Too much salt can do terrible things to a plant. Dissolved in solution, salt reduces the availability of water to the plant. Also, excess Na in high-salinity soil competes with K for uptake across the plasma membranes of plant cells, reducing plant growth and causing cellular injury and even death. Indeed, salinity tolerance is correlated with the ability of a plant to retain proper tissue K levels. In addition, soil salinity stress causes in-planta accumulation of reactive oxygen species (ROS), which can result in oxidative stress and cellular damage. Conversely, ROS also mediate plant salinity tolerance. For example, the ROS-producing protein RESPIRATORY BURST OXIDASE HOMOLOG F (RBOHF) plays a key role in soil salinity tolerance (Jiang et al., 2012). Under saline-soil conditions, Arabidopsis thaliana plants lacking RBOHF function exhibit high Na levels in their root stelar cells and xylem sap, enhanced root-to-shoot Na delivery, and high Na accumulation in shoots, adding up to soil salinity hypersensitivity. Other factors also regulate ionic homeostasis and plant salt tolerance. For example, salt-activated ethylene signaling regulates plant growth and development by altering the properties of the growth-repressing DELLA proteins (Achard et al., 2006). DELLA-independent pathways likely function in salt tolerance as well, but these pathways have remained unclear.
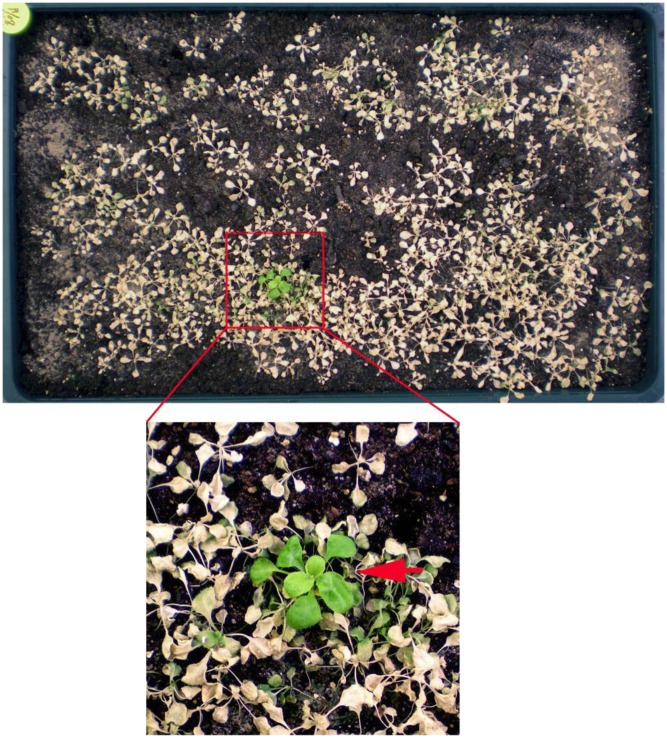
Jiang et al. (pages 3535–3552) aimed to uncover these DELLA-independent pathways by isolating and analyzing salt-tolerant Arabidopsis mutants. To this end, the authors screened mutagenized progeny of a mutant progenitor line devoid of all five DELLAs encoded by the Arabidopsis genome. Importantly, instead of screening for salt-tolerant mutants using a conventional in vitro culture system with plants enclosed in Petri dishes, which essentially blocks transpirational flow (which is highly important for delivery of Na to shoots) due to the lack of evaporation, they screened for mutants tolerant to soil salinity under conditions of active transpiration (i.e., in soil). Three weeks after watering mutagenized seedlings with NaCl solution, they selected a soil salinity–tolerant mutant (see figure). Tests revealed that this mutant exhibits enhanced ethylene production via loss of function of ETHYLENE OVERPRODUCER1 (ETO1; whose normal function is to reduce ethylene production by degrading ethylene biosynthetic enzymes); this mutant was designated eto1-15. Thus, salinity-induced ethylene is a potent promoter of salt tolerance. Salinity-treated eto1-15 plants exhibit reduced root Na influx and low root stelar and xylem-sap Na concentrations, leading to restricted root-to-shoot delivery of Na, along with high xylem-sap K concentrations. In addition, eto1-15 has constitutively elevated levels of ROS, which further increase in response to salinity treatment. Since these effects are similar to those produced by RBOHF under high-salinity conditions, the authors examined the behavior of an eto1-15 rbohf-4 (loss of function of RBOHF) double mutant. This double mutant is not salt tolerant, indicating that soil salinity tolerance conferred by ethylene requires RBOHF function. Moreover, the double mutant has wild-type ROS levels under normal conditions and fails to respond to salinity, suggesting that both salinity-induced and ethylene-induced ROS accumulation depend on the function of RBOHF. Finally, the authors found that while the regulation of Na accumulation is mediated by RBOHF, the lack of ETO1 function leads to significant increases in tissue K status via an RBOHF-independent mechanism.
Identification of a soil salinity–tolerant mutant. The highlighted section shows a green soil salinity–tolerant mutant (arrow) among bleached, dead, and dying plants. (Reprinted from Jiang et al. [2013], Supplemental Figure 1.)
Interestingly, the salt tolerance of eto1-15 plants is a conditional phenotype that is only apparent when plants are exposed to salinity when growing in soil and not when grown in vitro in Petri dishes on high-salt medium. Therefore, the authors would not have found this highly informative mutant using standard in vitro screening techniques.
References
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van Der Straeten D., Peng J., Harberd N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Jiang C., Belfield E.J., Mithani A., Visscher A., Ragoussis J., Mott R., Smith J.A.C., Harberd N.P. (2012). ROS-mediated vascular homeostatic control of root-to-shoot soil Na delivery in Arabidopsis. EMBO J. 31: 4359–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Belfield E.J., Cao Y., Smith J.A.C, Harberd N.P. (2013). An Arabidopsis soil-salinity tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. Plant Cell 25: 3535–3552 [DOI] [PMC free article] [PubMed] [Google Scholar]