Abstract
Background
Circadian clocks coordinate an organism’s activities and regulate metabolic homeostasis in relation to daily environmental changes, most notably light/dark cycles. As in other organisms, the timekeeping mechanism in mammals depends on a self-sustaining transcriptional negative feedback loop with a built-in time delay in feedback inhibition. Although the time delay is essential for generating a slow, self-sustaining negative feedback loop with a period close to 24 hours, the exact mechanisms underlying the time delay are not known.
Results
We show here that RNA interference mediated by microRNAs (miRNAs) is an essential mechanism in generating the time delay. In Dicer-deficient (and thus miRNA-deficient) cells and mice, circadian rhythms were dramatically shortened (by ~2 hours), although the rhythms remained robust. The period shortening was caused by faster PER1 and PER2 translation in the Dicer-deficient cells. We also identified three specific miRNAs that regulate Per expression, and showed that knockdown of these miRNAs in wild-type cells also shortened the circadian period.
Conclusions
Consistent with the canonical function of miRNAs as translational modulators of target genes and their widespread roles in cell physiology, circadian rhythms are also modulated by miRNA-mediated RNA interference acting on posttranscriptional regulation of key clock genes. Our present study definitively shows that RNA interference is an important modulator of circadian rhythms by controlling the pace of PER synthesis, and presents a novel layer of regulation for the clock.
Introduction
In mammals, a master clock located in the suprachiasmatic nucleus (SCN) of the hypothalamus drives circadian rhythms, adjusts itself according to light input, and synchronizes clocks in peripheral tissues such as liver and kidney [1–3]. It is now known that in addition to the SCN, peripheral tissues and even cultured cells have self-sustaining clocks whose molecular mechanism is very similar to that of the master clock [4–8]. Findings from studying cultured cells are faithfully reproduced in intact animals [6, 8–10].
As in model organisms such as Neurospora and Drosophila, the circadian clock in mammals depends on interacting transcriptional/translational feedback loops [11–13]. The backbone of the clock is a negative transcriptional feedback loop with delayed feedback inhibition. Within this feedback loop, the heterodimer of CLOCK (or its paralog NPAS2) and BMAL1 activates transcription of the negative elements, Per and Cry, as well as circadian output genes. As PER1 and PER2 levels rise in the cytoplasm, PER associates with CRY (1 and 2) and is progressively phosphorylated by PER kinases CK1δ/ε. Then the complexes translocate into or accumulate in the nucleus, where PER:CRY binds directly to CLOCK:BMAL1 to inhibit transcription [8, 14–16]. Additional feedback loops are generated by nuclear receptors of the REV-ERB and ROR families, which are responsible for generating Bmal1 oscillations and may also be involved in direct regulation of other clock genes [17–20]. Although PER, CRY, CLOCK (or NPAS2), and BMAL1 are all essential components of the main feedback loop, PER is of special importance for clock regulation. PER is the rate-limiting component involved in period and phase determination and in phase resetting of the circadian oscillator [21–24]. PER levels vary dramatically over the course of a circadian cycle [18, 24], and the endogenous rhythm in PER levels is required for clock function [8].
The time delay between generation of the transcriptional inhibitors (PER and CRY) and execution of the feedback inhibition is critical for the circadian feedback loop. Although the mechanisms for the time delay remain unclear, the time delay seems to be mainly mediated by slow accumulation of PER in the cytoplasm, as has been shown in flies [24–28]. In both mouse and Drosophila, PER is targeted for degradation following phosphorylation by CK1δ/ε and PER becomes more stable in the cytoplasm in the absence of CK1δ/ε [25, 29]. However, it has been shown that mouse PER accumulation in the cytoplasm is only modestly altered in CK1δ/ε-deficient cells, but PER is constitutively cytoplasmic in the cells, suggesting that PER phosphorylation by CK1δ/ε in the cytoplasm may be more important for timing of nuclear entry [29]. Considering that the accumulation of PER proteins is delayed several hours relative to Per mRNA, in both liver and SCN [24, 30], other mechanisms must also contribute to the delayed accumulation of PER in the cytoplasm and delayed nuclear entry for feedback inhibition.
MicroRNAs (miRNAs) are small, noncoding RNAs (~22 nucleotides) that normally inhibit translation of target genes by base-pairing with the 3′-untranslated region (3′-UTR) of the target gene mRNAs as part of RNA-induced silencing complexes (RISCs) [31, 32]. Since it has been proposed that more than one third of human genes are regulated by miRNAs in diverse aspects of cell physiology [33, 34], and multiple miRNA species have been implicated in the molecular clock and output pathways in several model organisms [35–41], it is highly likely that RNAi regulation plays critical roles in the mammalian clock. In an extensive in vivo study, Cheng et al. implicated miR-219 and 132 in the time-keeping mechanism: brain-specific miRNA219 was implicated in period determination, while miRNA132 was implicated in photic entrainment [36]. However, direct target genes for these miRNAs have not been identified. Other studies implicated different specific miRNAs in the regulation of clock genes such as Per and Bmal1, but it has not been shown how these miRNAs are integrated into the circadian system and the function of these miRNAs remains to be tested in vivo systems [38, 39]. In another extensive study searching for clock-relevant miRNAs in Drosophila, Kadener et al. demonstrated that a developmental regulator, bantam, can affect the circadian clock through translational regulation of clock (clk) [37]. Although it is imperative to understand how RNAi is involved in the time keeping mechanism, previous studies concerning RNAi in the clock mechanism have focused on specific miRNA species, and their roles in the clock mechanism were as subtle modulators rather than significant regulators. To systematically study how the miRNA-mediated regulation is integrated with the timekeeping mechanism, we employed a decisive and fundamental approach using Dicer mutant mice in which miRNA processing is globally compromised. Our current studies using Dicer mutant cells demonstrate that RNA interference mediated by miRNAs primarily affects the clock through translational control of Per in the cytoplasm, which delays cytoplasmic PER accumulation and thus generates a time delay in the circadian feedback inhibition. Our in vivo and in vitro studies have identified three miRNAs, miR-24, 29a and 30a, that affect the circadian clock through regulation of Per1 and 2 mRNA stability and translation. Thus, our studies exposed a novel mechanism for regulation of the pacemaker genes, Per1 and Per2, and for generating the time delay crucial for the circadian feedback loop.
Results
Period of the circadian clock is shortened in the absence of Dicer
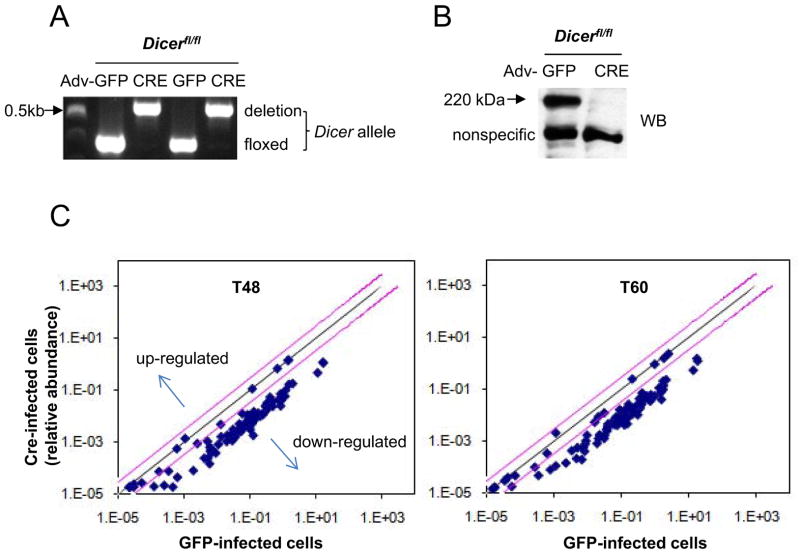
To begin to determine if RNA interference mediated by microRNAs is involved in the time-keeping mechanism, we measured the circadian clock in a Dicer mutant mouse in which biogenesis of mature miRNAs is globally disturbed. A conditional Dicer mutant mouse was generated by Harfe et al. [42] so that Dicer could be retained during early development, when it is essential, and disrupted later to enable researchers to study its roles in later development and adulthood. While we were generating mice combining the floxed Dicer mutation with different cre drivers, to study miRNA involvement in the clock in whole animals, we tested the roles of miRNA in the clock mechanism in a simpler system, Dicerflox/flox; Per2Luc mouse embryonic fibroblasts (MEFs). The Per2Luc allele involves a knockin of luciferase into the 3′ end of the Per2 gene coding sequence [5]. The resulting Per2:Luciferase fusion RNA retains the full length 3′-UTR of Per2 (which may be a target of miRNA), and the fusion protein retains the circadian function of PER2 while serving as a bioluminescent reporter. We deleted the floxed Dicer in the MEFs by introducing cre using an adenoviral vector. This method of transgene delivery is highly efficient (>95% delivery) [8, 29]. The adenovirus-cre-infected cells were grown for 5–6 days with two passages to deplete miRNAs in the Dicer-deleted cells. During this time, cells grew a little slower than adenovirus-gfp-infected control cells, but remained viable and morphologically normal as described previously by Harfe et al [42]. Dicer deletion was verified by PCR-genotyping and immunoblotting of these cells (Fig 1A, 1B, S1A).
Fig 1.
Deletion of Dicer in Dicerflox/flox MEFs by adenovirus-cre. Cells were infected with adenovirus expressing GFP or CRE and harvested 5 days later. (A) PCR-genotyping was performed using three primers described in Harfe et al. [42]. Two independent experiments are shown. (B) Representative immunoblots of the cell extracts. Dicer, indicated by the arrow, was eliminated in >95% of CRE-expressing cells, based on comparison to GFP cells. More immunoblots from different experiments are shown in Fig S1A. (C) Depletion of miRNAs in Dicer-deficient cells. Cells were infected with CRE- or GFP-expressing adenovirus, maintained for 5 days, given a 2-hr serum shock, and harvested 48 or 60 hours later to mimic the conditions for recording bioluminescence rhythms as we have done previously [8, 29]. The miRNA levels at T48 and T60 were normalized to control RNAs (Rnu6 and snoRNA202) according to the manufacturer’s protocol, and each point on the graph shows the average of 6 samples (duplicate samples in 3 independent experiments). The black line indicates no difference between GFP- and CRE-expressing cells. The pink lines indicate a 3-fold difference. Levels of all miRNAs except 8 species were significantly reduced between GFP and CRE cells (p<0.05). See also Table S1.
Depletion of most miRNAs was confirmed by quantitative real-time PCR using an miRNA PCR array which includes 88 of the most abundantly expressed mouse miRNAs (Fig 1C, Table S1). In cells collected 7–8 days after adenovirus-cre infection, most miRNA levels were reduced more than 10-fold, while small control RNAs (like Rnu6) were unaffected by loss of Dicer.
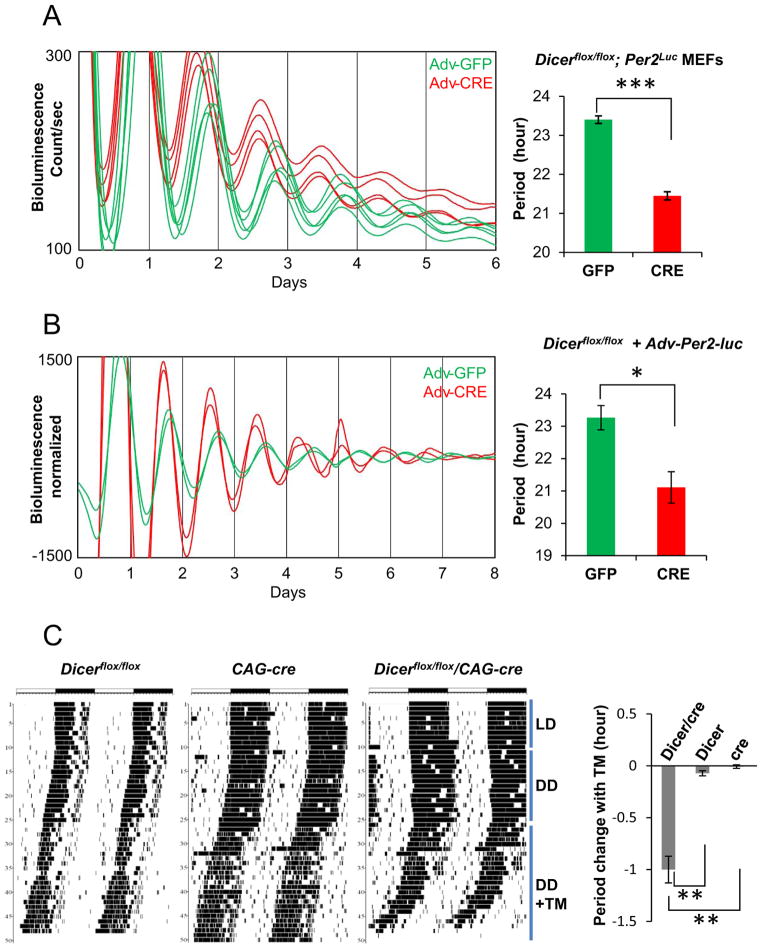
In terms of circadian rhythms in bioluminescence, CRE-expressing (i.e., Dicer-deficient) cells exhibited robust rhythms but a dramatically shorter period (by ~2 hrs) than GFP (Dicer-intact) cells (Fig 2A, S1B). These data suggest that miRNA-mediated RNAi may not be essential for the circadian feedback loop, but does strongly regulate the pace of the clock. The baseline of the bioluminescence rhythms was higher in Dicer mutant cells compared to control cells (Fig 2A), which is striking considering that there were fewer cells per dish due to the slow growth rate of Dicer mutant cells (Fig S1C). Clearly, PER2-LUC expression levels per cell were substantially increased when Dicer was disrupted.
Fig 2.
Circadian period is dramatically shortened in Dicer-deficient cells and mice. (A) The period of bioluminescence rhythms in CRE-expressing (Adv-CRE) Dicerflox/flox;mPer2Luc MEFs is shorter than that in control cells expressing GFP (Adv-GFP). Note that the basal lines are higher in CRE-MEFs than in GFP-MEFs. Quantitation of circadian period is shown in the graph. Mean+/−SEM; n=14 for each sample; p<0.001. See also Fig S1B and S1C. (B) Period is similarly shortened in Adv-CRE Dicerflox/flox MEFs without Per2Luc. Bioluminescence was produced from an exogenous Per2 promoter-driven luciferase reporter introduced into the MEFs using the adenoviral vector (Adv-Per2-Luc). Mean+/−SEM; n=5 for GFP, n=3 for CRE; p<0.05. (C) Period is also shortened in whole mice when Dicer is deleted using tamoxifen-dependent activation of CRE. Representative double-plotted actograms are shown for three different genotypes: Dicerflox/flox, CAG-cre-Esr1 and Dicerflox/flox/CAG-cre-Esr1 mice (n=5 each). Black markings indicate wheel running, and recordings from consecutive days are stacked in rows. Mice were entrained in a 12:12 LD cycle (12 hrs light and 12 hrs dark) for 2 weeks followed by 2 weeks in constant darkness (DD). On the 15th day, the food was switched to tamoxifen (TM)-containing food at 0.4mg/g food. All Dicerflox/CAG-cre-Esr1 mice were found dead at the end of the recording. In these Dicer-deficient mice, the period was ~1 hr shorter in the last 7 days before death when compared to the period in DD before tamoxifen treatment. However, there was no significant difference in period before and after tamoxifen treatment in control (Dicerflox/flox or CAG-cre-Esr1) mice. See also Fig S2.
The shortened period must be due to Dicer deletion by CRE, not by CRE itself, because CRE-expressing wild-type (wt) cells do not show a significant difference in circadian period compared to GFP-expressing wt cells, as shown previously [29]. Comparable period shortening was also observed in Dicer-deficient MEFs without the Per2Luc knockin but infected with adenovirus-Per2 promoter-Luc (a transient transgenic reporter) (Fig 2B), confirming that the effect is not due to abnormalities caused by insertion of luciferase into the Per2 locus.
Using mice with a transgenic cre driver, we determined whether Dicer deficiency shortens the circadian period not just in dispersed cells, but also in whole animals. Dicerflox/flox/CAG-cre-Esr1 mice were given mouse chow mixed with tamoxifen to trigger Dicer deletion [43], while their daily behavior was being monitored. The circadian period gradually shortened, and the mice died within 1–5 weeks after receiving tamoxifen (Fig 2C, S2A). However, the same treatment did not affect viability or circadian period in control mice with only the Dicerflox/flox allele or only the CAG-cre-Esr1 transgene, demonstrating that Dicer deletion produced the effects. The degree of period shortening— calculated by phase angle change in the last several days —was ~1 hr, half of that observed in Dicer-mutant MEFs. This modest effect is probably due to incomplete deletion of floxed Dicer in the SCN, because deletion of floxed genes in vivo was less efficient than in vitro. This is supported by genotyping PCR of brain tissue, performed when we tested two different cre drivers before selecting CAG-cre-Esr1. The CAG-cre-Esr1 transgenic mouse produced greater period shortening and more efficient deletion of the floxed Dicer allele than Scg2:tTA/tetO:cre transgenic mice based on PCR-genotyping of brain tissue, but deletion of Dicer in the brain occurred in only about half of the cells (Fig S2B). The period shortening is not a non-specific consequence of deleting essential genes: deletion of CK1δ affected viability but induced period lengthening, not shortening [29, 44].
The period shortening is caused by faster accumulation of PER in cytoplasm
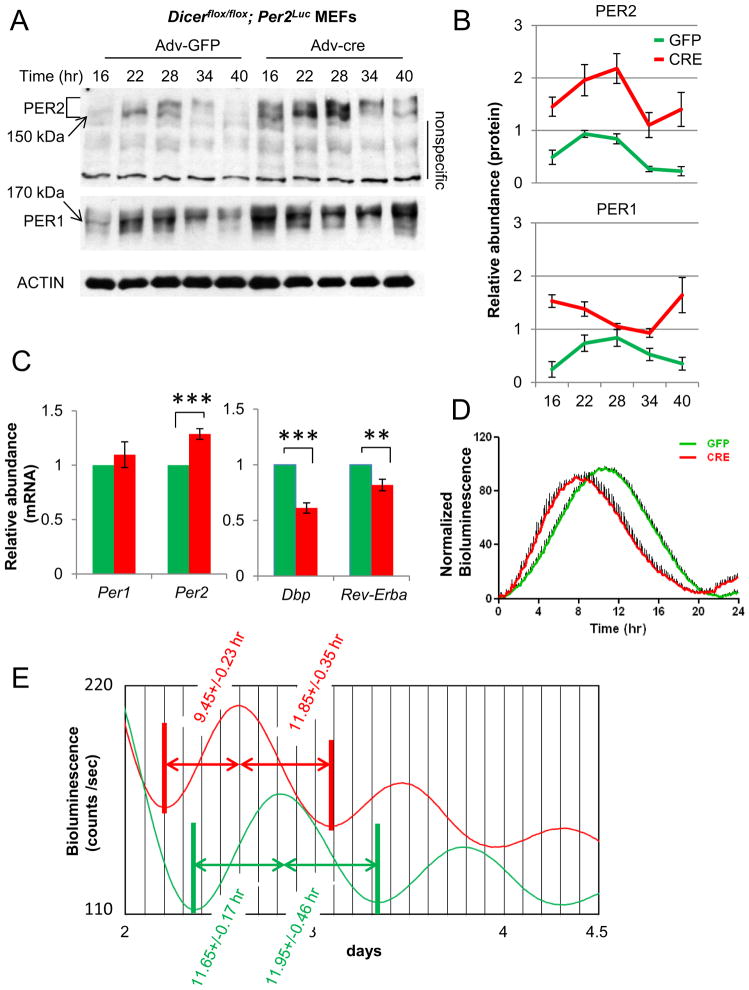
Since the circadian oscillator runs substantially faster in the Dicer mutant cells, some of the core clock components must have been affected by disrupted miRNA biogenesis in the mutant cells. We hypothesized that protein levels of the affected core clock components would be elevated in the Dicer mutant cells if they are direct targets of certain miRNAs. Among all the core clock proteins (CLOCK, BMAL1, PER1, PER2, CRY1, CRY2, CK1δ, and CK1ε), we found that only PER1 and PER2 levels were significantly elevated in the mutant cells (Fig 3A, 3B, S3A), suggesting that the mRNA of these two genes may be the direct targets of the miRNAs whose biogenesis is disrupted in the absence of Dicer. Per1 mRNA levels were not affected while Per2 mRNA levels were only slightly elevated in the Dicer mutant cells (Fig 3C). Since Per mRNA levels are down-regulated when PER protein is overexpressed [8], these data (high or unchanged mRNA levels in the face of higher levels of inhibitory protein) suggest that miRNAs probably affect Per1 and Per2 mRNA through both translational inhibition and mRNA degradation. Consistent with elevated levels of the limiting circadian inhibitors, mRNA levels of clock-controlled genes such as Dbp and Rev-Erbα were significantly reduced (Fig 3C).
Fig 3.
PER protein levels are elevated and bioluminescence (PER2-LUC) rhythms accumulate faster and peak earlier in Dicer-deficient cells (CRE) compared to Dicer-intact cells (GFP). (A) PER protein levels are higher in Dicer-deficient cells than control cells. The samples were harvested at the indicated times after a 2-hr serum shock. See also Fig S3A. (B) Quantitation is shown as mean+/−SEM of three independent experiments. (C) mRNA levels of Per1 and Per2 are only modestly increased in Dicer-deficient cells, less substantially than their protein levels. mRNA levels of clock-controlled genes Dbp and Rev-erbα are significantly lowered. mRNA levels were measured by quantitative RT-PCR from 6 samples harvested at several times over one circadian cycle. The data are shown as mean+/−SEM. (D) In the circadian rhythm of PER2-LUC expression, the upswing is shortened while the downswing is not affected. Dicerflox/flox;Per2Luc MEFs were treated with 40 ug/ml cycloheximide (CHX) for 10 hrs followed by washoff, and then bioluminescence was measured. We showed that endogenous PER1/2 can be almost completely depleted by CHX treatment for 10 hrs [29]. The peak of bioluminescence was set at 100 in each case, as has been done by Meng et al. and Etchegaray et al. [44, 59]. Note that the ~2-hr period shortening mostly comes from the accumulation (upswing) phase. The results are shown as mean+/−SEM of 10 samples. (E) Similar asymmetrical bioluminescence profiles are observed in Fig 2A. Single representative traces for GFP and Cre-MEFs are shown, but the numbers in mean+/−SEM were calculated from 10 samples each. Note that the period difference resulted from the shortened rising phase of the curves (p<0.01). Times from trough to peak and from peak to trough were calculated using Clocklab software after applying smoothing.
To test if mRNAs of Per1/2 genes are direct targets of miRNAs, we indirectly assessed the translation rate of Per2-Luc by measuring real-time accumulation of PER2-LUC between control and Dicer-deleted cells, after depletion of existing PER2-LUC by cycloheximide (CHX) treatment. PER2-LUC accumulates faster and peaks earlier in Dicer-deficient cells (Fig 3D), suggesting that the pace of PER2 cytoplasmic accumulation is regulated by miRNA. In Fig 3D, while the upswing of the bioluminescence rhythms was shortened in the Dicer mutant cells, the downswing was not significantly affected, suggesting that the period shortening in Dicer mutant cells is generated almost exclusively from the accumulation phase of the inhibitor in the feedback loop. This is consistent with the asymmetric bioluminescence traces for Dicer mutant cells in Fig 2A and S1B. In all of the traces for Dicer mutant cells, the upswing is shorter than the downswing, while the traces for the control cells are symmetric (Fig 3E).
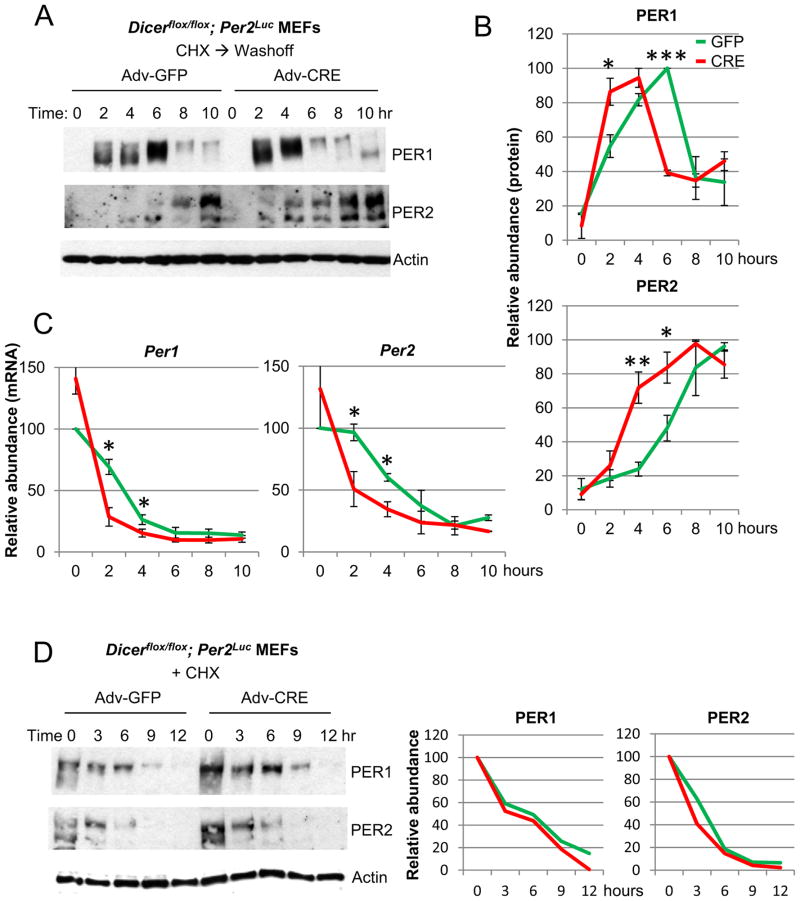
If the period shortening is indeed caused by a reduced time delay between transcription and feedback inhibition by the protein product, faster accumulation of the protein product and resulting earlier feedback inhibition should be observed in Dicer mutant cells. We measured Per mRNA and protein after CHX treatment, under the same conditions used for Fig 3D; this approach creates similar starting points for comparing control and Dicer mutant cells: PER and its inhibitory action on Per transcription have been depleted, and thus when CHX is removed, the pace of PER accumulation and Per inhibition can be readily compared. In Dicer-mutant cells, PER1 and PER2 protein accumulated and peaked faster (Fig 4A, B), and feedback inhibition was accelerated based on earlier down-regulation of mRNA levels in Dicer mutant cells (Fig 4C). However, consistent with the bioluminescence rhythms, PER degradation rates were not significantly affected in Dicer mutant cells (Fig 4D). These results further support that the period shortening is caused by faster accumulation of the limiting clock component PER rather than even compression of the whole cycle or faster relief of feedback inhibition through accelerated degradation of PER.
Fig 4.
Accumulation of PER proteins and feedback inhibition of Per are accelerated in Dicer-deficient cells. (A), (B) PER accumulates faster in Dicer-deficient cells. Cells were harvested after CHX treatment followed by washoff as described for Fig 3D and subjected to imunoblotting for PER1 and 2. The mean+/−SEM of 3 and 4 experiments is shown for PER1 and 2, respectively. (C) Feedback occurs earlier in Dicer mutant cells. Cells were also harvested for mRNA analysis under the same condition. Data are shown as mean+/−SEM of three experiments. (D) PER degradation rates are comparable between control and Dicer mutant cells. As has been done previously [29], cells were treated with CHX and harvested at the indicated times. Results are representative of two experiments.
Overexpression of 3′-untranslated regions (3′-UTR) of Per1 and Per2 mRNA causes period shortening
Target sites for miRNAs are highly conserved across different species and generally found in the 3′-UTR of mRNAs [33, 45, 46]. If certain miRNAs interfere with translation of Per1 and Per2 genes through their interaction with the 3′-UTR of these genes, and disruption of this mechanism caused the period shortening in the Dicer mutant cells, as suggested by the data above, then, we hypothesized, overexpression of the full-length 3′-UTRs of the Per1 and Per2 genes would produce similar results as in Dicer mutant cells. Since miRNAs bind target mRNAs stably through the protein complex, RISC, the overexpressed Per1 or Per2 3′-UTR would deplete endogenous miRNA molecules specific to the UTRs, similar to dominant negative approaches, which have been successfully used to study the function of many clock genes [8, 47–49].
When Per1 or Per2 3′-UTR was overexpressed in wt Per2Luc MEFs, we observed bioluminescence rhythms with a shortened period, as seen in Dicer mutant cells, suggesting that these UTRs indeed contain target sites for miRNA-induced translational regulation of Per1 and Per2 (Fig 5). The period shortening caused by overexpression of either Per1 or Per2 3′-UTRs was a little less than that in Dicer mutant cells. Overexpression of both UTRs also did not cause as much period shortening as in Dicer mutant cells, probably because the dominant negative approach is less efficient in disrupting the relevant miRNAs than the loss of Dicer function. However, we cannot rule out the possibilities that clock-relevant miRNA target sites may also reside in the coding region of Per1/2 or in the mRNA of other clock genes. Due to high transduction rates of the adenoviral vector, the levels of exogenous 3′-UTRs were hundreds of times higher than those of endogenous 3′-UTR and remained high 7 days after the infection (Fig S3B).
Fig 5.
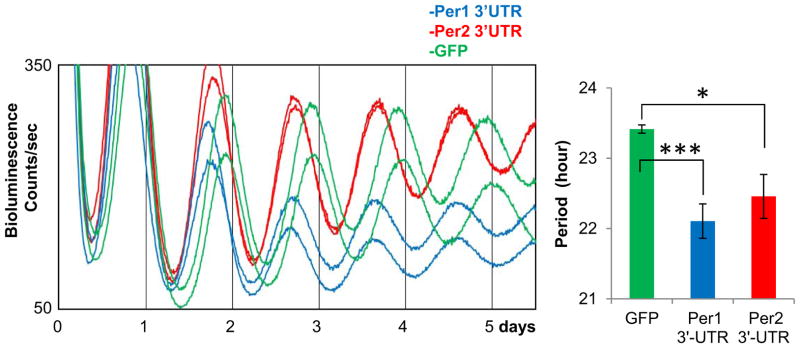
Overexpression of Per1 or Per2 3′-UTR in wt Per2Luc MEFs induces period shortening similar to Dicer-deficient cells. The 3′-UTR adenoviral constructs include a partial luciferase coding sequence (to increase stability of mRNA) followed by a full-length Per1 or Per2 3′-UTR. Data are shown as mean+/−SEM. n=6 for GFP; 8 for Per1 3′-UTR; 8 for Per2 3′-UTR. See also Fig S3B.
miR-24, 29a and 30a regulate the circadian clock by interfering with translation of Per1 and Per2
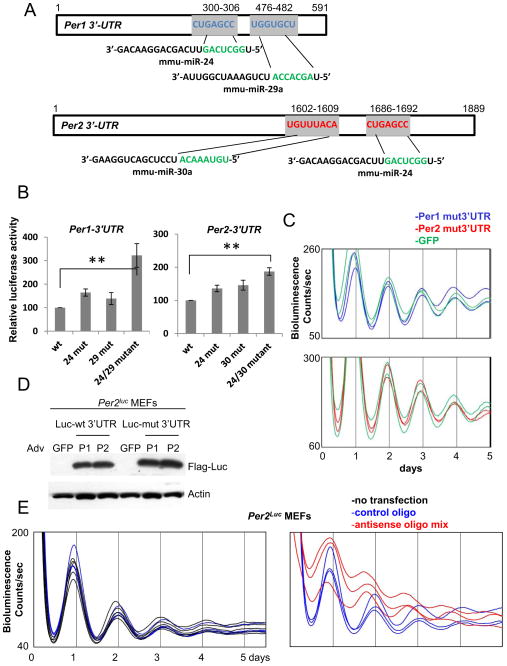
Since our data strongly indicate that Per1 and Per2 genes are regulated by miRNAs at the posttranscriptional level, we sought to determine which miRNAs are specifically involved in the timekeeping mechanism through interaction with the 3′-UTRs of Per1 and Per2. To identify Per-targeting miRNAs, we used the TargetScanMouse algorithm developed by the Bartel laboratory, which predicts miRNA:mRNA pairs by searching 3′-UTRs with conserved complementary sequence to the seed sequence (nucleotides 2–7) of the miRNAs [33]. The accuracy is greatly improved if the Watson-Crick base-pairing is extended immediately downstream (nucleotide 1) or upstream (nucleotide 8) of the miRNAs beyond the seed match. Using the algorithm, we identified several matching miRNAs, of which the three most promising species are miR-24, which could target both Per1 3′-UTR and Per2 3′-UTR; miR-29a, which could target Per1 3′UTR; and miR-30a, which could target Per2 3′-UTR (Fig 6A). All of these miRNAs exhibit perfect 7mer or 8mer matches to the target sites in the 3′UTR of Per1/2, and the target sites in the 3′UTRs were conserved across most mammalian species (Fig S4). These miRNAs were included in the miRNA PCR array described above, and their levels were reduced by >10-fold in Dicer knockout cells (see Table S1).
Fig 6.
miR-24/29a and 24/30a post-transcriptionally regulate Per1 and Per2, respectively. (A) Per1 and Per2 3′-UTRs have conserved target sites for miR-24/29a and 24/30a, respectively. The target sites were identified by TargetScanMouse 5.2. Note that these target sites have perfect 7 or 8 nucleotide matches. See also Fig S4. (B) Luciferase expression is higher when the target sites are mutated in transient transfection-based reporter assays. The mutant 3′-UTRs in the pGL3 basic plasmid were expressed in NIH3T3 cells and their expression levels were compared. Single mutations only modestly increased luciferase expression, but double mutations increased the luciferase expression significantly higher compared to wt UTRs. See also Fig S5A. (C) Overexpression of mutant miR-24/29a Per1 3′-UTR or mutant miR-24/30a Per2 3′-UTR does not induce period shortening. The mutations in the miRNA target sites were introduced into the adenoviral 3′-UTR constructs used in Fig 4. (D) Expression levels of these mutant UTRs were comparable to those of wt UTRs when expression levels of the partial luciferase attached to the UTRs were compared. (E) Circadian period is shortened in wt Per2Luc cells treated with antisense oligonucleotides to miR-24, 29a and 30a. A negative control oligonucleotide did not affect circadian period compared to non-treated control cells (left panel). The periods of bioluminescence rhythms in control and specific oligonucleotide-treated cells were 24.25+/−0.08 and 22.88+/−0.10, respectively (mean+/−SEM of 8 traces each; p<0.001). See also Fig S5B.
The first functional tests were luciferase reporter assays in transiently transfected NIH 3T3 cells. To test if these sites are regulated by miRNAs, these sites were mutated in the UTRs and the effect of the mutations on expression of the luciferase reporter was measured. Mutations of either miR-24 or miR-29a target sites in Per1 3′-UTR only increased luciferase activity slightly, but mutation of both target sites dramatically increased the luciferase activity, suggesting that Per1 is regulated by these two miRNAs in a combinatorial manner (Fig 6B). Similar combinatorial regulation was also observed for Per2 3′-UTR and the corresponding miRNAs (Fig 6B). The increase in expression of luciferase associated with mutant Per 3′-UTR was greater in NIH 3T3 cells compared to MEFs, probably because endogenous levels of the relevant miRNAs are higher in NIH 3T3 cells (Fig S5A).
To test further if these sites are major target sites, we overexpressed Per1 3′-UTR with mutated miR-24 and 29a sites and Per2 3′-UTR with mutated miR-24 and 30a sites in wt Per2Luc MEFs. Our data showed that overexpression of Per1 or Per2 wt 3′-UTR induces significant period shortening (Fig 5); if the UTRs mediate this effect by sequestering endogenous Per-targeting miRNAs, then overexpression of the mutant 3′-UTRs that cannot bind the specific miRNAs would not induce period shortening. Indeed, although expression levels of these mutant 3′-UTR were comparable to those of wt 3′-UTRs in the cells, the period was not significantly altered (Fig 6C, D) indicating that these sites are functional and clock-relevant miRNA target sites in vivo.
Finally, to test if miR-24, 29a and 30a are involved in the period determination, we knocked down these miRNAs in wt Per2Luc cells using Locked Nucleic Acid (LNA)-antisense oligonucleotides [50] and measured the period of the bioluminescence rhythms. As in Dicer mutant cells, the period of the circadian clock was significantly shortened in wt Per2Luc cells treated with the antisense oligonucleotides to the miRNAs (Fig 6E), demonstrating that these miRNAs are indeed involved in the timekeeping mechanism. The antisense oligonucleotide-treated cells, like Dicer mutant cells, had elevated basal levels of PER2-LUC, consistent with a role for the miRNAs in modulating PER synthesis. miR-24, 29a and 30a are likely to be involved in the timekeeping mechanism in most, if not all tissues, since they are widely expressed in fibroblasts, U2OS cells, and liver (liver data are shown in Fig S5B). These miRNAs did not exhibit significant circadian oscillations in liver, suggesting that miRNA-induced Per regulation is not time-gated (Fig S5B).
Discussion
The time delay, between transcription of Per and Cry and PER:CRY-mediated feedback inhibition, is one of the main features of the circadian feedback loop, but the underlying mechanism is not fully understood. In mammals and flies, it has been proposed that accumulation of the limiting negative regulator, PER, is somehow delayed because protein oscillations are delayed several hours relative to those of mRNA oscillations [24–26]. Here we reveal that RNAi is a crucial mechanism contributing to the time delay by interfering with translation of Per1 and Per2. We provide multiple lines of evidence supporting the mechanism. For example, the asymmetric shortening of the bioluminescence rhythms in Dicer mutant cells shows that the period of the rhythm is shortened mostly by accelerated upswing of PER (or PER2-LUC) synthesis, with little effect on the downswing, or PER turnover. This is further supported by our analysis of bioluminescence (PER2-LUC) and PER accumulation data after existing PER proteins are removed by CHX treatment, by the shortening of circadian period by overexpression of Per 3′-UTR, and by period shortening in wt cells by knockdown of Per-targeting miRNAs. The direct inhibition of PER accumulation in the cytoplasm by miRNAs provides a very intuitive mechanism for how miRNAs are integrated into the timekeeping mechanism, since most miRNAs function in the cytoplasm by inhibiting the translation of specific mRNA species.
The behavioral analysis of our conditional Dicer mutant mice confirmed that RNAi is important for circadian period in the SCN in vivo, not only in cultured MEFs. However, the mice showed milder period shortening compared to MEFs. We attribute this finding to the fact that Dicer was deleted only in half of brain cells and thus probably in only a subset of SCN cells, and behavioral phenotype is determined by the combined activities of all the SCN cells, as shown previously [51]. Kadener et al. previously studied how the Drosophila clock is affected by the knockdown of dicer-1 expression and found that dicer-1 knockdown flies did not show any significant circadian phenotype [37]. Furthermore, in Drosophila, TIMELESS (TIM) is critical for accumulation of dPER in the cytoplasm and delayed nuclear entry, whereas in mammals, mTIMELESS is irrelevant to the mammalian clock mechanism [52]. We believe that RNAi is a novel and alternative means of generating the time gap in mammals that is not conserved in flies.
In Dicer mutant cells and mice, where basic cell physiology is sufficiently disrupted to affect viability and/or cell growth, the circadian clock shows robust oscillations albeit with altered pace. The same is true of CK1δ mutant cells [29]. These data suggest that the clock mechanism is independent of the basic physiology related to cell survival, which may explain the resilience of the clock to physiological fluctuations in cells under different growth conditions and throughout the cell cycle.
Although our data implicate three specific Per-targeting miRNAs in the mammalian clock mechanism, we cannot rule out the possibility that other miRNAs are also important. First, our Dicer deletion did not eliminate all small, noncoding RNAs, so there may be Dicer-independent, RNA species involved in the clock mechanism. Second, we may have missed subtle dysregulation of other clock genes besides Per1 and Per2 in the Dicer mutant cells. As with the general function of miRNAs in gene expression, modulation of clock gene expression by miRNAs may not be dramatic. Even with Per1/2, mRNA and protein expression levels were only mildly affected in Dicer mutant cells. We believe that these small changes were able to produce a robust phenotype because they only had to alter the phase of PER oscillations, rather than absolute levels of Per expression. As long as PER shows normal oscillations, PER levels have only a subtle effect on period, as shown in Per transgenic mice and cells [8, 53]. Third, there may be additional miRNAs targeting Per1/2. Each miRNA may have multiple target genes and each gene can be targeted by multiple miRNAs. It is possible that miRNAs other than miR-24, 29a, and 30a can target Per1/2 genes. Because the seed pairing does not have to match perfectly and allows wobbles— as shown in let-7:lin41 and miR-196:HoxB8 pairings [54, 55]—there may be more clock-relevant miRNAs that cannot be detected by the current prediction algorithms. However, even if other miRNAs contribute to the circadian feedback loop’s time delay, our data clearly demonstrated that Per expression is modulated by miRNA interaction with the 3′-UTR of Per1/2 genes.
Experimental Procedures
Dicer mutant, cre driver transgenic mice
The floxed Dicer mutant (Dicer fl/fl) and Per2Luc mice were described previously [5, 42]. The conditional transgenic mice expressing the brain-specific Tetracycline Activator (Scg2-tTA) were described previously [56]. When these mice are mated with a second transgenic mouse that carries transgenic cre, controlled by a tetracycline-responsive promoter element (TRE or tetO), expression of cre is conditionally regulated by the presence or absence of doxycycline (Dox) in the drinking water [57]. In parallel, Dicerflox was combined with the CAG-cre-Esr1 transgene to activate CRE recombinase in the presence of tamoxifen [43]. This cre driver mouse was also used by the Evans lab recently to delete Rev-erbα and β genes and study the behavioral rhythms of the mutant mice [17]. Both tetO-cre and CAG-cre-Esr1 transgenic mice are commercially available from The Jackson Laboratory (stock # 006234 and 004682). All of these mice are in or close to the genetic background of the C57BL/6J strain, which exhibits robust locomotor activity rhythms. All animals were maintained in a climate-controlled room and used according to the FSU Animal Care and Use Committee’s guidelines.
RT-qPCR analysis for miRNAs
miRNAs were extracted from the samples by miRNeasy Mini kit (Qiagen, Germantown, MD, USA). For Fig 1C, first-strand cDNA was generated with the RT2miRNA First Stand Kit (Qiagen) and subjected to RT-PCR using the mouse miRNA Finder kit, which includes the 88 most abundant mouse miRNAs (Qiagen). Data were calculated according to the manufacturer’s protocol and presented in Table S1. For individual miRNAs, the same primers used in Fig 1C were ordered from Qiagen and used for RT-qPCR.
Bioluminescence and behavioral rhythms
For Dicer mutant cells, bioluminescence rhythms were measured 5 days after adenoviral cre or GFP infection as described previously [29]. For Fig 6E, miR-24, 29a and 30a were inhibited by commercial antisense oligonucleotides (miRCURY power inhibitors; #426983-08 for miR-24, #460039-1 for miR-29a and #460026-1 for miR-30a; Exiqon; Woburn, MA). The oligonucleotides were mixed at 50 nM (final concentration) each and introduced using Lipofectamine RNAi Max (Invitrogen, Grand Island, NY) into wt Per2Luc cells grown in 24-well plates. Bioluminescence rhythms were measured by a real-time luminometer that can accommodate 24-well plates (Actimetrics, Wilmette, IL). To confirm high transfection efficiency, miRCURY-24 antisense oligonucleotide was labeled with TexasRed and the efficiency was estimated by counting fluorescent cells. Knockdown efficiency for the miRNAs was measured by RT-qPCR. In two separate experiments, miR-24, 29a and 30a were knocked-down more than 90% compared to control cells transfected with the same concentration of negative control miRCURY power oligonucleotide (#199004-08). Period was calculated by the periodogram function in the Clocklab software using the 2nd and 3rd peaks.
Locomotor activity rhythms were measured as described previously [58] using the Stanford Software System (Santa Cruz, CA, USA). The mice were entrained in 12-hr light and 12-hr dark (LD) cycles for 2 weeks before being released into constant darkness (DD). After 2 weeks in DD, regular mouse chow was switched to tamoxifen-containing food (400 mg tamoxifen + 0.8% sucrose/kg food; Harlan laboratories, Inc.) for CAG-cre-Esr1 mice, and Dox water was switched to regular water for Scg2-tTA;tetO mice. When judged by genotyping PCR of brain tissue, the CAG-cre-Esr1 transgene was much more efficient in deleting Dicer in vivo and caused larger period shortening. Period was calculated using the χ2-periodogram in the Stanford Software System. In Dicer mutant mice (Dicerflox/flox/CAG-cre-Esr1) after the tamoxifen treatment, the last several days of behavioral data were used to calculate period by hand drawing a best-fit line across several activity onsets.
DNA constructs
Adenoviral GFP, Per2 promoter-luciferase and cre constructs have been described previously [8, 29]. The full-length 3′-UTRs of Per1 and Per2 were amplified from the pCMV-sport6-Per1 and Per2 plasmids (Addgene #4133009 and #3964321) using the following primers. The fragments were cloned into XbaI/SalI (for the Per1 3′-UTR) and XbaI/BamHI sites (Per2 3′-UTR) of the pGL3 basic plasmid.
Per1-3′UTR Fwd
5- ATC CTC TAG AACTCC ATT TTG GGG CCG CTT ACA GCA G -3
Per1-3′UTR Rev
5- ATC CGT CGA CCG AGG GAT ACA CTA GAG CGG CCG C -3
Per2-3′UTR Fwd
5- ATC CTC TAG ACC CTG TCC CCC AGC CAG AGGTC -3
Per2-3′UTR Rev
5- ATC CGG ATC CCG AGG GAT ACA CTA GAG CGG CCG C -3
The following mutations for the miR target sites were introduced into the constructs using the Quickchange II XL Site-Directed Mutagenesis kit (Agilent technologies, Santa Clara, CA, USA).
Per1 miR-24: CUGAGCC → GTCGCAG
Per1 miR-29a: UGGUGCU → GGTACCT
Per2 miR-24: CUGAGCCU → CCGCGCCU
Per2 miR-30a: UGUUUACA → CGCUUACA
To generate Flag tag-partial length luciferase-Per 3′-UTR in an adenoviral vector, the following primers were used to amplify the UTRs from the pGL3-Per 3′-UTR plasmids.
pGL3-basic-luc Fwd (with Flag tag)
5′-ATC CCT CGA GGC CAC CAT GGA TTA CAA GGA TGA CGA TGA CAA GTC CGG TTA TGT AAA CAA TCC GGA AGC G -3′
pGL3-basic-luc_Rev
5′-TAG GGA TAT CGG AAG CGG AAG AGC GCT CCC GGC A -3′
Lentiviruses expressing miR-24, 29a and 30a were purchased from the Open Biosystem (miRIDIAN shMIMIC microRNAs). Expression of these miRs was confirmed by RT-PCR as described above.
Real-time qPCR for clock genes, luciferase reporter assays and immunoblotting
Real time qPCR for clock genes was performed as described previously using the same primers [8]. Immunoblotting was performed using the antibodies described previously [14, 24]. PER1-1-R and PER2-1-R were used for PER1 and 2 immunoblotting, respectively. Novel anti-Dicer antibodies were generated using the N-terminal region of Dicer protein (amino acids 241–426) by Cocalico Biologicals, Inc (Reamstown, PA) and thoroughly characterized using in vitro and in vivo samples. Dicer GP47 was used for this study. Luciferase reporter assays were performed as described previously [8].
Supplementary Material
Highlights.
The period of the circadian clock is shortened in Dicer-mutant cells and mice.
The time delay in the feedback loop is compromised in the Dicer-mutant cells.
RNAi contributes to the time delay in the clock by targeting PER translation.
Three miRNAs regulate the accumulation of PER1 and PER2 in the cytoplasm.
Acknowledgments
We thank Dennis Chang for assistance with manuscript revisions. This work was supported by NIH grant NS-053616 awarded to Choogon Lee. Rongmin Chen was partially supported by DK-090730.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Schibler U. The daily rhythms of genes, cells and organs. Biological clocks and circadian timing in cells. EMBO reports. 2005;6(Spec No):S9–13. doi: 10.1038/sj.embor.7400424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annual review of neuroscience. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Current biology: CB. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yagita K, Tamanini F, van Der Horst GT, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 7.Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen R, Schirmer A, Lee Y, Lee H, Kumar V, Yoo SH, Takahashi JS, Lee C. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Molecular cell. 2009;36:417–430. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW, 3rd, Janes J, et al. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 11.Allada R, Emery P, Takahashi JS, Rosbash M. Stopping time: the genetics of fly and mouse circadian clocks. Annual review of neuroscience. 2001;24:1091–1119. doi: 10.1146/annurev.neuro.24.1.1091. [DOI] [PubMed] [Google Scholar]
- 12.Yu W, Hardin PE. Circadian oscillators of Drosophila and mammals. Journal of cell science. 2006;119:4793–4795. doi: 10.1242/jcs.03174. [DOI] [PubMed] [Google Scholar]
- 13.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 14.Lee C, Weaver DR, Reppert SM. Direct association between mouse PERIOD and CKIepsilon is critical for a functioning circadian clock. Molecular and cellular biology. 2004;24:584–594. doi: 10.1128/MCB.24.2.584-594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 16.Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 17.Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 19.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 20.Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 21.Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, et al. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 22.Shearman LP, Zylka MJ, Weaver DR, Kolakowski LF, Jr, Reppert SM. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 23.Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 24.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 25.Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 26.Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiu JC, Vanselow JT, Kramer A, Edery I. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes & development. 2008;22:1758–1772. doi: 10.1101/gad.1682708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ko HW, Jiang J, Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- 29.Lee HM, Chen R, Kim H, Etchegaray JP, Weaver DR, Lee C. The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16451–16456. doi: 10.1073/pnas.1107178108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annual review of physiology. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 31.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 32.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 34.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nature reviews. Molecular cell biology. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 35.Hansen KF, Sakamoto K, Obrietan K. MicroRNAs: a potential interface between the circadian clock and human health. Genome medicine. 2011;3:10. doi: 10.1186/gm224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, Nawathean P, Vagin VV, Zamore PD, Nelson SB, Rosbash M. A role for microRNAs in the Drosophila circadian clock. Genes & development. 2009;23:2179–2191. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shende VR, Goldrick MM, Ramani S, Earnest DJ. Expression and rhythmic modulation of circulating microRNAs targeting the clock gene Bmal1 in mice. PloS one. 2011;6:e22586. doi: 10.1371/journal.pone.0022586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagel R, Clijsters L, Agami R. The miRNA-192/194 cluster regulates the Period gene family and the circadian clock. The FEBS journal. 2009;276:5447–5455. doi: 10.1111/j.1742-4658.2009.07229.x. [DOI] [PubMed] [Google Scholar]
- 40.Sire C, Moreno AB, Garcia-Chapa M, Lopez-Moya JJ, San Segundo B. Diurnal oscillation in the accumulation of Arabidopsis microRNAs, miR167, miR168, miR171 and miR398. FEBS letters. 2009;583:1039–1044. doi: 10.1016/j.febslet.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 41.Luo W, Sehgal A. Regulation of circadian behavioral output via a MicroRNA-JAK/STAT circuit. Cell. 2012;148:765–779. doi: 10.1016/j.cell.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Developmental biology. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 44.Etchegaray JP, Machida KK, Noton E, Constance CM, Dallmann R, Di Napoli MN, DeBruyne JP, Lambert CM, Yu EA, Reppert SM, et al. Casein kinase 1 delta regulates the pace of the mammalian circadian clock. Molecular and cellular biology. 2009;29:3853–3866. doi: 10.1128/MCB.00338-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 46.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee H, Chen R, Lee Y, Yoo S, Lee C. Essential roles of CKIdelta and CKIepsilon in the mammalian circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21359–21364. doi: 10.1073/pnas.0906651106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muskus MJ, Preuss F, Fan JY, Bjes ES, Price JL. Drosophila DBT lacking protein kinase activity produces long-period and arrhythmic circadian behavioral and molecular rhythms. Molecular and cellular biology. 2007;27:8049–8064. doi: 10.1128/MCB.00680-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith EM, Lin JM, Meissner RA, Allada R. Dominant-negative CK2alpha induces potent effects on circadian rhythmicity. PLoS genetics. 2008;4:e12. doi: 10.1371/journal.pgen.0040012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petersen M, Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends in biotechnology. 2003;21:74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 51.Low-Zeddies SS, Takahashi JS. Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell. 2001;105:25–42. doi: 10.1016/s0092-8674(01)00294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hardin PE. The circadian timekeeping system of Drosophila. Current biology: CB. 2005;15:R714–722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 53.Xu Y, Toh KL, Jones CR, Shin JY, Fu YH, Ptacek LJ. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 55.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 56.Hong HK, Chong JL, Song W, Song EJ, Jyawook AA, Schook AC, Ko CH, Takahashi JS. Inducible and reversible Clock gene expression in brain using the tTA system for the study of circadian behavior. PLoS genetics. 2007;3:e33. doi: 10.1371/journal.pgen.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corbel SY, Rossi FM. Latest developments and in vivo use of the Tet system: ex vivo and in vivo delivery of tetracycline-regulated genes. Current opinion in biotechnology. 2002;13:448–452. doi: 10.1016/s0958-1669(02)00361-0. [DOI] [PubMed] [Google Scholar]
- 58.Chen R, Seo DO, Bell E, von Gall C, Lee C. Strong resetting of the mammalian clock by constant light followed by constant darkness. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2008;28:11839–11847. doi: 10.1523/JNEUROSCI.2191-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meng QJ, Logunova L, Maywood ES, Gallego M, Lebiecki J, Brown TM, Sladek M, Semikhodskii AS, Glossop NR, Piggins HD, et al. Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58:78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.