This work identifies TED, an autonomous maize transposable element related to the well-studied MuDR Mutator elements, but with some differences, including low copy number and a strong tendency to excise during the mitotic divisions of the gametophyte.
Abstract
Mutator (Mu) elements, one of the most diverse superfamilies of DNA transposons, are found in all eukaryotic kingdoms, but are particularly numerous in plants. Most of the present knowledge on the transposition behavior of this superfamily comes from studies of the maize (Zea mays) Mu elements, whose transposition is mediated by the autonomous Mutator-Don Robertson (MuDR) element. Here, we describe the maize element TED (for Transposon Ellen Dempsey), an autonomous cousin that differs significantly from MuDR. Element excision and reinsertion appear to require both proteins encoded by MuDR, but only the single protein encoded by TED. Germinal excisions, rare with MuDR, are common with TED, but arise in one of the mitotic divisions of the gametophyte, rather than at meiosis. Instead, transposition-deficient elements arise at meiosis, suggesting that the double-strand breaks produced by element excision are repaired differently in mitosis and meiosis. Unlike MuDR, TED is a very low-copy transposon whose number and activity do not undergo dramatic changes upon inbreeding or outcrossing. Like MuDR, TED transposes mostly to unlinked sites and can form circular transposition products. Sequences closer to TED than to MuDR were detected only in the grasses, suggesting a rather recent evolutionary split from a common ancestor.
INTRODUCTION
Though transposable elements (TEs) occupy a large fraction of most eukaryotic genomes and are major contributors to genome size evolution, the amplification of different TE superfamilies varies greatly from species to species (Kidwell, 2002; Wessler, 2006; Feschotte and Pritham, 2007; Ågren and Wright, 2011; Fedoroff, 2012). Among class 2 or DNA TEs (Wicker et al., 2007), the Mutator (Mu) superfamily has been particularly successful in colonizing plant genomes (Jiang et al., 2004; Holligan et al., 2006; Delseny et al., 2010; González and Deyholos, 2012), yet practically all knowledge of its transposition and regulation traces to genetic studies of the maize (Zea mays) Mu elements, the founding members of the superfamily (Robertson, 1978; Robertson and Stinard, 1989; Bennetzen, 1996; Walbot and Rudenko, 2002; Lisch and Jiang, 2009).
The autonomous maize Mu element MuDR (Chomet et al., 1991; Hershberger et al., 1991; Qin et al., 1991) controls the transposition of a diverse family of elements that share terminal inverted repeats (TIRs) but vary in internal sequence. MuDR encodes a Mutator Regulator A (MURA) protein with a highly conserved transposase domain (Hua-Van and Capy, 2008) and a MURB protein that has been implicated in element reinsertion, but whose function remains uncertain at this time (Lisch and Jiang, 2009). Mu-like elements (MULEs) have been reported in a number of other species besides maize (Lisch, 2002). Most are defective elements that would be incapable of autonomous transposition, but some encode intact proteins homologous to the MURA transposase, and a few have been shown to transpose autonomously: Hop1 in Fusarium oxysporum (Chalvet et al., 2003), Jittery in maize (Xu et al., 2004), and Os3378 in rice (Oryza sativa; Gao, 2012). Unlike MuDR, these autonomous elements do not encode a B function and, peculiarly, lack defective close relatives in their respective genomes. Here, we describe TED (for Transposon Ellen Dempsey), an autonomous maize MULE captured in the bz locus of a transposon-laden line (Rhoades and Dempsey, 1982). The resulting bz-m175 mutable allele has enabled a detailed analysis of the transposition properties and behavior of this novel autonomous MULE, which differs from MuDR in a number of ways.
TED encodes a protein closely related to MURA that we have called TEDA, but no other protein, so all its trans-acting transposition properties are dependent on this one protein. Transposition-deficient derivatives that lose part or all of TEDA arise with a high frequency at meiosis. Unlike MuDR, TED is a very-low-copy-number element that is neither silenced by inbreeding nor amplified by outcrossing. TED’s unusually high transposition frequency in the gametophytic generation, particularly in the embryo sac, can account for most recovered Bz’ germinal reversions. Like MuDR, TED transposes mostly to unlinked sites and forms circular transposition products of as yet unclear significance. Based on our reversion analysis of bz-m175, we infer that the double-strand breaks (DSBs) produced by TED excision are repaired by nonhomologous end joining in the mitotic divisions of the sporophyte and gametophyte, but not at meiosis, where they appear to be repaired by DNA synthesis using the sister chromatid as template. Sequences more similar to TED than to MuDR were detected only in grass genomes, suggesting that these elements diverged only recently from a common ancestor.
RESULTS
Isolation and Genetic Behavior of bz-m175
The bz-m175 mutable allele arose in an experiment designed to capture transposons in the maize stocks High Loss and High Knob (Rhoades and Dempsey, 1982), known to carry active elements from different superfamilies (Dennis et al., 1988; Shepherd et al., 1989). It conditions a uniformly fine-spotted bronze mutable (bz-mF) phenotype (Figure 1A), a telltale sign of a transposon mutation at bz that reverts late in aleurone development. In testcrosses of Sh bz-m175/sh-bz-X3 hemizygotes, the bz-mF phenotype cosegregates with the plump phenotype conditioned by the Sh allele (see Supplemental Table 1 online), suggesting that the transposon in bz-m175 acts autonomously.
Figure 1.
bz-m175 Phenotypes.
The bz-m175 allele conditions a uniformly fine-spotted mutable phenotype, as illustrated by the kernels in (A). This ear sector also shows a rare cluster of premeiotic germinal Bz’ purple revertants. Occasional large somatic sectors can also be seen in sporophytic tissues: leaf blade and midrib ([B] and [C]); husk (D); culm and leaf sheath (E); and tassel (F).
The bz-m175 allele has an obvious positive dosage effect on kernel spot number since the number of purple spots is twice as high when bz-m175 is crossed as the maternal parent to a bz tester than in the reciprocal cross (see Supplemental Figures 1A and 1B online). Large somatic revertant sectors are sometimes seen in adult plant tissues, indicating that the transposon at bz also reverts late in sporophytic development (Figures 1B to 1F). Occasionally, very heavily spotted kernels can be found in segregating populations of bz-m175 (see Supplemental Figure 1C online), but the phenotype is not heritable. The bz-m175 allele has been backcrossed four times to a W22 sh-bz-X3 stock without any obvious change in phenotype or apparent loss of activity.
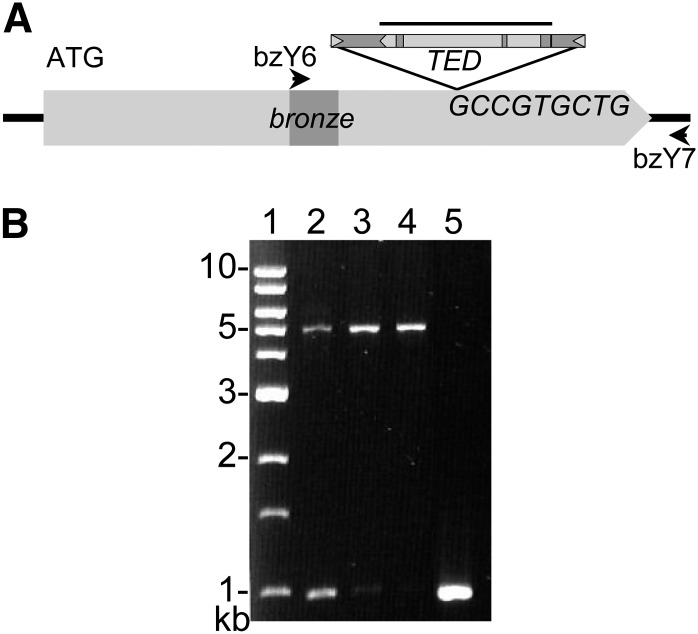
The mutable bz-m175 allele was isolated by PCR amplification from leaf DNA. Two PCR products were amplified from bz-m175/bz-R heterozygotes using primers spanning the bz coding region (Figure 2). The larger band was ∼4 kb longer than either the progenitor band or somatic excision products. Sequence analysis of the large PCR product showed that bz-m175 had resulted from the insertion of a novel TE in the second bz exon. The element was named TED, for Transposon Ellen Dempsey, in honor of one of the discoverers of the High Loss stock.
Figure 2.
The bz-m175 Allele.
(A) Structure of bz-m175, showing the TED element insertion in the bz second exon, the sequence of the 9-bp insertion site, and the location of primers bzY6 and bzY7 (arrows) and the TEDA hybridization probe (bar). bz exons and intron are light and dark gray, respectively. TED TIRs are triangles, and its coding and noncoding sequences are light and dark gray, respectively.
(B) Amplifications with bz-Y6 and bz-Y7 of the parental TED band and of somatic excision products from bz-m175. Lane 1, 1-kb DNA ladder; lane 2, bz-m175 seeding leaf; lanes 3 and 4, bz-m175 adult leaves; lane 5, Bz-McC wild-type control.
Structure of TED and TED-Like Elements in Maize
TED is 3960- bp long, causes a 9-bp target site duplication (TSD) at the insertion site, and ends in 191-bp TIRs, which are 92.1% identical to each other. It is predicted to encode a 785–amino acid protein that we have denominated TEDA because of its homology to MURA, the MuDR transposase (51% identity; 67% similarity over 742 amino acids). TED does not share significant nucleotide sequence similarity with either MUDR or other known elements in the Mu superfamily, except over the third and largest exon of the transposase gene. Not surprisingly, the TED TIRs lack a MURA binding site sequence (Benito and Walbot, 1997).
No sequences with significant similarity to the TED TIRs were found in the B73 maize genome, which contains 155 MULE families (Schnable and al., 2009). However, B73 sequences distantly related to the TEDA coding region and flanked by distinct TIRs were detected by tBLASTn searches using the TEDA transposase as query. Homology of these sequences to TED was confirmed by DNA gel blot hybridization (see Supplemental Figure 2 online). Unlike most known MULEs other than Jittery (Xu et al., 2004), TED is an extremely low-copy TE. Among the 11 inbred lines examined, only H99, I137TN, and W22 contain a single copy of a TED-like element, but none of them have TED activity (see later section). TEDA-hybridizing bands with TED activity were only detected in the bz-m175 progenitor line and in the original High Loss stocks of Rhoades and Dempsey.
Germinal Transposition of TED in bz-m175
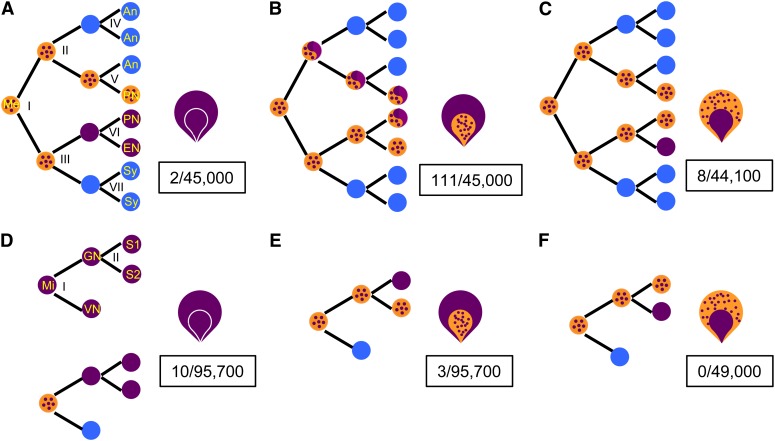
In crosses of bz-m175 females to a bz tester, purple kernels representing putative revertants (Bz’) are recovered at a frequency of around one per 400 gametes (113/45,000). However, only two showed the concordant Bz endosperm and embryo phenotypes expected of meiotic reversion events (Figure 3A). One of them occurred as a single purple kernel on the ear and the other as a cluster of four kernels, reflecting an early premeiotic event. The majority (111/113) were class 1 nonconcordant reversion events (Bz endosperm; bz-m embryo), indicating a high TED excision activity in the female gametophyte. These reversions can arise in any one of four possible non-egg cell lineages (Figure 3B).
Figure 3.
bz-m175 Reversion in Female and Male.
Mitotic divisions and differentiated cells of the female and male gametophytes are identified in (A) and (D), respectively. The exceptional kernel phenotypes produced in testcrosses of bz-m175 as either maternal or paternal parent are inferred to arise by the reversion events diagrammed to their left. The frequencies of each kernel class are boxed. Light-blue closed circles are nuclei that do not affect these kernel phenotypes. Adapted from Dooner and Belachew (1989).
(A) to (C) Female gametophyte (embryo sac): Me, megaspore; An, antipodals; PN, polar nuclei; EN, egg nucleus; Sy, synergids.
(A) Concordant kernel (Bz endosperm and embryo) from Bz’ reversion at megagametophytic mitosis I or at meiosis (data not shown).
(B) Class 1 nonconcordant kernel (Bz’ endosperm and bz-m embryo) from postmeiotic Bz’ reversion in four possible non-egg cell lineages.
(C) Class 2 nonconcordant kernel (bz-m endosperm and Bz’ embryo) from postmeiotic Bz’ reversion only in egg cell lineage.
(D) to (F) Male gametophyte (pollen): Mi, microspore; GN, generative nucleus; VN, vegetative (tube) nucleus; S1 and S2, sperm nuclei.
(D) Concordant kernels from meiotic Bz’ reversion (top) or postmeiotic Bz’ reversion in generative nucleus (bottom).
(E) Class 1 nonconcordant kernels from postmeiotic Bz’ reversion in sperm fertilizing the central cell.
(F) Class 2 nonconcordant kernels from postmeiotic Bz’ reversion in sperm fertilizing the egg.
The high frequency of class 1 nonconcordant reversions (2.5 × 10−3) prompted us to screen for the reciprocal nonconcordant reversions having a bz-m endosperm and a Bz embryo (class 2) among the same testcross progeny. This class can only arise from a TED excision that occurs at the last female gametophytic mitosis and is transmitted to the egg cell (Figure 3C), so its frequency is expected to be lower than that of class 1. The entire population of spotted kernels was germinated and screened for purple (Bz’) seedlings. Eight Bz’ revertants were identified and their heritability was confirmed by genetic test crosses. Their frequency, 1.8 × 10−4, is considerably lower than that of the reciprocal nonconcordant class and four times greater than that of the concordant class, though the numbers are too low to establish significance.
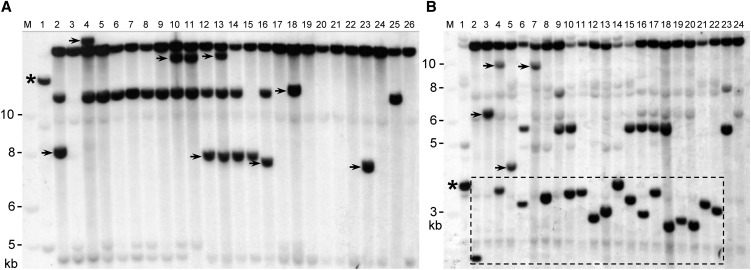
The occurrence of TED excisions in the female gametophyte allows us to analyze the role in reinsertion of the single gene encoding the TEDA transposase. We found that TED reinsertion was high among Bz’ germinal revertants (Figure 4A). Three new TED-hybridizing bands were detected among the two concordant excision events. The four clustered Bz’ revertants share a reinsertion event (lanes 12 to 15), as expected from a premeiotic TED transposition, but one has an additional unique reinsertion that must have occurred at or after meiosis (lane 13). Two of the nonconcordant class 2 reversions showed new TED bands (lanes 18 and 23), indicating that TED reinserted in a fraction of the egg cells after excising in the last megagametophytic mitosis. The DNA gel blot observations were confirmed by isolating and sequencing the nine TED reinsertion sites (see Supplemental Table 2 online). Our female parent reversion data indicate that TED excision and reinsertion are much higher in the postmeiotic gametophytic mitoses than at meiosis or the premeiotic mitoses, though they appear to be differentially regulated in the different cell lineages of the embryo sac.
Figure 4.
Detection of TED Transpositions.
(A) Bz’ revertants of bz-m175. Lane 1, bz-m175; lanes 2 to 24, Bz’ revertants; lane 25, sh-bz-X2; lane 26, sh-bz-X3. HindIII digest, TEDA probe.
(B) bz-s derivatives of bz-m175 (dashed rectangle). Lane 1, bz-m175; lanes 2 to 22, bz-s derivatives with a defective or fractured TED; lane 23, sh-bz-X2; lane 24, sh-bz-X3. SacI digest, TEDA probe.
Neither restriction enzyme cuts within the probe. trTED bands are marked with arrows and parental bz-m175 bands with asterisks. The nonsegregating high molecular weight band is present in all W22 stocks and the segregating lower band (∼12 kb in [A] and 5.5 kb in [B]) derives from the sh-bz-X2 parent.
By contrast, purple kernel Bz’ revertants are rarely recovered using bz-m175 as the pollen parent. In a population of almost 100,000 gametes, only 13 single-kernel Bz’ revertants were obtained. Thus, the overall reversion frequency of bz-m175 on the male side (approximately one per 10,000 gametes) is 25-fold lower than on the female side. Ten revertants were concordant and could have arisen during microsporogenesis or at the first pollen mitosis (Figure 3D). Four reinsertion events were detected among them (Figure 4A), a comparable reinsertion fraction to that observed in the egg cell. The remaining three revertants were nonconcordant, having originated most likely from the fertilization of the central cell of the embryo sac by a Bz’ sperm cell that arose from a TED excision in the second pollen mitosis (Figure 3E). The reciprocal class 2 nonconcordant revertants (Figure 3F) were searched for in the same testcross progeny. Among 50,000 seedlings germinated from spotted seed, no such revertants were found, a result in agreement with the low frequency of the reciprocal nonconcordant class in the same cross. Taken together, the bz-m175 reversion data indicate that TED transposition is much higher in the megagametophyte than in the microgametophyte.
All the Bz’ revertants had the unique single-nucleotide and indel polymorphisms of the Bz progenitor of bz-m175 (GenBank accession number KF279655), which was absent from the field of the reversion experiments, arguing that they did not arise from pollen contamination. None of the Bz’ revertants from either reciprocal cross bore a transposon footprint, but unselected somatic excision products revealed multiple footprints (see below). This observation underscores the importance of the highly conserved UDP-Glc binding domain disrupted by TED for enzymatic activity of the bz-encoded flavonoid 3-O-glucosyltransferase (Offen et al., 2006).
Stable bz (bz-s) mutations are recovered about three times more frequently than Bz’ revertants in crosses of bz-m175 females to a bz pollen tester (48/7300). These derivatives most likely arise at or around meiosis because they occur as single kernels in the ear and their recessive bronze endosperm phenotype can only be produced if the two nuclei in the central cell, whose lineages separate at the first postmeiotic mitosis, are mutant (Dooner and Belachew, 1989). All 48 bz-s mutations arose from deletions in the bz-m175 allele. Thirty (63%) carried internal TED deletions at the original site (Figure 5, 1 to 30). These are defective TED (dTED) nonautonomous elements, which can be activated by autonomous TED elements present elsewhere in the genome (see below). Because this class would not have been selected if a transposed TED had been retained in the genome, the true frequency of bz-s mutations from bz-m175 is even higher. Three (6%) carried a fractured TED (fTED) element lacking one of the two ends and 15 (31%) carried bronze deletions of unknown size.
Figure 5.
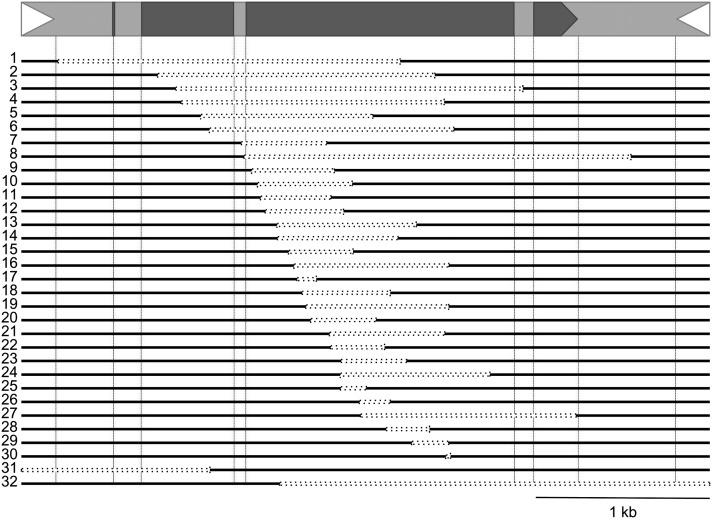
Structure of dTED and fTED Elements.
bz-s derivatives from bz-m175. 1 to 30, dTEDs; 31 and 32, fTEDs. Deletions are shown as dashed open bars. TIR locations and TEDA exon-intron junctions are indicated by vertical dashed lines. The dTED20 derivative was used to assay the activity of trTED elements in segregation analyses (see Supplemental Figure 4 online).
Different internal sequences have been deleted in the dTED elements. The deletions vary in size from 15 to 2240 bp and always include part of the TEDA coding region (210 to 3520), but never extend into the TIRs. In all cases, 2- to 8-bp short repeats flank the deletion endpoints (see Supplemental Table 3 online). Most deletions create premature stop codons that lead to nonfunctional transposases lacking the region coding for the conserved DDE triad domain (Hua-Van and Capy, 2008). Some deletions had filler DNA (4/30), as have been found in deletions from other transposons (Rubin and Levy, 1997; Yan et al., 1999; Raizada et al., 2001; Conrad et al., 2007; Huang and Dooner, 2012) and in spontaneous deletions from different organisms (Roth et al., 1985; Wessler et al., 1990). The filler DNA varies in size from 8 to 66 bp and originates from nearby sequences located 23 to 109 bp from the deletion junction. The largest filler DNA, found in dTED element No. 5 consists of two unequal fragments, 30 bp apart in the original TED element and flanked by different short repeats.
Either end can be lost in the fTED elements. One of the two 5′ TIR deletions removes 1084 bp from the TED 5′ end and 34 bp of adjacent bz sequence and is flanked by a perfect 8-bp direct repeat in the bz-m175 progenitor allele (see Supplemental Table 3 online, No. 31). A genetically active transposed TED (trTED) element was also detected in this line (data not shown). The other 5′ TIR deletion is much larger and extends into the distal bz-stc1 intergenic region. The 3039-bp 3′ TIR deletion removes 2437 bp from the TED 3′ terminus and 602 bp of adjacent bz sequence (Figure 5, No. 32). Much larger deletions account for the remaining 15 bz deletion derivatives from bz-m175. Eleven of them contain a trTED element elsewhere in the genome (Figure 6), suggesting that TED often reinserts in the genome after germinal excision. The genetic analysis of bz-s derivatives is described in greater detail in the next section.
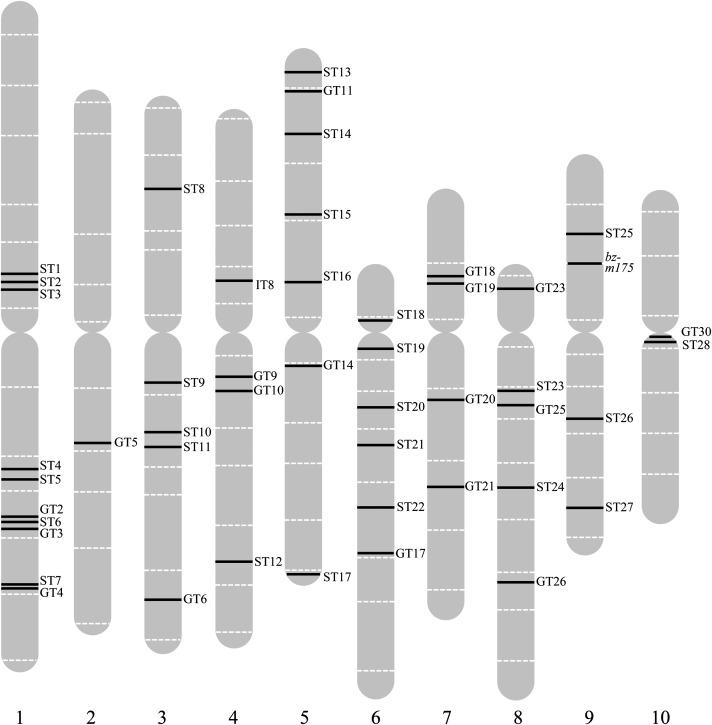
Figure 6.
Location of trTEDs in the Maize Genome.
trTEDs were mapped to one of the maize pseudomolecules by BLAST searches of the B73 genome sequence with TED-adjacent sequences. ST, somatic transpositions; GT, germinal transpositions.
As with Bz’ selections, a few rare premeiotic events were detected among the bz-s selections. More frequently, the multiple bz-s kernels of the same cob arose from independent excision events, as evidenced by the different alleles carried by different kernels. In one case, five different dTED elements (see Supplemental Table 3 online, bz-s derivatives 5, 8, 11, 13, and 16) were found among six bz-s kernels from the same cob, the remaining one carrying a large bz deletion and a trTED element. The lack of perfect adjacent deletions (deletions immediately next to TED) and the particularly large size of some deletions distinguish TED from most other DNA transposon families but are reminiscent of changes brought about by MuDR (Hsia and Schnable, 1996).
Genetic Interaction of trTEDs and dTEDs
The bz-s selections containing dTED elements (Figure 5) can serve as reporter lines to test the transposition activities of the trTED elements identified molecularly among both Bz’ and bz-s selections from bz-m175. A 20 × 30 grid was set up to test simultaneously the respective abilities of the 20 trTED elements to reactivate dTED excision from bz and of the 30 dTED elements to be mobilized by a TEDA transposase provided in trans. The results of all hybrid combinations were uniformly positive: The 20 trTED elements were capable of trans-activating excision of the 30 new dTED elements at bz, producing in each case a spotting pattern indistinguishable from that of the bz-m175 allele (see Supplemental Figure 3 online). trTED elements originally detected as new TEDA-hybridizing bands in a DNA gel blot (Figure 4) were always found to cosegregate with the spotting phenotype in crosses to bz-m(dTED) reporter lines (see Supplemental Figure 4 online). This indicates that TED generally retains its activity after transposing to new sites in the genome.
Nature and Distribution of TED Reinsertion Sites
To determine the nature of TED reinsertion sites, the DNA adjacent to the trTED elements was isolated and sequenced (see Supplemental Table 2 online). TED shows a strong preference for reinsertion in genic regions (16/18), as do other active MULEs (Fernandes et al., 2004; McCarty et al., 2005) and Ac/Ds (Cowperthwaite et al., 2002; Vollbrecht et al., 2010). Like its Mu cousin (Dietrich et al., 2002; May et al., 2003), TED displays a strong affinity for the 5′ end of genes (see Supplemental Figure 5 online). However, unlike Mu elements, which insert into a weak consensus sequence (Raizada et al., 2001), TED displays no obvious sequence preference. Occasionally, it transposes into low-copy repetitive sequences, yet it remains active. Since all trTEDs were first identified molecularly and then tested genetically, the latter observation suggests that trTEDs retain their activity regardless of the nature of the reinsertion site.
Although most TED reinsertions were flanked by the 9-bp TSDs characteristic of MULEs (Walbot and Rudenko, 2002), some were not (see Supplemental Table 2 online). Two TSDs were 10 bp long and one was 6 bp long, sizes within the range of MULE TSDs that have been identified computationally (Singer et al., 2001). Germinal transpositions were mostly to unlinked sites distributed throughout the genome (Figure 6; see Supplemental Table 2 online), as are those of MuDR (Lisch et al., 1995) and RescueMu (Raizada et al., 2001) in maize and of At-Mu1 in Arabidopsis thaliana ddm1 mutants (Singer et al., 2001).
Somatic Transposition of TED in bz-m175
The rarity of large revertant sectors in the endosperm and sporophyte would suggest that TED excises only late in somatic development. However, a prominent PCR band corresponding in size to TED excision products was detected in bz-m175 seedling leaves (Figure 2B). The wild-type-sized PCR products were cloned and sequenced to analyze the nature of the footprints resulting from somatic excision events. Fourteen different excision events (SE1 to SE14) were detected among 33 sequenced clones (see Supplemental Figure 6 online). Except for one +0 or no-footprint event (SE 9) and one 124-bp single-ended or fractured TED (fTED) element (SE 1), the majority were imperfect excisions of TED that left variable footprints, ranging from remnants of the transposon’s TIRs and the host TSD (SE2 to SE8) to small deletions from either or both sides of the bz host sequence (SE10 to SE14). The different somatic excision events were recovered in widely variable frequencies, attesting to the heterogeneity of their timing. Most somatic excision sites did not show stretches of microhomology at the deletion junctions.
Nonstoichiometric TEDA-hybridizing bands, indicative of early somatic reinsertion events, were readily detected in DNA gel blots of bz-m175 seedlings (see Supplemental Figure 7 online). A total of 29 somatic reinsertion sites were isolated from seedling DNA, sequenced, and mapped (see Supplemental Figure 5A and Supplemental Table 4 online). As in the germline, TED transposes preferentially into genic regions (23/29) scattered throughout the genome (Figure 6).
Molecular Characterization and Activity of TED-Like Elements in Other Maize Lines and Relatives
The Bz progenitor of bz-m175 shares a prominent TED-hybridizing band with the High Loss ancestral lines (see Supplemental Figure 2 online). This band corresponds to a TED-like element inserted in the promoter region of a gene encoding a bHLH domain–like protein (see Supplemental Table 5 online, IT16). Two lines of evidence argue that this is the TED element that generated the bz-m175 allele. First, the disappearance of this band in the original bz-m175 isolate is coupled with the appearance of a new band corresponding to the TED insertion at bz (see Supplemental Figure 2 online). Second, the High Loss ancestor lines were shown to carry an active TED element in genetic trans-activation tests with the bz-m(dTED) tester lines described above.
Some of the maize inbred lines examined, such as W22, H99, and I137TN, show a strong TED-hybridizing band (see Supplemental Figure 2 online, lanes 5, 9, 10, and 11) that lacks genetic activity in crosses to bz-m(dTED) reporter lines. The TED-related elements had inserted in a genic sequence in H99, a conserved noncoding sequence in I137TN, and a MITE in W22 (see Supplemental Table 5 online). Their loss of activity may have resulted from either mutations in the element or from silencing by DNA methylation. Among maize relatives, Zea nicaraguensis (Iltis and Benz, 2000) shows the largest number of TEDA-hybridizing bands in DNA gel blots (see Supplemental Figure 2 online). Although TED activity of these elements was not tested, sequence comparison showed that they are highly similar to the maize TED element (see Supplemental Figure 8 online) so they may be transpositionally competent. As with the trTED elements from bz-m175, insertion sites in Z. nicaraguensis tend to correspond to genic regions (see Supplemental Table 5 online).
Duplicative Transposition
Some plants carrying a tr-TED (bz-s/sh-bz-X3; tr-TED/+) produced 50% more spotted kernels than expected in crosses with plants carrying a bz-dTED reporter (bz-dTED/sh-bz-X3). This observation suggests that a second unlinked TED element is present in the genome, most likely from duplicative secondary transposition of the tr-TED. The analysis of one example is shown in Supplemental Figure 9 online. GT20, a line bearing one tr-TED element, was crossed with two different bz-dTED lines, dTED2 and dTED17. The progeny from the GT20 × dTED2 cross shows the expected cosegregation of the spotted phenotype with both tr-TED with dTED (lanes 1 to 9). By contrast, in the GT20 × dTED17 cross (lanes 10 to 18), a second TED-hybridizing band segregates among the spotted kernels (lanes 10 to 15). Only this band, but not the original tr-TED, is present in two spotted individuals (lanes 11 and 12), indicating that this is an active newly tr-TED element, arisen by duplicative transposition. Moreover, polymorphic, weaker hybridizing bands are observed in progenies from both crosses (12 bands among 18 individuals), suggesting that these tr-TEDs can also undergo somatic duplicative transpositions.
Different duplicative transposition frequencies have been reported for members of the Mu family in maize, as high as 100% for Mu elements in a standard Mu line (Alleman and Freeling, 1986) and as low as ∼6% for Mu1 and 51.5% for MuDR-1 in a Minimal Line (Lisch et al., 1995). To determine the duplicative transposition frequency of TED, we crossed 28 different plants of the GT20 line carrying a trTED to a bz-dTED reporter line. Fifteen of the crosses gave the expected 1/4 spotted kernels. However, the remaining 13 ears segregated 3/8 spotted kernels, a result consistent with two unlinked TED elements (see Supplemental Table 6 online). These data establish that the duplicative transposition frequency of TED is very comparable to that of MuDR-1 elements (46% versus 51%) and that TED duplicative transpositions, like excisive ones, tend to be to unlinked sites.
Circular TED Elements
Maize Mu elements can exist in a circular extrachromosomal form (Sundaresan and Freeling, 1987). Circles covalently closed at the ends of the inverted repeats are also a feature of several other TEs (Curcio and Derbyshire, 2003). Although the Mu circles are not cleanly conjoined at the TIR ends and may contain deletions (V. Sundaresan, unpublished data; cited in Walbot and Rudenko, 2002), their presence exclusively in Mu-active lines suggests that they originate during the transposition process.
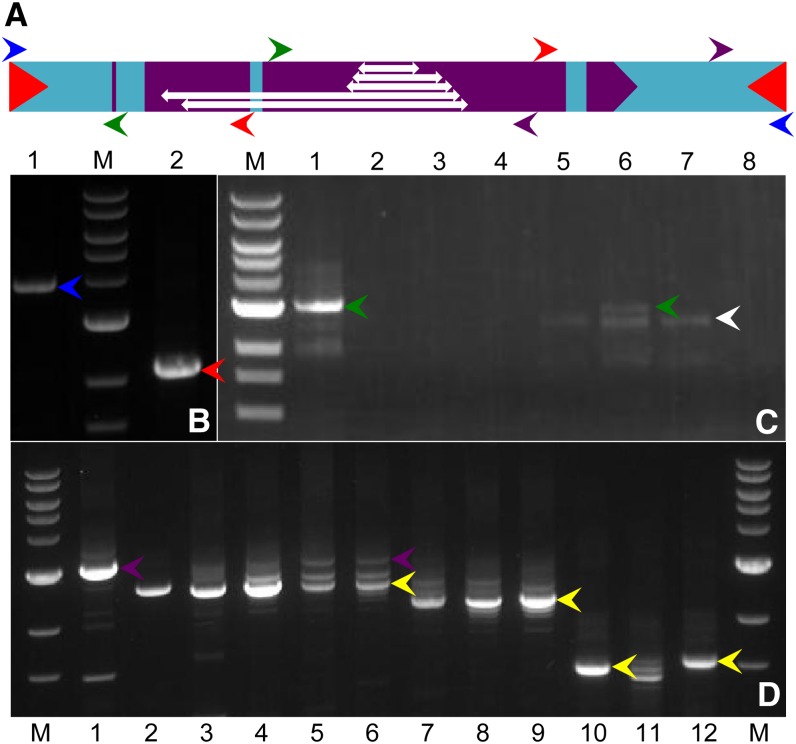
The similar preference of Mu and TED to transpose to unlinked genic sequences both germinally and somatically prompted us to search for circular-TED (cTED) DNA molecules in bz-m175 somatic tissues. We performed PCR on TED-active lines with divergent TED internal primers that would generate a product only if a circular DNA molecule was present (Figure 7A). In all cases, we readily detected PCR bands of the size expected of a TED element circularized by the end joining of its TIRs (Figures 7B to 7D) and cloned them for sequence analysis.
Figure 7.
PCR Amplification of cTED and dTED from bz-m175.
(A) Color-coded PCR primers in the parental TED sequence. dTED deletions are shown as double-headed arrows (from top to bottom, dTED20, dTED18, dTED13, dTED2, and dTED4).
(B) Amplification of bz-m175 DNA. Lane 1, a TIR single primer (blue arrows in [A]) amplifies the full-length element (blue arrowhead); lane 2, divergent internal primers (red arrows in [A]) produce a band of the expected size for a circular element (red arrowhead).
(C) Amplification with TED divergent primers (green arrows in [A]) of cTED and dTED elements (green and white arrowheads, respectively) in the trTED line GT30 (lane1) and in the bz-m progeny from a GT30 × dTED20 cross (lanes 5 to 7). No products are amplified in the TED-minus bz progeny from the same cross (lanes 2 to 4) or in the dTED20 line (lane 8).
(D) Amplifications with TED primers (purple arrows in [A]) of cTED (purple arrowheads) and corresponding dTED products (yellow arrowheads) from bz-m175/bz-R (lane 1) and four different trTED/dTED segregating lines. Lanes 2 to 6, dTED18; lanes 7 to 9, dTED13; lane 10, dTED2; lanes 11 and 12, dTED4; M, 1-kb DNA ladder.
Simple head-to-head fusions of the two TIRs would create new palindromic restriction sites for AvaI, XhoI, and BcgI, but these were not found. This indicates that the fusion of the two ends was more complex, as had been found for Mu circles (Sundaresan and Freeling, 1987). Sequencing of the PCR products revealed that all had the expected inverted configuration of 5′ and 3′ends fused head-to-head. However, due to the high sequence identity of the TIRs, mixed reads from both sequencing directions were obtained in the TIR regions. With the aim of sequencing one TIR end at a time, the cloned PCR products were digested at sites unique to each TIR. This allowed us, in most cases, to read the sequences to within 80 bp from the TIR ends (see Supplemental Figure 10 online).
PCR products of sizes other than expected, indicative of rearrangements, were also sequenced. Some had truncations of either or both ends at the junctions, the deletions ranging from <100 to >2 kb (see Supplemental Table 7 online). Others had internal deletions, flanked by imperfect direct repeats, suggesting possible microhomology-mediated deletions (McVey and Lee, 2008) prior to circularization.
dTED elements can form circular molecules, as well, but only if an active TED element is present. A population segregating for both trTED and dTED elements (GT30 and dTED20) was analyzed by inverse PCR using primers spanning the deletion in dTED (Figure 7A). These primers give rise to TED and dTED PCR products of different size, consistent with the internal 378-bp deletion that distinguishes the active and defective TED elements. We detected the predicted circular TED and dTED PCR products in active TED lines, but not in lines without TED activity (Figure 7C). Sequence analysis confirmed that the amplified PCR products were cTED and cdTED elements because they differed precisely at the site of the dTED20 deletion junction. As with cTED elements, end deletions of various sizes were found among cdTED20 elements (see Supplemental Table 7 online). Circular excision products were also formed in four other populations segregating dTED and trTED elements (Figure 7D; see Supplemental Table 7 online).
TED-Like Elements in Sequenced Plant Genomes
Five autonomous Mu transposons have been identified so far: MUDR (Chomet et al., 1991; Hershberger et al., 1991; Qin et al., 1991), Jittery (Xu et al., 2004), Hop (Chalvet et al., 2003), Os3378 (Gao, 2012), and TED. To investigate the distribution of TED-related elements in other plant genomes, we performed BLAST searches of plant sequence databases using the TED sequence as query.
Six sequences were found in the B73 maize genome. These probably correspond to the weakly hybridizing TEDA bands seen in DNA gel blots (see Supplemental Figure 2 online). All six sequences have typical MULE transposon features: They end in TIRs that vary in length from 115 to 186 bp and are flanked by 9-to 10-bp TSDs (see Supplemental Table 8 online). TIR identities vary from 82 to 99% within elements and from 77 to 99% between elements. However, the identities of these TIRs with TED’s were lower (75 to 79%), being restricted to a <90-bp region. Some of them are truncated copies of others (for instance, TEDLZm4 derives from TEDLZm2 by two separate short deletions; TEDLZm5 derives from TEDLZm2 by a single long deletion).
TEDA-hybridizing bands were also detected in sorghum (Sorghum bicolor), which is not surprising given its close relationship with maize. A BLASTN search of the sorghum genome sequence with TEDA as query identified five MULEs encoding transposases more similar to TEDA than to MURA (see Supplemental Figure 11 online). All five elements were closer to each other than to TED, indicating that they amplified after the sorghum-maize split 11.9 million years ago (Swigonová et al., 2004). The sorghum TED-like elements measure ∼4.6 kb, end in ∼200-bp TIRs of ∼95% sequence identity, and are flanked by 9- to 10-bp TSDs (see Supplemental Table 8 online). None of them encodes a MURB-like protein. The Brachypodium genome also contains a MULE element more closely related to TED than to MuDR. However, in phylogenetic comparisons with other plant MULE transposases, TEDA was found to be closer to MURA than to most other annotated plant sequences (see Supplemental Figure 11 online).
DISCUSSION
Relationship to MuDR and Mu System
Here, we describe the TED element of maize, the founding member of a novel family of very-low-copy-number transposons within the Mu superfamily. TED was recovered from the bz-m175 mutable allele in a High Loss stock known to undergo frequent chromosome breaks most likely because of its high content of active transposons (Rhoades and Dempsey, 1982). bz-m175 was subsequently introgressed by repeated backcrossing into the genetic background of the inbred W22 without any change in phenotype, loss of activity, or large increase in TED copy number. In this respect, TED resembles the minimal Mu line (Lisch, 2002) rather than the original higher MuDR copy number lines, where Mu activity was silenced by inbreeding (Robertson, 1986) and Mu1 copies greatly amplified by outcrossing (Alleman and Freeling, 1986).
TED and TED-like elements occur in the genus Zea and in close grass relatives, such as Tripsacum, Sorghum, and Brachypodium. These elements have the typical characteristics of a MULE: They encode transposases with homology to the Mu MURA protein, end in long inverted repeats of ∼200 bp, and cause a 9-bp duplication of the host insertion sites (Figure 2). TED is the third characterized autonomous transposon of the Mu superfamily in maize, a model system unique in its number of genetically defined elements. Based on its transposase homology, TED is much closer to MuDR (Eisen et al., 1994; Hershberger et al., 1995) than to Jittery (Xu et al., 2004), an active MULE related to the phytochrome A transcription factors FHY3/FAR1 (Hudson et al., 2003). A phylogenetic comparison of TED-related elements in maize and other sequenced grasses reveals a disproportionate abundance of TED-like elements in S. bicolor and Panicum virgatum, suggesting that the TED family has amplified differentially in different plant genomes (see Supplemental Figure 11 online). Similar observations have been made for retrotransposons (Vitte and Bennetzen, 2006) and other DNA transposons, such as PACK-MULEs (Jiang et al., 2004) and Helitrons (Du et al., 2009; Li and Dooner, 2012).
Absence of a B Function
Despite the similarities among members of the Mu superfamily from different plant species, only MuDR encodes a MURB protein. The initial analysis of MuDR deletion derivatives suggested that this protein may be required for somatic and germinal insertion (Lisch et al., 1999), but to this day its function remains elusive (Lisch and Jiang, 2009). TED is the closest autonomous relative of MuDR found to date (see Supplemental Figure 11 online). It is fully transposition competent, being capable of both excision and reinsertion in somatic and germinal tissues, but it lacks a B function, like other autonomous MULEs of widely divergent phylogenetic origins: Jittery of maize (Xu et al., 2004), Os3378 of rice (Gao, 2012), and Hop of Fusarium (Chalvet et al., 2003). These data argue that, if MURB is indeed required for MuDR reinsertion, that requirement evolved recently, after the separation of the MURA and TEDA clades.
Transposition in Sporophyte and Gametophyte
In contrast with Mu elements, which excise only late in development (McCarty et al., 1989; Raizada et al., 2001), TED can excise in young sporophytic tissues. PCR somatic excision products were more abundant in seedling leaves than in mature leaves (Figure 2). Although large revertant sectors are occasionally observed (Figures 1B to 1D), indicating an excision early in development, most reversions occur in the last few cell divisions, as in the MuDR system. However, whereas the timing of somatic excisions is not affected by dosage of the autonomous element in either system, the frequency of somatic excisions in the aleurone shows a positive response to TED dosage (see Supplemental Figures 1A and 1B online).
Our analysis of putative germinal revertants of bz-m175 uncovered an unusual TED feature, a high transposition frequency in the haploid gametophyte that can account for the majority of heritable excisions. In this respect, TED resembles the hobo-Activator-Tam2 (hAT) elements Inhibitor of R of maize and Tag1 of Arabidopsis thaliana, which excise in developing gametophytes, leading to a high frequency of germinal reversions (Williams et al., 1984; Eggleston et al., 1995; Galli et al., 2003), but differs from the majority of Arabidopsis transposons, which are transcribed and transpose in the pollen vegetative nucleus, but not in the sperm nuclei (Slotkin et al., 2009). Even though plants alternate between a diploid and a haploid phase, most genetic analysis in higher plants is performed with the dominant diploid sporophytic generation. Two properties of the bz-m175 mutable allele allow our genetic analysis to extend to the haploid gametophytic generation. First, the bz gene is expressed both in the endosperm and the embryo/seedling, plant parts produced from the separate fertilization events of the central cell and egg cell in the female gametophyte by the two sperm in the male gametophyte. Second, bz affects anthocyanin pigmentation, a most readily scorable trait in these tissues.
Putative germinal revertants can be identified as fully purple kernels in bz-m175 testcross ears (Figure 1A). The frequency of such kernels when using bz-m175 as female parent (2.5 × 10−3) is ∼100-fold higher than the reversion frequency of Mu1-containing mutable alleles (Bennetzen, 1996; Walbot and Rudenko, 2002). However, most of these Bz kernels (111/113) have a bz-m embryo (i.e., they do not represent germinal events transmissible to the next generation). These class 1 nonconcordant kernels can arise from reversion events in four of the seven mitoses that lead to the formation of the mature haploid female gametophyte or embryo sac: divisions I, II, V, or VI (Figure 3B). We infer that reversions occur at the two- or four-nucleate stage of the embryo sac because concordant kernels would have been more numerous if reversions had occurred at the one-nucleate stage (division I) and had an equal probability of being incorporated into either daughter cell. The reciprocal class 2 nonconcordant kernels, with a bz-m endosperm and a Bz embryo (Figure 3C), can only arise from a reversion event that is transmitted to the egg cell in the last embryo sac mitosis (division VI) and was recovered at a substantially lower frequency (1.8 × 10−4 versus 2.5 × 10−3). If a Bz’ reversion at this mitotic division has an equal probability of being incorporated into the sib polar nucleus, then it can account for only eight of the 111 class 1 nonconcordant kernels (Bz’ endosperm/bz-m embryo). This suggests that most reversions that produce class 1 nonconcordant kernels occur at mitotic divisions II or V in the embryo sac (Figure 3B) and, therefore, that TED excision in the haploid female gametophyte is under tight developmental control, as it is in the diploid sporophyte.
The frequency of concordant revertant kernels obtained in testcrosses using bz-m175 as pollen parent is 1 × 10−4, of the same order of magnitude as in the reciprocal cross. In the male gametophyte, the two sperm that participate in the double fertilization of the egg and central cell of the embryo sac share a common lineage until the last mitosis. Hence, concordant kernels can arise from reversion events either at meiosis or at the first pollen mitosis (Figure 3D). The reciprocal classes of nonconcordant kernels arise from reversion events at the second pollen mitosis and would be expected in roughly equal numbers, assuming no preferential fertilization by either sperm. As seen in Figures 3E and 3F, they occur in very low frequencies that do not differ significantly from each other.
All germinal Bz’ revertants alleles, regardless of parent of origin, carry a +0 footprint, reflecting the importance of the TED insertion site for bz gene function, as TED disrupts the sugar nucleotide binding site deemed essential for the glucosyltransferase activity of the BZ protein (Hughes and Hughes, 1994; Offen et al., 2006). No simple excisions were detected among stable bronze alleles. In this class, dTED elements of different sizes predominate, followed by bz deletions large enough to remove at least one of the PCR priming sites flanking bz and by a small fraction of fTED elements resulting from a deletion of one end of the inserted TED element plus adjacent bz sequences.
Like MuDR and nonautonomous Mu elements (Lisch et al., 1995), TED is capable of somatic as well as germinal reinsertion. In this, it differs from Jittery, an unusual MULE that can excise but not reinsert in the maize genome (Xu et al., 2004). Both somatic and germinal transpositions are mainly to unlinked sites and show preference for the 5′ end of targeted genes, including the upstream promoter region and the first few exons (see Supplemental Tables 2 and 4 and Supplemental Figure 5 online; Figure 6), properties also evidenced by Mu elements (Dietrich et al., 2002; May et al., 2003; McCarty et al., 2005).
Excisive versus Duplicative Transposition during Development
Like other plant transposons (Walbot and Rudenko, 2002; Li and Dooner, 2009; Lisch and Jiang, 2009; Huang and Dooner, 2012), TED can undergo both excisive and duplicative transposition. Appearance of the transposon at a new site is accompanied in the former by complete loss of the transposon from the original site and in the latter by retention of the transposon in whole or in part at the original site. Thus, the two types of transposition can be empirically distinguished by the nature of the novel products formed at the original insertion site: simple excision footprints in the former and defective elements in the latter.
Simple excision footprints contain remnants of the TSD generated by transposon insertion. Most transposon excision footprints in plants lack stretches of microhomology greater than 3 bp at the new deletion junction and are taken to arise by nonhomologous end joining of the DSB produced by element excision (Daley et al., 2005). A few show longer stretches of microhomology (≥ 5 bp) with filler DNA at the deletion junction sometimes and may arise by the rarer microhomology-mediated end joining repair of DSBs (McVey and Lee, 2008). Duplicative transpositions have been postulated to arise from replication repair of the DSB at the transposon excision site using either a sister or nonsister chromatid as template (Nassif et al., 1994). In this mechanism, incomplete repair would lead to variable deletions of the element at the donor site. Internally or terminally deleted elements found at the previous insertion site of autonomous elements often have microhomology and filler DNA at the deletion junction. Such elements occur at a high frequency in the progeny of bz-m175 plants.
Recessive stable mutations from unstable alleles occurring as single kernels in an ear can only arise at or around meiosis (Dooner and Belachew, 1989). Down-selection for stable bronze (bz-s) derivatives from bz-m175 revealed an unexpectedly high frequency of different classes of deletion mutations resulting from TED transposition. Defective TED (dTED) elements (i.e., TED internal deletions that did not alter the host gene) were the most frequent class (62.5% of all mutations), large deletions extending bidirectionally into the host gene for at least 6 kb were half as frequent, and fractured TED (fTED) elements or deletions of one end of TED that extended unidirectionally into the host gene were one-tenth as frequent.
Strikingly, no simple excision footprints were recovered among the 48 bz-s germinal derivatives. Given that TED excision in somatic tissues results in simple footprints and that practically all of them would have produced a stable bronze phenotype, we conclude that, unlike in somatic tissues, double-stranded gap repair is most probably the only mechanism for repairing TED-induced DSBs in meiotic cells. The short direct repeats flanking the deletion junctions in dTED and fTED elements and the filler DNA sequences in a subset of them can be best explained by repair synthesis using the sister chromatid as template (Nassif et al., 1994). The deletion of the entire bz region in the sh-bz-X2 homolog precludes the use of a nonsister chromatid as alternative template. The large bidirectional deletions of the host gene that can arise from TED excision have not been reported among mutable alleles carrying Mu elements, although in this study they were relatively frequent (2 × 10−3). Their absence from Mu alleles could be simply due to the fact that only a few loss-of-function mutations have been studied. However, many down-selections of mutable alleles carrying Ac/Ds transposons have been analyzed (e.g., Huang and Dooner, 2012) without uncovering large deletions. Since the predominant mechanisms of DSB repair in germinal tissues differ between hAT and Mu elements, it is conceivable that large bidirectional deletions may be a general feature distinguishing these two transposon superfamilies.
Plant DNA transposons can also differ in the relative frequencies with which the two types of transposition occur in different tissues. Whereas hAT elements, like Ac, exhibit predominantly excisive transposition in both somatic (Kunze and Weil, 2002) and meiotic (Huang and Dooner, 2012) tissues, Mu elements undergo mainly excisive transposition in somatic cells (Britt and Walbot, 1991; Doseff et al., 1991) and duplicative transposition in meiotic cells (Schnable et al., 1989; Britt and Walbot, 1991; Doseff et al., 1991; Lisch and Jiang, 2009). In this respect, TED shows a similar behavior to Mu (Figures 3 and 6). However, the frequency with which defective elements are formed at a specific site appears to be higher for TED than for Mu (Hsia and Schnable, 1996), even though Mu duplicative transposition can be as high as one event per element per generation (Alleman and Freeling, 1986). This observation suggests that replication repair of the DSB induced by element excision aborts prematurely, leading to the synthesis of an incomplete element, more frequently with TED than with Mu.
Circular Transposition Products
Putative extrachromosomal transposition intermediates in the form of linear or circular DNA have been reported for the Tc3 and Tc1 elements of Caenorhabditis elegans (Radice and Emmons, 1993; van Luenen et al., 1993). In maize, extrachromosomal Mu1 circular molecules were detected more than two decades ago (Sundaresan and Freeling, 1987), but their detailed structure was not resolved and, to this day, their biological significance remains unclear. Circularized Ds elements have also been reported in maize and in transgenic tobacco (Nicotiana tabacum; Gorbunova and Levy, 1997, 2000). On the other hand, no extrachromosomal circles were detected in ddm1 mutants of Arabidopsis, in which normally quiescent Mu elements were demethylated and activated (Singer et al., 2001), leading the authors to suggest that circular forms could be transposition intermediates that require the MURB function found only in maize. Here, we provide evidence for the formation of circular cTED molecules in bz-m175 plants and show that the formation of circular cdTED molecules occurs only in the presence of an autonomous TED element.
Using a PCR strategy, we have shown that TED/dTED excisions are accompanied by the formation of products with the expected size and joined TIR structure of extrachromosomal circular forms (Figure 7). Sequence analysis confirmed the head-to-head fusion structure and restriction analysis of the PCR products established that the fusions were not perfect, as the fragments lacked the restriction site at the junction predicted from precise joining of the transposon ends. Sequencing of PCR products with a shorter than expected length revealed deletions of variable sizes at the TIR joints (see Supplemental Table 7 online), in agreement with earlier findings in the Tc1 and Ac/Ds systems. The TIR head-to-head fusions are most likely free of adjacent TSD sequences, as the junction sites of the circularized elements share no sequence similarity with the TSD sequences at the donor site.
The formation of circular extrachromosomal transposon molecules only in the presence of an autonomous TED element establishes that these molecules are indeed the products of transposition reactions. Whether they are true transposition intermediates can only be conjectural at this time. The observation that TED, like Mu elements, transposes mostly to unlinked sites, in contrast with Ac/Ds, argues that the TED donor and recipient sites do not associate physically during the transposition reaction and favors the view of an extrachromosomal transposition intermediate. However, none of the observed products had a perfect head-to-head fusion structure, suggesting that they may represent abortive transposition intermediates that are incapable of reinsertion and accumulate in the cell. Possibly, the true transposition intermediates are short-lived linear TED structures with TIRs bound by transposase molecules that protect them from exonucleolytic degradation.
METHODS
Genetic Stocks
The following genetic stocks were used: bz-m175, a mutable allele with a fine-spotted phenotype arisen in a High Loss × High Knob genetic background (Rhoades and Dempsey, 1982); sh-bz-X2 and sh-bz-X3 (shrunken, bronze), x-ray–induced deletions of large chromosomal fragments extending beyond the 2-centimorgan sh-bz interval (Mottinger, 1973); and bz-R, the stable reference allele harboring a 340-bp deletion of bz coding sequences (Ralston et al., 1987).
Selection and Analysis of Bz′ and bz-s Derivatives
Bz′ germinal revertants were isolated from reciprocal testcrosses of Sh bz-m175/sh-bz-X2 hemizygotes to sh-bz-X3. In these crosses, exceptional plump, purple kernels represent Bz′ derivatives from bz-m175. In the same populations, nonconcordant Bz’ germinal revertants were screened by seedling phenotype (red coleoptile and leaf tips) after planting spotted kernels in sand benches in the greenhouse. All selections were self-pollinated and backcrossed to sh-bz-X3. bz-s stable bronze derivatives were obtained from testcrosses of Sh bz-m175/sh-bz-X2 hemizygotes to sh-bz-X3. Putative bz-s derivatives were selected as plump, stable bronze kernels, self-pollinated, and backcrossed to sh-bz-X3.
DNA Isolation and DNA Gel Blot Analysis
Maize (Zea mays) genomic DNA was isolated from seedling, adult leaf tissues, and pollen as described (Li et al., 2013). Restriction digested DNA (10 μg) was resolved on 0.6% agarose gels and transferred to Hybond XL nylon membranes (Amersham Biosciences). 32P-labeled probes were generated with Ready-To-Go DNA labeling beads (Amersham Biosciences). Hybridization and washing were performed according to the manufacturer’s instructions. The 2814-bp TEDA probe was amplified from a genomic clone using primers spanning the coding region of the TEDA gene. The 1157-bp bz probe was amplified from a genomic clone using primers spanning the bz coding region upstream of the TED insertion site.
PCR Amplifications and DNA Sequencing
PCR was performed according to the protocol of Expand High Fidelity PCR system (Hoffmann-La Roche). PCR products were cloned into pGEM-T Easy Vector (Promega) and transformed into XL-Blue–competent cells. Plasmids were purified with a Qiagen Spin Miniprep kit. DNA sequencing was performed in an ABI 3730 sequencer (Applied Biosciences) following the manufacturer’s instructions. Primers used in this work are shown in Supplemental Table 9 online.
Sequence Analysis and Mapping of TED Insertion Sites
Sequences flanking trTED elements were isolated by iPCR and sequenced. In brief, genomic DNA was digested with BamHI, EcoRI, HincII, HindIII, SacI, or SspI and then ligated under dilute conditions to favor intramolecular ligation. PCR was performed as described above using appropriate TED primers. Unique PCR bands were directly sequenced without additional purification. Sequences flanking both ends were assembled into parental sequences by merging the identical 9-bp TSDs on either side. The assembled sequences were queried against the maize B73 RefGen_v2 (MGSC) database by BLASTN (Altschul et al., 1997) and manually annotated after downloading the BAC contig from the database and analyzing the coding capacity of sequences 10 kb upstream and downstream of the insertion by BLASTX.
TED-Like Transposon Annotation and Phylogenetic Analysis
The entire TED sequence was queried against the maize B73 RefGen_v2 (MGSC) database by BLASTN. Hits from the transposase coding region were chosen for further analysis. To define the TIRs of the putative transposons, flanking sequences 10 kb upstream and downstream of the matching region were retrieved from the B73 database and aligned against each other using NCBI-BLAST 2. MULE TIR sequences were identified from pairwise alignments and subsequently refined from the flanking TSDs. The newly identified TED-like elements were then subjected to the same sequence analyses as described above. The TED sequence was also queried against the Sorghum and Brachypodium databases at Phytozome (http://www.phytozome.net). Sequence features of TED-like elements were manually annotated to define TIRs, TSDs, and the coding capacity of the insertion sites as described above for maize. Transposases encoded by these elements were predicted with Fgenesh++ (http://linux1.softberry.com). The DDE domain was extracted from the predicted transposase by comparison with MURA and TEDA and used for phylogenetic analysis together with other reported DDE domain transposases (Yuan and Wessler, 2011). A phylogenetic tree was constructed using neighbor joining in MEGA version 5.05 (http://www.megasoftware.net/), with 1000 bootstrap replicates and the pairwise deletion option for handling gaps.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: TED sequence (GenBank KF287636), bz-m175 allele (KF279654), Bz progenitor allele of bz-m175 (KF279655), Bz High Loss allele (KF279653), and Bz High Knob allele (KF279652).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Positive Dosage Effect of the bz-m175 Allele.
Supplemental Figure 2. TED Is a Low Copy Transposon in Maize and Its Grass Relatives.
Supplemental Figure 3. TED Dosage Effect.
Supplemental Figure 4. trTED Elements Maintain Their Transposase Activity.
Supplemental Figure 5. Sequence Features of TED Reinsertion Sites.
Supplemental Figure 6. Footprint Analysis of Somatic Excision Products from bz-m175.
Supplemental Figure 7. Detection of TED Somatic Reinsertions.
Supplemental Figure 8. Sequence Alignment of TIRs from TED at bz-m175 and TED-Like Elements Isolated from Zea nicaraguensis.
Supplemental Figure 9. Duplicative Transposition of TED Elements.
Supplemental Figure 10. Sequence Analysis of Circular TED Elements.
Supplemental Figure 11. Phylogenetic Tree of MULE Transposases.
Supplemental Table 1. Segregation in Testcrosses of Sh bz-m175/sh-bz-X3 Hemizygotes.
Supplemental Table 2. Germinal TED Transposition Sites Recovered via Inverse PCR Amplification.
Supplemental Table 3. Sequence Features of dTED and fTED Elements in bz-s Derivatives from bz-m175.
Supplemental Table 4. Somatic TED Reinsertion Sites Isolated by iPCR in Genomic DNA of Leaf Tissues.
Supplemental Table 5. TED-Related Elements Recovered via iPCR Amplification from Different Maize Lines and Teosinte.
Supplemental Table 6. Frequencies of trTED Duplications in Female Parent.
Supplemental Table 7. Sequence Features of cTED and cdTED Elements in TED-Active Lines.
Supplemental Table 8. MULEs Detected with a TEDA Query in Representative Plant Genomes.
Supplemental Table 9. Oligonucleotide Primers for PCR and iPCR Analysis.
Acknowledgments
We thank Qinghua Wang, Limei He, and Jun Huang for comments on the article, Xiuzhi Liu and Xiaofang Xue for help in the greenhouse and laboratory, and Marc Probasco for plant care. This article is dedicated to Manford Goh, one of our enthusiastic undergraduate field helpers, who passed away suddenly this year. Funding for this project was provided by the National Science Foundation (DBI-0929350) and the Waksman Institute.
AUTHOR CONTRIBUTIONS
Y.L. and H.K.D. designed the work, analyzed the data, and wrote the article. Y.L. and L.H. performed the research.
Glossary
- TE
transposable element
- TIR
terminal inverted repeat
- TSD
target site duplication
- DSB
double-strand break
References
- Ågren J.A., Wright S.I. (2011). Co-evolution between transposable elements and their hosts: A major factor in genome size evolution? Chromosome Res. 19: 777–786 [DOI] [PubMed] [Google Scholar]
- Alleman M., Freeling M. (1986). The Mu transposable elements of maize: Evidence for transposition and copy number regulation during development. Genetics 112: 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito M.I., Walbot V. (1997). Characterization of the maize Mutator transposable element MURA transposase as a DNA-binding protein. Mol. Cell. Biol. 17: 5165–5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J.L. (1996). The Mutator transposable element system of maize. Curr. Top. Microbiol. Immunol. 204: 195–229 [DOI] [PubMed] [Google Scholar]
- Britt A.B., Walbot V. (1991). Germinal and somatic products of Mu1 excision from the Bronze-1 gene of Zea mays. Mol. Gen. Genet. 227: 267–276 [DOI] [PubMed] [Google Scholar]
- Chalvet F., Grimaldi C., Kaper F., Langin T., Daboussi M.J. (2003). Hop, an active Mutator-like element in the genome of the fungus Fusarium oxysporum. Mol. Biol. Evol. 20: 1362–1375 [DOI] [PubMed] [Google Scholar]
- Chomet P., Lisch D., Hardeman K.J., Chandler V.L., Freeling M. (1991). Identification of a regulatory transposon that controls the Mutator transposable element system in maize. Genetics 129: 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad L.J., Bai L., Ahern K., Dusinberre K., Kane D.P., Brutnell T.P. (2007). State II dissociation element formation following activator excision in maize. Genetics 177: 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowperthwaite M., Park W., Xu Z., Yan X., Maurais S.C., Dooner H.K. (2002). Use of the transposon Ac as a gene-searching engine in the maize genome. Plant Cell 14: 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio M.J., Derbyshire K.M. (2003). The outs and ins of transposition: From mu to kangaroo. Nat. Rev. Mol. Cell Biol. 4: 865–877 [DOI] [PubMed] [Google Scholar]
- Daley J.M., Palmbos P.L., Wu D., Wilson T.E. (2005). Nonhomologous end joining in yeast. Annu. Rev. Genet. 39: 431–451 [DOI] [PubMed] [Google Scholar]
- Delseny M., Han B., Hsing Y.Ie. (2010). High throughput DNA sequencing: The new sequencing revolution. Plant Sci. 179: 407–422 [DOI] [PubMed] [Google Scholar]
- Dennis, E.S., Finnegan, E.J., Taylor, B.H., Peterson, T.A., Walker, A.R., and Peacock, W.J. (1988). Maize transposable elements: Structure, function, and regulation. In Plant Transposable Elements, O.E. Nelson, ed (New York: Plenum Press), pp. 101–113. [Google Scholar]
- Dietrich C.R., Cui F., Packila M.L., Li J., Ashlock D.A., Nikolau B.J., Schnable P.S. (2002). Maize Mu transposons are targeted to the 5′ untranslated region of the gl8 gene and sequences flanking Mu target-site duplications exhibit nonrandom nucleotide composition throughout the genome. Genetics 160: 697–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner H.K., Belachew A. (1989). Transposition pattern of the maize element Ac from the bz-m2(Ac) allele. Genetics 122: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doseff A., Martienssen R., Sundaresan V. (1991). Somatic excision of the Mu1 transposable element of maize. Nucleic Acids Res. 19: 579–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C., Fefelova N., Caronna J., He L., Dooner H.K. (2009). The polychromatic Helitron landscape of the maize genome. Proc. Natl. Acad. Sci. USA 106: 19916–19921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggleston W.B., Alleman M., Kermicle J.L. (1995). Molecular organization and germinal instability of R-stippled maize. Genetics 141: 347–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J.A., Benito M.I., Walbot V. (1994). Sequence similarity of putative transposases links the maize Mutator autonomous element and a group of bacterial insertion sequences. Nucleic Acids Res. 22: 2634–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N.V. (2012). Presidential address. Transposable elements, epigenetics, and genome evolution. Science 338: 758–767 [DOI] [PubMed] [Google Scholar]
- Fernandes J., Dong Q., Schneider B., Morrow D.J., Nan G.L., Brendel V., Walbot V. (2004). Genome-wide mutagenesis of Zea mays L. using RescueMu transposons. Genome Biol. 5: R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C., Pritham E.J. (2007). DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41: 331–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli M., Theriault A., Liu D., Crawford N.M. (2003). Expression of the Arabidopsis transposable element Tag1 is targeted to developing gametophytes. Genetics 165: 2093–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D. (2012). Identification of an active Mutator-like element (MULE) in rice (Oryza sativa). Mol. Genet. Genomics 287: 261–271 [DOI] [PubMed] [Google Scholar]
- González L.G., Deyholos M.K. (2012). Identification, characterization and distribution of transposable elements in the flax (Linum usitatissimum L.) genome. BMC Genomics 13: 644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V., Levy A.A. (1997). Circularized Ac/Ds transposons: Formation, structure and fate. Genetics 145: 1161–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V., Levy A.A. (2000). Analysis of extrachromosomal Ac/Ds transposable elements. Genetics 155: 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger R.J., Benito M.I., Hardeman K.J., Warren C., Chandler V.L., Walbot V. (1995). Characterization of the major transcripts encoded by the regulatory MuDR transposable element of maize. Genetics 140: 1087–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger R.J., Warren C.A., Walbot V. (1991). Mutator activity in maize correlates with the presence and expression of the Mu transposable element Mu9. Proc. Natl. Acad. Sci. USA 88: 10198–10202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holligan D., Zhang X., Jiang N., Pritham E.J., Wessler S.R. (2006). The transposable element landscape of the model legume Lotus japonicus. Genetics 174: 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia A.P., Schnable P.S. (1996). DNA sequence analyses support the role of interrupted gap repair in the origin of internal deletions of the maize transposon, MuDR. Genetics 142: 603–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua-Van A., Capy P. (2008). Analysis of the DDE motif in the Mutator superfamily. J. Mol. Evol. 67: 670–681 [DOI] [PubMed] [Google Scholar]
- Huang J.T., Dooner H.K. (2012). The spectrum and frequency of self-inflicted and host gene mutations produced by the transposon Ac in maize. Plant Cell 24: 4149–4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson M.E., Lisch D.R., Quail P.H. (2003). The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 34: 453–471 [DOI] [PubMed] [Google Scholar]
- Hughes J., Hughes M.A. (1994). Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (Manihot esculenta Crantz) cotyledons. DNA Seq. 5: 41–49 [DOI] [PubMed] [Google Scholar]
- Iltis H.H., Benz B.F. (2000). Zea nicaraguensis (Poaceae), a new teosinte from Pacific coast Nicaragua. Novon 10: 382–390 [Google Scholar]
- Jiang N., Bao Z., Zhang X., Eddy S.R., Wessler S.R. (2004). Pack-MULE transposable elements mediate gene evolution in plants. Nature 431: 569–573 [DOI] [PubMed] [Google Scholar]
- Kidwell M.G. (2002). Transposable elements and the evolution of genome size in eukaryotes. Genetica 115: 49–63 [DOI] [PubMed] [Google Scholar]
- Kunze, R., and Weil, C.F. (2002). The hAT and CACTA superfamilies of plant transposons. In Mobile DNA II, N.L. Craig, R. Craigie, M. Gellert, and A.M. Lambowitz, eds (Washington, D.C.: ASM Press), pp. 565–610. [Google Scholar]
- Li Y., Dooner H.K. (2009). Excision of Helitron transposons in maize. Genetics 182: 399–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., and Dooner, H.K. (2012). Helitron proliferation and gene-fragment capture. In Plant Transposable Elements: Impact on Genome Structure and Function, M.A. Gransbastien and J.M. Casacuberta, eds (Berlin: Springer-Verlag), pp. 193–227. [Google Scholar]
- Li, Y., Wang, Q., Segal, G., and Dooner, H.K. (2013). Gene tagging with engineered Ds elements in maize. In Methods in Molecular Biology: Plant Transposable Elements, T. Peterson, ed (New York: Springer Science+Business Media), pp. 83–99. [DOI] [PubMed] [Google Scholar]
- Lisch D. (2002). Mutator transposons. Trends Plant Sci. 7: 498–504 [DOI] [PubMed] [Google Scholar]
- Lisch D., Chomet P., Freeling M. (1995). Genetic characterization of the Mutator system in maize: Behavior and regulation of Mu transposons in a minimal line. Genetics 139: 1777–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D., Girard L., Donlin M., Freeling M. (1999). Functional analysis of deletion derivatives of the maize transposon MuDR delineates roles for the MURA and MURB proteins. Genetics 151: 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch, D., and Jiang, N. (2009). Mutator and MULE transposons. In Handbook of Maize: Genetics and Genomics, J.L. Bennetzen and S. Hake, eds (New York: Springer), pp. 277–306. [Google Scholar]
- May B.P., Liu H., Vollbrecht E., Senior L., Rabinowicz P.D., Roh D., Pan X., Stein L., Freeling M., Alexander D., Martienssen R. (2003). Maize-targeted mutagenesis: A knockout resource for maize. Proc. Natl. Acad. Sci. USA 100: 11541–11546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty D.R., Carson C.B., Stinard P.S., Robertson D.S. (1989). Molecular analysis of viviparous-1: An abscisic acid-insensitive mutant of maize. Plant Cell 1: 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty D.R., et al. (2005). Steady-state transposon mutagenesis in inbred maize. Plant J. 44: 52–61 [DOI] [PubMed] [Google Scholar]
- McVey M., Lee S.E. (2008). MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet. 24: 529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottinger J. (1973). Unstable mutants of bronze induced by premeiotic X-ray treatment in maize. Theor. Appl. Genet. 43: 190–195 [DOI] [PubMed] [Google Scholar]
- Nassif N., Penney J., Pal S., Engels W.R., Gloor G.B. (1994). Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14: 1613–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offen W., Martinez-Fleites C., Yang M., Kiat-Lim E., Davis B.G., Tarling C.A., Ford C.M., Bowles D.J., Davies G.J. (2006). Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 25: 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin M.M., Robertson D.S., Ellingboe A.H. (1991). Cloning of the Mutator transposable element MuA2, a putative regulator of somatic mutability of the a1-Mum2 allele in maize. Genetics 129: 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radice A.D., Emmons S.W. (1993). Extrachromosomal circular copies of the transposon Tc1. Nucleic Acids Res. 21: 2663–2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizada M.N., Nan G.L., Walbot V. (2001). Somatic and germinal mobility of the RescueMu transposon in transgenic maize. Plant Cell 13: 1587–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston E.J., English J., Dooner H.K. (1987). Stability of deletion, insertion and point mutations at the bronze locus in maize. Theor. Appl. Genet. 74: 471–475 [DOI] [PubMed] [Google Scholar]
- Rhoades M.M., Dempsey E. (1982). The induction of mutable systems in plants with the high-loss mechanism. Maize Genetics Coop. Newslet. 56: 21–26 [Google Scholar]
- Robertson D.S. (1978). Characterization of a Mutator system in maize. Mutat. Res. 51: 21–28 [Google Scholar]
- Robertson D.S. (1986). Genetic studies on the loss of mu mutator activity in maize. Genetics 113: 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D.S., Stinard P.S. (1989). Genetic analyses of putative two-element systems regulating somatic mutability in Mutator-induced aleurone mutants of maize. Dev. Genet. 10: 482–506 [Google Scholar]
- Roth D.B., Porter T.N., Wilson J.H. (1985). Mechanisms of nonhomologous recombination in mammalian cells. Mol. Cell. Biol. 5: 2599–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin E., Levy A.A. (1997). Abortive gap repair: Underlying mechanism for Ds element formation. Mol. Cell. Biol. 17: 6294–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., Peterson P.A., Saedler H. (1989). The bz-rcy allele of the Cy transposable element system of Zea mays contains a Mu-like element insertion. Mol. Gen. Genet. 217: 459–463 [DOI] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Shepherd N.S., Rhoades M.M., Dempsey E. (1989). Genetic and molecular characterization of a-mrh-Mrh, a new mutable system of Zea mays. Dev. Genet. 10: 507–519 [DOI] [PubMed] [Google Scholar]
- Singer T., Yordan C., Martienssen R.A. (2001). Robertson’s Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev. 15: 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R.K., Vaughn M., Borges F., Tanurdzić M., Becker J.D., Feijó J.A., Martienssen R.A. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V., Freeling M. (1987). An extrachromosomal form of the Mu transposons of maize. Proc. Natl. Acad. Sci. USA 84: 4924–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonová Z., Lai J., Ma J., Ramakrishna W., Llaca V., Bennetzen J.L., Messing J. (2004). Close split of sorghum and maize genome progenitors. Genome Res. 14 (10A): 1916–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Luenen H.G., Colloms S.D., Plasterk R.H. (1993). Mobilization of quiet, endogenous Tc3 transposons of Caenorhabditis elegans by forced expression of Tc3 transposase. EMBO J. 12: 2513–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte C., Bennetzen J.L. (2006). Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc. Natl. Acad. Sci. USA 103: 17638–17643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E., et al. (2010). Genome-wide distribution of transposed dissociation elements in maize. Plant Cell 22: 1667–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot, V., and Rudenko, G.N. (2002). MuDR/Mu transposable elements of maize. In Mobile DNA II, N.L. Craig, R. Craigie, M. Gellert, and A.M. Lambowitz, eds (Washington, D.C.: ASM Press), pp. 533–564. [Google Scholar]
- Wessler S., Tarpley A., Purugganan M., Spell M., Okagaki R. (1990). Filler DNA is associated with spontaneous deletions in maize. Proc. Natl. Acad. Sci. USA 87: 8731–8735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler S.R. (2006). Transposable elements and the evolution of eukaryotic genomes. Proc. Natl. Acad. Sci. USA 103: 17600–17601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T., et al. (2007). A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8: 973–982 [DOI] [PubMed] [Google Scholar]
- Williams W.M., Satyanarayana K.V., Kermicle J.L. (1984). R-stippled maize as a transposable element system. Genetics 107: 477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Yan X., Maurais S., Fu H., O’Brien D.G., Mottinger J., Dooner H.K. (2004). Jittery, a Mutator distant relative with a paradoxical mobile behavior: Excision without reinsertion. Plant Cell 16: 1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Martínez-Férez I.M., Kavchok S., Dooner H.K. (1999). Origination of Ds elements from Ac elements in maize: Evidence for rare repair synthesis at the site of Ac excision. Genetics 152: 1733–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y.W., Wessler S.R. (2011). The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc. Natl. Acad. Sci. USA 108: 7884–7889 [DOI] [PMC free article] [PubMed] [Google Scholar]