HD-ZIPIII proteins and KANADI proteins are responsible for tailoring transcription in cells that will make up different parts of the leaf and in the self-renewing stem cells. This article reports which genes are turned on and off by the HD-ZIPIII and KANADI proteins and, therefore, how these proteins act to regulate the growth of new leaves and branches.
Abstract
The broadly conserved Class III HOMEODOMAIN LEUCINE ZIPPER (HD-ZIPIII) and KANADI transcription factors have opposing and transformational effects on polarity and growth in all tissues and stages of the plant's life. To obtain a comprehensive understanding of how these factors work, we have identified transcripts that change in response to induced HD-ZIPIII or KANADI function. Additional criteria used to identify high-confidence targets among this set were presence of an adjacent HD-ZIPIII binding site, expression enriched within a subdomain of the shoot apical meristem, mutant phenotype showing defect in polar leaf and/or meristem development, physical interaction between target gene product and HD-ZIPIII protein, opposite regulation by HD-ZIPIII and KANADI, and evolutionary conservation of the regulator–target relationship. We find that HD-ZIPIII and KANADI regulate tissue-specific transcription factors involved in subsidiary developmental decisions, nearly all major hormone pathways, and new actors (such as INDETERMINATE DOMAIN4) in the ad/abaxial regulatory network. Multiple feedback loops regulating HD-ZIPIII and KANADI are identified, as are mechanisms through which HD-ZIPIII and KANADI oppose each other. This work lays the foundation needed to understand the components, structure, and workings of the ad/abaxial regulatory network directing basic plant growth and development.
INTRODUCTION
Class III HOMEODOMAIN LEUCINE ZIPPER (HD-ZIPIII) and KANADI (KAN) transcription factors play opposing roles in defining polarity along the ad/abaxial axis of the Arabidopsis thaliana leaf. HD-ZIPIII factors promote the development of adaxial (upper) characteristics and KAN1 factors promote the development of abaxial (lower) characteristics in the leaf (Eshed et al., 2001; Kerstetter et al., 2001; McConnell et al., 2001; Prigge et al., 2005). The “axis” runs from shoot to root down the middle of the plant. Leaf primordia form with their future upper, or adaxial, domain toward the center of the plant and their future lower, or abaxial, domain toward the periphery of the plant. In the stem, root, and early embryo, the ad/abaxial dimension is called the radial dimension.
The development of the leaf blade into distinct upper (adaxial) and lower (abaxial) domains is vital to its function as a photosynthetic organ. Chloroplast-rich palisade cells pack together tightly in the upper domain, facilitating light capture, while loosely arranged cells in the lower domain facilitate the exchange of CO2 and oxygen. Cells in vascular bundles are arranged with xylem, the conduit for water and minerals, located adaxial to phloem, the conduit for sugars. Other characteristics distinguishing ad- and abaxial domains include trichome density and type, stomata density, cell shape and size, and the presence of secondary metabolites. Strikingly, formation of the leaf blade requires the establishment of polarity in the leaf primordium: Juxtaposition of adaxial and abaxial cells in the young leaf primordium is required for, and defines the site of, blade outgrowth (Waites and Hudson, 1995; McConnell and Barton, 1998; Evans, 2007). The balance of ad- and abaxializing activities is also important for coordinating growth between the upper and lower leaf domains; adaxialized leaves curl up, while abaxialized leaves curl down (Ochando et al., 2006; Wenkel et al., 2007).
Branching of the shoot is dependent on the ad/abaxial polarity system: Establishment of new stem cell centers, or meristems, depends on it. New shoot apical meristems (or buds) form only within the adaxial domain of the embryo and on the adaxial bases of leaf primordia. Failure to establish an adaxial domain results in failure of new shoot apical meristems to form, whereas the development of ectopic adaxial tissue results in ectopic shoot apical meristem formation (Talbert et al., 1995; McConnell and Barton, 1998; Kerstetter et al., 2001; Otsuga et al., 2001). This is one of several developmental processes in which HD-ZIPIII genes promote growth while KAN genes inhibit growth (summarized in Liu et al., 2012).
Additional studies have broadened our understanding of the role of HD-ZIPIII and KAN1 proteins to include regulation of seed coat development (Leon-Kloosterziel et al., 1994; Eshed et al., 2001; Kelley et al., 2009; McAbee et al., 2006), polarity along the apical basal (root-shoot) axis of the embryo (Smith and Long, 2012), patterning of the vasculature (Zhong and Ye, 2001; Carlsbecker et al., 2010; Ilegems et al., 2010), outgrowth and positioning of leaf primordia (Emery et al., 2003; Izhaki and Bowman, 2007), lateral root production (Hawker and Bowman, 2004), and shoot apical meristem size (Green et al., 2005; Williams et al., 2005).
HD-ZIPIII and KAN genes encode two distinct types of transcription factors. The five Class III HD-ZIP proteins of Arabidopsis, REVOLUTA (REV), PHABULOSA (PHB), PHAVOLUTA, ARABIDOPSIS THALIANA HOMEOBOX GENE 8 (ATHB8), and INCURVATA/CORONA, have a DNA binding homeodomain followed by a Leu zipper domain at the N terminus. C-terminal to the Leu zipper is a START domain, predicted to bind an as yet unknown hydrophobic ligand, and a PER-ARNT-SIM (PAS) domain (Mukherjee et al., 2009). In vitro, HD-ZIPIII proteins bind to a palindromic DNA sequence as dimers, with dimerization occurring between the Leu zipper domains (Sessa et al., 1998). Dimerization of HD-ZIPIII proteins is negatively regulated intramolecularly by the PAS domain and intermolecularly by the LITTLE ZIPPER (ZPR) proteins (Wenkel et al., 2007; Magnani and Barton, 2011). The four closely related KAN genes encode members of the GARP family of transcription factors (Kerstetter et al., 2001). The GARP domain is a myb-like DNA binding domain (Hosoda et al., 2002).
HD-ZIPIII and KAN transcription factors have perhaps the broadest ranging effects of any developmental regulators known in plants, yet only a small number of HD-ZIPIII and KAN1 regulatory targets are known. REV activates transcription of the four ZPR (ZPR1-4) genes, which in turn inhibit dimerization of HD-ZIPIII proteins, forming a negative feedback loop (Wenkel et al., 2007; Kim et al., 2008; Magnani and Barton, 2011). Similarly, KAN1 directly represses the ASYMMETRIC LEAVES2 (AS2) gene in abaxial domains, and AS2 in turn represses KAN1 (Wu et al., 2008). There are likely many more targets of HD-ZIPIII and KAN, and, since they act oppositely to one another, it is likely that some genes will be oppositely regulated by these factors.
To better understand how the HD-ZIPIII and KAN1 proteins affect development, we identified a high-confidence set of REV and KAN targets. These targets include genes encoding components of the major plant hormone pathways and transcription factors that regulate subsidiary developmental processes, such as meristem formation or polarity within the vasculature. The discovery of this set of targets, and the elucidation of their interactions with REV and KAN (several target genes are oppositely regulated by REV and KAN), establish a foundation for defining and ultimately understanding the regulatory network controlling ad/abaxial development in the plant.
RESULTS
Identification of Transcripts Regulated by REV and KAN1
To identify genes regulated by the HD-ZIPIII REV and KAN1 transcription factors, we analyzed whole-genome transcriptional responses to dexamethasone-inducible REV and KAN proteins fused to a glucocorticoid response (GR) domain (Wenkel et al., 2007; Wu et al., 2008). Addition of dexamethasone releases the GR-transcription factor fusion protein from a cytoplasmic HEAT SHOCK PROTEIN90 complex, allowing nuclear translocation and synchronous activation of the transcription factor (Schena et al., 1991).
Dexamethasone addition to GR-REV transgenic lines causes phenotypic adaxialization of leaves and rapid upregulation of known targets ZPR1, ZPR2, ZPR3, and ZPR4, indicating that GR-REV protein fusions retain function (Wenkel et al., 2007; Magnani and Barton, 2011). Similarly, dexamethasone addition to KAN1-GR transgenic lines causes phenotypic abaxialization of leaves and rapid downregulation of known target AS2, indicating that the KAN1-GR fusion protein retains function (Wu et al., 2008).
35S:GR-REV* (resistant to microRNAs 165 and 166), 35S:KAN1-GR, or wild-type Columbia (Col) week-old seedlings were treated with dexamethasone for 0, 30, or 60 min, and transcript levels across the entire genome were measured by hybridization to ATH1 microarrays. In addition, a 120-min time point was included for GR-REV.
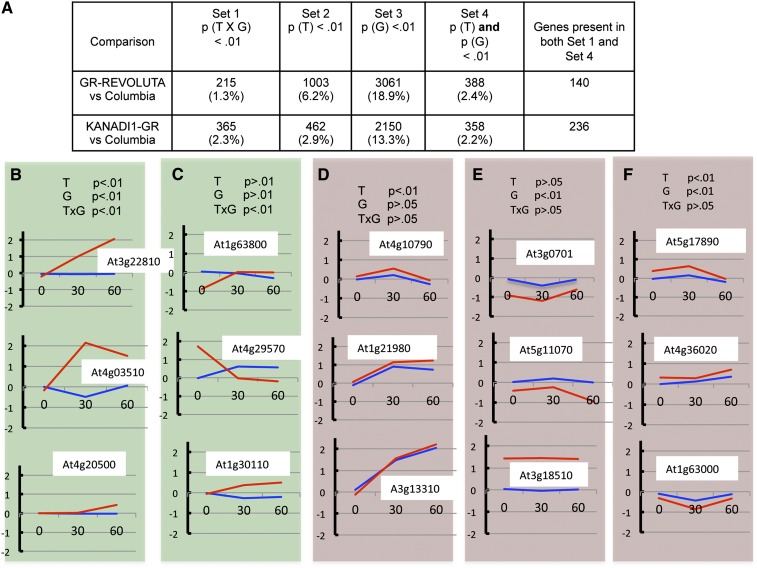
By concentrating on genes that show a statistically significant interaction between time of treatment and genotype, we can both increase our ability to find genuine targets (i.e., decrease false negatives) and reduce the background of false positives (Nettleton, 2006). Inspection of the behavior of transcripts significant for time (t), genotype (g), and/or the interaction between the two (txg) illustrates this (Figures 1B to 1F). The examples in Figure 1B were significant for both t and g as well as for txg, while the examples in 1C were significant for only txg. In both, transcripts showed different patterns in response to dexamethasone treatment in the transgenic and wild-type lines and were therefore included in our set of candidate targets (Set 1, Figure 1A). This approach identified 215 candidate target genes in the REV versus the wild type (Col) analysis and 365 genes in the KAN1 versus the wild type (Col) analysis (see Supplemental Data Sets 1A to 1D online).
Figure 1.
Two-Way ANOVA of Whole-Genome Transcript Responses to Activation of GR-REV or KAN1-GR.
(A) The number (%) of genes whose transcript levels showed significant changes over treatment time (T), genotype (G), or time by genotype interaction (TxG). Percentages given are percentage of total genes (n = 16164) that passed the filter for gene expression level.
(B) Examples of time courses for three genes whose transcripts show significant TxG interactions.
(C) Examples of time courses for three genes whose transcripts show significant TxG interactions but where the two-way ANOVA does not detect significant variation over time or genotype individually.
(D) Examples of time courses for three genes whose transcripts show significant variation over time only.
(E) Examples of time courses for three genes whose transcripts show significant variation over genotype only.
(F) Examples of time courses for three genes whose transcripts show significant variation over time and genotype but no significant TxG interaction.
Red curve = transcript levels in GR-REV; blue curve = transcript levels in wild-type Col ecotype. The y axis shows transcript levels in log base 2 units (i.e., a value of 1 indicates a doubling of transcript levels). The x axis shows time in minutes.
By contrast, a large number of transcripts showed significant differences for t and/or g but not for txg. This was due to differences in baseline levels of transcripts in the two genotypes (transcripts significant for g; Figure 1E) or to dexamethasone response in both the wild-type and the GR-REV line (transcripts significant for t; Figure 1D). Finally, transcripts may display both different baselines between genotypes and similar responses to dexamethasone (transcripts significant for t and for g but not txg; Figure 1F). As a consequence, the “overlap” set of genes that differed over both time and genotype (Set 4) includes many false positives and is not equivalent to the set of genes that showed an interaction between time and genotype (Set 1).
Note that some transcripts in the transgenic lines showed altered levels at time 0 (Figure 1 and other figures throughout this article). This may be due to “leakiness” of the transgene or to some other alteration of the transcriptional steady state of the cell in the transgenic line.
Comparison of Genes Regulated by Inducible REV and KAN1
The fraction of up-regulated genes was higher in the REV versus the wild type (Col) analysis than in the KAN1 versus the wild type (Col) analysis: 56.7% compared with 33.4% (see Supplemental Figure 1A online; P < 0.0001, Fisher exact test). This is consistent with the evidence to date that REV is an activator of transcription, whereas KAN1 is a repressor of transcription (Wenkel et al., 2007; Wu et al., 2008; Magnani and Barton, 2011).
For both REV and KAN1, transcripts generally showed larger fold changes at 60 min than at 30 min. The average change for upregulated genes was approximately twofold at 30 min and 2.8-fold at 60 min (see Supplemental Figure 1A online; 1.0 and 1.5 in log base 2) and for downregulated genes was ∼0.5-fold at 30 min and 0.35-fold at 60 min (approximately −1.0 and −1.5 in log base 2).
The range of gene regulation and the distribution of two-way analysis of variance (ANOVA) P values were similar in the GR-REV and KAN1-GR experiments (see Supplemental Figure 1 and Supplemental Data Sets 1A to 1D online). Genes upregulated in the GR-REV experiment varied from 1.3- to 14.4-fold, while genes upregulated in KAN1-GR varied from 1.26- to 22.0-fold. Genes downregulated in the GR-REV experiment varied from 0.08 to 0.69, while genes downregulated by KAN1-GR varied from 0.03 to 0.80. In both experiments, up- and downregulated genes were similarly distributed according to statistical significance varying from 1 × 10−11 to the cutoff of 0.01 (see Supplemental Figures 1B and 1C online). When plotted against the max fold change observed for each gene, the corresponding P value showed only a modest correlation (see Supplemental Figures 1A and 1D to 1G online), indicating that ranking by fold change may not be a reliable way to assign confidence to a target.
A P-SCAN analysis of 1000 bp upstream of the candidate targets (Zambelli et al., 2009) showed that REV upregulated genes, but not REV downregulated genes, were enriched in the Class III HD-ZIP binding sites identified in vitro by Sessa et al. (1998) (P = 0.04; Table 1). A similar P-SCAN analysis showed that KAN1 downregulated genes, but not upregulated genes, were enriched in the KAN1 binding site upstream of AS2 (Wu et al., 2008) (P = 0.0021 to 0.012). Again, this is consistent with previous observations showing that REV is a transcriptional activator and KAN1 is a repressor.
Table 1. Probability of Observed HD-ZIPIII and KAN Binding Sites Upstream of GR-REV– and KAN1-GR–Regulated Genes Occurring by Chance Alonea.
| Binding Site | GR-REV |
KAN1-GR |
||
|---|---|---|---|---|
| Up | Down | Up | Down | |
| HD-ZIPIIIb | 0.04 | 0.64 | 0.17 | 0.06(*) |
| (ATHB-9) | ||||
| KAN1(A)c | 0.45 | 0.35 | 0.79 | 0.012* |
| KAN1(B) | 0.83 | 0.47 | 0.99 | 0.0021* |
| KAN1(C) | 0.052(*) | 0.82 | 0.80 | 0.0052* |
Significant at P < 0.05. (*)Significant at P < 0.10.
One kilobase upstream of each regulated gene was surveyed.
ATHB9 binding site is GTAATG/CATTAC (Sessa et al., 1998).
KAN1 binding sites are three overlapping single KAN binding sites from the dimeric site defined by Wu et al. (2008). KAN1(A), AAGAAT; KAN1(B), AGAATA; KAN1(C), GAATAA.
Interestingly, at a less stringent threshold (P < 0.10), KAN1 (C) binding sites were overrepresented in the promoters of REV upregulated genes, while HD-ZIPIII binding sites were overrepresented in the promoters of KAN1 downregulated genes. This hints that a large fraction of genes upregulated by REV are regulated by KAN1 and a large fraction of genes downregulated by KAN1 are regulated by REV.
To ensure the identification of biologically significant targets of REV and KAN1 regulation, we performed additional analyses and experiments and identified seven supported subsets of genes targeted by REV and/or KAN1.
Supported Subset One: Genes with Adjacent REV Binding Sites
Transcripts regulated by GR-REV or KAN1-GR may be directly regulated by these factors or they may be regulated by intermediary factors. While both are of interest, identification of genes directly regulated by REV or KAN provides important mechanistic information and adds experimental support to target gene identification.
We compared the genes we identified as targets of REV in our time-course experiments (Set 1 in Figure 1A; see Supplemental Data Sets 1A and 1B online) with potential REV targets identified by Brandt et al. (2012) who identified DNA to which REV binds by performing chromatin immunoprecipitation (ChIP)-sequencing on dexamethasone-treated GR-FLG-REV plants from the same transgenic line. Of the 143 binding sites (286 genes = 1.0% of protein coding genes in the genome as a whole) identified in that work, we found 41 located next to 39 genes or 18% from the REV-regulated set (see Supplemental Data Set 1E online). This corresponds to an enrichment of 14-fold in the REV-regulated set. A total of 77% of the binding sites were 5′ to the regulated transcript, ranging from immediately upstream to 7500 bp away. A total of 23% percent of binding sites were 3′ to the regulated locus, ranging from immediately downstream to 4800 bp away. In most cases, only one gene neighboring a binding site showed significant regulation. The set includes the known REV target ZPR1 (Wenkel et al., 2007) but fails to include ZPR2, ZPR3, and 4 genes, also known REV targets, from the list of genes with binding sites, indicating that the list of binding sites identified by ChIP-sequencing is incomplete.
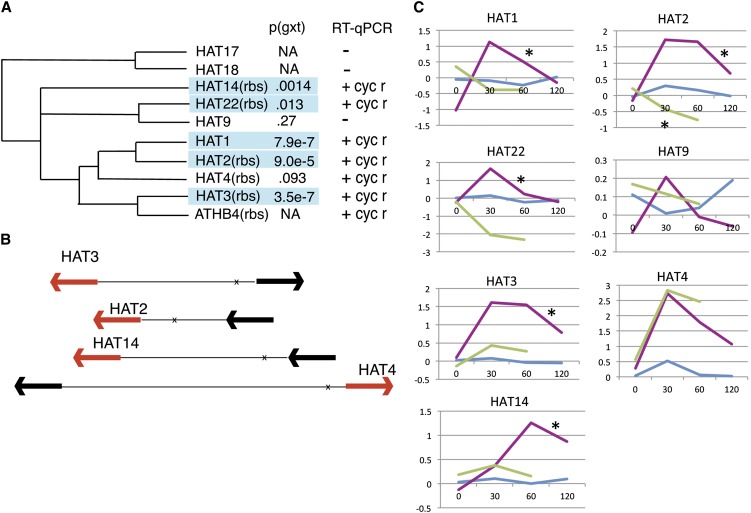
Genes whose transcripts are regulated by GR-REV and that are flanked by REV binding sites (see Supplemental Data Set 1E online) include six genes regulated by both KAN1-GR and GR-REV (see Subset 2 below): These encode the zinc finger protein INDETERMINATE DOMAIN4 (IDD4), the ZINC FINGER PROTEIN8 (ZFP8) regulator of epidermal cell fates (Gan et al., 2007), the SNF1-related protein kinase 3 (SnrK3) CBL-Interacting Protein Kinase12 (CIPK12), and the class II HD-ZIP genes HAT2, HAT3, and HAT14.
Quantitative RT-PCR (qRT-PCR) experiments on independent samples established that all Class II HD-ZIP genes except HAT9, HAT17, and HAT18 were activated by GR-REV (Figure 2; see Supplemental Data Set 1F online). Activation occurred in the presence and absence of the protein synthesis inhibitor cycloheximide, indicating no new protein synthesis was required to activate the transcription of these genes and they are therefore likely direct targets. In all cases where this could be examined, the time-course data for the Class II genes showed a spike in transcript levels followed by a decrease (Figure 2). The decrease may indicate negative feedback. Indeed, in many cases (see Supplemental Figure 2 online), the presence of cycloheximide in the experiment allowed higher levels of transcripts to accumulate at 1 h, indicating that the negative feedback requires new protein synthesis. Since Class II HD-ZIP proteins have been shown to negatively regulate their own transcription (Ciarbelli et al., 2008), they are good candidates for mediating this feedback step.
Figure 2.
Regulation of Class II HD-ZIP Gene Family Members by REV and KAN1.
(A) Phylogenetic tree of HD-ZIPII genes (after Ciarbelli et al., 2008) showing relationship between family members and P value (time by genotype interaction) for regulation of transcript in GR-REV time-course experiment (comparison is to wild-type Col). rbs, adjacent to REV binding site; +, transcript was REV regulated in independent qRT-PCR experiments; -, transcript was not REV regulated in independent qRT-PCR experiments; cyc r, regulation was resistant to presence of cycloheximide; NA, not assayable by microarray.
(B) Location of REV binding sites (identified in Brandt et al., 2012; marked with an x) relative to closest flanking coding sequences (arrows). Transcripts that show regulation by REV are indicated with red arrows.
(C) Microarray time-course data for Class II HD-ZIP genes. x axis, time points are 0, 30, 60, and 120 min after dexamethasone addition to the medium; y axis, transcript level normalized to mean value of untreated Col transcripts (log base 2). Magenta line represents GR-REV; blue line represents wild-type Col; green line represents KAN1-GR. No time-course data are given for ATHB4 since it is not represented on the ATH1 array. Asterisk indicates behavior significant from the wild type (P < .01).
These experiments extend observations by Brandt et al. (2012) who found that four Class II HD-ZIP genes are transcriptionally activated by REV (HAT2, HAT3, HAT4, and ATHB4). Strengthening the importance of their regulation by GR-REV, recent mutant analysis shows that HAT3, HAT4, and ATHB4 are required for normal ad/abaxial leaf development (Bou-Torrent et al., 2012; Turchi et al., 2013).
Of the seven Class II HD-ZIP genes upregulated by REV, four were regulated by KAN1: HAT2 and HAT22 downregulated and HAT3 and ATHB4 upregulated (Figure 2; see Supplemental Data Set 1F online). Downregulation of HAT2 and HAT22 by KAN1 was resistant to cycloheximide, indicating that these genes are direct regulatory targets of REV and KAN1 acting in opposition.
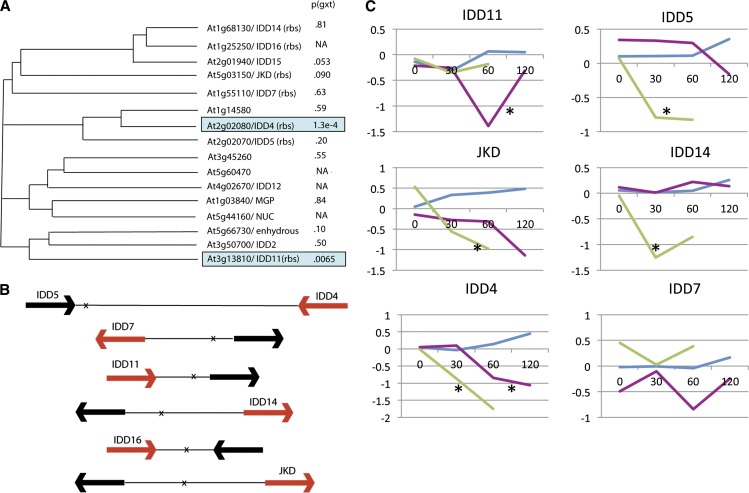
The set of GR-REV–regulated genes with adjacent REV binding sites also contained two related IDD Cys2His2 zinc finger protein genes (IDD4 and IDD11). Furthermore, of the 16 genes in the IDD gene family, REV binding sites (as defined by ChIP-sequencing in Brandt et al., 2012) flanked seven of them (Figures 3A and 3B), and, of the 12 family members for which transcript levels could be measured, four were downregulated by KAN1 (IDD4, IDD5, JACKDAW, and IDD14; P < 0.01; Figure 3). Thus, IDD family genes are enriched for genes with adjacent REV binding sites (58% as opposed to 1.0% for the genome as a whole) and for KAN regulation (33% versus 2.3% for the genome as a whole). In contrast with previously identified targets of REV that are upregulated, IDD4 and several other IDD family members showed a pattern of late (60 min) downregulation by REV (Figure 3). The presence of a REV binding site next to IDD4 strongly suggests that REV can act as a transcriptional repressor at a subset of genes. If true, this would be a previously unknown role for REV.
Figure 3.
Regulation of IDD C2H2 Zinc Finger Gene Family Members by REV and KAN1.
(A) Phylogenetic tree of group a and b IDD proteins (after Englbrecht et al., 2004) and their regulation by REV. P values are for interaction between time and genotype in comparison between GR-REV and the wild type. rbs indicates presence of REV binding site in vicinity of gene as determined by Brandt et al. (2012).
(B) Schematic showing position of REV binding sites (identified in Brandt et al., 2012; marked with an x) relative to closest flanking coding sequences (arrows). Transcripts that show regulation by REV are indicated with red arrows.
(C) Graphs showing time-course behavior of regulated IDD transcripts. Magenta represents GR-REV line; blue represents wild-type Col; Green represents KAN1-GR. x axis, time points are 0, 30, 60, and 120 min after dexamethasone addition to the medium; y axis, transcript level normalized to mean value of untreated Col transcripts (log base 2). Asterisk indicates behavior significant from the wild type (P < .01).
Supported Subset 2: Genes Regulated by Both REV and KAN1
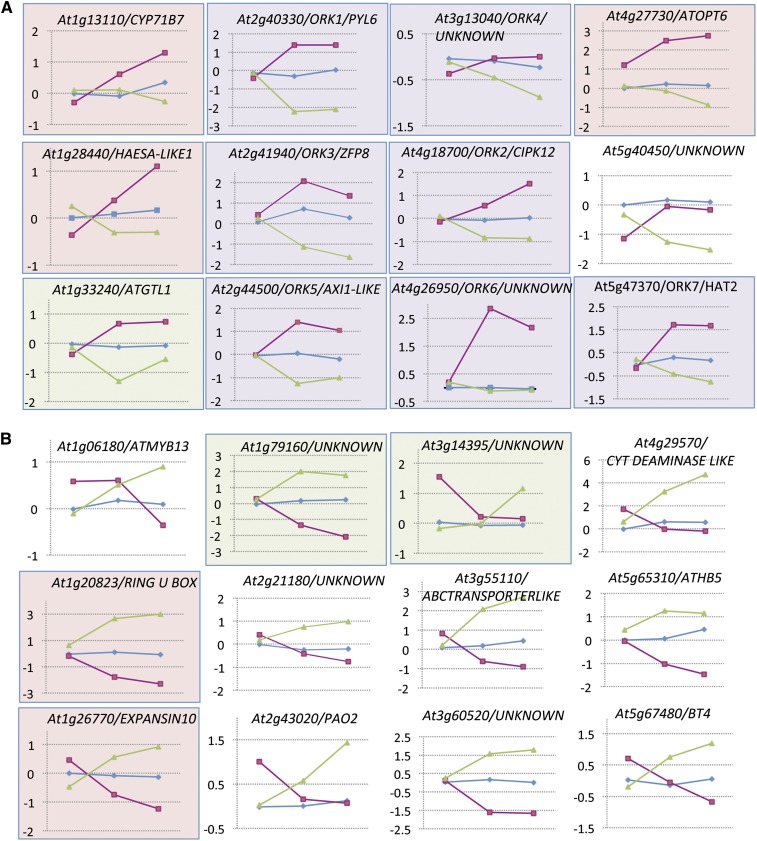
Analysis of the IDD and Class II HD-ZIP gene families shows that genes regulated by REV have a high probability of being regulated by KAN1 as well. Given the opposing roles of REV and KAN1, the set of genes regulated by both of these factors, especially those that are oppositely regulated, should identify a high confidence set of genes involved in ad/abaxial development. Of the 215 REV and 365 KAN1 candidate-regulated genes, 42 were common to both lists (see Supplemental Data Set 1G online). Eighteen genes in the overlap set were regulated in the same direction by the two transcription factors, 11 down and seven up (see Supplemental Figure 3 online). Twenty-four of the 42 genes were regulated in opposite directions in the two experiments: 12 were upregulated by GR-REV and downregulated by KAN1-GR (Table 2, Figure 4A), while the remaining 12 were downregulated by GR-REV and upregulated by KAN1-GR (Table 2, Figure 4B). We refer to the oppositely regulated genes as ORK genes (for oppositely regulated by REV and KAN1).
Table 2. Genes Oppositely Regulated by REV and KAN1.
| AGI | GR-REV p(TxG)a | GR-KAN p(TxG)a | Regulation by REV/KAN | Gene Name/Target Description |
|---|---|---|---|---|
| AT5G65310 | 3.06E-07 | 6.40E-04 | Down/up | ATHB5 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN5) |
| AT1G79160 | 6.66E-05 | 7.52E-05 | Down/up | Protein of unknown function |
| AT3G60520 | 1.10E-04 | 1.97E-04 | Down/up | Putative protein |
| AT5G67480 | 5.78E-03 | 2.62E-05 | Down/up | BT4 (BTB AND TAZ DOMAIN PROTEIN4); protein binding |
| AT1G20823 | 8.83E-04 | 2.77E-04 | Down/up | Ring U-box family protein |
| AT1G26770 | 1.41E-03 | 3.39E-04 | Down/up | ATEXPA10 (ARABIDOPSIS THALIANA EXPANSIN A10) |
| AT2G43018 /// AT2G43020 | 2.16E-03 | 2.61E-04 | Down/up | PAO2 polyamine oxidase 2 |
| AT4G29570 | 2.93E-03 | 5.77E-04 | Down/up | Cytidine deaminase-like protein |
| AT1G06180 | 7.03E-04 | 3.89E-03 | Down/up | ATMYB13 (myb domain protein13); DNA binding/transcription factor |
| AT2G21180 | 1.89E-03 | 1.60E-03 | Down/up | Hypothetical protein |
| AT3G14395 | 7.90E-04 | 9.13E-03 | Down/up | Expressed protein |
| AT3G55110 | 6.92E-03 | 1.68E-03 | Down/up | ABC transporter like |
| AT2G40330 | 4.50E-06 | 9.09E-06 | Up/down | PYR1-like 6 |
| AT1G33240 | 4.78E-06 | 1.85E-04 | Up/down | AT-GTL1 (Arabidopsis thaliana GT2-like1); transcription factor |
| AT4G26950 | 3.51E-07 | 9.95E-03 | Up/down | Putative protein |
| AT1G28440 | 4.53E-06 | 1.87E-03 | Up/down | HSL1 (HAESA-LIKE1); ATP binding kinase/protein Ser/Thr kinase |
| AT4G18700 | 6.51E-06 | 1.64E-03 | Up/down | CIPK12; kinase |
| AT2G41940 | 8.40E-03 | 3.41E-06 | Up/down | ZFP8 (ZINC FINGER PROTEIN8); nucleic acid binding/transcription factor/zinc ion binding |
| AT5G47370 | 9.03E-05 | 2.10E-03 | Up/down | HAT2; Class II HD-ZIP transcription factor |
| AT1G13110 | 3.65E-05 | 7.25E-03 | Up/down | CYP71B7 (cytochrome P450, family 71, subfamily B, polypeptide 7); oxygen binding |
| AT2G44500 | 1.79E-03 | 1.70E-04 | Up/down | AXI1-like |
| AT4G27730 | 7.74E-04 | 2.26E-03 | Up/down | ATOPT6 (oligopeptide transporter 6); oligopeptide transporter |
| AT3G13040 | 4.85E-04 | 4.36E-03 | Up/down | Hypothetical protein |
| AT5G40450 | 6.92E-03 | 5.45E-03 | Up/down | Putative protein microtubule-associated protein homolog Drosophila melanogaster |
Figure 4.
Microarray Time-Course Data for Genes Oppositely Regulated by REV and KAN1 (ORKs).
(A) ORKs upregulated in GR-REV and downregulated by KAN1-GR.
(B) ORKs downregulated in GR-REV and upregulated by KAN1-GR.
x axis, time points are 0, 30, and 60 min after dexamethasone addition to the medium; y axis, transcript level normalized to mean value of untreated Col transcripts (log base 2). Magenta line represents GR-REV; blue line represents wild-type Col; green line represents KAN1-GR. Genes in pink-shaded boxes retested significant for REV regulation (P < 0.05) by qRT-PCR. Genes in green-shaded boxes retested significant for KAN1 regulation (P < 0.05) by qRT-PCR. Genes in purple shaded boxes retested significant for KAN1 and REV regulation (P < 0.05) by qRT-PCR.
To validate that the ORK loci are regulated by REV and KAN1, we performed qRT-PCR on an independent set of samples treated with dexamethasone for 60 min, the time point with the largest average fold change in our microarray experiment (see Supplemental Figure 1A online). Genes with transcript level changes resistant to cycloheximide were candidates for genes directly regulated by either GR-REV or GR-KAN (see Supplemental Figures 4 and 5 and Supplemental Data Set 1F online). (Note, however, that the microarray-based experiment included six wild-type controls for each of three time points [total sample number = 18] while the qRT-PCR experiment included three wild-type controls for each of two time points [total sample number = 6]). Failure to validate targets by qRT-PCR may reflect this difference in sampling.)
The qRT-PCR experiments showed that candidate ORK genes upregulated by GR-REV and downregulated by GR-KAN were more likely to be validated by qRT-PCR and were more likely to be direct targets than the inversely regulated set (Figure 4). Direct targets of both REV and KAN based on cycloheximide resistance included: ORK1/PYRABACTIN RESISTANCE1 LIKE6 (PYL6), a member of the family of ABA receptors (At2g40330); ORK2/CIPK12, a SnRK3 kinase (At4g18700); ORK6, a member of a 15-member family of genes containing a DOMAIN OF UNKNOWN FUNCTION 581 (At4g26950); ORK3/ZFP8, a zinc finger protein (At2g41940); and ORK7/HAT2 (At5g47370). Of these, ORK2/CIPK12, ORK7/HAT2, and ORK3/ZFP8 have ChIP-identified REV binding sites (Brandt et al., 2012; see Supplemental Data Set 1E online).
We reanalyzed the data using two-way ANOVA to compare transcription in GR-REV plants directly to that in KAN1-GR plants (= REV versus KAN two-way ANOVA). Because this compares transcript changes in GR- REV directly to changes in KAN1-GR, this is a more sensitive way to identify genes oppositely regulated by REV and KAN1 but a less sensitive way to identify genes regulated in the same direction. (Note that this analysis will include genes regulated by REV only or KAN1 only as well.) In this analysis, we identified 812 genes (Figure 5; see Supplemental Data Set 1H online), 461 of which were upregulated in dexamethasone-treated GR-REV lines relative to dexamethasone-treated KAN1-GR lines. Of these genes, 274 were new to this analysis. An analysis of the gene ontology terms associated with these 461 genes revealed terms related to (1) protein modification and degradation, (2) regulation of transcription, and (3) signaling to be the top three categories (see Supplemental Data Set 1I online).
Figure 5.
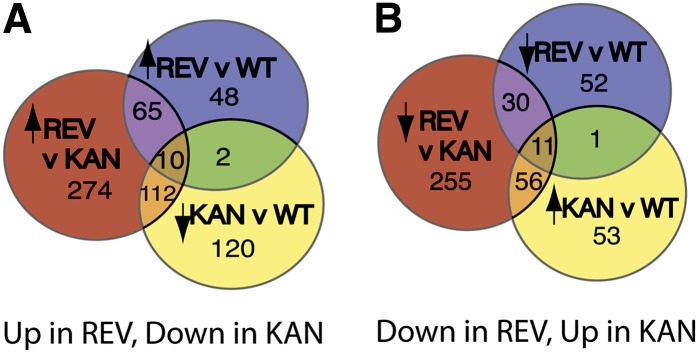
Overlap between Sets of Transcripts Identified as Regulated by GR-REV and KAN1-GR.
(A) Summary of genes upregulated by REV and downregulated by KAN.
(B) Summary of genes downregulated by REV and upregulated by KAN.
REV v WT indicates transcripts significant (P < 0.01) for the interaction between time and genotype compared between a GR-REV line and a wild-type Col line, both treated with dexamethasone at time 0. KAN v WT is the analogous comparison between a KAN1-GR line and a wild-type Col line. KAN v REV is the analogous comparison between a GR-REV line and a KAN1-GR line. Arrows indicate direction of regulation at 30 min.
We queried the REV versus KAN analysis and found that additional family members among the five ORK genes show evidence of regulation: 5/9 PYL, 9/17 CIPK12, 5/7 HD-ZIPII, 3/6 ZFP8, and 3/8 ORK6 family members assayed showed evidence of differential regulation by REV and KAN1 (see Supplemental Figure 6 online for CIPK and PYL loci and Figure 6C for ORK6 and relatives).
Figure 6.
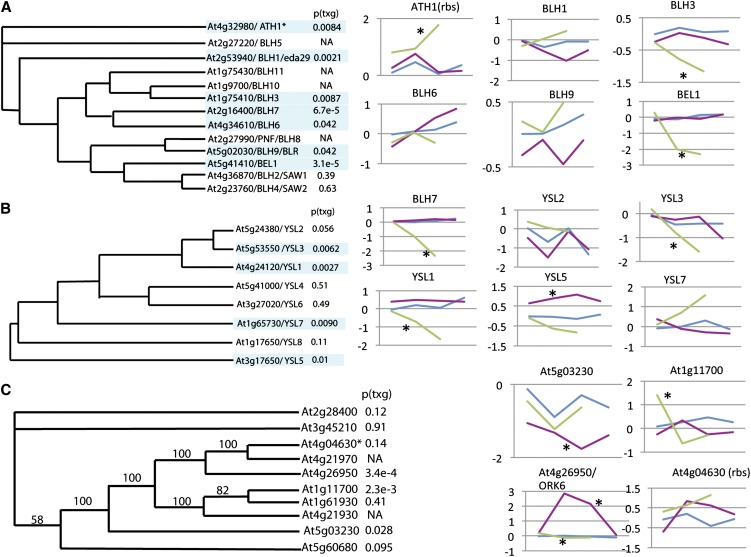
Multiple Members of the BEL, YSL, and ORK6 Gene Families Are Regulated by REV and/or KAN1.
BEL1 gene family (A), YSL gene family (B), and ORK6 gene family (C). P values next to phylogenetic trees are for genotype by time interaction in a comparison of GR-REV and KAN1-GR treated with dexamethasone. Magenta lines represent GR-REV; blue lines represent wild-type Col; green lines represent KAN1. Time points are 0, 30, 60, and 120 min after treatment. Black asterisk indicates significant P value (genotype by time < 0.05) in comparison to wild-type Col. BEL1 tree is described by Mukherjee et al. (2009). YSL tree is described by Chu (2010). ORK6 tree was generated by first aligning sequences in Clustal (see Supplemental Data Set 2 online) and then entering this alignment into MrBayes version 3.2 (900,000 trials).
Supported Subset 3: Genes with Several REV- and/or KAN1-Regulated Family Members
As shown above, multiple members of at least six gene families, all five ORK gene families and the IDD family, are targets of REV regulation. Thus, REV’s connection to its target genes appears to be, at least in some cases, evolutionarily ancient.
Two other gene families showed extensive regulation. Seven of nine BEL homeobox and four of eight YSL metal transporter family members showed evidence of regulation (P values are for comparison of REV versus KAN; Figures 6A and 6B). Whereas most of the regulation seems to occur through KAN1 for the BEL and YSL family genes (KAN versus the wild type was statistically significant, whereas REV versus the wild type was not), there is a reported REV binding site in the vicinity of ATH1 (Brandt et al., 2012). YSL proteins are metal transporters associated with the vascular strand (Chu et al., 2010). BEL1 is required for ovule development, and other BEL genes are required for shoot apical meristem formation (Rutjens et al., 2009; discussed below); both processes are influenced by KAN1.
Thus, many of the genes regulated by REV and KAN1 are clustered in gene families. This suggests the evolution of the regulator/target relationship arose early in the evolution of the land plant lineage.
Supported Subset 4: Genes Previously Known to Regulate Meristem and Leaf Development
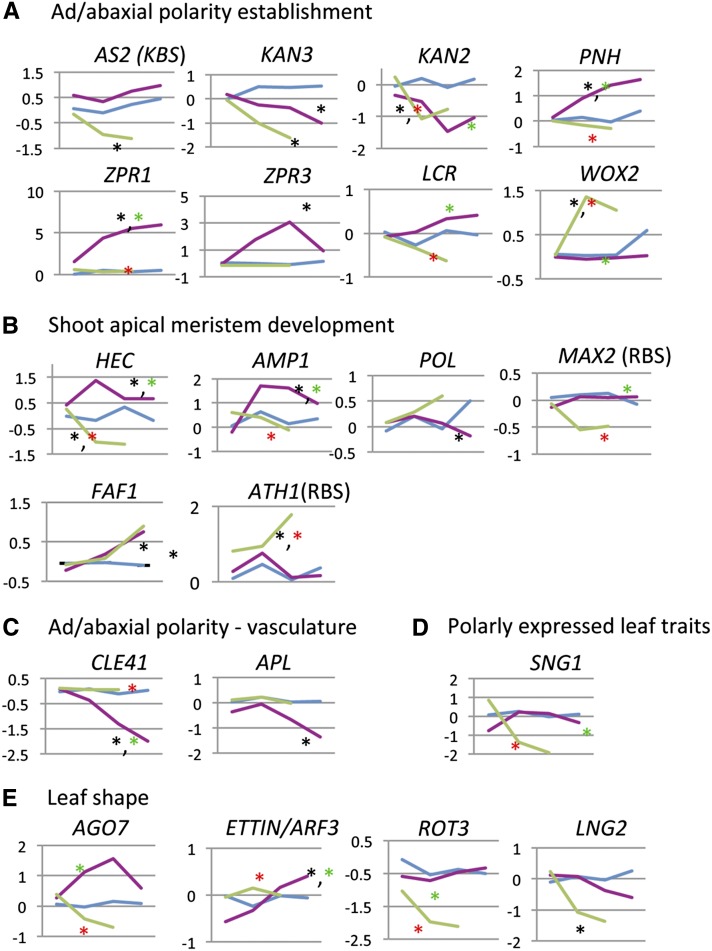
REV and KAN1 regulate organ polarity and meristem formation and maintenance. For this reason, we identified known regulators of organ polarity, meristem formation, and leaf development within the set of KAN1- and REV-regulated transcripts (Table 3, Figure 7).
Table 3. Genes Known to Regulate Meristem and Leaf Development That Are Regulated by REV and KAN1.
| P (Genotype by Time Interaction)a |
|||
|---|---|---|---|
| Locus | REV versus Col | KAN versus Col | REV versus KAN |
| Ad/abaxial polarity | |||
| AS2 (At1g65620) | 0.99 | 0.0034 | 0.10 |
| KAN3 (At4g17695) | 0.032 | 1.6E-04 | 0.11 |
| KAN2 (At1g32240) | 0.16 | 0.023 | 0.013 |
| PNH (At5g43810) | 0.0043 | 0.857 | 0.0061 |
| LCR (At1g27340) | 0.24 | 0.074 | 0.0031 |
| ZPR1 (At2g45450) | 7.3E-08 | 0.61 | 3.0E-05 |
| ZPR3 (At3g52770) | 0.0033 | 0.89 | 0.067 |
| Regulation of shoot meristem formation and function | |||
| WUSCHEL-LIKE HOMEOBOX2 (At5g59340) | 0.21 | 1.5E-06 | 1.7E-05 |
| POL (At2g46920) | 0.034 | 0.33 | 0.35 |
| AMP (At3g54720) | 0.0036 | 0.24 | 3.0E-05 |
| HECATE (At5g67060) | 0.014 | 0.0041 | 0.023 |
| FAF1 (At4g02810) | 0.010 | 0.0059 | 0.62 |
| MORE AXILLARY MERISTEMS2 (At2g42620) | 0.84 | 0.39 | 0.044 |
| ARABIDOPSIS THALIANA HOMEOBOX GENE1 (At4g32980) | 0.81 | 0.049 | 0.0084 |
| Ad/abaxial polarity in vascular strands | |||
| CLE41 (At3g24770) | 9.8E-06 | 0.84 | 0.0062 |
| APL (At1g79430) | 7.4E-04 | 0.75 | 0.45 |
| Leaf shape | |||
| ARGONAUTE7 (At1g69440) | 0.062 | 0.13 | 0.0029 |
| ETTIN/ARF3 (At2g33860) | 0.0345 | 0.448 | 0.031 |
| ROT3 (At4g36380) | 0.42 | 0.15 | 1.9E-04 |
| LNG2 (At3g02170) | 0.36 | 0.027 | 0.070 |
| Adaxial limited character | |||
| SNG1 (At2g22990) | 0.13 | 0.16 | 0.0021 |
Values in bold indicate significance at <0.05.
Figure 7.
REV- and KAN1-Regulated Loci with Known Roles in Shoot Development.
REV- and KAN-regulated transcripts previously shown to play a role in ad/abaxial polarity (A), shoot apical meristem formation and function (B), development of ad/abaxial polarity in the vasculature (C), the biosynthesis of secondary metabolites that are polarly localized (D), and leaf shape (E). x axis is time after dexamethasone induction. Time points are 0, 30, 60, and 120 min. Magenta represents GR-REV; blue represents Col wild type; green represents KAN1-GR. Black asterisk indicates statistically significant difference (P < 0.05) relative to the wild type. Red and green asterisks indicate statistically significant difference relative to GR-REV and KAN-1GR, respectively. Associated P values can be found in Table 3. KBS, possesses adjacent KAN binding site (genetically defined; Wu et al., 2008); RBS, possesses adjacent REV binding site (ChIP defined; Brandt et al., 2012).
Genes That Regulate Ad/abaxial Polarity
Genes in this subgroup affect polar development of the leaf and the ability of the plant to form and/or maintain shoot apical meristems.
ASYMMETRIC2
A transcription factor required for adaxial development, ASYMMETRIC2 (AS2) is a direct target of KAN1 repression (Wu et al., 2008). AS2 levels were reduced rapidly by KAN1-GR but were unaffected by GR-REV induction (Figure 7).
KANADI2 and KANADI3
The four KAN genes act redundantly to promote abaxial leaf fate, prevent organ outgrowth, and for seed coat development (Leon-Kloosterziel et al., 1994; Kerstetter et al., 2001; Eshed et al., 2004; Izhaki and Bowman, 2007). KAN2 and KAN3 expression decreased in response to KAN1-GR induction, indicating negative feedback between the KAN1 gene family members (Table 3, Figure 7). Induction of GR-REV also downregulated KAN2 transcripts (P < 0.05).
PINHEAD/ZWILLE
PINHEAD (PNH), a member of the ARGONAUTE family of silencing genes, is expressed in the adaxial domain of cotyledons and leaves and in vascular tissue and is required for meristem formation (McConnell and Barton, 1995; Lynn et al., 1999). Gain-of-function mutations cause upward leaf curling, a hallmark of adaxialized mutants (Lynn et al., 1999; Newman et al., 2002). PNH was upregulated following GR-REV induction (Table 3, Figure 7).
WUSCHEL-LIKE HOMEOBOX2
The homeobox gene WUSCHEL-LIKE HOMEOBOX2 (WOX2) is expressed in the apical cell of the two-cell embryo and later becomes apically and adaxially restricted (Haecker et al., 2004). Thus, the expression domain of WOX2 falls within that of HD-ZIPIII mRNA and is excluded from the domain expressing KAN1. WOX2, together with other WOX loci, is required for shoot apical meristem and cotyledon formation, and embryos lacking WOX activity are phenotypically similar to HD-ZIPIII loss-of-function mutants (Breuniger et al., 2008). WOX2 was upregulated by KAN1 (Table 3, Figure 7). This is likely to be an indirect effect and, given the WOX2 expression pattern, likely represents the nonautonomous action of KAN1 on adjacent, adaxially located cells.
LEAF CURLING RESPONSIVENESS
Loss-of-function mutations in the LEAF CURLING RESPONSIVENESS (LCR) gene cause upward leaf curling, while increased expression of LCR causes downward leaf curling (Song et al., 2012) and meristem termination (Knauer et al., 2013). Thus, LCR behaves like an abaxializing agent. LCR encodes an F-box protein whose target is unknown. LCR was upregulated in GR-REV relative to KAN1-GR (Table 3, Figure 7). Since this is counter to the direction of action of these genes (GR-REV is adaxializing while LCR is abaxializing), this regulation is likely part of a negative feedback mechanism.
LITTLE ZIPPER1 and LITTLE ZIPPER3
LITTLE ZIPPER (ZPR) genes encode short proteins that bind to and inhibit HD-ZIPIII proteins (Wenkel et al., 2007; Kim et al., 2008). As in earlier experiments, we found upregulation of both ZPR1 and ZPR3 by REV in our extended time-course experiment. The two genes differed slightly in their behavior, with ZPR1 levels remaining high, while ZPR3 mRNA levels peaked and had begun to decline by 2 h (Figure 7). No regulation by KAN1-GR of either locus was detected.
Genes Required for Shoot Apical Meristem Formation and Function
Genes in this category affect the ability of the plant to form and/or maintain shoot meristems but do not grossly affect leaf polarity.
BEL-1-LIKE Homeodomain Genes
BEL-1 LIKE homeodomain proteins form heterodimers with Class I KNOTTED-LIKE HOMEOBOX proteins. Together, these are required for shoot apical meristem formation and maintenance (Long et al., 1996; Kanrar et al., 2006; Rutjens et al., 2009). We found no Class I KNOX genes among the REV- or KAN-regulated loci. However, several members of the BEL family were affected in the time-course experiments (Table 3, Figures 6 and 8). Among the BEL family of genes, ATH1 together with POUNDFOOLISH/BEL1 LIKE HOMEODOMAIN8 BLH8 and PENNYWISE/BEL1 LIKE HOMEODOMAIN9 are required for shoot apical meristem formation (Rutjens et al., 2009). ATH1 was upregulated by KAN1-GR (P = 0.049 GR-KAN versus Col). No significant effect of GR-REV induction on ATH1 was observed in the time course even though a REV binding site was identified adjacent to ATH1 (Brandt et al., 2012).
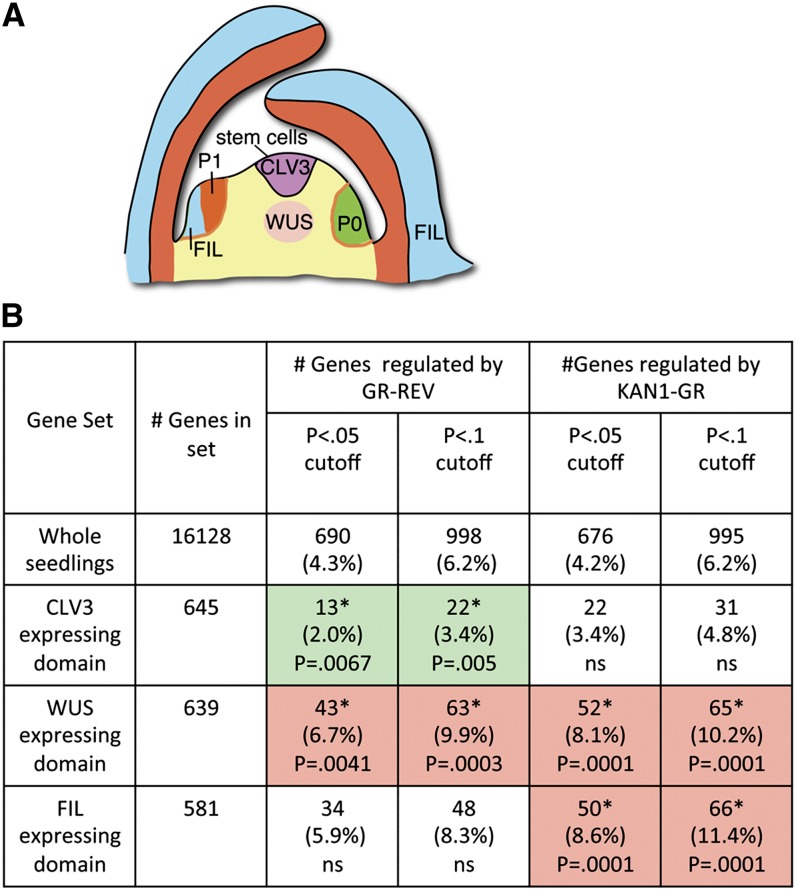
Figure 8.
Evidence for Spatial Regulation of REV- and KAN-Regulated Transcripts.
(A) Diagram of vegetative shoot apical meristem. The CLV3, WUS, and FIL domains are color coded purple, pink, and blue, respectively. Cells in the CLV3 domain are predominantly the stem cells that divide to produce descendants that are pushed into the peripheral and rib zones to produce differentiated leaf and stem segments, respectively.
(B) Table showing fraction of genes expressed specifically in CLV3, WUS, and FIL domains that are regulated by either REV or KAN1. Gene lists for GR-REV or KAN1-GR were generated using cutoffs of either P < 0.05 or P < 0.1. Pink shading indicates sets in which more genes than expected by chance alone are regulated. Green shading indicates where fewer genes than expected by chance alone are regulated. Asterisk indicates difference from expected number at P < 0.01.
POLTERGEIST
The POLTERGEIST (POL) locus encodes a phosphatase required for stem cell activity in the shoot apical meristem (Yu et al., 2003). POL regulates cellular polarity in the embryo (Song et al., 2008) and was downregulated toward the end of the time course by GR-REV (Table 3, Figure 7).
MORE AXILLARY BRANCHES2
MORE AXILLARY BRANCHES2 (MAX2) encodes an F-box protein responsible for inhibiting axillary meristem outgrowth in response to a signal transported from base to tip of the plant (Stirnberg et al., 2007). MAX2 was upregulated by both REV and KAN (Table 3, Figure 7) and is adjacent to a ChIP-defined REV binding site (see Supplemental Data Set 1E online).
FANTASTIC FOUR1
The FANTASTIC FOUR1 (FAF1), FAF2, FAF3, and FAF4 genes encode plant-specific proteins of unknown function that when overexpressed cause shoot apical meristem arrest (Wahl et al., 2010). The FAF1 gene is expressed at the adaxial base of leaf primordia and in the developing vasculature. FAF1 (but not FAF2, FAF3, or FAF4) responded to both GR-REV and KAN1-GR induction with an increase in transcript levels (Table 3, Figure 7).
ALTERED MERISTEM PROGRAM
Mutants defective for the ALTERED MERISTEM PROGRAM (AMP) gene make an enlarged shoot apical meristem and produce more closely spaced organ primordia (Chaudhury et al., 1993). Tricotyledonous plants are common. This is similar to what is seen when HD-ZIPIII activity is increased (Kim et al., 2008). AMP was upregulated by REV. This is opposite to that expected from their inferred biological roles and may therefore reflect a negative feedback step.
Genes That Regulate Vascular Polarity
CLAVATA3/ESR RELATED41
CLAVATA3/ESR RELATED41(CLE41) is a small peptide signal synthesized in the presumptive phloem of the developing vasculature (Etchells et al., 2012). It is required to control cell division in the adjacent cambium and to repress xylem cell fate. Asymmetric expression of CLE41 is important to its function. It was downregulated by REV (Table 3, Figure 7), consistent with the known role for REV in promoting adaxial vascular strand development (i.e., xylem).
ALTERED PHLOEM
ALTERED PHLOEM (APL) is a MYB-like transcription factor required for phloem identity (Bonke et al., 2003). APL was downregulated by REV, consistent with our understanding that REV promotes adaxial vascular strand development (xylem) and represses abaxial vascular strand development (phloem).
Genes That Regulate Leaf Shape
ZIPPY
ZIPPY is a member of the ARGONAUTE family of silencing genes. Loss-of-function mutations in ZIPPY cause downward curled leaves, a trait shared with many abaxialized mutants (Hunter et al., 2003). Abaxialization of ZIPPY mutants can be explained by inappropriate persistence of its target, ETTIN/AUXIN RESPONSE FACTOR (ARF3), which is thought to be important in abaxial leaf development. ZIPPY is expressed in the adaxial medial domain of leaf primordia (Chitwood et al., 2009) and is upregulated by REV (Table 3, Figure 7).
AUXIN RESPONSE FACTOR3
ZIPPY targets trans-acting siRNAs which in turn target a subset of ARF mRNAs. One of those is ETTIN/ARF3 which has been interpreted as promoting abaxial development. Furthermore, ETTIN has been reported to physically associate with KAN1 (Kelley et al., 2012). Surprisingly, ETTIN/ARF3 mRNA increased in response to GR-REV activation (Table 3, Figure 7), opposite to what was expected, as increased ZIPPY is predicted to result in increased trans-acting siRNAs production and, therefore, decreased ETTIN/ARF3 levels.
LONGIFOLIA2
Mutations in the LONGIFOLIA (LNG1) and LNG2 genes cause leaves to develop longer, narrower blades through increased polar cell expansion (Lee et al., 2006). KAN1 reduced LNG2 levels (Figure 7). Examination of expression data from Yadav et al. (2009) shows that LNG2 is enriched in the WUSCHEL (WUS)–expressing domain of the shoot apical meristem, a domain enriched for both REV- and KAN1-regulated genes (see next section).
ROTUNDIFOLIA3
ROTUNDIFOLIA3 (ROT3) encodes an enzyme in the brassinolide biosynthesis pathway (Ohnishi et al., 2006). Like the LNG1 and LNG2 genes, ROT3 is required for the proper development of an elongated leaf shape but operates independently of them. Similar to those genes, ROT3 was downregulated by KAN1-GR (Figure 7).
Genes That Regulate Leaf Cell Differentiation
SINAPOYL GLUCOSE1
SINAPOYL GLUCOSE1 (SNG1) encodes an enzyme that catalyzes a step in the biosynthesis of synapoylmalate, a compound found primarily in adaxial leaf cells, where it acts to protect the cells from UV radiation (Lehfeldt et al., 2000). SNG1 was downregulated by KAN1 (Table 3, Figure 7), suggesting the mechanism for localized sinapoylmalate accumulation involves repression of gene expression for the SNG1 enzyme in the abaxial domains of the leaf.
Supported Subset 5: Genes Expressed in FIL, CLAVATA, and WUS Domains Regulated by REV and/or KAN1
The HD-ZIPIII genes are expressed in the adaxial domains of early-stage leaf primordia and in the shoot apical meristem to varying extents (McConnell et al., 2001; Prigge et al., 2005). KAN1 genes, on the other hand, are expressed in the abaxial leaf domain in leaf primordia (Eshed et al., 2004; Izhaki and Bowman, 2007). Whereas these regulators show quite distinct patterns of expression, their targets need not do so; quantitative differences in target gene between REV- and KAN-expressing tissues could be sufficient to provide a difference in cell type and/or behavior. Nevertheless, it is likely that at least some downstream targets show distinct ad- and abaxial expression patterns. Whereas carrying out in situ hybridization experiments on the regulated targets is beyond the scope of this work, we compared the expression of the predicted REV and KAN1 targets to transcripts enriched in subdomains of the shoot apical meristem as determined by Yadav et al. (2009) (Figure 8). These are the CLAVATA3 (CLV3)–expressing domain (central domain including the uppermost layers of the shoot apical meristem); the WUS-expressing domain (central domain just underneath the CLV3-expressing domain); and the FILAMENTOUS FLOWER (FIL)-expressing domain (abaxial domain of primordia; diagrammed in Figure 8A). REV is expressed in the CLV3 and WUS domains, while KAN1 is primarily expressed in the FIL domain.
Both REV-regulated and KAN1-regulated transcripts were statistically overrepresented in the WUS domain (Figure 8B). KAN1-regulated, but not REV-regulated, transcripts were overrepresented in the FIL domain. Somewhat surprisingly, REV-regulated targets were underrepresented in the CLV3 domain. REV/KAN-regulated genes expressed in these domains are listed in Supplemental Data Sets 1J to 1M online.
Supported Subset 6: Genes Encoding Proteins That Physically Interact with REV
To determine if any of the other genes regulated by GR-REV might be part of feedback loops similar to ZPR proteins, we screened for proteins that physically interact with REV in yeast. To increase the likelihood of finding genes expressed in few cells, the library we screened was generated from RNA made from inflorescence meristems. Two separate screens were done: one using the full-length REV protein and one using REV lacking the terminal MEKHLA domain. The truncated REV construct was used because the MEKHLA domain inhibits both homodimerization of REV as well as the ability of REV to interact with the ZPR proteins (Magnani and Barton, 2011). Table 4 shows the proteins that were isolated more than once in these screens and that retested positive in a follow-up test. The majority (10/12) of the interacting proteins were transcription factors.
Table 4. REV-Interacting Proteins and Their Regulation.
| Gene | Interacts with Full-Length REV in Retest? (No. Times Isolated in Y2H) | Interacts with REV-MEKHLA in Retest? (No. Times Isolated in Y2H) | Transcript Regulated by GR-REV? | Biological Function |
|---|---|---|---|---|
| TCP8 (At1g58100) | Yes (2) | Yes (2) | No | Unknown |
| TCP14 (At3g47620) | Yes (0) | Yes (28) | Upregulated, P = 0.028 | Loss-of-function mutants have upward curled leaves,1 delayed germination, hypersensitivity to abscisic acid2 |
| TIFY8 (At4g32570) | Yes (8) | Yes (2) | No | Unknown |
| IDD4 (At2g02080) | Yes (8) | Yes (5) | Downregulated, P = 0.045 REV binding site adjacent | Overexpression causes downward curled leaves (Figure 10) |
| IDD5 (At2g02070) | ND (0) | Yes (2) | REV binding site adjacent | Unknown |
| ID4-like (At1g14580) | ND (4) | ND (0) | No | Unknown |
| ARR2 (At4g16110) | Yes (0) | Yes (2) | No | Cytokinin-mediated transcriptional regulation. Cytokinin interacts with PHABULOSA in regulating root growth3 |
| Zinc finger (At1g53190) | ND (0) | Yes (8) | NA | Unknown |
| Ring finger (At4g01290) | ND (0) | Yes (2) | No | Unknown |
| CDF3 At3g47500 | ND (0) | Yes (2) | No | Transcriptional repressor of flowering regulator CO |
| Curculin-like At1g78820 | Yes (2) | ND (0) | No | Unknown |
| Major Facilitator Superfamily protein (At5g05310) | Yes (2) | ND (0) | No | Unknown |
NA, not assayable in microarray study; ND, not determined; Y2H, yeast two-hybrid assay.
Two members of the TCP family of transcription factors were isolated. The gene encoding one of these, TCP14, was transcriptionally upregulated by GR-REV. In tcp14 loss-of-function mutants, leaves curl upward (Kieffer et al., 2011). This suggests the wild-type role of TCP14 is to physically interact with and inhibit, or alter, the ability of REV to activate genes that coordinate leaf growth on upper and lower sides of the leaf.
Three members of the IDD family of zinc finger–containing transcription factors were identified. Strikingly, among these was IDD4, which was identified in earlier sections of this work. This suggests that IDD4 acts together with GR-REV to promote and coordinate outgrowth of a flattened leaf blade.
The HD-ZIPIII gene PHB interacts genetically with the cytokinin pathway, in part by activating transcription of a cytokinin biosynthesis enzyme (Dello Ioio et al., 2012). It is therefore significant that ARABIDOPSIS RESPONSE REGULATOR2 (ARR2), a member of the ARR gene family of cytokinin-responsive factors, was identified in the screen. Three others that were tested, ARR1, ARR12, and ARR14, all interacted physically with REV. ARR12 and ARR14 interacted only with the MEKHLA-less REV construct. No evidence for transcriptional regulation of ARR factor-encoding genes by either REV or by KAN1 was observed.
Supported Subset 7: Genes That Cause Alterations in Leaf Development When Overexpressed
To determine if the genes we identified in the time-course experiments as regulated by REV and/or KAN1 were sufficient to cause changes in leaf morphology, we systematically overexpressed the corresponding full-length cDNAs for 27 genes (listed in Supplemental Data Set 1N online) regulated by REV, KAN, or both under the control of the 35S promoter and examined the phenotypes of T1 seedlings. T1 seedlings were grown on sterile medium (including 30 µM hygromycin) so that plants with subviable morphologies could be observed. T2 plants were grown without selection. Genes were scored for having an effect on leaf morphology if at least four T1 plants showed a similar alteration in leaf morphology. In addition, phenotypes were validated in the T2 generation for four of the lines shown in Figure 9 (IDD4, TCP-DOMAIN PROTEIN 9 [TCP9], TETRATRICOPEPTIDE-REPEAT THIOREDOXIN-LIKE 4 [TTL4], and At2g44500). T2 plants were not tested for At2g44500 and could not be tested for SCHLAFMUTZE (SMZ) because the T1 plants were sterile.
Figure 9.
REV- and KAN1-Regulated Genes That Cause Changes in Leaf Morphology When Overexpressed.
(A) and (B) Adaxial (upper) (A) and abaxial (B) views of wild-type Col leaf.
(C) and (D) Adaxial (C) and abaxial (D) views of leaf from plant overexpressing IDD4/At2g02080. Note curled under leaf blade.
(E) and (F) Adaxial (E) and abaxial (F) views of leaf from plant overexpressing SCHLAFMÜTZE/At3g54990. Leaves are narrow, asymmetric, and lobed.
(G) View from above of leaf from plant overexpressing TCP9/At2g45680 showing a duplicated, fused leaf.
(H) View from above of leaf from plant overexpressing TTL4/At3g58620 showing a duplicated, fused leaf.
(I) and (J) Adaxial (I) and abaxial (J) view of leaf from plant overexpressing HLS/At4g37580 showing a duplicated, fused leaf.
(K) and (L) Adaxial (K) and abaxial (L) view of leaf from plant overexpressing At2g44500. Leaf is folded up at the basal end of the leaf blade.
IDD4 was found to cause downward leaf curling in this assay (Figures 9C and 9D). SMZ, an AP2-like protein (Yant et al., 2010), caused the formation of narrow, asymmetric, and lobed leaves (Figures 9E and 9F). The TCP9 protein At2g45680 caused the production of duplicated, fused leaves as did the TTL4 and HLS proteins. Upward leaf curling was observed when At2g44500, a predicted O-fucosyltransferase gene, was overexpressed. Thus, At2g45680, TTL4, and HOOKLESS (HLS) are candidates for establishing overall ad/abaxial polarity of the leaf primordium, while SMZ, IDD4, and At2g44500 are candidates for regulating and/or coordinating leaf blade outgrowth.
DISCUSSION
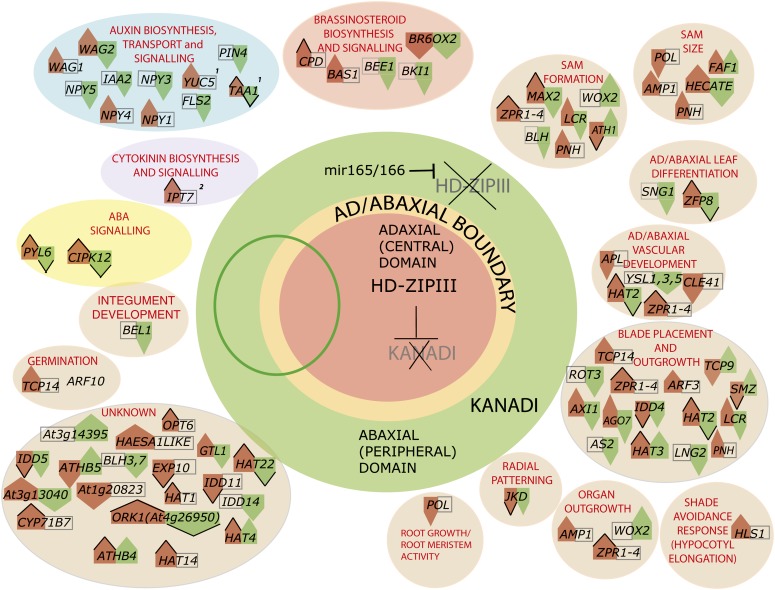
REV and KAN1 exert dramatic effects on plant development: They affect the top/bottom (or ad/abaxial) polarity of leaves, the formation of new meristems, and the placement and formation of leaf blades. Figure 10 summarizes the REV/KAN regulation of many target genes identified in this study and the processes and tissues they are known, or hypothesized, to act in. This bird's eye view of REV and KAN action shows they act at the top of a hierarchy that includes many genes involved in signal transduction, especially in hormone signaling, as well as many transcription factors that play subsidiary roles in leaf polarity and meristem formation.
Figure 10.
Model for Transcriptional Regulation of Ad/abaxial Polarity by REV and KAN1.
Schematic of plant from above, looking down the long axis of the plant. REV and other HD-ZIPIII genes are expressed in the central, or adaxial, domain of the plant. Much of the shoot apical meristem resides within this domain. Expression of HD-ZIPIII mRNAs is limited to the adaxial domain, at least in part, through the action of microRNAs 165 and 166, which cause degradation of HD-ZIPIII RNA in the abaxial domain. It is not known what controls mir165 and mir166 expression patterns. The leaf primordium (shown schematically as an oval) develops from the flanks of the apical meristem with its adaxial (upper) side located closer to the center of the plant and its abaxial (lower) side located toward the periphery. The leaf blade grows out at the junction of ad- and abaxial domains. Processes affected by the ad/abaxial network, specifically by HD-ZIPIII and KAN1 proteins, are indicated in bubbles surrounding the schematic. Genes targeted by REV and/or KAN1 are shown in bubbles corresponding to the processes in which they are hypothesized to take part. Red arrows (left side of each double arrow) indicate up- or downregulation by REV. Green arrows (right side of each double arrow) indicate up- or downregulation by KAN1. Arrowheads outlined in black indicate that the interaction is likely direct, based either on the presence of the corresponding binding site in the target gene or on the observation that regulation is resistant to cycloheximide. (1) Reported by Brandt et al. (2012). (2) Reported by Dello Ioio et al. (2012).
Our observations support the primary function of HD-ZIPIII proteins as activators, and KAN1 proteins as repressors, of gene expression. First, a majority of the candidate REV target genes were upregulated, while a majority of the KAN1 candidate target genes were downregulated. Second, of genes that behave as expected for direct targets of REV, all but two (EXPANSIN10 and IDD4) were upregulated. This parallels transient expression experiments in tobacco (Nicotiana tabacum) in which a ZPR3–β-glucuronidase reporter is activated in the presence of REV (Magnani and Barton, 2011). All genes that behaved as likely direct targets of KAN1 were downregulated, consistent with the known requirement for the KAN binding site in the negative regulation of AS2 (Wu et al., 2008).
Caveats: False Negatives and False Positives
Although this system is effective at identifying genes regulated by REV and KAN1, we have likely missed many target genes. First, if tissue-specific cofactors are necessary for regulating a subset of targets in a small number of cells, we might not detect those targets since the cofactor would be missing in many cells overexpressing the transcription factor. Second, ∼40% of protein coding genes cannot be examined in our survey since they are either not on the ATH1 array or are expressed at insufficient levels for comparison using microarrays. Third, downregulated genes that encode stable mRNAs might not be identified in a short time course such as the one followed here. However, negatively regulated genes showed similar fold changes to the positively regulated genes over the 1-h time course, indicating many negatively regulated genes have unstable mRNAs. The most strongly negatively regulated gene reached 3% of starting mRNA levels by 1 h after induction. Assuming complete inhibition of new mRNA synthesis by the transcription factor, this corresponds to an mRNA with a half-life of roughly 12 min. Thus, negative regulators such as KAN1 have the potential to substantially clear the cell of particular transcripts over the course of 1 to 2 h.
It is also possible that we have identified genes that are not true targets of REV or KAN. First, we may have identified loci that do not normally respond to REV but to other members of the HD-ZIPIII gene family since these proteins share similar DNA binding domains. Second, the GR-REV and KAN1-GR proteins are expressed at high levels in the transgenic plants and may bind to sites not normally bound at the HD-ZIPIII or KAN concentrations normally found in the cell. Additional genetic and gene expression studies will be required to sort this out, but the identified targets provide good starting points for such studies.
The IDD4 Gene, a New Actor in the Ad/abaxial Pathway
Multiple lines of evidence support a role for the IDD4 transcription factor in ad/abaxial leaf development. First, IDD4 is downregulated by both REV and KAN1. While REV primarily has been observed to act a transcriptional regulator, the IDD4 gene is adjacent to a REV binding site, strongly suggesting that in some cases REV acts as a transcriptional repressor. Given the ability of REV to interact with several other types of transcription factors in yeast two-hybrid experiments, a reasonable model is that REV interacts with factors that turn it into a repressor at some loci.
IDD4 is an a1-type zinc finger protein that contains four tandemly arranged zinc fingers (Englbrecht et al., 2004). It is named after the Indeterminate gene of maize (Zea mays) that controls flowering time (Colasanti et al., 2006). In addition to identifying the IDD4 locus as a target of REV and KAN1 regulation, we also isolated the IDD4 protein as a physical interactor with REV in a yeast two-hybrid assay. When expressed under the control of the constitutive 35S promoter, we find that IDD4 causes downward leaf curling. Since this is what happens when HD-ZIPIII function is reduced, we hypothesize that IDD4 acts to counter HD-ZIPIII activity in the leaf by physically altering REV activity.
Multiple members of the IDD family of transcription factor genes are regulated by REV and KAN1 and/or have adjacent REV binding sites. Of these, biological function has only been reported for JACKDAW. JACKDAW is most strongly expressed at the boundary of adaxial and abaxial cells in the hypocotyl/root of the embryo (Welch et al., 2007). Liu et al. (2012) proposed the hypothesis that a boundary expression pattern for IDD loci could be established in part by negative regulation of IDD transcription by REV and KAN1 in the respective adaxial (REV) and abaxial (KAN1) domains but not along the boundary between these domains. The gradient of HD-ZIPIII expression in the embryo (McConnell et al., 2001) together with the mutually antagonistic regulation between REV and IDD would be responsible for low levels of IDD mRNA in the central most region of the embryo and high levels of IDD at the boundary.
Identification of Feedback Loops
Several of the genes regulated by REV and KAN1 play a part in feedback loops (see Supplemental Figure 7 online). Indeed, the first target genes of REV identified, those encoding the ZPR family of proteins, appear to be part of such a feedback loop (Wenkel et al., 2007; Kim et al., 2008). The ZPR proteins consist primarily of a Leu zipper that is similar to and can dimerize with the larger HD-ZIPIII proteins. In vitro, this small protein prevents the HD-ZIPIII protein from binding to DNA and in vitro overexpression of ZPR proteins results in loss of HD-ZIPIII function (Wenkel et al., 2007). Seedlings carrying mutations in loss of function mutants for both ZPR3 and ZPR4 display a subset of phenotypes seen in HD-ZIPIII gain-of-function mutants: multiple cotyledons and an enlarged shoot apical meristem (Kim et al., 2008).
Similar to the ZPR proteins, the transcription factors IDD4 and TCP14 were both found in this study to physically interact with REV and are regulated by REV (TCP14 up and IDD4 down). Both appear to act oppositely to REV in their effect on leaf development; TCP14 loss-of-function mutants have upward curled leaves and IDD4 overexpressors have downward curled leaves. REV loss-of-function mutants have downward curled leaves and REV overexpressors have upward curled leaves. Thus, regulation of TCP14 and IDD4 transcription by REV may be part of additional feedback loops.
Another feedback loop links HD-ZIPIII transcription factors with control of synthesis and/or action of the microRNAs that destroy the HD-ZIPIII mRNAs. REV increases transcript levels of the PNH gene. PNH transcripts are located in the adaxial and central domains of the embryo (Lynn et al., 1999). PNH encodes an ARGONAUTE family member. Unlike other family members that degrade the mRNAs they are targeted to, PNH has been proposed to lack slicing activity in vivo and associate specifically with microRNA165/166 to protect HD-ZIPIII mRNAs (Zhu et al., 2011). If true, this would indicate that REV and PNH act together in a feed forward loop.
Models for Antagonistic Action of HD-ZIPIII and KAN1 Factors
REV and KAN1 promote opposite polar fates in the leaf. The mRNAs for REV and KAN are localized to opposite sides of the leaf primordium. In plants where HD-ZIPIII genes are ectopically expressed in the abaxial leaf domain, KAN1 expression is reduced and vice versa. The results from our studies indicate at least two ways this antagonism works. First, REV downregulates at least one of the KAN1 genes, KAN3. It is unknown whether this is a direct effect on KAN3 transcription or an indirect effect. We found no evidence of the opposite effect, KAN1 negatively regulating HD-ZIPIII transcription. This is consistent with findings by Gillmor et al. (2010); mutations in subunits of the MEDIATOR complex cause reduced expression of KAN1 and KAN2 but have no effect on expression of the HD-ZIPIII PHABULOSA gene. KAN1 also downregulates expression of KAN2 and KAN3 gene family members in what appears to be a negative feedback loop (see Supplemental Figure 7 online).
Second, REV and KAN1 have opposite effects on a set of promoters. The PYL6, CIPK12, ZFP8, HAT2, and ORK6 (At4g26950) loci all appear to be directly and oppositely targeted with REV activating and KAN1 repressing them. Kelley et al. (2009) proposed a balance model to explain the opposing interactions of KAN1 and HD-ZIPIII genes in ovule development. They observed that KAN4 is expressed normally in ovules with ectopic HD-ZIPIII expression. Based on this as well as on the observations of phenotypes of multiple mutant combinations, they proposed a “balance” model where the amount of HD-ZIPIII activity relative to KAN1 activity is weighed in determining adaxial versus abaxial fates. The discovery of genes regulated by both REV and KAN1 provide support for a balance model. The responses of such common targets are expected to depend on the relative amounts of HD-ZIPIII and KAN activity and/or on the relative timing of regulation (i.e., which factor is first to act). Future use of direct target genes as reporters should enable testing of the balance model and, if it holds, to determine what concentrations of REV relative to KAN1 allow expression versus repression of the locus.
REV Interacts Physically with Different Types of Transcription Factors
Several different types of transcription factors interact physically with REV. These include IDD4 (a C2H2 zinc finger protein), TCP14 (contains a TCP DNA binding domain), ARR2 (contains a MYB-like DNA binding domain), and TIFY DOMAIN 8 PROTEIN (contains the plant-specific TIFY domain). For two of these, IDD4 and TCP14, we found evidence of REV regulation of the corresponding loci; TCP14 is upregulated by REV, whereas IDD4 is downregulated. In addition, IDD4 has an adjacent REV binding domain (Brandt et al., 2012). For three of these, there is phenotypic evidence that the proteins play a role in ad/abaxial development. We observed downward curled leaves in IDD4 overexpressors, a phenotype seen in abaxialized mutants. Plants with reduced TCP14 function are reported to have upward curled leaves (Kieffer et al., 2011). Upward curling is seen in adaxialized mutants and is increased by HD-ZIPIII activity. Mutants in ARR genes are defective in cytokinin responses and interact genetically with mutants in the HD-ZIPIII gene PHABULOSA (Dello Ioio et al., 2012).
These newly identified interacting proteins are in addition to previously identified proteins that include the ZPR proteins and the AP2 domain–containing proteins DÖRNRÖSCHEN and DÖRNRÖSCHEN-LIKE (Chandler et al., 2007; Wenkel et al., 2007; Kim et al., 2008). For both ZPR proteins and DÖRNRÖSCHEN proteins, there is evidence of genetic interaction to support the significance of the physical interaction.
The identification of many types of interacting transcription factors suggests either that the interacting transcription factors contain a common HD-ZIPIII interaction domain that has yet to be identified or that HD-ZIPIII proteins contain many different interaction surfaces that can be exploited by the various interactors. The first alternative, the presence of a domain common to all these proteins, includes not only domains formed by primary amino acid sequence but also domains involving protein modifications. The protein modifications would then be recognized by REV, perhaps by the StAR-related lipid transfer domain, which is known to recognize ligands.
REV's interaction with multiple transcription factor types suggests REV acts in combination with additional factors to achieve tissue and stage specificity in regulation. It is known that the presence of REV alone cannot explain the specificity of ZPR expression patterns as these genes are expressed in only a subset of REV-expressing cells (Wenkel et al., 2007). This is one context where additional factors must exist to allow the observed cell specificity of REV action.
Evolution of HD-ZIPIII KAN1 Regulation of Targets
In several gene families, we found many family members to be regulated by REV and/or KAN1. This was true for the YSL, BEL, IDD4, HD-ZIPII, PYL, and CIPK families. The large number of related loci that are under REV and/or KAN1 control suggest that the targets came under REV and or KAN1 control early in the evolution of land plants. Conversely, “solitary” loci such as SNG1 may be relatively new evolutionary recruits to the ad/abaxial pathway. Sinapoylglucosides are specific to the Brassicaceae where they act as UV shields. In Arabidopsis, they are expressed where they would be expected to be most useful: on the upper side of the leaf. Downregulation by KAN1 may explain their location in the leaf.
Unexpected and Novel Targets
We identified unexpected targets using this approach. Although the ABA signaling pathway has not been previously implicated in the regulation of ad/abaxial development, regulation of this pathway by REV and KAN is striking. The PYL6 ABA receptor and the CIPK12 signaling kinase genes are both oppositely regulated by REV (up) and KAN (down). Examination of the response of other members of these families shows that regulation of family members by REV and/or KAN is extensive. These findings suggest the importance of future experiments to determine what the role of ABA might be in ad/abaxial plant development and, conversely, the effects the ad/abaxial pathway might have on ABA signaling in the plant.
Also among the oppositely regulated genes is ORK6, a gene encoding a protein of unknown function. The fact that this locus is highly regulated by REV and KAN suggests that ORK6 is involved in leaf and or meristem development and will help guide future experiments on ORK6 function. This is an example of how network analysis can aid in the understanding of function for proteins where the amino acid sequence does not provide helpful clues.
METHODS
Plant Growth and Transgenic Lines
The GR-REV and KAN1-GR transgenic lines used in this study have been previously described (Wenkel et al., 2007; Wu et al., 2008). A Col control was grown simultaneously with each of the transgenic lines. To minimize noise caused by differences in treatment, we grew seedlings in liquid culture in flasks. Seeds were sterilized and cold treated (4° for 48 h), and seedlings were grown in Murashige and Skoog liquid media with 1% Suc (50 seedlings/50 mL) at 20°C in constant light for 7 d before induction with 50 µm dexamethasone (1:1000 dilution from a 50 mM stock dissolved in 95% ethanol). Plants were treated with dexamethasone at mid-day (∼11 am to 1 pm). At times 0, 30, 60, and 120 min, plants were removed from flasks, media were drained, and tissue was flash frozen in liquid nitrogen.
For cycloheximide treatment, lines carrying GR-REV or KAN1-GR transgenes and Col were grown as above. At 7 d of growth, all samples were pretreated with either 50 µM cycloheximide (1:1000 dilution from a 50 mM stock dissolved in 95% ethanol) or mock treated with ethanol alone (1:1000 dilution of 95% ethanol). After 20 min, all samples were treated with either 50 µM dexamethasone or mock treated with ethanol alone (1:1000 dilution of 95% ethanol) for 60 min before freezing in liquid nitrogen.
Microarray and Data Analyses
Total RNA was prepared using the Qiagen RNeasy kit according to the manufacturer, and genomic DNA was removed from the samples with the Ambion DNA free kit. cRNA was prepared and hybridized to Affymetrix ATH1 GeneChips according to the manufacturer’s instructions, then washed and labeled on the GeneChip Fluidics Station 400. Three biological replicates for each time point were used for the transgenic lines, and six replicates were used for each wild-type time point (except for 120 min, where only three were used). Chips were scanned on an Agilent GeneArray Scanner G2500A, and images were analyzed with Affymetrix GeneChip operating software.
Data analysis was performed using the GeneSpring 11 software package. CEL files were loaded as a group, normalized with the GC-RMA algorithm, and filtered on level of expression to remove the lowest 30% in order to minimize variation in the expression signals. To identify genes with significant changes in expression in response to DEX induction of GR-REV or GR-KAN, two-way ANOVA was conducted on the filtered data set with asymptotic P value computation and Benjamini-Hochberg multiple testing correction. CEL files are available at the Gene Expression Omnibus database under number GSE30702.
Overexpression of REV- or KAN-Regulated Genes
Clones with the corresponding Arabidopsis thaliana cDNA flanked by Gateway sequences in pENTR223 (Yamada et al., 2003) were obtained from the ABRC. The cDNAs were moved into the pJAN35S or pMDC32 vector using the Gateway system according to the manufacturer’s instructions. The resulting overexpression constructs were transformed into Col plants using the floral dip method. Transformants were identified with BASTA (pJAN35S vector) or hygromycin (pMDC32 vector) selection on sterile media to allow the recovery of weak or morphologically abnormal seedlings. Resistant plants were transplanted to soil and self-progeny seeds harvested.
qRT-PCR
Biological triplicates of seedlings were grown in sterile liquid culture and treated as described above. Two micrograms of total RNA (Qiagen RNeasy kit) was used to make cDNA using an oligo(dT) primer and M-MLV reverse transcriptase according to the manufacturer’s instructions (Applied Biosystems/Ambion). PCR was done using gene-specific primers (see Supplemental Data Set 1O online) in technical triplicates on a LightCycler 480 system using the LightCycler 480 DNA SYBR Green I Master mix (Roche Applied Science). The ratio of experimental target mRNA to an ACTIN control for each sample was calculated by Applied Biosystems software. An average for the biological replicates and standard deviation were calculated in Excel.
Yeast Two-Hybrid Library Screening
Bait proteins were fused to the GAL4 DNA binding domain in pDEST32 (Invitrogen) and used to screen two Arabidopsis prey libraries constructed in pEXP-AD502 (Invitrogen), which contains the GAL4 activation domain. Screens were performed using His selection in the presence of 3-amino-1,2,4-triazole, according to the manufacturer’s instructions. One library was constructed from RNA isolated from Col Arabidopsis inflorescence tissue (meristem through first open flower). The second library was constructed from vegetative apex tissue of 4-week-old late-flowering FRI-Col plants (meristems plus leaves smaller than 3 mm) (Lee et al., 1994). Positive clones were rescreened for interaction and scored as positive if they matched or exceeded the interaction strength of the intermediate control standards pPC97-CYH2-dDP/pPC86-dE2F (Magnani and Barton, 2011).
Accession Numbers
CEL files with data from the microarray experiments are available at the Gene Expression Omnibus database under the number GSE30702.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Direction and Amplitude of Transcript Level Changes in GR-REV and KAN1-GR Lines Treated with Dexamethasone.
Supplemental Figure 2. Microarray-Based Time-Course Data for Genes Regulated in the Same Direction by Both GR-REV and KAN1-GR (P < 0.01).
Supplemental Figure 3. qRT-PCR Data for ORKs Upregulated in GR-REV and Downregulated in KAN1-GR.
Supplemental Figure 4. qRT-PCR Data for ORKs Downregulated in GR-REV and Upregulated in KAN1-GR.
Supplemental Figure 5. qRT-PCR Data for Class II HD-ZIP Loci.
Supplemental Figure 6. Phylogenetic Trees of the PYL Family of ABA Receptor-Coding Genes and the SnRK3 Family of Signaling Kinases.
Supplemental Figure 7. Identification of REV and KAN1 Targets Uncover Multiple Feedback Loops.
Supplemental Data Set 1A. Genes Upregulated by GR-REV.
Supplemental Data Set 1B. Genes Downregulated by GR-REV.
Supplemental Data Set 1C. Genes Upregulated by KAN1-GR.
Supplemental Data Set 1D. Genes Downregulated by KAN1-GR.
Supplemental Data Set 1E. Genes with Transcripts That Change upon GR-REV Induction That Also Have Adjacent REV Binding Sites.
Supplemental Data Set 1F. Fold Changes in Transcript Levels of ORKs and Class II HD-ZIPs in Response to 60-min Dexamethasone Treatment with and without Cycloheximide as Measured by qRT-PCR.
Supplemental Data Set 1G. Genes with Transcripts Regulated by Both REV and KAN1: Overlap Set of GR-REV versus Col and KAN-GR versus Col.
Supplemental Data Set 1H. Genes Regulated by Both GR-REV and KAN1-GR: Set Significant in Two-Way ANOVA Comparing GR-REV versus KAN1-GR Directly.
Supplemental Data Set 1I. Overrepresented GO Categories among REV- versus KAN1-Regulated Genes.
Supplemental Data Set 1J. GR-REV–Regulated Genes Found in the WUS Domain.
Supplemental Data Set 1K. GR-REV–Regulated Genes Found in the CLAVATA Domain.
Supplemental Data Set 1L. KAN1-GR–Regulated Genes Found in the WUSCHEL Domain.
Supplemental Data Set 1M. KAN1-GR–Regulated Genes Found in the FIL Domain.
Supplemental Data Set 1N. Genes Screened for Defects in Leaf Morphology in the Overexpression Assay.
Supplemental Data Set 1O. Primers Used in qRT-PCR Experiments.
Supplemental Data Set 2. Text File of the Alignment of ORK6 Proteins Used for the Phylogenetic Analysis Shown in Figure 6.
Acknowledgments
We thank our colleagues David Ehrhardt and Matthew Evans for critical comments on the article. This work was funded by Grant RO1 GM 069293 from the National Institutes of Health and IOS-0929413 from the National Science Foundation.
AUTHOR CONTRIBUTIONS
B.J.R. performed microarray experiments, assisted in writing, and analyzed data. T.L. performed and analyzed overexpression experiments. N.R.N. performed qRT-PCR experiments. E.M. generated clones for the two-hybrid screens and performed the retests of positive clones. T.H. and R.K. generated and tested the KAN1-GR lines. S.M. performed the two-hybrid screen. M.K.B. analyzed data and wrote the article.
Glossary
- Col
Columbia
- ANOVA
analysis of variance
- ChIP
chromatin immunoprecipitation
- qRT-PCR
quantitative RT-PCR
References
- Bonke M., Thitamadee S., Mähönen A.P., Hauser M.-T., Helariutta Y. (2003). APL regulates vascular tissue identity in Arabidopsis. Nature 426: 181–186 [DOI] [PubMed] [Google Scholar]
- Bou-Torrent J., Salla-Martret M., Brandt R., Musielak T., Palauqui J.C., Martínez-García J.F., Wenkel S. (2012). ATHB4 and HAT3, two class II HD-ZIP transcription factors, control leaf development in Arabidopsis. Plant Signal. Behav. 7: 1382–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt R., et al. (2012). Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. 72: 31–42 [DOI] [PubMed] [Google Scholar]
- Breuninger H., Rikirsch E., Hermann M., Ueda M., Laux T. (2008). Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 14: 867–876 [DOI] [PubMed] [Google Scholar]
- Carlsbecker A., et al. (2010) Cell signalling by microRNA 165/6 directs gene dose-dependent root cell fate. Nature 465: 316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, J.W., Cole, M., Flier, A, Grewe, B. and Werr, W. (2007). The AP2 transcription factors DORNROSCHEN and DORNROSCHEN-LIKE redundantly control embryo patterning via interaction with PHAVOLUTA. Development 134: 1653–1662. [DOI] [PubMed]
- Chaudhury A.M., Letham S., Craig S., Dennis E.S. (1993). amp1 - A mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J. 4: 907–916 [Google Scholar]
- Chitwood D.H., Nogueira F.T., Howell M.D., Montgomery T.A., Carrington J.C., Timmermans M.C. (2009). Pattern formation via small RNA mobility. Genes Dev. 23: 549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, H.H. 2010, Analyses of Arabidopsis Yellow Stripe-like (YSL) Family of Metal Transporters. PhD dissertation (Amherst, MA: University of Massachusetts). [Google Scholar]
- Chu H.H., Chiecko J., Punshon T., Lanzirotti A., Lahner B., Salt D.E., Walker E.L. (2010). Successful reproduction requires the function of Arabidopsis Yellow Stripe-Like1 and Yellow Stripe-Like3 metal-nicotianamine transporters in both vegetative and reproductive structures. Plant Physiol. 154: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarbelli A.R., Ciolfi A., Salvucci S., Ruzza V., Possenti M., Carabelli M., Fruscalzo A., Sessa G., Morelli G., Ruberti I. (2008). The Arabidopsis homeodomain-leucine zipper II gene family: Diversity and redundancy. Plant Mol. Biol. 68: 465–478 [DOI] [PubMed] [Google Scholar]
- Colasanti J., Tremblay R., Wong A.Y., Coneva V., Kozaki A., Mable B.K. (2006). The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R., Galinha C., Fletcher A.G., Grigg S.P., Molnar A., Willemsen V., Scheres B., Sabatini S., Baulcombe D., Maini P.K., Tsiantis M. (2012). A PHABULOSA/cytokinin feedback loop controls root growth in Arabidopsis. Curr. Biol. 22: 1699–1704 [DOI] [PubMed] [Google Scholar]
- Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Englbrecht C.C., Schoof H., Böhm S. (2004). Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y., Baum S.F., Perea J.V., Bowman J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Izhaki A., Baum S.F., Floyd S.K., Bowman J.L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Etchells J.P., Provost C.M., Turner S.R. (2012). Plant vascular cell division is maintained by an interaction between PXY and ethylene signalling. PLoS Genet. 8: e1002997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.M.S. (2007). The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo Sac and leaf development. Plant Cell 19: 46–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y., Liu C., Yu H., Broun P. (2007). Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 134: 2073–2081 [DOI] [PubMed] [Google Scholar]
- Gillmor C.S., Park M.Y., Smith M.R., Pepitone R., Kerstetter R.A., Poethig R.S. (2010). The MED12-MED13 module of Mediator regulates the timing of embryo patterning in Arabidopsis. Development 137: 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K.A., Prigge M.J., Katzman R.B., Clark S.E. (2005). CORONA, a member of the Class III Homeodomain Leucine Zipper gene family in Arabidopsis, regulates stem cell specification and organogenesis. Plant Cell 17: 691–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker A., Gross-Hardt R., Geiges B., Sarkar A., Breuninger H., Herrmann M., Laux T. (2004). Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668 [DOI] [PubMed] [Google Scholar]
- Hawker N.P., Bowman J.L. (2004). Roles for Class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol. 135: 2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda K., Imamura A., Katoh E., Hatta T., Tachiki M., Yamada H., Mizuno T., Yamazaki T. (2002). Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14: 2015–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C., Sun H., Poethig R.S. (2003). The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 13: 1734–1739 [DOI] [PubMed] [Google Scholar]
- Ilegems M., Douet V., Meylan-Bettex M., Uyttewaal M., Brand L., Bowman J.L., Stieger P.A. (2010). Interplay of auxin, KANADI and Class III HD-ZIP transcription factors in vascular tissue formation. Development 137: 975–984 [DOI] [PubMed] [Google Scholar]
- Izhaki A., Bowman J.L. (2007). KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19: 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanrar S., Onguka O., Smith H.M. (2006). Arabidopsis inflorescence architecture requires the activities of KNOX-BELL homeodomain heterodimers. Planta 224: 1163–1173 [DOI] [PubMed] [Google Scholar]
- Kelley D.R., Arreola A., Gallagher T.L., Gasser C.S. (2012). ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 139: 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D.R., Skinner D.J., Gasser C.S. (2009). Roles of polarity determinants in ovule development. Plant J. 57: 1054–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter R.A., Bollman K., Taylor R.A., Bomblies K., Poethig R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411: 706–709 [DOI] [PubMed] [Google Scholar]
- Kieffer M., Master V., Waites R., Davies B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.S., Kim S.G., Lee M., Lee I., Park H.Y., Seo P.J., Jung J.H., Kwon E.J., Suh S.W., Paek K.H., Park C.M. (2008). HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell 20: 920–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer S., Holt A.L., Rubio-Somoza I., Tucker E.J., Hinze A., Pisch M., Javelle M., Timmermans M.C., Tucker M.R., Laux T. (2013). A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell 24: 125–132 [DOI] [PubMed] [Google Scholar]
- Lee I., Michaels S.D., Masshardt A.S., Amasino R.M. (1994). The late-flowering phenotype of FRIGIDA and mutations in LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J. 6: 903–909 [Google Scholar]
- Lee Y.K., Kim G.-T., Kim I.-J., Park J., Kwak S.-S., Choi G., Chung W.-I. (2006). LONGIFOLIA1 and LONGIFOLIA2, two homologous genes, regulate longitudinal cell elongation in Arabidopsis. Development 133: 4305–4314 [DOI] [PubMed] [Google Scholar]
- Lehfeldt C., Shirley A.M., Meyer K., Ruegger M.O., Cusumano J.C., Viitanen P.V., Strack D., Chapple C. (2000). Cloning of the SNG1 gene of Arabidopsis reveals a role for a serine carboxypeptidase-like protein as an acyltransferase in secondary metabolism. Plant Cell 12: 1295–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Kloosterziel K.M., Keijzer C.J., Koornneef M. (1994). A seed shape mutant of Arabidopsis that is affected in integument development. Plant Cell 6: 385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Reinhart B.J., Magnani E., Huang T., Kerstetter R.A., Barton M.K. (2012). Of blades and branches: Understanding and expanding the Arabidopsis ad/abaxial regulatory network through target gene identification. Cold Spring Harb. Symp. Quant. Biol. 77: 31–45 [DOI] [PubMed] [Google Scholar]
- Long J., Moan E., Medford J., Barton M.K. (1996). The Arabidopsis SHOOTMERISTEMLESS gene encodes a member of the KNOTTED class of homeodomain proteins. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Lynn K., Fernandez A., Aida M., Sedbrook J., Tasaka M., Masson P., Barton M.K. (1999). The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126: 469–481 [DOI] [PubMed] [Google Scholar]
- Magnani E., Barton M.K. (2011). A per-ARNT-sim-like sensor domain uniquely regulates the activity of the homeodomain leucine zipper transcription factor REVOLUTA in Arabidopsis. Plant Cell 23: 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAbee J. M., Hill T. A., Skinner D. J., Izhaki A., Hauser B. A., Meister R. J., Venugopala Reddy G., Meyerowitz E. M., Bowman J. L., Gasser C. S. (2006). ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant J. 46: 522–531 [DOI] [PubMed] [Google Scholar]
- McConnell J.R., Barton M.K. (1995). Effect of mutations in the PINHEAD gene of Arabidopsis on the formation of the shoot apical meristem. Dev. Genet. 16: 358–366 [Google Scholar]
- McConnell J.R., Barton M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942 [DOI] [PubMed] [Google Scholar]
- McConnell J.R., Emery J., Eshed Y., Bao N., Bowman J., Barton M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- Mukherjee K., Brocchieri L., Bürglin T.R. (2009). A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 26: 2775–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettleton D. (2006). A discussion of statistical methods for design and analysis of microarray experiments for plant scientists. Plant Cell 18: 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman K.L., Fernandez A.G., Barton M.K. (2002). Regulation of axis determinacy by the Arabidopsis PINHEAD gene. Plant Cell 14: 3029–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochando I., Jover-Gil S., Ripoll J.J., Candela H., Vera A., Ponce M.R., Martínez-Laborda A., Micol J.L. (2006). Mutations in the microRNA complementarity site of the INCURVATA4 gene perturb meristem function and adaxialize lateral organs in Arabidopsis. Plant Physiol. 141: 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Szatmari A.M., Watanabe B., Fujita S., BAncos S., Koncz C., Lafos M., Shibata K., Yokota T., Sakata K., Szekeres M., Mizutani M. (2006). C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 18: 3275–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D., DeGuzman B., Prigge M.J., Drews G.N., Clark S.E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25: 223–236 [DOI] [PubMed] [Google Scholar]
- Prigge M.J., Otsuga D., Alonso J.M., Ecker J.R., Drews G.N., Clark S.E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17: 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutjens B., Bao D., van Eck-Stouten E., Brand M., Smeekens S., Proveniers M. (2009). Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J. 58: 641–654 [DOI] [PubMed] [Google Scholar]
- Schena M., Lloyd A.M., Davis R.W. (1991). A steroid-inducible gene expression system for plant cells. Proc. Natl. Acad. Sci. USA 88: 10421–10425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G., Steindler C., Morelli G., Ruberti I. (1998). The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol. Biol. 38: 609–622 [DOI] [PubMed] [Google Scholar]
- Smith Z.R., Long J.A. (2010). Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 464: 423–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.B., Huang S.Q., Dalmay T., Yang Z.M. (2012). Regulation of leaf morphology by microRNA394 and its target LEAF CURLING RESPONSIVENESS. Plant Cell Physiol. 53: 1283–1294 [DOI] [PubMed] [Google Scholar]
- Song S.-K., Hofhuis H., Lee M.M., Clark S.E. (2008). Key divisions in the early Arabidopsis embryo require POL and PLL1 phosphatases to establish the root stem cell organizer and vascular axis. Dev. Cell 15: 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P., Furner I.J., Ottoline Leyser H.M. (2007). MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 50: 80–94 [DOI] [PubMed] [Google Scholar]
- Talbert P.B., Adler H.T., Parks D.W., Comai L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121: 2723–2735 [DOI] [PubMed] [Google Scholar]
- Tatematsu K., Nakabayashi K., Kamiya Y., Nambara E. (2008). Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 53: 42–52 [DOI] [PubMed] [Google Scholar]
- Turchi L., Carabelli M., Ruzza V., Possenti M., Sassi M., Peñalosa A., Sessa G., Salvi S., Forte V., Morelli G., Ruberti I. (2013). Arabidopsis HD-Zip II transcription factors control apical embryo development and meristem function. Development 140: 2118–2129 [DOI] [PubMed] [Google Scholar]
- Wahl V., Brand L.H., Guo Y.-L., Schmid M. (2010). The FANTASTIC FOUR proteins influence shoot meristem size in Arabidopsis thaliana. BMC Plant Biol. 10: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites R., Hudson A. (1995). phantastica: A gene required for dorsiventrality of leaves in Antirrhinum majus. Development 121: 2143–2154 [Google Scholar]
- Welch D., Hassan H., Blilou I., Immink R., Heidstra R., Scheres B. (2007). Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 21: 2196–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S., Emery J., Hou B.H., Evans M.M.S., Barton M.K. (2007). A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell 19: 3379–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L., Grigg S.P., Xie M., Christensen S., Fletcher J.C. (2005). Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132: 3657–3658 [DOI] [PubMed] [Google Scholar]
- Wu G., Lin W.C., Huang T., Poethig R.S., Springer P.S., Kerstetter R.A. (2008). KANADI1 regulates adaxial-abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2. Proc. Natl. Acad. Sci. USA 105: 16392–16397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R.K., Girke T., Pasala S., Xie M., Reddy G.V. (2009). Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc. Natl. Acad. Sci. USA 106: 4941–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., et al. (2003). Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Dinh T.T., Ott F., Lanz C., Wollmann H., Chen X., Schmid M. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L.P., Miller A.K., Clark S.E. (2003). POLTERGEIST encodes a protein phosphatase 2C that regulates CLAVATA pathways controlling stem cell identity at Arabidopsis shoot and flower meristems. Curr. Biol. 13: 179–188 [DOI] [PubMed] [Google Scholar]
- Zambelli F., Pesole G., Pavesi G. (2009). Pscan: Finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 37 (Web Server issue): W247–W252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R. Q., Ye Z. H. (2001). Alteration of auxin polar transport in the Arabidopsis ifl1 mutants. Plant Physiol. 126: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Hu F., Wang R., Zhou X., Sze S.-H., Liou L.W., Barefoot A., Dickman M., Zhang X. (2011). Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145: 242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]