This work examines the transcriptomic responses of Chlamydomonas to the combined conditions of anoxia and darkness, finding that the cells implement acclimation strategies to save and generate energy using all available resources and that COPPER RESPONSE REGULATOR1, a transcription factor involved in signaling in hypoxia and copper deficiency, plays an important role in these responses.
Abstract
Anaerobiosis is a stress condition for aerobic organisms and requires extensive acclimation responses. We used RNA-Seq for a whole-genome view of the acclimation of Chlamydomonas reinhardtii to anoxic conditions imposed simultaneously with transfer to the dark. Nearly 1.4 × 103 genes were affected by hypoxia. Comparing transcript profiles from early (hypoxic) with those from late (anoxic) time points indicated that cells activate oxidative energy generation pathways before employing fermentation. Probable substrates include amino acids and fatty acids (FAs). Lipid profiling of the C. reinhardtii cells revealed that they degraded FAs but also accumulated triacylglycerols (TAGs). In contrast with N-deprived cells, the TAGs in hypoxic cells were enriched in desaturated FAs, suggesting a distinct pathway for TAG accumulation. To distinguish transcriptional responses dependent on COPPER RESPONSE REGULATOR1 (CRR1), which is also involved in hypoxic gene regulation, we compared the transcriptomes of crr1 mutants and complemented strains. In crr1 mutants, ∼40 genes were aberrantly regulated, reaffirming the importance of CRR1 for the hypoxic response, but indicating also the contribution of additional signaling strategies to account for the remaining differentially regulated transcripts. Based on transcript patterns and previous results, we conclude that nitric oxide–dependent signaling cascades operate in anoxic C. reinhardtii cells.
INTRODUCTION
In aerobic organisms, the production of energy by oxidative phosphorylation requires oxygen (O2). Additionally, many biosynthetic pathways use O2 as an oxidant or reagent (Raymond and Segrè, 2006), and the presence of O2 influences the bioavailability of metals (Anbar, 2008). O2 deficiency confronts aerobic organisms with the challenge of producing sufficient energy and cell components to allow growth, or at least survival. Acclimation to O2 limitation thus requires the adjustment of nearly all cellular pathways; this adjustment mostly occurs by differential gene expression, often at the level of transcription (Mustroph et al., 2010). Biological responses to the absence of O2 (anoxia) or limitations in O2 (hypoxia) have been examined in many organisms, including those that carry out oxygenic photosynthesis. The reactions of crops to flooding and the consequent O2 depletion in the roots, are intensively studied (Bailey-Serres and Voesenek, 2008; Bailey-Serres et al., 2012). In the unicellular green alga Chlamydomonas reinhardtii, hypoxic conditions have been investigated primarily in the context of the production of molecular hydrogen (H2) (Hemschemeier and Happe, 2011; Catalanotti et al., 2013).
C. reinhardtii is a common reference organism for studying plant-specific processes such as photosynthesis or inorganic nutrient assimilation (Grossman, 2000; Rochaix, 2002; Merchant et al., 2006). However, this alga has retained many genes from the common ancestor of both plants and animals (Merchant et al., 2007), which has made it a valuable model for studying animal-specific pathways, such as the biology of cilia (Marshall, 2008). C. reinhardtii makes its natural habitat in soil and fresh water environments, which frequently become anoxic because of respiratory activity during growth of organisms. This environmental variability may explain the extensive metabolic flexibility of the alga (Grossman et al., 2007). Of interest in terms of anaerobic metabolism, C. reinhardtii was also reported to have enzymes typically found in prokaryotes. It has two molecular H2-producing [FeFe]-hydrogenases, HYDA1 and HYDA2 (Stripp and Happe, 2009). In the light, these enzymes generate H2 using photosynthetically provided electrons (Ghirardi et al., 2009; Hemschemeier and Happe, 2011). C. reinhardtii also employs a pyruvate:formate lyase (PFL1) and the enzymes involved in the PFL pathway, which form the backbone of the fermenting metabolism in (facultative) anaerobic bacteria like Escherichia coli (Atteia et al., 2006; Hemschemeier et al., 2008; Philipps et al., 2011; Magneschi et al., 2012). Additionally, the alga has a pyruvate:ferredoxin oxidoreductase (PFR1) (Mus et al., 2007; Hemschemeier et al., 2008; Terashima et al., 2010; van Lis et al., 2013; Noth et al., 2013).
The signaling cascades operative in C. reinhardtii under anaerobic conditions show some overlap with signaling cascades operating in the copper deficiency response. The COPPER RESPONSE REGULATOR1 (CRR1) transcription factor activates a subset of genes as a response to hypoxia. CRR1 is an important regulator of the acclimation of C. reinhardtii to Cu deficiency (Eriksson et al., 2004; Kropat et al., 2005; Sommer et al., 2010), and several genes that are upregulated in Cu-deficient conditions are also upregulated in hypoxia (Quinn et al., 2000, 2002). The hypoxic response of CRR1 target genes is vital for cells, as crr1 mutants have a severe growth defect in hypoxic conditions in the light (Eriksson et al., 2004). Genes known to be important for the Cu deficiency response of C. reinhardtii are activated in hypoxia, and genes known to be responsive to O2 limitation, such as HYDA1 and PFR1, are also induced in Cu-deficient cells (Castruita et al., 2011). The CRR1 transcriptional activator is also involved in HYDA1 regulation (Pape et al., 2012). However, in contrast with all other CRR1 targets identified so far, expression of HYDA1 is not completely dependent on CRR1, as crr1 mutants still induce HYDA1 gene expression (Quinn et al., 2002; Pape et al., 2012). Thus, other factors must contribute to HYDA1 promoter activity.
CRR1 is a multidomain 1232–amino acid protein that binds to the promoter of its target genes via a SQUAMOSA-PROMOTER BINDING PROTEIN DNA binding domain. CRR1 has ankyrin repeats, which are known protein–protein interaction modules, and a C-terminal Cys-rich region similar to metallothionein (Kropat et al., 2005). Notably, both modules are needed for the transcriptional activation of the Cu-responsive cytochrome c6 (CYC6) gene upon hypoxia, but not upon Cu depletion (Sommer et al., 2010).
This study aimed to identify the physiological responses and putative signaling pathways of C. reinhardtii subjected to anaerobiosis by generating whole-genome transcript profiles. In particular, we sought to gain deeper insights into the role of CRR1 in the hypoxic response and the modulating activity of the C-terminal metallothionein-like domain of CRR1. For this purpose, wild-type cultures, crr1 mutants, and strains containing a CRR1 protein lacking the Cys-rich C terminus were transferred to anaerobic conditions in the dark. This setup was chosen to mimic natural conditions, in which the O2 concentration in the medium probably decreases gradually in the night as O2 evolution shuts off and cellular respiratory activity consumes O2 (Quinn et al., 2002). The transcript profiles indicated that the cells implement various strategies to acclimate to the absence of light and O2; thus, the absence of all means to efficiently generate energy and cell components. Besides predictable patterns regarding cell growth and photosynthesis, one of the patterns that emerged was a complex strategy of energy generation via the utilization of all reserves available. Our data also supported the close connection between Cu and O2 deficiency and the important role of CRR1 under both conditions.
RESULTS
Defining a Setup Simulating Natural Anoxic Conditions
Various experimental approaches have been used to analyze the hypoxic or anaerobic response of C. reinhardtii. Cells incubated in dim light without aeration become hypoxic over time because of respiratory activity and decreased oxygen evolution (Quinn et al., 2000, 2002). Hypoxia can also be achieved in sealed cultures under standard light intensities in the absence of sulfur or nitrogen, which results in loss of photosystem II (PSII) activity and, hence, decreased photosynthetic O2 evolution and finally net consumption of O2 by respiration (Melis et al., 2000; Philipps et al., 2012). These approaches have in common that the onset of anaerobiosis is accompanied by substantial metabolic (Doebbe et al., 2010) and transcriptional (Nguyen et al., 2008; González-Ballester et al., 2010) changes, many of which are related to nutrient depletion. Another approach is to transfer concentrated cell suspensions to the dark while they are purged with O2-free inert gas (Happe and Kaminski, 2002; Mus et al., 2007). This approach was used in a prior study using a microarray platform (Mus et al., 2007).
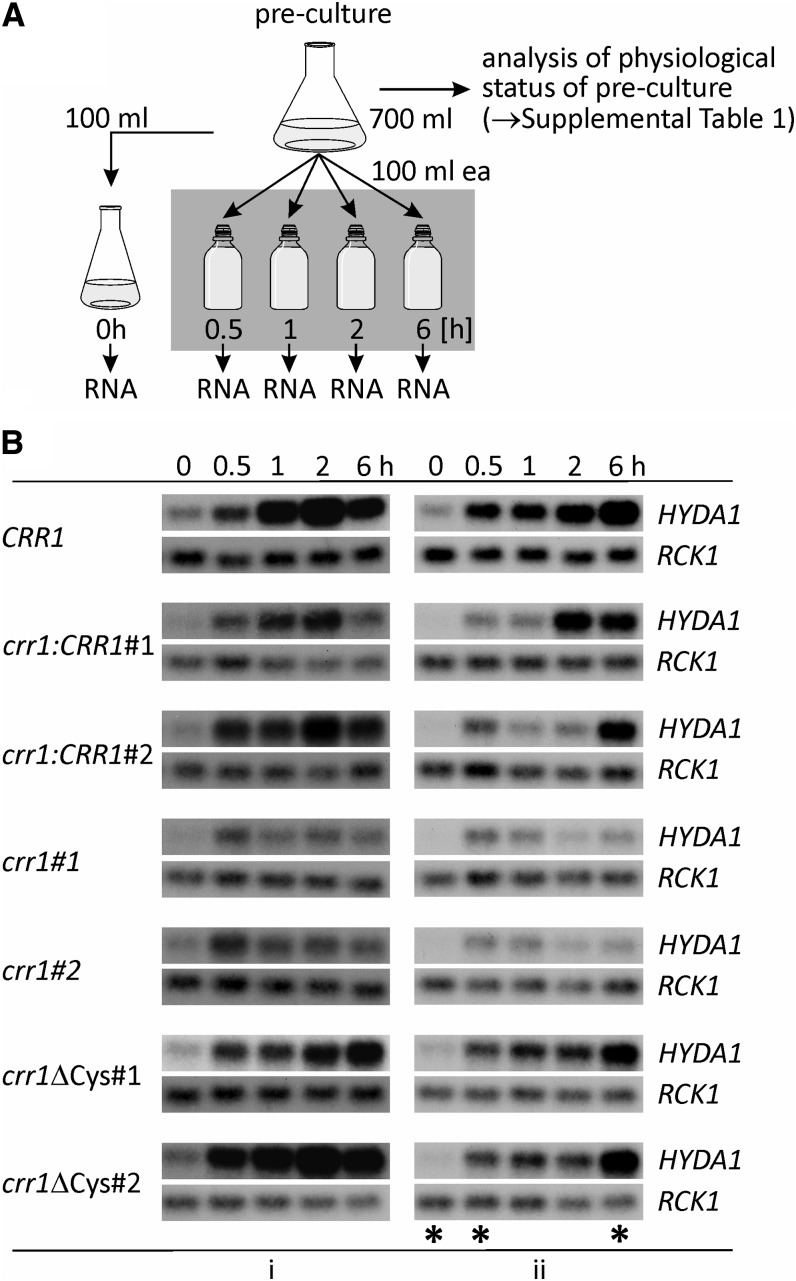
In this work, we sought to generate hypoxia in a situation that might reflect the natural onset in the environment; therefore, we used a protocol in which photomixotrophically grown cells are transferred to sealed flasks in the dark, allowing a gradual removal of dissolved O2 by respiratory activity (self-anaerobization) (Figure 1A). The strains used in this work include the usual C. reinhardtii laboratory strain CC-124 wild-type mt− [137c] (CRR1 nit1 nit2), crr1, a mutant defective in the Cu deficiency response and hypoxic signaling (Kropat et al., 2005), and complemented derivatives of crr1. These were crr1:CRR1 representing the rescued strain and strains containing a C terminally truncated CRR1 protein (termed crr1ΔCys). The latter represents a strain fully rescued for the Cu nutrition response but partially defective in hypoxic signaling because of a deletion of the C-terminal metallothionein-like domain that has been implicated in O2-responsive signaling through CRR1 (Sommer et al., 2010). We analyzed two clones for each of the respective transformants (Figure 1B), which are labeled #1 and #2 in this work. Note that because of differences in the rate of O2 consumption, the time course of anaerobization is prone to variation. Therefore, we monitored respiratory O2 consumption rates as well as the acetate content of the spent medium in each preculture in order to define conditions for the best time points to initiate self-anaerobization (see Supplemental Table 1 online). Acetate concentrations were comparable, but C. reinhardtii crr1 and crr1ΔCys mutants exhibited lower respiratory rates, which were ∼60 to 80% of those of the rescued clones (crr1:CRR1#1 in Supplemental Table 1 online), which is consistent with an impact of the crr1 mutation on cytochrome oxidase function (Eriksson et al., 2004). This must result in a slower progress of the establishment of anaerobic conditions. Therefore, the HYDA1 transcript, known from previous work to be a sensitive sentinel gene for anaerobiosis (Happe and Kaminski, 2002), was used to assess the onset of hypoxia at various time points after the transfer of the cells to sealed flasks in the dark (0.5, 1, 2, and 6 h). Since the cells can get anaerobic during collection, it is relevant to demonstrate also that the untreated (aerated) cells do not express HYDA1. RNA hybridization analyses showed no or low amounts of HYDA1 transcript in the 0-h samples, while HYDA1 mRNA accumulated in all strains during the time course of the experiments (Figure 1B). In RNA samples of both crr1 mutants, HYDA1 signals were weaker than in the other strains. This was consistent with CRR1 being involved in the transcriptional regulation of HYDA1 (Pape et al., 2012).
Figure 1.
Experimental Setup and Sample Evaluation by RNA Gel Blot Hybridization.
(A) Schematic of the experimental proceeding applied for establishing anoxic conditions in algal cultures. Time points of RNA sampling are indicated.
(B) HYDA1 RNA hybridization in samples isolated according to the scheme shown in (A). Shown are results from two independent experiments (i and ii) of all C. reinhardtii clones analyzed in this study. Wild-type strain CC-124 is indicated as CRR1. Transcript levels of RCK1 (CBLP, Cre13.g599400) served as loading control. Asterisks in the right panel (ii) mark the RNA samples that were sequenced.
In addition to HYDA1, changes of transcript levels of other sentinel genes were analyzed by quantitative RT-PCR (qRT-PCR) in order to verify the expected status of the cells, among them PFR1 and CYC6 (oligonucleotides used for qRT-PCR are listed in Supplemental Table 2 online). The relative fold changes of all chosen marker transcripts were as expected (see Supplemental Table 3 online), showing that the chosen setup was suitable to analyze the anaerobic response of C. reinhardtii. For RNA sequencing, we chose samples that would reflect early and late responses. Therefore, RNA isolated at 0.5 and 6 h of self-anaerobization from experiment labeled ii (Figure 1B; see Supplemental Tables 1 and 3 online) were sequenced.
Quality and Statistics of RNA-Seq Data
Samples isolated from independent clones of each genotype were analyzed as replicates to ensure the reproducibility of the data. Statistical analyses of both the expression estimates obtained for each individual sample in units of reads per kilobase of mappable transcript length per million mapped reads (RPKM) and the fold changes of filtered transcripts showed that the data obtained from each pair of clones correlated well (R2 values between 0.89 and 0.97), while the correlation between the rescued strains and C. reinhardtii wild-type CC-124 was lower (R2 values between 0.63 and 0.74) (see Supplemental Figure 1 online). The quality of the RNA-Seq data was additionally validated by analyzing the fold changes of 33 transcripts in the same RNA samples by qRT-PCR (R2 values between 0.83 and 0.97) (see Supplemental Figure 2 online).
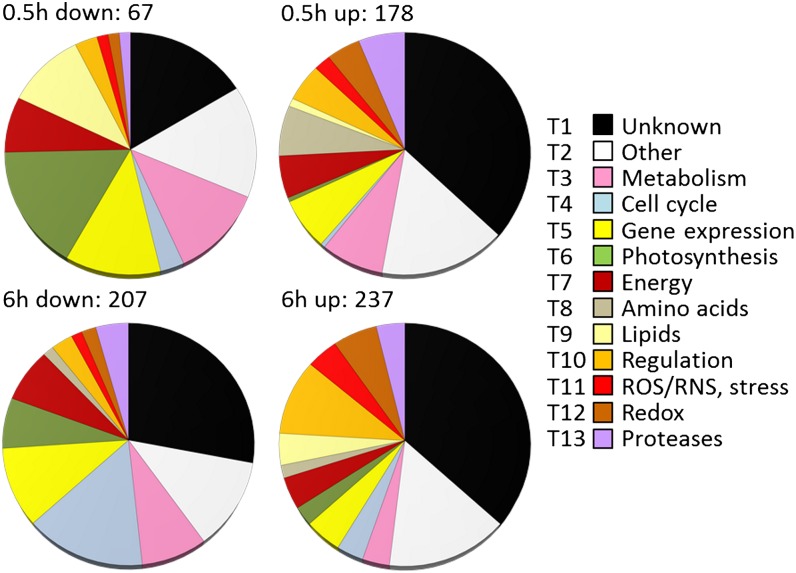
Including all mRNAs whose abundance changed more than fourfold in at least one of the pairwise comparisons (0.5 versus 0 h and 6 versus 0 h) with a false discovery rate (FDR) ≤ 0.05 resulted in a list of 1376 significantly regulated transcripts. As the annotations and gene ontology (GO) terms available for C. reinhardtii are still limited, the deduced proteins were manually annotated (see Supplemental Methods 1 online) and grouped into functional categories (see Supplemental Data Set 1 online). A total of 439 proteins had no known motifs or domains of unknown function and were classified as “unknown” (see Supplemental Data Set 1, T1, online). A total of 198 proteins were grouped as “other” (see Supplemental Data Set 1, T2, online) as their functions could not be specifically categorized. Proteins that could be assigned to specific functions or processes were grouped accordingly (see Supplemental Data Set 1, T3 to T13, online). Figure 2 shows an overview of the categories and their representation by transcripts, respectively, deduced from the data of the C. reinhardtii wild-type CC-124. The pattern differed moderately from that observed in the rescued strain, but the trends were similar (see Supplemental Figure 3 online). The patterns were basically reflected by GO enrichment analyses (see Supplemental Table 4 online).
Figure 2.
Functional Annotation of Differentially Accumulating Transcripts in Dark-Anoxic Wild-Type C. reinhardtii.
Transcripts whose abundance changed at least fourfold (FDR ≤ 5%) in cells incubated for 0.5 or 6 h under dark-anoxic conditions were selected. Numbers show the total amount of transcripts down- or upregulated at each time point. Deduced proteins were grouped according to functional categories (described in Supplemental Methods 1 online). T1 to T13 indicate the sheet number in Supplemental Data Set 1 online in which individual transcripts are listed. ROS, reactive oxygen species; RNS, reactive nitrogen species.
Putative CRR1 Targets
In view of the known connections between Cu and O2 limitation in C. reinhardtii, we first compared the data obtained in this study with a previous RNA-Seq analysis of Cu-deprived algae (Castruita et al., 2011). Eighty-three anoxia-responsive transcripts are also differentially regulated in Cu-deficient C. reinhardtii cells (Cu-responsive transcripts are marked in Supplemental Data Sets 1 and 2 online and summarized in Supplemental Data Set 2, T2, online). From the group of intersecting transcripts, 46 were assigned CRR1 targets upon Cu deficiency (Castruita et al., 2011). Calculated from the total number of C. reinhardtii genes and the total number of anoxia-responsive transcripts from the 6-h versus 0-h comparisons, the overlap between these previously identified CRR1 targets and the set of anoxia-responsive genes identified in this study was significant (hypergeometric P value = 1.7e−38), indicating that the overlap was not merely due to the large number of anoxia-responsive transcripts. In the latter study, the cutoff for defining CRR1 targets was set to a twofold difference, but we chose to apply a more stringent filter in this work because the much shorter time frame (6 h versus days) was likely to cause higher variations. To determine if the expression of a gene is dependent on CRR1 and, if so, if the Cys-rich C terminus is involved, the fold changes after 6-h anoxia in crr1 and crr1ΔCys mutants were compared with those of the rescued strain (crr1:CRR1). From all annotated C. reinhardtii v4.3 gene models, transcripts whose fold changes after 6-h anoxia had an FDR < 0.05 in C. reinhardtii crr1:CRR1 were used as the basis for the comparisons. Transcripts were considered as CRR1 targets when their fold changes after 6-h anoxia were at least fourfold different from those in the rescued strain in each both mutant colonies (see Supplemental Data Set 2, T1, online). Though this approach resulted in loss of the HYDA1 gene in the list (Pape et al., 2012) and possibly other genes whose expression is only partially dependent on CRR1, it was chosen to prevent assigning too many false positives.
Under the previous constraints, 19 genes were identified that were misregulated in both crr1 and crr1ΔCys mutants (Figures 3A and 3B). Twenty-three genes were only affected in the crr1 colonies, indicating that the Cys-rich C terminus was not involved in their regulation. In crr1ΔCys colonies, 16 transcripts were deregulated but showed nearly normal expression patterns in crr1 colonies (Figures 3A and 3B, Table 1; see Supplemental Table 5 online). From the 42 transcripts that showed abnormal fold changes in crr1 colonies, 20 were also identified as CRR1 targets in Cu-deficient C. reinhardtii cells (Castruita et al., 2011) (Figure 3C; see Supplemental Data Set 2, T2, online).
Figure 3.
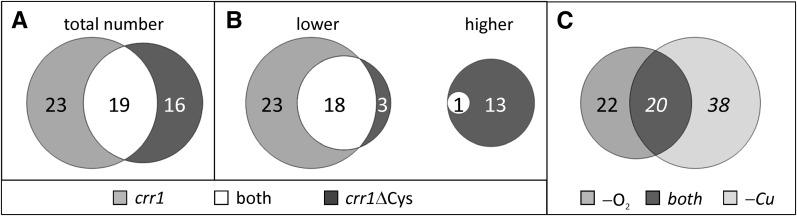
A Subset of CRR1-Responsive Genes Is Regulated by Both Cu Nutrition and Hypoxia.
CRR1 targets were determined in the subset of transcripts whose fold changes in the 6-h versus 0-h comparison had an FDR < 0.05 in the rescued strain crr1:CRR1. A transcript was defined as CRR1 target when its fold change after 6-h anoxia was at least fourfold different from the fold changes in the rescued strain.
(A) Total number of anoxic CRR1 targets.
(B) Anoxic CRR1 targets differentiated between transcripts whose fold changes after 6-h anoxia were lower in the mutants compared with the rescued strain (lower) and those whose fold changes were higher (higher).
(C) Intersection of CRR1 targets defined in anoxia and in Cu deficiency. Anoxic CRR1 targets identified in crr1 mutants were compared with the CRR1 targets identified in Cu deficiency by Castruita et al. (2011).
Table 1. Putative CRR1 Targets.
| 6-h versus 0-h Fold Changes | ||||||||
|---|---|---|---|---|---|---|---|---|
|
crr1ΔCys |
crr1 |
|||||||
| Locus ID | Gene name | Description | Cua | crr1:CRR1 | #1 | #2 | #1 | #2 |
| CRR1 targets depending on Cys-rich C terminus | ||||||||
| Cre17.g700950 | FDX5 | Ferredoxin 5 | Y | 1238.1 | 21.2 | 13.4 | 0.27 | 0.26 |
| Cre16.g651050 | CYC6 | Cytochrome c6 | Y | 152.5 | 0.44 | 2.2 | 1.4 | 1.4 |
| Cre02.g087073 | Unknown | 110.7 | 5.5 | 6.3 | 12.2 | 1.4 | ||
| Cre02.g084400 | * | Prolyl-4-hydroxylase with ShK domain | Y | 68.3 (137.8) | 20.6 (19.7) | 12.4 (13.2) | 3.4 | 1.4 |
| Cre01.g039450 | DUF3611, transmembrane | 55.0 | 2.4 | 5.2 | 1.4 | 1.4 | ||
| Cre02.g087050 | Unknown | 24.9 | 5.1 | 4.0 | 2.3 | 1.7 | ||
| Cre44.g787700 | *GOX7 | Glyoxal or galactose oxidase | Y | 22.4 | 3.6 | 3.5 | 2.9 | 3.2 |
| Cre10.g426800 | Chloramphenicol acetyltransferase-like domain | Yb | 20.0 | 0.49 | 0.52 | 0.41 | 0.72 | |
| Cre10.g444200 | Unknown, transmembrane | 18.9 | 2.4 | 2.7 | 2.9 | 5.7 | ||
| Cre06.g265300 | WD40/YVTN repeat-like-containing domain | Yb | 15.5 | 0.8 | 1.0 | 1.4 | 1.4 | |
| Cre18.g750700 | * | Glyoxal or galactose oxidase | 12.3 | 1.6 | 1.0 | 1.4 | 1.4 | |
| Cre01.g029700 | Unknown, transmembrane | 8.7 (328.7) | 0.5 | 1.2 | 3.1 (60.4) | 2.7 (50.7) | ||
| Cre10.g434350 | CTR2 | CTR type copper ion transporter | Y | 5.7 (35.3) | 0.8 | 0.7 | 3.4 (0.4) | 0.6 (0.1) |
| Cre03.g191250 | LCI34 | Unknown; gene is low-CO2-inducible | Yb | 4.3 (174.5) | 0.6 (37.6) | 1.0 (58.0) | 1.3 (58.1) | 1.0 (39.1) |
| Cre12.g512400 | Protein related to nitrogen assimilation regulatory proteins | Y | 3.8 (37.7) | 1.1 (10.8) | 1.0 (11.1) | 0.8 (9.1) | 0.8 (9.3) | |
| Cre10.g434650 | CTR3 | Copper transport accessory protein | Y | 3.6 | 0.3 | 0.5 | 0.4 | 0.7 |
| Cre15.g645100 | * | Squalene epoxidase/monooxygenase | 3.5 | 0.8 | 0.8 | 0.7 | 0.8 | |
| Cre16.g693050 | SIR2 | Nitrite/Sulfite reductase ferredoxin-like domain | 1.7 (15.6) | 0.3 | 0.3 | 0.3 (3.9) | 0.6 (4.4) | |
| CRR1 targets independent from Cys-rich C terminus | ||||||||
| Cre02.g137700 | Unknown | 143.9 (9.9) | 21.0 (20.8) | 29.4 (17.5) | 4.1 (0.3) | 7.1 (0.2) | ||
| Cre12.g510058 | Unknown | 69.7 | 31.4 | 3.9 | 0.9 | 0.7 | ||
| Cre08.g372200 | Unknown | Y | 64.9 | 31.9 | 143.7 | 5.2 | 2.5 | |
| Cre17.g730100 | RSE1 | Intramembrane metalloprotease (peptidase M50) | Y | 40.2 | 14.9 | 33.3 | 6.8 | 5.6 |
| Cre13.g585050 | WD40/YVTN repeat-like-containing domain, transmembrane | 20.5 | 54.2 | 38.6 | 2.3 | 2.8 | ||
| Cre03.g179500 | * | Prolyl-4-hydroxylase, transmembrane | Y | 19.4 | 11.0 | 13.3 | 1.3 | 3.3 |
| Cre18.g750800 | *GOX8 | Glyoxal or galactose oxidase, transmembrane | 15.8 | 8.9 | 10.2 | 2.0 | 2.9 | |
| Cre07.g318800 | HSP22A | Small heat shock protein 22A | 12.4 | 9.3 | 22.5 | 1.7 | 3.1 | |
| Cre10.g444250 | CRAL/TRIO domain | 9.8 | 3.0 | 2.7 | 1.2 | 1.5 | ||
| Cre07.g346050 | *CRD1 | Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase | Y | 8.9 | 5.7 | 8.0 | 0.7 | 0.7 |
| Cre10.g443050 | * | Prolyl-4-hydroxylase with ShK domain | Y | 8.0 | 4.8 | 10.4 | 1.0 | 0.9 |
| Cre07.g346900 | Unknown, transmembrane | Y | 7.1 (557.0) | 0.6 (548.7) | 1.4 (281.0) | 0.2 (2.0) | 0.2 (0.6) | |
| Cre12.g507800 | Unknown, transmembrane | Y | 6.0 | 2.7 | 4.1 | 0.4 | 0.5 | |
| Cre12.g490477 | WD40 domain | 5.5 | 9.7 | 18.5 | 0.7 | 0.4 | ||
| Cre16.g686450 | α/β-hydrolase superfamily | 5.2 | 1.0 | 1.7 | 0.4 | 0.6 | ||
| Cre02.g084450 | * | Prolyl-4-hydroxylase with ShK domain | Y | 5.2 | 1.7 | 2.1 | 0.9 | 0.6 |
| Cre13.g570600 | CTR1 | CTR-type copper ion transporter | Y | 5.1 | 3.1 | 3.9 | 1.2 | 1.2 |
| Cre12.g530350 | IRT2 | Iron-nutrition responsive ZIP (zinc/iron permease) family transporter | Y | 4.3 | 1.5 | 0.8 | 0.5 | 0.6 |
| Cre12.g490500 | CGL78 | DUF2488/Ycf54 | Y | 3.7 | 7.1 | 9.4 | 0.5 | 0.5 |
| Cre02.g085450 | *CPX1 | Coproporphyrinogen III oxidase | Y | 3.6 | 6.9 | 2.8 | 0.3 | 0.5 |
| Cre02.g136050 | *HPD1 | 4-hydroxyphenylpyruvate dioxygenase | Y | 2.1 | 1.0 | 0.7 | 0.4 | 0.2 |
| Cre08.g367400 | LHCSR3 | Stress-related Chl a/b binding protein | 0.26 (6.6) | 0.04 (2.7) | 0.02 (3.1) | 0.06 (2.0) | 0.06 (1.6) | |
| Cre03.g162800 | LCI1 | Unknown membrane protein; gene is low-CO2-inducible | 0.04 (0.52) | 0.004 (0.11) | 0.004 (0.47) | 0.004 (0.07) | 0.004 (0) | |
| Only misregulated in crr1ΔCys colonies | ||||||||
| Cre10.g433800 | Unknown | 21.5 | 2.6 | 3.7 | 6.2 | 5.6 | ||
| Cre12.g488450 | VSP3 | Extracellular matrix protein, transmembrane | 2.5 | 0.4 | 0.4 | 2.2 | 2.2 | |
| Cre16.g681750 | Plasma-membrane Ca2+-translocating P-type ATPase | 0.3 | 0.05 | 0.04 | 0.2 | 0.2 | ||
The entries in this table were selected from automated lists generated by comparisons of the transcriptomes and contain transcripts whose fold changes after 6-h anoxia were at least fourfold lower in crr1 and/or crr1ΔCys mutants than in the rescued strain crr1:CRR1. The list was manually curated with respect to aberrant RPKM values and known CRR1 targets (see Methods). In case putative targets were manually curated due to extraordinary RPKM values in the 6-h samples, these are indicated next to the fold changes in parentheses and italics. See Supplemental Data Set 2 online for more details, such as estimated expression in RPKM and FDR. Locus IDs are from the C. reinhardtii v4.3 genome annotation on Phytozome v8.0. An asterisk in the gene name column indicates that the gene encodes an O2-dependent enzyme. Y (yes) in the column labeled Cu shows that a transcript is a CRR1 target in Cu deficiency as shown by Castruita et al. (2011). Blank spaces in the second column indicate that the gene is unnamed.
Deduced from Castruita et al. (2011). Blank spaces indicate that the gene was not regulated in Castruita et al. (2011).
The gene is Cu responsive but not defined as a CRR1 target in Cu deficiency.
In agreement with known CRR1 targets, most of the targets assigned here exhibited lower fold changes in the mutants than in the rescued strain (Figure 3B, Table 1). Transcripts that were deregulated in both crr1 and crr1ΔCys mutants included the previously characterized CRR1 targets CYC6, FDX5 (encoding ferredoxin 5), CTR2, and CTR3 (coding for Copper Transport proteins 2 and 3) (Table 1; Quinn et al., 2002; Page et al., 2009; Terauchi et al., 2009; Lambertz et al., 2010). Also, new putative CRR1 targets identified by Castruita et al. (2011), in Cu-deficient C. reinhardtii cultures, appeared in this set, such as Cre02.g084400, encoding a prolyl-4-hydroxylase (P4H), GOX7 (glyoxal or galactose oxidase), or Cre12.g512400, encoding a protein related to nitrogen assimilation regulatory proteins (Table 1). GO analyses indicated an enrichment of three encoded proteins having electron carrier activity (CYC6, FDX5, and Cre12.g512400, P value 4.7 × 10−5; the latter two were also represented by the category of metal cluster binding proteins, P value 0.002).
Putative targets only identified in the crr1 mutant colonies, but not in the crr1ΔCys mutants, contained characterized CRR1 targets, too. These were CRD1, CPX1, and CTR1 (Table 1) (Quinn et al., 2002; Allen et al., 2008; Page et al., 2009). Several of the genes in this set were also defined as CRR1 targets by Castruita et al. (2011), such as three genes encoding P4Hs (Cre03.g179500, Cre10.g443050, and Cre02.g084450), IRT2 (iron nutrition-responsive ZIP family transporter), HPD1 (4-hydroxyphenylpyruvate dioxygenase), and RSEP1 (intramembrane metalloprotease). Proteins involved in the biological processes of porphyrin biosynthesis (CPX1 and CRD1; P value 9.8 × 10−4) and metal ion transport (IRT2 and CTR1; P value 0.012) or having oxidoreductase activity (CPX1, CRD1, HPD1, Cre03.g179500, Cre10.g443050, and Cre02.g084450; P value 1.3 × 10−4) were identified as enriched. The latter six enzymes require O2 as a substrate, and the P4Hs were represented by the GO molecular function category “oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen” (P value 2.1 × 10−4). Our manual annotation revealed 11 transcripts encoding O2-dependent enzymes to be misregulated in crr1 mutants, four of these also in the crr1ΔCys mutants (Table 1).
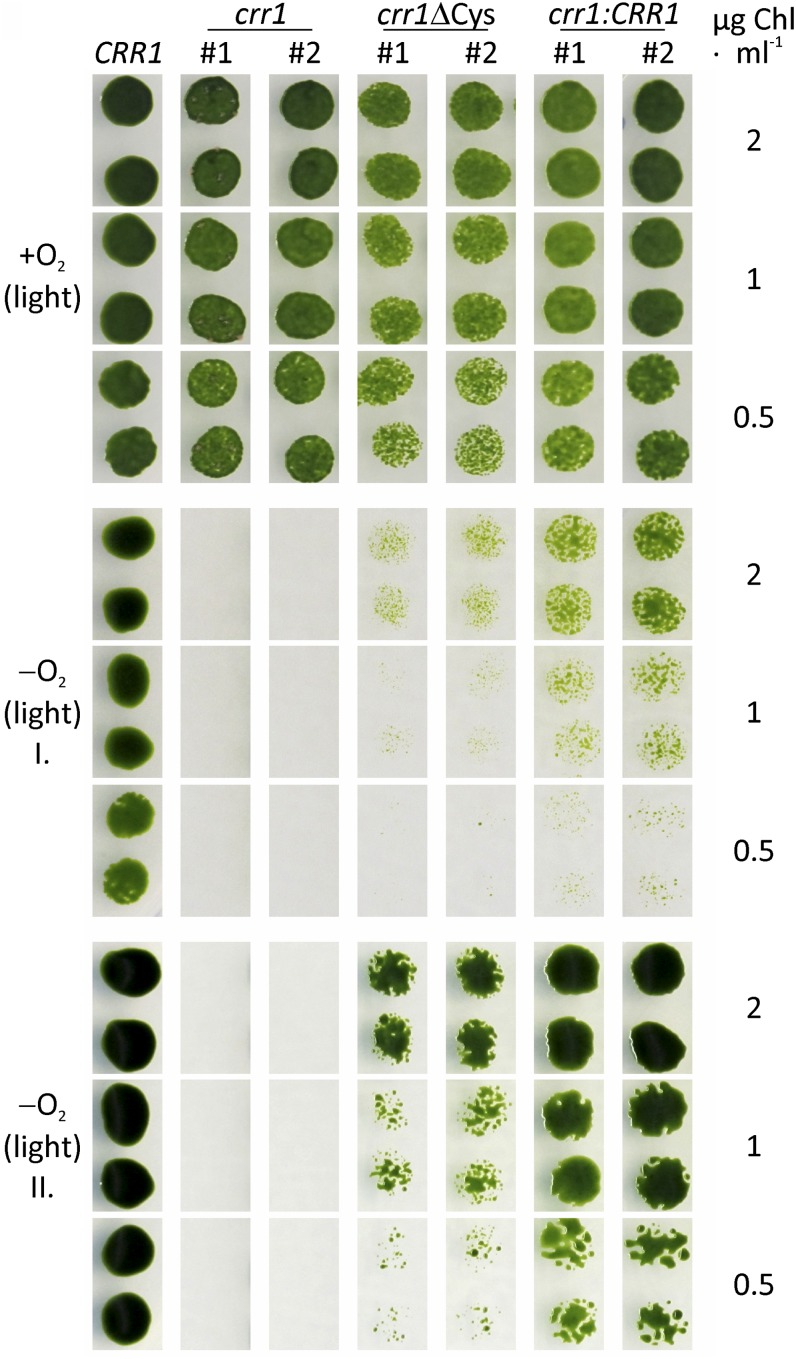
Because the regulation of several CRR1 targets appeared to be independent from the Cys-rich C terminus of CRR1, we tested if crr1ΔCys mutants are able to grow in an anaerobic atmosphere in the light. While crr1 mutants do not grow under this condition (Eriksson et al., 2004), both crr1ΔCys colonies grew, albeit slower than the rescued strains (Figure 4).
Figure 4.
An Anaerobic Atmosphere Impacts the Growth of the C. reinhardtii crr1 Mutant.
The indicated strains (wild-type CC-124 is labeled by CRR1) were grown to the midexponential growth phase under standard conditions. Then they were diluted to chlorophyll concentrations of 2, 1, and 0.5 µg mL−1 and spotted on two TAP agar plates. Each one of the plates was incubated aerobically (+O2) or anaerobically (−O2) in the light (50 µmol photons m−2 s−1). Photographs were taken after 5 (+O2 and −O2 I) and 8 d of growth (−O2 II).
From the list of genes that were significantly misregulated only in crr1ΔCys colonies, 13 were not as strongly downregulated as in C. reinhardtii crr1:CRR1 (Figure 3B; see Supplemental Table 5 online). GO analyses indicated a single enrichment in the molecular function of nucleic acid binding (P value 0.007). Our own annotation indicated that transcripts coding for proteins involved in gene expression, cell cycle, and cell structure were enriched (see Supplemental Table 5 online).
Downregulation of Growth and Gene Expression
C. reinhardtii grows much slower in the dark, even in the presence of O2 and acetate (Harris, 2001). In sealed flasks in the dark, cell division of wild-type CC-124 and rescued strain crr1:CRR1 was even more impaired (see Supplemental Figure 4A online). This was reflected by reduced levels of transcripts coding for proteins with known or proposed functions in cell cycle, DNA synthesis, and gene expression (see Supplemental Data Set 1, T4 and T5, online). Among those were several transcripts probably involved in plastid division, specifically, FTSZ1, FTSZ2, MIND1, and MINE1 (Wang et al., 2003; Maple and Møller, 2007; Adams et al., 2008) (Table 2). Only few transcripts assigned to cell cycle regulation, circadian rhythm, or gametogenesis accumulated (see Supplemental Data Set 1, T4, online).
Table 2. Differentially Accumulating Transcripts in Dark-Anoxic versus Illuminated and Aerated C. reinhardtii Strains.
|
CRR1 |
crr1:CRR1 |
crr1ΔCys |
crr1 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus ID | Name | Description | RPKM | Fold Change |
RPKM | Fold Change |
RPKM | Fold Change |
RPKM | Fold Change |
||||
| 0 h | 0.5 h | 6 h | 0 h | 0.5 h | 6 h | 0 h | 0.5 h | 6 h | 0 h | 0.5 h | 6 h | |||
| Sentinel genes | ||||||||||||||
| Cre03.g199800 | HYDA1 | [FeFe]-hydrogenase 1 | 76.8 | 3.1 | 8.1 | 54.8 | 3.6 | 8.7 | 140.9 | 2.8 | 5.2 | 45.5 | 5.0 | 4.3 |
| Cre09.g396600 | HYDA2 | [FeFe]-hydrogenase 2 | 19.0 | 5.7 | 3.6 | 20.1 | 4.4 | 2.1 | 19.0 | 5.1 | 1.5 | 18.9 | 5.3 | 2.4 |
| Cre06.g296750 | HYDEF | [FeFe]-hydrogenase maturation factor | 8.4 | 4.8 | 7.3 | 1.7 | 10.3 | 11.2 | 2.7 | 11.0 | 2.6 | 3.0 | 10.5 | 4.3 |
| Cre06.g296700 | HYDG | [FeFe]-hydrogenase maturation factor | 55.8 | 4.9 | 4.2 | 13.0 | 10.0 | 7.1 | 20.8 | 9.3 | 2.9 | 22.4 | 11.0 | 4.4 |
| Cre11.g473950 | PFR1 | Pyruvate:ferredoxin oxidoreductase | 4.6 | 8.9 | 31.5 | 6.3 | 6.5 | 21.0 | 7.8 | 5.6 | 4.2 | 3.0 | 8.8 | 8.7 |
| Cre17.g700950 | FDX5 | Ferredoxin 5 | 29.0 | 0.3 | 310.0 | 1.3 | 0.4 | 1238 | 1.9 | 0.3 | 21.2 | 0.3 | 0.2 | 0.3 |
| Cre16.g651050 | CYC6 | Cytochrome c6 | 0.1 | 1.3 | 419.3 | 0 | 1.0 | 152.5 | 86.9 | 0.6 | 0.4 | 0.1 | 1.0 | 1.4 |
| Cre02.g085450 | CPX1 | Coproporphyrinogen III oxidase | 103.0 | 0.4 | 23.7 | 83.5 | 0.3 | 3.6 | 119.3 | 0.3 | 6.9 | 125.2 | 0.3 | 0.3 |
| Cre07.g346050 | CRD1 | Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase | 203.9 | 0.6 | 9.6 | 58.1 | 0.7 | 8.9 | 110.2 | 0.6 | 5.7 | 49.6 | 0.8 | 0.7 |
| Cell division and cell cycle | ||||||||||||||
| Cre02.g118600 | FTSZ1 | Plastid division protein | 28.1 | 0.7 | 0.1 | 22.5 | 1.1 | 0.1 | 36.1 | 1.0 | 0.4 | 19.3 | 0.8 | 0.2 |
| Cre15.g646150 | FTSZ2 | Plastid division protein | 15.6 | 0.5 | 0.1 | 15.1 | 1.1 | 0.1 | 18.6 | 1.2 | 0.3 | 11.7 | 1.0 | 0.1 |
| Cre12.g522950 | MIND1 | Chloroplast septum site determinant | 44.2 | 0.6 | 0.1 | 41.9 | 1.2 | 0.1 | 45.5 | 1.3 | 0.5 | 27.7 | 0.9 | 0.3 |
| Cre17.g720350 | MINE1 | Chloroplast division site determinant | 26.7 | 0.6 | 0.1 | 21.1 | 0.9 | 0.2 | 19.8 | 1.1 | 0.8 | 13.2 | 1.0 | 0.4 |
| Photosynthesis | ||||||||||||||
| Cre07.g325500 | CHLH1 | Mg-chelatase, subunit H | 121.2 | 0.1 | 0.7 | 65.8 | 0.2 | 0.9 | 99.1 | 0.2 | 0.8 | 115.2 | 0.2 | 0.6 |
| Cre05.g246800 | GUN4 | Tetrapyrrole binding protein involved in plastid-to-nucleus signaling | 52.2 | 0.1 | 0.6 | 38.8 | 0.2 | 1.0 | 43.1 | 0.3 | 1.2 | 57.4 | 0.1 | 0.6 |
| Cre12.g517700 | NYC1 | Putative chlorophyll b reductase homologous to NYC1 | 1.6 | 2.2 | 5.1 | 4.5 | 2.4 | 6.5 | 5.3 | 3.5 | 2.9 | 4.8 | 4.0 | 6.6 |
| Cre05.g231450 | Putative chlorophyllide-a oxygenase | 2.3 | 4.6 | 2.7 | 4.5 | 5.6 | 8.0 | 4.9 | 5.2 | 3.6 | 5.7 | 7.3 | 6.9 | |
| Energy metabolism | ||||||||||||||
| Cre03.g185250 | SSS2 | Soluble starch synthase II | 13.9 | 6.5 | 5.3 | 21.9 | 4.6 | 8.5 | 23.3 | 6.2 | 4.8 | 40.5 | 4.1 | 4.4 |
| Cre01.g032700 | GLK1 | Glucokinase | 29.9 | 0.8 | 0.4 | 24.3 | 0.9 | 0.2 | 32.3 | 0.9 | 0.5 | 23.7 | 0.9 | 0.4 |
| Cre10.g424750 | PPD1 | Pyruvate phosphate dikinase | 0.6 | 12.3 | 58.6 | 0.1 | 1.1 | 10.4 | 0.1 | 2.2 | 18.6 | 0.1 | 0.5 | 6.2 |
| Cre14.g629750 | MME2 | NADP-dependent malic enzyme | 12.6 | 1.7 | 6.2 | 12.2 | 1.6 | 2.9 | 13.7 | 1.9 | 10.4 | 15.9 | 2.0 | 1.7 |
| Cre04.g228350 | Lipoamide acyltransferase of branched-chain α-keto acid dehydrogenase complex | 1.4 | 7.6 | 2.7 | 6.0 | 6.7 | 7.9 | 7.2 | 6.1 | 2.9 | 7.0 | 6.4 | 5.6 | |
| Cre12.g539900 | 3-Methyl-2-oxobutanate dehydrogenase E1, α | 1.0 | 7.1 | 2.3 | 1.8 | 5.4 | 7.9 | 3.1 | 5.1 | 3.2 | 3.0 | 3.9 | 4.8 | |
| Cre06.g311050 | 3-Methyl-2-oxobutanate dehydrogenase E1, β | 2.8 | 3.2 | 2.5 | 5.0 | 4.3 | 5.5 | 6.9 | 4.3 | 2.9 | 7.0 | 3.7 | 3.9 | |
| Cre06.g296400 | Putative isovaleryl-CoA dehydrogenase | 4.7 | 7.5 | 3.5 | 9.2 | 4.4 | 5.8 | 14.1 | 3.7 | 2.9 | 13.6 | 4.4 | 4.2 | |
| Cre13.g593500 | MCC1 | Methylcrotonoyl-CoA carboxylase α | 1.9 | 8.0 | 1.6 | 3.4 | 5.2 | 7.3 | 6.3 | 3.6 | 1.7 | 6.2 | 4.0 | 3.8 |
| Cre03.g181200 | Methylcrotonoyl-CoA carboxylase β | 2.0 | 10.2 | 4.6 | 6.2 | 6.3 | 7.7 | 11.8 | 4.9 | 2.8 | 14.5 | 4.8 | 4.8 | |
| Cre16.g687950 | ETF1 | Electron transfer flavoprotein α-subunit | 2.7 | 4.0 | 1.6 | 3.2 | 5.6 | 6.3 | 4.5 | 6.8 | 3.3 | 4.8 | 5.5 | 5.1 |
| Cre27.g775600 | ETF2 | Electron transfer flavoprotein β-subunit | 3.4 | 3.5 | 1.4 | 7.9 | 3.5 | 3.6 | 7.3 | 3.4 | 2.0 | 8.0 | 3.5 | 3.1 |
| Lipids and FAs | ||||||||||||||
| Cre16.g687350 | Acyl-CoA oxidase | 11.0 | 8.9 | 1.8 | 21.1 | 6.0 | 3.9 | 19.9 | 5.9 | 1.5 | 28.4 | 5.0 | 2.7 | |
| Cre16.g695100 | Acyl-CoA oxidase | 27.6 | 2.6 | 1.2 | 15.4 | 3.6 | 1.5 | 19.8 | 3.4 | 0.7 | 20.9 | 2.4 | 1.0 | |
| Cre05.g231950 | Acyl-CoA oxidase, C terminal | 19.6 | 4.3 | 0.8 | 17.9 | 5.5 | 4.1 | 18.5 | 6.0 | 1.6 | 23.9 | 4.5 | 2.8 | |
| Cre11.g467350 | Acyl-CoA oxidase | 37.8 | 3.7 | 0.6 | 27.0 | 4.5 | 1.8 | 25.4 | 5.1 | 1.2 | 32.6 | 4.8 | 2.2 | |
| Cre12.g557750 | DGTT1 | Diacylglycerol O-acyltransferase | 2.0 | 1.0 | 2.8 | 1.9 | 1.3 | 3.2 | 1.4 | 0.7 | 4.0 | 1.0 | 1.3 | 7.0 |
| Cre12.g498750 | LIP2 | TAG lipase | 1.7 | 1.7 | 4.4 | 2.2 | 3.8 | 6.6 | 2.1 | 4.7 | 5.2 | 2.1 | 4.0 | 6.1 |
| Cre17.g698600 | LIP3 | TAG lipase | 1.1 | 1.3 | 4.7 | 1.2 | 2.1 | 6.8 | 1.7 | 1.2 | 1.8 | 1.7 | 0.8 | 2.6 |
| Cre03.g193500 | PGD1 | Galactoglycerolipid lipase | 2.5 | 1.1 | 1.6 | 3.1 | 2.5 | 4.2 | 4.2 | 2.8 | 2.0 | 5.1 | 2.3 | 3.0 |
| Cre11.g480250 | TAG lipase | 6.1 | 2.9 | 1.5 | 8.3 | 2.9 | 2.5 | 4.9 | 2.7 | 2.8 | 6.4 | 2.9 | 3.8 | |
| Cre06.g287000 | Caleosin-related protein | 2.3 | 1.5 | 2.8 | 0.2 | 2.0 | 20.7 | 0.7 | 1.0 | 10.0 | 0.7 | 0.5 | 6.9 | |
| Cre13.g590500 | DES6 | Plastid ω6 FA desaturase | 160.3 | 1.0 | 13.7 | 476.2 | 1.1 | 4.2 | 908.6 | 1.2 | 2.2 | 883.3 | 1.2 | 2.3 |
| Cre17.g701700 | FAB2 | Plastid Δ9 stearate desaturase | 98.4 | 0.7 | 6.8 | 92.1 | 0.8 | 3.8 | 120.1 | 0.8 | 2.1 | 118.3 | 0.8 | 1.1 |
| Cre09.g397250 | FAD5A | MGDG-specific palmitateΔ7 desaturase | 52.2 | 0.2 | 5.0 | 63.7 | 0.2 | 2.0 | 74.2 | 0.2 | 1.0 | 87.5 | 0.3 | 0.7 |
| Cre01.g037700 | FAD3 | Putative linoleate desaturase | 40.8 | 0.2 | 2.5 | 43.4 | 0.2 | 1.0 | 60.5 | 0.1 | 1.5 | 58.6 | 0.1 | 0.5 |
| ROS/RNS | ||||||||||||||
| Cre07.g320750 | CYG12 | Heme-NO binding guanylate cyclase | 0.3 | 8.5 | 23.9 | 3.5 | 1.7 | 3.1 | 3.0 | 2.3 | 2.7 | 4.5 | 2.4 | 3.6 |
| Cre07.g320700 | CYG11 | Heme-NO binding guanylate cyclase | 0.3 | 17.5 | 0.7 | 3.9 | 2.1 | 1.5 | 1.2 | 3.5 | 1.3 | 1.4 | 5.7 | 2.5 |
| Cre11.g474800 | OTA1 | Orn transaminase | 17.5 | 1.5 | 6.6 | 18.5 | 1.5 | 1.5 | 27.2 | 1.7 | 1.3 | 25.2 | 1.8 | 1.0 |
| Cre16.g648350 | Pro oxidase | 3.7 | 6.8 | 2.1 | 4.1 | 3.1 | 4.8 | 7.1 | 2.4 | 1.4 | 6.0 | 1.5 | 2.8 | |
| Cre05.g232150 | GDH2 | Glu dehydrogenase | 26.8 | 2.6 | 1.8 | 22.3 | 3.7 | 10.0 | 31.1 | 3.7 | 2.4 | 37.5 | 3.5 | 5.4 |
| Cre09.g389250 | AOF1 | Flavin-containing amine oxidase | 29.6 | 0.3 | 5.8 | 18.0 | 0.2 | 2.1 | 17.6 | 0.2 | 1.8 | 17.6 | 0.2 | 0.8 |
| Cre10.g447750 | AOF2 | Flavin-containing amine oxidase | 3.5 | 3.1 | 0.6 | 1.7 | 4.2 | 3.2 | 3.4 | 3.7 | 1.1 | 3.4 | 3.5 | 2.5 |
| Cre01.g000650 | AMX2 | Copper amine oxidase | 0.7 | 2.2 | 4.1 | 1.0 | 3.8 | 8.8 | 1.6 | 2.8 | 5.2 | 1.0 | 2.3 | 14.0 |
| Cre08.g360550 | ERM3 | NO-forming nitrite reductase | 1.1 | 6.5 | 1.6 | 3.7 | 2.5 | 0.8 | 5.4 | 2.6 | 0.5 | 5.2 | 2.8 | 1.1 |
| Cre01.g007950 | CYP55B1 | Cytochrome P450, putative NO reductase | 5.6 | 7.3 | 1.8 | 1.2 | 4.0 | 6.1 | 2.1 | 3.9 | 2.1 | 1.2 | 3.2 | 2.8 |
| Cre16.g661200 | THB8 | Truncated hemoglobin | 0.01 | 2.9 | 1690 | 0 | 11.8 | 837.5 | 0 | 48.2 | 1295 | 0 | 56.1 | 4926 |
| Cre16.g661000 | THB7 | Truncated hemoglobin | 2.3 | 3.2 | 9.9 | 3.0 | 3.1 | 8.2 | 2.5 | 5.2 | 6.6 | 8.8 | 3.3 | 4.7 |
| Cre16.g663000 | THB12 | Truncated hemoglobin | 3.1 | 4.1 | 3.5 | 2.1 | 3.3 | 4.3 | 2.0 | 4.8 | 2.3 | 3.1 | 4.2 | 2.9 |
| Cre03.g188400 | RBO2 | Respiratory burst oxidase | 3.4 | 3.2 | 7.7 | 1.8 | 2.1 | 1.7 | 0.9 | 2.8 | 1.2 | 0.6 | 3.9 | 2.7 |
| Cre01.g010500 | Mitochondrial ROS modulator 1-like | 25.5 | 0.7 | 0.3 | 20.9 | 0.7 | 0.3 | 30.2 | 0.4 | 0.3 | 25.2 | 0.5 | 0.2 | |
| Cre07.g346100 | Endonuclease V | 4.4 | 5.7 | 1.0 | 2.7 | 1.9 | 1.5 | 5.3 | 1.3 | 1.0 | 3.2 | 0.7 | 0.6 | |
Genes shown are discussed in the text and in the order of appearance. Blank spaces in second column indicate unnamed genes. Locus IDs are from the C. reinhardtii v4.3 genome annotation on Phytozome v8.0. Functional annotations were manually curated when required, as described in the Supplemental Methods 1 online. Estimated expression levels (in units of RPKM) in the illuminated and aerated precultures (0 h) as well as fold changes after 0.5 and 6 h of dark-anoxic incubation relative to the 0-h time point are shown for each clone #1 of the analyzed strains. CRR1 stands for the wild-type CC-124. When a control expression value is depicted as zero, the transcript was not detectable at the sequencing level used in this work. In such cases, the fold changes were computed after imputation of missing values. Supplemental Data Set 1 (T1 to 13) online shows expression values for all samples and FDRs for all comparisons.
Decreased Photosynthetic Activity
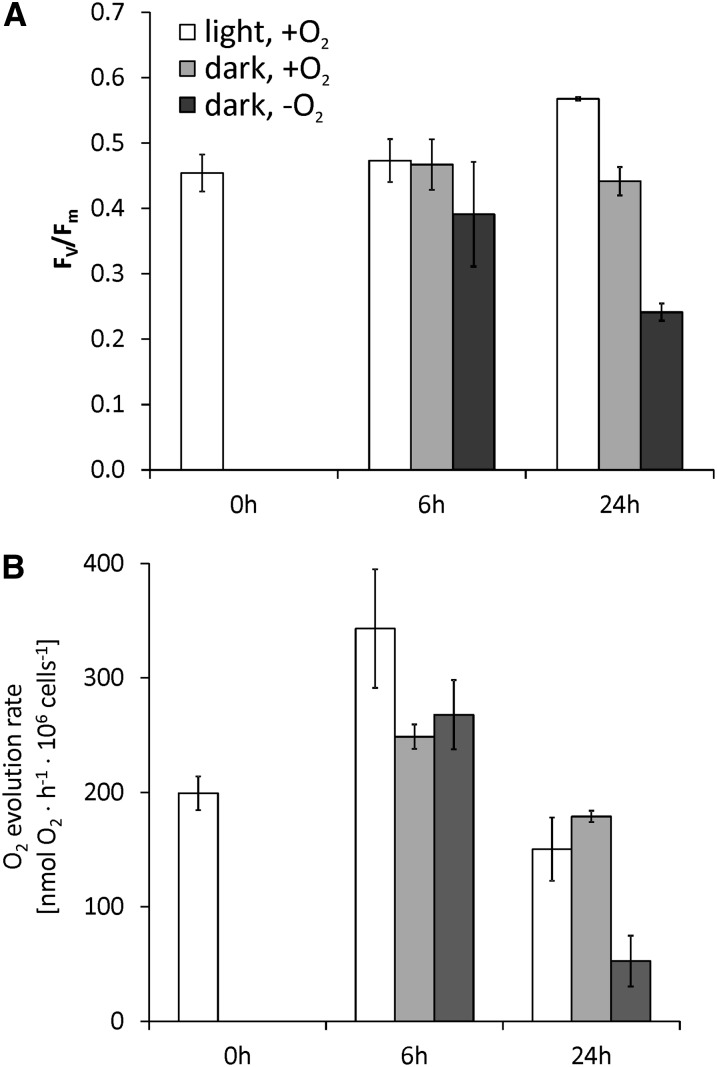
The amounts of various transcripts related to photosynthesis and the carbon concentrating mechanism decreased in the dark-anoxic C. reinhardtii strains (see Supplemental Data Set 1, T6, online). In total, 20 transcripts encoding light-harvesting or other chlorophyll binding proteins were reduced in abundance. Also, several mRNAs involved in tetrapyrrole and chlorophyll biosynthesis were downregulated especially at the early time point, such as CHLH1, encoding Mg-chelatase H subunit, or GUN4, coding for a regulatory subunit of Mg-chelatase involved in the regulation of chlorophyll biosynthesis and plastid-to-nucleus signaling (Formighieri et al., 2012) (Table 2). The general downregulation of genes associated with chlorophyll binding or biosynthesis is supported by literature showing that dark-grown C. reinhardtii cells have lower chlorophyll contents (Chekounova et al., 2001; Duanmu et al., 2013) and by our own analyses of dark-anoxic CC-124 and crr1:CRR1 cultures (see Supplemental Figure 4B online). We analyzed if the reduced abundances of transcripts related to photosynthesis would be reflected by the status of the photosynthetic apparatus. PSII pulse amplitude modulated chlorophyll fluorescence measurements (Baker, 2008) showed that the maximum PSII quantum efficiency Fv/Fm did not decrease significantly after 6 h of self-anaerobization. However, after 24 h, Fv/Fm of dark-anoxic cells was over 50% reduced compared with cells cultivated in standard growth conditions, while algae incubated under oxic conditions in the dark did only show a 20% decrease (Figure 5A). The lowered PSII quantum yield was reflected by similarly decreased photosynthetic O2 evolution rates (Figure 5B).
Figure 5.
Impact of Light and Oxygen on Photosynthetic Parameters of the C. reinhardtii Wild Type.
(A) Maximum PSII quantum efficiency determined in culture aliquots incubated in the measuring cuvette in the dark for 15 min.
(B) O2 evolution rates determined from the same cultures analyzed in (A) prior to the chlorophyll fluorescence measurements.
Cultures had a cell density of 3 × 106 cells mL−1 at 0 h. Values of three independent experiments are shown. Error bars = sd.
There were few photosynthesis-related transcripts whose amounts increased, among those CRR1 targets known to accumulate in anaerobiosis (CPX1, CRD1, and CYC6) and transcripts encoding a putative chlorophyll b reductase (NON-YELLOW COLORING1 [NYC1]) and a possible chlorophyllide-a oxygenase (Cre05.g231450).
Activation of Heterotrophic Energy Generation
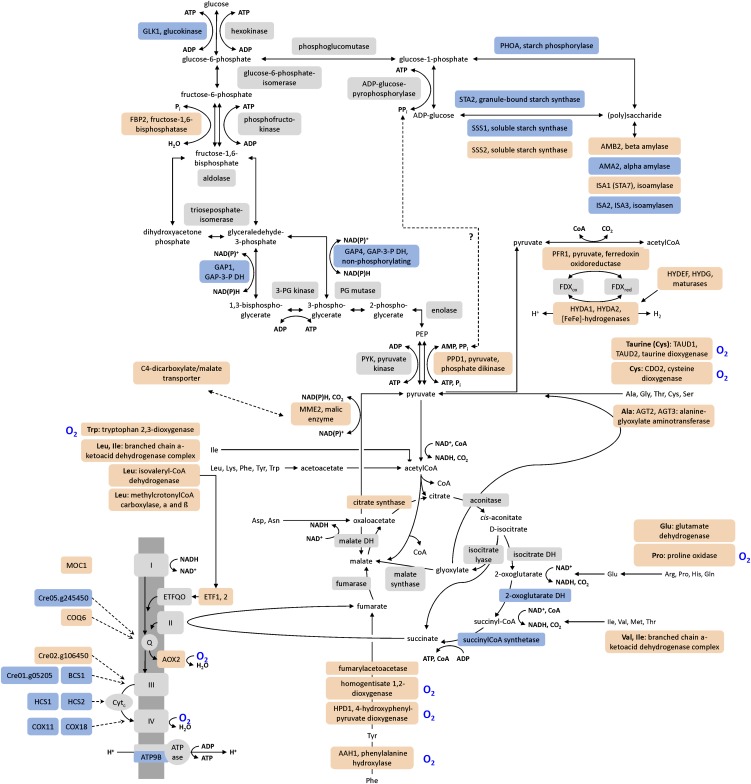
Starch is degraded in anaerobic C. reinhardtii cells and, therefore, the most likely substrate for glycolysis and fermentation (Gfeller and Gibbs, 1984; Kreuzberg, 1984). In the strains analyzed here, four transcripts related to starch metabolism (encoding soluble starch synthase II [SSS2], isoamylase [ISA1/STA7], β-amylase [AMYB2], and a putative α-glucosidase [AGL1]) accumulated (see Supplemental Data Set 1, T7, online). Transcripts associated with glycolysis were not significantly affected, except GLK1, encoding glucokinase (Table 2). However, PPD1 and MME2, encoding Pyruvate phosphate dikinase1 and NADP malic enzyme2, possibly involved in gluconeogenesis, accumulated (Table 2).
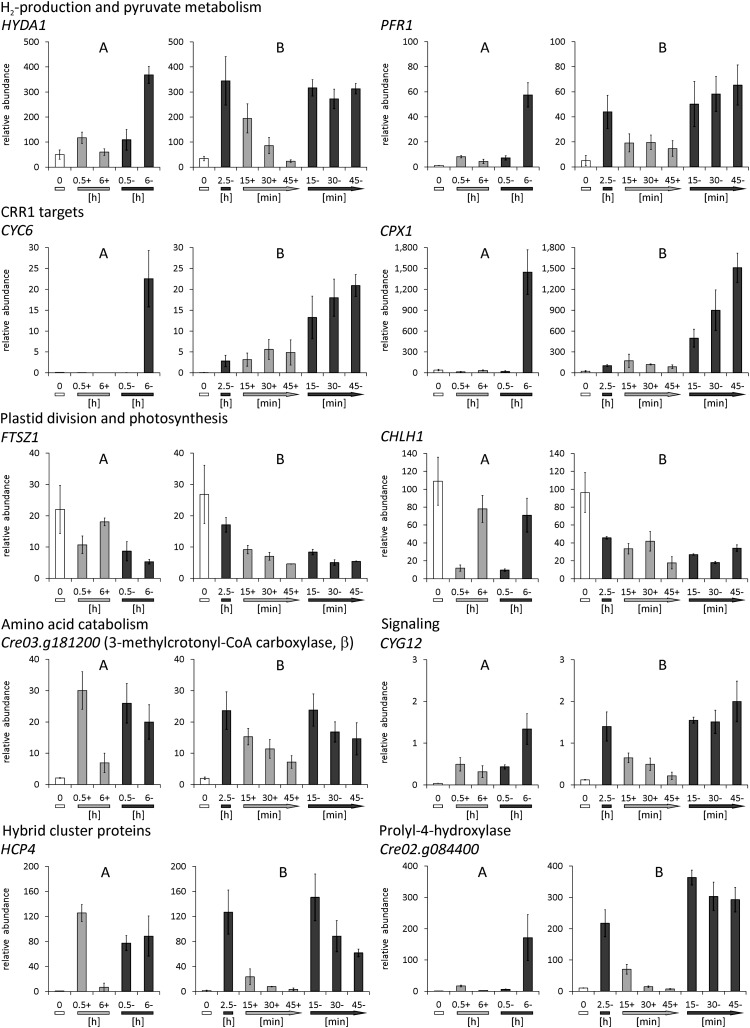
Especially at the early time point (0.5 h), the amounts of transcripts encoding amino acid catabolic enzymes increased (see Supplemental Data Set 1, T8, online; Figure 6). For example, in all C. reinhardtii strains, transcripts involved in the degradation of branched-chain amino acids were upregulated (Table 2). These were Cre04.g228350, Cre12.g539900, and Cre06.g311050, encoding the lipoamide acyltransferase component and the α- and β-subunits of the E1 component of 3-methyl-2-oxobutanate dehydrogenase (branched-chain keto acid dehydrogenase), as well as Cre06.g296400, coding for a putative isovaleryl-CoA dehydrogenase (IVD), MCC1, and Cre03.g181200, encoding the α- and β-subunits of methylcrotonoyl-CoA carboxylase (Table 2, Figure 6). Also, ETF1 and ETF2 mRNAs accumulated in all strains transiently in the 0.5-h samples (Table 2). ETF1 and ETF2 encode the α- and β-subunits of electron-transfer flavoprotein (ETF). ETF accepts electrons from mitochondrial dehydrogenases and transfers them to electron-transfer flavoprotein:ubiquinone oxidoreductase (ETFQO). Thereby, the ETF/ETFQO system allows cells to use electrons derived from diverse catabolic pathways for respiratory energy generation.
Figure 6.
Regulation of Genes Involved in Energy-Generating Pathways.
The figure summarizes pathways employed for energy generation and indicates proteins whose transcript amounts increased (red), decreased (blue), or stayed unchanged (light gray) in anoxic C. reinhardtii CC-124 (CRR1) cells. O2-dependent enzymes are marked by a blue O2 label. The accumulation of various transcripts involved in amino acid catabolism indicated that, in parallel to starch, the cells used amino acids as a substrate for heterotrophic ATP synthesis, possibly via gluconeogenesis and the glyoxylate cycle.
Anaerobic C. reinhardtii Cells Degrade Fatty Acids and Accumulate Triacylglycerol
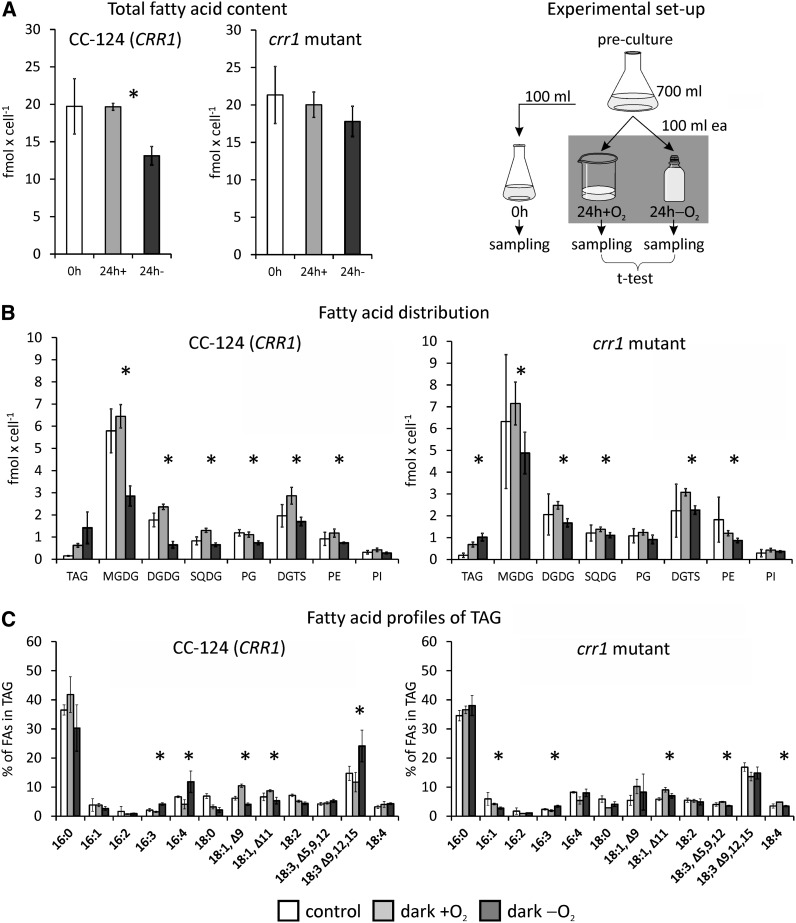
The early and transient accumulation of transcripts encoding four acyl-CoA oxidases and two putative peroxisomal biogenesis factors (PEX11) (Cre12.g540500 and Cre06.g263300) in all strains pointed to a degradation of fatty acids (FAs) (Table 2; see Supplemental Data Set 1, T4 and T9, online). Furthermore, the amount of several transcripts related to FA, membrane lipid, or triacylglycerol (TAG) metabolism changed (Table 2; see Supplemental Data Set 1, T9, online). Therefore, we examined the FA and lipid profiles of the C. reinhardtii wild type (CC-124) and the crr1 mutant after 24 h of incubation under aerobic or anaerobic conditions in the dark. The amount of total FAs per cell decreased by ∼30% in the wild type and by 16% in the crr1 mutant incubated in sealed flasks. By contrast, no (minus 0.34% in strain CC-124) or smaller (minus 6% in the mutant) changes of the total FA content were observed in aerobically incubated cultures (Figure 7A). The lipid analysis also showed changes in the profiles of lipid classes. Amounts of the plastid membrane lipids monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG) decreased in anaerobic cells, while TAG accumulated (Figure 7B). In both C. reinhardtii wild type and crr1 mutants, the TAG content increased approximately fourfold in dark-aerobic versus illuminated and aerobic cells (from 0.15 ± 0.02 to 0.63 ± 0.08 fmol cell−1 in the wild type and from 0.19 ± 0.10 to 0.68 ± 0.11 fmol cell−1 in the mutant). In anaerobic cultures, the TAG amounts reached 1.42 ± 0.71 and 1.03 ± 0.17 fmol cell−1 in strains CC-124 and crr1, respectively. Enhanced TAG amounts were paralleled by increased abundances of DGTT1 transcript, encoding a type-2 diacylglycerol acyltransferase, Cre06.g287000, encoding a caleosin-like (calcium binding lipid body) protein, as well as LIPG2, LIPG3, CGLD15/PGD1, and Cre11.g480250 putative lipase transcripts (Table 2). Notably, the TAGs in dark-anaerobic wild-type cells were enriched in 16:4 and 18:3Δ9,12,15 (Figure 7C). Both are the main FAs associated with MGDG, and 18:3Δ9,12,15 is also a major FA species esterified to DGDG (Giroud et al., 1988) (see Supplemental Figure 5 online).
Figure 7.
Profiles of FAs and Lipids.
The total content of FAs (A), the amounts of FAs associated with TAG or membrane lipids (B), and profile of FA species associated with TAG (C) were analyzed in C. reinhardtii wild-type CC-124 (CRR1) and crr1 mutant cells grown aerated in the light (white bars) and then transferred to open beakers (light gray bars, dark +O2) or sealed flasks (dark gray bars, dark −O2) in the dark for 24 h. Values shown are the averages of biological triplicates. Error bars indicate sd. Asterisks indicate that the difference between samples incubated anaerobically versus aerobically in the dark was significant (P value < 0.05). SQDG, sulfoquinovosyldiacylglycerol; PG, phosphatidylglycerol; DGTS, diacylglyceryl-N,N,N-trimethylhomoserine; PE, phosphatidylethanolamine; PI, phosphatidylinositol.
Several FA desaturase transcripts accumulated to a greater extent in the 6-h samples of the dark-anoxic C. reinhardtii wild type, specifically DES6, FAB2, FAD5A, and FAD3 (Table 2). Except FAD3, these transcripts also increase in Cu-deprived C. reinhardtii cultures, and FAB2 and FAD5A were described as CRR1 targets (Castruita et al., 2011). Despite the changed abundances of FA desaturase encoding transcripts, the desaturation status of FAs did not change significantly in anaerobic algae (see Supplemental Figure 5 online).
Transcripts Involved in ROS/RNS-Based Signaling
The functional category for genes involved in regulation and signaling was one of the largest. This was not unexpected, as the transition to anaerobiosis in the dark requires drastic metabolic rearrangements in photosynthetic aerobic organisms. In one category, 37 protein kinase encoding transcripts appeared as hypoxia targets (see Supplemental Data Set 1, T10, online). In C. reinhardtii, components of hypoxia-specific signal transduction pathways are largely unknown, except for the transcription factor CRR1. A recent study reported nine homologs of the PAS domain/heme based O2 sensor of N2-fixing rhizobia, FixL, in C. reinhardtii (Murthy et al., 2012). In the anoxic algae analyzed here, six transcripts encoding PAS domain–containing transmembrane proteins accumulated, among them Cre13.g586700, which was already identified by Mus et al. (2007), as an anoxia target, and Cre08.g373200, which was named FXL5 and shown to bind heme and O2 by Murthy et al. (2012) (see Supplemental Data Set 1, T10, online). These proteins might therefore be candidates for O2 sensors in C. reinhardtii.
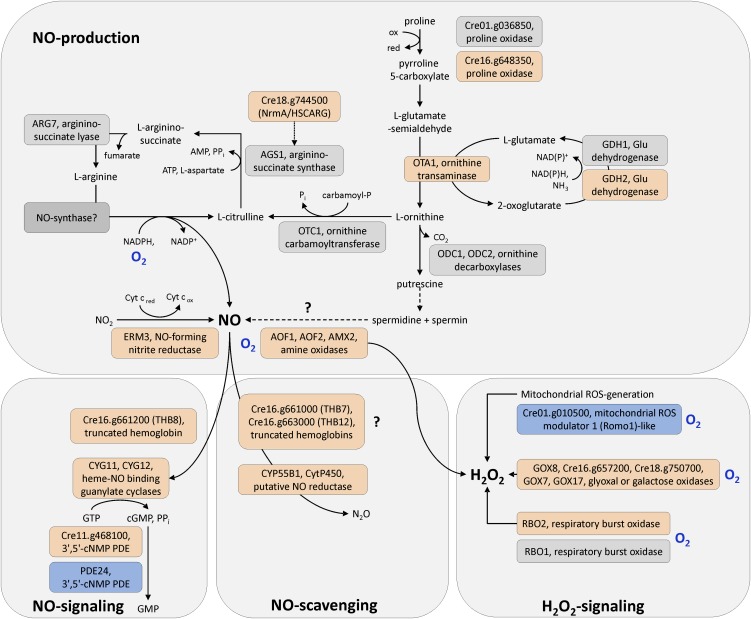
As one outcome of this study, we recently showed that nitric oxide (NO) plays a role in hypoxic growth and gene expression in the microalga (Hemschemeier et al., 2013). Especially the accumulation of two transcripts encoding heme/NO binding guanylate cyclases, CYG11 and CYG12 (Table 2, Figure 8), was an indicator for NO as a signaling molecule in anoxic C. reinhardtii cells. These enzymes, also called soluble guanylate cyclases, mediate NO signals in mammals by generating cGMP upon NO binding to their heme groups (Koesling et al., 2004). In the strains analyzed here, CYG11 mRNA abundance increased predominantly at the early time point (0.5 h), while CYG12 was upregulated early and increased to higher amounts in the 6-h samples.
Figure 8.
ROS/NO Production, Signaling, and Detoxification Pathways.
Upregulated transcripts and the encoded proteins, respectively, are shown in red, and downregulated transcripts (proteins) are colored in blue. Not significantly regulated transcripts (enzymes) are indicated by gray boxes. Blue O2 labels mark O2-dependent steps. Question marks indicate uncertain pathways.
In animals, NO is generated from Arg during the NO-Arg cycle (Bryan et al., 2009). In our data, OTA1, Cre16.g648350, and GDH2 appeared as hypoxia targets (Table 2). These transcripts encode Orn aminotransferase, Pro oxidase, and Glu dehydrogenase, respectively, which generate Orn or deliver intermediates of the Orn biosynthesis pathway (Figure 8). Orn is a precursor of citrulline, which is part of the NO-Arg cycle.
Further candidates that, according to parallels in the literature, might be involved in NO generation were the flavin-containing amine oxidases AOF1 and AOF2 and the Cu-amine oxidase AMX2, whose transcripts accumulated (Table 2, Figure 8). Amine oxidases were shown to be involved in polyamine induced NO formation in Arabidopsis thaliana (Wimalasekera et al., 2011). Another NO producer might be Cre08.g360550, a homolog of fungal Cu-containing denitrifying or respiratory nitrite reductases (Kim et al., 2009). Cre08.g360550 was upregulated in all 0.5-h samples (Table 2, Figure 8). Notably, also a transcript encoding a cytochrome P450 protein homologous to NO reductases (P450nor), Cre01.g007950 (CYP55B1), accumulated (Table 2, Figure 8).
A specific downstream target of the NO-based signaling cascade could not be identified. However, a novel component of the regulatory network is a truncated hemoglobin (2/2Hb) encoded by transcript Cre16.g661200, which accumulated 103-fold from hardly detectable levels in the noninduced state (Table 2, Figure 8) and which we termed TRUNCATED HEMOGLOBIN8 (THB8; Hemschemeier et al., 2013). Two additional 2/2Hb transcripts accumulated in all strains, namely, Cre16.g661000 (THB7) and Cre16.g663000 (THB12), but the fold changes were much lower (fivefold to 10-fold) (Table 2).
Besides NO, reactive oxygen species (ROS) are important second messengers of the response to hypoxia in plants (Bailey-Serres and Chang, 2005; Blokhina and Fagerstedt, 2010; Licausi, 2011) and mammals (Brüne and Zhou, 2003; Fuchs et al., 2010; Ho et al., 2012). Our transcript profiles indicated that ROS might play a role in dark-anoxic C. reinhardtii cultures (Figure 8) (see Supplemental Data Set 1, T11, online). The RBO2 transcript, which encodes one of two isoforms of respiratory burst oxidases in C. reinhardtii, accumulated after 0.5 h (Table 2). Additionally, amounts of transcript Cre01.g010500, encoding a mitochondrial ROS modulator 1 (Romo 1)-like transmembrane protein, decreased three- to fourfold (Table 2, Figure 8). Patterns of several transcripts in anoxic C. reinhardtii strains indicated that the cells activated stress defense systems. For example, an endonuclease V encoding transcript was upregulated (Table 2). The latter protects E. coli specifically against DNA- damage caused by NO and nitrous acid (Schouten and Weiss, 1999; Weiss, 2006). Also, four hybrid cluster/prismane protein (HCP) encoding transcripts (HCP1-HCP4) accumulated in the anoxic algae (see Supplemental Data Set 1, T12, online) as has been shown before (Mus et al., 2007). HCP2 and HCP3 are also upregulated in Cu-deficient cells (Castruita et al., 2011). E. coli HCP has peroxidase activity and protects the cells from oxidative stress (Almeida et al., 2006). It might also be involved in NO defense, together with respiratory nitrite reductase (Filenko et al., 2007).
Darkness as Regulatory Stimulus
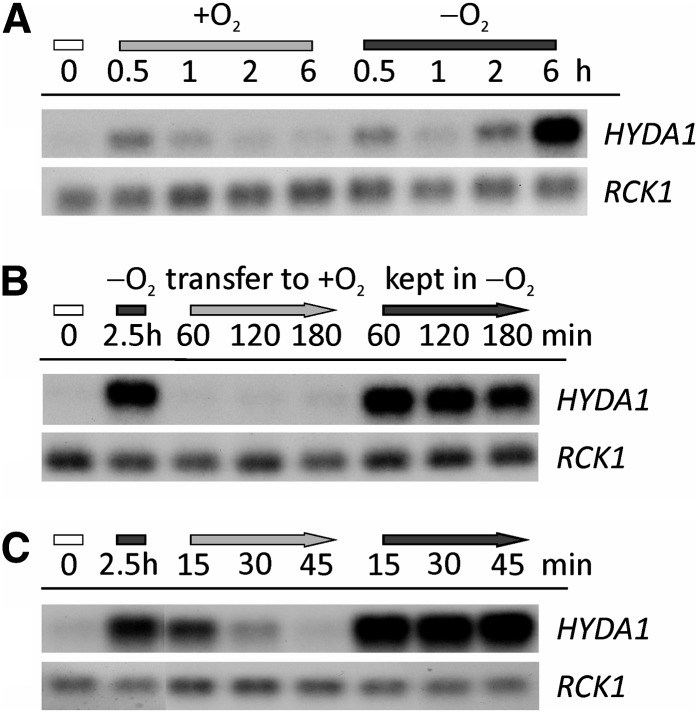
In Arabidopsis, transcriptional and posttranscriptional responses to darkness versus O2 deprivation overlap significantly (Juntawong and Bailey-Serres, 2012). We tested the effect of a light-dark shift on transcripts selected from several functional groups described above by two types of control experiments. First, light-grown cultures of the C. reinhardtii wild-type CC-124 were transferred to open beakers in the dark and stirred well to ensure efficient aeration (dark-oxic incubation) (see Supplemental Figure 6A online for a scheme of the experimental setup). Second, cells were incubated in sealed flasks in the dark for 2.5 h to establish anaerobiosis before transferring these cells to open beakers in the dark (reaeration) (see Supplemental Figure 6B online). In both cases, control cells were also kept in sealed flasks in the dark. RNA was isolated after various time points and analyzed by HYDA1 RNA hybridization (Figure 9) and qRT-PCR analyses. The HYDA1 RNA hybridizations showed that HYDA1 transcript amounts moderately increased after 0.5 h also in cells incubated in open beakers but the signal decreased afterwards (Figure 9A). This pattern was confirmed by qRT-PCR analyses on the same samples (Figure 10). Upon reaeration, HYDA1 transcript levels in anaerobic C. reinhardtii cells decreased rapidly: After 60 min, HYDA1 levels were as low as in the control sample (0 h) (Figure 9B). In a refined setup, HYDA1 amounts decreased already after 15 min of reaeration (Figures 9C and 10). A similar pattern was observed analyzing other transcripts related to H2 production, HYDA2, HYDEF, and HYDG (the latter two encoding specific [FeFe]-hydrogenase maturases; Posewitz et al., 2004) (see Supplemental Figure 7 online) and cDNAs involved in pyruvate metabolism, PFR1 (Figure 10) and PPD1 (see Supplemental Figure 7 online). In case of known or putative CRR1 targets, no or only a moderate increase of transcript amounts was observed in dark-oxic cells. However, the abundance of all analyzed CRR1 targets decreased upon reaeration, though not as fast as HYDA1 (see CYC6 and CPX1 in Figure 10 or Cre12.g512400 and FDX5 in Supplemental Figure 7 online).
Figure 9.
Impact of O2 on HYDA1 Transcript in the Dark.
(A) Cells were incubated for the indicated time points in open (+O2, light-gray bar) or sealed (−O2, dark-gray bar) flasks in the dark. Cell concentrations at 0 h (white bar) were 2.2 × 106 cells mL−1.
(B) and (C) Light-grown cells (0 h, white bar) were incubated for 2.5 h in sealed flasks in the dark (dark-gray bar) and then transferred to open beakers in the dark (+O2, light-gray arrow) or kept in sealed flasks (−O2, dark-gray arrow) for the indicated time points. Cell numbers of the precultures were 2.7 (B) and 2.2 × 106 cells mL−1 (C).
RNA was isolated at the depicted time points. HYDA1 RNA hybridization was conducted with a HYDA1-specific probe. RCK1 (CBLP, Cre13.g599400) served as the reference transcript. Supplemental Figures 6A and 6B online provide schemes of the experimental setups.
Figure 10.
Effect of O2 on Transcript Abundances Estimated by qRT-PCR.
(A) Cells were incubated aerobically (+; light-gray bars) or anaerobically (–; dark-gray bars) in the dark (see Supplemental Figure 6A online).
(B) Cell suspensions were first incubated for 2.5 h in sealed flasks in the dark (2.5-; dark-gray bar) and then transferred to open beakers in the dark (+; light gray arrows) or kept in sealed flasks (–; dark gray arrows) (see Supplemental Figure 6B online).
In all cases, RNA was isolated also from precultures grown aerated in the light (0; white bars). Relative transcript abundances were calculated relative to the RCK1 transcript (CBLP, Cre13.g599400). Values shown are from biological triplicates, analyzed in technical triplicates. Error bars = sd.
In contrast with the transcripts described above, cDNAs involved in growth or photosynthesis did not respond differently to dark-oxic or dark-anoxic conditions. Abundances of the transcript encoding Plastid division protein1 (FTSZ1) and Mg-chelatase subunit H (CHLH1) (Figure 10) as well as LHCSR1 (coding for a stress-related chlorophyll a/b binding protein) and CAH1 (carbonic anhydrase) (see Supplemental Figure 7 online) decreased independently from the presence or absence of O2, and they showed no significant response to reaeration.
An intermediate response was observed for transcripts encoding enzymes involved in amino acid catabolism. Cre03.g181200 (coding for methylcrotonoyl-CoA carboxylase, β-subunit) (Figure 10) and Cre06.g296400 (encoding a putative isovaleryl-CoA dehydrogenase) (see Supplemental Figure 7 online) had significantly increased amounts under both dark-oxic and dark-anoxic conditions, but their abundances in anaerobiosis were moderately higher. A similar pattern was observed for ETF1 (electron transfer flavoprotein, α-subunit) (see Supplemental Figure 7 online).
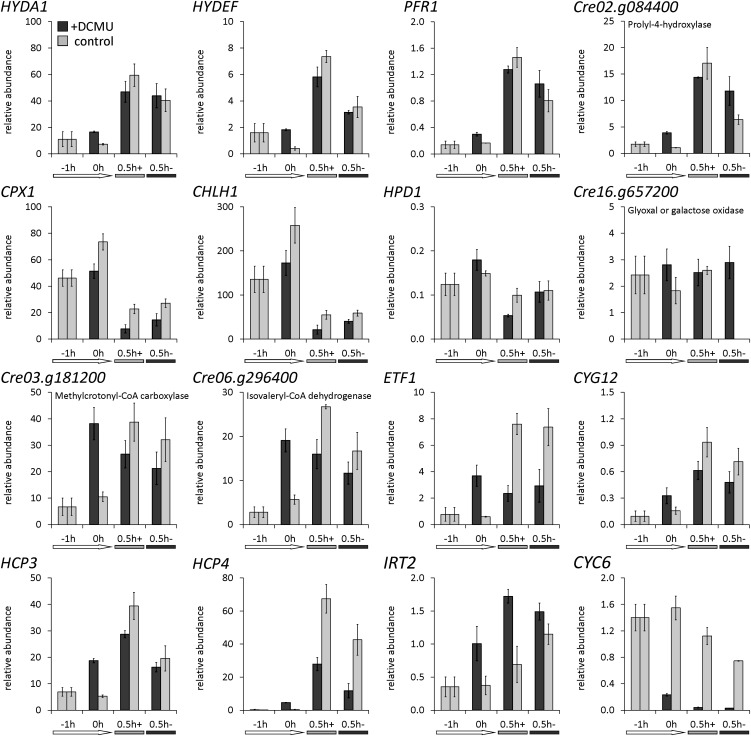
Transcripts from functional categories that we thought were interesting in the context of O2 sensing and signaling, such as CYG12 (heme/NO binding guanylate cyclase) (Figure 10) and two transcripts encoding proteins with putative functions in regulatory steps (Cre16.g668550, RING-type Zn-finger protein; Cre03.g209200, protein with RNA recognition motif) (see Supplemental Figure 7 online) responded moderately to dark-oxic conditions, but increased significantly only in dark-anoxic cells and decreased upon reaeration. Both HCP4 and HCP3 cDNAs, coding for hybrid cluster proteins, accumulated strongly but transiently in dark-oxic C. reinhardtii cells, while their amounts decreased upon longer incubation in the presence of O2 or upon reaeration (Figure 10; see Supplemental Figure 7 online). Selected transcripts encoding O2-dependent enzymes were Cre02.g084400 (P4H) (Figure 10), HPD1 (4-hydroxyphenylpyruvate dioxygenase), and Cre16.g657200 (glyoxal/galactose oxidase) (see Supplemental Figure 7 online). These three transcripts responded only moderately to darkness when air was present and their abundances in anaerobic algal cells decreased when air was reintroduced, confirming that their expression is dependent on O2 rather than darkness.
We examined if the observed effects of darkness were due to the abrupt stop of photosynthetic activity or to a sudden change of intracellular O2 concentrations, respectively. This was done by analyzing transcript abundances in C. reinhardtii cells pretreated with the specific PSII inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) in the light before they were transferred to dark-oxic or dark-anoxic conditions (Figure 11). RNA was first isolated from the algae grown aerated in the light, then from cells incubated for 1 h in the presence of DCMU in the light and finally after 0.5-h dark-oxic or -anoxic conditions (see Supplemental Figure 6C online for a scheme of the experimental setup). Some of the analyzed transcripts did not or not strongly respond to DCMU in the light and still responded to darkness. These were HYDA1, HYDEF, PFR1, and Cre02.g084400 (P4H), which were identified to be upregulated upon the transfer to darkness before, as well as CPX1 and CHLH1, whose amounts decreased in cells incubated in the dark for 0.5 h (Figure 11). Some of the transcripts that had not shown a significant reaction to darkness before were also not influenced by DCMU, such as HPD1 and Cre16.g657200 (glyoxal/galactose oxidase) (Figure 11). The abundances of transcripts associated with amino acid catabolism, by contrast, increased significantly already in the light when photosynthetic O2 evolution was blocked by DCMU: Cre03.g181200 (methylcrotonoyl-CoA carboxylase, β-subunit), Cre06.g296400 (putative isovaleryl-CoA dehydrogenase), and ETF1, and their amounts did not further increase upon the transfer to darkness (Figure 11). An intermediary response was observed for CYG12, HCP3, HCP4, and IRT2, whose amounts were enhanced in illuminated, DCMU-treated cells and increased further upon a transfer to darkness (Figure 11). Notably, the relative amounts of CYC6 (cytochrome c6), which are quite low in C. reinhardtii incubated in the presence of Cu and O2, decreased significantly in DCMU-treated cells and then further when these cells were transferred to darkness (Figure 11).
Figure 11.
Preincubation with DCMU Affects Dark-Induced Transcript Accumulation.
C. reinhardtii wild-type CC-124 was grown under standard conditions in the light until a cell density of 3 × 106 cells mL−1. Then, the culture was split in two and 2.5 µM of the specific photosystem II inhibitor DCMU was added to one aliquot, while an equivalent volume of ethanol was added to the other. After further incubation in the light for 1 h, each cell suspension was transferred to either open beakers (+) or sealed flasks (–) in the dark for 0.5 h. RNA was isolated before the preculture was split (indicated by −1h), after 1-h incubation in the light with (dark-gray bars) or without (light-gray bars) DCMU (0h) and after 0.5 h of incubation in the dark (0.5h+ and 0.5h−). Transcript abundances were calculated relative to the RCK1 transcript (CBLP, Cre13.g599400). Values shown are averages of biological replicates. Error bars = sd. See Supplemental Figure 6C online for a scheme of the experimental setup.
DISCUSSION
We applied RNA-Seq technology to analyze the role of the CRR1 transcription factor and its Cys-rich C terminus in the hypoxic response of C. reinhardtii. Furthermore, we aimed to reveal transcriptional patterns that would allow us to define the strategies that wild-type cells use to deal with limitations in O2. The experimental setup of self-anaerobization was chosen to mimic natural conditions, as one might envision a gradual decrease of O2 concentrations in shallow waters or soil by respiratory activity in the dark. Thereby, this study differed from former microarray-based transcriptome analyses, which examined concentrated cell suspensions resuspended in buffer and purged with argon in the dark (Mus et al., 2007). This difference was reflected in that only ∼40 transcripts indicated as targets in the latter study were also regulated here. Even though both the C. reinhardtii strains (CC-425; cw15, sr-u-60, arg7-8, mt+) used by Mus et al. (2007) and the methodology of the transcriptome analyses differ and prevent a proper comparison between both studies, one can imagine that the microalgae will react quite differently when O2 is removed abruptly. Furthermore, Mus et al. (2007), resuspended the cells in buffer, so that the cells might have also reacted to the absence of nutrients and acetate. By contrast, the system used here was inherently more prone to variability because of its dependence on the metabolic activity of the culture. This can be assessed from the HYDA1 RNA hybridization analyses (Figure 1) and the standard deviations of the qRT-PCR analyses shown in Figures 10 and 11 and Supplemental Figure 7 online. Nevertheless, the chosen setup of self-anaerobization resulted in the expected responses of all known and well-studied hypoxia and CRR1 targets and several of the enriched functional categories reflected physiological acclimation strategies supported by the literature and our own analyses.
Dark-Anoxic C. reinhardtii Cells Downregulate Growth and Photosynthesis
The response of plants, mammals, and bacteria to low-O2 stress has been analyzed by whole-genome approaches before. The public availability of the data sets led to a comparative study evaluating common and plant-specific responses (Mustroph et al., 2010). Conserved responses include the downregulation of genes associated with protein synthesis and oxidative metabolism and the accumulation of transcripts involved in primary carbon metabolism and stress defense (Mustroph et al., 2010). The transcript profiles obtained from the C. reinhardtii strains analyzed here reflected these conserved strategies. The expected patterns included an overall decrease of transcripts involved in cell growth (see Supplemental Data Set 1, T4 and T5, online) and photosynthesis (see Supplemental Data Set 1, T6, online). Both trends were supported by physiological analyses showing that C. reinhardtii cells transferred to dark anoxia grew much slower, had lower chlorophyll contents, and exhibited reduced PSII quantum yields. In view of these data, it can be assumed that the few cell division–related transcripts whose amounts increased exert negative control functions and that the photosynthetic transcripts that accumulated have a role in the acclimation of a photosynthetic organism to prolonged darkness. For example, the increased amounts of transcripts encoding a putative chlorophyll b reductase homologous to NYC1 and a putative chlorophyllide-a oxygenase (Cre05.g231450) in all examined strains indicated an activation of the chlorophyll cycle. This cycle, which interconverts chlorophyll a and chlorophyll b, and the chlorophyll b reductase NYC1 in particular, have been shown to be important regulating factors of chlorophyll and light-harvesting complex degradation during senescence in higher plants (Horie et al., 2009; Sato et al., 2009; Tanaka and Tanaka, 2011).
Amino Acids and FAs Might Contribute to Energy Generation in Anoxia
In agreement with the conserved responses to anaerobiosis defined by Mustroph et al. (2010), the RNA-Seq data of dark-anoxic C. reinhardtii cells showed increased abundances of transcripts related to heterotrophic energy generation and electron disposal (see Supplemental Data Set 1, T7 to T9, online). In the absence of O2, the generation of sufficient energy is a major challenge for aerobes. Our transcriptome data indicated that C. reinhardtii resorts to all available energy sources when transferred to darkness and O2 limitation. The presence of transcript data from an early time point after the transfer to darkness (0.5-h samples) made it possible to distinguish between early and late responses. The early accumulation of transcripts encoding amino acid catabolic enzymes suggests that C. reinhardtii resorts first to amino acid degrading pathways (Figure 6). Especially transcripts related to branched-chain amino acid degradation accumulated, but also transcripts coding for Tyr, Trp, and Phe catabolic enzymes. Together with the parallel and transient upregulation of ETF1 and ETF2 (encoding the α- and β-subunit of electron transfer flavoprotein), this transcription pattern was reminiscent of Arabidopsis, in which genes involved in protein and amino acid degradation are upregulated when the plants are subjected to darkness and/or Suc starvation (Buchanan-Wollaston et al., 2005; Caldana et al., 2011). In Arabidopsis, genetic and metabolic data indicate that the plants oxidatively degrade mainly Leu, Ile, Val, Phe, Trp, and Tyr and transfer the electrons to the mitochondrial electron transport chain via electron-transfer flavoprotein and electron-transfer flavoprotein:ubiquinone oxidoreductase (Ishizaki et al., 2005; Ishizaki et al., 2006).
In Arabidopsis, the catabolism of amino acids was also proposed to fuel the tricarboxylic acid (TCA) cycle (Caldana et al., 2011). In anaerobiosis, a condition marked by low redox potential, feeding a pathway that generates reducing equivalents might be counterproductive. However, the only TCA cycle–related transcripts whose amounts were reduced in the 6-h samples of most C. reinhardtii strains analyzed here were OGD1 and OGD2, encoding the E1 and E2 subunits of 2-oxoglutarate dehydrogenase, SCL1 (succinyl-CoA synthetase α-subunit) and CIS2 (peroxisomal citrate synthase) (Figure 6; see Supplemental Data Set 1, T7, online). 2-Oxoglutarate dehydrogenase and succinyl-CoA synthetase are specific for the TCA cycle. Transcripts coding for enzymes shared by the TCA cycle and the glyoxylate cycle were not significantly regulated (Figure 6). It might be proposed that the glyoxylate cycle was employed to convert amino acids to fermentable substrates also after the onset of anaerobiosis. The cycle is active in anaerobic C. reinhardtii cells (Gibbs et al., 1986; Willeford and Gibbs, 1989) and dark-anoxic algae excrete ammonium (Aparicio et al., 1985). The products of amino acid degradation might be used for gluconeogenesis, whose induction was indicated by increased amounts of the pyruvate:phosphate dikinase transcript PPD1 and MME2, a transcript encoding an NADP-dependent malic enzyme. The fermentative enzyme pyruvate:ferredoxin oxidoreductase (PFR1), the transcript of which was the only one involved in pyruvate fermentation which accumulated significantly, might help to maintain redox balance. We showed recently that PFR1 not only oxidizes pyruvate but also oxaloacetate (Noth et al., 2013). PFR1 provides reduced ferredoxin and thereby allows H2 generation by the [FeFe]-hydrogenases. The generation of H2 has a physiological advantage because reducing equivalents are reoxidized upon the generation of a diffusible and nontoxic fermentative end product.
A rather unexpected observation was the early (0.5 h) and transient accumulation of acyl-CoA oxidase transcripts and the degradation of FAs in dark-anoxic C. reinhardtii cells. Acyl-CoA oxidases are O2-dependent enzymes; yet, the decrease of FA content was more pronounced in anaerobic cultures (Figure 7A). Anaerobic oxidation of FAs occurs in bacteria (Campbell et al., 2003) and might be possible in C. reinhardtii.
Dark-Anoxic C. reinhardtii Cells Employ a Distinct Pathway of TAG Biosynthesis
Despite the decrease of total FAs, the dark-anoxic algal cells accumulated TAGs. The amount of TAGs (1 to 2 fmol cell−1) was considerably lower than that observed in N-deprived cells (∼20 fmol cell−1 after 48 h), but in the range of TAG accumulation reported for other nutritional stress conditions, like phosphorous (P) or Zn deficiency (Boyle et al., 2012). Under the conditions applied here, TAG might accumulate as a general reaction to stress (Kropat et al., 2011; Liu and Benning, 2013; Merchant et al., 2012) or, as discussed below, serve as storage for certain FA species.
TAG accumulation was accompanied by increased abundances of DGTT1, encoding diacylglycerol O-acyltransferase. According to its transcriptional upregulation in N-starved C. reinhardtii cells, DGTT1 has been implicated in TAG biosynthesis (Miller et al., 2010; Boyle et al., 2012). DGTT1 transcription is enhanced in several nutritional stress conditions (-Fe, -Zn, -S, and -P) in addition to N starvation (Boyle et al., 2012), indicating that DGTT1 is involved in stress-triggered TAG accumulation in general. The putative lipases whose transcripts accumulated to various degrees in all examined strains (LIPG2, LIPG3, CGLD15/PGD1, and Cre11.g480250) might be responsible for releasing FAs for oxidative degradation or TAG synthesis. In the case of CGLD15/PGD1, its role in TAG accumulation has been established (Li et al., 2012). LIPG2 and two further putative TAG lipase encoding genes (Cre02.g127300 and Cre07.g350000) are induced upon N starvation and were also discussed to be involved in TAG synthesis (Boyle et al., 2012). Lipid droplet formation in anoxic C. reinhardtii cells might be supported by the caleosin-like (calcium binding lipid-body) protein Cre06.g287000, whose transcript increased threefold to 20-fold. Cre09.g405500, encoding the major lipid droplet protein of the lipid droplet proteome and involved in lipid droplet size regulation (Moellering and Benning, 2010), increased moderately (approximately twofold) in all strains, too (see Supplemental Data Set 3 online).
The overlap between TAG biosynthesis–related transcripts accumulating upon N and O2 deprivation was relatively small. Moreover, both the significantly smaller amounts of TAG produced in dark-anaerobic C. reinhardtii cultures and the profile of FAs associated with TAG suggest that the mechanism of lipid production differs under both conditions. While in TAG in N-deprived cells, the amount of de novo–synthesized FA 18:1Δ9 increases (Boyle et al., 2012), the TAGs in dark-anaerobic wild type cells were enriched in 16:4 and 18:3Δ9,12,15 instead (Figure 7C). Both are the main FAs associated with MGDG, and 18:3Δ9,12,15 is also a major FA species of DGDG (Giroud et al., 1988) (see Supplemental Figure 5 online). In view of the decrease of MGDG and DGDG amounts in the anoxic algae, a redistribution of the polyunsaturated FAs from the major plastid lipids MGDG and DGDG to TAGs seems likely, reflecting disassembly of the photosynthetic machinery. The storage of FAs typically associated with major plastid membranes might be a strategy for the rapid resynthesis of thylakoid membranes upon reillumination.
CRR1 Might Integrate Metal Availability and O2 Sensing
We compared the transcript profiles of C. reinhardtii crr1 mutants to those of the rescued strain because CRR1 is the only transcription factor known so far that is involved in hypoxic gene expression. About 40 genes were misregulated in the mutants, and 20 of these are also CRR1 dependent in Cu deficiency (Castruita et al., 2011). Together with the observation that 83 of all anoxia-responsive transcripts were differently expressed also in Cu deprivation, these results show that the group of genes sensitive to both stimuli and putatively regulated by CRR1 under both conditions is larger than previously realized.
The physiological reason for the close interaction of Cu and O2 limitation signaling networks is still unclear. One hypothesis suggests that, as Cu becomes insoluble under anaerobic conditions, Cu-deficient algae might anticipate hypoxia and vice versa. However, this model could not be confirmed, as hypoxic algae still contain the holo-form of the Cu-containing plastocyanin protein, which is a marker for the bioavailable Cu status (Merchant and Bogorad, 1986; Quinn et al., 2002). Still, as the O2 concentration generally influences the bioavailability of metals (Anbar, 2008) and pathways regulating metal homeostasis interact frequently (Merchant et al., 2006), a role of CRR1 and its subdomains, respectively, in governing reactions to stimuli altering metal homeostasis may be suggested.
CRR1 is necessary also for C. reinhardtii to grow in Zn deficiency (Malasarn et al., 2013) and deletion of its C terminus results in extraordinary Zn accumulation and increased transcript abundances of Zn-responsive genes in replete medium (Sommer et al., 2010). CRR1 is also required for growth in low Fe (Eriksson et al., 2004), and this might be attributed to loss of IRT2 expression, encoding a ZIP (zinc/iron permease) family transporter, in the crr1 mutant but not in wild-type cells or the rescued strain (Table 1) (Chen et al., 2008). The concept of anaerobiosis changing mechanisms of metal assimilation and/or the demand for certain metal species in C. reinhardtii may additionally be deduced from the fact that 17 transcripts related to metal transport or distribution changed in the anoxic strains analyzed here (see Supplemental Data Set 1, T3, online).
We noticed that several putative CRR1 targets in anoxic algae encode O2-dependent enzymes, such as CPX1 or CRD1. CRD1 codes for an isoform of the chlorophyll biosynthetic enzyme Mg-protoporphyrin IX monomethyl ester aerobic oxidative cyclase. CRD1 and the isoform CTH1 are reciprocally expressed in Cu or O2 deficiency in a CRR1-dependent manner (Moseley et al., 2000, 2002; Kropat et al., 2005; Allen et al., 2008). CRD1 is the only CRR1 target for which a role under both Cu and O2 deficiency has been demonstrated so far. C. reinhardtii crd1 mutants become chlorotic under both conditions because a functional CRD1 isoform is then vital for chlorophyll biosynthesis and PSI accumulation (Moseley et al., 2000). Thus, though the two O2-dependent enzymes CRD1 and CTH1 have homologous primary structures, they fulfill specialized functions, possibly via different affinities toward O2. It might be speculated that a similar kind of isoform specialization is repeatedly operative in Cu- and O2-deficient C. reinhardtii. The transcripts that were upregulated in the anaerobic cultures analyzed here might well encode O2-dependent enzymes needed in hypoxic or Cu-limited cells in the light.
Our analyses of the C. reinhardtii crr1ΔCys mutant allowed us to examine gene regulatory responses that might depend on the C terminus of CRR1. For example, while the hypoxic induction of the CYC6 gene needs the Cys-rich C terminus (Sommer et al., 2010), the known Cu-responsive CRR1 targets CRD1, CPX1, and CTR1 were upregulated in the crr1ΔCys strains. In contrast with crr1 mutants, the latter were able to grow under hypoxic conditions in the light, showing that the Cys-rich C terminus of CRR1 has no essential role in hypoxic C. reinhardtii cells. This was a further indicator that CRR1 fulfills modulated functions, probably integrating various stimuli related to metal and O2 homeostasis.
Oxygen Sensing via O2-Dependent Enzymes?
A total of 30 transcripts encoding O2-dependent enzymes accumulated in the anoxic C. reinhardtii cells, and 11 of these transcripts are putative CRR1 targets (summarized in Supplemental Data Set 1, T14, online). Among the former were several FA desaturase encoding genes. The upregulation of the desaturase transcripts may represent a mechanism to compensate for the reduced activity of the enzymes when one of the substrates (O2) is present at low concentration. Alternatively, it may be the case that the membranes become more desaturated as part of an acclimation mechanism. In Cu-deficient C. reinhardtii cells, which also accumulate desaturase transcripts, the desaturation level of several FA species indeed increases (Castruita et al., 2011). Our FA profiling of anoxic cells indicates that the former is more likely, demonstrating the importance of thylakoid membrane lipid composition in this organism. The upregulation of transcripts encoding O2-dependent enzymes indicates that the cells sought to maintain O2-dependent biosynthetic pathways, which, in turn, might be a form of O2 sensing. For example, both tetrapyrrole and sterol biosynthetic pathways include several O2-requiring catalytic steps. In yeast, information on the O2 status is mediated via heme and sterol concentrations (Hon et al., 2003; Todd et al., 2006; Bien and Espenshade, 2010). Another form of sensing the O2 status via enzymatic activity is the posttranscriptional regulation of hypoxia inducible factor 1α (HIF1α), which is central to the hypoxic response of mammals. HIF1α-specific P4Hs hydroxylate HIF1α under normoxic conditions, and hydroxylated HIF1α becomes ubiquitinylated and degraded by the proteasome (Semenza, 2004; Kapitsinou and Haase, 2008). In C. reinhardtii, several P4H encoding transcripts were shown to accumulate under anaerobic conditions (Mus et al., 2007), and we also noted five P4H encoding transcripts whose amounts increased in the anoxic cells analyzed here. Four of the P4H encoding genes appeared to be hypoxic CRR1 targets (Table 1) and are also upregulated dependent on CRR1 in Cu-deficient algae (Castruita et al., 2011).
Is the Photosynthetic Apparatus Involved in Anoxia-Responsive Gene Expression?
The decreased amounts of transcripts associated with photosynthesis in all C. reinhardtii strains and mutants analyzed here indicated a significant influence of the absence of light. This was confirmed by our analyses of control samples of the wild type incubated under dark-oxic conditions or reaerated after anaerobic induction and by the effect of DCMU on various transcripts. Plastid-to-nucleus signaling is an established concept and employs versatile regulatory routes, such as intermediates of tetrapyrrole synthesis, the redox state of the plastoquinone pool, or metabolites (Pfannschmidt et al., 2003; Strand, 2004; Kleine et al., 2009). Most of the originating processes will be affected under dark-anoxic conditions. Transcripts that were selected from different functional categories, such as photosynthesis or fermentation, showed varying patterns in response to darkness or DCMU treatment, indicating that the stimulus for their regulation differed. With regard to O2 sensing, a sudden intracellular drop of O2 concentrations in the absence of photosynthetic O2 evolution might be one of these triggers. It was noticeable, however, that especially transcripts involved in the anaerobic metabolism of C. reinhardtii (HYDA1, HYDEF, and PFR1) were hardly affected by DCMU. By contrast, HYDA1 amounts increased in dark-oxic algae treated with the reducing agent β-mercaptoethanol despite the presence of O2 (see Supplemental Figure 8 online), indicating that reducing conditions may play a role in HYDA1 regulation. Acetate-fed algae show a highly reduced plastoquinone pool in the dark (Wollman and Delepelaire, 1984; Finazzi et al., 1999). By contrast, transcripts associated with amino acid catabolism were strongly affected by DCMU already in illuminated and aerated cell suspensions. As noted above, amino acids are used in Arabidopsis as energy sources in the dark and upon Suc starvation (Buchanan-Wollaston et al., 2005; Caldana et al., 2011). Genes involved in this metabolism might be regulated by plastid-to-nucleus signaling, which conveys some signal of starvation via metabolites (Kleine et al., 2009). In the natural habitats of C. reinhardtii, darkness is probably often accompanied by anaerobiosis or microaerobiosis. It might be speculated that the algae use the status of the photosynthetic apparatus as an early warning system to anticipate anaerobiosis.
NO Plays a Role in the Hypoxic Response of C. reinhardtii
We showed recently that NO is essential for hypoxic growth in C. reinhardtii and that the NO molecule, applied to aerobic cells, can elicit the accumulation of the otherwise anaerobically inducible transcripts HYDA1 and HYDA2 (Hemschemeier et al., 2013). NO is an important second messenger in all organisms. In plants, NO influences various processes, including responses to hypoxia (Dordas et al., 2003). NO generation in C. reinhardtii has not been elucidated yet. In higher plants, one of several NO sources is nitrate reductase (NR) (Blokhina and Fagerstedt, 2010, and references therein), and nitrite- and NR-dependent NO generation occurs in C. reinhardtii (Sakihama et al., 2002). However, the CC-124 wild type, which we used here, is NR deficient. In animals, NO is produced by NO synthase (NOS) (described in Bryan et al., 2009). In C. reinhardtii, no NOS homolog has been detected yet, but NOS activity has been inferred from the inhibition of NO synthesis by the NOS inhibitor l-NAME (de Montaigu et al., 2010). In our RNA-Seq data, we noticed the accumulation of transcripts putatively encoding a NO-forming copper-containing nitrite reductase (Cre08.g360550) and a NO reductase belonging to the cytochrome P450 family. Though speculative, the encoded enzymes might be involved in NO generation and detoxification, given an intracellular source of nitrite. Protection against NO has been reported for NO reductase of Pseudomonas syringae (Kakishima et al., 2007).
The NO signal might be forwarded by one of the six isoforms of heme/NO binding or so-called soluble guanylate cyclases of C. reinhardtii (de Montaigu et al., 2010), of which CYG12 and CYG11 transcripts accumulated in dark-anoxic algae. These enzymes, well studied in mammals (Koesling et al., 2004), indeed play a role in the green alga: The CYG56 isoform of C. reinhardtii as well as NO, cyclic guanosine monophosphate (cGMP), and Ca2+ regulate the ammonium-induced repression of nitrate assimilation genes (de Montaigu et al., 2010). A further component of the NO-based signaling cascade in C. reinhardtii is a 2-on-2-helices or truncated hemoglobin (Wittenberg et al., 2002). Transcript Cre16.g661200, coding for isoform THB8, was one of the most strongly induced transcripts in the dark-anoxic cells examined here, and we analyzed THB8 as a candidate protein important in anoxic acclimation. Algal strains with posttranscriptionally reduced THB8 transcript levels exhibit a severe growth defect under anaerobic conditions in the light (Hemschemeier et al., 2013). The hypoxic growth of the knockdown strains is not restored, but completely abolished by a chemical NO scavenger, and growth of the wild type is strongly impaired. We therefore postulated that the NO binding THB8 protein might be involved in a signaling cascade itself (Hemschemeier et al., 2013). Notably, THB8 is one of 12 putative truncated hemoglobins in the green alga. The function of the remaining 11 proteins remains elusive, and a function of THB7 and THB12, whose transcripts moderately increased in hypoxic cells, too, has to be proven.
In summary, the results we obtained through RNA-Seq and the accompanying physiological analyses gave insights into how C. reinhardtii acclimates to the absence of O2 in the dark. In accordance with known strategies of other organisms, the metabolic response of the algae was obviously aimed at saving, diverting, and generating energy, reflected by reduced growth and photosynthetic activity and the transcriptional activation of pathways of heterotrophic energy production. Several of the transcriptional responses were dependent on CRR1, but a major part of the transcriptional responses was CRR1 independent. Sensing of the O2 status might be accomplished via different routes, such as the activity of O2-dependent biosynthetic pathways, the redox potential, and the status of the photosynthetic apparatus. Regardless of the specific nature of the O2 sensor(s), NO is one of the second messengers involved in the hypoxic response of C. reinhardtii.
METHODS
Strains, Culture Conditions, and Analysis of the Physiological Status of the Cells
The wild type used in this work was Chlamydomonas reinhardtii strain CC-124 mt− [137c] (CRR1 nit1 nit2). CRR1-deficient C. reinhardtii strains were derivatives of CC-3960 crr1-2 arg7 (Eriksson et al., 2004; Kropat et al., 2005) transformed with plasmid pARG7.8 (Debuchy et al., 1989). The complemented strain (crr1:CRR1) was analyzed as the wild type genetically related to the crr1 mutant (Kropat et al., 2005; Castruita et al., 2011). Furthermore, we included a partially complemented strain, crr1ΔCys. This was created by transforming the crr1 mutant with a truncated CRR1 gene encoding a CRR1 protein that lacks the Cys-rich C terminus (amino acids 1127 to 1230) (Sommer et al., 2010). From each strain library, two individual clones, termed #1 and #2, served as replicates.
Liquid cultures of all strains were grown in Tris acetate phosphate (TAP) medium (Harris, 1989) on a rotary shaker (180 rpm) at 24°C and bottom-down illumination of 80 µmol photons m−2 s−1 (warm white light). Cell numbers were determined using a hemocytometer, and the chlorophyll content was analyzed in 80% acetone as reported before (Porra, 2002). Acetate in the medium was quantified in eightfold diluted supernatants of culture aliquots as described previously (Philipps et al., 2011), using a UV test for acetic acid from R-Biopharm. The pH value of the medium was measured in the spent medium. Respiratory and photosynthetic rates of C. reinhardtii cells were determined using the OxyLab system from Hansatech Instruments. The electrode was calibrated setting the O2 content of air saturated water as 100% and by adding sodium dithionite to achieve 0% oxygen. O2 consumption rates of 1-mL cell samples were recorded for 3 min in the dark before measuring O2 evolution rates for 5 min in the light (80 µmol photons m−2 s−1), followed by another 5-min incubation in the dark.
Pulse amplitude modulated chlorophyll fluorescence measurements were conducted using a DUAL-PAM-100 measuring device (Walz). Ground fluorescence F0 was determined in aliquots of C. reinhardtii cultures incubated in the measuring cuvette in the dark for 15 min. The intensity of the measuring beam was 0.2 μmol of photons m−2 s−1. Maximum fluorescence Fm was determined by application of a 600-ms saturating pulse of 10,000 μmol of photons m−2 s−1. Fv/Fm as indicator of the maximum quantum efficiency of PSII was calculated according to the formula Fv/Fm = (Fm − F0)/Fm.
Establishment of Anaerobic Conditions in C. reinhardtii Cultures
Cultures were grown in 700 mL of TAP medium in 2-liter flasks until they reached a cell density of 2.2 to 2.7 × 106 cells mL−1. Then, 100-mL culture aliquots were transferred to square glass bottles sealed with Red Rubber Suba Seals 37 (Sigma-Aldrich), which had a total volume of 118 mL after insertion of the Suba Seals. The flasks were subsequently incubated in a darkroom on magnetic stirrers. Reference cultures were 100-mL culture aliquots removed from the main cultures, transferred to 500-mL Erlenmeyer flasks and placed back to growth conditions until RNA extraction. Dark-aerobic conditions were established by incubating 100 mL of algal suspension in 500-mL glass beakers loosely covered with aluminum foil. When indicated, a stock solution of 25 mM DCMU in ethanol was added to the cultures to reach a final concentration of 2.5 µM.
RNA Isolation and Hybridization
Total RNA was isolated at time points indicated in the text according to the method described before (Hill et al., 1991) with the following modifications: 100 mL of cell suspension was harvested by centrifugation (3440g) at 4°C and resuspended in 1.6 mL of ice-cold lysis buffer (Schmidt et al., 1984) without SDS. After addition of 0.4 mL 10% (w/v) SDS, cells were vigorously vortexed and RNA was extracted and precipitated. RNA integrity was checked visually after formaldehyde gel electrophoresis, by Agilent 2100 Bioanalyzer analysis and by RNA hybridization using a 32P-labeled RCK1- (CBLP-) specific probe derived from plasmid pcf8-13 (Schloss, 1990) by EcoRI digestion. RNA gel electrophoresis and blotting were conducted according to standard techniques (Maniatis et al., 1982) using 10 µg of total RNA. 32P-labeling was done as described before (Feinberg and Vogelstein, 1983, 1984) using [α-32P]-deoxycytidine triphosphate (-dCTP) from Perkin-Elmer. RNA samples were tested for HYDA1 transcript amounts using a probe sequence derived from the 3′-untranslated region and amplified by the following oligonucleotides: 5′-GAGGAGAAGGACGAGAAGAA-3′ and 5′-TGCCAGTACCTACGCCTAA-3′.
DNase Treatment, cDNA Synthesis, and qRT-PCR
DNA was removed from total RNA using TURBO DNase from Ambion/Applied Biosystems. cDNA for qRT-PCR analyses was synthesized using M-MLV reverse transcriptase from Invitrogen according to the manual in the presence of RNase inhibitor (RNasin from Promega) and by priming with oligo(dT18) oligonucleotides. cDNA was diluted 10-fold before use. qRT-PCR reactions performed at UCLA contained 4 μL of cDNA, 6 pmol of each forward and reverse oligonucleotide (see Supplemental Table 2 online), 2 μL Taq-polymerase, 0.5 μL 10 mM deoxynucleotide triphosphate (New England Biolabs), 2 μL 10× Ex Taq buffer (Mg2+ plus) (TaKaRa), 2 μL 10× SYBR mix (0.1% [v/v] SYBR Green 1 Nucleic Acid Gel Stain from Cambrex, 1% [v/v] Tween 20, 1 mg mL−1 BSA, and 50% [v/v] DMSO) in a 20-μL volume. Each sample was analyzed in technical triplicates using DNA Engine Opticon 2 from MJ Research. The PCR program included the following steps: 95°C for 5 min, followed by 40 cycles of 95°C for 15 s, 65°C for 20 s, and 72°C for 20 s. Fluorescence was measured after each cycle at 72°C. A melting curve was performed afterwards from 70 to 95°C with plate reads every 0.5°C. Relative fold changes were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001), and relative abundances were calculated according to Pfaffl (2001). RCK1 (CBLP, Cre13.g599400) served as reference transcript. The qRT-PCR setup at Ruhr University was as described above, except that the peqGOLDTaq DNA polymerase kit (PeqLab) was applied for the PCR reaction using Buffer S and 5× Enhancer solution included in the kit and that extension was performed at 63°C. Comparability of both qRT-PCR setups was checked by comparing the cycle threshold values and fold changes of marker transcripts (HYDA1, HYDEF, PFR1, FDX5, CPX1, and CYC6) obtained at either UCLA or Ruhr University using the same RNA preparations (data not shown). Oligonucleotides are listed in Supplemental Table 2 online.
RNA Sequencing and Data Analysis
Total RNA was treated with DNase as described above and sequenced by the Joint Genome Institute using the Illumina platform. Around 20 million 76-bp reads were obtained per sample. All sequence files are publicly available at the National Center for Biotechnology Information’s Gene Expression Omnibus (accession number GSE42035). Reads were aligned to the Au10.2 transcripts sequences (version 4.3 assembly of the C. reinhardtii genome) using Bowtie (Langmead et al., 2009) in single-end mode and with a maximum tolerance of three mismatches. Expression estimates were obtained for each individual sample in units of RPKM (Mortazavi et al., 2008) after normalization by the number of aligned reads and transcript mappable length (computed for 76-mers) according to Castruita et al. (2011). Expression fold changes were computed from the average between replicates of the same experiment. Differential expression analysis was performed using the DESeq package (Anders and Huber, 2010), and FDRs were estimated in R with the Benjamini-Hochberg correction (Benjamini and Hochberg, 1995). For each pairwise comparison, we selected genes showing a fold regulation greater than or equal to four, while keeping FDR below 5% (see Supplemental Data Set 1, T1 to T13, online). Functional analysis of the data was done by manually annotating the deduced proteins and sorting them according to known or putative functions as described in detail in the Supplemental Methods 1 online. In parallel, GO enrichment analyses were performed using the Algal Functional Annotation Tool at http://pathways.mcdb.ucla.edu/algal/index.html (Lopez et al., 2011). Only biological processes or molecular functions that contained at least two members and a hypergeometric P value <0.05 were considered.
To determine if the expression of a gene is dependent on CRR1 and, if so, if the Cys-rich C terminus is involved, the fold changes after 6-h anoxia in crr1 and crr1ΔCys mutants were compared with those of the rescued strain (crr1:CRR1). From all annotated C. reinhardtii v4.3 gene models, transcripts whose fold changes after 6-h anoxia had an FDR < 0.05 in C. reinhardtii crr1:CRR1 (3157 in total) were used as the basis for the comparisons, irrespective of the extent of the fold change. Transcripts were considered as CRR1 targets when their fold changes after 6-h anoxia were at least fourfold different from those in the rescued strain in each both mutant colonies. The computed lists of putative CRR1 targets were manually curated, taking estimated expression values into account. Especially in crr1ΔCys mutants, many computed targets were due to abnormal RPKM values in the 0-h samples and, therefore, probably unrelated to the anoxic treatment. Note that CYC6 would have been deleted during manual curation because its estimated expression was abnormally high in the control sample of one of the crr1ΔCys colonies and higher than in the rescued strain in the 6-h sample of the same colony (see Supplemental Data Set 2, T1, online). However, CYC6 is one of the strongly regulated genes (∼150-fold upregulated in C. reinhardtii crr1:CRR1), and the twofold downregulation and only threefold upregulation in crr1ΔCys colony #1 and #2, respectively, show a marked misregulation of CYC6 in this mutant type, as has been reported before (Sommer et al., 2010).
Lipid and FA Analysis
Lipid and FA profiles were analyzed from biological triplicates and technical duplicates from wild-type CC-124 and mutant crr1 (clone #1). Samples were taken from control (illuminated and aerated) cultures grown to ∼3 × 106 cells mL−1 and from cells incubated for 24 h in sealed flasks or in open beakers in the dark as described above. For total FA quantification, 2 mL of cell suspension was vacuum filtered on Whatman GF/A 25 mm circles. The filter was transferred to a 2-mL screw cap tube and immediately frozen in liquid nitrogen. For analyzing the relative contents of FAs and lipids, 90 mL of cells was collected by centrifugation at 4°C and the pellet was resuspended in 1.5 mL of ice-cold water. The suspension was transferred to 2-mL tubes and centrifuged at maximum speed for 2 min at 4°C. The cell pellet was frozen in liquid nitrogen. Both filters and cell pellets were lyophilized and stored at −80°C until the analysis. Then, filtered cells were transferred to fatty acid methyl ester (FAME) reaction tubes and subjected to FAME and gas chromatography as described below. Cell pellets were resuspended in 1 mL of methanol:chloroform:formic acid (2:1:0.1 [v/v/v]), followed by adding 0.5 mL of extraction solution (1 M KCl and 0.2 M H3PO4) and vigorous vortexing. The lipid phase was extracted after centrifugation for 3 min at 13,780g. Lipids were developed by thin layer chromatography separation with petroleum ether:diethylether:acetic acid (8:2:0.1 [v/v/v]) as solvent. Lipid samples were spotted on baked Si60 silica plates (EMD Chemicals). After separation, lipids were reversibly stained by exposure to iodine vapor and quantitatively recovered by scraping the corresponding spots with a razor blade and transferring the material to a FAME reaction tube. One milliliter of methanolic HCl (1 n) was added to each FAME reaction tube, followed by loading 5 µg of the internal standard pentadecanoic acid (15:0, 50 µg mL−1 in methanol). The head space was purged with nitrogen gas and the cap was tightly sealed. Samples were incubated at 80°C for 25 min and allowed to cool to room temperature. Aqueous NaCl (0.9%, 1 mL) and n-hexane (1 mL) were added and the samples were vigorously shaken. After centrifugation at 2170g for 3 min, the hexane layer on the top was removed, transferred to a new tube, and dried under nitrogen stream. The resulting FAMEs were dissolved in 100 μL of hexane. FA content and composition of the extracts was determined by gas chromatography–flame ionization detection as described before (Rossak et al., 1997). The capillary DB-23 column (Agilent) was operated as follows: initial temperature 140°C, increased by 25°C min−1 to 160°C, then by 8°C min−1 to 250°C, and held at 250°C for 4 min.
Accession Numbers
Sequence data from this article can be found in the C. reinhardtii v5.3.1 genome assembly at Phytozome v9.1 (http://www.phytozome.net/chlamy) under the accession numbers (locus IDs) provided in Tables 1 and 2, except for the following: g9657 (CRR1; GenBank AAY33924) and Cre01.g044800 (PFL1). The following are deleted from Phytozome v9.1: Cre02.g087073 (unknown), Cre12.g510058 (unknown), and Cre12.g490477 (WD40 domain). The following are renamed in Phytozome v9.1: Cre11.g473950 (PFR1, g1910), Cre44.g787700 (GOX7, g11470), Cre18.g750700 (glyoxal or Gal oxidase, g11468), Cre15.g645100 (squalene epoxidase/monooxygenase, g17770), Cre16.g693050 (SIR2, g15615), Cre02.g137700 (unknown, g9962), Cre18.g750800 (GOX8, g11468), Cre16.g686450 (α/β-hydrolase, g16365), Cre02.g136050 (HPD1, g9907), Cre15.g646150 (FTSZ2, g2757), Cre05.g231450 (putative chlorophyllide-a oxygenase, g18260), Cre04.g228350 (lipoamide acyltransferase of branched-chain α-keto acid dehydrogenase complex, g4996), Cre13.g593500 (MCC1, g6242), Cre03.g181200 (methylcrotonoyl-CoA carboxylase β, g3755), Cre27.g775600 (ETF2, g13818), Cre05.g231950 (acyl-CoA oxidase, C terminal, g5140), Cre16.g648350 (Pro oxidase, g15692), Cre10.g447750 (AOF2, g11012), and Cre02.g129500 (HCP3, g9756).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Correlations of Expression and Fold-Change Values between Replicates.
Supplemental Figure 2. Correlation between Fold-Changes Obtained by RNA-Seq and qRT-PCR.
Supplemental Figure 3. Functional Annotation of Differentially Expressed Genes in the C. reinhardtii Wild Type and in Rescued Strain crr1:CRR1.
Supplemental Figure 4. Cell Numbers and Chlorophyll Content in the Dark.
Supplemental Figure 5. Fatty Acid Profiles of Dark-Incubated C. reinhardtii Cultures.
Supplemental Figure 6. Schemes of Experimental Setups Used to Analyze the Effect of Dark-Aerobic versus Dark-Anaerobic Conditions.
Supplemental Figure 7. Additional qRT-PCR Analyses Examining the Effect of O2 on Transcript Abundances.
Supplemental Figure 8. Effect of β-Mercaptoethanol on HYDA1 Transcript Amounts under Dark-Oxic Conditions.
Supplemental Table 1. Assessment of the Physiological State of Cultures Used for Transcriptome Profiling.
Supplemental Table 2. Oligonucleotides Used for Quantitative Real-Time PCR.
Supplemental Table 3. Fold-Changes of Marker Transcripts under Dark-Anoxic Conditions Analyzed by qRT-PCR.
Supplemental Table 4. Gene Ontology Enrichment of Dark-Anoxia Targets in the C. reinhardtii Wild-Type CC-124 (CRR1) and the Rescued Strain crr1:CRR1 #1.
Supplemental Table 5. Putative CRR1 Targets Showing Higher Fold Changes in the Mutants Compared with the Rescued Strain.
Supplemental Methods 1. Functional Analysis of Gene Products and Categorization Principles.
Supplemental Data Set 1. Tables 1 to 14 (T1 to T14): Anoxia-Responsive Genes Grouped into Functional Categories.
Supplemental Data Set 2. Tables 1 and 2 (T1 and T2): Hypoxic CRR1 Targets and Comparison to Transcriptome Data of Cu-Deficient Cells.
Supplemental Data Set 3. Expression Estimates and Fold Changes of All Chlamydomonas reinhardtii v4.3 Gene Models.
Acknowledgments
RNA samples were sequenced by the U.S. Department of Energy Joint Genome Institute, which is supported by the Office of Science of the U.S. Department of Energy under Contract DE-AC02-05CH11231. A.H. was funded by the Deutsche Forschungsgemeinschaft (He 5790/1-1 and -2 and HE 5790/3-1). The work was further supported by the Deutsches Zentrum für Luft- und Raumfahrt (DLR) (ModuLES to T.H.). D.C. and M.P. are supported by a grant from the National Institutes of Health (GM092473). Research was also supported in part by a grant to C.B. from the U.S. Air Force Office of Scientific Research (FA9550-11-1-0264). S.S.M. acknowledges support from GM42143.
AUTHOR CONTRIBUTIONS
A.H. designed and performed research. T.H. and S.S.M. contributed to the design of the experiments. D.C. provided computational tools and performed the bioinformatic analyses of RNA-Seq data. M.P. supervised the bioinformatic analysis. B.L. performed the lipid analyses, which was supervised by C.B. A.H., T.H., and S.S.M. wrote the article.
Glossary
- PSII
photosystem II
- qRT-PCR
quantitative RT-PCR
- RPKM
reads per kilobase of mappable transcript length per million mapped reads
- FDR
false discovery rate
- GO
gene ontology
- ETF
electron-transfer flavoprotein
- ETFQO
electron-transfer flavoprotein:ubiquinone oxidoreductase
- FA
fatty acid
- TAG
triacylglycerol
- MGDG
monogalactosyldiacylglycerol
- DGDG
digalactosyldiacylglycerol
- NO
nitric oxide
- ROS
reactive oxygen species
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
- TCA
tricarboxylic acid
- P4H
prolyl-4-hydroxylase
- NR
nitrate reductase
- NOS
NO synthase
- TAP
Tris acetate phosphate
- FAME
fatty acid methyl ester
References
- Adams S., Maple J., Møller S.G. (2008). Functional conservation of the MIN plastid division homologues of Chlamydomonas reinhardtii. Planta 227: 1199–1211 [DOI] [PubMed] [Google Scholar]
- Allen M.D., Kropat J., Merchant S.S. (2008). Regulation and localization of isoforms of the aerobic oxidative cyclase in Chlamydomonas reinhardtii. Photochem. Photobiol. 84: 1336–1342 [DOI] [PubMed] [Google Scholar]
- Almeida C.C., Romão C.V., Lindley P.F., Teixeira M., Saraiva L.M. (2006). The role of the hybrid cluster protein in oxidative stress defense. J. Biol. Chem. 281: 32445–32450 [DOI] [PubMed] [Google Scholar]
- Anbar A.D. (2008). Oceans. Elements and evolution. Science 322: 1481–1483 [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio P.J., Azuara M.P., Ballesteros A., Fernández V.M. (1985). Effects of light intensity and oxidized nitrogen sources on hydrogen production by Chlamydomonas reinhardii. Plant Physiol. 78: 803–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atteia A., van Lis R., Gelius-Dietrich G., Adrait A., Garin J., Joyard J., Rolland N., Martin W. (2006). Pyruvate formate-lyase and a novel route of eukaryotic ATP synthesis in Chlamydomonas mitochondria. J. Biol. Chem. 281: 9909–9918 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J., Chang R. (2005). Sensing and signalling in response to oxygen deprivation in plants and other organisms. Ann. Bot. (Lond.) 96: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J., Fukao T., Gibbs D.J., Holdsworth M.J., Lee S.C., Licausi F., Perata P., Voesenek L.A., van Dongen J.T. (2012). Making sense of low oxygen sensing. Trends Plant Sci. 17: 129–138 [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J., Voesenek L.A. (2008). Flooding stress: Acclimations and genetic diversity. Annu. Rev. Plant Biol. 59: 313–339 [DOI] [PubMed] [Google Scholar]
- Baker N.R. (2008). Chlorophyll fluorescence: A probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59: 89–113 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57: 289–300 [Google Scholar]
- Bien C.M., Espenshade P.J. (2010). Sterol regulatory element binding proteins in fungi: Hypoxic transcription factors linked to pathogenesis. Eukaryot. Cell 9: 352–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokhina O., Fagerstedt K.V. (2010). Oxidative metabolism, ROS and NO under oxygen deprivation. Plant Physiol. Biochem. 48: 359–373 [DOI] [PubMed] [Google Scholar]
- Boyle N.R., et al. (2012). Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 287: 15811–15825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüne B., Zhou J. (2003). The role of nitric oxide (NO) in stability regulation of hypoxia inducible factor-1alpha (HIF-1alpha). Curr. Med. Chem. 10: 845–855 [DOI] [PubMed] [Google Scholar]
- Bryan N.S., Bian K., Murad F. (2009). Discovery of the nitric oxide signaling pathway and targets for drug development. Front Biosci (Landmark Ed) 14: 1–18 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P.O., Nam H.G., Lin J.F., Wu S.H., Swidzinski J., Ishizaki K., Leaver C.J. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Caldana C., Degenkolbe T., Cuadros-Inostroza A., Klie S., Sulpice R., Leisse A., Steinhauser D., Fernie A.R., Willmitzer L., Hannah M.A. (2011). High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J. 67: 869–884 [DOI] [PubMed] [Google Scholar]
- Campbell J.W., Morgan-Kiss R.M., Cronan J.E., Jr. (2003). A new Escherichia coli metabolic competency: Growth on fatty acids by a novel anaerobic beta-oxidation pathway. Mol. Microbiol. 47: 793–805 [DOI] [PubMed] [Google Scholar]
- Castruita M., Casero D., Karpowicz S.J., Kropat J., Vieler A., Hsieh S.I., Yan W., Cokus S., Loo J.A., Benning C., Pellegrini M., Merchant S.S. (2011). Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23: 1273–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotti C., Yang W., Posewitz M.C., Grossman A.R. (2013). Fermentation metabolism and its evolution in algae. Front. Plant Sci. 4: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekounova E., Voronetskaya V., Papenbrock J., Grimm B., Beck C.F. (2001). Characterization of Chlamydomonas mutants defective in the H subunit of Mg-chelatase. Mol. Genet. Genomics 266: 363–373 [DOI] [PubMed] [Google Scholar]
- Chen J.C., Hsieh S.I., Kropat J., Merchant S.S. (2008). A ferroxidase encoded by FOX1 contributes to iron assimilation under conditions of poor iron nutrition in Chlamydomonas. Eukaryot. Cell 7: 541–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy R., Purton S., Rochaix J.D. (1989). The argininosuccinate lyase gene of Chlamydomonas reinhardtii: An important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 8: 2803–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Montaigu A., Sanz-Luque E., Galván A., Fernández E. (2010). A soluble guanylate cyclase mediates negative signaling by ammonium on expression of nitrate reductase in Chlamydomonas. Plant Cell 22: 1532–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebbe A., Keck M., La Russa M., Mussgnug J.H., Hankamer B., Tekçe E., Niehaus K., Kruse O. (2010). The interplay of proton, electron, and metabolite supply for photosynthetic H2 production in Chlamydomonas reinhardtii. J. Biol. Chem. 285: 30247–30260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dordas C., Rivoal J., Hill R.D. (2003). Plant haemoglobins, nitric oxide and hypoxic stress. Ann. Bot. (Lond.) 91 (Spec No): 173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu D., Casero D., Dent R.M., Gallaher S., Yang W., Rockwell N.C., Martin S.S., Pellegrini M., Niyogi K.K., Merchant S.S., Grossman A.R., Lagarias J.C. (2013). Retrograde bilin signaling enables Chlamydomonas greening and phototrophic survival. Proc. Natl. Acad. Sci. USA 110: 3621–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M., Moseley J.L., Tottey S., Del Campo J.A., Quinn J., Kim Y., Merchant S. (2004). Genetic dissection of nutritional copper signaling in Chlamydomonas distinguishes regulatory and target genes. Genetics 168: 795–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A.P., Vogelstein B. (1983). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132: 6–13 [DOI] [PubMed] [Google Scholar]
- Feinberg A.P., Vogelstein B. (1984). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Addendum. Anal. Biochem. 137: 266–267 [DOI] [PubMed] [Google Scholar]
- Filenko N., Spiro S., Browning D.F., Squire D., Overton T.W., Cole J., Constantinidou C. (2007). The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. J. Bacteriol. 189: 4410–4417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finazzi G., Furia A., Barbagallo R.P., Forti G. (1999). State transitions, cyclic and linear electron transport and photophosphorylation in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1413: 117–129 [DOI] [PubMed] [Google Scholar]
- Formighieri C., Ceol M., Bonente G., Rochaix J.D., Bassi R. (2012). Retrograde signaling and photoprotection in a gun4 mutant of Chlamydomonas reinhardtii. Mol. Plant 5: 1242–1262 [DOI] [PubMed] [Google Scholar]
- Fuchs B., Sommer N., Dietrich A., Schermuly R.T., Ghofrani H.A., Grimminger F., Seeger W., Gudermann T., Weissmann N. (2010). Redox signaling and reactive oxygen species in hypoxic pulmonary vasoconstriction. Respir. Physiol. Neurobiol. 174: 282–291 [DOI] [PubMed] [Google Scholar]
- Gfeller R.P., Gibbs M. (1984). Fermentative metabolism of Chlamydomonas reinhardtii: I. Analysis of fermentative products from starch in dark and light. Plant Physiol. 75: 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardi M.L., Dubini A., Yu J., Maness P.C. (2009). Photobiological hydrogen-producing systems. Chem. Soc. Rev. 38: 52–61 [DOI] [PubMed] [Google Scholar]
- Gibbs M., Gfeller R.P., Chen C. (1986). Fermentative metabolism of Chlamydomonas reinhardii: III. Photoassimilation of acetate. Plant Physiol. 82: 160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroud C., Gerber A., Eichenberger W. (1988). Lipids of Chlamydomonas reinhardtii - Analysis of molecular species and intracellular site(s) of biosynthesis. Plant Cell Physiol. 29: 587–595 [Google Scholar]
- González-Ballester D., Casero D., Cokus S., Pellegrini M., Merchant S.S., Grossman A.R. (2010). RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 22: 2058–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A. (2000). Acclimation of Chlamydomonas reinhardtii to its nutrient environment. Protist 151: 201–224 [DOI] [PubMed] [Google Scholar]
- Grossman A.R., Croft M., Gladyshev V.N., Merchant S.S., Posewitz M.C., Prochnik S., Spalding M.H. (2007). Novel metabolism in Chlamydomonas through the lens of genomics. Curr. Opin. Plant Biol. 10: 190–198 [DOI] [PubMed] [Google Scholar]
- Happe T., Kaminski A. (2002). Differential regulation of the Fe-hydrogenase during anaerobic adaptation in the green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 269: 1022–1032 [DOI] [PubMed] [Google Scholar]
- Harris, E.H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. (San Diego, CA: Academic Press). [DOI] [PubMed] [Google Scholar]
- Harris E.H. (2001). Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 363–406 [DOI] [PubMed] [Google Scholar]
- Hemschemeier A., Düner M., Casero D., Merchant S.S., Winkler M., Happe T. (2013). Hypoxic survival requires a 2-on-2 hemoglobin in a process involving nitric oxide. Proc. Natl. Acad. Sci. USA 110: 10854–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemschemeier A., Happe T. (2011). Alternative photosynthetic electron transport pathways during anaerobiosis in the green alga Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1807: 919–926 [DOI] [PubMed] [Google Scholar]
- Hemschemeier A., Jacobs J., Happe T. (2008). Biochemical and physiological characterization of the pyruvate formate-lyase Pfl1 of Chlamydomonas reinhardtii, a typically bacterial enzyme in a eukaryotic alga. Eukaryot. Cell 7: 518–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K.L., Li H.H., Singer J., Merchant S. (1991). Isolation and structural characterization of the Chlamydomonas reinhardtii gene for cytochrome c6. Analysis of the kinetics and metal specificity of its copper-responsive expression. J. Biol. Chem. 266: 15060–15067 [PubMed] [Google Scholar]
- Ho J.J., Man H.S., Marsden P.A. (2012). Nitric oxide signaling in hypoxia. J. Mol. Med. 90: 217–231 [DOI] [PubMed] [Google Scholar]
- Hon T., Dodd A., Dirmeier R., Gorman N., Sinclair P.R., Zhang L., Poyton R.O. (2003). A mechanism of oxygen sensing in yeast. Multiple oxygen-responsive steps in the heme biosynthetic pathway affect Hap1 activity. J. Biol. Chem. 278: 50771–50780 [DOI] [PubMed] [Google Scholar]
- Horie Y., Ito H., Kusaba M., Tanaka R., Tanaka A. (2009). Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J. Biol. Chem. 284: 17449–17456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K., Larson T.R., Schauer N., Fernie A.R., Graham I.A., Leaver C.J. (2005). The critical role of Arabidopsis electron-transfer flavoprotein: Ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17: 2587–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K., Schauer N., Larson T.R., Graham I.A., Fernie A.R., Leaver C.J. (2006). The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J. 47: 751–760 [DOI] [PubMed] [Google Scholar]
- Juntawong P., Bailey-Serres J. (2012). Dynamic light regulation of translation status in Arabidopsis thaliana. Front. Plant Sci. 3: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakishima K., Shiratsuchi A., Taoka A., Nakanishi Y., Fukumori Y. (2007). Participation of nitric oxide reductase in survival of Pseudomonas aeruginosa in LPS-activated macrophages. Biochem. Biophys. Res. Commun. 355: 587–591 [DOI] [PubMed] [Google Scholar]
- Kapitsinou P.P., Haase V.H. (2008). The VHL tumor suppressor and HIF: Insights from genetic studies in mice. Cell Death Differ. 15: 650–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.W., Fushinobu S., Zhou S., Wakagi T., Shoun H. (2009). Eukaryotic nirK genes encoding copper-containing nitrite reductase: Originating from the protomitochondrion? Appl. Environ. Microbiol. 75: 2652–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T., Voigt C., Leister D. (2009). Plastid signalling to the nucleus: Messengers still lost in the mists? Trends Genet. 25: 185–192 [DOI] [PubMed] [Google Scholar]
- Koesling D., Russwurm M., Mergia E., Mullershausen F., Friebe A. (2004). Nitric oxide-sensitive guanylyl cyclase: Structure and regulation. Neurochem. Int. 45: 813–819 [DOI] [PubMed] [Google Scholar]
- Kreuzberg K. (1984). Starch fermentation via a formate producing pathway in Chlamydomonas reinhardii, Chlorogonium elongatum and Chlorella fusca. Physiol. Plant. 61: 87–94 [Google Scholar]
- Kropat J., Hong-Hermesdorf A., Casero D., Ent P., Castruita M., Pellegrini M., Merchant S.S., Malasarn D. (2011). A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 66: 770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J., Tottey S., Birkenbihl R.P., Depège N., Huijser P., Merchant S. (2005). A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA 102: 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertz C., Hemschemeier A., Happe T. (2010). Anaerobic expression of the ferredoxin-encoding FDX5 gene of Chlamydomonas reinhardtii is regulated by the Crr1 transcription factor. Eukaryot. Cell 9: 1747–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Moellering E.R., Liu B., Johnny C., Fedewa M., Sears B.B., Kuo M.H., Benning C. (2012). A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 24: 4670–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F. (2011). Regulation of the molecular response to oxygen limitations in plants. New Phytol. 190: 550–555 [DOI] [PubMed] [Google Scholar]
- Liu B., Benning C. (2013). Lipid metabolism in microalgae distinguishes itself. Curr. Opin. Biotechnol. 24: 300–309 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lopez D., Casero D., Cokus S.J., Merchant S.S., Pellegrini M. (2011). Algal Functional Annotation Tool: A web-based analysis suite to functionally interpret large gene lists using integrated annotation and expression data. BMC Bioinformatics 12: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magneschi L., Catalanotti C., Subramanian V., Dubini A., Yang W., Mus F., Posewitz M.C., Seibert M., Perata P., Grossman A.R. (2012). A mutant in the ADH1 gene of Chlamydomonas reinhardtii elicits metabolic restructuring during anaerobiosis. Plant Physiol. 158: 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malasarn D., Kropat J., Hsieh S.I., Finazzi G., Casero D., Loo J.A., Pellegrini M., Wollman F.A., Merchant S.S. (2013). Zinc deficiency impacts CO2 assimilation and disrupts copper homeostasis in Chlamydomonas reinhardtii. J. Biol. Chem. 288: 10672–10683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., Fritsch, E.F., and Sambrook, J.(1982). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory). [Google Scholar]
- Maple J., Møller S.G. (2007). Plastid division: Evolution, mechanism and complexity. Ann. Bot. (Lond.) 99: 565–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W.F. (2008). Basal bodies platforms for building cilia. Curr. Top. Dev. Biol. 85: 1–22 [DOI] [PubMed] [Google Scholar]
- Melis A., Zhang L., Forestier M., Ghirardi M.L., Seibert M. (2000). Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol. 122: 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S., Bogorad L. (1986). Regulation by copper of the expression of plastocyanin and cytochrome c552 in Chlamydomonas reinhardi. Mol. Cell. Biol. 6: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S.S., Allen M.D., Kropat J., Moseley J.L., Long J.C., Tottey S., Terauchi A.M. (2006). Between a rock and a hard place: Trace element nutrition in Chlamydomonas. Biochim. Biophys. Acta 1763: 578–594 [DOI] [PubMed] [Google Scholar]
- Merchant S.S., Kropat J., Liu B., Shaw J., Warakanont J. (2012). TAG, you’re it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr. Opin. Biotechnol. 23: 352–363 [DOI] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R., et al. (2010). Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 154: 1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering E.R., Benning C. (2010). RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell 9: 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Moseley J., Quinn J., Eriksson M., Merchant S. (2000). The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19: 2139–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.L., Page M.D., Alder N.P., Eriksson M., Quinn J., Soto F., Theg S.M., Hippler M., Merchant S. (2002). Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14: 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy U.M., Wecker M.S., Posewitz M.C., Gilles-Gonzalez M.A., Ghirardi M.L. (2012). Novel FixL homologues in Chlamydomonas reinhardtii bind heme and O(2). FEBS Lett. 586: 4282–4288 [DOI] [PubMed] [Google Scholar]
- Mus F., Dubini A., Seibert M., Posewitz M.C., Grossman A.R. (2007). Anaerobic acclimation in Chlamydomonas reinhardtii: Anoxic gene expression, hydrogenase induction, and metabolic pathways. J. Biol. Chem. 282: 25475–25486 [DOI] [PubMed] [Google Scholar]
- Mustroph A., Lee S.C., Oosumi T., Zanetti M.E., Yang H., Ma K., Yaghoubi-Masihi A., Fukao T., Bailey-Serres J. (2010). Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 152: 1484–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A.V., Thomas-Hall S.R., Malnoë A., Timmins M., Mussgnug J.H., Rupprecht J., Kruse O., Hankamer B., Schenk P.M. (2008). Transcriptome for photobiological hydrogen production induced by sulfur deprivation in the green alga Chlamydomonas reinhardtii. Eukaryot. Cell 7: 1965–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noth J., Krawietz D., Hemschemeier A., Happe T. (2013). Pyruvate:ferredoxin oxidoreductase is coupled to light-independent hydrogen production in Chlamydomonas reinhardtii. J. Biol. Chem. 288: 4368–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M.D., Kropat J., Hamel P.P., Merchant S.S. (2009). Two Chlamydomonas CTR copper transporters with a novel cys-met motif are localized to the plasma membrane and function in copper assimilation. Plant Cell 21: 928–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape M., Lambertz C., Happe T., Hemschemeier A. (2012). Differential expression of the Chlamydomonas [FeFe]-hydrogenase-encoding HYDA1 gene is regulated by the copper response regulator1. Plant Physiol. 159: 1700–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T., Schütze K., Fey V., Sherameti I., Oelmüller R. (2003). Chloroplast redox control of nuclear gene expression—A new class of plastid signals in interorganellar communication. Antioxid. Redox Signal. 5: 95–101 [DOI] [PubMed] [Google Scholar]
- Philipps G., Happe T., Hemschemeier A. (2012). Nitrogen deprivation results in photosynthetic hydrogen production in Chlamydomonas reinhardtii. Planta 235: 729–745 [DOI] [PubMed] [Google Scholar]
- Philipps G., Krawietz D., Hemschemeier A., Happe T. (2011). A pyruvate formate lyase-deficient Chlamydomonas reinhardtii strain provides evidence for a link between fermentation and hydrogen production in green algae. Plant J. 66: 330–340 [DOI] [PubMed] [Google Scholar]
- Porra R.J. (2002). The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 73: 149–156 [DOI] [PubMed] [Google Scholar]
- Posewitz M.C., King P.W., Smolinski S.L., Zhang L., Seibert M., Ghirardi M.L. (2004). Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J. Biol. Chem. 279: 25711–25720 [DOI] [PubMed] [Google Scholar]
- Quinn J.M., Barraco P., Eriksson M., Merchant S. (2000). Coordinate copper- and oxygen-responsive Cyc6 and Cpx1 expression in Chlamydomonas is mediated by the same element. J. Biol. Chem. 275: 6080–6089 [DOI] [PubMed] [Google Scholar]
- Quinn J.M., Eriksson M., Moseley J.L., Merchant S. (2002). Oxygen deficiency responsive gene expression in Chlamydomonas reinhardtii through a copper-sensing signal transduction pathway. Plant Physiol. 128: 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond J., Segrè D. (2006). The effect of oxygen on biochemical networks and the evolution of complex life. Science 311: 1764–1767 [DOI] [PubMed] [Google Scholar]
- Rochaix J.D. (2002). Chlamydomonas, a model system for studying the assembly and dynamics of photosynthetic complexes. FEBS Lett. 529: 34–38 [DOI] [PubMed] [Google Scholar]
- Rossak M., Schäfer A., Xu N., Gage D.A., Benning C. (1997). Accumulation of sulfoquinovosyl-1-O-dihydroxyacetone in a sulfolipid-deficient mutant of Rhodobacter sphaeroides inactivated in sqdC. Arch. Biochem. Biophys. 340: 219–230 [DOI] [PubMed] [Google Scholar]
- Sakihama Y., Nakamura S., Yamasaki H. (2002). Nitric oxide production mediated by nitrate reductase in the green alga Chlamydomonas reinhardtii: An alternative NO production pathway in photosynthetic organisms. Plant Cell Physiol. 43: 290–297 [DOI] [PubMed] [Google Scholar]
- Sato Y., Morita R., Katsuma S., Nishimura M., Tanaka A., Kusaba M. (2009). Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 57: 120–131 [DOI] [PubMed] [Google Scholar]
- Schloss J.A. (1990). A Chlamydomonas gene encodes a G protein beta subunit-like polypeptide. Mol. Gen. Genet. 221: 443–452 [DOI] [PubMed] [Google Scholar]
- Schmidt R.J., Myers A.M., Gillham N.W., Boynton J.E. (1984). Chloroplast ribosomal proteins of Chlamydomonas synthesized in the cytoplasm are made as precursors. J. Cell Biol. 98: 2011–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten K.A., Weiss B. (1999). Endonuclease V protects Escherichia coli against specific mutations caused by nitrous acid. Mutat. Res. 435: 245–254 [DOI] [PubMed] [Google Scholar]
- Semenza G.L. (2004). Hydroxylation of HIF-1: Oxygen sensing at the molecular level. Physiology (Bethesda) 19: 176–182 [DOI] [PubMed] [Google Scholar]
- Sommer F., Kropat J., Malasarn D., Grossoehme N.E., Chen X., Giedroc D.P., Merchant S.S. (2010). The CRR1 nutritional copper sensor in Chlamydomonas contains two distinct metal-responsive domains. Plant Cell 22: 4098–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand A. (2004). Plastid-to-nucleus signalling. Curr. Opin. Plant Biol. 7: 621–625 [DOI] [PubMed] [Google Scholar]
- Stripp S.T., Happe T. (2009). How algae produce hydrogen—News from the photosynthetic hydrogenase. Dalton Trans. 45: 9960–9969 [DOI] [PubMed] [Google Scholar]
- Tanaka R., Tanaka A. (2011). Chlorophyll cycle regulates the construction and destruction of the light-harvesting complexes. Biochim. Biophys. Acta 1807: 968–976 [DOI] [PubMed] [Google Scholar]
- Terashima M., Specht M., Naumann B., Hippler M. (2010). Characterizing the anaerobic response of Chlamydomonas reinhardtii by quantitative proteomics. Mol. Cell. Proteomics 9: 1514–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terauchi A.M., Lu S.F., Zaffagnini M., Tappa S., Hirasawa M., Tripathy J.N., Knaff D.B., Farmer P.J., Lemaire S.D., Hase T., Merchant S.S. (2009). Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii. J. Biol. Chem. 284: 25867–25878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd B.L., Stewart E.V., Burg J.S., Hughes A.L., Espenshade P.J. (2006). Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol. Cell. Biol. 26: 2817–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lis R., Baffert C., Couté Y., Nitschke W., Atteia A. (2013). Chlamydomonas reinhardtii chloroplasts contain a homodimeric pyruvate: Ferredoxin oxidoreductase that functions with FDX1. Plant Physiol. 161: 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Kong D., Wang Y., Hu Y., He Y., Sun J. (2003). Isolation of two plastid division ftsZ genes from Chlamydomonas reinhardtii and its evolutionary implication for the role of FtsZ in plastid division. J. Exp. Bot. 54: 1115–1116 [DOI] [PubMed] [Google Scholar]
- Weiss B. (2006). Evidence for mutagenesis by nitric oxide during nitrate metabolism in Escherichia coli. J. Bacteriol. 188: 829–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willeford K.O., Gibbs M. (1989). Localization of the enzymes involved in the photoevolution of H(2) from acetate in Chlamydomonas reinhardtii. Plant Physiol. 90: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimalasekera R., Villar C., Begum T., Scherer G.F. (2011). COPPER AMINE OXIDASE1 (CuAO1) of Arabidopsis thaliana contributes to abscisic acid- and polyamine-induced nitric oxide biosynthesis and abscisic acid signal transduction. Mol. Plant 4: 663–678 [DOI] [PubMed] [Google Scholar]
- Wittenberg J.B., Bolognesi M., Wittenberg B.A., Guertin M. (2002). Truncated hemoglobins: A new family of hemoglobins widely distributed in bacteria, unicellular eukaryotes, and plants. J. Biol. Chem. 277: 871–874 [DOI] [PubMed] [Google Scholar]
- Wollman F.A., Delepelaire P. (1984). Correlation between changes in light energy distribution and changes in thylakoid membrane polypeptide phosphorylation in Chlamydomonas reinhardtii. J. Cell Biol. 98: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]