Abstract
Effector proteins represent a refined mechanism of bacterial pathogens to overcome plants’ innate immune systems. These modular proteins often manipulate host physiology by directly interfering with immune signaling of plant cells. Even if host cells have developed efficient strategies to perceive the presence of pathogenic microbes and to recognize intracellular effector activity, it remains an open question why only few effectors are recognized directly by plant resistance proteins. Based on in-silico genome-wide surveys and a reevaluation of published structural data, we estimated that bacterial effectors of phytopathogens are highly enriched in long-disordered regions (>50 residues). These structurally flexible segments have no secondary structure under physiological conditions but can fold in a stimulus-dependent manner (e.g., during protein–protein interactions). The high abundance of intrinsic disorder in effectors strongly suggests positive evolutionary selection of this structural feature and highlights the dynamic nature of these proteins. We postulate that such structural flexibility may be essential for (1) effector translocation, (2) evasion of the innate immune system, and (3) host function mimicry. The study of these dynamical regions will greatly complement current structural approaches to understand the molecular mechanisms of these proteins and may help in the prediction of new effectors.
Plants and pathogens are entangled in a continual arms race. While host organisms have developed complex and dynamic immune systems able to recognize a wide range of pathogens and to discriminate them from beneficial microbes (Jones and Dangl, 2006; Medzhitov, 2007), bacterial pathogens have evolved refined adaptation strategies to overcome the plant’s innate immune system. Among these ingenious adaptations are effector proteins. Most of these proteins are secreted via the type III secretion system (TTSS) into the host cytoplasm, where they manipulate the immune signaling and the physiology of plant cells and thereby improve bacterial fitness within the host (Dean, 2011).
Plant–pathogen interactions are highly dynamic processes, both from the evolutionary and the physiological point of view. Here, we postulate that they are equally dynamic at the protein-structure level. This is based on our finding that numerous effector proteins are predicted to be intrinsically disordered (ID) and that this feature may be essential for (1) effector translocation, (2) evasion of the innate immune system, and (3) host function mimicry. Intrinsic disorder has so far been postulated to preferentially occur in eukaryotic proteins. While on average ∼20% of the eukaryotic proteome harbors long (>50 residues) ID segments, these regions are only predicted at low abundance (8% on average) in bacterial proteomes (Dunker et al., 2000). The most likely reason for this discrepancy is the lack of efficient mechanisms to protect unfolded proteins from degradation (Ward et al., 2004). However, when surveying genomes of pathogenic bacteria with the widely used PONDR VL-XT program (Romero et al., 2001), we observed that not only the average percentage of sequence disorder, but most strikingly long (>50 residues) stretches of intrinsic disorder are highly overrepresented in secreted effectors, with especially high levels in phytopathogenic bacteria (Pseudomonas syringae, ∼39%; Ralstonia solanacearum, ∼70%; Xanthomonas spp, ∼77%) (Table 1; see Supplemental Table 1 online). This striking enrichment of unstructured regions strongly suggests positive evolutionary selection of intrinsic disorder in effector proteins and highlights their dynamic nature.
Table 1. Predictions of Intrinsic Disorder in Effectors and Whole Proteomes of Different Bacterial Species.
| Organism | Average Percentage of Disordered Residues |
Percentage of Proteins Harboring ID Regions >50 Residues |
||
|---|---|---|---|---|
| All Proteins | TTSS Effectors | All Proteins | TTSS Effectors | |
| P. syringae | 38.6 | 35.6 | ||
| phaseolicola 1448A | 26.1 | 42.0 | 10.1 | 52.4 |
| syringae B728a | 26.2 | 41.4 | 10.7 | 57.1 |
| tomato DC3000 | 26.4 | 39.7 | 10.2 | 34.4 |
| R. solanacearum | 42.6 | 69.6 | ||
| GMI1000 | 29.2 | 43.5 | 11.9 | 66.7 |
| Xanthomonas sp | 49.2 | 75.7 | ||
| X. campestris pv vesicatoria 85-10 | 29.6 | 50.9 | 13.5 | 69.6 |
| X. oryzae pv oryzae KACC10331 | 29.7 | 46.3 | 12.5 | 82.3 |
| X. campestris pv campestris ATCC 33913 | 29.1 | 44.6 | 13.3 | 68.9 |
| S. enterica | 22.1 | 18.5 | ||
| enterica ser. typhimurium LT2 | 23.0 | 21.5 | 7.0 | 19.2 |
Disorder parameters of representative effectors (see Supplemental Table 1 online) were calculated per species (highlighted in bold) and were compared to the values calculated for the proteomes from which the majority of the effectors were extracted. For completeness, effectors belonging to protein families absent in these strains were extracted from closely related strains (see Supplemental Table 1 online). Proteomes of P. syringae pv phaseolicola (strain 1448A; 5170 proteins), P. syringae pv syringae (strain B728a; 5088 proteins), P. syringae pv tomato (strain DC3000; 5618 proteins), R. solanacearum (strain GMI1000; 5108 proteins), X. campestris pv vesicatoria (strain 85-10; 4726 proteins), X. oryzae pv oryzae (strain KACC10331; 4065 proteins), X. campestris pv campestris (strain ATCC 33913; 4178 proteins), and S. typhimurium (strain LT2 ; 4555 proteins) were downloaded from the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov/genome/). Additionally, parameters were individually calculated for the different strains. Intrinsic disorder predictions were calculated with the PONDR VL-XT program (Romero et al., 2001). Here, scores below and above 0.5 indicate residues predicted to be ordered and disordered, respectively. The average percentage of sequence disordered was calculated as the mean value of the percentage of disordered residues (PONDR score > 0.5) per protein from all proteins. The percentage of long ID regions was calculated as the percentage of proteins harboring ID regions >50 residues.
WHAT EXACTLY IS INTRINSIC DISORDER?
Per definition, ID regions are flexible protein segments that have no ordered secondary or tertiary structure under physiological conditions in vitro. Historically, unfolded proteins have been considered to be a nuisance and an artifact of recombinant overexpression. However, the recognition that this state is of biological relevance led to a revision of the structure-function paradigm, which claimed that the function of a protein is defined by an ordered structure (Dunker and Obradovic, 2001). Disordered regions show lower sequence complexity and exhibit large net charges at neutral pH as a consequence of harboring many noninteracting charged groups and few hydrophobic residues (Uversky et al., 2000; Uversky, 2002). These properties result in high sequential, structural, and spatiotemporal heterogeneity of disordered proteins and regions, where intrinsic disorder can have multiple faces, can affect different levels of protein structural organization, and whole proteins, or various protein regions can be disordered to a different degree (Uversky, 2013a). ID proteins often adapt diverse bound structures as a result of interaction with structurally different partners with low affinity but in a specific manner (Dyson and Wright, 2005; Oldfield et al., 2008; Hsu et al., 2013). Such low-affinity and high-specificity binding is essential for efficient signal transduction. Furthermore, phosphorylation (Iakoucheva et al., 2004; Marín and Ott, 2012) and other posttranslational modifications sites locate preferentially within ID regions (Uversky, 2013b). Altogether, this may explain why ∼70% of eukaryotic signaling proteins are predicted to harbor long disordered regions (Iakoucheva et al., 2002; Marín et al., 2012). Prominent examples of plant proteins harboring long disordered regions are the Arabidopsis thaliana bZIP transcription factor long hypocotyl5 (HY5) and the cryptochrome CRY1 involved in photomorphogenic development (Partch et al., 2005; Yoon et al., 2006; Sun et al., 2013).
FUNCTIONAL RELEVANCE OF INTRINSIC DISORDER IN EFFECTOR PROTEINS
In the case of effector proteins, most of the available structural data refer to folded domains, but there is increasing experimental evidence for the occurrence of intrinsic disorder in these proteins. For example, it has been shown by NMR spectroscopy that the N-terminal region of the Yersinia pseudotuberculosis YopE effector is ID and undergoes partial induced folding upon association to its cognate chaperone SycE (Rodgers et al., 2008). Similar observations were made for the P. syringae effectors AvrPto and AvrRpt2, where partial disorder was also demonstrated by NMR spectroscopy. AvrPto consists of a core that is composed of a three-helix bundle motif and long ID N and C termini (Wulf et al., 2004). The protein core exists in a slowly exchanging equilibrium between folded and unfolded states in the bacterial cytoplasm. However, upon entrance to the host cytoplasm, the unfolded fraction undergoes induced folding in a pH-dependent manner and the folded form becomes stable (Dawson et al., 2009). Prior to the delivery into plant cells, AvrRpt2 remains in a mostly unfolded form. However, upon interaction with the Rotamase CYP1 peptidyl-prolyl isomerase, it undergoes induced folding, activation, and autoproteolytic cleavage of the 71 N-terminal residues, which are predicted to be disordered (Coaker et al., 2005, 2006). Another prominent example is the P. syringae effector HopPmaL that harbors two folded domains, while the majority of the protein is ID, as shown by NMR spectroscopy (Figure 1, inset) (Singer et al., 2012). However, the molecular function of these flexible regions is thus far unknown, which highlights the need for functional studies on disordered regions in general. The lack of structure-function studies for ID regions is likely exacerbated by experimental difficulties, such as insolubility and proclivity to degradation.
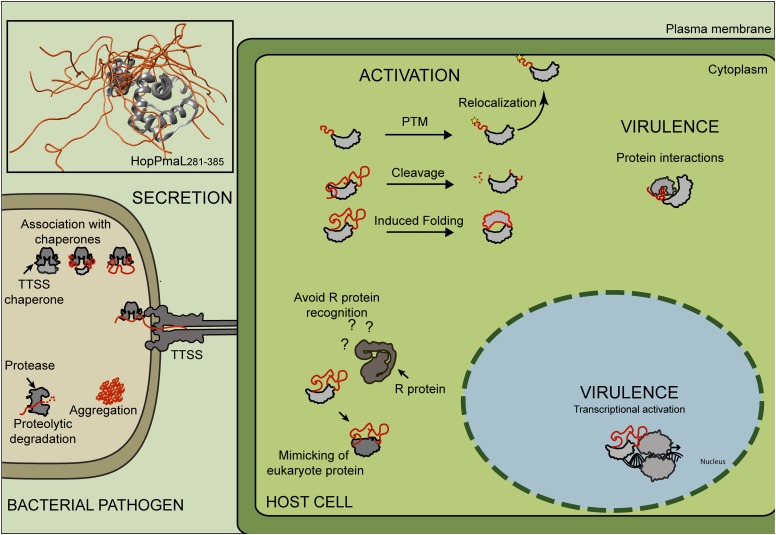
Figure 1.
Intrinsic Disorder in Effector Proteins.
Bacterial effector proteins are secreted via TTSS into the host cell cytoplasm, where they manipulate host cell immune signaling and physiology. Structural flexibility is required for efficient secretion via TTSS, as proteins can only be secreted in an unfolded state. Posttranslational modifications (PTM) can be added to residues within ID segments to determine subcellular localization of the protein inside the host cell. This structural signature may also contribute to effector virulence via mimicking or interactions with signaling proteins and evasion of immune recognition by Resistance (R) proteins. Top left box: The structural model of HopPmaL281-385 (PDBID: 2LF3; inset top left corner) illustrates the structural flexibility of ID regions. The unfolded region is depicted in orange.
Although most of the available structural models for effectors support induced folding at some point, we postulate that the disordered state is essential for central aspects of effector biology. Secretion of disordered regions would be of inherent advantage, as it would not require active unfolding, which is required prior to insertion into the narrow channel formed by the TTSS apparatus (Stebbins and Galán, 2001). To protect these proteins from degradation prior to host–pathogen contact and secretion, some effector proteins are stored in the microbial cytoplasm in complex with cognate chaperones, which bind their N-terminal regions. A number of crystal structures of effector–chaperone complexes support this assumption (Stebbins and Galán, 2001; Zheng et al., 2012). These ATP-independent chaperones most likely protect ID effectors from degradation and aggregation and could have arisen as a secondary secretion signal in later stages of evolution. This idea is supported by the above-mentioned example of YopE-induced folding upon association with its cognate chaperone (Rodgers et al., 2008). Moreover, numerous chaperones seem to be required for substrate stabilization (Figure 1) (Losada and Hutcheson, 2005).
However, the importance of structural flexibility in effectors is not restricted to secretion. A large proportion of effectors have long disordered regions in the middle and/or in C-terminal regions (Salmonella enterica, ∼60%; ∼P. syringae, ∼52%; R. solanacearum, ∼63%; Xanthomonas spp, ∼71% of the total number of effectors harboring long disordered regions). As ID regions are often associated with signaling and transcriptional regulation functions, they could mimic plant immune signaling components. Furthermore, the ability of ID proteins to interact with multiple protein partners could enable them to interfere with host–protein interaction networks. A prominent example of such an interaction is the viral ID protein E1A, which interferes with host cell signaling by recruiting regulatory proteins such as CBP and pRb (Berk, 2005). Whether the interaction of E1A with these proteins results in positive or negative cooperativity depends on the presence of E1A interaction sites (Ferreon et al., 2013). Such differential allosteric behavior may enable context-specific fine-tuning of protein interactions and thereby enable viruses to maximize functional complexity with a reduced proteome (Ferreon et al., 2013). A similar scenario can be envisioned for bacterial pathogens, which rely on a limited set of secreted proteins to manipulate host signaling.
However, the most far-reaching hypothesis is that intrinsic disorder represents a key structural feature that helps to avoid direct recognition by cognate resistance proteins in plants (Figure 1). This hypothesis is supported by the observation that ID proteins generally evolve faster than ordered proteins (Brown et al., 2011). They exhibit different evolutionary patterns in comparison to folded proteins, accept diverse point mutations, and show higher rates of insertions, deletions, and repeat expansions. This receptivity toward mutations is likely to facilitate evasion of immune recognition. Thus, ID may represent the hypothesized mechanism that allows pathogens to overcome effector-triggered immunity as previously suggested (Jones and Dangl, 2006; Medzhitov, 2007). In return, indirect recognition of effector proteins may have evolved to overcome the evolution of ID segments.
Taken together, studies on the dynamics of ID regions will open new avenues to explore novel effector mechanisms, as in other systems these regions have been shown to be key for protein function. If we want to understand how effectors as a whole manipulate plant immune components, we need to understand the molecular function of ID regions and their contribution to virulence. Additionally, the study of these regions may aid the prediction of new bacterial effectors. These ideas may also be applicable to fungal and oomycete effectors given that ID regions have also been identified in effectors from these eukaryotic pathogens (Schneider et al., 2010; Yaeno et al., 2011).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. List of TTSS Effectors Used for ID Predictions.
Acknowledgments
We thank Orlando De Lange for critically reading the article and Sophien Kamoun and Benjamin Petre for fruitful discussions on the project. Financial support was provided by the Collaborative Research Center (Sonderforschungsbereich) SFB924 and the Emmy Noether Program (Grant OT423/2-1) funded by the German Research Foundation (Deutsche Forschungsgemeinschaft).
AUTHOR CONTRIBUTIONS
M.M. designed the study. M.M and V.N.U. performed the in-silico analyses. All authors discussed the results. M.M. and T.O. wrote the article.
References
- Berk A.J. (2005). Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 24: 7673–7685 [DOI] [PubMed] [Google Scholar]
- Brown C.J., Johnson A.K., Dunker A.K., Daughdrill G.W. (2011). Evolution and disorder. Curr. Opin. Struct. Biol. 21: 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coaker G., Falick A., Staskawicz B. (2005). Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science 308: 548–550 [DOI] [PubMed] [Google Scholar]
- Coaker G., Zhu G., Ding Z., Van Doren S.R., Staskawicz B. (2006). Eukaryotic cyclophilin as a molecular switch for effector activation. Mol. Microbiol. 61: 1485–1496 [DOI] [PubMed] [Google Scholar]
- Dawson J.E., Seckute J., De S., Schueler S.A., Oswald A.B., Nicholson L.K. (2009). Elucidation of a pH-folding switch in the Pseudomonas syringae effector protein AvrPto. Proc. Natl. Acad. Sci. USA 106: 8543–8548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean P. (2011). Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol. Rev. 35: 1100–1125 [DOI] [PubMed] [Google Scholar]
- Dunker A.K., Obradovic Z. (2001). The protein trinity—Linking function and disorder. Nat. Biotechnol. 19: 805–806 [DOI] [PubMed] [Google Scholar]
- Dunker A.K., Obradovic Z., Romero P., Garner E.C., Brown C.J. (2000). Intrinsic protein disorder in complete genomes. Genome Inform. Ser. Workshop Genome Inform. 11: 161–171 [PubMed] [Google Scholar]
- Dyson H.J., Wright P.E. (2005). Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6: 197–208 [DOI] [PubMed] [Google Scholar]
- Ferreon A.C., Ferreon J.C., Wright P.E., Deniz A.A. (2013). Modulation of allostery by protein intrinsic disorder. Nature 498: 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu W.L., Oldfield C.J., Xue B., Meng J., Huang F., Romero P., Uversky V.N., Dunker A.K. (2013). Exploring the binding diversity of intrinsically disordered proteins involved in one-to-many binding. Protein Sci. 22: 258–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva L.M., Brown C.J., Lawson J.D., Obradović Z., Dunker A.K. (2002). Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 323: 573–584 [DOI] [PubMed] [Google Scholar]
- Iakoucheva L.M., Radivojac P., Brown C.J., O’Connor T.R., Sikes J.G., Obradovic Z., Dunker A.K. (2004). The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 32: 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Losada L.C., Hutcheson S.W. (2005). Type III secretion chaperones of Pseudomonas syringae protect effectors from Lon-associated degradation. Mol. Microbiol. 55: 941–953 [DOI] [PubMed] [Google Scholar]
- Marín M., Ott T. (2012). Phosphorylation of intrinsically disordered regions in remorin proteins. Front. Plant Sci. 3: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín M., Thallmair V., Ott T. (2012). The intrinsically disordered N-terminal region of AtREM1.3 remorin protein mediates protein—protein interactions. J. Biol. Chem. 287: 39982–39991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. (2007). Recognition of microorganisms and activation of the immune response. Nature 449: 819–826 [DOI] [PubMed] [Google Scholar]
- Oldfield C.J., Meng J., Yang J.Y., Yang M.Q., Uversky V.N., Dunker A.K. (2008). Flexible nets: Disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics 9 (suppl. 1): S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch C.L., Clarkson M.W., Ozgür S., Lee A.L., Sancar A. (2005). Role of structural plasticity in signal transduction by the cryptochrome blue-light photoreceptor. Biochemistry 44: 3795–3805 [DOI] [PubMed] [Google Scholar]
- Rodgers L., Gamez A., Riek R., Ghosh P. (2008). The type III secretion chaperone SycE promotes a localized disorder-to-order transition in the natively unfolded effector YopE. J. Biol. Chem. 283: 20857–20863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P., Obradovic Z., Li X., Garner E.C., Brown C.J., Dunker A.K. (2001). Sequence complexity of disordered protein. Proteins 42: 38–48 [DOI] [PubMed] [Google Scholar]
- Schneider D.R., Saraiva A.M., Azzoni A.R., Miranda H.R., de Toledo M.A., Pelloso A.C., Souza A.P. (2010). Overexpression and purification of PWL2D, a mutant of the effector protein PWL2 from Magnaporthe grisea. Protein Expr. Purif. 74: 24–31 [DOI] [PubMed] [Google Scholar]
- Singer A.U., Wu B., Yee A., Houliston S., Xu X., Cui H., Skarina T., Garcia M., Semesi A., Arrowsmith C.H., Savchenko A. (2012). Structural analysis of HopPmaL reveals the presence of a second adaptor domain common to the HopAB family of Pseudomonas syringae type III effectors. Biochemistry 51: 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins C.E., Galán J.E. (2001). Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414: 77–81 [DOI] [PubMed] [Google Scholar]
- Sun X., Rikkerink E.H., Jones W.T., Uversky V.N. (2013). Multifarious roles of intrinsic disorder in proteins illustrate its broad impact on plant biology. Plant Cell 25: 38–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V.N. (2002). Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 11: 739–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V.N. (2013a). Unusual biophysics of intrinsically disordered proteins. Biochim. Biophys. Acta 1834: 932–951 [DOI] [PubMed] [Google Scholar]
- Uversky V.N. (2013b). Intrinsic disorder-based protein interactions and their modulators. Curr. Pharm. Des. 19: 4191–4213 [DOI] [PubMed] [Google Scholar]
- Uversky V.N., Gillespie J.R., Fink A.L. (2000). Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 41: 415–427 [DOI] [PubMed] [Google Scholar]
- Ward J.J., Sodhi J.S., McGuffin L.J., Buxton B.F., Jones D.T. (2004). Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337: 635–645 [DOI] [PubMed] [Google Scholar]
- Wulf J., Pascuzzi P.E., Fahmy A., Martin G.B., Nicholson L.K. (2004). The solution structure of type III effector protein AvrPto reveals conformational and dynamic features important for plant pathogenesis. Structure 12: 1257–1268 [DOI] [PubMed] [Google Scholar]
- Yaeno T., Li H., Chaparro-Garcia A., Schornack S., Koshiba S., Watanabe S., Kigawa T., Kamoun S., Shirasu K. (2011). Phosphatidylinositol monophosphate-binding interface in the oomycete RXLR effector AVR3a is required for its stability in host cells to modulate plant immunity. Proc. Natl. Acad. Sci. USA 108: 14682–14687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M.K., Shin J., Choi G., Choi B.S. (2006). Intrinsically unstructured N-terminal domain of bZIP transcription factor HY5. Proteins 65: 856–866 [DOI] [PubMed] [Google Scholar]
- Zheng Z., Ma D., Yahr T.L., Chen L. (2012). The transiently ordered regions in intrinsically disordered ExsE are correlated with structural elements involved in chaperone binding. Biochem. Biophys. Res. Commun. 417: 129–134 [DOI] [PMC free article] [PubMed] [Google Scholar]