Abstract
Objective: We conducted a meta-analysis to compare the EBV DNA and VCA-IgA in diagnosis of Nasopharyngeal Carcinoma, and provide important evidence for screening method of NPC.
Methodology: Three databases, Medline (from Jan. 1966 to Jan. 2012), EMBASE (from January 1988 to Jan. 2012) and Chinese Biomedical Database (from January 1980 to Jan. 2012) were used to detect the role of EBV DNA and VCA-IgA in diagnosis of NPC. Meta-DiSc statistical software was used for analysis.
Results: Twenty seven case-control and cohort studies were included in final analysis. A total of 1554 cases and 2932 controls were included in our meta-analysis. The Sensitivity specificity, positive likelihood (+LR) and likelihood negative (-LR) of EBV-DNA in diagnosis of NPC were 0.75(0.72-0.76), 0.87(0.85-0.88), 6.98(4.50-10.83) and 0.18(0.11-0.29), respectively, and they were 0.83(0.81-0.85), 0.85(0.83-0.86), 10.89(5.41-21.93) and 0.20(0.14-0.29) for VCA-IgA. The SROC for EBV DNA detection was 0.939, while this was 0.936 for VCA-IgA detection. The subgroup analysis showed EBV-DNA had larger areas under the summary receiver operator curve when compared with VCA-IgA in high quality and low quality studies.
Conclusion: Our meta-analysis indicated the EBV DNA had higher sensitivity and specificity in diagnosis of NPC.
Key Words: Epstein-Barr virus (EBV), DNA, VCA-IgA, Diagnosis, Nasopharyngeal Carcinoma
INTRODUCTION
Nasopharyngeal carcinoma (NPC) is a rare disease on a world scale, and it accounted for 0.7% of all cancers, and ranked the 23rd most common new cancer in the world.1 However, it is endemic in some specific areas, such as in Hong Kong, and south of China.1 The intermediate rates are observed in several indigenous populations in South East Asia and in natives of the Arctic region, North Africa and the Middle East.1 Epstein-Barr Virus (EBV) is a well known risk factor for NPC. Patients with NPC are noted to have high levels of EBV antibodies.2,3 The infection of EBV is not associated directly in inducing by the tumor, but infection in the healthy individuals means increased risk of cancer.4-6
Several diagnostic methods are used for NPC detection, but the EBV serology examinations test IgA antibodies against viral capsid antigen (VCA) to IgA to early antigen are the most common detection methods for diagnosis of NPC.2 This method is cheap and non- invasive, and therefore, it is acceptable for patients and could be widely used in clinics. Quantitative EBV DNA and VCA-IgA analysis has been reported to be a sensitive detection tool in diagnosis of NPC.7,8
Recent studies indicated the cell-free EBV DNA had high detection rate in the plasma and serum among patients with NPC.9,10 Recently several studies have showed plasma EBV-DNA and VCA-IgA level might be a sensitive and reliable biomarker for the diagnosis of NPC at a molecular level in clinical practice.11-14 However, the there is no consensus yet which is a better test for the early diagnosis of nasopharyngeal carcinoma. Reasons may include the different sources of EBV antigens, different antibody assays and the selection of cases from different geographic origins. Therefore, we conducted a meta-analysis to evaluate which EBV serology examination had the better sensitivity and specificity in the diagnosis of NPC.
METHODOLOGY
Searching strategy: Three databases, Medline (from Jan. 1966 to Jan. 2012), EMBASE (from January 1988 to Jan. 2012) and Chinese Biomedical Database (from January 1980 to Jan. 2012), were systematically searched by using related terms (‘Epstein-Barr Virus’, ‘EBV’, ‘DNA’, ‘VCA-IgA’, ‘serological test’, ‘nasopharyngeal carcinoma’ (NPC). There was no restriction on the language of the papers. References cited in retrieved studies were reviewed for more eligible studies. The criteria used for including studies were (1) Case-control or cohort studies on the role of EBV-DNA and VCA-IgA in diagnosis of NPC; (2) identification of NPC was confirmed histologically/pathologically; (3) Available data regarding sensitivity and specificity of EBV-DNA and VCA-IgA in diagnosis of NPC; If the authors reported more than once the data on publication papers, we only included the complete data into our review. The exclusion criteria were case only study, reviews, and overlapping studies.
Data extraction: Two reviewers independently reviewed the final abstracts of all potential articles, and decided one should be included into final meta-analysis. In case there was any disagreement, it was resolved by discussion. If the data were missing in the included studies, we attempted to contact the authors by emails or telephones in order to include complete data. From these finally selected studies, we included author’s names, location, study type, number of participants of studies in terms of EBV-DNA and VCA-IgA (Table-I).
Table-I.
Characteristics of included studies
| Study ID | Location |
Sample size
|
Method | Study design | Score of bias | |
|---|---|---|---|---|---|---|
| Case | Control | |||||
| Zhang 2012[15] | Mainland China | 40 | 50 | EBA DNA | Case-control | 6 |
| Zhu 2012[16] | Mainland China | 168 | 60 | EBA DNA and VCA-IgA | Case-control | 6 |
| Feng 2009 [17] | Mainland China | 65 | 29 | EBA DNA and VCA-IgA | Case-control | 7 |
| Kong 2010 [18] | Mainland China | 56 | 60 | EBA DNA | Case-control | 9 |
| Liao 2010 [19] | Mainland China | 34 | 30 | EBA DNA and VCA-IgA | Case-control | 4 |
| Sun 2010 [20] | Mainland China | 62 | 62 | EBA DNA and VCA-IgA | Case-control | 5 |
| Tan 2010[21] | Mainland China | 12 | 40 | EBA DNA and VCA-IgA | Case-control | 3 |
| Wai 2010[22] | Hong Kong | 18 | 1181 | EBA DNA | Case-control | 8 |
| Luo 2009 [23] | Mainland China | 160 | 76 | EBA DNA and VCA-IgA | Case-control | 5 |
| Chang 2008 [24] | Mainland China | 156 | 265 | EBA DNA | Cohort | 5 |
| Sun 2008[25] | Mainland China | 68 | 90 | EBA DNA and VCA-IgA | Case-control | 5 |
| Ozyar 2007[26] | Turkey | 24 | 29 | EBA DNA | Case-control | 7 |
| O 2007[27] | United State | 24 | 84 | EBA DNA and VCA-IgA | Case-control | 8 |
| Li 2007[28] | China | 781 | 171 | VCA-IgA | Case-control | 5 |
| Huang 2006[29] | China | 184 | 80 | VCA-IgA | Case-control | 5 |
| Leung 2004 [30] | Hong Kong | 139 | 178 | EBA DNA and VCA-IgA | Case-control | 7 |
| Shao 2004[31] | Mainland China | 147 | 78 | EBA DNA | Case-control | 6 |
| Fan 2004[32] | Mainland China | 65 | 68 | EBA DNA and VCA-IgA | Case-control | 5 |
| Krishna 2004[33] | India | 17 | 15 | EBA DNA | Case-control | 6 |
| Chan 2003[34] | Mainland China | 55 | 163 | EBA DNA and VCA-IgA | Case-control | 5 |
| Pratesi 2003[35] | Italy | 15 | 32 | EBA DNA | Case-control | 7 |
| Fang 2003[36] | China | 114 | 842 | VCA-IgA | Case-control | 5 |
| Huang 2003[37] | China | 84 | 60 | VCA-IgA | Case-control | 4 |
| Mai 2002[38] | Mainland China | 66 | 58 | EBA DNA | Case-control | 5 |
| Mutirangura 1998[8] | Thailand | 13 | 111 | EBA DNA | Case-control | 7 |
| Shotelersuk 2000[39] | Thailand | 93 | 130 | EBA DNA | Case-control | 6 |
| Lo 1999[9] | Hong Kong | 57 | 43 | EBA DNA | Case-control | 8 |
| Total | 1554 | 2932 | ||||
Quality of study: The quality of included studies was according to the Cochrane Handbook for diagnostic test accuracy review. The criteria included sampling, data collection, design of study, detection application and selection bias. The quality scores ranged from 0 to 10. Score<6 was defined as low quality, and score≥6 was defined as high quality.
Statistical analysis: Statistical analysis was conducted by using Meta-DiSc statistical software version 1.4 (Unit of Clinical Biostatistics, Ramony Cajal Hospital, Madrid, Spain). The accuracy indexes of EBV-DNA and VCA-IgA was pooled by meta-analysis, such as sensitivity, specificity, positive likelihood ratio (LR+) and negative likelihood ratio (LR–). The heterogeneity was evaluated by I2 with p-values < 0.1. The I2 value of 25%, 50% and 75% were regarded as low, moderate and high heterogeneity, respectively (Higgins et al., 2003)39 and its possible sources of heterogeneity were evaluated by subgroup analysis. If moderate or high heterogeneity existed, the random effects model was used. Otherwise, a fixed-effect model was used for pooled results. Summary receiver operating characteristic (SROC) curve was used for evaluating the global summary of test performance, and the area under the SROC curve presents the overall performance of the detection method. The area under the curve of 1 presents perfect discriminatory ability. All P values are two sides and P<0.05 was regarded as statistical significant.
RESULTS
Characteristics of studies: A total of 758 records were selected by searching the databases. After excluding the overlapping studies and those which were not in line with the inclusion criteria. A total of 29 studies were included and assessed for meta-analysis. After reviewing the original paper, we excluded 2 studies. Finally, 27 case-control and cohort studies were included in final analysis. A total of 2717 cases and 4085 controls were included in our meta-analysis (Table-I).
We analyzed the pooled sensitivity, specificity, positive likelihood (+LR) and likelihood negative (-LR) of EBV-DNA and VCA-IgA (Table II and III). The Sensitivity specificity, positive likelihood (+LR) and likelihood negative (-LR) of EBV-DNA in diagnosis of NPC were 0.75(0.72-0.76), 0.87(0.85-0.88), 6.98(4.50-10.83) and 0.18(0.11-0.29), respectively, and they were 0.83(0.81-0.85), 0.85(0.83-0.86), 10.89(5.41-21.93) and 0.20(0.14-0.29) for VCA-IgA.
Table-II.
The diagnostic characteristics of included studies in terms of EBV-DNA
| Study ID | TP | FP | FN | TN | Sensitivity(95% CI) | Specificity(95% CI) | +LR(95% CI) | -LR(95% CI) |
|---|---|---|---|---|---|---|---|---|
| Zhang 2012 | 27 | 10 | 13 | 40 | 0.68(0.51-0.81) | 0.80(0.66-0.90) | 3.38(1.86-6.12) | 0.41(0.26-0.65) |
| Zhu 2012 | 58 | 2 | 110 | 58 | 0.35(0.27-0.42) | 0.97(0.88-1.0) | 10.36(2.61-41.1) | 0.68(0.60-0.76) |
| Kong 2010 | 41 | 7 | 15 | 53 | 0.73(0.60-0.84) | 0.88(0.77-0.95) | 6.28(2.07-12.82) | 0.30(0.20-0.47) |
| Liao 2010 | 20 | 3 | 14 | 27 | 0.59(0.41-0.75) | 0.90(0.74-0.98) | 5.88(1.94-17.85) | 0.46(0.30-0.70) |
| Sun 2010 | 59 | 4 | 3 | 58 | 0.94(0.86-0.99) | 0.94(0.84-0.98) | 14.75(5.71-38.12) | 0.05(0.02-0.16) |
| Tan 2010 | 33 | 0 | 90 | 40 | 0.27(0.91-1.0) | 1.00(0.91-1.0) | 22.15(1.39-353.54) | 0.74(0.66-0.83) |
| Wai 2010 | 15 | 153 | 3 | 1028 | 0.83(0.57-0.96) | 0.87(0.85-0.89) | 6.43(4.99-8.29) | 0.19(0.07-0.54) |
| Feng 2009 | 45 | 1 | 20 | 28 | 0.69(0.57-0.80) | 0.97(0.82-0.99) | 20.08(2.91-138.69) | 0.32(0.22-0.46) |
| Luo 2009 | 110 | 9 | 50 | 67 | 0.69(0.61-0.76) | 0.88(0.79-0.94) | 5.81(3.12-10.82) | 0.35(0.28-0.45) |
| Chang 2008 | 127 | 9 | 29 | 255 | 0.81(0.74-0.87) | 0.97(0.94-0.98) | 23.88(12.51-45.58) | 0.19(0.14-0.27) |
| Sun 2008 | 65 | 6 | 3 | 84 | 0.96(0.88-0.99) | 0.93(0.86-0.98) | 14.34(6.61-31.11) | 0.05(0.02-0.14) |
| Ozyar 2007 | 24 | 10 | 0 | 19 | 1.00(0.86-1.00) | 0.66(0.46-0.82) | 2.8(1.71-4.57) | 0.03(0.01-0.48) |
| O 2007 | 17 | 7 | 5 | 79 | 0.77(0.55-0.92) | 0.92(0.84-0.97) | 9.49(4.51-20.0) | 0.25(0.11-0.54) |
| Leung 2004 | 132 | 4 | 7 | 174 | 0.95(0.90-0.98) | 0.98(0.94-0.99) | 42.26(16.02-111.44) | 0.05(0.03-0.11) |
| Fan 2004 | 64 | 29 | 1 | 39 | 0.99(0.92-1.0) | 0.57(0.45-0.69) | 2.31(1.75-3.05) | 0.03(0.01-0.19) |
| Shao 2004 | 138 | 12 | 9 | 66 | 0.94(0.88-0.97) | 0.85(0.75-0.92) | 6.10(3.62-10.29) | 0.07(0.04-0.14) |
| Krishna 2004 | 15 | 2 | 5 | 10 | 0.75(0.51-0.91) | 0.83(0.52-0.98) | 4.5(1.24-16.35) | 0.3(0.14-0.67) |
| Chan 2003 | 31 | 3 | 24 | 160 | 0.56(0.42-0.70) | 0.98(0.95-0.99) | 30.62(9.75-96.23) | 0.45(0.33-0.60) |
| Pratesi 2003 | 15 | 20 | 0 | 12 | 1.0(0.78-1.0) | 0.38(0.21-0.56) | 1.56(1.18-2.07) | 0.08(0.01-1.31) |
| Mai 2002 | 56 | 6 | 10 | 52 | 0.85(0.74-0.93) | 0.90(0.79-0.96) | 8.20(3.82-17.62) | 0.17(0.10-0.30) |
| Mutirangura 1998 | 13 | 29 | 0 | 82 | 1.0(0.75-0.82) | 0.74(0.65-0.82) | 3.66(2.64-5.07) | 0.05(0.003-0.74) |
| Shotelersuk 2000 | 83 | 63 | 10 | 67 | 0.89(0.81-0.94) | 0.52(0.43-0.60) | 1.84(1.52-2.23) | 0.21(0.11-0.38) |
| Lo 1999 | 55 | 3 | 2 | 40 | 0.97(0.88-0.95) | 0.93(0.81-0.99) | 13.83(4.64-41.24) | 0.04(0.01-0.15) |
| Pooled results | 1243 | 392 | 423 | 2538 | 0.75(0.72-0.76) | 0.87(0.85-0.88) | 6.98(4.50-10.83) | 0.18(0.11-0.29) |
Table-III.
The diagnostic characteristics of included studies in terms of VCA-IgA
| Study ID | TP | FP | FN | TN | Sensitivity | Specificity | +LR(95% CI) | -LR(95% CI) |
|---|---|---|---|---|---|---|---|---|
| Zhu 2012 | 105 | 2 | 53 | 28 | 0.67(0.58-0.74) | 0.93(0.78-0.99) | 9.97(2.60-38.20) | 0.36(0.28-0.46) |
| Liao 2010 | 15 | 1 | 19 | 29 | 0.44(0.27-0.61) | 0.97(0.83-0.99) | 13.24(1.86-94.32) | 0.58(0.43-0.79) |
| Sun 2010 | 58 | 32 | 5 | 58 | 0.92(0.82-0.97) | 0.64(0.54-0.74) | 2.59(1.94-3.45) | 0.12(0.05-0.29) |
| Tan 2010 | 88 | 1 | 35 | 39 | 0.72(0.63-0.79) | 0.98(0.87-1.0) | 28.62(4.12-198.85) | 0.29(0.22-0.39) |
| Luo 2009 | 120 | 4 | 40 | 72 | 0.75(0.68-0.82) | 0.95(0.87-0.89) | 14.25(5.47-37.14) | 0.26(0.20-0.35) |
| Sun 2008 | 15 | 153 | 3 | 1028 | 0.83(0.59-0.96) | 0.87(0.85-0.89) | 6.43(4.99-8.29) | 0.19(0.07-0.54) |
| O 2007 | 29 | 57 | 3 | 66 | 0.91(0.75-0.98) | 0.54(0.44-0.63) | 1.96(1.57-2.44) | 0.18(0.06-0.52) |
| Li 2007 | 704 | 0 | 77 | 171 | 0.90(0.88-0.92) | 1.0(0.98-1.0) | 309.91(19.46-4935.2) | 0.10(0.08-0.12) |
| Huang 2006 | 146 | 2 | 38 | 78 | 0.79(0.73-0.85) | 0.98(0.91-0.99) | 31.74(8.06-125.96) | 0.21(0.16-0.28) |
| Leung 2004 | 112 | 8 | 27 | 170 | 0.81(0.73-0.87) | 0.96(0.91-0.98) | 17.93(9.06-35.46) | 0.20(0.15-0.29) |
| Chan 2003 | 40 | 5 | 4 | 94 | 0.91(0.78-0.76) | 0.95(0.89-0.98) | 18.0(7.62-42.50) | 0.10(0.04-0.24) |
| Fang 2003 | 107 | 193 | 7 | 649 | 0.94(0.88-0.96) | 0.77(0.74-0.80) | 4.10(3.59-4.68) | 0.08(0.04-0.16) |
| Huang 2003 | 146 | 2 | 38 | 78 | 0.79(0.73-0.85) | 0.98(0.91-1.0) | 31.74(8.06-124.96) | 0.21(0.16-0.28) |
| Pooled results | 1685 | 460 | 349 | 2560 | 0.83(0.81-0.85) | 0.85(0.83-0.86) | 10.89(5.41-21.93) | 0.20(0.14-0.29) |
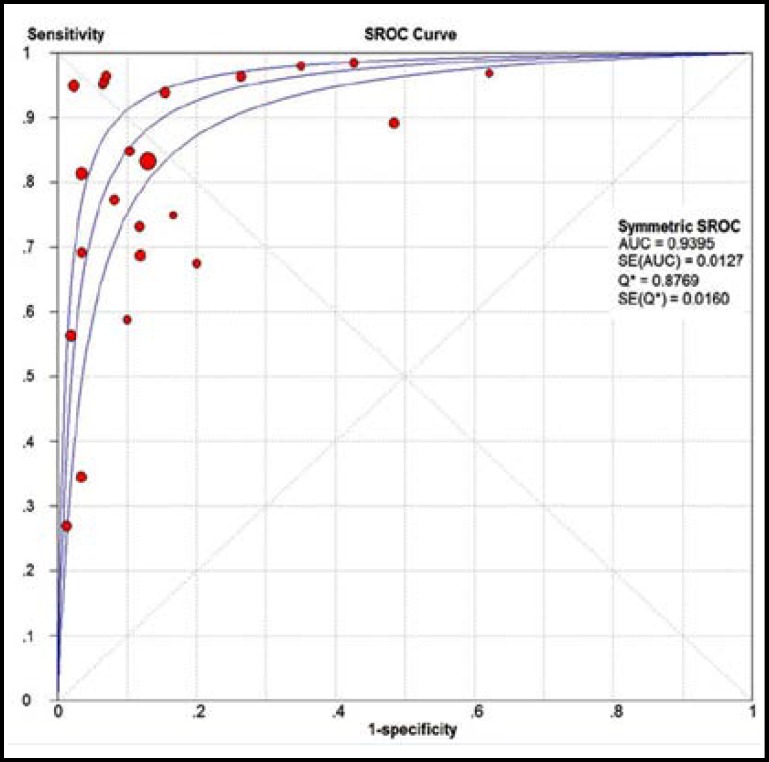
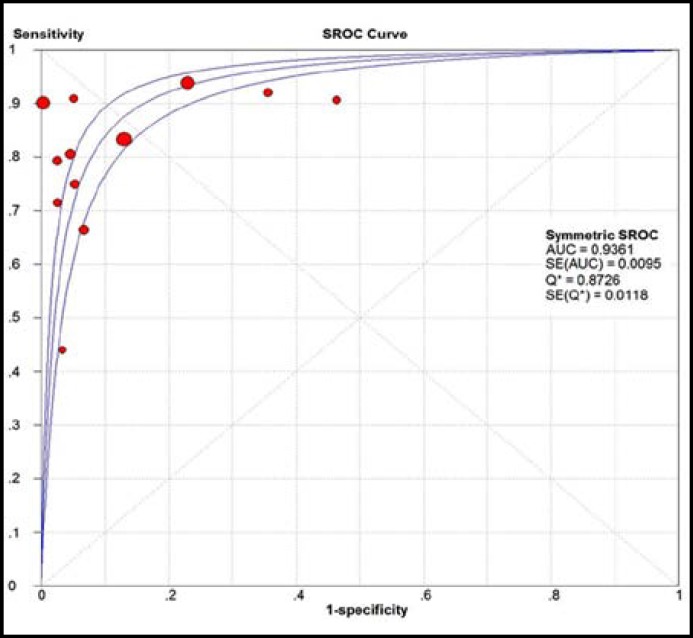
The largest area of diagnosis under the summary receiver operator curve (AUC) for NPC by overall EBV DNA detection was 0.939, while the SROC was 0.936 for VCA-IgA detection (Fig. 1 and 2). In the pooled analysis for EBV-DNA, there was significant heterogeneity across studies (p<0.05, I2>50%). While, no significant heterogeneity was found between studies in terms of VCA-IgA.
Fig.1.
SROC for the pooled accuracy of EBV-DNA for NPC detection
Fig.2.
SROC for the pooled accuracy of VCA-IgA for NPC detection
Subgroup analysis was taken according to the quality of studies to investigate the heterogeneity within the included studies (Table-IV), which indicated studies with low quality had lower sensitivity, specificity, +LR and -LR for both EBV-DNA and VCA-IgA detection. We could find the EBV-DNA had larger areas under the summary receiver operator curve when compared with VCA-IgA in high quality and low quality studies. The subgroup analysis significantly decreases the heterogeneity among studies, with the p value of 0.12 for EBV-DNA and 0.31 for VCA-IgA methods.
Table-IV.
The diagnostic characteristics of EBV DNA in plasma and serum
| Subgroup | TP | FP | FN | TN | Pooled Sensitivity | Pooled Specificity | Pooled +LR(95% CI) | Pooled -LR(95% CI) | SROC |
|---|---|---|---|---|---|---|---|---|---|
| High quality of studies | |||||||||
| EBV-DNA | 678 | 323 | 199 | 1756 | 0.77(0.74-0.80) | 0.85(0.83-0.86) | 5.54(2.25-9.16) | 0.16(0.07-0.37) | 0.93 |
| VCA-IgA | 246 | 67 | 83 | 264 | 0.75(0.69-0.79) | 0.80(0.75-0.84) | 6.94(0.43-111.52) | 0.25(0.13-0.47) | 0.89 |
| Low qu ality of studies | |||||||||
| EBV-DNA | 565 | 69 | 224 | 782 | 0.72(0.68-0.75) | 0.92(0.90-0.94) | 10.20(4.27-24.36) | 0.20(0.09-0.43) | 0.96 |
| VCA-IgA | 1439 | 393 | 266 | 2296 | 0.84(0.82-0.86) | 0.85(0.84-0.87) | 13.05(5.69-29.93) | 0.19(0.12-0.29) | 0.944 |
A single study in our meta-analysis was removed each time to analyze the robust of the pooled results, and the results did not greatly changed (Data not shown). The Egger’s test were used to assess the publication bias, and no significant publication bias was found in our meta-analysis.
DISCUSSION
Meta-analysis has been regarded as an important tool to more precisely define the effect of treatment for diseases and to identify potentially important sources of between-study heterogeneity. There is no systematic review to compare the EBV DNA and VCA-IgA in diagnosis of NPC. Only one previous study showed the sensitivity and specificity of EBV DNA in diagnosis of NPC38, but it could not reach a conclusive result whether EBV DNA is better for VCA-IgA. Hence, our study included 27 recently published studies comparing the effectiveness EBV DNA and VCA-IgA in diagnosis of NPC. Our meta-analysis involved 2757 cases and 4085 controls. Finally, we found EBV DNA had a higher accuracy than VCA-IgA in diagnosis of NPC. The EBV DNA had large SROC of 0.94, while the VCA-IgA had SROC of 0.936. Morever, the high quality of studies in terms of EBV DNA detection had high accuracy in diagnosis of NPC when compared with VCA-IgA (AUC of EBV DNA: 0.93; AUC of VCA-IgA: 0.89).
Heterogeneity is a potential problem in explaining the results of meta-analysis, and identifying the sources of heterogeneity is an important goals of meta-analysis.39 In our study, we assessed the between-study heterogeneity by using the I2 statistic to quantify the between-study heterogeneity39, and the results suggested great heterogeneity between studies in terms of EBV-DNA. Therefore, we further performed subgroup analysis by risk of bias. The results showed that risk of bias was an main source of heterogeneity.
There are two possible limitations in our meta-analysis which mainly influence the explanation of the results. Firstly, there might be publication bias in our study. All the studies included into meta-analysis were published paper; however, there might be many unfavorable results which may not have been published. We plan to include more studies in clinical trials registration and paper presented in conferences. Secondly, there might be selection bias in our study. Secondly as most of the studies included the NPC cases and controls in the same hospital or places, which could influence the results of study.
In conclusion, our results demonstrated the EBV DNA and VCA-IgA detection methods had better effect in diagnosis of NPC. However, EBV DNA detection method had high accuracy in diagnosis of NPC.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–08. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Henle G, Henle W. Epstein-Barr virus-specific IgA serum antibodies as an outstanding feature of nasopharyngeal carcinoma. Int J Cancer. 1976;17:1–7. doi: 10.1002/ijc.2910170102. [DOI] [PubMed] [Google Scholar]
- 3.Old LJ, Boyse EA, Oettgen HF, de Harven E, Geering G, Williamson B, Clifford P. Precipitating antibody in human serum to an antigen present in cultured Burkitt’s lymphoma cells. Proc. Natl. Acad. Sci USA. 1966:1699–1740. doi: 10.1073/pnas.56.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng Y, Zhang LG, Wu YC, Huang YS, Huang NQ, Li JY, et al. Prospective studies on nasopharyngeal carcinoma in Epstein-Barr virus IgA/VCA antibody-positive persons in Wuzhou city, China. Int J Cancer. 1985;36(5):545–547. doi: 10.1002/ijc.2910360505. [DOI] [PubMed] [Google Scholar]
- 5.Cheng WM, Chan KH, Chen HL, Luo RX, Ng SP, Luk W, et al. Assessing the risk of nasopharyngeal carcinoma on the basis of EBV antibody spectrum. Int J Cancer. 2002;97(4):489–492. doi: 10.1002/ijc.1641. [DOI] [PubMed] [Google Scholar]
- 6.Chien YC, Chen JY, Liu MY, Yang HI, Hsu MM, Chen CJ, Yang CS. Serologic markers of Epstein-Barr virus infection and nasopharyngeal carcinoma in Taiwanese men. N Engl J Med. 2001;345:1877–1882. doi: 10.1056/NEJMoa011610. [DOI] [PubMed] [Google Scholar]
- 7.Teo P, Yu P, Lee WY, Leung SF, Kwan WH, Yu KH, et al. Significant prognosticators after primary radiotherapy in 903 nondisseminated nasopharyngeal carcinoma evaluated by computer tomography. Int J Radiat Oncol Biol Phys. 1996;36(2):291–304. doi: 10.1016/s0360-3016(96)00323-9. [DOI] [PubMed] [Google Scholar]
- 8.Mutirangura A, Pornthanakanem W, Theamboonlers A, Sriuranpong V, Lertsanguansinchi P, Yenrudi S, et al. Epstein–Barr viral DNA in serum of patients with nasopharyngeal carcinoma. Clin Cancer Res. 1998;4:665–669. [PubMed] [Google Scholar]
- 9.Lo YM, Chan LY, Chan AT, Leung SF, Lo KW, Zhang J, et al. Quantitative and temporal correlation between circulating cell-free Epstein–Barr virus DNA and tumor recurrence in nasopharyngeal carcinoma. Cancer Res. 1999;59:5452–5455. [PubMed] [Google Scholar]
- 10.Feinmesser R, Miyazaki I, Cheung R, Freeman JL, Noyek AM, Dosch HM. Diagnosis of nasopharyngeal carcinoma by DNA amplification of tissue obtained by fine-needle aspiration. N Engl J Med. 1992;326:7–21. doi: 10.1056/NEJM199201023260103. [DOI] [PubMed] [Google Scholar]
- 11.Yang X, Goldstein AM, Chen CJ. Distribution of Epstein-Barr viral load in serum of individuals from nasopharyngeal carcinoma high-risk families in Taiwan. Int J Cancer. 2006;118(3):780–784. doi: 10.1002/ijc.21396. [DOI] [PubMed] [Google Scholar]
- 12.Shao JY, Zhang Y, Li YH, Gao HY, Feng HX, Wu QL, et al. Comparison of Epstein-Barr virus DNA level in plasma, peripheral blood cell and tumor tissue in nasopharyngeal carcinoma. Anticancer Res. 2004;24:4059–4066. [PubMed] [Google Scholar]
- 13.Fan H, Nicholls J, Chua D, Chan KH, Sham J, Lee S, et al. Laboratory markers of tumor burden in nasopharyngeal carcinoma: a comparison of viral load and serologic tests for Epstein-Barr virus. Int J Cancer. 2004;112:1036–1041. doi: 10.1002/ijc.20520. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Gao HY, Feng HX, Deng L, Huang MY, Hu B, et al. Quantitative analysis of Epstein-Barr virus DNA in plasma and peripheral blood cells in patients with nasopharyngeal Carcinoma. Natl Med J Chin (Chin) 2004;84:982–986. [PubMed] [Google Scholar]
- 15.Zhang LW, Luo BQ, Dou XQ, Deng YH, Zeng ZM, Li JZ, et al. Quantitative analysis of Epstein-Barr virus DNA in saliva, blood serum and peripheral blood cells in patients with nasopharyngeal carcinoma. Chinese Journal of Otorhinolaryngology-Skull Base Surg. 2012;18(1):24–27. [Google Scholar]
- 16.Zhu HF, He X. Significance of Detecting Plasma EBV DNA and VCA-IgA in Patients with Nasopharyngeal Carcinoma. J Chinese Oncology. 2012;18(2):111–113. [Google Scholar]
- 17.Feng HY, Huang YJ, Zhou XY, Mo YX, Huang GW. Diagnosis of NPC with Plasma EBV DNA. J Tropical Med. 2009;9(8):913–915. [Google Scholar]
- 18.Kong P. The clinical value of the quantitative determination of EBV-DNA in blood serum and PBMC patients with nasopharyngeal carcinoma. J Shandong Med Coll. 2010;32(5):321–324. [Google Scholar]
- 19.Liao LY, Kong M, Liu CJ. Comparison of detection of white blood cell’s EBV DNA and plasma’s VCA-IgA in nasopharyngeal carcinoma patients. Med Innovation China. 2010;7(32):145–146. [Google Scholar]
- 20.Sun JG, Wang HX, Xiao FG, Liu YL. Clinical application of plasma ell free EBV DNA, serum CYFR A21-1 and VCA-IgA in patients with nasopharyngeal carcinoma. Modern Oncology. 2010;18(10):1930–1932. [Google Scholar]
- 21.Tan YJ, Su XK, Cui JH. Comparison of detection of serum VCA—lgA,EA-IgA and EBV-DNA in nasopharyngeal carcinoma patient. Chongqing Med J. 2010;39(6):703–706. [Google Scholar]
- 22.Ng WT, Choi CW, Lee MC, Law LY, Yau TK, Lee AW. Outcomes of nasopharyngeal carcinoma screening for high risk family members in Hong Kong. Fam Cancer. 2010;9(2):221–228. doi: 10.1007/s10689-009-9296-y. [DOI] [PubMed] [Google Scholar]
- 23.Luo YL, Ou GP, Chi PD, Liang YN, Liu YH, Huang MY. Combined determination of Epstein-barr virus related antibodies and antigens for diagnosis of nasopharyngeal carcinoma. Chinese J China. 2009;28(1):96–99. [PubMed] [Google Scholar]
- 24.Chang KP, Hsu CL, Chang YL, Tsang NM, Chen CK, Lee TJ, et al. Complementary serum test of antibodies to Epstein-Barr virus nuclear antigen-1 and early antigen: a possible alternative for primary screening of nasopharyngeal carcinoma. Oral Oncol. 2008;44:784–792. doi: 10.1016/j.oraloncology.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Sun JG, Zheng AP. Clinical significance of plasma EBV DNA and VCA-IgA for nasopharyngeal carcinoma. Modern Oncology. 2008;16(12):2086–2087. [Google Scholar]
- 26.Ozyar E, Gültekin M, Alp A, Hasçelik G, Ugur O, Atahan IL. Use of plasma Epstein- Barr virus DNA monitoring as a tumor marker in follow-up of patients with nasopharyngeal carcinoma: preliminary results and report of two cases. Int J Biol Markers. 2007;22:194–199. doi: 10.1177/172460080702200305. [DOI] [PubMed] [Google Scholar]
- 27.O TM, Yu G, Hu K, Li JC. Plasma Epstein-Barr virus immunoglobulin A and DNA for nasopharyngeal carcinoma screening in the United States. Otolaryngol Head Neck Surg. 2007;136:992–997. doi: 10.1016/j.otohns.2006.11.053. [DOI] [PubMed] [Google Scholar]
- 28.Li Q. The relationship between EB virus VCA-IgA antibody and the diagnosis of nasopharyngeal carcinoma. J Hainan Med College (Chin) 2007;13:180–183. [Google Scholar]
- 29.Huang LS, Zhu B, Chen YH, Zhao HL. Significance of combined detection of serum tumor makers in the diagnosis of nasopharyngeal carcinoma. Lab Med (Chin) 2006;21:472–474. [Google Scholar]
- 30.Leung SF, Tam JS, Chan AT, Zee B, Chan LY, Huang DP, et al. Improved accuracy of detection of nasopharyngeal carcinoma by combined application of circulating Epstein-Barr virus DNA and anti-Epstein-Barr viral capsid antigen IgA antibody. Clin Chem. 2004;50(2):339–345. doi: 10.1373/clinchem.2003.022426. [DOI] [PubMed] [Google Scholar]
- 31.Krishna SM, James S, Kattoor J. Serum EBV DNA as a biomarker in primary nasopharyngeal carcinoma of Indian origin. Jpn J Clin Oncol. 2004;34(6):307–311. doi: 10.1093/jjco/hyh055. [DOI] [PubMed] [Google Scholar]
- 32.Chan KH, Gu YL, Ng F, Ng PS, Seto WH, Sham JS, et al. EBV specific antibody-based and DNA-based assays in serologic diagnosis of nasopharyngeal carcinoma. Int J Cancer. 2003;105:706–709. doi: 10.1002/ijc.11130. [DOI] [PubMed] [Google Scholar]
- 33.Pratesi C, Bortolin MT, D’Andrea M. Quantitative plasma/serum EBV DNA load by LMP2A determination in an Italian cohort of NPC patients. J Clin Virol. 2003;28:155–164. doi: 10.1016/s1386-6532(02)00276-7. [DOI] [PubMed] [Google Scholar]
- 34.Fang QQ. Application of EB virus detection and the needle aspiration of neck lymph node in the diagnosis of nasopharyngeal carcinoma. Fujian Med J (Chin) 2003;25:54–55. [Google Scholar]
- 35.Huang WG, Lin J, Chen RC. Detection of diagnosis about nasopharyngeal carcinoma combined with EBVCA-IgA and EBVA-IgA in serum. J Pract Med Tech (Chin) 2003;10:1235–1236. [Google Scholar]
- 36.Mai S, Zong Y, Zhang M, Zhong B, Lin S. Detection of Epstein-Barr virus DNA in plasma/serum: a useful serological indicator for diagnosis of nasopharyngeal carcinoma. Chin Med J (Engl) 2002;115(12):1895–1897. [PubMed] [Google Scholar]
- 37.Shotelersuk K, Khorprasert C, Sakdikul S. Epstein-Barr virus DNA in serum/plasma as a tumor marker for nasopharyngeal cancer. Clin Cancer Res. 2000;6:1046–1051. [PubMed] [Google Scholar]
- 38.Liu Y, Fang Z, Liu L, Yang S, Zhang L. Detection of Epstein-Barr virus DNA in serum or plasma for nasopharyngeal cancer: a meta-analysis. Genetic Testing and Molecular Biomarkers. 2011;15(7-8):495–502. doi: 10.1089/gtmb.2011.0012. [DOI] [PubMed] [Google Scholar]
- 39.Higgins JP, Thompson SG, Deeks JJ. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]