Abstract
Objective: The study aim was to explore the role of DEK in tumor progression and prognostic of hepatocellular carcinoma (HCC).
Methodology: DEK protein in 178 samples of HCC was evaluated by immunohistochemical method. Additionally, the correlation between DEK expression and the clinicopathological features was evaluated by x2 test or Fisher’s exact test, the survival rates were calculated by the Kaplan-Meier method, and the relationship between prognostic factors and patient survival was also by the Cox analysis.
Results: DEK protein expression was noted in 86 cases of HCC, and 61 cases of normal liver tissues. DEK positive rate were closely correlated with the tumor size, grade, AJCC stage and survival rate (P<0.05, respectively). HCC with large tumor, lower grade, and late-stage, concomitant with DEK expression, had the lowest 5-years survival rate than HCC with above factors but without DEK expression (P<0.01, respectively). DEK expression emerged as significant independent hazard factors for survival in HCC (P<0.01).
Conclusions: DEK could promote aggressiveness of cancer behavior, and hence poor prognosis of the HCC. It might be an independent poor prognostic factor and can serve as a useful new therapeutic biomarker.
Key Words: Carcinoma, DEK, Hepatocellular, Immubohistochemistry, Prognosis
INTRODUCTION
Hepatocellular carcinoma (HCC) is the most common primary hepatic malignancy, especially in Africa and Asia.1,2 Despite many years of sustained efforts, the long-term outcome employing current therapies remain dismal as both recurrences of the lesions within the liver and distant metastases continue to increase. Therefore, both understanding and identification of useful biomarker for disease progression are of central importance for optimizing treatment outcome.
The proto-oncogene DEK protein was originally identified as a fusion with the CAN/NUP214 nucleoporin in a subset of acute myeloid leukemia patients.3,4 In many human aggressive tumors such as neuroblastoma, bladder cancer, cervical cancer, melanoma and the acute leukemia of adults, DEK was subsequently reported as a gene that is frequently upregulated.5-9 The available evidence suggests that DEK protein is closely related to apoptosis by its ability to inhibit p53-mediated apoptosis, cooperate with the viral oncogenes E6 and E7 to overcome senescence, promote HRAS-driven keratinocyte transformation, and promote cellular motility and invasion.4,10-12
Previous studies13 have showed that DEK protein was closely related with the proliferation of serous ovarian tumor cell, and DEK overexpression was significantly correlated with the increased proliferating index of Ki-67 in cervical cancer. Furthermore, it was found that DEK expression is correlated with the prognosis of a variety of human tumors such as breast cancer and ovarian cancer, is expected to become the new molecular targets of the cancer treatment.14 However, there is no study which has addressed to support increased DEK protein levels in HCC patients. This study was conducted to investigate the relationship of DEK expression with clinical pathologic parameters and prognosis in HCC patients.
METHODOLOGY
Samples: All the 178 cases were routinely processed and diagnosed HCC with strict follow-up were randomly selected from the patients undergoing surgery between 2004 and 2007 in the Liaoning and Jilin regions of China. The pathological parameters, including age, gender, size, grade, the presence of cirrhosis and nodal metastasis, portal vein tumor thrombus (PVTT), clinical stage, and survival data were carefully reviewed in all above cases. The patients’ age ranged from 32 to 73 years with a mean age of 55.4yr. The male to female ratio was 116:62. Tumor was staged according to the 7th edition of the American Joint Committee on Cancer (AJCC).15 Of the 178 HCC encompassed 100 cases as early-stage (I-II) and 78 cases as late-stage (III-IV). In which, 38 cases as well differentiated and 140 cases as worse differentiated cancers (grade). All the adjacent benign liver tissue from cancer resection margin were also included, and none of these patients had received chemotherapy before surgery.
All samples were approved by the Institutional Review Board, and informed consent from all patients were obtained before sample collection.
Immunohistochemistry for DEK in paraffin-embedded tissues: Tissue sections were deparaffinized, rehydrated and incubated with 3% H2O2 in methanol for 15 minutes at room temperature to eliminate endogenous peroxidase activity. The antigen was retrieved at 95°C for 20 minutes by placing the slides in 0.01 M sodium citrate buffer (pH6.0). The slides were then incubated with primary antibody DEK (1:50, BD Biosciences Pharmingen, CA, USA) at 4°C overnight. After incubation at room temperature for 30 minutes with biotinylated secondary antibody, the slides were incubated with streptavidin-peroxidase complex at room temperature for 30 minutes. Immunostaining was developed by using chromogen, 3,3′-diaminobenzidine and counterstained with Mayer’s hematoxylin. We used the Mouse IgG isotope controls, which showed negative staining. Also, the positive tissue sections were processed omitting the primary antibody (mouse anti-DEK) as negative controls.
The interpretation criteria were described previously.[6] Briefly, the immunostaining for DEK was semi-quantitatively scored as negative ("-", no or less than 5% positive cells), "+" (5~50% positive cells), and "++" (more than 50% positive cells was). Only the nuclear expression pattern was considered as positive staining which included the weakly(+) and strongly (++) positive cells.
Follow-up observation: All 178 patients had been followed-up for 5 years or until death. At the end of follow-up, 94 patients remained alive.
Statistical analysis: Statistical analyses were performed using the SPSS 17.0. Correlation between DEK expression and clinicopathological characteristics was evaluated by x2 test and Fisher’s exact test. The survival rates after tumor removal were calculated by the Kaplan-Meier method, and difference in survival curves was analyzed by the log rank test. Multivariate survival analysis was performed on all the significant characteristics measured by univariate survival analysis through the Cox proportional hazard regression model. P<0.05 was considered as statistical significance.
RESULTS
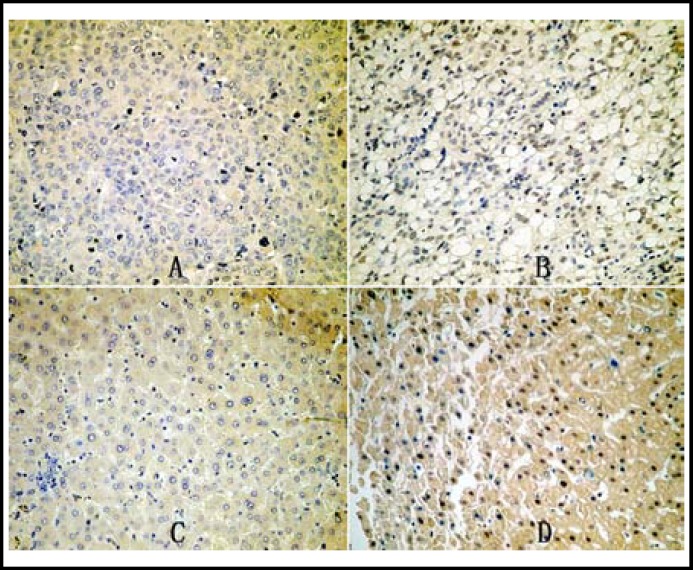
Expression of DEK in HCC: DEK expression showed nucleus immunohistochemical staining pattern in HCC. There was statistical significance between the expression (+and ++) in the HCCs (48.31%, 86/178) and adjacent benign liver tissues (25.28%, 45/178) (P<0.001). (Fig.1)
Fig.1.
Immunohistochemical staining of DEK in HCC and benign liver tissue. DEK is negative (A) and positive (B) in HCC; DEK is negative (C) and positive (D) in benign liver tissue. (Original magnification, ×200
Clinicopathological and prognostic significance of DEK expression: To evaluate the role of DEK in HCC progression, we correlated DEK expression with major clinicopathological features, Table-I and Fig.3 shows that the DEK expression rate in the large tumors (>3cm), lower grade tumors (moderate of poor differentiation), and late-stage tumors (III-IV) was significantly higher than the smaller (≤3cm), higher grade (well differentiation), and early-stage (I-II) cases (P=0.023, 0.007, and 0.005, respectively). However, no statistical difference was found between DEK expression and age, gender, cirrhosis status, lymph node metastasis (LN), and PVTT (P>0.05, respectively).
Table-I.
Relationship between DEK expression and clinicpathological factors
| Characteristic | Cases | Positive Cases | P value | ||||
|---|---|---|---|---|---|---|---|
| Gender | 0.521 | ||||||
| Male | 116 | 54 | |||||
| Female | 62 | 32 | |||||
| Age (Y) | 0.426 | ||||||
| ≥55 | 99 | 51 | |||||
| <55 | 77 | 35 | |||||
| Tumor size (cm) | 0.023* | ||||||
| ≤3 | 67 | 25 | |||||
| >3 | 111 | 61 | |||||
| Tumor grade | 0.007** | ||||||
| higher | 38 | 11 | |||||
| lower | 140 | 75 | |||||
| Cirrhosis | 0.587 | ||||||
| - | 51 | 23 | |||||
| + | 127 | 63 | |||||
| LN | 0.159 | ||||||
| - | 61 | 25 | |||||
| + | 117 | 61 | |||||
| PVTT | 0.113 | ||||||
| - | 103 | 55 | |||||
| + | 75 | 31 | |||||
| TNM | 0.005** | ||||||
| Early | 102 | 40 | |||||
| Late | 76 | 46 |
*P < 0.05, **P< 0.01
The correlation between the survival status and DEK expression by Kaplan-Meier method:
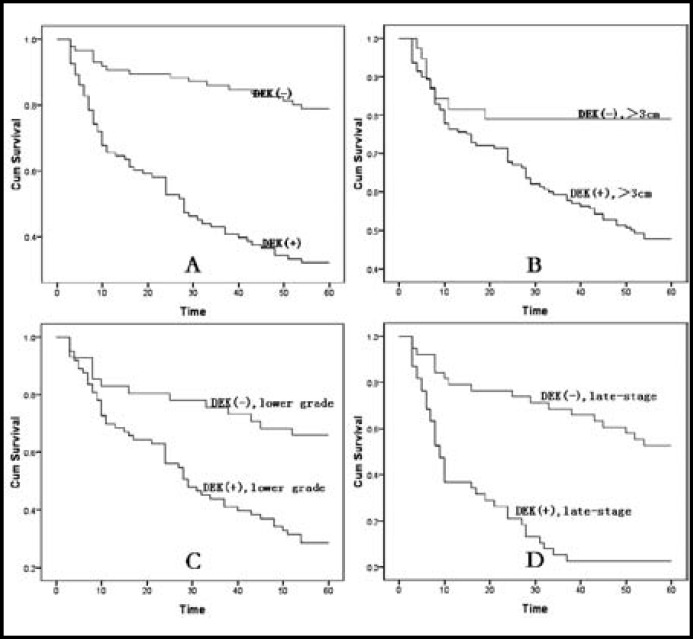
To further confirm the role of DEK expression in HCC progression, we analyzed the 5-years survival rate of 178 HCC cases by Kaplan-meier, and found that HCC patients with DEK expression had a lower 5-years survival rate than those without DEK expression, P<0.001 (Fig. 2A). Meanwhile, we also analyzed the correlation between other factors and the 5-years survival rate in HCC, and found that age, size, grade, PVTT, and AJCC stage were the key factors associated with 5-years survival rate. By combination analysis, we found that HCC with large tumor, lower grade, and tumor stage concomitant with DEK expression showed significantly lower 5-years survival rate than HCC with above factors but without DEK expression (Fig. 2B-D, P=0.007, P=0.001, and P<0.001, respectively), but no statistical relationship is seen between the case with and without LN and PVTT (P>0.05).
Fig.2.
Kaplan-Meier analysis of 5-years survival in 178 HCC patients in relation to DEK protein expression. (2A) Patients with DEK expression had a significantly Lower 5-years survival rate (P<0.001). (2B) HCC with larger tumor concomitant with DEK expression were associated with the worst 5-years survival, significantly worse than HCC with larger tumor only (P=0.007). (2C) HCC with lower grade concomitant with DEK expression had the lower 5-years survival than HCC with lower grade only (P=0.001). (2D) HCC with later-stage concomitant with DEK expression were associated with the worst 5-years survival, significantly worse than HCC with later-stage only (P<0.001).
DEK expression is an independent prognostic factor in HCC by Cox proportional hazard regression model: On univariate analysis all significant variables in Kaplan-Meier test, patients of HCC tumors with DEK expression had a significantly lower 5-years survival than those without DEK expression tumors (HR: 0.516, 95% CI: 0.383-0.694, P<0.001). Meanwhile, age, tumor size, tumor grade, LN, PVTT, and stage, were also associated with 5-years survival rate. Additionally, multivariate analysis was performed using the Cox proportional hazards model. We found that, larger tumor, lower grade, PVTT positive, and late-stage of HCC proved to be independent prognostic factors for survival in HCC. Importantly, DEK expression emerged as significant independent prognostic factors in HCC (HR: 0.161, 95%CI: 0.102-0.256, P<0.001) (Table-II).
Table-II.
Univariate survival analyses (Cox regression model) of various factors in 178 patients
| Factors | B | SE | Wald | HR(95%CI) | P value | |
|---|---|---|---|---|---|---|
| Univariate analyses | ||||||
| Age | 0.333 | 0.151 | 4.844 | 1.396(1.037-1.878) | 0.028* | |
| Gender | 0.007 | 0.155 | 0.003 | 0.987(0.737-1.355) | 0.897 | |
| Tumor size | -0.304 | -0.155 | 3.840 | 0.738(0.545-1.000) | 0.049* | |
| Tumor grade | -0.386 | -0.157 | 6.077 | 0.680(0.500-0.924) | 0.013* | |
| Cirrhosis | 0.003 | 0.167 | 0.000 | 1.003(0.723-1.390) | 0.987 | |
| LN | 0.473 | 0.159 | 8.863 | 1.604(1.175-2.190) | 0.003** | |
| PVTT | -0.307 | 0.152 | 4.075 | 0.736(0.546-0.991) | 0.043* | |
| TNM | -0.763 | 0.154 | 24.498 | 0.466(0.345-0.631) | 0.000** | |
| DEK | -0.766 | 0.153 | 19.011 | 0.516(0.383-0.694) | 0.000** | |
| Multivariant analyses | ||||||
| Age | 0.258 | 0.157 | 2.687 | 1.294(0.951-1.762) | 0.101 | |
| Tumor size | -1.192 | 0.243 | 24.022 | 0.304(0.188-0.489) | 0.000** | |
| Tumor grade | -0.323 | 0.169 | 3.643 | 0.727(0.519-1.009) | 0.056 | |
| LN | 0.318 | 0.168 | 3.589 | 1.374(0.989-1.908) | 0.058 | |
| PVTT | -0.380 | 0.173 | 4.804 | 0.684(0.487-0.961) | 0.028* | |
| TNM | -0.814 | 0.188 | 18.745 | 0.443(0.307-0.641) | 0.000** | |
| DEK | -1.825 | 0.236 | 59.803 | 0.161(0.102-0.256) | 0.000** | |
*P < 0.05, **P < 0.01
DISCUSSION
DEK is located on chromosome 6p22.3, initially described as the target of a recurrent t(6;9) translocation in a subset of acute myeloid leukemia (AML) patients. The human DEK protein consists of 375 amino acids with four distinct tretches of acidic amino acids. It has a central SAP box DNA-binding domainandan additional carboxy-terminal DNA-binding region that partially overlaps with a multimerization domain.16
DEK protein function in the cells has not been clarified. Researches has shown that DEK is a phospho-protein with several phosphorylation sites of which most are clustered in the carboxy terminal region.17 The protein is localized strictly in the nucleus and occurs in copy numbers ranging from about one million copies/nucleus in proliferating lymphocytes to 4–6 million copies/nucleus in cultured HeLa cells. However, the abundance of DEK changes with the physiology of the cell, it is high in proliferating cells, and low in resting and terminally differentiated cells.16
Subsequently, DEK has been shown to promote tumorigenesis in a variety of cancer cell types, at least in part by its ability to interfere with cell division or DNA repair, inhibit cell differentiation, senescence and apoptosis, and cooperate with transforming oncogenes. Kavanaugh GM, et al.18 reported that DEK overexpression promotes the transformation of human keratinocytes, and that DEK knockout mice are partially resistant to chemically induced papilloma formation. Moreover, Trisha, et al5 used littermate DEK knockout, heterozygous and wild type mice for their experiments, and found that there was a significant delay in the formation of papillomas in DEK knockout mice compared to wild type and heterozygous mice. Papillomas ultimately formed in the DEK knockout mice, suggesting a role for DEK in tumor initiation in this model. Shibata, et al19 also showed that DEK overexpression, partly through an increase in its gene dose, mediates the activity of global transcriptional regulators and is associated with tumor initiation activity and poor prognosis in high-grade neuroendocrine carcinoma. Interestingly, some reports have indicated that DEK expression correlates with resistance to chemotherapeutic drugs like camptothecin, etoposide, neocarzinostatin, and doxorubicin, which is often used to treat breast cancer.20-22 In addition, recent reports have shown that DEK mRNA expression is up-regulated in invasive ductal breast cancers with particularly strong gene expression in high grade and late stage breast cancers, making it a potential new target in the fight against recurrence.5,20,23,24
At present, the reports of relationship of HCC and DEK has not been retrived yet. In this study, we performed immunohistochemical staining and analysis in 178 cases of HCC and their adjacent benign tissues, and found that DEK played important roles in the progression of HCC potentially. HCC with DEK expression correlated with large, high grade, late-stage tumors. Additionally, the combination of these indicators are more conductive to the HCC prognosis assessment. By combination analysis, DEK expression significantly affect 5-years survival rate of HCC, and the HCC with larger tumors, lower differentiation, and later-stage, concomitant with DEK expression, had the significantly lower 5-years survival rate than HCC with the factors but without DEK expression. Further analysis showed that DEK expression emerged as significantly independent hazard factors for survival in HCC. These findings led us to suggest that DEK promoted aggressiveness of cancer behavior, and hence poor prognosis of the HCC.
Our findings are perhaps the first to report on the prognostic significance of DEK in HCC, and proved DEK as a potential biomarker for HCC to evaluate its role in tumor progression and prognosis. DEK expression was more commonly seen in cases having poor prognostic factors of HCC, leading to bad differentiation, late-stage, and poor prognosis. DEK may serve as a useful new therapeutic biomarker. However, further studies are needed to confirm these observations.
Authors Contributors
LLJ conceived the study. CLT performed research and wrote the first draft. All authors contributed to the design and interpretation of the study and approved the final version of the manuscript.
References
- 1.Parkin DM, Pisani P, Ferlay J. Global cancer statistics. CA Cancer J Clin. 1999;49(1):33–64. doi: 10.3322/canjclin.49.1.33. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka M, Katayama F, Kato H, Tanaka H, Wang J, Qiao YL, Inoue M. Hepatitis B and C virus infection and hepatocellular carcinoma in China: a review of epidemiology and control measures. J Epidemiol. 2011;21(6):401–416. doi: 10.2188/jea.JE20100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Von Lindern M, Fornerod M, Van Baal S. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol. 1992;12(4):1687–1697. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wise-Draper TM, Allen HV, Jones EE. Apoptosis inhibition by the human DEK oncoprotein involves interference with p53 functions. Mol Cell Biol. 2006;26(20):7506–7519. doi: 10.1128/MCB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wise-Draper TM, Mintz-Cole RA, Morris TA. Overexpression of the cellular DEK protein promotes epithelial transformation in vitro and in vivo. Cancer Res. 2009;69(5):1792–1799. doi: 10.1158/0008-5472.CAN-08-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Q, Li Z, Lin H, Han L. DEK over expression in uterine cervical cancers. Pathol Int. 2008;58(6):378–382. doi: 10.1111/j.1440-1827.2008.02239.x. [DOI] [PubMed] [Google Scholar]
- 7.Orlic M, Spencer CE, Wang L. Expression analysis of 6p22 genomic gain in retinoblastoma. Genes Chromosomes Cancer. 2006;45(1):72–82. doi: 10.1002/gcc.20263. [DOI] [PubMed] [Google Scholar]
- 8.Carro M S, Spiga F M, Quarto M. DEK Expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle. 2006;5(11):1202–1207. doi: 10.4161/cc.5.11.2801. [DOI] [PubMed] [Google Scholar]
- 9.Datta A, Adelson M E, Mogilevkin Y. Oncoprotein DEK as a tissue and urinary biomarker for bladder cancer. BMC Cancer. 2011;11 doi: 10.1186/1471-2407-11-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kappes F, Khodadoust MS, Yu L. DEK expression in melanocytic lesions. Hum Pathol. 2011;42(7):932–938. doi: 10.1016/j.humpath.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel RM, Walters LL, Kappes F. DEK expression in Merkel cell carcinoma and small cell carcinoma. J Cutan Pathol. 2012;39(8):753–757. doi: 10.1111/j.1600-0560.2012.01941.x. [DOI] [PubMed] [Google Scholar]
- 12.Lupescu A, Jilani K, Zbidah M. Induction of apoptotic erythrocyte death by rotenone. Toxicology. 2012;300(3):132–137. doi: 10.1016/j.tox.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Han S, Xuan Y, Liu S, Zhang M. Clinicopathological significance of DEK overexpression in serous ovarian tumors. Pathol Int. 2009;59(7):443–447. doi: 10.1111/j.1440-1827.2009.02392.x. [DOI] [PubMed] [Google Scholar]
- 14.Liu S, Wang X, Sun F, Kong J. DEK over expression is correlated with the clinical features of breast cancer. Pathol Int. 2012;62(3):176–181. doi: 10.1111/j.1440-1827.2011.02775.x. [DOI] [PubMed] [Google Scholar]
- 15.Cheng CH, Lee CF, Wu TH. Evaluation of the new AJCC staging system for resectable hepatocellular carcinoma. World J Surg Oncol. 2011;9 doi: 10.1186/1477-7819-9-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kappes F, Damoc C, Knippers R. Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK. Mol Cell Biol. 2004;24(13):6011–6020. doi: 10.1128/MCB.24.13.6011-6020.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fornerod M, Boer J, van Baal S. Relocation of the carboxyterminal part of CAN from the nuclear envelope to the nucleus as a result of leukemia-specific chromosome rearrangements. Oncogene. 1995;10(9):1739–1748. [PubMed] [Google Scholar]
- 18.Kavanaugh GM, Wise-Draper TM, Morreale RJ. The human DEK oncogene regulates DNA damage response signaling and repair. Nucleic Acids Res. 2011;39(17):7465–7476. doi: 10.1093/nar/gkr454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata T, Kokubu A, Miyamoto M. DEK oncoprotein regulates transcriptional modifiers and sustains tumor initiation activity in high-grade neuroendocrine carcinoma of the lung. Oncogene. 2010;29(33):4671–4681. doi: 10.1038/onc.2010.217. [DOI] [PubMed] [Google Scholar]
- 20.Privette Vinnedge LM, McClaine R, Wagh PK. The human DEK oncogene stimulates β-catenin signaling, invasion and mammosphere formation in breast cancer. Oncogene. 2011;30(24):2741–2752. doi: 10.1038/onc.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kappes F, Fahrer J, Khodadoust MS. DEK is a poly(ADP-ribose) acceptor in apoptosis and mediates resistance to genotoxic stress. Mol Cell Biol. 2008;28(10):3245–3257. doi: 10.1128/MCB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khodadoust M S, Verhaegen M, Kappes F. Melanoma proliferation and chemoresistance controlled by the DEK oncogene. Cancer Res. 2009;69(16):6405–6413. doi: 10.1158/0008-5472.CAN-09-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abba MC, Sun H, Hawkins KA. Breast cancer molecular signatures as determined by SAGE: correlation with lymph node status. Mol Cancer Res. 2007;5(9):881–890. doi: 10.1158/1541-7786.MCR-07-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodes DR, Yu J, Shanker K. Oncomine: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]