Allopolyploids usually arise by hybridization of two species. This work examines the evolutionary role of a triploid bitter-cress hybrid (Cardamine × insueta) in the origin of trigenome hybrids (Cardamine × schulzii) and elucidates the 110-year history of interspecies hybridization in Cardamine, demonstrating that semifertile triploid hybrids may facilitate the origin of new trigenome allopolyploids.
Abstract
This article describes the use of cytogenomic and molecular approaches to explore the origin and evolution of Cardamine schulzii, a textbook example of a recent allopolyploid, in its ∼110-year history of human-induced hybridization and allopolyploidy in the Swiss Alps. Triploids are typically viewed as bridges between diploids and tetraploids but rarely as parental genomes of high-level hybrids and polyploids. The genome of the triploid semifertile hybrid Cardamine × insueta (2n = 24, RRA) was shown to combine the parental genomes of two diploid (2n = 2x = 16) species, Cardamine amara (AA) and Cardamine rivularis (RR). These parental genomes have remained structurally stable within the triploid genome over the >100 years since its origin. Furthermore, we provide compelling evidence that the alleged recent polyploid C. schulzii is not an autohexaploid derivative of C. × insueta. Instead, at least two hybridization events involving C. × insueta and the hypotetraploid Cardamine pratensis (PPPP, 2n = 4x−2 = 30) have resulted in the origin of the trigenomic hypopentaploid (2n = 5x−2 = 38, PPRRA) and hypohexaploid (2n = 6x−2 = 46, PPPPRA). These data show that the semifertile triploid hybrid can promote a merger of three different genomes and demonstrate how important it is to reexamine the routinely repeated textbook examples using modern techniques.
INTRODUCTION
Hybrid speciation on the same ploidal level (homoploid hybridization) or via hybridization and genome doubling (allopolyploidy) are relatively common evolutionary processes in land plants. It is estimated that ∼15% of angiosperm speciation events have resulted from recent polyploidy (Wood et al., 2009) and the legacy of ancient polyploidy (whole-genome duplication) is retained in all angiosperm genomes so far analyzed (Soltis et al., 2009; Jiao et al., 2011; Arrigo and Barker, 2012). Stable allopolyploidy requires a hybridization event, genome doubling, reproductive isolation, genome diploidization, and, eventually, long-term survival or extinction. Although the formation of allopolyploid species is ongoing, there are only a few documented examples of recently (approximately <150 years old) formed allopolyploid species. Such allopolyploids are Spartina anglica (Ainouche et al., 2004), Senecio cambrensis/Senecio eboracensis (Abbott and Lowe, 2004), Tragopogon mirus and Tragopogon miscellus (Ownbey, 1950; Soltis et al., 2004), Mimulus peregrinus (Vallejo-Marín, 2012), and Cardamine schulzii (Urbanska-Worytkiewicz, 1977b; Urbanska et al., 1997).
Because the parental genomes and the time of origin of recently formed allopolyploid species are usually known, they represent exciting models to elucidate the immediate consequences of hybridization and polyploidy. Indeed, recently formed allopolyploids have been analyzed from various perspectives, including fertility, mode of reproduction, gene expression, chromosome pairing, epigenetic reprogramming, chromosome repatterning, or ecological preferences (reviewed in Doyle et al., 2008; Hegarty and Hiscock, 2008; Soltis and Soltis, 2009; Abbott and Rieseberg, 2012). Dozens of studies have focused on Spartina, Senecio, and Tragopogon allopolyploids, but only three molecular studies have focused on the genome evolution of C. schulzii since its discovery in 1974 (Urbanska et al., 1997; Neuffer and Jahncke, 1997; Franzke and Mummenhoff, 1999).
The bitter-cress species C. schulzii Urbanska occurs in the alpine meadows of Urnerboden in the Swiss Alps and is reported to be a recent autoallohexaploid species (2n = 6x = 48; RRRRAA genome) formed by a genome doubling of the triploid Cardamine × insueta Urbanska shortly after the origin of C. × insueta around the turn of the 20th century (Zimmerli, 1986) or during the 1950s (Urbanska and Landolt, 1999). C. × insueta (2n = 3x = 24, RRA) is a partially fertile triploid hybrid between two diploid species (2n = 2x = 16), Cardamine amara (AA) and Cardamine rivularis (RR; for species identity, see Marhold, 1995), presumably formed at Urnerboden some 100 to 150 years ago (Urbanska-Worytkiewicz and Landolt, 1972; Urbanska et al., 1997). Although the two parental species have different ecological preferences (C. amara prefers wetter habitats than C. rivularis), C. × insueta probably arose when the two parental species came into direct contact due to the intensifying exploitation of the Alpine meadows of Urnerboden at the turn of the 20th century. The year-round inhabitation of the Urnerboden alp (since 1877), marked by forest clearing, drainage, grazing, and/or mowing, established new habitats that became colonized by the parental species and particularly by their triploid hybrid. Continuing agricultural activities ensured the long-term survival of C. × insueta and the later origin of the hexaploid C. schulzii (Urbanska-Worytkiewicz, 1977b). Since its description (Urbanska-Worytkiewicz, 1977b), C. schulzii has been reported to represent an example of a very recent neoallopolyploid species, established in a human-influenced habitat (Grant, 1981; Abbott and Lowe, 2004; Soltis et al., 2004; Soltis and Soltis, 2009; Hegarty et al., 2012). Human-related introductions are also responsible for the recent occurrence of the hypotetraploid Cardamine pratensis (2n = 4x−2 = 30, PPPP), a species that is closely related to C. rivularis, at Urnerboden (Urbanska et al., 1997).
Here, we aimed to gain insights into the origin and genome stability of the triploid C. × insueta and alleged autoallohexaploid C. schulzii using newly available molecular approaches and Arabidopsis thaliana–related genomic resources. Using a unique combination of comparative chromosome painting (CCP) and genomic in situ hybridization (GISH), we were able to identify the entire parental genomes and individual chromosomes in the allopolyploid genomes. Furthermore, the use of genome size estimates, the retrieval of species-specific repeats from next-generation sequencing data, the analysis of chloroplast DNA (cpDNA) variation, and the quantification of single nucleotide polymorphism (SNP) ratio by PyroMark enabled us to corroborate and expand the conclusions based on comparative cytogenetics. Our multidisciplinary study illuminates genome evolution in the hybrid Cardamine populations and demonstrates the evolutionary potential of triploid hybrids to promote further hybridization and polyploid speciation. Furthermore, this work demonstrates how important it is to reexamine textbook examples of alleged allopolyploid species using state-of-the-art techniques.
RESULTS
Diploid Parental Species Display Contrasting Genome Features
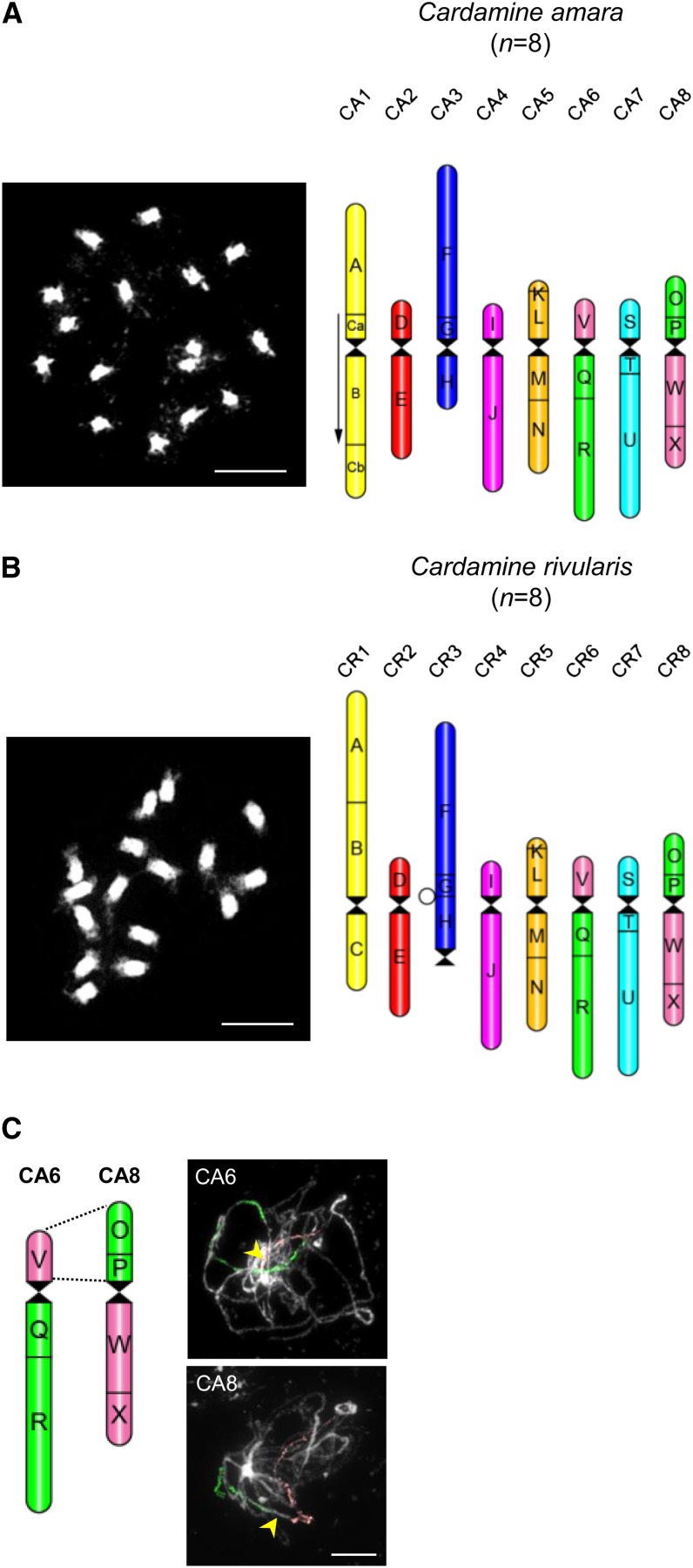
We counted chromosomes and/or estimated ploidal levels using flow cytometry in 65 and 134 individuals of C. amara and C. rivularis, respectively, across 12 subpopulations at Urnerboden (see Supplemental Figure 1 online). In C. amara, 59 plants were diploid (AA, 2n = 2x = 16), two were triploid (AAA, 2n = 3x = 24), and four were tetraploid (AAAA, 2n = 4x = 32). As only diploid plants contributed to the origin of C. × insueta (RRA), all further analyzes focused on this cytotype. All plants of C. rivularis (RR) were diploid. The parental species differ by genome size, whereby the C. rivularis genome (380 Mb) is 43% bigger than that of C. amara (217 Mb) (see Supplemental Table 1 online). This difference is reflected by smaller chromosomes (∼2 µm) and less pericentromeric heterochromatin in C. amara compared with larger chromosomes (∼4 µm) and prominent pericentromeric heterochromatin (built up by the 290-bp Carcen tandem repeat) in C. rivularis (Figures 1A and 1B; see Supplemental Figure 2A online).
Figure 1.
Reconstructed Comparative Karyotypes in C. amara and C. rivularis (2n = 2x = 16).
(A) and (B) DAPI-stained mitotic chromosomes and comparative ideograms of C. amara (A) and C. rivularis (B). The arrow indicates the 2.5-Mb inversion on chromosome CA1, and the open circle shows the ancestral position of the relocated centromere on CR3.
(C) Identification of a translocation between chromosomes CA6 and CA8 by CCP of pachytene spreads in C. amara. Arrowheads indicate centromere regions.
Bars = 5 µm
For 11 individuals of the diploid C. amara (11 subpopulations) and 16 individuals of C. rivularis (seven subpopulations) (see Supplemental Figure 1 online), a comparative cytogenetic map was constructed by CCP with 644 chromosome-specific BAC clones of Arabidopsis used as painting probes. Arabidopsis BAC contigs were arranged and fluorescently labeled on the premise that the genus Cardamine (tribe Cardamineae) is embedded within the crucifer Lineage I (Franzke et al., 2011) whose genomes most likely descended from the ancestral crucifer karyotype (ACK) with eight chromosomes (n = 8, AK1 to AK8) and 24 conserved genomic blocks (A to X) (Schranz et al., 2006). CCP analysis showed that structurally the karyotypes of both Cardamine species closely resembled the ACK (Figures 1A and 1B; see Supplemental Figures 3A and 3D online) and were stable in all 10 and 16 analyzed individuals, respectively. Genomes of the two parental species are largely collinear, including the shared whole-arm translocation between the ancestral chromosomes AK6 and AK8 (Figures 1A to 1C), but differ by two species-specific rearrangements (Figures 1A and 1B; see Supplemental Figures 3A to 3E online). In C. rivularis, the CR1 chromosome has the ancestral block association (Figure 1B; see Supplemental Figures 3D and 3E online), whereas in C. amara, chromosome CA1 bears a 13-Mb pericentric inversion with breakpoints between blocks A and B (Arabidopsis BAC clones T29M8/F6F9) and C (F9I5/F6D8) (Figure 1A; see Supplemental Figures 3A to 3C online). This inversion was identified in all three cytotypes of C. amara (see Supplemental Figure 3C online). In addition, whereas CA3 in C. amara is a submetacentric homoeolog of the ancestral chromosome AK3 (Figure 1A; see Supplemental Figure 3A online), the C. rivularis homoeolog CR3 is telocentric presumably due to centromere repositioning or to two consecutive inversions, one pericentric and the other paracentric (Figure 1B; see Supplemental Figures 3D and 3E online).
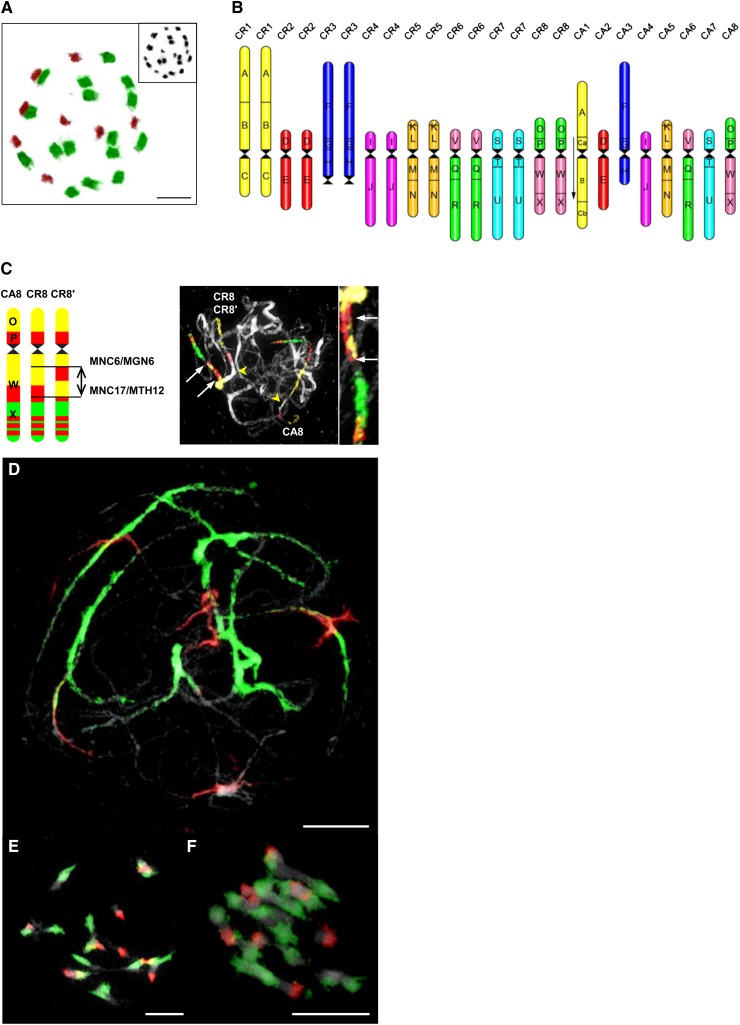
CCP/GISH Analysis of the Triploid Hybrid C. × insueta (RRA)
The highest abundance of triploid plants occurred at the locus classicus (subpopulation #3) and in neighboring meadows (subpopulations #8 and #10) (see Supplemental Figure 1 online). In seven subpopulations, 87 individuals were verified as C. × insueta (2n = 3x = 24) by chromosome counting and/or flow cytometry. Male-sterile plants with indehiscent anthers (i.e., anthers not releasing pollen; see Supplemental Figure 4A online) dominated all subpopulations. Only a single plant with dehiscent anthers (i.e., anthers releasing pollen; see Supplemental Figure 4B online) was found. In the male-fertile hybrid plants, the pollen fertility reached 22% and pollen size ranged from 26.4 to 33.5 μm (n = 4193). The estimated 2C-value genome size of C. × insueta (969.76 Mb) differed from the theoretical sum of parental C-values (i.e., RR + A, 2C of C. rivularis + 1C of C. amara) only by 1% (see Supplemental Table 1 online). In chromosome analyzes, the eight smaller (A) chromosomes of C. amara were readily discernible from the 16 bigger (RR) chromosomes of C. rivularis, and in 32 plants, the RRA genome composition was confirmed by GISH using genomic DNA (gDNA) probes of C. amara and C. rivularis (Figure 2A). The combined CCP/GISH analysis of all chromosomes in 32 plants revealed the additivity of the parental chromosome complements (Figure 2B). In 27 analyzed plants from six subpopulations, no evidence for intergenomic translocations or other major rearrangements was found. In all five analyzed plants from the subpopulation #4, a 2.4-Mb paracentric inversion on one homolog of chromosome CR8 was detected. The breakpoints occurred within the genomic block W, corresponding to Arabidopsis BAC clones MNC6/MGN6 and MNC17/MTH12 (Figure 2C).
Figure 2.
Genome Structure of the Triploid Hybrid C. × insueta (2n = 3x = 24).
(A) GISH revealing eight chromosomes of C. amara (red fluorescence) and 16 chromosomes of C. rivularis (green fluorescence). DAPI-stained chromosome spread shown in the inset.
(B) Comparative ideogram of C. × insueta based on comparison with the eight ancestral chromosomes and 24 genomic blocks (A to X) of the ACK (Schranz et al. 2006) and the reconstructed karyotypes of C. amara (chromosomes CA1 to CA8) and C. rivularis (CR1 to CR8).
(C) The 2.4-Mb paracentric inversion on the CR8' homolog (blocks O, P, W, and X) in plants from subpopulation #4. Arrows point to the inverted region, and arrowheads indicate centromeres.
(D) to (F) GISH revealing chromosomes of C. amara (red) and C. rivularis (green) at pachytene (D), diakinesis (E), and metaphase I (F). Bars = 5 µm
The semifertile triploid C. × insueta has been reported to breed by producing RA and R gametes, a process called polarized meiosis (Urbanska-Worytkiewicz, 1977a, 1978). Using GISH, we investigated male meiosis in five plants with indehiscent anthers and in the single plant having dehiscent anthers. At pachytene, RR bivalents and A univalents were observed in 62.5% of cells, RRA trivalents were less frequent (25%), and RA bivalents and R univalents were observed only in 12.5% of cells (Figures 2D to 2F). At anaphase I, the irregular segregation of varying numbers of A and R chromosomes distributed to daughter cells was most frequently observed (92.3%). In total, 39 different combinations of segregating R and A chromosomes were observed. However, neither polarized genome segregation (8R8A:8A) nor unreduced cells (RRA) were observed in any of the analyzed plants. The irregular meiotic segregation is reflected by the presence of anaphase bridges between A and R chromosomes (14%; see Supplemental Figure 5C online) and one to three A-genome laggards in anaphase I (57%; see Supplemental Figure 5A online). In metaphase and anaphase I, small chromosome-like segments labeled by gDNA of C. amara were often located close to the forming phragmoplast between the two daughter cells (see Supplemental Figure 5B online). We assume that these are chromosome fragments or sister chromatids of the A genome univalents simultaneously pulled to opposite poles of the spindle. Consequently in anaphase I and II, micronuclei labeled by gDNA of C. amara (see Supplemental Figure 5D online) are three times more frequent than R genome micronuclei (75.6% versus 24.4%).
The Genome of C. schulzii
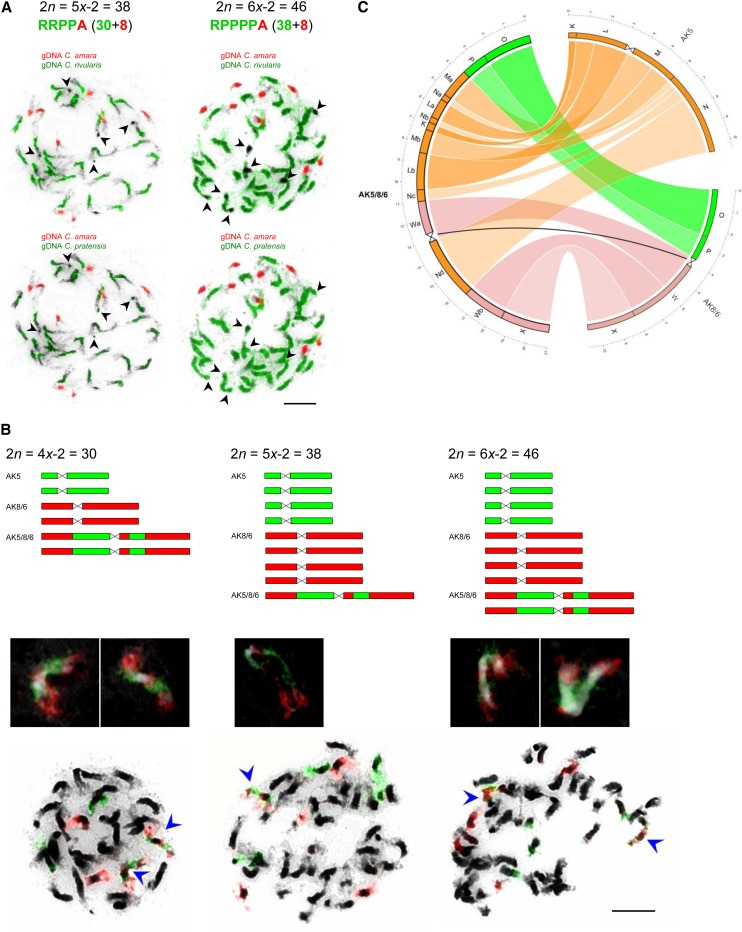
The presumably hexaploid species C. schulzii with the RRRRAA genome composition and 48 chromosomes was first described as an autopolyploid derivative of the triploid hybrid C. × insueta (Urbanska-Worytkiewicz, 1977b). However, despite thorough and intensive investigations, hexaploid RRRRAA plants have not been found at Urnerboden. Instead, 27 individuals with 38 (six plants) or 46 (21 plants) chromosomes fitting the original description of C. schulzii were identified at its locus classicus (subpopulations #3) and in adjacent areas (subpopulations #8 and #10) (see Supplemental Figure 1 online). GISH analysis of the hypopentaploid (2n = 5x−2 = 38) and hypohexaploid (2n = 6x−2 = 46) plants showed that in both genomes eight smaller chromosomes were labeled by gDNA of C. amara, whereas 30 or 38 chromosomes were positively identified using gDNA of C. rivularis (Figure 3A). Among the 30 or 38 chromosomes, six chromosomes bore terminal heterochromatic knobs not labeled by gDNA of C. rivularis (Figure 3A). To investigate the origin of these knob-bearing chromosomes by GISH, we used gDNA of the hypotetraploid C. pratensis (PPPP, 2n = 4x−2 = 30), a species closely related to C. rivularis and also occurring at Urnerboden (see Supplemental Figure 1 online). gDNA of C. pratensis labeled all 30 and 38 chromosomes in hypopentaploid and hypohexaploid hybrids, respectively, including all heterochromatic knobs (Figure 3A). These results indicated that the knob-bearing chromosomes most likely originate from C. pratensis. This was further confirmed through a 177-bp satellite repeat (Prasat) retrieved from 454 sequence reads of C. pratensis (∼0.12% of genome) and localized to four or five terminal knobs in this species (see Supplemental Figure 6A online). In both hypopentaploid and hypohexaploid accessions, the repeat is localized to four to six terminal heterochromatic knobs (see Supplemental Figures 6B and 6C online), whereas it is absent in C. rivularis, C. amara, and C. × insueta (data not shown). Although the interpretation of the CCP/GISH patterns in hypopentaploid/hexaploid plants was hampered by multivalents formed by homologous and homoeologous chromosomes at pachytene, and by the inability of GISH to discriminate chromosomes of C. rivularis and C. pratensis, we were able to infer the purported genome compositions through CCP/GISH with probes for chromosomes CA1/CR1 and CA3/CR3. As the two chromosomes structurally differ between C. amara and C. rivularis/C. pratensis (Figure 1; see Supplemental Figures 3A to 3E online), one copy of CA1 and CA3 versus four or five copies of the two chromosomes can be recognized in the hypopentaploid and hypohexaploid genomes by CCP/GISH. Meiotic chromosome pairing was analyzed in five 2n = 46 individuals from subpopulation #3. At pachytene, various chromosome exclusions resulting in micronuclei of different sizes dominated (see Supplemental Figure 7A online), and chromosome segregation at later meiotic stages can be interpreted only with difficulty. The meiotic irregularities were mirrored by the production of pollen grains of different sizes and low fertility (4.0 to 6.6%) (see Supplemental Figure 7B online). Interestingly, more than 90% of pollen (n = 42 only) was stained as fertile in a single hypopentaploid plant analyzed.
Figure 3.
Genome Composition of C. × schulzii.
(A) GISH in the hypopentaploid (2n = 5x−2 = 38) and hypohexaploid (2n = 6x−2 = 46) accessions. Mitotic chromosome spreads were labeled by gDNA of C. amara (red) and C. rivularis (green) and reprobed by gDNA of C. amara (red) and C. pratensis (green). Arrowheads indicate terminal heterochromatic knobs.
(B) Frequency of AK5 and AK8/6 homoeologs and the fusion AK5/8/6 chromosome in genomes of C. pratensis and C. × schulzii. The same chromosomes identified by comparative painting with BAC contig probes for AK5 (green) and AK8/6 (red) homoeologs. Insets and arrowheads show the AK5/8/6 fusion chromosome. Bar = 5 µm.
(C) Circos plot showing the collinearity between AK5 and AK8/6 homoeologs in C. amara and C. rivularis and the fusion chromosome AK5/8/6 in the hypotetraploid C. pratensis and in C. × schulzii. Capital letters refer to genomic blocks of the ACK (Schranz et al., 2006), and broken blocks are indexed by small letters.
As these results suggest a hybrid origin of C. schulzii, we refer to these plants as C. × schulzii thereafter.
Descending Dysploidy in C. pratensis
To further elucidate the role of the hypotetraploid C. pratensis in the formation of the two C. × schulzii hybrids, we first tested whether or not the 30 chromosomes of C. pratensis resulted from descending dysploidy (Lawrence, 1931) or otherwise. In C. pratensis, CCP with painting probes corresponding to AK5 and AK8/6 homoeologs in C. amara and C. rivularis, labeled two AK5 and AK8/6 homologs, as well as a chromosome pair with a reduced pericentromere and longer chromosome arms (Figure 3B). The detailed analysis of the long chromosome pair revealed that AK5 and AK8/6 homologs had participated in a so-called nested chromosome fusion (NCF), a translocation event involving the “insertion” of chromosome AK5 into the pericentromere of AK8/6. This rearrangement involves three breakpoints, one in the pericentromeric region of the recipient chromosome (AK8/6) and two in the subtelomeric regions of the insertion chromosome (AK5). This occurred simultaneously with the NCF event or shortly after the AK5 centromere was inactivated or deleted. The NCF was followed by a pericentric inversion with breakpoints within genomic blocks N and W, and two subsequent paracentric inversions within blocks originating from AK5 (Figure 3C; see Supplemental Figure 6 and Supplemental Movie 1 online). The origin of the fusion chromosome AK5/8/6 was responsible for the reduction of chromosome number from 2n = 4x = 32 to 2n = 4x−2 = 30.
Homologs of chromosomes AK5/8/6, AK5, and AK8/6 paired as bivalents in 71% of analyzed diakineses in pollen mother cells (PMCs). Quadrivalents of AK5/8/6 and AK8/6 chromosomes were formed in 29% of analyzed cells, while pairing of AK5/8/6 and AK5 was not observed.
The Fusion Chromosome AK5/8/6 Is Present in Both C. × schulzii Hybrids
The fusion AK5/8/6 chromosome found in the complement of hypotetraploid C. pratensis was also identified in both hybrids referred to as C. × schulzii (see above). There was only one AK5/8/6 homolog in the hypopentaploid hybrid, whereas two homologs were present in plants with 2n = 46 (Figure 3B). These data showed that in addition to C. amara and C. rivularis, the hypotetraploid C. pratensis is also one of the parental species of both C. × schulzii hybrids. The origin of the hybrid genomes have been inferred from the number of AK5/8/6 homologs. One homolog in the hypopentaploid genome suggests that a reduced gamete of C. pratensis (n = 15) contributed to the origin of the hypopentaploid hybrid with the RRPPA genome. By contrast, the presence of two AK5/8/6 homologs in the hypohexaploid hybrid with the RPPPPA genome implies that an unreduced gamete of C. pratensis (n = 30) was involved in its origin. Genome size data confirm the purported genome composition of both hypopentaploid and hypohexaploid hybrids, as the estimated and theoretical C-values differ by only by 1.02 and 1.03%, respectively (see Supplemental Table 1 online).
Crambo Tandem Repeat Elucidates the Genome Composition of Herbarium Specimens of C. × schulzii
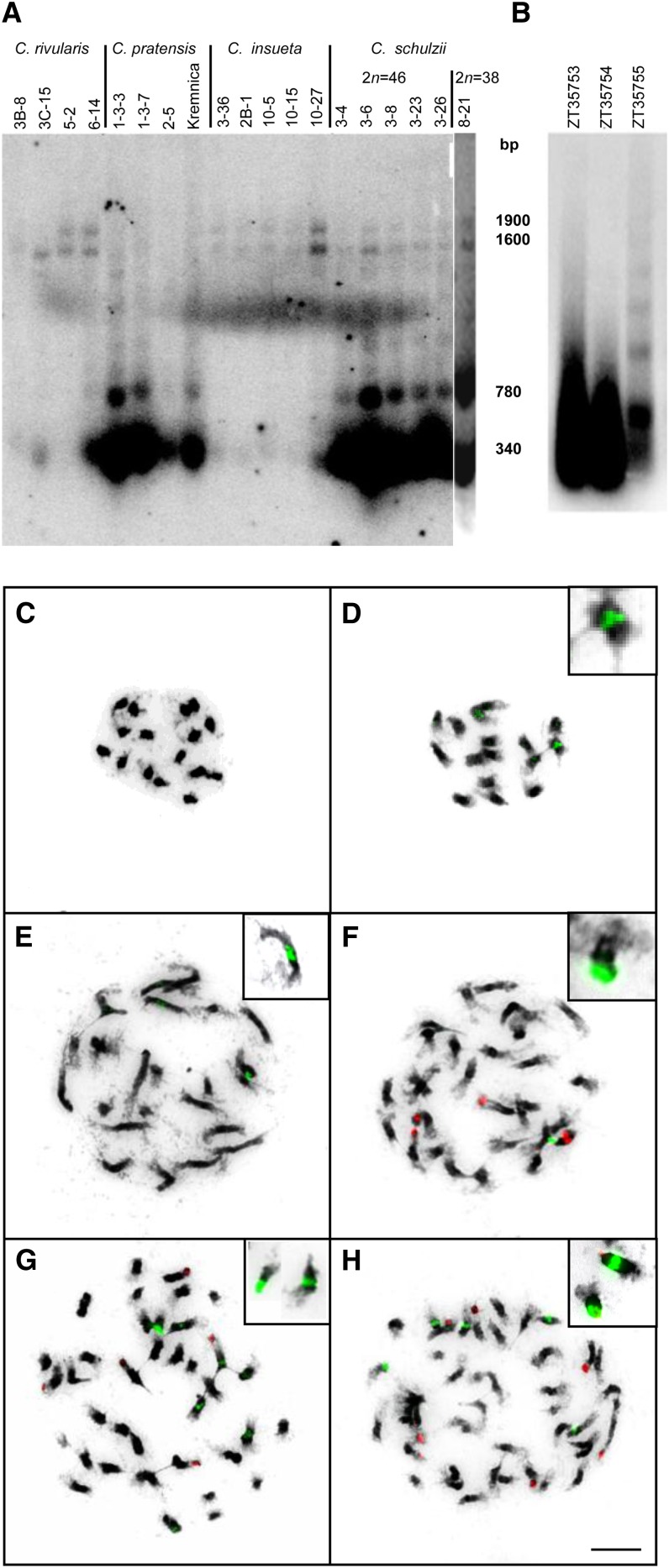
Restriction enzyme analysis in C. pratensis identified a new tandem repeat, Crambo, which was also shown to be present in the C. rivularis genome but absent in C. amara (Figure 4). Additional low-pass 454 sequencing of all three parental genomes (C. amara, C. rivularis, and C. pratensis) confirmed that the repeat is abundant in C. pratensis (0.158% of genome), underrepresented in C. rivularis (0.036%), and nearly absent in C. amara (<0.001%). In C. pratensis, the Crambo repeat is present in high copy number in a highly homogenous 340- and 780-bp MboI fraction, whereas in C. rivularis, low copy numbers of Crambo are represented by a high molecular weight 1600- and 1900-bp MboI fraction (Figure 4A). As expected, C. × insueta inherited the 1600- and 1900-bp fraction from C. rivularis, while there was no or weak signal in the 340- and 780-bp fractions. In both C. × schulzii hybrids, the 340- and 780-bp as well as 1600- and 1900-bp fraction were detected (Figure 4A). To investigate whether the extant C. × schulzii plants are identical to those originally described as C. schulzii collected at the locus classicus more than 35 years ago (Urbanska-Worytkiewicz, 1977b), selected herbarium specimens were analyzed for the Crambo repeat. The low molecular weight fractions typical for C. pratensis were amplified in all three herbarium vouchers to the similar extent as in 2n = 38 and 46 hybrids collected by us (Figure 4B). This is strong evidence that plants described as C. schulzii (Urbanska-Worytkiewicz, 1977b) are identical to the C. × schulzii plants analyzed by us. Chromosome in situ localization of Crambo differs among the analyzed taxa (Figures 4C to 4H). Whereas one large and one minor interstitially positioned loci are characteristic for C. rivularis and C. × insueta (Figures 4D and 4E), a single large terminal locus was observed in C. pratensis (Figure 4F). Interstitial and terminal Crambo loci in both C. × schulzii hybrids testify to the parentage of both C. pratensis and C. × insueta (Figures 4G and 4H). No Crambo-related fluorescence in situ hybridization (FISH) signals were detected in C. amara (Figure 4C).
Figure 4.
Genome-Specific Variation of Satellite Repeats in Cardamine Accessions.
(A) DNA gel blot hybridization showing structural polymorphisms of the Crambo repeats in Cardamine accessions collected at Urnerboden in the course of this study (Kremnica refers to C. pratensis from Slovakia, 2n = 44).
(B) DNA gel blot hybridization shows amplification of Crambo repeats in herbarium specimens collected as C. schulzii at the time of its discovery. The absence of signal in the high molecular fraction is due to significant degradation of DNA.
(C) to (H) Chromosome localization of Crambo (green fluorescence) and Prasat (red fluorescence) satellites in Cardamine taxa from Urnerboden.
(C) C. amara, no hybridization signals.
(D) C. rivularis, two interstitial loci of Crambo.
(E) C. × insueta, two interstitial loci of Crambo.
(F) to (H) Interstitial and/or terminal loci of Crambo (green) and Prasat (red) hybridizing to heterochromatic knobs in the hypotetraploid C. pratensis (F), hypopentaploid C. × schulzii (G), and hypohexaploid C. × schulzii (H). Insets display examples of Crambo-bearing chromosomes. Bar = 5 µm.
Quantitative SNP Detection by PyroMark Confirms the Composition of Hybrid Genomes
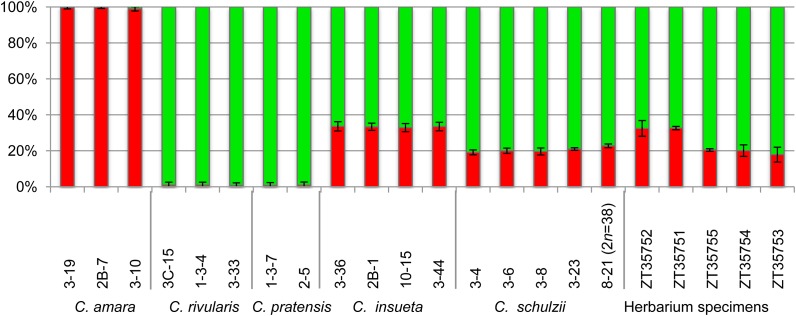
To further verify whether C. × schulzii had only one (A) genome derived from C. amara, and whether the extant C. × schulzii hybrids have the same genome composition as the herbarium specimens determined as C. schulzii, we used pyrosequencing to distinguish SNPs divergent between C. amara and C. rivularis/pratensis (Figure 5). PyroMark 96 ID provides information on the relative abundance of SNPs by comparing the intensity of light emission generated by the incorporation of allele- or SNP-specific bases during the sequencing reaction by polymerase. We measured the SNP ratio at four SNP positions in two genes (see Methods for details) to assess the genome contribution of each gene. The SNPs ratio shared between C. rivularis and C. pratensis, but absent in C. amara, was analyzed in 22 individuals of C. amara, C. rivularis, C. × insueta, C. pratensis, and C. × schulzii, as well as in five herbarium specimens determined as C. × insueta (ZT35751 and ZT35752) or C. schulzii (ZT35753, ZT35754, and ZT35755). The C. amara–type SNP ratio (A-percentage) was always around 100% in C. amara, and the C. rivularis/pratensis–type SNP ratio was close to 100% in C. rivularis and C. pratensis. The A-percentage in C. × insueta plants collected by us was around 33%. This value is in accordance with the expected value (33.3%) based on the verified genome ratio (RRA). The A-percentage of the hypohexaploid hybrids (RPPPPA) varied between 19 and 21%. The hypopentaploid accession (8-21; RRPPA) showed slightly higher A-percentage than the hypohexaploid ones (22.8%), reflecting the 1 (A):4 (R/P) genome composition (Figure 3A). Next, we examined five herbarium specimens collected and identified as C. × insueta and C. schulzii in the 1970s and 1980s. Two C. × insueta specimens (ZT35751 and ZT35752) showed comparable A-percentages to the extant samples of C. × insueta. Three herbarium specimens of C. schulzii (ZT35753, ZT35754, and ZT35755) showed a similar A-ratio as observed for the extant plants of C. × schulzii. These results suggest that the analyzed herbarium specimens of C. schulzii had only a single A genome, and their genome composition was similar to the extant hypohexaploid plants of C. × schulzii.
Figure 5.
Ratio of Gene Copy Numbers in Cardamine Accessions from Urnerboden Detected by Pyrosequencing (PyroMark).
The red and green bars show the average ratio of four SNP sites between C. amara and C. rivularis/C. pratensis, respectively; error bars indicate the standard deviations based on the C. amara percentage. The last five columns show the analyzed herbarium specimens of C. × insueta (ZT35752 and ZT35751) and C. × schulzii (ZT35755, ZT35754, and ZT35753).
cpDNA Haplotype Variation Elucidates Maternal Parentage of Hybrid Genomes
The near complete plastome genomes of progenitor species were assembled from 454 reads. The lengths of consensus plastomes were 154,367 bp (C. amara), 154,378 bp (C. rivularis), and 154,629 bp (C. pratensis). Pairwise comparisons showed the highest sequence identity for the species pair C. pratensis and C. rivularis (99.51%), followed by C. rivularis and C. amara (99.06%), and with the lowest value obtained for C. pratensis and C. amara (98.88%). The Cardamine plastomes differed from that of Arabidopsis in ∼10% of nucleotides. To clarify the direction of the interspecific crosses resulting in the origin of C. × insueta and C. × schulzii hybrids, cpDNA variation (Sanger sequencing of the RNA polymerase beta subunit [rpoB]-tRNA-Cys [trnCGCA] intergenic spacer) was screened in 109 individuals, including all taxa from Urnerboden and the parental species also from additional populations. Both the maximum likelihood tree and parsimony network showed a clear distinction between the haplotypes retrieved from C. amara (C. amara clade; haplotypes A1 to A5 in Figure 6) and those from C. rivularis and C. pratensis (C. rivularis/C. pratensis clade; haplotypes R1-R3 and P1-P6, respectively), as well as the close relatedness of the latter two species (Figure 6; see Supplemental Data Set 1 online). We determined intraindividual haplotypic variation in the rpoB-trnCGCA subregion using mapped 454 reads (see Supplemental Data Set 2 online). The abundant (>10% reads) SNPs were not recovered from any of the sequenced genomes, and most of the low abundant SNPs could be attributed to sequencing errors. We conclude that the intraindividual haplotypic variation was low in that region. However, variation was found among the individuals and/or populations of the progenitor species, but not among the hybrid individuals. All C. rivularis plants from Urnerboden (22 individuals) bore a single, specific haplotype (R1), which was present in all analyzed accessions of C. × insueta (I1, 19 individuals) and C. × schulzii (S1, five hypopentaploid and 14 hypohexaploid individuals). This provides conclusive evidence that C. rivularis from the local Urnerboden population was the maternal progenitor of all C. × insueta plants and that R subgenomes within the RRPPA and RPPPPA hybrid genomes of C. × schulzii were maternally inherited (see also the SNP at the alignment position 76 in Supplemental Data Set 2 online).
Figure 6.
Phylogenetic Analyses of cpDNA Sequence Data (rpoB-trnC Intergenic Spacer) of Cardamine Accessions.
(A) Maximum likelihood phylogenetic tree, and (B) maximum parsimony network Terminal labels (A) include the accession code and species abbreviation: am, C. amara (green; haplotypes denoted as A1 to A5); riv, C. rivularis (red; haplotypes R1 to R3); prat, C. pratensis (blue; haplotypes P1 to P6); ins, C. × insueta (brown; haplotype I1); sch5x and sch6x, hypopentaploid and hypohexaploid C. × schulzii (brown; haplotype S1). The capital letter U indicates accessions from the Urnerboden site. Values above branches show bootstrap support. Interrupted branches were shortened by half. In the network (B), the circles represent haplotypes (the circle sizes are proportional to haplotype frequencies), the lines indicate mutational steps, and the black dots are unsampled haplotypes.
DISCUSSION
Ancestral and Genome-Specific Chromosome Rearrangements in the Diploid Cardamine Species
Comparative cytogenetic maps of the two diploid species, C. amara and C. rivularis, have shed light on genome evolution in Cardamine (∼200 species) when compared with the ACK, and these new insights will contribute to understanding the genomic processes and evolution across the monophyletic tribe Cardamineae as a whole (∼337 species). As the tribe comprises some important vegetable (water-cress and horseradish), weedy, and model species (e.g., C. hirsuta; Canales et al., 2010), the established karyotypes provide a useful genomic resource for this crucifer group. As the two Cardamine genomes closely resemble the eight chromosomes of the ACK (Schranz et al., 2006), we conclude that the progenitor genome of Cardamineae has descended from the ACK. The whole-arm translocation between ancestral chromosomes AK6 and AK8 shared by both species most likely predates the generic and species diversification in Cardamine/Cardamineae (T. Mandáková and M.A. Lysak, unpublished data). Nevertheless, although the genomes of C. amara and C. rivularis are largely collinear, two homoeologous chromosomes differ in structure. In particular, the origin of the telocentric chromosome CR3 in C. rivularis (homoeolog of AK3 in ACK and of CA3 in C. amara) is unique in the context of genome evolution in Brassicaceae. Whereas this chromosome has retained the ancestral organization of genomic blocks, its centromere has relocated to one of the chromosome ends. The only other example where centromere repositioning in crucifers is inferred is in the telocentric homoeolog of AK8 in Neslia paniculata (Lysak et al., 2006). However, in both instances, a double inversion restoring the ancestral collinearity cannot be ruled out as an alternative mechanism (Lysak et al., 2006), and more data are needed to resolve the issue.
The Triploid Genome of C. × insueta: Its Origin, Stability, and Long-Term Survival
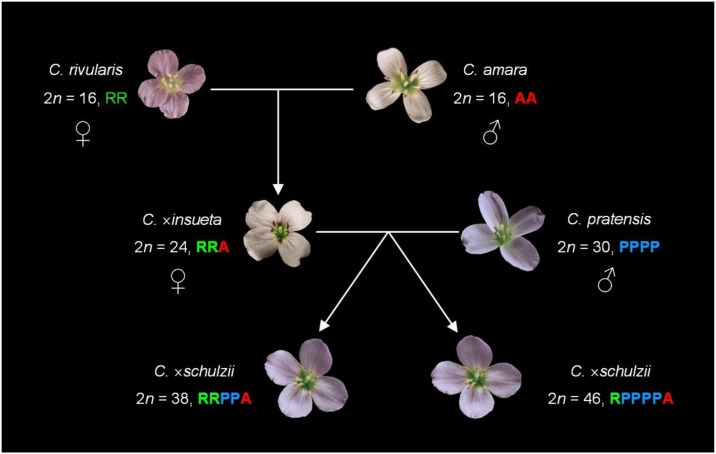
Combined CCP/GISH analysis of C. × insueta confirmed its triploid RRA genome constitution (Urbanska-Worytkiewicz and Landolt, 1972), and cpDNA analysis corroborated C. rivularis as the maternal parent (Figure 7), as proposed earlier by Urbanska et al. (1997). Although the presence of triploid and tetraploid plants of C. amara at Urnerboden prove that unreduced gametes can be produced, in accordance with Urbanska et al. (1997), triploid hybrids with the AAR genome have not been found. Our study has demonstrated the stability of the hybrid genome throughout the Urnerboden plateau since its probable origin more than 110 years ago. Despite the potential for meiotic recombination between paternal (A) and maternal (R) homoeologs given the largely conserved collinearity, CCP/GISH failed to detect any large-scale translocation or insertion/deletion events between the two parental genomes. Only five plants of subpopulation #4 were heterozygous for a 2.4-Mb paracentric inversion within the rivularis subgenome. Phenotypes, chromosome pairing, anther indehiscence, and pollen fertility of subpopulation #4 plants were comparable with other C. × insueta populations. The overall genome stability of the semifertile hybrid is to a large extent determined by the prevailing vegetative propagation by stolones, fragments, and plantlets in leaf axils (Urbanska-Worytkiewicz, 1977a, 1980; our observation). Sexual reproduction is of limited importance as a consequence of the odd-numbered genome composition and prevalent anther indehiscence (Urbanska-Worytkiewicz, 1977a). The triploid hybrid’s adaptation to a wider spectrum of ecological niches along with vegetative propagation has likely contributed to its successful colonization of human-influenced and managed habitats (Urbanska-Worytkiewicz, 1980) as well as its long-term survival. The triploid C. × insueta coexisting with its parental species since the time of its formation mirrors the coexistence of the hexaploid hybrid Spartina × townsendii (2n = 60) and the derived allododecaploid S. anglica (2n = 120, 122, 124) at the site of their formation at the end of the 19th century (Renny-Byfield et al., 2010).
Figure 7.
Inferred Parentage of C. × insueta and C. × schulzii.
C. schulzii Is a Trigenomic Hybrid
The neohexaploid C. schulzii with 48 chromosomes was first reported from Urnerboden in 1974 (Urbanska-Worytkiewicz and Landolt, 1974) and formally described as an autoallohexaploid species (RRRRAA) derived from the triploid hybrid C. ×insueta (RRA) 3 years later (Urbanska-Worytkiewicz, 1977b). Despite our intensive screening of Urnerboden populations, hexaploid RRRRAA plants have not been found. Instead, all plants morphologically fitting the species description and occurring at the locus classicus (Urbanska-Worytkiewicz, 1977b) were hypohexaploid (2n = 6x−2 = 46) or hypopentaploid (2n = 5x−2 = 38), with hypohexaploids being 3.5 times more frequent. Our multidisciplinary approach has unequivocally proved that these plants represent trigenome hybrids between the triploid C. × insueta and hypotetraploid C. pratensis (2n = 4x−2 = 30, PPPP). In all hypopentaploid (RRPPA) and hypohexaploid (RPPPPA) plants, eight chromosomes originate from C. amara, whereas the 30 or 38 remaining chromosomes come from C. rivularis/C. pratensis. The differential amplification and chromosomal localization of the Crambo tandem repeat in C. rivularis and C. pratensis, and the C. pratensis–specific satellite repeat Prasat allowed us to conclude that both species participated at the origin of C. × schulzii. Furthermore, the identification of only one fusion chromosome (AK5/8/6), typical for the hypotetraploid C. pratensis, in hypopentaploids suggested that two (PP) genomes of C. pratensis were involved in the origin of this hybrid genome. Consequently, two fusion chromosomes in the hypohexaploid complement reflect the participation of four C. pratensis genomes (PPPP). As both hybrids share the cpDNA haplotype with C. rivularis and C. × insueta, containing only R genomes, the most likely scenario of the origin of the hypopentaploid hybrids (RRPPA) is via the fertilization of an unreduced egg cell of C. × insueta (RRA) with a reduced pollen of C. pratensis (PP) (Figure 7). As the hypopentaploid hybrid has only 38 chromosomes (instead of the expected 2n = 39), either an aneuploid unreduced egg cell (n = 23) of C. × insueta or an aneuploid pollen (n = 14) of C. pratensis is inferred as contributing to its origin. In hypohexaploids (RPPPPA), an egg cell of C. × insueta with the RA genome was presumably pollinated by unreduced pollen of C. pratensis (PPPP) (Figure 7). Nevertheless, are these proposed scenarios also true for plants originally described as the hexaploid C. schulzii? We have shown that the more than 35-year-old herbarium specimens of C. schulzii contain the low molecular weight fraction of the Crambo tandem repeat typical for C. pratensis. Moreover, pyrosequencing performed on the same herbarium specimens showed genome ratios comparable with those obtained for the present-day plants of C. × schulzii from the same locality. These results demonstrate how historical DNA sequences retrieved from herbarium specimens can be useful. For example, they have been used to monitor the genetic diversity of endangered species over time (Cozzolino et al., 2007) or to compare the dynamics of rRNA genes between extant and 80-year-old populations of Tragopogon allopolyploids (Kovarik et al., 2005). Recently, next-generation sequencing has been employed to sequence the entire nuclear genome of a 43-year-old herbarium specimen of Arabidopsis (Staats et al., 2013). This opens new possibilities for a wider utilization of herbarium specimens in genetic and phylogenetic studies.
The production of unreduced RRA gametes in C. × insueta was observed to be rather frequent (Urbanska et al., 1997) and has been proven experimentally (Urbanska-Worytkiewicz, 1977a). It has previously been shown in Brassica that unreduced gamete production is much higher in interspecies hybrids than in the parental genotypes, especially at cold temperatures (Mason et al., 2011). Our data also indirectly confirm the proposed polarized chromosome assortment (8R8A:8A) in C. × insueta, at least in the maternal triploid plant of the hypohexaploid C. × schulzii (the RA subgenome of the RPPPPA genome; Figure 7). Although polarized chromosome segregation in PMCs of the single C. × insueta plant with dehiscent anthers was not observed, the process was proven by cytological analyses and through experimental crossing experiments (Urbanska-Worytkiewicz, 1977a, 1978). An in-depth analysis is required to test whether the production of RA and R gametes in triploids is indeed the result of polarized meiosis or whether such gametes are produced due to the random assortment of meiotic chromosomes in embryo-sac mother cells and PMCs in C. × insueta.
The available literature records and herbarium collections do not document the occurrence of C. pratensis at the Urnerboden alp until 1994 (Urbanska et al., 1997). Nevertheless, our results demonstrate that the hybridization between C. × insueta and the hypotetraploid C. pratensis occurred prior to 1974 (Urbanska-Worytkiewicz and Landolt, 1974; this article). The legacy of C. pratensis in the hybrid genomes of C. × schulzii argues for a much earlier introduction of C. pratensis to Urnerboden from lower altitudes, most likely through hay transport and/or agricultural machinery (Urbanska et al., 1997; Neuffer et al., 2009). The available data imply recurrent invasions of C. pratensis to Urnerboden, followed by the periodical disappearance of the species, up to its present occurrence. As shown for Senecio, Spartina, or Tragopogon, hybridization and allopolyploidy often occur between invading and native species (Abbott and Lowe, 2004; Ainouche et al., 2004), and both processes are closely interlinked with human activities (Abbott and Rieseberg, 2012).
To sum up, we could not confirm the proposed origin of C. schulzii as an autopolyploid derivative of the triploid C. × insueta, as originally proposed. Instead, the plants described as a neohexaploid species represent largely sterile trigenomic hybrids between the triploid C. × insueta and hypotetraploid C. pratensis. We argue that the 46 small chromosomes were originally counted as 48 due to the limited power of early genome analysis techniques, and the bona fide hexaploid chromosome number was interpreted as the duplicated 24-chromosome complement of C. × insueta (Urbanska-Worytkiewicz, 1977b). Apparently, the hybrid nature of C. × schulzii was also unintentionally revealed by Neuffer and Jahncke (1997) when these authors found 28 nonparental random amplified polymorphic DNA marker bands in the bona fide C. schulzii plants, and from these bands only four were detected in C. × insueta.
Descending Dysploidy in C. pratensis
C. pratensis exhibits extensive variation in chromosome number from 2n = 16 up to 2n = 56 (Kučera et al., 2005). The nature of this variation remains obscure, although polyploidy and hybridization are suggested to be the underlying mechanisms (Lövkvist, 1956, Lihová and Marhold, 2006). Lawrence (1931) was the first to suggest that the hypotetraploid complement (2n = 30) resulted from a fusion between two nonhomologous chromosomes within the eutetraploid chromosome set. Here, we find evidence to support this hypothesis and we elucidate the origin of the fusion chromosome in the hypotetraploid. As argued by Lawrence (1931), it remains to be clarified whether the chromosome fusion occurred in one of the diploid (2n = 16) progenitors of the tetraploid C. pratensis or within the tetraploid genome itself. The origin of fusion chromosomes via NCF events (i.e., through the breakage in a centromere region of one chromosome and the insertion of an entire other chromosome) has been described as a prominent mechanism of karyotype evolution in several grass species (Luo et al., 2009; International Brachypodium Initiative, 2010; Murat et al., 2010). Although principally similar, the NCF detected in C. pratensis differs from NCFs described in grasses. Whereas the centromere of the insertion chromosome is maintained and that of the recipient chromosome is inactivated in grasses, in C. pratensis, the centromere of the insertion chromosome is lost and the centromere of the recipient chromosome remains functional. In Brassicaceae, NCFs have been previously documented as mediating chromosome number decreases in the ancestral genomes of Pachycladon (Mandáková et al., 2010a) and Hornungia (Lysak et al., 2006) but not as a mechanism responsible for intraspecific chromosome number variation. Our data explain the extensive intraspecific karyological variation in the C. pratensis species complex (Lövkvist, 1956; Kučera et al., 2005) as the combination of intraspecific descending dysploidy and hybridization resulting in series of dysploid chromosome numbers deviating from the multiples of the ancestral base number (x = 8). Chromosome number variation observed in C. pratensis further illustrates a caveat of using somatic (2n) or base (x) chromosome numbers to interpret genome evolution. The recently established techniques, such as massive parallel sequencing and CCP, as well as detailed FISH/GISH analyses have highlighted several cases where the examination of the allopolyploid genomes revealed their complex origin and extensive intraspecific variability (Kantama et al., 2007; Xiong et al., 2011; Chester et al., 2012). Chromosome numbers increase due to repetitive cycles of whole-genome duplication (i.e., auto- and allopolyploidy) and decrease through fusion-like chromosome translocations during the genome diploidization process (Lysak et al., 2006; Arrigo and Barker, 2012; Mandáková et al., 2010a, 2010b). Multispecies hybridization, exemplified here by Cardamine, and aneuploidy (Comai, 2005; Henry et al., 2005; Considine et al., 2012) may further modify chromosome numbers. Hence, simple algebraic inferences of gametic and base chromosome numbers without reflecting species’ genome histories often can be misleading (Cusimano et al., 2012).
C. × insueta as a Mediator of Three-Species Genome Merger
Although triploids are generally sterile, some can be semifertile and produce aneuploid and euploid gametes (Ramsey and Schemske, 1998; Burton and Husband, 2001; Husband, 2004; Comai, 2005; Henry et al., 2005; Mason et al., 2011). Such triploids producing reduced and unreduced gametes and hybridizing with either diploids or triploids play the role of a bridge in the two-step formation of tetraploids or hexaploids (Ramsey and Schemske, 1998; Comai, 2005). Although triploid C. × insueta plants with dehiscent anthers occur only with a frequency of ∼1 to 1.5% (Urbanska-Worytkiewicz, 1977a; this study), their pollen fertility may exceed 20%. Three euploid types of pollen are believed to be produced by these plants: R, RA, and RRA. Comparable gametes were inferred to occur during macrosporogenesis (Urbanska-Worytkiewicz, 1977a). The semifertile C. × insueta, producing polarized, triploid, and aneuploid gametes, is acting as a triploid bridge, however, not by mating with its diploid parents or other triploids (Husband, 2004) but by mediating a merger of three different genomes through a cross with another species. The hybrid vigor, together with vegetative and sexual reproduction, ensure the long-term stability of triploid populations at Urnerboden and are an ideal prerequisite for ongoing recurrent hybridization between C. × insueta with its parental and nonparental species to form new hybrids and allopolyploids.
METHODS
Study Site and Plant Collections
The village of Urnerboden (canton Uri, Switzerland) is situated within Switzerland’s largest cattle alp of the same name at ∼1350 m above sea level. The high-mountain plateau is ∼6 km long and ∼0.5 km wide; high-mountain temperatures and snow cover (∼6 months a year) are important ecological factors. The year-round inhabitation and subsequent intensive grazing, mowing, and drainage dates back to 1877. Cardamine populations at Urnerboden were visited during three successive vegetation seasons. Leaf material from individual plants for flow-cytometric ploidy estimation and molecular analyses were collected in silica gel, and flower buds were fixed in freshly prepared 3:1 (ethanol:acetic acid) fixative and stored in 70% ethanol at −20°C. Some plants were transferred and grown in greenhouses of Central European Institute of Technology, Masaryk University. Herbarium vouchers of representative individuals of the studied taxa collected in the field or in the experimental greenhouse were deposited in the Herbarium of the Institute of Botany, Slovak Academy of Sciences in Bratislava (SAV). Herbarium vouchers of Cardamine × insueta and Cardamine × schulzii were obtained on loan from the Herbarium of the University of Zurich (Z): Cardamine amara (ZT 35743, collected in 1977; ZT 35744, collected in 1983), Cardamine rivularis (ZT 35745, collected in 1977), C. × insueta (ZT 35751, collected in 1981; ZT 35752, collected in 1977), C. × schulzii (ZT 35753, collected in 1977; ZT 35754, collected in 1974; ZT 35755, collected in 1974).
Flow Cytometry and Chromosome Counting
All silica gel–dried samples analyzed in this study were checked for ploidal level based on the fluorescence intensity of nuclei stained using 4′,6-diamidino-2-phenylindole (DAPI) estimated by a CyFlow ML flow cytometer (Partec), following the protocol described by Suda and Trávníček (2006). Genome size and guanine-cytosine (GC) content were estimated at the Department of Botany and Zoology, Masaryk University. The estimates were performed on a CyFlow ML flow cytometer. Propidium iodide was used for genome size estimates and DAPI for calculating GC content. Nuclei of rice (Oryza sativa subsp japonica ‘Nipponbare’) (1C = 388.82 Mb, GC content 43.6%; International Rice Genome Sequencing Project, 2005) were used as a primary standard for genome size measurements. Mitotic and meiotic chromosome numbers were counted from young anthers of inflorescences fixed in ethanol:acetic acid (3:1) fixative and stored in 70% ethanol at −20°C. Spread-like chromosome preparations were prepared following Lysak and Mandáková (2013) and stained by DAPI (2 μg/mL) in Vectashield (Vector Laboratories). Chromosome figures were photographed with an Olympus BX-61 epifluorescence microscope and AxioCam CCD camera (Zeiss).
454 Sequencing and Tandem Repeat Identification
Total gDNA for sequencing was extracted from healthy young leaves according to Dellaporta et al. (1983) followed by RNase treatment (50 µg/mL). DNA samples of the three parental Cardamine species were sequenced using the Roche 454 GS-FLX Titanium sequencing instrument at the Functional Genomics Center Zurich (C. amara and C. rivularis) and at Microsynth (Cardamine pratensis). In total, 217,107,224 bp was sequenced in C. amara (0.8× genome coverage), 197,571,466 bp of sequence data were obtained for C. rivularis (0.45× genome coverage), and 167,047,291 bp was sequenced in C. pratensis (0.2× genome coverage). To identify repeat content, we used the clustering-based repeat identification pipeline developed by Novák et al. (2010, 2013), performing all-to-all similarity comparisons of 454 reads followed by their graph-based clustering, to identify groups of reads (clusters) derived from repetitive elements. To identify potential tandem repeats, selected cluster contigs were analyzed by Tandem Repeat Finder (Benson, 1999) and Dotter (Sonnhammer and Durbin, 1995).
Tandem Repeat Probes
Oligonucleotides were designed directly from DNA alignments. Probe sequences (70 to 85 nucleotides) were manually selected to obtain a high level of sequence complexity to maximize probe specificity and a GC content of 30 to 50%. The sequence was checked to minimize self-annealing, and the complementary sequence was generated and checked using the programs OligoCalc and OligoAnalyzer. Unmodified lyophilized DNA oligonucleotides were purchased from Sigma-Aldrich at the 25 nM scale and resuspended in water to a final concentration of 100 μM. Equal volumes of complementary oligonucleotides were combined in a 0.2-mL tube and heated in a PCR machine to 95°C for 5 min. The tube was immediately transferred to a beaker containing 400 mL of 90°C water and left to cool slowly until the water temperature reached ∼35°C. Concentrations of the resulting double-stranded DNA templates were quantified using a NanoDrop spectrophotometer and labeled by nick translation.
DNA Labeling and Fluorescence in Situ Hybridization
Whole inflorescences stored in 70% ethanol were digested in the pectolytic enzyme mixture, and meiotic and mitotic chromosome preparations were prepared from young anthers as described by Lysak and Mandáková (2013). Four types of FISH experiments were performed: (1) GISH with labeled gDNA of C. amara, C. rivularis, and C. pratensis as probes, (2) CCP with chromosome-specific BAC contigs of Arabidopsis thaliana arranged according to the 24 genomic blocks building up the eight ancestral chromosomes of the ACK (Schranz et al., 2006), (3) CCP/GISH (i.e., simultaneous hybridization of labeled gDNA probes together with BAC contig painting probes), and (4) FISH with labeled tandem repeat probes. All probes were labeled with biotin-deoxyuridine triphosphate (dUTP), digoxigenin-dUTP, or Cy3-dUTP by nick translation following Lysak and Mandáková (2013). Labeled probes were ethanol precipitated and resuspended in 20 μL of hybridization mixture containing 50% formamide and 10% dextrane sulfate in 2× SSC (1× SSC is 0.15 M sodium chloride and 0.015 M sodium citrate) buffer. The probes were applied to suitable chromosome preparations and denaturated together on a hot plate at 80°C for 2 min. After the overnight (12 to 16 h) hybridization at 37°C, posthybridization washing was conducted in 20% formamide in 2× SSC at 42°C, and hapten-labeled probes were immunodetected as described by Lysak and Mandáková (2013). Slides were counterstained with DAPI (2 μg/mL) in Vectashield and photographed as described above. Suitable images were pseudocolored and merged in Adobe CS Photoshop. Schematic intergenome and interchromosome comparisons based on CCP data and the ACK concept (Schranz et al., 2006) were prepared using Circos (Krzywinski et al., 2009).
Cloning of Crambo Repeats and DNA Gel Blot Hybridization
The conventional approach to satellite repeat isolation based on restriction enzyme digestion of gDNA was applied to isolate Crambo repeats (Hemleben et al., 2007). Briefly, digestion of gDNA from C. pratensis with BstNI yielded a prominent high molecular weight relic of >10 kb. The relic was excised, and the embedded DNA was digested with MboI at 37°C overnight. DNA was then purified using a kit (Macherey-Nagel). Before cloning, a single adenine was added to both ends of DNA using DyNAzyme II DNA polymerase (Finnzymes). The purified deoxyadenosine-tailed BstNI-MboI fragment was cloned into the pDrive Cloning vector (Qiagen PCR cloning kit). Inserts that strongly hybridized on blots with the 32P-labeled gDNA probe from C. pratensis were isolated and sequenced. Probes were labeled with the [α-32P]deoxycytidine triphosphate using a random prime kit (Dekaprime; Fermentas). DNA gel blot hybridization was performed as described by Matyášek et al. (2011). About 2 μg of gDNA digested with MboI was loaded per electrophoretic lane and hybridized with the 32P-labeled Crambo clone 7. The radioactive signals were visualized using a Typhoon phosphor imager and ImageQuant software (both GE Healthcare). The copy number of the satellite repeat units was determined using slot blot hybridization (Hemleben et al., 2007).
Quantification of SNP Ratio by Pyrosequencing (PyroMark)
The gene copy number ratio was verified using the pyrosequencing technique. We chose two genes, CHALCONE SYNTHASE (CHS) and APETALA3 (AP3), for this analysis, as these genes are not duplicated in diploid Cardamine species (Koch et al., 2001; Lihová et al., 2006). The set of gene-specific (PCR amplification) primers and sequencing primer(s) was designed by PyroMark Assay Design v2.0 software (Qiagen). The gene-specific amplification primers were designed at the conserved regions between C. amara and C. rivularis/C. pratensis within each gene, so that they included SNP positions within the amplified fragment. The sequencing primer was also nested within the conserved region to a targeted SNP position(s). Two and one sequencing primers were designed for CHS and AP3, respectively (see Supplemental Table 2 online). DNA was extracted from leaf tissue using DNeasy kit (Qiagen). DNA fragments were amplified with the amplification primers by Ex Taq polymerase (Takara). The amplified DNA fragments were sequenced using the PyroMark ID system (Qiagen) with the sequencing primers at the Genetic Diversity Center (Zurich). The obtained peak was automatically analyzed using the PyroMark system using allele quantification mode to determine the SNP ratio at the targeted position. The multiple SNP ratios in two gene fragments were averaged to determine the A:R ratio of each individual.
cpDNA Analysis
cpDNA variation was assessed to identify the maternal progenitors of the studied hybrids C. × insueta and C. × schulzii (maternal inheritance of chloroplasts has been proven for Brassicaceae [e.g., see Johannessen et al., 2005]). 454 Sequence reads obtained from three individuals of the parental species (C. amara, C. rivularis, and C. pratensis) were assembled and mapped to the cpDNA genome of Arabidopsis (closely related to Cardamine) and used for pairwise comparisons. The bioinformatic analysis was performed using the CLC Genomics Workbench software (version 6.0.4; CLC Bio). Between ∼28,000 and 53,000 reads (5 to 10% of total reads) were mapped to the Arabidopsis reference plastome (NC_000932), providing relatively deep (>100×) sequencing depths. However, several regions of low complexity and high sequence divergence were less deeply covered. Plastome consensus sequences were created from sequence builds using default program parameters (Ns were inserted in positions with no reads). The sequence identity divergence was calculated as a percentage of overlapping alignments where the two sequences agree. The rpoB-trnCGCA intergenic spacer, a representative of the highly variable noncoding cpDNA regions (Shaw et al., 2005), was chosen for PCR amplifications and Sanger sequencing to screen variation in multiple individuals (38 hybrid individuals and 71 individuals of the parental species from Urnerboden and additional populations), following the protocol used by Španiel et al. (2011). Four indels identified in the sequence alignment were coded as additional binary characters (see Supplemental Data Set 1 online). A phylogenetic tree was inferred using maximum likelihood approach with Arabidopsis as an outgroup. A best-fit model of nucleotide substitutions was selected in jModelTest v0.1.1 (Darriba et al., 2012) based on the Akaike information criterion. The maximum likelihood analyses were computed in GARLI v2.0 (Zwickl 2006), implementing the optimal model, setting five million generations, and with multiple runs using different tree search strategies. Bootstrap analysis with 500 replicates was performed to assess the branch support. The method of statistical parsimony was also employed to identify haplotypes and infer their relationships (TCS version 1.18; Clement et al., 2000). The rpoB-trnCGCA intergenic spacer sequences were also extracted from 454 reads obtained for the three progenitor species. The coverage of the ∼1-kb rpoB-trnCGCA region was 65 to 120×. The haplotypes obtained from Sanger and 454 sequences were in good agreement except for a low complexity adenine thymine–rich region (position 338 to 388 of the alignment) that could not be correctly assembled (see Supplemental Data Set 2 online).
Pollen Fertility
Selected inflorescences were stored in 10% ethanol at room temperature for several days. Anthers from mature flowers were stained by Alexander stain under a cover slip and observed under a light microscope. Stained pollen grains were photographed using an Olympus BX-61 epifluorescence microscope and AxioCam charge-coupled device camera and their size measured using the ImageJ program (National Institutes of Health).
Accession Numbers
The rpoB-trnCGCA sequence data from this article can be found in GenBank under accession numbers KC902641 to KC902749 (altogether 109 accessions). Three clones of the Crambo repeat correspond to GenBank accession numbers JQ412178 to JQ412180.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Location of 12 Analyzed Subpopulations and Distribution of the Analyzed Cardamine Taxa at the Urnerboden Plateau.
Supplemental Figure 2. Centromere Localization of the Carcen Satellite Repeat in C. rivularis and the Hypotetraploid C. pratensis.
Supplemental Figure 3. Comparative Genome Structure of C. amara and C. rivularis.
Supplemental Figure 4. Anthers in C. × insueta (2n = 24).
Supplemental Figure 5. Irregular Meiotic Segregation and Pollen Fertility in C. × insueta.
Supplemental Figure 6. Chromosome Localization of Prasat Satellite Repeat in C. pratensis, Hypopentaploid C. × schulzii, and Hypohexaploid C. × schulzii.
Supplemental Figure 7. Male Meiosis and Pollen Fertility in the Hypohexaploid C. × schulzii (2n = 46).
Supplemental Table 1. Genome Size and GC Content in the Analyzed Cardamine Taxa.
Supplemental Table 2. Primers Used for Pyrosequencing.
Supplemental Data Set 1. Sequence Alignment of the rpoB-trnCGCA Intergenic Spacer Based on Sanger Sequencing, Corresponding to the Phylogenetic Analysis in Figure 6.
Supplemental Data Set 2. Sequence Alignment of the rpoB-trnCGCA Intergenic Spacer, Including Sanger Sequence Data and the Data Extracted from the Plastomes Assembled from 454 Reads of the Progenitor Species.
Supplemental Movie 1. Reconstruction of the Origin of the AK5/8/6 Chromosome in the Hypotetetraploid C. pratensis (2n = 30).
Acknowledgments
We thank Petr Bureš for genome size estimates and Ilia J. Leitch and Ingo Schubert for a critical reading of the article. The assistance of Jiří Macas, Petra Hloušková, Elias Landolt, Milan Pouch, Stanislav Španiel, Martin Krzywinski, and Herbert Hurka is greatly acknowledged. We thank the Genetic Diversity Center Zurich and Functional Genomic Center Zurich for their technical support. This work was supported by research grants from the Czech Science Foundation to M.A.L., K.Ma., and K.Mu. (P501/10/1014) and to A.K. (P501/13/10057S) by the European Regional Development Fund (CZ.1.05/1.1.00/02.0068) and the European Social Fund (CZ.1.07/2.3.00/20.0189). K.K.S. was funded by Young Investigator Award of Human Frontier Science Program and Swiss National Foundation (31003A_140917 and Sinergia AVE CRSI33_127155) and the University Research Priority Programs of the University of Zurich. A Marie Heim-Vögtlin grant of the Swiss National Science Foundation to R.S.-I. is acknowledged.
AUTHOR CONTRIBUTIONS
M.A.L., K.Ma., K.Mu., A.K., and K.K.S. designed the research. T.M., J.Z.-L., A.K., and R.S.-I. performed research. M.A.L., T.M., J.Z.-L., A.K., R.S.-I., and K.K.S. wrote the article.
Glossary
- CCP
comparative chromosome painting
- GISH
genomic in situ hybridization
- SNP
single nucleotide polymorphism
- ACK
ancestral crucifer karyotype
- gDNA
genomic DNA
- NCF
nested chromosome fusion
- PMC
pollen mother cells
- FISH
fluorescence in situ hybridization
- DAPI
4′,6-diamidino-2-phenylindole
- cpDNA
chloroplast DNA
References
- Abbott R.J., Lowe A.J. (2004). Origins, establishment and evolution of new polyploidy species: Senecio cambrensis and S. eboracensis in the British Isles. Biol. J. Linn. Soc. Lond. 82: 467–474 [Google Scholar]
- Abbott, R.J., and Rieseberg, L.H. (2012). Hybrid speciation. In Encyclopedia of Life Sciences (eLS, online). (Chichester, UK: John Wiley & Sons). doi: 10.1002/9780470015902.a0001753.pub2 [Google Scholar]
- Ainouche M.L., Baumel A., Salmon A. (2004). Spartina anglica C. E. Hubbard: A natural model system for analysing early evolutionary changes that affect allopolyploid genomes. Biol. J. Linn. Soc. Lond. 82: 475–484 [Google Scholar]
- Arrigo N., Barker M.S. (2012). Rarely successful polyploids and their legacy in plant genomes. Curr. Opin. Plant Biol. 15: 140–146 [DOI] [PubMed] [Google Scholar]
- Benson G. (1999). Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton T.L., Husband B.C. (2001). Fecundity and offspring ploidy in matings among diploid, triploid and tetraploid Chamerion angustifolium (Onagraceae): Consequences for tetraploid establishment. Heredity (Edinb) 87: 573–582 [DOI] [PubMed] [Google Scholar]
- Canales C., Barkoulas M., Galinha C., Tsiantis M. (2010). Weeds of change: Cardamine hirsuta as a new model system for studying dissected leaf development. J. Plant Res. 123: 25–33 [DOI] [PubMed] [Google Scholar]
- Chester M., Gallagher J.P., Symonds V.V., Cruz da Silva A.V., Mavrodiev E.V., Leitch A.R., Soltis P.S., Soltis D.E. (2012). Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc. Natl. Acad. Sci. USA 109: 1176–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement M., Posada D., Crandall K.A. (2000). TCS: A computer program to estimate gene genealogies. Mol. Ecol. 9: 1657–1659 [DOI] [PubMed] [Google Scholar]
- Comai L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6: 836–846 [DOI] [PubMed] [Google Scholar]
- Considine M.J., Wan Y., D’Antuono M.F., Zhou Q., Han M., Gao H., Wang M. (2012). Molecular genetic features of polyploidization and aneuploidization reveal unique patterns for genome duplication in diploid Malus. PLoS ONE 7: e29449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzolino S., Cafasso D., Pellegrino G., Musacchio A., Widmer A. (2007). Genetic variation in time and space: the use of herbarium specimens to reconstruct patterns of genetic variation in the endangered orchid Anacamptis palustris. Conserv. Genet. 8: 629–639 [Google Scholar]
- Cusimano N., Sousa A., Renner S.S. (2012). Maximum likelihood inference implies a high, not a low, ancestral haploid chromosome number in Araceae, with a critique of the bias introduced by 'x'. Ann. Bot. (Lond.) 109: 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Taboada G.L., Doallo R., Posada D. (2012). jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta S.L., Wood J., Hicks J.B. (1983). A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1: 19–21 [Google Scholar]
- Doyle J.J., Flagel L.E., Paterson A.H., Rapp R.A., Soltis D.E., Soltis P.S., Wendel J.F. (2008). Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 42: 443–461 [DOI] [PubMed] [Google Scholar]
- Franzke A., Lysak M.A., Al-Shehbaz I.A., Koch M.A., Mummenhoff K. (2011). Cabbage family affairs: The evolutionary history of Brassicaceae. Trends Plant Sci. 16: 108–116 [DOI] [PubMed] [Google Scholar]
- Franzke A., Mummenhoff K. (1999). Recent hybrid speciation in Cardamine (Brassicaceae). Conversion of nuclear ribosomal ITS sequences in statu nascendi. Theor. Appl. Genet. 98: 831–834 [Google Scholar]
- Grant, V. (1981). Plant Speciation, 2nd ed. (New York: Columbia University Press). [Google Scholar]
- Hegarty, M.J., Abott, R.J., and Hiscock, S.J. (2012). Allopolyploid speciation in action: The origins and evolution of Senecio cambrensis In Polyploidy and Genome Evolution, P.S. Soltis and D.E. Soltis, eds (Berlin, Heidelberg, Germany: Springer-Verlag), pp. 245–270. [Google Scholar]
- Hegarty M.J., Hiscock S.J. (2008). Genomic clues to the evolutionary success of polyploid plants. Curr. Biol. 18: R435–R444 [DOI] [PubMed] [Google Scholar]
- Hemleben V., Kovarik A., Torres-Ruiz R.A., Volkov R.A., Beridze T. (2007). Plant highly repeated satellite DNA: Molecular evolution, distribution, and use for identification of hybrids. Syst. Biodivers. 5: 277–289 [Google Scholar]
- Henry I.M., Dilkes B.P., Young K., Watson B., Wu H., Comai L. (2005). Aneuploidy and genetic variation in the Arabidopsis thaliana triploid response. Genetics 170: 1979–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husband B.C. (2004). The role of triploid hybrids in the evolutionary dynamics of mixed-ploidy populations. Biol. J. Linn. Soc. Lond. 82: 537–546 [Google Scholar]
- International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jiao Y., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Johannessen M.M., Andersen B.A., Damgaard C., Jørgensen R.B. (2005). Maternal inheritance of chloroplasts between Brassica rapa and F1-hybrids demonstrated by cpDNA markers specific to oilseed rape and B. rapa. Mol. Breed. 16: 271–278 [Google Scholar]
- Kantama L., Sharbel T.F., Schranz M.E., Mitchell-Olds T., de Vries S., de Jong H. (2007). Diploid apomicts of the Boechera holboellii complex display large-scale chromosome substitutions and aberrant chromosomes. Proc. Natl. Acad. Sci. USA 104: 14026–14031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M.A., Weisshaar B., Kroymann J., Haubold B., Mitchell-Olds T. (2001). Comparative genomics and regulatory evolution: Conservation and function of the Chs and Apetala3 promoters. Mol. Biol. Evol. 18: 1882–1891 [DOI] [PubMed] [Google Scholar]
- Kovarik A., Pires J.C., Leitch A.R., Lim K.Y., Sherwood A.M., Matyasek R., Rocca J., Soltis D.E., Soltis P.S. (2005). Rapid concerted evolution of nuclear ribosomal DNA in two Tragopogon allopolyploids of recent and recurrent origin. Genetics 169: 931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kučera J., Valko I., Marhold K. (2005). On-line database of the chromosome numbers of the genus Cardamine (Brassicaceae). Biologia 60: 473–476 [Google Scholar]
- Lawrence W.J.C. (1931). The chromosome constitution of Cardamine pratensis and Verbascum phoeniceum. Genetica 13: 183–208 [Google Scholar]
- Lihová, J., and Marhold, K. (2006). Phylogenetic and diversity patterns in Cardamine (Brassicaceae) – A genus with conspicuous polyploid and reticulate evolution. In Plant Genome: Biodiversity and Evolution: Phanerogams (Angiosperm -Dicotyledons), Vol. 1C, A.K. Sharma and A. Sharma, eds (Enfield, NH: Science Publishers), pp. 149–186 [Google Scholar]
- Lihová J., Shimizu K.K., Marhold K. (2006). Allopolyploid origin of Cardamine asarifolia (Brassicaceae): Incongruence between plastid and nuclear ribosomal DNA sequences solved by a single-copy nuclear gene. Mol. Phylogenet. Evol. 39: 759–786 [DOI] [PubMed] [Google Scholar]
- Lövkvist B. (1956). The Cardamine pratensis complex. Outline of its cytogenetics and taxonomy. Symb. Bot. Ups. 14: 1–131 [Google Scholar]
- Luo M.C., et al. (2009). Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc. Natl. Acad. Sci. USA 106: 15780–15785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M.A., Berr A., Pecinka A., Schmidt R., McBreen K., Schubert I. (2006). Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl. Acad. Sci. USA 103: 5224–5229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M.A., Mandáková T. (2013). Analysis of plant meiotic chromosomes by chromosome painting. Methods Mol. Biol. 990: 13–24 [DOI] [PubMed] [Google Scholar]
- Mandáková T., Heenan P.B., Lysak M.A. (2010a). Island species radiation and karyotypic stasis in Pachycladon allopolyploids. BMC Evol. Biol. 10: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T., Joly S., Krzywinski M., Mummenhoff K., Lysak M.A. (2010b). Fast diploidization in close mesopolyploid relatives of Arabidopsis. Plant Cell 22: 2277–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhold, K. (1995). Cardamine rivularis auct. non Schur in the Eastern Alps. Carinthia II 53 (Sonderheft): 101–102.
- Mason A.S., Nelson M.N., Yan G., Cowling W.A. (2011). Production of viable male unreduced gametes in Brassica interspecific hybrids is genotype specific and stimulated by cold temperatures. BMC Plant Biol. 11: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyášek R., Fulneček J., Leitch A.R., Kovařík A. (2011). Analysis of two abundant, highly related satellites in the allotetraploid Nicotiana arentsii using double-strand conformation polymorphism analysis and sequencing. New Phytol. 192: 747–759 [DOI] [PubMed] [Google Scholar]
- Murat F., Xu J.H., Tannier E., Abrouk M., Guilhot N., Pont C., Messing J., Salse J. (2010). Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution. Genome Res. 20: 1545–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer B., Jahncke P. (1997). RAPD analyses of recent hybrid speciation events in Cardamine (Brassicaceae). Folia Geobot. Phytotaxon. 32: 57–67 [Google Scholar]
- Neuffer, B., Mönninghoff, U., and Hurka, H. (2009). Arealausbreitung entlang von Höhengradienten – Wiesenschaumkraut am Urnerboden. In Biologische Invasionen und Phytodiversität. Auswirkungen und Handlungsoptionen, W. Bleeker and H. Hurka, eds (Osnabrück, Germany: Universität Osnabrück), pp. 20–21. [Google Scholar]
- Novák P., Neumann P., Macas J. (2010). Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 11: 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák P., Neumann P., Pech J., Steinhaisl J., Macas J. (2013). RepeatExplorer: A Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29: 792–793 [DOI] [PubMed] [Google Scholar]
- Ownbey M. (1950). Natural hybridization and amphiploidy in the genus Tragopogon. Am. J. Bot. 37: 487–499 [Google Scholar]
- Ramsey J., Schemske D.W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29: 467–501 [Google Scholar]
- Renny-Byfield S., Ainouche M., Leitch I.J., Lim K.Y., Le Comber S.C., Leitch A.R. (2010). Flow cytometry and GISH reveal mixed ploidy populations and Spartina nonaploids with genomes of S. alterniflora and S. maritima origin. Ann. Bot. (Lond.) 105: 527–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz M.E., Lysak M.A., Mitchell-Olds T. (2006). The ABC’s of comparative genomics in the Brassicaceae: Building blocks of crucifer genomes. Trends Plant Sci. 11: 535–542 [DOI] [PubMed] [Google Scholar]
- Shaw J., Lickey E.B., Beck J.T., Farmer S.B., Liu W., Miller J., Siripun K.C., Winder C.T., Schilling E.E., Small R.L. (2005). The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 92: 142–166 [DOI] [PubMed] [Google Scholar]
- Soltis D.E., Soltis P.S., Pires J.C., Kovarik A., Tate J., Mavrodiev E. (2004). Recent and recurrent polyploidy in Tragopogon (Asteraceae): Cytogenetic, genomic and genetic comparisons. Biol. J. Linn. Soc. Lond. 82: 485–501 [Google Scholar]
- Soltis D.E., Albert V.A., Leebens-Mack J., Bell C.D., Paterson A.H., Zheng C., Sankoff D., Depamphilis C.W., Wall P.K., Soltis P.S. (2009). Polyploidy and angiosperm diversification. Am. J. Bot. 96: 336–348 [DOI] [PubMed] [Google Scholar]
- Soltis P.S., Soltis D.E. (2009). The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 60: 561–588 [DOI] [PubMed] [Google Scholar]
- Sonnhammer E.L., Durbin R. (1995). A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167: GC1–GC10 [DOI] [PubMed] [Google Scholar]
- Staats M., Erkens R.H.J., van de Vossenberg B., Wieringa J.J., Ken Kraaijeveld K., Stielow B., Geml J., Richardson J.E., Bakker F.T. (2013). Genomic treasure troves: Complete genome sequencing of herbarium and insect museum specimens. PLoS ONE 8: e69189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda J., Trávníček P. (2006). Reliable DNA ploidy determination in dehydrated tissues of vascular plants by DAPI flow cytometry—New prospects for plant research. Cytometry A 69: 273–280 [DOI] [PubMed] [Google Scholar]
- Španiel S., Marhold K., Passalacqua N.G., Zozomová-Lihová J. (2011). Intricate variation patterns in the diploid-polyploid complex of Alyssum montanum-A. repens (Brassicaceae) in the Apennine Peninsula: Evidence for long-term persistence and diversification. Am. J. Bot. 98: 1887–1904 [DOI] [PubMed] [Google Scholar]
- Urbanska K.M., Hurka H., Landolt E., Neuffer B., Mummenhoff K. (1997). Hybridization and evolution in Cardamine (Brassicaceae) at Urnerboden, central Switzerland: Biosystematic and molecular evidence. Plant Syst. Evol. 204: 233–256 [Google Scholar]
- Urbanska, K.M., and Landolt, E. (1999). Patterns and processes of man-influenced hybridisation in Cardamine L. In Plant Evolution in Man-made Habitats, L.W.D. van Raamsdonk and H.C.M. den Nijs, eds (Amsterdam: Hugo de Vries Laboratory), pp. 29–47. [Google Scholar]
- Urbanska-Worytkiewicz K. (1977a). Reproduction in natural triploid hybrids (2n = 24) between Cardamine rivularis Schur and C. amara L. Ber. Geobot. Inst. ETH. Stiftung Rübel 44: 42–85 [Google Scholar]
- Urbanska-Worytkiewicz K. (1977b). An autoallohexaploid in Cardamine L., new to the Swiss flora. Ber. Geobot. Inst. ETH. Stiftung Rübel 44: 86–103 [Google Scholar]
- Urbanska-Worytkiewicz K.M. (1978). Ségrégation polarisée chez les hybrides naturels triploïdes (2n = 24) entre Cardamine rivularis Schur (2n = 16) et C. amara L. (2n = 16). Bull. Soc. Bot. Franc. Actual Bot. 1–2: 91–93 [Google Scholar]
- Urbanska-Worytkiewicz, K.M. (1980). Reproductive strategies in a hybridogenous population of Cardamine L. Acta Oecol., Oecol. Pl. 1: 137–150.
- Urbanska-Worytkiewicz K., Landolt E. (1972). Natürliche Bastarde zwischen Cardamine amara L. und C. rivularis Schur aus den Schweizer Alpen. Ber. Geobot. Inst. ETH. Stiftung Rübel 41: 88–101 [Google Scholar]
- Urbanska-Worytkiewicz K., Landolt E. (1974). Hybridation naturelle entre Cardamine rivularis Schur et C. amara L., ses aspects cytologiques et écologiques. Act. Soc. Helv. Sci. Nat. 1974: 89–90 [Google Scholar]
- Vallejo-Marín M. (2012). Mimulus peregrinus (Phrymaceae): A new British allopolyploid species. PhytoKeys 14: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood T.E., Takebayashi N., Barker M.S., Mayrose I., Greenspoon P.B., Rieseberg L.H. (2009). The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. USA 106: 13875–13879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z., Gaeta R.T., Pires J.C. (2011). Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. USA 108: 7908–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli S. (1986). Einfluss der Bewirtschaftung auf die Entstehung und Struktur der Cardamine-Populationen auf dem Urnerboden. Veröff. Geobot. Inst. ETH. Stiftung Rübel 87: 141–154 [Google Scholar]
- Zwickl, D.J. (2006). Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets under the Maximum Likelihood Criterion. PhD dissertation (Austin, TX: University of Texas). [Google Scholar]