queried for predictive biomarkers of systemic and renal diseases [40]. The power of label-free quantitation has even been applied to the discovery of personalized immunosuppressive therapies for kidney transplant patients [41]. Likewise, comparative profiling of serum and plasma peptidomes has gained momentum as these samples are widely used as sources of biomarkers of various cancers [42,43] and other diseases [44,45]. The salivary peptidome has also drawn attention recently for the diagnosis and treatment of oral diseases, and as a potential tool for the diagnosis of systemic diseases [46].
As proteolytic events generate much of the complex peptidome in biological fluids and cells, high throughput MS-based methods are well suited for quantitative analysis of peptides resulting from proteolysis [47] and have been extensively used for understanding the regulation of bioactive peptides [7,48–50]. Although MS-based quantitation methods aid the effort to characterize peptides and determine their functions and interactions, the functional follow-up studies represent major efforts because newly discovered peptide targets require validation to establish their diagnostic or therapeutic value.
Conclusions
Mass spectrometry has revolutionized the way we characterize biologically important molecules. This versatile technology multiplies the descriptive power and throughput of chemical analysis, brings molecular characterization to a new level of confidence, and opens new opportunities for inquiry across disciplinary boundaries in synergy with other “omics” technologies. Numerous MS-based strategies used to determine endogenous peptide abundance have matured enough to allow their transfer from specialized applications in fundamental research to diagnostic peptide marker discovery and the development of peptide therapeutics. Considering the wide availability of high-resolution instruments, a desired shift toward versatile and cost-effective label-free quantitative peptidomics is inevitable. At present, implementation of quantitative MS methods in peptidomics is slow relative to proteomics, but we are optimistic that acquiring valuable quantitative knowledge from mass spectrometric information will become faster and easier in the future. Continued progress will focus on sensitive and reliable methods for absolute quantitation, and on the development of rigorous and user-friendly statistical tools for the assessment and reporting of quantitative data.
Highlights.
Mass spectrometry can measure relative and absolute levels of endogenous peptides.
Stable isotope labels provide relative changes in peptide levels between treatments.
Label-free approaches quantify peptide signal intensities or sequencing events.
Quantitative peptidomics facilitate functional bioactive peptide characterization.
Quantitative peptidomics is used in biomarker and peptide therapeutics research.
Acknowledgments
This work was supported by Award Number P30 DA018310 from the National Institute on Drug Abuse and by National Science Foundation Division of Chemistry under grant CHE-11-11705 (with co-funding from the Division of Biological Infrastructure). The content is solely the responsibility of the authors and does not necessarily represent the official views of the award agencies.
Footnotes
Conflict of interest
The authors declare they have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Elena V. Romanova, Email: romanova@illinois.edu.
Sarah E. Dowd, Email: dowd2@illinois.edu.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Hou X, Xie F, Sweedler JV. Relative quantitation of neuropeptides over a thousand-fold concentration range. J Am Soc Mass Spectrom. 2012;23:2083–2093. doi: 10.1007/s13361-012-0481-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kusmierz JJ, Sumrada R, Desiderio DM. Fast atom bombardment mass spectrometric quantitative analysis of methionine-enkephalin in human pituitary tissues. Anal Chem. 1990;62:2395–2400. doi: 10.1021/ac00220a026. [DOI] [PubMed] [Google Scholar]
- 3.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 4.Larance M, Bailly AP, Pourkarimi E, Hay RT, Buchanan G, Coulthurst S, Xirodimas DP, Gartner A, Lamond AI. Stable-isotope labeling with amino acids in nematodes. Nat Methods. 2011;8:849–851. doi: 10.1038/nmeth.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5*.Yin P, Hou X, Romanova EV, Sweedler JV. Neuropeptidomics: mass spectrometry-based qualitative and quantitative analysis. Methods Mol Biol. 2011;789:223–236. doi: 10.1007/978-1-61779-310-3_14. Detailed protocols for LC/MS based experiments on identification, characterization, and quantitation of neuropeptides with the emphasis on the sample preparation steps so integral to experimental success. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gelman JS, Wardman J, Bhat VB, Gozzo FC, Fricker LD. Quantitative peptidomics to measure neuropeptide levels in animal models relevant to psychiatric disorders. Methods Mol Biol. 2012;829:487–503. doi: 10.1007/978-1-61779-458-2_31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wardman JH, Zhang X, Gagnon S, Castro LM, Zhu X, Steiner DF, Day R, Fricker LD. Analysis of peptides in prohormone convertase 1/3 null mouse brain using quantitative peptidomics. J Neurochem. 2010;114:215–225. doi: 10.1111/j.1471-4159.2010.06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Pan H, Peng B, Steiner DF, Pintar JE, Fricker LD. Neuropeptidomic analysis establishes a major role for prohormone convertase-2 in neuropeptide biosynthesis. J Neurochem. 2010;112:1168–1179. doi: 10.1111/j.1471-4159.2009.06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelman JS, Sironi J, Castro LM, Ferro ES, Fricker LD. Peptidomic analysis of human cell lines. J Proteome Res. 2011;10:1583–1592. doi: 10.1021/pr100952f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gelman JS, Dasgupta S, Berezniuk I, Fricker LD. Analysis of peptides secreted from cultured mouse brain tissue. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbapap.2013.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubakhin SS, Sweedler JV. Quantitative measurements of cell-cell signaling peptides with single-cell MALDI MS. Anal Chem. 2008;80:7128–7136. doi: 10.1021/ac8010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12*.Brockmann A, Annangudi SP, Richmond TA, Ament SA, Xie F, Southey BR, Rodriguez-Zas SR, Robinson GE, Sweedler JV. Quantitative peptidomics reveal brain peptide signatures of behavior. Proc Natl Acad Sci U S A. 2009;106:2383–2388. doi: 10.1073/pnas.0813021106. Example of stable isotopic labeling for functional characterization of bioactive neuropeptides in the regulation of honey bee behavior. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller LK, Hou X, Rodriguiz RM, Gagnidze K, Sweedler JV, Wetsel WC, Devi LA. Mice deficient in endothelin-converting enzyme-2 exhibit abnormal responses to morphine and altered peptide levels in the spinal cord. J Neurochem. 2011;119:1074–1085. doi: 10.1111/j.1471-4159.2011.07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin P, Bousquet-Moore D, Annangudi SP, Southey BR, Mains RE, Eipper BA, Sweedler JV. Probing the production of amidated peptides following genetic and dietary copper manipulations. PLoS ONE. 2011;6:e28679. doi: 10.1371/journal.pone.0028679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sterkel M, Urlaub H, Rivera-Pomar R, Ons S. Functional proteomics of neuropeptidome dynamics during the feeding process of Rhodnius prolixus. J Proteome Res. 2011;10:3363–3371. doi: 10.1021/pr2001012. [DOI] [PubMed] [Google Scholar]
- 16.Gregorius B, Jakoby T, Schaumloffel D, Tholey A. Metal labeling for accurate multiplexed peptide quantification via matrix-assisted laser desorption/ionization mass spectrometry. Anal Bioanal Chem. 2013;405:2735–2741. doi: 10.1007/s00216-012-6686-z. [DOI] [PubMed] [Google Scholar]
- 17**.Neilson KA, Ali NA, Muralidharan S, Mirzaei M, Mariani M, Assadourian G, Lee A, van Sluyter SC, Haynes PA. Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics. 2011;11:535–553. doi: 10.1002/pmic.201000553. Review article about the label-free methods. Includes examples of quantitative analysis using two methods and highlights available software packages. [DOI] [PubMed] [Google Scholar]
- 18*.Lee JE, Zamdborg L, Southey BR, Atkins N, Jr, Mitchell JW, Li M, Gillette MU, Kelleher NL, Sweedler JV. Quantitative peptidomics for discovery of circadian-related peptides from the rat suprachiasmatic nucleus. J Proteome Res. 2013;12:585–593. doi: 10.1021/pr300605p. Application of the label-free spectral counting approach in conjunction with high-resolution LC-MS for quantitiative measurements of bioactive peptides. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scholz B, Alm H, Mattsson A, Nilsson A, Kultima K, Savitski MM, Falth M, Skold K, Brunstrom B, Andren PE, et al. Neuropeptidomic analysis of the embryonic Japanese quail diencephalon. BMC Dev Biol. 2010:10. doi: 10.1186/1471-213X-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossbach U, Nilsson A, Falth M, Kultima K, Zhou Q, Hallberg M, Gordh T, Andren PE, Nyberg F. A quantitative peptidomic analysis of peptides related to the endogenous opioid and tachykinin systems in nucleus accumbens of rats following naloxone-precipitated morphine withdrawal. J Proteome Res. 2009;8:1091–1098. doi: 10.1021/pr800669g. [DOI] [PubMed] [Google Scholar]
- 21.Gallien S, Duriez E, Domon B. Selected reaction monitoring applied to proteomics. J Mass Spectrom. 2011;46:298–312. doi: 10.1002/jms.1895. [DOI] [PubMed] [Google Scholar]
- 22.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012;9:555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 23.Kosanam H, Ramagiri S, Dass C. Quantification of endogenous alpha- and gamma-endorphins in rat brain by liquid chromatography-tandem mass spectrometry. Anal Biochem. 2009;392:83–89. doi: 10.1016/j.ab.2009.05.041. [DOI] [PubMed] [Google Scholar]
- 24.Perroud B, Alvarado R, Espinal G, Morado A, Phinney B, Warden C. In vivo multiplex quantitative analysis of 3 forms of alpha melanocyte stimulating hormone in pituitary of prolyl endopeptidase deficient mice. Molecular Brain. 2009;2:14. doi: 10.1186/1756-6606-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xi L, Jin YP, Parker EA, Josh P, Jones A, Wijffels G, Colgrave ML. Challenges in mass spectrometry-based quantification of bioactive peptides: A case study exploring the neuropeptide Y family. Biopolymers. 2012;98:357–366. doi: 10.1002/bip.22109. [DOI] [PubMed] [Google Scholar]
- 26*.Filiou MD, Martins-de-Souza D, Guest PC, Bahn S, Turck CW. To label or not to label: applications of quantitative proteomics in neuroscience research. Proteomics. 2012;12:736–747. doi: 10.1002/pmic.201100350. An insightful review on quantitative mass spectrometry workflows, feasibility, advantages, and disadvantages of the available techniques for the analysis of the central nervous system specimens from model organisms and human subjects. [DOI] [PubMed] [Google Scholar]
- 27.Kim KY, Joo HJ, Kwon HW, Kim H, Hancock WS, Paik YK. Development of a method to quantitate nematode pheromone for study of small-molecule metabolism in Caenorhabditis elegans. Anal Chem. 2013;85:2681–2688. doi: 10.1021/ac4001964. [DOI] [PubMed] [Google Scholar]
- 28.Romanova EV, Rubakhin SS, Sweedler JV. One-step sampling, extraction, and storage protocol for peptidomics using dihydroxybenzoic acid. Anal Chem. 2008;80:3379–3386. doi: 10.1021/ac7026047. [DOI] [PubMed] [Google Scholar]
- 29.Wang JH, Jiang XY, Sturm RM, Li LJ. Combining tissue extraction and off-line capillary electrophoresis matrix-assisted laser desorption/ionization Fourier transform mass spectrometry for neuropeptide analysis in individual neuronal organs using 2,5-dihydroxybenzoic acid as a multi-functional agent. J Chromatogr. 2009;1216:8283–8288. doi: 10.1016/j.chroma.2009.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang XZ, Petruzziello F, Zani F, Fouillen L, Andren PE, Solinas G, Rainer G. High identification rates of endogenous neuropeptides from mouse brain. J Proteome Res. 2012;11:2819–2827. doi: 10.1021/pr3001699. [DOI] [PubMed] [Google Scholar]
- 31.Ahmed MM, Gardiner KJ. Preserving protein profiles in tissue samples: Differing outcomes with and without heat stabilization. J Neurosci Methods. 2011;196:99–106. doi: 10.1016/j.jneumeth.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svensson M, Boren M, Skold K, Falth M, Sjogren B, Andersson M, Svenningsson P, Andren PE. Heat stabilization of the tissue proteome: a new technology for improved proteomics. J Proteome Res. 2009;8:974–981. doi: 10.1021/pr8006446. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez-Galarza FF, Lawless C, Hubbard SJ, Fan J, Bessant C, Hermjakob H, Jones AR. A critical appraisal of techniques, software packages, and standards for quantitative proteomic analysis. Omics. 2012;16:431–442. doi: 10.1089/omi.2012.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cappadona S, Baker PR, Cutillas PR, Heck AJ, van Breukelen B. Current challenges in software solutions for mass spectrometry-based quantitative proteomics. Amino Acids. 2012;43:1087–1108. doi: 10.1007/s00726-012-1289-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colaert N, Vandekerckhove J, Martens L, Gevaert K. A case study on the comparison of different software tools for automated quantification of peptides. Gel-Free Proteomics: Methods and Protocols. 2011;753:373–398. doi: 10.1007/978-1-61779-148-2_25. [DOI] [PubMed] [Google Scholar]
- 36.Romanova EV, Lee JE, Kelleher NL, Sweedler JV, Gulley JM. Mass spectrometry screening reveals peptides modulated differentially in the medial prefrontal cortex of rats with disparate initial sensitivity to cocaine. AAPS Journal. 2010;12:443–454. doi: 10.1208/s12248-010-9204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romanova EV, Lee JE, Kelleher NL, Sweedler JV, Gulley JM. Comparative peptidomics analysis of neural adaptations in rats repeatedly exposed to amphetamine. J Neurochem. 2012;123:276–287. doi: 10.1111/j.1471-4159.2012.07912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fricker LD. Neuropeptidomics to study peptide processing in animal models of obesity. Endocrinology. 2007;148:4185–4190. doi: 10.1210/en.2007-0123. [DOI] [PubMed] [Google Scholar]
- 39.Gruson D, Bodovitz S. Rapid emergence of multimarker strategies in laboratory medicine. Biomarker. 2010;15:289–296. doi: 10.3109/13547500903560065. [DOI] [PubMed] [Google Scholar]
- 40.Ling XB, Mellins ED, Sylvester KG, Cohen HJ. Urine peptidomics for clinical biomarker discovery. Adv Clin Chem. 2010;51:181–213. doi: 10.1016/s0065-2423(10)51007-2. [DOI] [PubMed] [Google Scholar]
- 41.Quintana LF, Campistol JM, Alcolea MP, Banon-Maneus E, Sol-Gonzalez A, Cutillas PR. Application of label-free quantitative peptidomics for the identification of urinary biomarkers of kidney chronic allograft dysfunction. Mol Cell Proteomics. 2009;8:1658–1673. doi: 10.1074/mcp.M900059-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pietrowska M, Polanska J, Suwinski R, Widel M, Rutkowski T, Marczyk M, Dominczyk I, Ponge L, Marczak L, Polanski A, et al. Comparison of peptide cancer signatures identified by mass spectrometry in serum of patients with head and neck, lung and colorectal cancers: association with tumor progression. Int J Oncol. 2012;40:148–156. doi: 10.3892/ijo.2011.1186. [DOI] [PubMed] [Google Scholar]
- 43.Shen Y, Tolic N, Liu T, Zhao R, Petritis BO, Gritsenko MA, Camp DG, Moore RJ, Purvine SO, Esteva FJ, et al. Blood peptidome-degradome profile of breast cancer. PLoS ONE. 2010;5:e13133. doi: 10.1371/journal.pone.0013133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terracciano R, Preiano M, Palladino GP, Carpagnano GE, Barbaro MP, Pelaia G, Savino R, Maselli R. Peptidome profiling of induced sputum by mesoporous silica beads and MALDI-TOF MS for non-invasive biomarker discovery of chronic inflammatory lung diseases. Proteomics. 2011;11:3402–3414. doi: 10.1002/pmic.201000828. [DOI] [PubMed] [Google Scholar]
- 45.Hansen HG, Overgaard J, Lajer M, Hubalek F, Hojrup P, Pedersen L, Tarnow L, Rossing P, Pociot F, McGuire JN. Finding diabetic nephropathy biomarkers in the plasma peptidome by high-throughput magnetic bead processing and MALDI-TOF-MS analysis. Proteomics Clin Appl. 2010;4:697–705. doi: 10.1002/prca.200900169. [DOI] [PubMed] [Google Scholar]
- 46.Amado F, Lobo MJ, Domingues P, Duarte JA, Vitorino R. Salivary peptidomics. Expert Rev Proteomics. 2010;7:709–721. doi: 10.1586/epr.10.48. [DOI] [PubMed] [Google Scholar]
- 47.Lone AM, Kim YG, Saghatelian A. Peptidomics methods for the identification of peptidase-substrate interactions. Curr Opin Chem Biol. 2013;17:83–89. doi: 10.1016/j.cbpa.2012.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gelman JS, Sironi J, Berezniuk I, Dasgupta S, Castro LM, Gozzo FC, Ferro ES, Fricker LD. Alterations of the intracellular peptidome in response to the proteasome inhibitor bortezomib. PLoS ONE. 2013;8:e53263. doi: 10.1371/journal.pone.0053263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tinoco AD, Kim YG, Tagore DM, Wiwczar J, Lane WS, Danial NN, Saghatelian A. A peptidomics strategy to elucidate the proteolytic pathways that inactivate peptide hormones. Biochemistry (Mosc) 2011;50:2213–2222. doi: 10.1021/bi2000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim YG, Lone AM, Nolte WM, Saghatelian A. Peptidomics approach to elucidate the proteolytic regulation of bioactive peptides. Proc Natl Acad Sci U S A. 2012;109:8523–8527. doi: 10.1073/pnas.1203195109. [DOI] [PMC free article] [PubMed] [Google Scholar]