Abstract
Despite intense interest in the proteolysis of the ß-Amyloid Precursor Protein (APP) in Alzheimer's disease (AD), how the normal processing of this type I receptor-like glycoprotein is physiologically regulated remains ill-defined. In recent years, several candidate protein ligands for APP, including F-spondin, Reelin, β1 Integrin, Contactins, Lingo-1 and Pancortin, have been reported. However, a cognate ligand for APP that regulates its processing by α- or β-secretase has yet to be widely confirmed in multiple laboratories. Here, we developed new assays in an effort to confirm a role for one or more of these candidate ligands in regulating APP ectodomain shedding in a biologically relevant context. A comprehensive quantification of APPsα and APPsβ, the immediate products of secretase processing, in both non-neuronal cell lines and primary neuronal cultures expressing endogenous APP yielded no evidence that any of these published candidate ligands stimulate ectodomain shedding. Rather, Reelin, Lingo-1 and Pancortin-1 emerged as the most consistent ligands for significantly inhibiting ectodomain shedding. These findings led us to conduct further detailed analyses of the interactions of Reelin and Lingo-1 with APP.
Keywords: Regulated intramembrane proteolysis, Amyloid Precursor Protein, ectodomain shedding, Lingo-1/Reelin, Alzheimer's disease
Although the stepwise proteolysis of the ß-Amyloid Precursor Protein (APP) to release amyloid ß-protein (Aß) has been central to the study of Alzheimer's disease (AD), how the normal processing of this conserved type I membrane glycoprotein is physiologically regulated remains poorly defined. In AD, amyloid (neuritic) plaques are principally composed of the potentially neurotoxic Aβ peptides, which are generated by the sequential proteolytic processing of APP (reviewed in (1)). Cleavage of APP by either α- or β- secretase results in the shedding of large extracellular portions of APP termed APPsα or APPsβ, respectively. The remaining C-terminal fragments (CTFα and CTFβ) are then cleaved intramembranously by γ-secretase. Cleavage of CTFβ by γ -secretase releases Aβ into the luminal/extracellular space and AICD into the cytoplasm. Cleavage of CTFα by γ-secretase releases the smaller p3 fragment into the lumen/extracellular space and AICD into the cytoplasm. The latter pathway, which begins with APP ectodomain shedding by α-secretase, predominates in almost all cell types and precludes Aβ production.
Experimental studies suggest several roles for APP in brain development, including migration of neuronal precursor cells (2, 3), neurite outgrowth (4-7), cell adhesion (8, 9) and synapse formation (10, 11). Since its initial discovery, APP has been hypothesized to be a cell surface receptor (12). In recent years, several candidate protein ligands for APP, including F-spondin (13, 14), Reelin (15, 16), β1 Integrin (4, 16), Contactins (17-19), Lingo-1 (19) and Pancortin (20), have been reported to interact physically with the ectodomain of APP and modulate APP processing and, in some cases, APP function in neurodevelopment.
F-spondin, a secreted extracellular matrix glycoprotein, was identified in unbiased screens for APP interactors (13, 19). Transfection of F-spondin was initially found to inhibit β-secretase cleavage of APP as measured by CTFP levels in human embryonic kidney (HEK) 293 cells also over-expressing APP and BACE1 (13). F-spondin inhibited AICD-dependent gene transactivation, suggesting that α-secretase cleavage was also inhibited by F-spondin (13). However, a subsequent study reported that F-spondin enhanced APPsα and CTFα in addition to reducing APPsβ in COS7 cells overexpressing APP (14).
β1 Integrin is a type I single transmembrane protein important for cell adhesion. We have found β1 Integrin to physically interact with APP and to be involved in APP-dependent neurite outgrowth (4). These findings were confirmed in a study by another group in which they also reported that β1 Integrin enhanced APPsα and CTFα levels in COS7 cells overexpressing APP (16).
The Contactins (CNTNs) are GPI-anchored neuronal-specific cell adhesion molecules (reviewed in (21)). CNTN4 was identified in a screen for extracellular APP binding partners in embryonic chick brain, and only CNTN3 and CNTN4 but not the remaining CNTN family members were found to directly bind APP in vitro (17). However, other groups reported evidence of a physical interaction of CNTN2 (18) and CNTN1(19) with APP. Expression of CNTN4 in HEK cells overexpressing APP led to an increase of CTFα levels in some experiments and a decrease in others (17). CNTN2 was reported to enhance AICD, CTFα and CTFβ in both overexpressed and endogenous assays (18). Functional interactions between CNTN4 and APP in neurite outgrowth (17) and CNTN2 and APP in neurogenesis (18) have been reported.
Lingo-1 (leucine rich repeat and Ig domain-containing Nogo receptor interacting protein-1), a single-transmembrane protein, is a member of the Nogo-66 receptor complex and negatively regulates axonal myelination and regeneration (reviewed in (22)). Lingo-1 was among the proteins identified (along with F-spondin) in the APP interactome study of intact mouse brain (19). This study reported a physical interaction between APP and Lingo-1 and showed that knockdown of Lingo-1 in HEK293 cells stably overexpressing APP bearing the “Swedish” AD mutation increased CTFα and lowered CTFβ, whereas overexpression of Lingo-1 increased CTFβ (19). A separate group confirmed a physical interaction in an overexpressed cell system and determined that the interaction occurs via the ectodomain of Lingo-1 (23).
Reelin, a large glycoprotein, is secreted from Cajal-Retzius cells in the embryonic cortex and regulates the migration of neuronal precursor cells (reviewed in (24)). In the adult cortex, Reelin is secreted by a subset of interneurons and plays a role in synaptic plasticity (reviewed in (25)). In two studies, Reelin was shown to physically interact with APP and enhance APPsα and CTFα levels in COS7 cells overexpressing APP (15, 16). Subsequently, another group showed that reduction of Reelin enhanced both CTFβ and Aβ in APP transgenic mice (26). A functional interaction between Reelin and APP in neurite outgrowth has been described (16).
We recently reported Pancortin, a secreted glycoprotein with multiple isoforms, as a candidate ligand for APP (20). Pancortin was identified in an unbiased screen of endogenous proteins from murine cortical slices that interacted with the APP ectodomain (20). Pancortin-1 and Pancortin-2 were found to specifically reduce β-secretase but not α-secretase cleavage of endogenous APP in HEK293 cells, while Pancortin-3 had no effect (20). We also uncovered a functional interaction of Pancortin isoforms with APP in regulating the entry of neuronal precursor cells into the cortical plate (20).
In the context of these numerous reports of candidate ligands with often variable individual results, a cognate ligand for APP that regulates its processing by α- or β- secretase has yet to be widely confirmed by multiple laboratories in biologically relevant systems. Here, in an effort to confirm a role for one or more reported candidate ligands in regulating α- or β- secretase cleavage of APP, we describe a systematic comparison of candidate ligands by directly quantifying APPsα and APPsβ, the immediate products of secretase processing, across multiple assays. First, we compare candidate APP ligands in a non-neuronal mammalian cell line with overexpression of APP, in keeping with virtually all of the above initial reports on these candidate ligands. Then, we compare the candidates in novel assays we have developed to measure proteolytic processing of endogenous APP in both non-neuronal and neuronal cell lines. From these studies, we do not confirm any candidates as triggering ectodomain shedding of APP. However, Reelin, Lingo-1 and Pancortin-1 emerge as the most consistent ligands that reduce α-and/or β- secretase cleavage of APP. Accordingly, we report further detailed analyses of the interactions of Reelin and Lingo-1 with APP.
Experimental Procedures
Plasmids
Plasmids utilized for transient and stable transfections include, pCAX-APP751(human) as described (2), pcDNA-APP695-swedish (human), as described (27), pCAX-β1 Integrin (mouse) as described (4), PCAX –Pancortin-1 and -4 (mouse) as described (20). pcDNA4-His/Myc-F-spondin(human) was kindly provided by T. Sudoff (13). Lingo-1 (human) was obtained from DF/HCC DNA Resource Core deposited by the Mammalian Gene Collection consortium and cloned into the pCDH vector. CNTN2-Fc(human) was kindly provided by J. Flanagan (17) and cloned into the pcDNA vector. pcDNA-Reelin (mouse) was kindly provided by T. Curran (28). Constructs for the Reelin fragments (N-R6, R3-8, 3-6, N-R2, and R7-8) (mouse) were provided by A. Goffinet (29). pcDNA-APOER2 were kindly provided by J. Herz. VLDLR-myc (mouse) was kindly provided by H-S Hoe (14). HEK293 cell lines stably expressing Reelin or vector control, CER or CEP4 respectively, were kindly provided by T. Curran (30).
Immunoprecipitation (IP) and Western blotting (WB)
For IP studies, HEK293 cells were transfected with the specified combinations of Reelin, APOER2, VLDLR, and APP. Cells were lysed in 1% NP40 STEN buffer (150 mM NaCl, 50 mM Tris, 2 mM EDTA, and 1.0% (v/v) NP-40). Lysates were IPed with either anti-Reelin (G10, Millipore), anti-APOER2 (Abcam, Cambridge, MA) anti-VLDLR (R&D Systems, Minneapolis, MN), or anti-APP (C9) and protein A and G agarose resin (Sigma, St. Louis, MO) overnight and washed 3X with 1% NP40 STEN buffer. Lysates, CM, and immunoprecipitations were electrophoresed on 10–20% Tricine or 4-12% Bis-Tris gels (Invitrogen, Carlsbad, CA) and transferred to nitrocellulose. Western blotting (WB) was performed with primary antibodies anti-APP (C9, 1:1,000; Selkoe lab), anti-APPsα (1736, 1:2,000; Selkoe lab), anti-rodent APP/APPsα (597,1:200; Immuno-biological Laboratories, Minneapolis, MN), anti-GAPDH (1:2,000; Millipore, Billerica, MA), anti-FLAG (M2; 1:1,000; Sigma, St. Louis, MO), anti-Lingo-1(1:1,000; Millipore, Billerica, MA), anti-β1 Integrin (1:1,000; Abcam, Cambrdige, MA), anti-F-spondin (1:1,000; Abcam, Cambrdige, MA), anti-Reelin (N-terminal, G10) (1:500, Millipore, Billerica, MA), anti-Reelin (mid-region, R4B) (1:1000, Developmental Studies Hybridoma Bank, Iowa City, IA) anti-Reelin (C-terminal) (1:1000, E5, Santa Cruz Biotechnology, Santa Cruz, CA), anti-Tau (1:2,000, Dako, Carpinteria, CA), anti-BACE1 (1:500, Millipore, Billerica, MA), anti-ADAM17 (1:500, Abcam, Cambridge, MA), each followed by IRDye800- or IRDye680-conjugated secondary antibodies (1:10,000; Rockland Immunochemicals, Gilbertsville, PA) and detection with the LICOR Odyssey detection system. For quantitative Western blots, Pancortin-3 (Rice et al., 2012), Reelin and F-spondin (R&D systems, Minneapolis, MN) recombinant proteins were utilized as standards.
APP processing assays in HEK293 cells
HEK293 cells were plated in 6 well plates at 1×10̂6 cells/well and transiently transfected with cDNA of each candidate ligand or empty vector (as control) using Fugene HD (Promega, Madison, WI). In the assay to examine effects on overexpressed APP, APP751 was co-transfected with the candidate ligand or empty vector. At 24 hr post-transfection, media (DMEM + 10% FBS) were replaced, and at 48 hr post-transfection, media were collected and centrifuged at 200 g for 5 min, and cells were lysed in 1% NP-40 STEN buffer. Human APPsα, APPsβ, Aβ40, and Aβ42 levels in the CM was quantified by multiplex ELISA kits (Meso Scale Discovery, Gaithersburg, MD) and normalized to holoAPP in the lysate (for the overexpressed APP assay) or normalized to total intracellular protein (for the endogenous APP assay). For DAPT treatment of Lingo-1 transfected HEK293 cells, cells were treated for 24 hrs with 5 μM DAPT in DMSO. One-way ANOVA tests were performed with the Bonferroni correction for multiple comparisons.
APP processing assays in rat primary cortical cultures
Cortical neurons from E1 8 Sprague Dawley rats were plated in 6-well poly-D-lysine coated plates at 7.5 × 10̂5 cells/well typically for 4 DIV (but but some experiments ranged for 2-12 DIV with similar effects). In our co-culture assay, The HEK293 stable cell lines expressing the ligand of interest were pelleted by centrifugation and then resuspended in neuronal medium. The HEK293 cells are then plated at 7.5 × 10̂5 cells/well overlying the neurons for 18-24 hr. Medium is then changed 4 hrs later to remove any unattached HEK293 cells. In our CM assay, neurons were treated for 18-24 hr with CM from the HEK293 cell lines stably expressing the ligand of interest. CM was obtained from stable cell lines conditioned for 24 hr in serum-free optiMEM concentrated 10× with Amicon Ultra 10K MWCO centrifugal filters (Millipore) and diluted to a 1× solution in Neurobasal media. After the 18-24 hr period of CM treatment or co-culture, CM was collected and centrifuged at 2000 rpm for 5 min. Cells were lysed in 1% NP-40 STEN. Endogenous rat neuronal APPsα was quantified by a rodent-specific ELISA kit (Immunobiological Laboratories). APPsα was normalized to a neuronal-specific protein, Tau, by WB analysis in the co-culture assay and to total intracellular protein by BCA assay (Fisher) in the CM assay. One-way ANOVA tests were performed with the Bonferroni correction for multiple comparisons.
Results
Effects of candidate ligands on APPsα and APPsβ levels in HEK293 cells
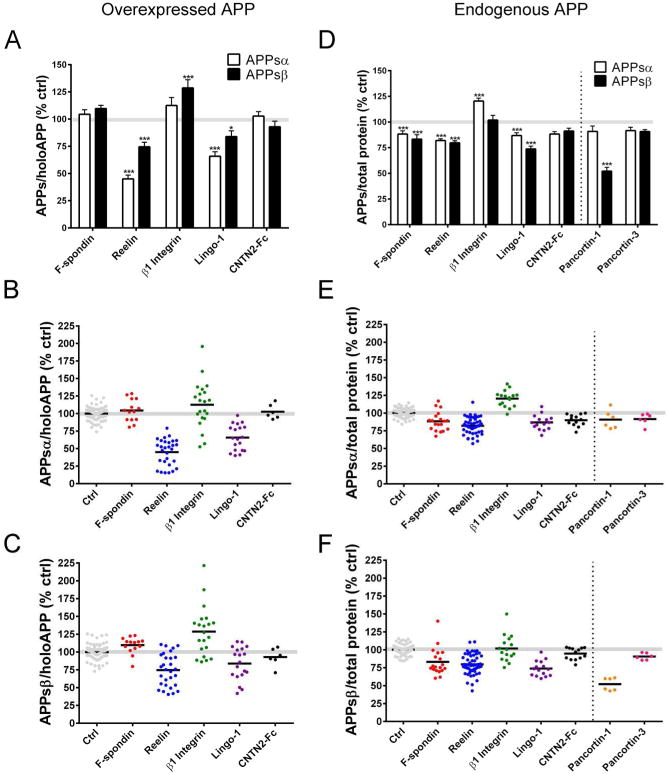
Candidate APP ligands were first examined in a non-neuronal mammalian cell line overexpressing APP, because this had been done in nearly all of the initial reports of these particular candidate ligands (reviewed in Introduction). In this assay, HEK 293 cells were transiently co-transfected with one of the putative ligands and human APP751, and the medium (DMEM with 10% FBS) was changed 24 hr after transfection. At 48 hr, the conditioned media (CM) were collected, and cells were lysed. APPsα and APPsβ levels in the CM were measured by a sensitive and highly reproducible MSD multiplex ELISA. APPsα and APPsβ levels were normalized to holoAPP, which was measured by WB of the respective cell lysates. Using this assay, F-spondin and CNTN2-Fc did not significantly modulate APPsα or APPsβ levels (Fig 1A-C). Expression of Reelin resulted in the greatest change of APPsα and APPsβ levels, with a decrease of 54.9 ± 3.5% (p<0.001) and 25.4 ± 4.2% (p<0.001), respectively (Fig 1A-C). Expression of Lingo-1 also significantly reduced levels of APPsα by 34.1 ± 4.2% (p<0.001) and APPsβ by 16.1 ± 5.3% (p<0.05) (Fig 1A-C). β1 Integrin increased APPsβ levels by 28.6% ± 7.5 (p<0.001) and did not significantly modulate APPsα levels (Fig 1A-C).
Figure 1. Effects of candidate ligands on APPsα and APPsβ levels in HEK293 cells.
(A-C) HEK293 cells were co-transfected with APP751 (human, wild-type) and candidate ligands or empty vector (as control). APPsα and APPsβ levels were quantified by ELISA and normalized to holoAPP by Western blot and shown as a percentage of control. (D-F) HEK293 cells (expressing only endogenous human APP) were transfected with candidate ligands or empty vector (as control) and APPsα and APPsβ levels were quantified by ELISA and normalized to total intracellular protein and shown as a percentage of control. (A,D) Bar graph showing average APPsα and APPsβ levels of all experiments. Scatter plots showing APPsα (B,E) and APPsβ (C,F) levels for each replicate of each experiment. Error bars represent s.e.m.; * p<.05, ***p<.001
Of note, these mean changes in soluble APP shedding were all determined by ELISAs on numerous individual samples performed over multiple experimental days (Fig. 1B, C). Reelin produced the most robust and consistent effect in this assay (see scatterplots in Fig. 1B-C). Across all replicates, Reelin overexpression resulted in a 25% or greater decrease in APPsα (Fig 1B). Transfection of β1 Integrin resulted in the highest variability across experiments, as APPsα and APPsβ levels were enhanced in some experiments but reduced in others (Figs 1B-C). These variable effects of β1 Integrin on APPsα and APPsβ shedding appeared to be due to differences in holoAPP expression levels. In this overexpression assay system, co-transfection of β1 Integrin with APP751 led to much greater percent changes in holoAPP levels than did co-transfection of the other 4 candidates (Fig S1). Even after normalization of the APPsα and APPsβ levels to holoAPP levels in each experiment, the effects of β1 Integrin on APPsα secretion were significantly correlated (R2 =.44, p<0.01) with differences in holoAPP expression (Fig S1). For example, when holoAPP levels were relatively high in the β1 Integrin co-transfectants compared to vector transfected controls, then relative APPsα/holoAPP levels were also high. In contrast, we observed no significant correlations between APPsα/holoAPP and changes in holoAPP expression levels for F-spondin, Reelin and Lingo-1 (Fig. S1).
Proteolytic processing of overexpressed APP can be quite different than that of endogenous APP. For example, we observed that the APPsα/APPsβ ratio in CM was 6.6 ± 0.7 in HEK293 cells expressing just endogenous human APP but was a remarkable 78.4 ± 8.8 in HEK293 cells overexpressing human APP (Fig S2). This striking many-fold difference highlights the non-physiological nature of the procesing of overexpressed APP, and we therefore developed assays to investigate the effects of candidate ligands on proteolytic processing of endogenous APP, something that has not typically been reported for potential APP ligands. These experiments were initially performed in HEK293 cells using the same methods as in the above assay with the exception that no co-transfection of APP occurred. WBs of cellular lysates confirmed that the transfection of the candidate ligands did not alter endogenous holoAPP levels (Fig 2B); therefore, endogenous APPsα and APPsβ levels in the CM were normalized to the more quantitative measure of total intracellular protein concentration in the lysate. Using this assay, we found that the expression of each candidate ligand resulted in a small but significant change in APPs level. APPsα and APPsβ levels were significantly reduced by the expression of F-spondin (APPsα: 11.7 ± 3.1%; p<0.001; APPsβ: 16.7 ± 4.3%; p<0.001), Reelin (APPsα: 18.0 ± 1.6%; p<0.001; APPsβ: 20.2 ± 2.0%; p<0.001), and Lingo-1 (APPsα: 13.1 ± 2.7%; p<0.001; APPsβ: 26.2 ± 2.8%; p<0.001) (Fig 1D-F). Expression of β1 Integrin significantly increased APPsα by 20.4 ± 2.9% (p<0.001) but did not significantly change APPsβ levels (Fig 1D). CNTN2 did not significantly affect APPsα or APPsβ levels. Thus, while Reelin and Lingo-1 strongly inhibited α- and β- secretase cleavage of overexpressed APP in HEK293 cells (Fig. 1A), cleavage of endogenous APP was more weakly – but still significantly -- inhibited by Reelin and Lingo-1 (Fig. 1D). As a comparative control in this same assay, we repeated experiments on the proteins Pancortin-1 and Pancortin-3 that we recently described as APP ectodomain ligands (20). In agreement with our previous report, Pancortin-1 significantly reduced β-secretase cleavage (47.9 ± 3.6%; p<0.001) without affecting α-secretase cleavage of endogenous APP, whereas the isoform Pancortin-3 had no significant effects on either α- or β- secretase cleavage. The effect of Pancortin-1 on APPsβ was more robust and less variable than any of the other candidate ligands we tested in this endogenous APP cleavage assay in 293 cells (Fig 1E-F).
Figure 2. Representative Western blots of APP shedding assays with HEK293 cells.
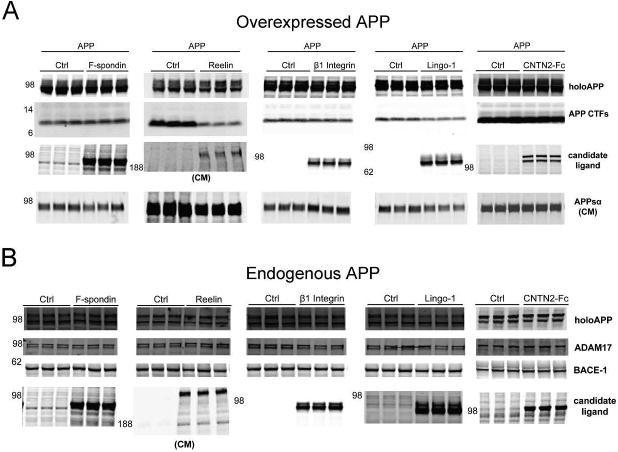
(A) Western blot of lysates (and CM where noted) showing expression levels of holoAPP, APP CTF, candidate ligands, and APPsα in a representative experiment with co-transfection of candidate ligands and APP751 into HEK293 cells. (B) Western blot of lysates (and CM where noted) showing expression levels of holoAPP, secretases, and candidate ligands from a representative experiment with transfection of candidate ligands into HEK293 cells with endogenous APP.
Western blots (WB) from representative experiments are shown for the HEK293 cell assays in which APP was either overexpressed (Fig 2A) or endogenous (Fig 2B), demonstrating the expression levels of both holoAPP and the candidate ligands. Expression levels were similar among the various candidate ligands, with an estimate of 5-10 μg/mg of cell lysate or 5-10 mg/mL secreted into the CM for those tested by quantitative Western blot (Fig S3). In the initial experiments where APP was overexpressed, CTFs and APPsα could be readily detected by WB, and these paralleled the changes in APPs by ELISA (Fig 2A, see e.g., Reelin and Lingo-1). Levels of ADAM17 (an α-secretase for APP) and BACE-1 (β-site APP cleaving enzyme-1, or β-secretase) were not changed by the expression of the candidate ligands (Fig 2B), suggesting that any effects on APP processing we observed were not due to changes in the levels of the secretases that cleave APP.
In both of these HEK293 cell assays (Fig. 1), we utilized DMEM media with 10% fetal bovine serum (FBS). Previous reports of these and other candidate APP ligands have used a variety of medium conditions, including medium with serum, serum-free (SF) medium, and SF medium supplemented with bovine serum albumin (BSA) as a carrier protein. This technical variability could help explain some of the different results obtained by different labs. We found that medium supplemented with either FBS or BSA enhanced the recovery and subsequent detection of APPsα in the CM by over 3-fold (Fig S4A). Moreover, transfected cells conditioned in media with serum were healthier than those with only BSA. Therefore, we chose to condition our cells in DMEM+10% FBS for all of the studies reported above, as this allows the best health of the transfected cells and the best recovery of APPsα. Importantly, we showed that the effect of Reelin on endogenous APPsα levels was similar across these three media conditions (Fig S4B).
Effects of candidate APP ligands on APPsα levels in primary cortical neuronal cultures
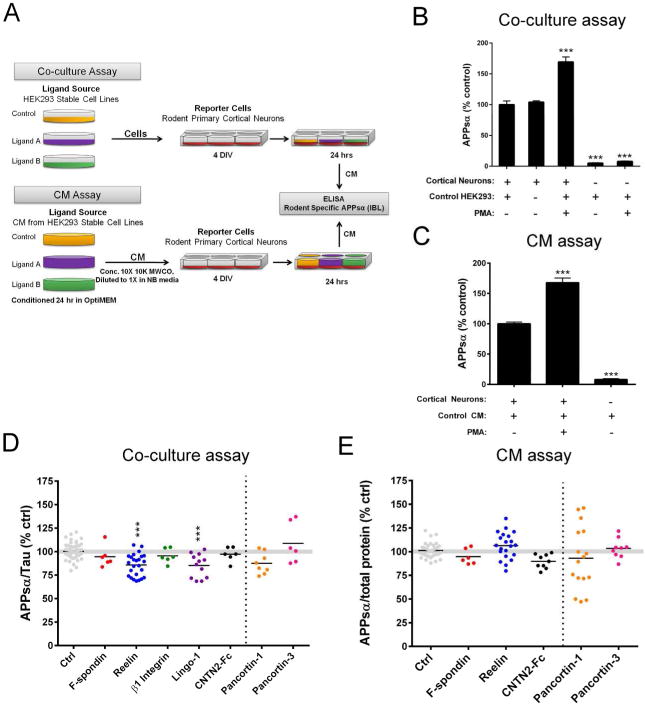
The most physiologically relevant culture system for analyzing putative ligands that regulate processing of APP in the central nervous system would assay the effects on endogenous APP in primary neurons with the ligands presented in trans. To this end, we developed both co-culture and conditioned medium (CM) assays in untransfected primary neuronal cultures (Fig. 3A). In both assays, E18 rat primary cortical neurons were utilized as the reporter cell. In our co-culture assay, stable HEK293 cells expressing the ligands of interest were co-cultured overlying the neurons for 18-24 hr (Fig. 3A). Alternatively, in our CM assay, neurons were treated with the CM of stably transfected HEK293 cells expressing the ligand of interest (Fig 3A). Endogenous APPsα produced from the neurons (but not from the human HEK293 cells) was detected by a rodent-specific APPsα ELISA. APPsα was normalized to a neuron-specific marker (Tau) in the lysate of our co-culture assay (in order to normalize to only the neuronal reporter cells but not the HEK293 ligand source) or to total intracellular protein in our CM assay. As an important negative control, we observed no significant difference in APPsα levels secreted from neurons cultured alone compared to those co-cultured with control (untransfected) HEK293 cells at the optimized densities of both cell types employed here (Fig 3B, first two bars). In both assays, an expected increase in neuronal APPsα could be detected in the CM upon treatment with PMA (phorbol-12-myristate-13-acetate) as a positive control (Fig 3B-C) (31, 32). Further, human APPsα from the HEK293 cells represents a negligible percentage of the total APPsα detected in both the co-culture and CM assays, confirming the specificity of our rodent-specific ELISA (Figs 3B-C).
Figure 3. Effects of candidate ligands on APPsα levels in primary cortical cultures.
(A) Schematic of methods used for co-culture and CM assays. (B-C) Positive controls (PMA treatment) and negative controls (neurons and HEK293 cells alone) for the co-culture (B) and CM (C) assays. (D) ELISA quantification of APPsα (endogenous, rodent) secreted from cortical neurons co-cultured with HEK293 cells stably expressing ligand of interest. (E) ELISA quantification of APPsα (endogenous, rodent) secreted from cortical neurons treated with CM from HEK293 cells expressing ligand of interest. Error bars represent s.e.m.; * p<.05
Reelin and Lingo-1 (which showed the most consistent effects in reducing APPsα in the HEK293 cell assays) significantly reduced neuronal APPsα in the co-culture assay (Fig 3D-E). Reelin reduced APPsα by 14.3 ± 2.4% (p<0.001), and Lingo-1 reduced APPsα by 14.7 ± 3.6% (p<0.001). F-spondin, β1 Integrin, and CNTN2-Fc did not significantly modulate APPsα levels in the neuronal co-culture assay (Fig 3D). Only those proteins that are secreted could be tested in the CM assay. As in the co-culture assay, treatment of neurons with F-spondin and CNTN2-Fc CM had no effect on APPsα levels (Fig 3E). In contrast to the neuronal co-culture assay and the two HEK293 assays, Reelin CM had no effect on neuronal APPsα levels (Fig 3E). For comparison, we performed experiments with Pancortin-1 and Pancortin-3 in both of these assays. We had previously reported a decrease in endogenous APPsβ but not APPsα levels by expressing Pancortin-1 in HEK293 cells. Here, we were only able to perform a rodent-specific ELISA for APPsα, since rodent-specific antibodies for APPsβ are not available, and we confirmed that there was no significant effect of either Pancortin isoform on APPsα secretion in neurons (Fig 3D,E).
Western blots from representative experiments are shown for both the co-culture (Fig 4A) and CM (Fig 4B) assays in rodent cortical neuronal cultures, demonstrating the expression levels of the candidate ligands and endogenous rat APP. Expression of endogenous holoAPP in the neuronal lysates was not affected by the candidate ligands in either assay (Fig 4A,B). Expression of Tau, which we used as a neuronal-specific protein for normalization, was relatively consistent across conditions (Fig 4A).
Figure 4. Representative Western blots of APP shedding assays with rat primary cortical cultures.
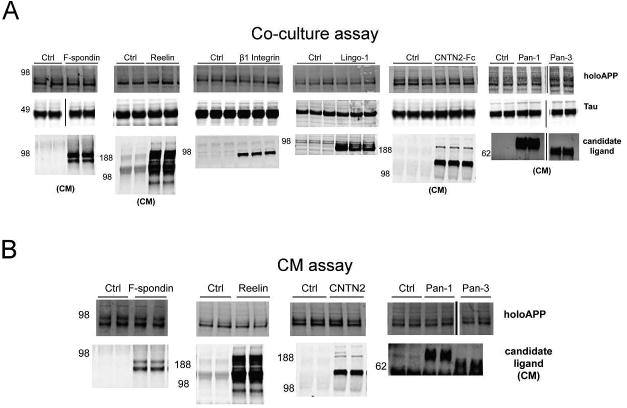
(A) Western blot of lysates (and CM where noted) showing expression levels of holoAPP, Tau, and candidate ligands in a representative experiment with co-culture of neurons with HEK293 cells expressing putative ligands. (B) Western blot of lysates (and CM where noted) showing expression levels of holoAPP and candidate ligands from a representative experiment with neurons treated with CM from HEK293 cells expressing putative ligands.
Addressing discrepancies in the effects of Reelin and Lingo-1 on APP cleavage
After testing these candidate ligands in assays on both overexpressed and endogenous APP and in both neuronal and non-neuronal cells, Reelin, Lingo-1 and Pancortin-1 emerged as the candidate APP ligands with the most consistent and quantitatively significant effects on the α- and/or β-secretase cleavages of APP (Table 1). However, the effects of Lingo-1 and Reelin in our assays were not identical to previous reports. We have already characterized in detail the interaction of the Pancortins with APP in a recent publication (20). Here, we attempt to reconcile experimentally the discrepancy between our data and previous reports for Reelin and Lingo-1.
Table 1. Summary of candidate ligands tested in multiple APP shedding assays.
The magnitude of statistically significant changes (p<.05) in APPsα or APPsβ are represented by arrows. ↑ or ↓ < 25% change relative to control; ↑↑ or ↓↓ = 25-50% change; ↑↑↑ or ↓↓↓ > 50% change. An equals sign (=) represents no significance difference. Gray boxes represent conditions not determined.
| Cell type | Ligand Source | APP | F-spondin | Reelin | β1 Integrin | Lingo1 | CNTN2-Fc | Pancortin-1 | Pancortin-3 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| APPsα | APPsβ | APPsα | APPsβ | APPsα | APPsβ | APPsα | APPsβ | APPsα | APPsβ | APPsα | APPsβ | APPsα | APPsβ | |||
| HEK293 | transient transfection | transient transfection | = | = | ↓↓↓ | ↓↓ | = | ↑↑ | ↓↓ | ↓ | = | = | ||||
| endogenous | ↓ | ↓ | ↓ | ↓ | ↑ | = | ↓ | ↓↓ | = | = | = | ↓↓↓ | = | = | ||
| primary rat cortical neurons | co-culture with HEK293 stables | endogenous | = | ↓ | = | ↓ | = | = | = | |||||||
| CM from HEK293 stables | endogenous | = | = | = | = | = | ||||||||||
For Lingo-1 in our HEK293 assay with overexpressed APP, the variability in the magnitude of reduction of APPsα and APPsβ (normalized to holoAPP) across experiments appears to be due to the expression levels of Lingo-1. The degree of reduction in APPsα and APPsβ was directly and significantly correlated with protein levels of Lingo-1 (APPsα: R2 =.69, p<.001; APPsβ: R2 =.76, p<.001) (Fig. 5A-B). The reduction of APPsβ by Lingo-1 was in contrast to a previous study (19) in which Lingo-1 enhanced P-secretase cleavage of APP. This discrepancy could be due to differences in the processing of wild-type APP, which we studied, and APP with the Swedish AD mutation that Bai et al (19) studied. Therefore, we tested the effects of Lingo-1 in HEK293 cells transfected with APP695 bearing the Swedish mutation (APPswe). Co-transfection of Lingo-1 with APPswe significantly reduced APPsα but, unlike with wild-type APP, had variable effects on APPsβ. Lingo-1 caused enhanced APPsβ in some experiments and reduced APPsβ levels in others and overall had no significant effect on APPsβ (Fig 5C-D). However, we noticed that co-transfection of the standard 1 μg of Lingo-1 cDNA with APPswe cDNA reduced APPswe expression. Therefore, we tested 0.1 μg of Lingo-1 cDNA in which APPswe expression was less affected, but Lingo-1 still had variable effects on APPsα and APPsβ, with no overall significant effect on either (Fig 5C, E). In our studies of Lingo-1, we also uncovered evidence of the γ-secretase-dependent intramembrane cleavage of Lingo-1. Upon transfection of Lingo-1 into HEK293 cells, we detected a ∼10 kDa fragment of Lingo-1 with a C-terminal Lingo-1 antibody, and the cellular levels of this CTF were enhanced 2-fold when the cells were treated with a γ-secretase inhibitor (DAPT) (Fig 5F-G). These data strongly suggest that Lingo-1 is processed by γ-secretase via the regulated intramembranous proteolysis mechanism, something which was not previously known. Overexpression of APP did not alter the production of the Lingo-1 CTF (Fig 5F-G).
Figure 5. Biochemical analysis of the interaction of Lingo-1 with APP.
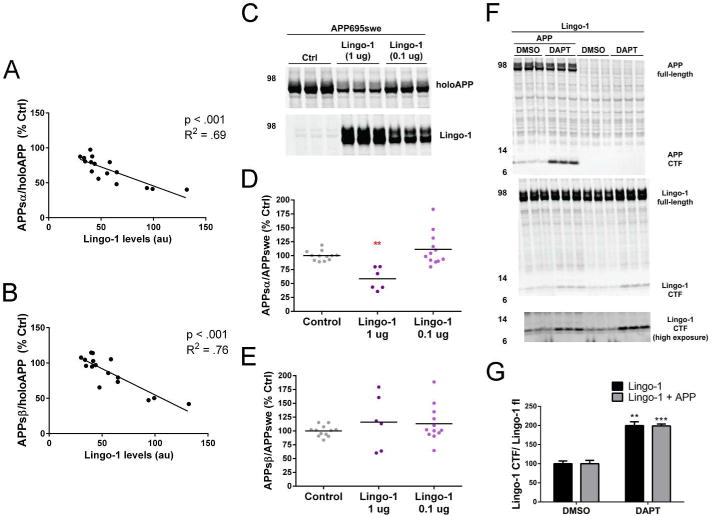
(A-B) For co-transfection of Lingo-1 and APP751 (human, wild-type) into HEK293 cells, the percent change in APPsα/holoAPP (A) or APPsβ/holoAPP (B) was graphed as a function of Lingo-1 expression. A regression correlation was performed, and p values represent statistical significance of the slope deviating from 0. (C-E) HEK293 cells were transfected with APP695-swedish (human) and either vector only (control) or Lingo-1 (with both 1.0 μg and 0.1 μg of DNA). (C) Western blot of lysates showing expression levels of holoAPP and Lingo-1. Quantification of APPsα (D) APPsβ (E) for each replicate of each experiment shown with scatterplots. (F-G) HEK293were transfected with Lingo-1 or Lingo-1 plus APP751 (human, wild-type) and treated for 24 hrs with DAPT or DMSO (as control). (F) Western blot of lysates showing expression levels of holoAPP, APP CTF, Lingo-1 (apparent full-length and CTF). (G) Quantification of Lingo-1 CTF/full-length Lingo-1 with and without expression of APP. **p<.01, ***p<.001
Reelin was previously reported to enhance APPsα and CTF and reduce Aβ levels (15, 16), whereas we found a reduction of APPsα and APPsβ levels by Reelin. First, we examined CTF and Aβ levels. Upon co-transfection of APP with Reelin in HEK293 cells, APP CTFs and Aβ40 and Aβ42 could be readily detected. Expression of Reelin not only decreased APPsα and APPsβ as documented above, but it also substantially reduced levels of the APP CTF, Aβ40, and Aβ42 (Figs. 2 and 6A). Next, we investigated whether the reduction in APPsα and APPsβ by Reelin expression was dose-dependent, or if differences in expression levels of Reelin might explain the conflicting results. Increasing concentrations of Reelin cDNA were transfected, leading to rising expression of Reelin secreted into the CM (Fig 6B) accompanied by a dose-dependent decrease in both APPsα and APPsβ (Fig 6C), consistent with our earlier Reelin results (Fig. 1A and B). Finally, the effect of Reelin on APPsα was investigated according to the methods in which Reelin was previously reported to enhance APPsα levels (15, 16). Here, Reelin CM was applied to COS7 cells transfected with APP751. In contrast to this prior study, we found no effect of Reelin when utilizing this method (Fig S5).
Figure 6. Biochemical analysis of the interaction of Reelin with APP.
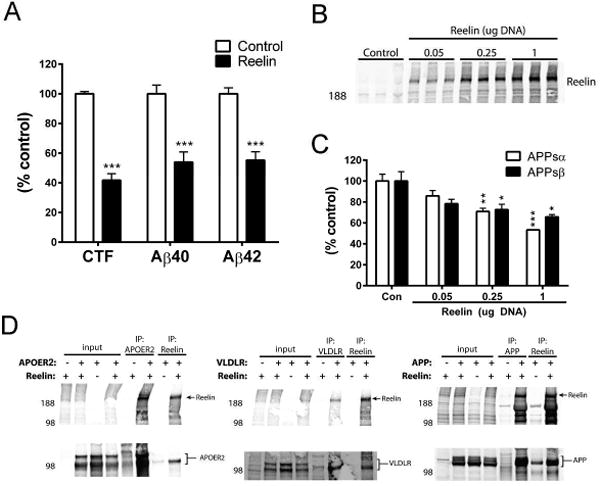
(A) Quantification of Aβ40, and Aβ42 (by ELISA) and CTF (by WB) in HEK293 assay with over-expression of APP751 (human, wild-type). (B) Reelin expression in CM of HEK293 cells co-transfected with APP751 (human, wild-type) and increased concentrations of Reelin cDNA. (C) Quantification of APPsα and APPsβ levels in response to increasing Reelin expression. (D) HEK293 cells were transfected with listed combinations of Reelin, APP, APOER2, and VLDLR, and co-IPs were performed for Reelin or else APP, APOER2, or VLDLR. Error bars represent s.e.m.; * p<.05, **p<.01, ***p<.001
Biochemical analysis of the interaction of Reelin with APP
Next, we sought to confirm whether Reelin and APP could physically interact. Reelin and APP were co-transfected into HEK293 cells, and lysates were immunoprecipitated (IP) for either Reelin or APP. Indeed, Reelin co-IPed with APP, and in the reverse direction, APP co-IPed with Reelin (Fig. 6D, right panel). As a negative control, IP with the APP antibody (C9) failed to co-IP Reelin in the absence of APP overexpression. IP with the Reelin antibody (G10) did IP detectable levels of APP in the absence of Reelin overexpression; however, overexpressing Reelin greatly enhanced the co-IP of APP with Reelin above this background endogenous level (Fig 5D). Importantly, we found that the co-IP of Reelin and APP was quantitatively comparable to the co-IP of Reelin with its canonical receptors, APOER2 and VLDLR (33-35) (Fig 5D).
Reelin undergoes proteolytic cleavages in primary neurons at both its C-terminal and N-terminal ends to generate five fragments (Fig 7A) (29, 36, 37). To determine which physiological fragments of Reelin may be sufficient for the inhibition of α-secretase cleavage of APP, HEK293 cells were co-transfected with APP751 and cDNAs for full-length Reelin or else each of the 5 known Reelin fragments. Reelin antibodies with epitopes towards the different regions of Reelin (G10, R4B, E5) (Fig 7A) were used to detect all of the Reelin fragments by Western blot (Fig 7B). Each fragment of Reelin significantly reduced α-secretase cleavage of APP compared to control (Fig 7C). However, expression of the fragments containing the N-terminal region of Reelin (N-R6 and N-R2) resulted in the greatest reduction in APPsα, and this lowering was not significantly different than that seen from full-length Reelin (Fig 7C). Conversely, expression of Reelin fragments containing the C-terminal region but lacking the N-terminal region (R3-8 and R7-8) led to relatively higher levels of APPsα, compared to the effect of full-length Reelin (Fig 7C). These differential effects due not appear to be due to differential expression levels. When the levels of each transfected Reelin fragment detected with single antibodies were compared to full-length Reelin, their expression levels were relatively similar to one another, with only R7-8 having higher expression levels but still less effect on APP shedding (Fig 7B). Thus, while each physiological proteolytic fragment of Reelin can inhibit α-secretase cleavage of APP to some extent, the N-terminal region of Reelin is the most active.
Figure 7. Effects of Reelin fragments on APPsα in HEK293 cells.
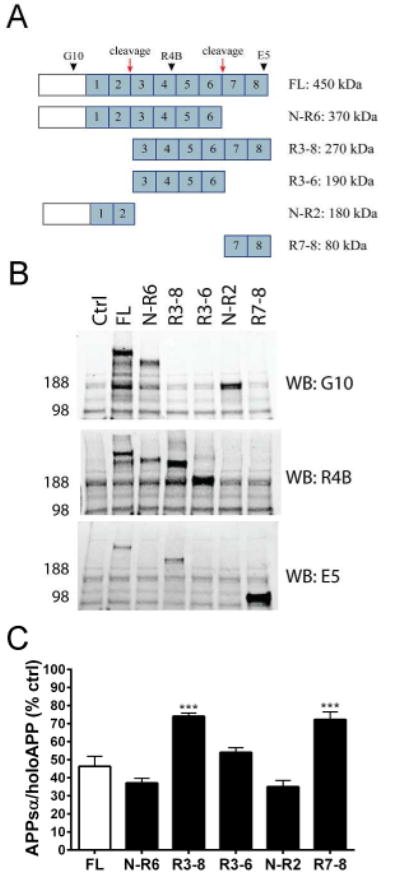
Schematic of Reelin fragments generated from proteolytic processing. Reelin repeat domains are numbered in blue. Red arrows represent cleavage sites. Black arrowheads represent antibody epitopes. (B) Western blots of cell lysates showing expression of Reelin fragments transfected into HEK293 cells. (C) ELISA quantification of APPsα levels in HEK293 cells co-transfected with APP751 (human, wild-type) and either full-length Reelin or individual Reelin fragments. Error bars represent s.e.m.; ***p<.001, relative to full-length Reelin (FL)
Discussion
Novel and systematic approaches to analyze the effects of candidate ligands on APP processing
Several extracellular and membrane-bound proteins have been proposed as candidate ligands that may modulate proteolytic processing of the ubiquitously expressed APP polypeptide. However, these candidates have not been validated by multiple laboratories and have often been examined solely with overexpressed APP and with one or two assay systems. In an effort to clarify a role for one or more of the reported ligands in regulating the processing of APP, we systematically and rigorously investigated the ability of these candidates to modulate α- and β-secretase cleavage of APP. In contrast to virtually all prior studies, we used multiplex ELISA-based assays to obtain quantitative measures of both APPsα and APPsβ, as opposed to solely relying on Western blotting. Further, many of the previous studies show data from a single “representative” experiment. We chose to show our data by scatter-plot analysis of all experiments, and we found that for most putative ligands, a single experiment could not adequately represent the complete data set of the range of effects on APP cleavage and would thus be misleading. Instead, we report a comprehensive quantification of data points across all experiments to capture the inherent biological variability of the effects of each ligand, as well as the technical variability for different assay types. Furthermore, the quantification of secreted APPsα/β we used provides a direct measure of the α- and β-secretase cleavages, as opposed to measuring only CTFα/β, the levels of which can be further complicated by the degree of γ-secretase activity.
Another important aspect to consider when performing assays to accurately measure APPsα/β generation in the CM is the appropriate normalization. Changes in cellular holoAPP levels can change APPsα and APPsβ levels independent of any effects of a ligand on α- and β- secretase cleavage per se. Because co-transfection of APP concurrent with the candidate ligands could lead to differences in APP levels due to technical rather than biological reasons, we normalized data in which APP is overexpressed to holoAPP levels in the lysates of the same cultures. However, because normalizing to holoAPP, which as an end-point measurement in lysates may not fully correct for variations in the levels of APPs that accumulate in the media, we also checked for correlations between the holoAPP cellular level and the APPsα/holoAPP ratio (Fig S1). For assays measuring cleavage of endogenous APP, we observed no detectable differences in holoAPP in the lysates by Western blot and therefore normalized to the more quantitative measure of total intracellular protein concentration in the lysate. For the co-cultures of neurons with HEK293 cells, we normalized to a neuron-specific marker (Tau) in the lysate, in order to normalize only to the neuronal reporter cells but not the HEK293 ligand source.
As a result of attention to these various technical factors and controls, a side-by-side comparison of candidate ligands across multiple assays is presented here for the first time. We initially used an assay similar to the original reports for each of these ligands, i.e., with non-neuronal immortalized mammalian cells overexpressing APP, to enable direct comparisons to those previous reports. We found that co-transfection of APP with certain candidate ligands can lead to the most dramatic effects on APPsα/p levels, perhaps due to a wider dynamic range inherent to the overexpression assay or to artifacts from supraphysiological levels of APP or non-biologically relevant changes in APP levels. For example, we found evidence that despite normalization to holoAPP, the effects of β1 Integrin in the overexpression assay were due to variations in APP expression levels (Fig S1). Furthermore, we found that APPsα/APPsβ ratios were more than 10-fold higher with overexpressed APP (∼75) than with endogenous APP (∼6), suggesting a fundamental alteration in the processing of overexpressed APP (Fig S2). A likely explanation is that β-secreatse (BACE1) is not highly expressed endogenously in these non-neuronal cells, and overexpressing APP leads to much greater processing in the α-secretase pathway. For these reasons and because the goal is to determine the in vivo neurobiological relevance of these ligands, it is critical to confirm any findings from APP-overexpressing systems in endogenous and, preferably, neuronal systems. In this context, we proceeded to develop novel assays to compare the candidate ligands in non-neuronal and neuronal cell lines relying on endogenous APP. In particular, we believe the co-culture assay using primary rat cortical neurons has advantages over other systems: 1) APP is endogenously expressed by the neuronal reporters; 2) necessary but unknown co-receptors/co-ligands also should be endogenously expressed in the neuronal reporters; 3) ligands are continuously produced by the co-cultured HEK cells (rather than requiring artificial pulse administration); 4) ligands that require expression on the plasma membrane for activity will be expressed in their natural state; and 5) effects on APP processing that could be relevant to AD are best studied in neurons.
F-spondin
While F-spondin was the first reported candidate APP ligand with perhaps the most evidence across laboratories for effects on APP cleavage (13, 14, 38), we observed little evidence of these effects in our assays. We found no significant changes in APPsα or APPsβ levels in HEK293 cells co-transfected with APP and F-spondin (Fig 1A) or in primary neurons co-cultured with F-spondin stable cells lines (Fig 3D) or treated with F-spondin-containing CM (Fig 3F). However, we did observe a subtle decrease of endogenous APPsα and APPsβ in plain HEK293 cells transfected with F-Spondin (Fig 1D). A potential underlying difference between our results and previous results is that in order to maintain a less artificial system, we did not overexpress BACE1 (as in (13)) or APOER2 (as in (14)). Perhaps the most direct contrast between our studies and previous studies was in the treatment of primary neurons with F-Spondin-containing CM. Previously, F-spondin CM was reported to enhance CTFα levels in primary neurons (14), but we failed to observe an effect on APPsα with a similar assay.
Integrin β1
Expression of Integrin β1 was previously reported in one study to enhance APPsα and APP CTF (16). Here, we found only minor evidence for a subtle overall enhancement of APPsα and APPsβ in HEK293 cells. However, this effect was not confirmed in primary neurons. Further, in contrast to the relative consistency of the rest of the candidate ligands, transfection of Integrin β1 in HEK293 cells overexpressing APP resulted in very high variability in APPs secretion. The effects ranged from very dramatic increases in APPsα and APPsβ to only subtle or no changes in APPsα/β levels or even reductions in APPsα/β levels in some experiments (Fig 1B-C). We found that this variability in APPsα was significantly correlated with the expression of holoAPP upon co-transfection of APP751 with β1 Integrin, even after normalization to holoAPP levels (Fig S1). The changes in holoAPP levels do not appear to represent a biologically relevant effect of β1 Integrin on APP expression, as β1 Integrin did not change expression of endogenous APP. Thus, changes in APPsα upon co-transfection of β1 Integrin with APP appear to be an artifact due to differences in APP co-transfection efficiency.
Contactin-2
CNTN2 has been reported to modulate APP processing by increasing AICD, CTFα and CTFβ levels in both over expressed and endogenous assays (18). However, our data did not confirm these findings. We found no effects of a soluble (Fc-tagged) form of CNTN2 on APPsα or APPsβ both in our endogenous and overexpressed assays.
Lingo-1
Knockdown of Lingo-1 has been reported to enhance CTFα and reduce CTFβ levels, while overexpression of Lingo-1 was reported to enhance CTFβ levels in HEK293 cells overexpressing the APPswe mutation (19). As predicted from Bai et al. (19), we found that Lingo-1 reduced APPsα levels in each of our assays (Figs 1 & 3), including primary neuronal cultures (Fig 3). However, instead of an enhancement in β-secretase cleavage of APP by Lingo-1 (19), we found that Lingo-1 reduced β-secretase cleavage. The discrepancy between these effects on β-secretase cleavage of APP may be due to differences in the processing of wild-type APP and APPswe. The Swedish mutation of APP markedly enhances β-secretase cleavage of APP (27) and modifies the principal subcellular loci for β-secretase cleavage (39). β-secretase cleavage of wild-type APP occurs in large part upon internalization and endosomal recycling of cell-surface APP, whereas the Swedish mutation causes APP to be cleaved in considerable part by β-secretase within the secretory pathway (39). In contrast to our results with wild-type APP, we found that Lingo-1 produced quite variable effects on β-secretase cleavage of APPswe (Fig 5E). Lingo-1 enhanced APPsβ-swe in some experiments (similar to (19)) but reduced APPsβ-swe in other experiments (similar to our data with wild-type APP, Fig 1C). Thus, the separate mechanisms of β-secretase cleavage of the two APP variants could explain the apparent differences in Lingo-1 effects on β-secretase cleavage of APP in these studies.
Reelin
The effects of Reelin on APP shedding was confirmed across our multiple assays, including with endogenous APP in neurons. However, in contrast to previous studies in which Reelin increased α-secretase cleavage of APP (15, 16), we observed that Reelin decreased α-secretase cleavage of APP (Figs. 1 & 3). We also found that Reelin reduced β-secretase cleavage of APP (Fig 1), which corroborates a previous study in which a reduction of Reelin enhanced Aβ and CTFβ levels in APP transgenic mouse brain (26). We solidified this evidence by showing that the effect of Reelin on APPsα is dose-dependent and that Reelin also decreases CTF, Aβ40, and Aβ42. Moreover, we confirmed reports of a physical interaction between Reelin and APP and showed a similar level of Reelin-APP co-IP as is seen with its canonical receptors, ApoER2 and VLDLR.
In an attempt to reconcile the opposing effects of Reelin on APPsα, we replicated as closely as we could the methods described previously that resulted in an increase in APPsα (15, 16). However, using this method we found no significant effect of Reelin on APPsα levels (Fig S5). These conflicting effects on APPsα do not appear to be due to differences in the concentrations of Reelin, as a range of Reelin concentrations resulted in a decrease of APPsα in our hands (Fig 6C). Because Reelin is cleaved to generate several fragments, it is possible that different cell types secrete alternate Reelin products. However, we found that expression of cDNAs encoding each physiological Reelin fragment reduced APPsα to some extent in our assay (Fig 7).
Pancortins
Pancortin-1 produced the most robust and consistent effects on cleavage of endogenous APP of any of the candidate ligands tested (Fig 1D-F). Pancortin-1 also was the only candidate ligand which specifically reduced β-secretase processing while having no effects on α-secretase processing of APP. With Pancortins being expressed not only in embryonic but also adult cortex (40, 41), regulation of β-secretase cleavage by Pancortin-1 could turn out to have important implications for the pathogenesis or treatment of Alzheimer's disease. Recently, Pancortin was shown to interact with members of the Lingo-1 signaling pathway and regulate axonal growth (42). As Pancortin and Lingo-1 were top APP ligands in our assays, future studies to determine how the Pancortin and Lingo-1 signaling pathways may intersect to regulate APP processing will be important.
A classic ligand for APP?
Since its cloning 25 years ago, APP has been intensively studied as regards its processing via regulated intramembrane proteolysis and the role of its Aß fragment in AD pathogenesis, but studies of its physiological function and processing have received less attention and led to an array of complex, sometimes conflicting findings. For example, analogous to the sizeable number of proteins purported to be candidate ligands for APP, a number of genes had been reported to be transcriptionally activated by the APP intracellular domain (AICD) (43-46). Like the candidate APP ligands, potential target genes had usually been reported by single labs, and attempts to confirm them had been largely unsuccessful (47, 48). One particularly clarifying study in this field published by De Strooper and colleagues systematically compared these target genes in the same assay system and found that each was at best indirectly and weakly influenced by APP processing or not at all (47). A central goal of our study was to provide similar clarity for most of the reported candidate ligands of APP.
Our study raises the central question of whether a classic ligand for APP that positively triggers processing by α- or β- secretase exists. While we did find effects of Reelin, Lingo-1 and Pancortin-1 on APP processing to be consistent across the multiple assays we used, the effects of Reelin and Lingo-1 were subtle in endogenous systems and not identical to previous reports (15, 16, 19). Further, each ligand we tested turned out to inhibit cleavage rather than stimulate α- or β-secretase processing. Whereas a larger portion of APP processing appears to be constitutive than regulated, in contrast to the ligand-regulated cleavage of Notch (49, 50), the ability of PMA to robustly stimulate α-secretase cleavage of APP (Fig 2 B-C and (31, 32, 51)) suggests that there is a cellular capacity for α-secretease cleavage of APP to be enhanced. On the other hand, it is possible that cognate ligand(s) for APP regulate neuronal functions of APP without significantly modulating its proteolytic processing. It is also possible that instead of a single protein ligand, several proteins and non-protein factors may have coordinated effects to regulate APP cleavage. Thus, each ectodomain-binding ligand may individually result in only subtle effects, particularly in the more biologically relevant context of endogenous, wild-type APP in neurons that we explored. Furthermore, it may be that apparent ligand effects are more indirect, perhaps through common intracellular adaptor proteins or signaling molecules or even through competition of common binding partners (14, 15, 52). The cellular context of APP may affect ligand binding and cleavage of APP, for example homo- or hetero- dimerization of APP (53) or the subcellullar localization and trafficking of APP (54, 55). Finally, the reported ability of the APP ectodomain to bind certain glycosaminoglycans and proteoglycans (56-58) may contribute to a multifactorial ligand regulation of APP secretory processing, and this should now be explored in the context of the ligands that most consistently affect the shedding of APP, such as Reelin, Pancortin-1 and Lingo-1. Such further research is needed to better define the basic functions of this conserved and ubiquitously expressed protein and to better understand the consequences of chronically altering its proteolytic processing in older humans with AD-type cognitive syndromes.
Supplementary Material
Acknowledgments
We thank T. Shin for technical assistance. We thank T. Sudoff, T. Curran, J. Flanagan, J. Herz, H.-S. Hoe and A. Goffinet for providing DNA constructs. The 54B Reelin antibody developed by A. Goffinet was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa Department of Biology, Iowa City, IA. We thank M. LaVoie for critical reading of the manuscript and members of the Selkoe and Young-Pearse laboratories for helpful discussions.
Funding source statement: This work was supported by National Institutes of Health grants R01 AG06173 (D.J.S.) and R00 MH085004 (T.L.Y-P.) and a Jerome L. Rappaport Fellowship (H.C.R.).
Abbreviations
- APP
ß-Amyloid Precursor Protein
- AD
Alzheimer's disease
- Aß
amyloid ß-protein
- CTF
C-terminal fragment
- HEK
human embryonic kidney
- BACE
β-site APP cleaving enzyme-1
- AICD
APP intracellular domain
- CNTN
Contactin
- Lingo-1
leucine rich repeat and Ig domain containing Nogo receptor interacting protein-1
- Pan-1/Pan-3
Pancortin-1/3
- CM
conditioned medium
- WB
Western blot
- ELISA
enzyme-linked immunosorbent assay
- ADAM
a disintegrin and metalloproteinase family
- SF
serum-free
- FBS
fetal bovine serum
- BSA
bovine serum albumin
- swe
Swedish
- PMA
phorbol-12-myristate-13-acetate
- IP
immunoprecipitation
Footnotes
Supporting Information: β1 Integrin shows a correlation between holoAPP and APPsα/holoAPP (Fig S1). APPsα/APPsβ is dramatically higher and more variable with overexpressed relative to endogenous APP (Fig S2). Quantitative Western blot analysis of candidate ligand expression levels (Fig S3). FBS and BSA increase the detection of APPsα (Fig S4). Effect of Reelin CM treatment on COS7 cells overexpressing APP751 (Fig S5). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Haass C, Kaether C, Thinakaran G, Sisodia S. Trafficking and Proteolytic Processing of APP. Cold Spring Harbor Perspectives in Medicine. 2012;2 doi: 10.1101/cshperspect.a006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young-Pearse TL, Bai J, Chang R, Zheng JB, Loturco JJ, Selkoe DJ. A Critical Function for beta-Amyloid Precursor Protein in Neuronal Migration Revealed by In Utero RNA Interference. J Neurosci. 2007;27:14459–14469. doi: 10.1523/JNEUROSCI.4701-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pramatarova A, Chen K, Howell BW. A genetic interaction between the APP and Dab1 genes influences brain development. Mol Cell Neurosci. 2008;37:178–186. doi: 10.1016/j.mcn.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young-Pearse TL, Chen A, Chang R, Marquez C, Selkoe DJ. Secreted APP regulates the function of full-length APP in neurite outgrowth through interaction with integrin beta1. Neural Development. 2008;3 doi: 10.1186/1749-8104-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rama N, Goldschneider D, Corset V, Lambert J, Pays L, Mehlen P. Amyloid Precursor Protein Regulates Netrin-1-mediated Commissural Axon Outgrowth. J Biol Chem. 2012;287:30014–30023. doi: 10.1074/jbc.M111.324780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perez RG, Zheng H, Van der Ploeg LH, Koo EH. The beta-amyloid precursor protein of Alzheimer's disease enhances neuron viability and modulates neuronal polarity. J Neurosci. 1997;17:9407–9414. doi: 10.1523/JNEUROSCI.17-24-09407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Araki W, Kitaguchi N, Tokushima Y, Ishii K, Aratake H, Shimohama S, Nakamura S, Kimura J. Trophic effect of b-amyloid precursor ptotein on cerebral cortical neurons in culture. Biochem Biophys Res Commun. 1991;181:265–271. doi: 10.1016/s0006-291x(05)81412-3. [DOI] [PubMed] [Google Scholar]
- 8.Ghiso J, Rostagno A, Gardella JE, Liem L, Gorevic PD, Frangione B. A 109-amino-acid C-terminal fragment of Alzheimer's-disease amyloid precursor protein contains a sequence, -RHDS-, that promotes cell adhesion. Biochem J. 1992;288:1053–1059. doi: 10.1042/bj2881053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soba P, Eggert S, Wagner K, Zentgraf H, Siehl K, Kreger S, Lower A, Langer A, Merdes G, Paro R, Masters CL, Muller U, Kins S, Beyreuther K. Homo- and heterodimerization of APP family members promotes intercellular adhesion. EMBO J. 2005;24:3624–3634. doi: 10.1038/sj.emboj.7600824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Priller C, Bauer T, Mitteregger G, Krebs B, Kretzschmar HA, Herms J. Synapse formation and function is modulated by the amyloid precursor protein. J Neurosci. 2006;26:7212–7221. doi: 10.1523/JNEUROSCI.1450-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Wang B, Yang L, Guo Q, Aithmitti N, Songyang Z, Zheng H. Presynaptic and postsynaptic interaction of the amyloid precursor protein promotes peripheral and central synaptogenesis. J Neurosci. 2009;29:10788–10801. doi: 10.1523/JNEUROSCI.2132-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Muller-Hill B. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 13.Ho A, Sudhof TC. Binding of F-spondin to amyloid-beta precursor protein: a candidate amyloid-beta precursor protein ligand that modulates amyloid-beta precursor protein cleavage. Proc Natl Acad Sci. 2004;101:2548–2553. doi: 10.1073/pnas.0308655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoe HS, Wessner D, Beffert U, Becker AG, Matsuoka Y, Rebeck GW. F-spondin interaction with the apolipoprotein E receptor ApoEr2 affects processing of amyloid precursor protein. Mol Cell Biol. 2005;25:9259–9268. doi: 10.1128/MCB.25.21.9259-9268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoe HS, Tran TS, Matsuoka Y, Howell BW, Rebeck GW. DAB1 and Reelin effects on amyloid precursor protein and ApoE receptor 2 trafficking and processing. J Biol Chem. 2006;281:35176–35185. doi: 10.1074/jbc.M602162200. [DOI] [PubMed] [Google Scholar]
- 16.Hoe HS, Lee K, Carney R, Lee J, Markova A, Lee JY, Howell B, Hyman B, Pak D, Bu G, Rebeck G. Interaction of reelin with amyloid precursor protein promotes neurite outgrowth. J Neurosci. 2009;29:7459–7532. doi: 10.1523/JNEUROSCI.4872-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osterfield M, Egelund R, Young LM, Flanagan JG. Interaction of amyloid precursor protein with contactins and NgCAM in the retinotectal system. Development. 2008;135:1189–1199. doi: 10.1242/dev.007401. [DOI] [PubMed] [Google Scholar]
- 18.Ma QH, Futagawa T, Yang WL, Jiang XD, Zeng L, Takeda Y, Xu RX, Bagnard D, Schachner M, Furley AJ, Karagogeos D, Watanabe K, Dawe GS, Xiao ZC. A TAG1-APP signalling pathway through Fe65 negatively modulates neurogenesis. Nat Cell Biol. 2008;10:283–294. doi: 10.1038/ncb1690. [DOI] [PubMed] [Google Scholar]
- 19.Bai Y, Markham K, Chen F, Weerasekera R, Watts J, Horne P, Wakutani Y, Bagshaw R, Mathews PM, Fraser PE, Westaway D, St George-Hyslop P, Schmitt-Ulms G. The in vivo brain interactome of the amyloid precursor protein. Mol Cell Proteomics. 2008;7:15–34. doi: 10.1074/mcp.M700077-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Rice H, Townsend M, Bai J, Suth S, Cavanaugh W, Selkoe D, Young-Pearse T. Pancortins interact with amyloid precursor protein and modulate cortical cell migration. Development. 2012;139:3986–3996. doi: 10.1242/dev.082909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimoda Y. Contactins: Emerging key roles in the development and function of the nervous system. Cell Adh Migr. 2009;3:64–70. doi: 10.4161/cam.3.1.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mi S, Sandrock A, Miller R. LINGO-1 and its role in CNS repair. Int J Biochem Cell Biol. 2008;40:1971–1978. doi: 10.1016/j.biocel.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 23.Stein T, Walmsley A. The leucine-rich repeats of LINGO-1 are not required for self-interaction or interaction with the amyloid precursor protein. Neuroscience Letters. 2012;509:9–12. doi: 10.1016/j.neulet.2011.11.029. [DOI] [PubMed] [Google Scholar]
- 24.Honda T, Kobayashi K, Mikoshiba K, Nakajima K. Regulation of cortical neuron migration by the Reelin signaling pathway. Neurochem Res. 2011;36:1270–1279. doi: 10.1007/s11064-011-0407-4. [DOI] [PubMed] [Google Scholar]
- 25.Förster E, Bock H, Herz J, Chai X, Frotscher M, Zhao S. Emerging topics in Reelin function. Eur J Neurosci. 2010;31:1511–1519. doi: 10.1111/j.1460-9568.2010.07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kocherhans S, Madhusudan A, Doehner J, Breu K, Nitsch R, Fritschy JM, Knuesel I. Reduced Reelin expression accelerates amyloid-beta plaque formation and tau pathology in transgenic Alzheimer's disease mice. J Neurosci. 2010;30:9228–9268. doi: 10.1523/JNEUROSCI.0418-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Citron M, Oltersdorf T, Haass C, McConlogue L, Hung AY, Seubert P, Vigo-Pelfrey C, Lieberburg I, Selkoe DJ. Mutation of the b-amyloid precursor protein in familial Alzheimer's disease increases b-protein production. Nature. 1992;360:672–674. doi: 10.1038/360672a0. [DOI] [PubMed] [Google Scholar]
- 28.D'Arcangelo G, Nakajima K, Miyata T, Ogawa M, Mikoshiba K, Curran T. Reelin is a secreted glycoprotein recognized by the CR-50 monoclonal antibody. J Neurosci. 1997;17:23–31. doi: 10.1523/JNEUROSCI.17-01-00023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jossin Y, Ignatova N, Hiesberger T, Herz J, Lambert de Rouvroit C, Goffinet A. The central fragment of Reelin, generated by proteolytic processing in vivo, is critical to its function during cortical plate development. J Neurosci. 2004;24:514–521. doi: 10.1523/JNEUROSCI.3408-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benhayon D, Magdaleno S, Curran T. Binding of purified Reelin to ApoER2 and VLDLR mediates tyrosine phosphorylation of Disabled-1. Brain Res Mol Brain Res. 2003;112:33–45. doi: 10.1016/s0169-328x(03)00032-9. [DOI] [PubMed] [Google Scholar]
- 31.Lammich S, Kojro E, Postina R, Gilbert S, Pfeiffer R, Jasionowski M, Haass C, Fahrenholz F. Constitutive and regulated alpha-secretase cleavage of Alzheimer's amyloid precursor protein by a disintegrin metalloprotease. PNAS. 1999;96:3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buxbaum J, Liu K, Luo Y, Slack J, Stocking K, Peschon J, Johnson R, Castner B, Cerretti D, Black R. Evidence that tumor necrosis factor alpha converting enzyme is involved in regulated alpha-secretase cleavage of the Alzheimer amyloid protein precursor. J Biol Chem. 1998;273:27765–27767. doi: 10.1074/jbc.273.43.27765. [DOI] [PubMed] [Google Scholar]
- 33.Hiesberger T, Trommsdorff M, Howell B, Goffinet A, Mumby M, Cooper J, Herz J. Direct binding of Reelin to VLDL receptor and ApoE receptor 2 induces tyrosine phosphorylation of disabled-1 and modulates tau phosphorylation. Neuron. 1999;24:481–489. doi: 10.1016/s0896-6273(00)80861-2. [DOI] [PubMed] [Google Scholar]
- 34.Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer R, Richardson J, Herz J. Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell. 1999;97:689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 35.D'Arcangelo G, Homayouni R, Keshvara L, Rice DS, Sheldon M, Curran T. Reelin is a ligand for lipoprotein receptors. Neuron. 1999;24:471–479. doi: 10.1016/s0896-6273(00)80860-0. [DOI] [PubMed] [Google Scholar]
- 36.Jossin Y, Gui L, Goffinet A. Processing of Reelin by embryonic neurons is important for function in tissue but not in dissociated cultured neurons. J Neurosci. 2007;27:4243–4295. doi: 10.1523/JNEUROSCI.0023-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambert de Rouvroit C, de Bergeyck V, Cortvrindt C, Bar I, Eeckhout Y, Goffinet A. Reelin, the extracellular matrix protein deficient in reeler mutant mice, is processed by a metalloproteinase. Exp Neurol. 1999;156:214–217. doi: 10.1006/exnr.1998.7007. [DOI] [PubMed] [Google Scholar]
- 38.Hafez D, Huang J, Richardson J, Masliah E, Peterson D, Marr R. F-spondin gene transfer improves memory performance and reduces amyloid-β levels in mice. Neuroscience. 2012;223:465–472. doi: 10.1016/j.neuroscience.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haass C, Lemere CA, Capell A, Citron M, Seubert P, Schenk D, Lannfelt L, Selkoe DJ. The Swedish mutation causes early-onset Alzheimer's disease by b-secretase cleavage within the secretory pathway. Nature Med. 1995;1:1291–1296. doi: 10.1038/nm1295-1291. [DOI] [PubMed] [Google Scholar]
- 40.Nagano T, Nakamura A, Konno D, Kurata M, Yagi H, Sato M. A2-Pancortins (Pancortin-3 and Pancortin-4) are the dominant Pancortins during neocortical development. J Neurochem. 2000:75. doi: 10.1046/j.1471-4159.2000.0750001.x. [DOI] [PubMed] [Google Scholar]
- 41.Danielson PE, Forss-Petter S, Battenberg EL, deLecea L, Bloom FE, Sutcliffe JG. Four structurally distinct neuron-specific olfactomedin-related glycoproteins produced by differential promoter utilization and alternative mRNA splicing from a single gene. J Neurosci Res. 1994;38:468–478. doi: 10.1002/jnr.490380413. [DOI] [PubMed] [Google Scholar]
- 42.Nakaya N, Sultana A, Lee HS, Tomarev S. Olfactomedin 1 interacts with the nogo a receptor complex to regulate axon growth. J Biol Chem. 2012;287:37171–37184. doi: 10.1074/jbc.M112.389916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- 44.Kim HS, Kim EM, Lee JP, Park CH, Kim S, Seo JH, Chang KA, Yu E, Jeong SJ, Chong YH, Suh YH. C-terminal fragments of amyloid precursor protein exert neurotoxicity by inducing glycogen synthase kinase-3beta expression. Faseb J. 2003;17:1951–1953. doi: 10.1096/fj.03-0106fje. [DOI] [PubMed] [Google Scholar]
- 45.von Rotz RC, Kohli BM, Bosset J, Meier M, Suzuki T, Nitsch RM, Konietzko U. The APP intracellular domain forms nuclear multiprotein complexes and regulates the transcription of its own precursor. J Cell Sci. 2004;117:4435–4448. doi: 10.1242/jcs.01323. [DOI] [PubMed] [Google Scholar]
- 46.Pardossi-Piquard R, Petit A, Kawarai T, Sunyach C, Alves da Costa C, Vincent B, Ring S, D'Adamio L, Shen J, Muller U, St George Hyslop P, Checler F. Presenilin-dependent transcriptional control of the Abeta-degrading enzyme neprilysin by intracellular domains of betaAPP and APLP. Neuron. 2005;46:541–554. doi: 10.1016/j.neuron.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 47.Hebert SS, Serneels L, Tolia A, Craessaerts K, Derks C, Filippov MA, Muller U, De Strooper B. Regulated intramembrane proteolysis of amyloid precursor protein and regulation of expression of putative target genes. EMBO Rep. 2006;7:739–745. doi: 10.1038/sj.embor.7400704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hass M, Yankner B. A {gamma}-secretase-independent mechanism of signal transduction by the amyloid precursor protein. J Biol Chem. 2005;280:36895–37799. doi: 10.1074/jbc.M502861200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mumm J, Schroeter E, Saxena M, Griesemer A, Tian X, Pan D, Ray W, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Molecular Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 50.Schroeter E, Kisslinger J, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 51.Hung AY, Haass C, Nitsch RM, Qiu WQ, Citron M, Wurtman RJ, Growdon JH, Selkoe DJ. Activation of protein kinase C inhibits cellular production of the amyloid b-protein. J Biol Chem. 1993;268:22959–22962. [PubMed] [Google Scholar]
- 52.Hoe HS, Rebeck G. Functional interactions of APP with the apoE receptor family. J Neurochem. 2008;106:2263–2271. doi: 10.1111/j.1471-4159.2008.05517.x. [DOI] [PubMed] [Google Scholar]
- 53.Libeu C, Descamps O, Zhang Q, John V, Bredesen D. Altering APP proteolysis: increasing sAPPalpha production by targeting dimerization of the APP ectodomain. PloS one. 2012;7 doi: 10.1371/journal.pone.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ehehalt R, Keller P, Haass C, Thiele C, Simons K. Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts. J Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haass C, Koo EH, Capell A, Teplow DB, Selkoe DJ. Polarized sorting of b-amyloid precursor protein and its proteolytic products in MDCK cells is regulated by two independent signals. J Cell Biol. 1995a;128:537–547. doi: 10.1083/jcb.128.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Narindrasorasak S, Lowery D, Gonzalez-DeWhitt P, Poorman R, Greenberg B, Kisilevsky R. High affinity interactions between the Alzheimer's beta-amyloid precursor proteins and the basement membrane form of heparan sulfate proteoglycan. J Biol Chem. 1991;266:12878–12961. [PubMed] [Google Scholar]
- 57.Clarris HJ, Cappai R, Heffernan D, Beyreuther K, Masters CL, Small DH. Identification of heparin-binding domains in the amyloid precursor protein of Alzheimer's disease by deletion mutagenesis and peptide mapping. J Neurochem. 1997;68:1164–1172. doi: 10.1046/j.1471-4159.1997.68031164.x. [DOI] [PubMed] [Google Scholar]
- 58.Mok S, Sberna G, Heffernan D, Cappai R, Galatis D, Clarris H, Sawyer W, Beyreuther K, Masters C, Small D. Expression and analysis of heparin-binding regions of the amyloid precursor protein of Alzheimer's disease. FEBS Lett. 1997;415:303–310. doi: 10.1016/s0014-5793(97)01146-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.