This work reveals that the woodland strawberry ortholog of the floral activator SOC1 (Fv SOC1) is the general regulator of the photoperiodic development in this perennial short-day plant. It suppresses photoperiodic flowering by activating a major floral repressor Fv TFL1 and mediates photoperiodic signaling to promote runner development through regulating gibberellin biosynthetic genes.
Abstract
In the annual long-day plant Arabidopsis thaliana, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) integrates endogenous and environmental signals to promote flowering. We analyzed the function and regulation of the SOC1 homolog (Fragaria vesca [Fv] SOC1) in the perennial short-day plant woodland strawberry (Fragaria vesca). We found that Fv SOC1 overexpression represses flower initiation under inductive short days, whereas its silencing causes continuous flowering in both short days and noninductive long days, similar to mutants in the floral repressor Fv TERMINAL FLOWER1 (Fv TFL1). Molecular analysis of these transgenic lines revealed that Fv SOC1 activates Fv TFL1 in the shoot apex, leading to the repression of flowering in strawberry. In parallel, Fv SOC1 regulates the differentiation of axillary buds to runners or axillary leaf rosettes, probably through the activation of gibberellin biosynthetic genes. We also demonstrated that Fv SOC1 is regulated by photoperiod and Fv FLOWERING LOCUS T1, suggesting that it plays a central role in the photoperiodic control of both generative and vegetative growth in strawberry. In conclusion, we propose that Fv SOC1 is a signaling hub that regulates yearly cycles of vegetative and generative development through separate genetic pathways.
INTRODUCTION
The molecular control of flowering has been studied in detail in the annual model plants Arabidopsis thaliana and rice (Oryza sativa; Kim et al., 2009; Tsuji et al., 2011; Turnbull, 2011), whereas less is known about perennial species. In Arabidopsis, four major genetic pathways (i.e., the photoperiodic, vernalization, autonomous, and gibberellin [GA] pathways) regulate flowering time (Simpson, 2004; Turck et al., 2008; Kim et al., 2009; Mutasa-Göttgens and Hedden, 2009). Many genes with sequence similarity to known flowering genes are found in different plant families, but their functions may differ between species (Suárez-López et al., 2001; Hayama et al., 2003; Hecht et al., 2005, 2011; Mouhu et al., 2009). In perennials, which undergo repeated cycles of vegetative and reproductive phases, flowering time is controlled by seasonal regulation of flowering genes (Böhlenius et al., 2006; Wang et al., 2009; Koskela et al., 2012). Moreover, homologous genes, or genes classified in the same gene family, may have roles during the seasonal cycle beyond those that are known in annual species (Böhlenius et al., 2006; Hsu et al., 2011). Therefore, careful analysis of gene functions in the different phases of seasonal cycles is required to better understand perennial growth.
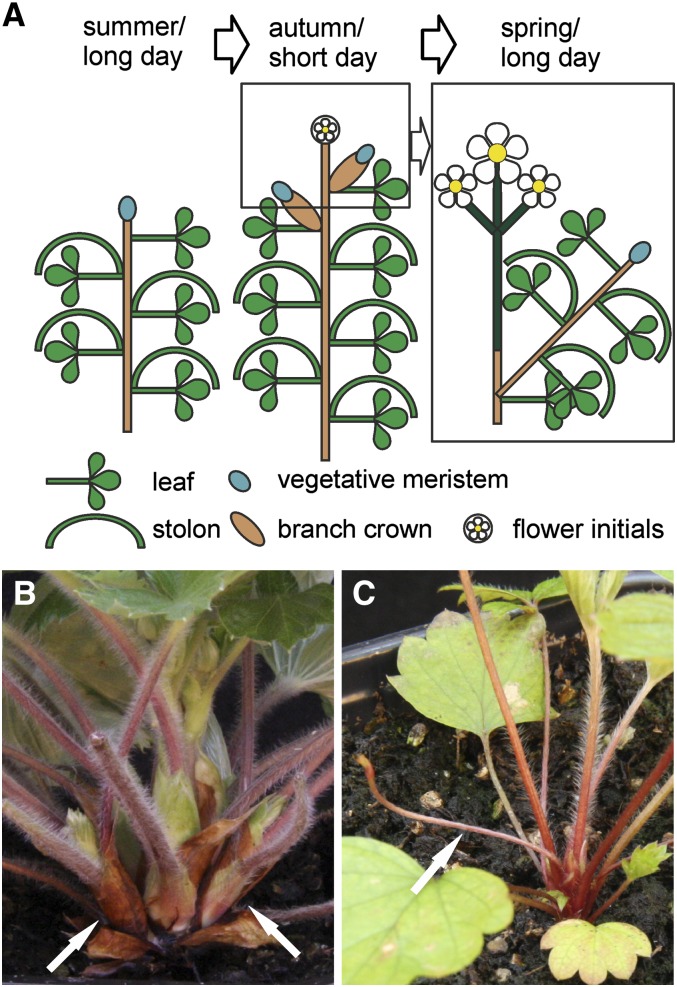
We use diploid woodland strawberry (Fragaria vesca) as a model to study the environmental control of perennial growth. Strawberry belongs to the Rosaceae family, which includes many economically important fruit crops, such as apple (Malus domestica), pear (Pyrus communis), peach (Prunus persica), plum (Prunus domestica), and cherry (Prunus avium), and ornamental genera, such as Rosa, Potentilla, and Spiraea (Potter et al., 2007). Woodland strawberry and other strawberry species are perennial rosette herbs. Most accessions of strawberry and cultivars of the garden strawberry (Fragaria × ananassa) are seasonal flowering short-day (SD) plants (Heide, 1977; Heide and Sønsteby, 2007). During the vegetative phase under long days (LDs), strawberries spread clonally through aboveground stolons called runners, which are formed from the axillary buds of the rosette stem, called the crown (Figure 1; Konsin et al., 2001; Hytönen et al., 2004; Heide and Sønsteby, 2007), and consist of two long internodes followed by a daughter plant. Under SDs in the autumn, runner formation ceases and the uppermost axillary buds differentiate to axillary leaf rosettes called branch crowns (Figure 1; Konsin et al., 2001; Hytönen et al., 2004), and vegetative growth is reduced as characterized by decreased petiole elongation (Guttridge and Thompson, 1964; Wiseman and Turnbull, 1999; Konsin et al., 2001). At the same time, SDs activate flower initiation in the shoot apex of the main crown as well as in branch crowns that have become competent for floral development before or during the inductive SDs (Figure 1; Hytönen et al., 2004). However, the youngest axillary shoots remain vegetative, enabling the next seasonal growth cycle.
Figure 1.
Development of Strawberry Shoot.
(A) Schematic representation of the shoot structure and development in seasonal flowering strawberry (SD F. vesca). Under LDs in summer, the plant grows vegetatively and axillary buds typically differentiate into runners (stolons). Autumn SDs cause flower initiation in the apical meristem and the development of axillary branch crowns. The terminal inflorescence emerges in the next season, and newly formed axillary branch crowns continue vegetative development. Note that branch crowns formed in the autumn often produce terminal inflorescences in the next spring.
(B) Close-up of a strawberry crown with the main crown in the middle and axillary branch crowns (arrows) in both sides. An Fv SOC1-RNAi line, which produces only a few runners, was photographed.
(C) Close-up of a young strawberry seedling with a newly emerged runner (arrow). Simple leaves in the figure are juvenile leaves.
Early genetic studies have shown that different single genes, SEASONAL FLOWERING LOCUS (SFL) and RUNNERING LOCUS (RL), regulate seasonal flowering and runner formation in strawberry, and recessive alleles of these loci cause perpetual (continuous) flowering and runner-less phenotypes, respectively (Brown and Wareing, 1965; Albani et al., 2004). RL has not been identified. However, it is known that the photoperiodic control of GA biosynthesis is involved in the axillary bud differentiation to runners and branch crowns (Hytönen et al., 2009). Iwata et al. (2012) showed that SFL and the RECURRENT BLOOMING locus in roses encode homologs of TERMINAL FLOWER1 (TFL1), a member of the phosphatidyl ethanolamide binding protein family. We confirmed the function of Fv TFL1 as a major floral repressor and showed that its photoperiodic control in the shoot apex explains seasonal flowering in strawberry (Koskela et al., 2012). LDs in summer activate Fv TFL1 mRNA expression, and flower initiation only occurs in autumn, when Fv TFL1 is downregulated by SDs. In spring, high Fv TFL1 expression is restored in the apices of new branch crowns to allow the production of new vegetative shoots (Koskela et al., 2012). Perpetual-flowering strawberry accessions, by contrast, do not require SDs for flower induction because of a frame shift mutation, which prevents the production of the functional Fv TFL1 (Iwata et al., 2012; Koskela et al., 2012). In these accessions, LD strongly advances flowering by activating another phosphatidyl ethanolamide binding protein homolog, Fv FLOWERING LOCUS T1 (Fv FT1) (Koskela et al., 2012). Also in seasonal flowering strawberry, Fv FT1 is expressed specifically under LDs, correlating negatively with the photoperiodic flower induction.
FT is a major activator of photoperiodic flowering in many LD and SD plants, including perennials (Turck et al., 2008; Pin and Nilsson, 2012). It is activated by CONSTANS (CO) in the leaf vascular tissues under flower-inductive conditions (Suárez-López et al., 2001; An et al., 2004). Consequently, FT protein relocates through phloem to the shoot apex, where it forms a complex with a bZIP protein FD and 14-3-3 proteins to induce APETALA1 (AP1) and flowering (Abe et al., 2005; Wigge et al., 2005; Corbesier et al., 2007; Tamaki et al., 2007; Taoka et al., 2011). In Arabidopsis, TFL1 has not been shown to control photoperiodic flowering, although it can also bind FD (Hanano and Goto, 2011). TFL1 functions to maintain an indeterminate inflorescence meristem by repressing floral meristem identity genes AP1 and LEAFY (LFY), whereas AP1 and LFY downregulate TFL1 in the flanks of the inflorescence meristem to specify floral meristems (Liljegren et al., 1999; Ratcliffe et al., 1999).
CO and FT have been shown to promote flowering also through the MADS box transcription factor SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1; Samach et al., 2000; Moon et al., 2005; Yoo et al., 2005). SOC1 may be the primary target of FT, since it is the first gene to be activated after a single inductive LD in the shoot apex (Torti et al., 2012). In addition to the photoperiodic pathway, SOC1, as well as FT, integrates signals from the vernalization, autonomous, and GA flowering pathways (Lee et al., 2000; Moon et al., 2003, 2005; Li et al., 2008; Jung et al., 2012). SOC1 interacts with multiple MADS box proteins, including AGAMOUS LIKE24 (AGL24), FRUITFUL (FUL), and AP1, and regulates the expression of several flowering genes, such as SHORT VEGETATIVE PHASE, AGL15, and AGL18, by directly binding to their regulatory sequences (de Folter et al., 2005; Lee et al., 2008; Seo et al., 2009; Immink et al., 2012; Tao et al., 2012).
Here, we report the functional analysis of the SOC1 homolog in the perennial SD plant strawberry. We show evidence that Fv FT1 may mediate the photoperiodic regulation of Fv SOC1. This regulation of Fv SOC1 plays an important role in the photoperiodic development of strawberry, since both overexpression and silencing of Fv SOC1 strongly compromise the regulation of both vegetative and generative development by daylength. We also demonstrate that Fv SOC1 activates the expression of the floral repressor Fv TFL1, which prevents flower induction under LD conditions. In addition, Fv SOC1 promotes vegetative development by activating the expression of several GA biosynthetic genes. Our results suggest that Fv SOC1 is a general regulator of photoperiodic development that mediates photoperiodic signaling to regulate flowering and vegetative growth through separate genetic pathways.
RESULTS
Fv SOC1 Is an Ortholog of SOC1
We previously cloned Fv SOC1, which is the closest strawberry homolog of Arabidopsis SOC1, sharing 66% identity at the amino acid sequence level (Mouhu et al., 2009; Shulaev et al., 2011). We performed a maximum likelihood phylogenetic analysis using randomized axelerated maximum likelihood (Stamatakis et al., 2008), which showed that Fv SOC1 groups with other known SOC1 homologs from Rosaceae (e.g., Trainin et al., 2013), within the same clade as SOC1 homologs from other rosids (e.g., Hecht et al., 2005; see Supplemental Figure 1 and Supplemental Data Set 1 online). Moreover, a syntenic view generated by CoGe (Lyons and Freeling, 2008) showed that the microsynteny around SOC1 and Fv SOC1 is conserved (see Supplemental Figure 2 online). The protein sequence alignment of Fv SOC1 and several SOC1-like proteins showed that Fv SOC1 contains the highly conserved MADS box domain, the K domain, and the SOC1 domain characteristic of these proteins (see Supplemental Figure 3 online; Riechmann and Meyerowitz, 1997; Vandenbussche et al., 2003; Nakamura et al., 2005). We also overexpressed Fv SOC1 in the Arabidopsis Columbia ecotype under the cauliflower mosaic virus 35S promoter and analyzed flowering time. Similarly to the constitutive expression of Arabidopsis SOC1 (Lee et al., 2000), heterologous overexpression of Fv SOC1 advanced flowering in both LD and SD conditions in Columbia-0 (see Supplemental Figure 4 online). Therefore, we conclude that Fv SOC1 is the ortholog of SOC1, acting as a floral activator in Arabidopsis.
Fv SOC1 Is under Photoperiodic Regulation in SD Strawberry
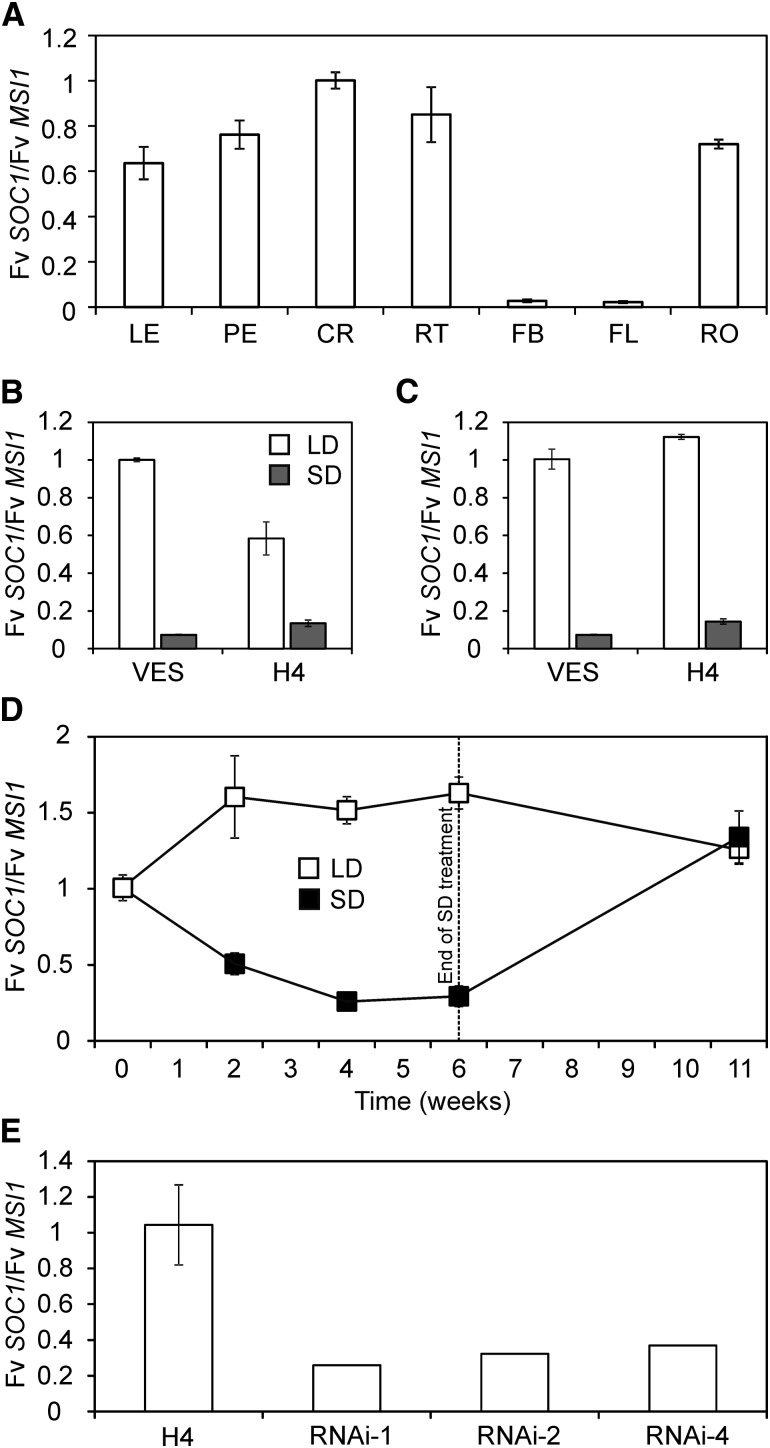
We first analyzed the daily rhythm of Fv SOC1 in seasonal flowering SD strawberry plants (PI551792; National Clonal Germplasm Repository, Corvallis, OR; abbreviated to VES in the figures). Results indicated that Fv SOC1 is slightly downregulated at ZT8 and ZT12 (zeitgeber time) compared with other time points under LDs, whereas no clear rhythm was found under SDs (see Supplemental Figure 5 online). Based on these data, all samples for gene expression analyses were collected at ZT4. To examine the spatial expression pattern of Fv SOC1, we collected tissue samples from plants exposed to a 6-week SD flower induction treatment followed by LD conditions. We found that Fv SOC1 was highly expressed in all tissues except flower buds and flowers (Figures 2A to 2C). In the shoot apex, Fv SOC1 mRNA was abundant in both apical and axillary meristems as well as in the vascular tissue and leaf primordia (see Supplemental Figure 6 online). To analyze if daylength has an effect on Fv SOC1 expression, we subjected young SD strawberry seedlings to photoperiodic treatments and collected leaf and shoot apex samples. We found that under LDs, the expression of Fv SOC1 was high in both shoot apices and leaves. However, strong downregulation of Fv SOC1 was found in both tissues of plants grown under flower-inductive SDs (Figures 2B and 2C). Time-course analysis showed that Fv SOC1 mRNA levels stayed at high levels in the shoot apex under LDs (Figure 2D). By contrast, during a 6-week SD treatment, Fv SOC1 expression gradually decreased and reached low levels after 4 weeks of SDs, similarly to the major floral repressor Fv TFL1 in the same sample set (as shown in Koskela et al., 2012). To see if exposure to LDs after SD treatment restores Fv SOC1 expression, we collected shoot apex samples from vegetative axillary shoots that developed after the SD treatment. In these samples, Fv SOC1 expression was increased to a level of expression similar to that detected before SD treatment (Figure 2D). Therefore, our results suggest that Fv SOC1 is seasonally regulated similarly to Fv TFL1, and its expression correlates negatively with flower initiation in SD strawberry.
Figure 2.
Spatial Expression Pattern and Regulation of Fv SOC1.
(A) Relative expression of Fv SOC1 in different plant organs. Clonally propagated 2-month-old SD strawberry plants were grown under SDs for 6 weeks followed by standard LDs until anthesis. LE, leaf; PE, petiole; CR, axillary branch crown; RT, runner tip; FB, flower bud; FL, opened flower; RO, root. Mean fold change was calculated relative to axillary branch crown. Error bars represent se; n = 2.
(B) and (C) Relative expression of Fv SOC1 in leaves (B) and shoot apices (C) of SD strawberry (VES) and Hawaii-4 (H4) seedlings. Samples were collected from plants grown under LDs or SDs until the three-leaf stage. Mean fold change was calculated relative to SD strawberry grown under LDs. Error bars represent se; n = 3 to 4.
(D) Time-course analysis of Fv SOC1 relative expression under SDs and LDs. Clonally propagated 5-week-old plants of SD strawberry were subjected to photoperiodic treatments for 6 weeks (weeks 0 to 6) followed by LDs for 5 weeks. Primary shoot apices were analyzed during weeks 0 to 6, and newly formed branch crowns were analyzed at week 11. Mean fold change was calculated relative to week 0. Error bars represent se; n = 3.
(E) Relative expression of Fv SOC1 in shoot apices of Fv FT1-RNAi lines in the H4 background. H4 and three independent Fv FT1-RNAi lines were grown under standard LD conditions and samples were collected at the three-leaf stage. Fold change was calculated relative to H4. Error bar represents se; n = 3 for H4, and n = 1 for transgenic lines, which are shown as biological replicates. See Koskela et al. (2012) for the Fv FT1 expression levels.
Regulation of Fv SOC1 in the Perpetual Flowering Mutant
We also analyzed the expression of Fv SOC1 in the Hawaii-4 accession (PI551572; National Clonal Germplasm Repository, Corvallis, OR; called H4 hereafter). H4 lacks functional Fv TFL1; therefore, it flowers perpetually after flower induction and its photoperiodic requirement for flower induction is reversed (i.e., it flowers earlier under LDs than SDs) (Koskela et al., 2012). Also in H4, Fv SOC1 was regulated by photoperiod. However, our data indicated that under LDs, the mRNA level of Fv SOC1 was slightly lower in H4 leaf samples compared with SD strawberry, and the downregulation of Fv SOC1 under SDs was less dramatic in H4 (Figures 2B and 2C). Therefore, we tested if Fv TFL1 regulates the expression of Fv SOC1 by analyzing previously reported Fv TFL1 transgenic lines in the SD strawberry background (Koskela et al., 2012). Neither RNA interference (RNAi) silencing nor overexpression of Fv TFL1 affected the expression of Fv SOC1, suggesting that Fv SOC1 is not regulated by Fv TFL1 (see Supplemental Figure 7 online). Therefore, Fv SOC1 may act upstream of Fv TFL1 or independently of it.
To examine if Fv FT1 regulates Fv SOC1, we studied previously reported Fv FT1-RNAi lines in the H4 background (Koskela et al., 2012). We found about a threefold downregulation of Fv SOC1 mRNA levels in the shoot apices of three independent RNAi lines (Figure 2E). Given that both Fv FT1 and Fv SOC1 are activated by LDs (Koskela et al., 2012), our results indicate that Fv FT1 is likely involved in the photoperiodic regulation of Fv SOC1 mRNA expression, which correlates positively with flowering in H4. In SD strawberry, these genes are similarly regulated, but their expression correlates negatively with flowering.
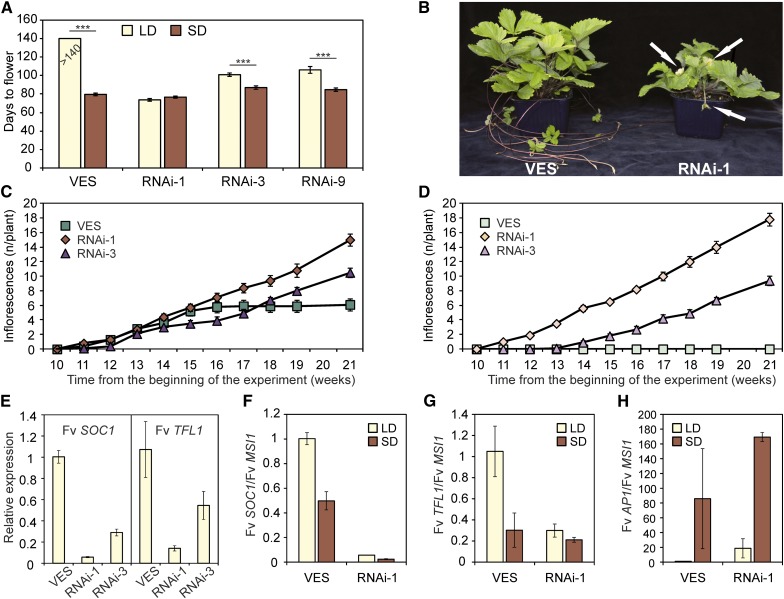
RNAi Silencing of Fv SOC1 Downregulates Fv TFL1 and Causes Day-Neutral Flowering in Strawberry
Since our real-time PCR and in situ hybridization analyses showed that Fv SOC1 has spatial and temporal expression patterns similar to the floral repressor Fv TFL1 in the shoot apex (Figures 2C and 2D; see Supplemental Figure 6 online; Koskela et al., 2012), we hypothesized that Fv SOC1 may regulate Fv TFL1 expression to repress flowering in strawberry. Therefore, we silenced Fv SOC1 in the SD strawberry background using an RNAi construct expressed under the 35S promoter. Analysis showed that Fv SOC1 was strongly downregulated in three independent transgenic lines (see Supplemental Table 1 online). We propagated these lines clonally from runner cuttings and subjected the plants and nontransgenic controls to LD and SD treatments for 6 weeks followed by standard LDs (see Methods). All plants flowered after inductive SD treatment, but only Fv SOC1-RNAi plants flowered under noninductive LD conditions (Figures 3A and 3B). Compared with LDs, SDs advanced flowering by 2 to 3 weeks in two Fv SOC1-RNAi lines, whereas the line Fv SOC1-RNAi#1 flowered at the same time in both photoperiods. SD-treated nontransgenic plants produced on average six terminal inflorescences on the top of the primary and axillary leaf rosettes during weeks 11 to 16 after the beginning of the photoperiodic treatment, before returning to the vegetative growth phase (Figure 3C). By contrast, RNAi#1 and RNA#3 lines continuously produced new inflorescences in both photoperiods (Figures 3C and 3D; see Supplemental Figure 8 online), similarly to perpetual-flowering H4, which lacks functional Fv TFL1 (Koskela et al., 2012).
Figure 3.
Silencing of Fv SOC1 Causes Day-Neutral Flowering in Strawberry.
(A) Flowering time of SD strawberry (VES) and Fv SOC1-RNAi lines #1, #3, and #9. Clonally propagated 5-week-old plants were subjected to photoperiodic treatments for 6 weeks followed by standard LD conditions. Time from the beginning of the SD treatment to the first open flower was observed. Horizontal bars represent statistically significant differences between photoperiodic treatments (separately for SD strawberry and transgenic lines); ***P < 0.001. Error bars represent se; n = 10.
(B) The phenotype of LD-grown SD strawberry and Fv SOC1-RNAi plants. SD strawberry is vegetative and produces runners, whereas the RNAi plant is flowering and has no runners. Arrows point to flowers.
(C) and (D) Time-course analysis of inflorescence formation in SD strawberry and Fv SOC1-RNAi lines #1 and #3. Cumulative number of inflorescences is shown. Clonally propagated 5-week-old plants were subjected to 6 weeks of SDs followed by LDs (C) or grown under continuous LDs (D). Error bars represent se; n = 10.
(E) The expression of Fv SOC1 and Fv TFL1 in 5-week-old clonally propagated SD strawberry and Fv SOC1-RNAi plants grown under LDs. Mean fold change was calculated relative to SD strawberry. Error bars represent se; n = 3.
(F) to (H) Relative expression of Fv SOC1 (F), Fv TFL1 (G), and Fv AP1 (H) in the shoot apices of SD strawberry and Fv SOC1-RNAi line #1. Clonally propagated 5-week-old plants were subjected to LDs or SDs for 4 weeks before sampling. Mean fold change was calculated relative to LD-grown SD strawberry. Error bars represent se; n = 3; for SD-grown strawberry, n = 2.
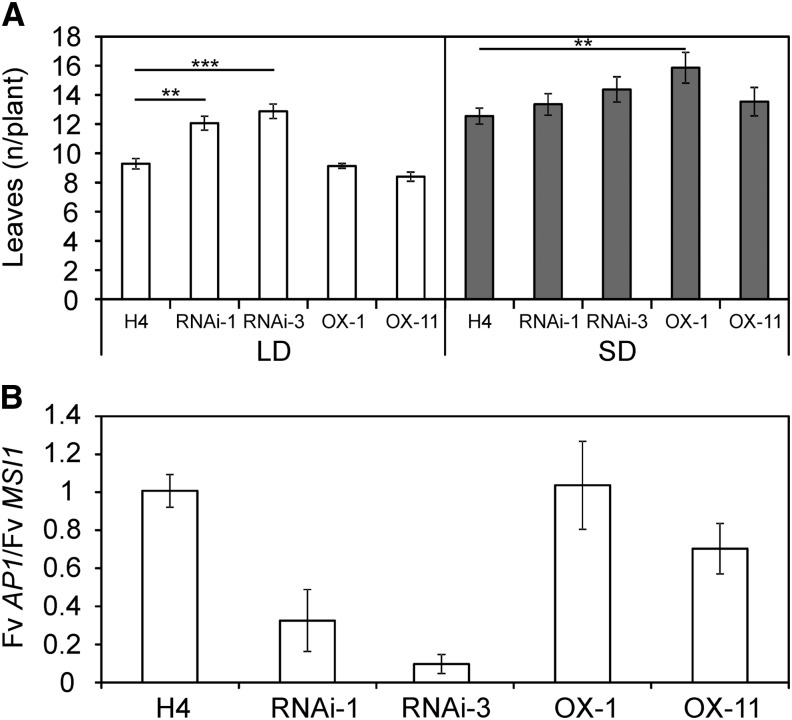
To test if Fv SOC1 regulates the expression of Fv TFL1 in the shoot apex, we analyzed its expression levels in young Fv SOC1-RNAi plants in the SD strawberry background. Fv TFL1 was downregulated in the shoot apices of LD-grown RNAi plants compared with nontransgenic SD strawberry. The level of downregulation correlated with the strength of Fv SOC1 RNAi silencing (Figure 3E). To confirm that these differences were not caused by changes in the meristem identity in these plants, we analyzed the expression of the floral meristem identity gene Fv AP1, which is strongly upregulated at the time of flower induction (Mouhu et al., 2009; Koskela et al., 2012). Fv AP1 was expressed at low levels in all RNAi plants compared with flower-induced SD-grown nontransgenic plants (see Supplemental Figure 9 online). This result confirmed that the apical meristems of LD-grown plants were vegetative at this stage and that the low Fv TFL1 expression levels in RNAi plants were not caused by the activation of Fv AP1.
Next, we studied gene expression levels in SD strawberry and in Fv SOC1-RNAi line #1 in the SD strawberry background after an additional 4 weeks of photoperiodic treatments. In SD strawberry, both Fv SOC1 and Fv TFL1 were downregulated under SDs compared with LDs, correlating negatively with the expression of Fv AP1, which was activated under SDs (Figures 3F to 3H). In RNAi plants, by contrast, downregulation of both Fv SOC1 and Fv TFL1 was detected in both photoperiods. In addition, compared with LD-grown nontransgenic plants, Fv AP1 was ∼20 and 160 times upregulated in LD- and SD-grown RNAi plants, respectively, and plants flowered in both photoperiods. These results strongly suggest that, under LDs, Fv SOC1 promotes the expression of Fv TFL1, which prevents the activation of Fv AP1 and flower initiation in SD strawberry.
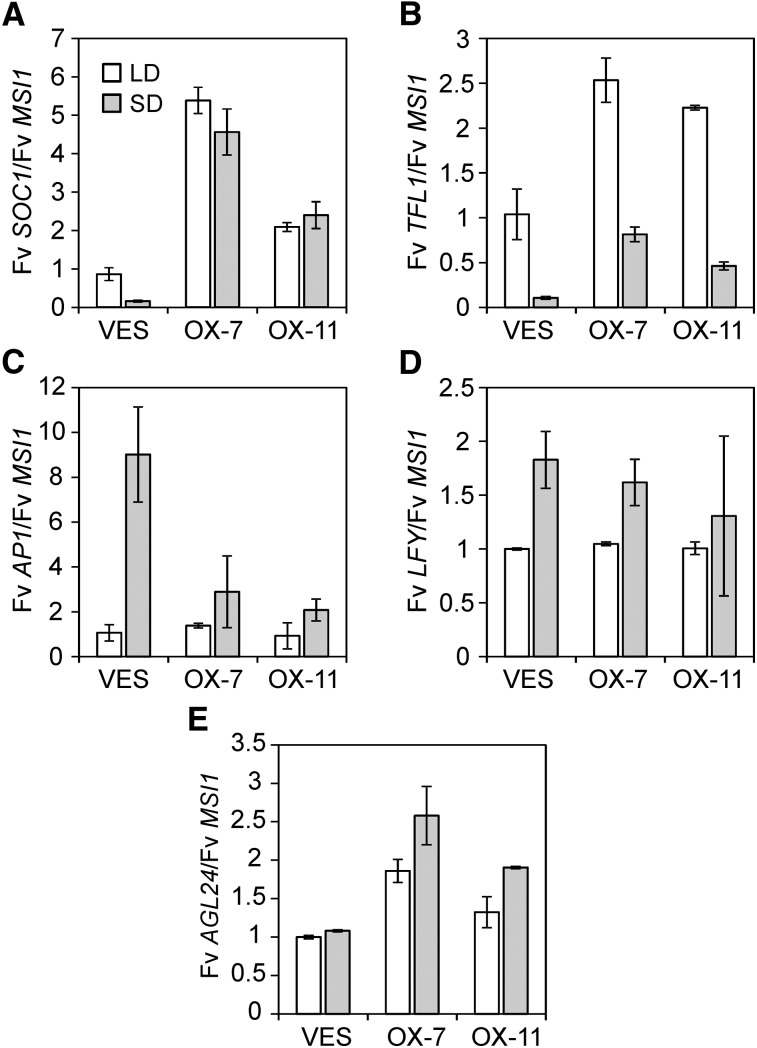
Fv SOC1 Overexpression Represses Flowering in Strawberry
We also overexpressed Fv SOC1 under the 35S promoter in SD strawberry (Fv SOC1-OX). We selected transgenic lines based on Fv SOC1 expression levels (see Supplemental Table 1 online) and subjected clonally propagated plants to SD treatment for 45 d followed by standard LDs. Only a few Fv SOC1-OX plants flowered after SD treatment, whereas all SD-grown nontransgenic control plants flowered and produced several inflorescences (Table 1). Moreover, opposite to Fv SOC1-RNAi plants, all LD-grown Fv SOC1-OX plants and nontransgenic control plants remained vegetative. SDs strongly repressed Fv SOC1 and Fv TFL1 expression in the shoot apices of nontransgenic control plants, whereas the expression of floral marker Fv AP1 was ninefold upregulated in SDs compared with LDs, correlating with flowering (Figures 4A to 4C). In Fv SOC1-OX lines, consistent with their flowering phenotypes, upregulation of Fv TFL1 was found in both photoperiods compared with control plants grown under the same photoperiods. However, in contrast with nontransgenic SD strawberry, Fv SOC1 and Fv TFL1 expression levels did not fully correlate in transgenic lines (Figures 4A and 4B). Although Fv SOC1 expression remained at high levels in Fv SOC1-OX lines in both photoperiods, clear downregulation of Fv TFL1 mRNA levels by SDs were still seen in these plants, suggesting the presence of both Fv SOC1-dependent and -independent regulatory mechanisms of Fv TFL1. In Arabidopsis, AP1 is known to downregulate TFL1 (Liljegren et al., 1999; Ratcliffe et al., 1999), and we observed mild increases of Fv AP1 mRNA levels in parallel with reduced Fv TFL1 expression levels in SD-grown Fv SOC1-OX lines compared with LD-grown plants. The analysis of Fv SOC1-OX plants further supported the hypothesis that Fv SOC1 activates Fv TFL1 to repress flowering in SD strawberry. However, based on these data, the role of Fv AP1 as a negative regulator of Fv TFL1 cannot be excluded.
Table 1. Overexpression of Fv SOC1 Represses Flowering in Strawberry.
| Transgenic Line | Flowering Plants (Total Plants) |
Inflorescences (n/Plant) |
||
|---|---|---|---|---|
| LD | SD | LD | SD | |
| VES | 0 (9) | 7 (7) | 0 | 3.7 ± 0.5 |
| OX-7 | 0 (10) | 1 (10) | 0 | 0.1 ± 0.1 |
| OX-10 | 0 (10) | 0 (10) | 0 | 0 |
| OX-11 | 0 (10) | 2 (10) | 0 | 0.3 ± 0.2 |
Five-week-old plants of SD strawberry (VES) and three independent Fv SOC1-OX lines (OX-7, OX-10, and OX-11) were subjected to SD or LD treatments for 6 weeks followed by standard LD growing conditions, and their flowering phenotypes were observed. The number of inflorescences (n/plant) is indicated as mean ± se.
Figure 4.
Overexpression of Fv SOC1 Activates Fv TFL1 and Represses Fv AP1.
Relative expression of Fv SOC1 (A), Fv TFL1 (B), Fv AP1 (C), Fv LFY (D), and Fv AGL24 (E) in shoot apices of SD strawberry (VES) and Fv SOC1-OX lines #7 and #11. Clonally propagated 5-week-old plants were grown for 4 weeks under LDs or SDs before sampling. Mean fold change was calculated relative to LD-grown SD strawberry. Error bars represent se; n = 2.
We also analyzed the mRNA levels of Fv LFY and Fv AGL24, since SOC1 has been reported to bind directly to the promoters of LFY and AGL24 to activate their expression and to advance flowering in Arabidopsis (Lee et al., 2008; Liu et al., 2008). Constitutive overexpression of Fv SOC1 did not affect Fv LFY mRNA expression in the SD strawberry background, whereas Fv AGL24 tended to be slightly upregulated in Fv SOC1-OX lines compared with nontransgenic SD strawberry (Figures 4D and 4E). However, gene expression analysis in Fv SOC1-RNAi plants suggested that Fv SOC1 does not regulate Fv AGL24 or Fv LFY (see Supplemental Figure 10 online).
Fv SOC1 Does Not Repress Flowering in Hawaii-4
Since our results indicated that Fv SOC1 regulates flowering through Fv TFL1, we explored whether Fv SOC1 regulates flowering time in H4, which lacks functional Fv TFL1 (Koskela et al., 2012). We transformed H4 plants with the Fv SOC1-RNAi or Fv SOC1-OX construct, selected transgenic lines according to Fv SOC1 expression levels (see Supplemental Table 1 online), and produced T1 seedlings by self-pollination. To observe the flowering time of T1 seedlings, the plants were subjected to SD and LD treatments for six weeks. As observed earlier by Koskela et al. (2012), H4 plants flowered earlier under LDs than under SDs. Fv SOC1-OX lines and H4 control plants produced equal number of leaves before developing a terminal inflorescence under LDs, whereas Fv SOC1-RNAi plants were late flowering compared with nontransgenic H4 (Figure 5A). Under SDs, however, no differences were found in the flowering time of Fv SOC1-RNAi lines and H4 control plants, whereas one overexpression line produced slightly more leaves than H4. Our results, which show that Fv SOC1 does not repress flowering under LDs in the absence of functional Fv TFL1 in H4, in contrast with SD strawberry, which contains functional Fv TFL1, support the hypothesis that Fv SOC1 regulates flowering upstream of Fv TFL1. Moreover, the late flowering of Fv SOC1-RNAi lines under LDs suggests that Fv SOC1 is required for the LD flowering response in H4.
Figure 5.
Silencing of Fv SOC1 Delays Flowering in Hawaii-4 under LDs.
(A) Flowering time of Fv SOC1-RNAi lines #1 and #3 and Fv SOC1-OX lines #1 and #11 in the Hawaii-4 (H4) background. T1 seedlings were subjected to LDs or SDs for 6 weeks followed by standard LD conditions. Flowering time was calculated as the total number of leaves formed in the main crown below the terminal inflorescence. Error bars represent se; n = 7 to 16. Horizontal bars represent statistically significant differences to H4 (separately for LD and SD); **P < 0.01 and ***P < 0.001.
(B) Relative expression of Fv AP1 in shoot apices of H4 and transgenic lines grown under LDs. Samples were collected at the three-leaf stage. Mean fold change was calculated relative to H4. Error bars represent se; n = 3.
To understand the molecular control of flowering time in H4 transgenic lines, we analyzed the expression of Fv AP1, Fv AGL24, and Fv LFY under LDs. Consistent with flowering time, we found similar Fv AP1 expression levels in the shoot apices of H4 and Fv SOC1-OX lines. However, Fv AP1 was downregulated in Fv SOC1-RNAi lines, indicating that Fv SOC1 may be required for the activation of Fv AP1 under LDs in H4 (Figure 5B). In contrast with Fv AP1, neither the expression of Fv AGL24 nor Fv LFY was affected in H4 transgenic lines (see Supplemental Figure 11 online). To verify that the mutation in H4 Fv TFL1 does not convert the repressor into an activator that is still upregulated by Fv SOC1, we analyzed transgenic lines overexpressing the mutated Fv TFL1 in SD strawberry and H4 backgrounds. Flowering was not advanced in these transgenic lines in either SD strawberry (see Supplemental Figure 12 online) or H4 (Koskela et al., 2012), ruling out the proposed new function of mutated Fv TFL1.
Fv SOC1 Regulates Vegetative Growth in Strawberry
We also monitored the vegetative development of Fv SOC1 transgenic lines in the SD strawberry background. Overexpression and RNAi silencing of Fv SOC1 affected vegetative vigor, as indicated by the length of the petioles (see Supplemental Figure 13 online). Moreover, Fv SOC1-OX plants grown in the greenhouse for more than a year became elongated, whereas Fv SOC1-RNAi plants had a bushy growth habit (see Supplemental Figure 8 online).
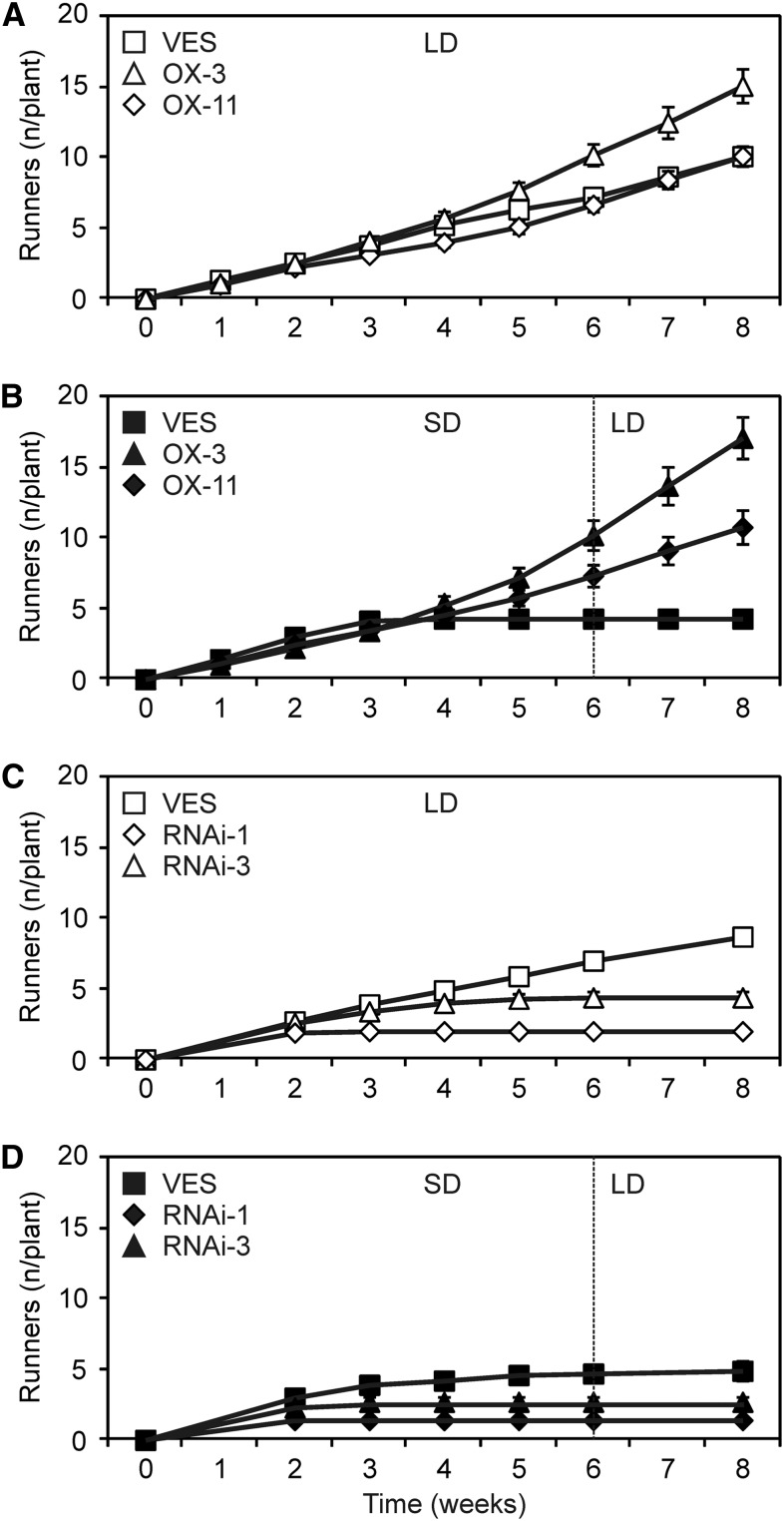
To examine the effect of Fv SOC1 on the photoperiodic regulation of vegetative development, we performed 6 weeks of photoperiodic treatment on clonally propagated plants of Fv SOC1-RNAi and Fv SOC1-OX lines in the SD strawberry background and observed runner formation. Nontransgenic control plants halted runner production after a few weeks under SDs, whereas Fv SOC1-OX plants formed runners continuously during the observation period, regardless of the photoperiod (Figures 6A and 6B). By contrast, plants of the strongest Fv SOC1-RNAi line stopped runner formation after 2 to 3 weeks in both SDs and LDs (Figures 6C and 6D). Also in the weaker Fv SOC1-RNAi line, runner formation ceased in both photoperiods, but this occurred a few weeks later in LDs compared with SDs. Consistent with reduced runner formation, most axillary buds developed into branch crowns in Fv SOC1-RNAi lines, and the plants became highly branched even under LDs (see Supplemental Figure 8 online). Moreover, Fv SOC1-OX lines produced only a few branch crowns even under SDs, where nontransgenic control plants formed several branch crowns (see Supplemental Figure 14 online). Taken together, in addition to the regulation of flowering, we conclude that Fv SOC1 is involved in the photoperiodic regulation of axillary bud differentiation into runners and branch crowns in SD strawberry.
Figure 6.
Fv SOC1 Enhances Runner Formation in Strawberry.
(A) and (B) Cumulative number of runners (n/plant) in clonally propagated plants of SD strawberry (VES) and Fv SOC1-OX lines #3 and #11. Five-week-old plants were grown under LDs (A) and under SDs for 6 weeks followed by LDs (B). Error bars represent se; n = 7 to 10.
(C) and (D) Cumulative number of runners in SD strawberry and Fv SOC1-RNAi lines #1 and #3. Five-week-old plants were grown under LDs (C) and under SDs for 6 weeks followed by LDs (D). Error bars represent se; n = 10.
Fv SOC1 Activates GA Biosynthesis
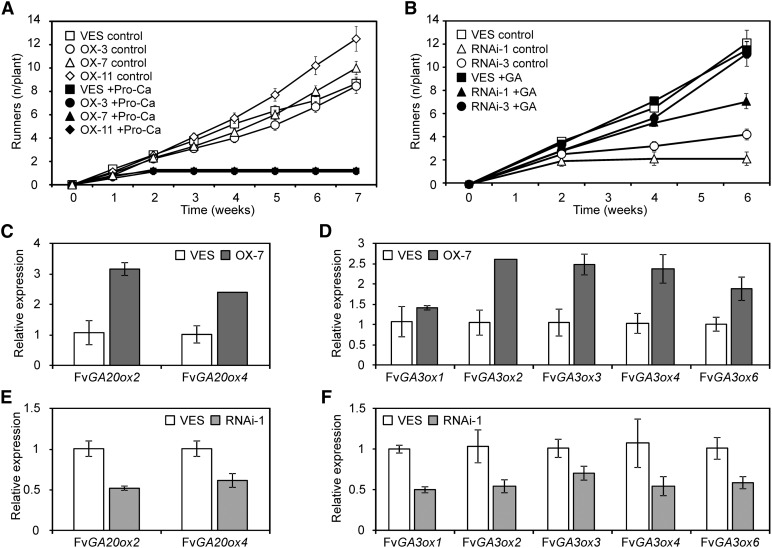
In strawberry, GA affects petiole elongation (Guttridge and Thompson, 1964; Wiseman and Turnbull, 1999) and promotes axillary bud differentiation into runners (Hytönen et al., 2009). To analyze if the enhanced vegetative growth of Fv SOC1-OX lines is GA dependent, we treated LD-grown plants with the inhibitor of GA biosynthesis prohexadione-calcium (Evans et al., 1999). We found that prohexadione-calcium completely inhibited runner formation in both SD strawberry and Fv SOC1-OX plants in the SD strawberry background (Figure 7A), confirming that a normal level of GA biosynthesis is also required for runner formation in Fv SOC1-OX lines. Next, we treated LD-grown Fv SOC1-RNAi and SD strawberry plants with GA3. Nontransgenic control plants produced runners continuously, and GA3 had no effect on runner formation (Figure 7B). However, runner formation ceased in nontreated RNAi plants within a few weeks, whereas GA3 strongly enhanced the production of new runners.
Figure 7.
Fv SOC1 Activates GA Biosynthesis to Promote Runner Growth in Strawberry.
(A) The effect of the inhibitor of GA biosynthesis, prohexadione-calcium (Pro-Ca), on runner formation in SD strawberry (VES) and Fv SOC1-OX lines. The cumulative number of runners (n/plant) is shown. Five-week-old plants were grown under LDs and treated with 100 ppm Pro-Ca or water (control) at week 0. Error bars represent se; n = 9 to 10.
(B) The effect of GA3 on runner formation in SD strawberry and Fv SOC1-RNAi lines. Cumulative number of runners is shown. Five-week-old plants were grown under LDs and treated with 50 ppm GA3 solution or mock treated (-GA) at weeks 0 and 2. Error bars represent se; n = 10 to 11; for RNAi #1, n = 5.
(C) and (D) Relative expression of Fv GA20ox (C) and Fv GA3ox (D) genes in the leaves of clonally propagated SD strawberry and Fv SOC1-OX line #7 grown under LDs. Mean fold change was calculated relative to the expression in SD strawberry separately for each gene. Error bars represent se; n = 2.
(E) and (F) Relative expression of Fv GA20ox (E) and Fv GA3ox (F) genes in the leaves of clonally propagated SD strawberry and Fv SOC1-RNAi line #1 grown under LDs. Mean fold change was calculated relative to the expression in SD strawberry separately for each gene. Error bars represent se; n = 3.
Growth regulator treatments of transgenic lines suggested that Fv SOC1 may activate GA biosynthesis to control vegetative development in SD strawberry. Therefore, we studied the mRNA levels of GA biosynthetic gene homologs (Hedden and Thomas, 2012; Kang et al., 2013). We found opposite changes in the expression of several genes in Fv SOC1-OX and Fv SOC1-RNAi lines in the SD strawberry background. Two putative GA20-oxidase (GA20ox) and four GA3ox homologs were activated in Fv SOC1-OX plants (Figures 7C and 7D). By contrast, an opposite trend in the expression of these genes was observed in the strong Fv SOC1-RNAi line #1 (Figures 7E and 7F). In addition, the mRNA level of the strawberry homolog of GA2ox, which encodes a GA degradation enzyme (Hedden and Thomas, 2012), was slightly increased in Fv SOC1-OX lines and decreased in the Fv SOC1-RNAi line (see Supplemental Figure 15 online).
Next, we analyzed the expression of the strawberry homolog of GIBBERELLIC ACID INSENSITIVE (GAI), which encodes a DELLA growth repressor in the GA pathway (Peng et al., 1997). Consistent with earlier results that genes encoding DELLA proteins are feed-forward regulated by GA (Hytönen et al., 2009; Hedden and Thomas, 2012), we found that Fv GAI was upregulated in Fv SOC1 overexpression plants compared with SD strawberry and, again, that the response was opposite in the RNAi line (see Supplemental Figure 15 online). Strawberry homologs of positive regulators of GA signaling, GIBBERELLIN INSENSITIVE DWARF1 and SLEEPY (Harberd et al., 2009), were not clearly affected in transgenic lines (see Supplemental Figure 15 online). In conclusion, our results indicate that Fv SOC1 may activate the expression of many genes of the GA biosynthetic pathway to regulate vegetative development in strawberry.
The Activation of Fv TFL1 by Fv SOC1 Is GA Independent
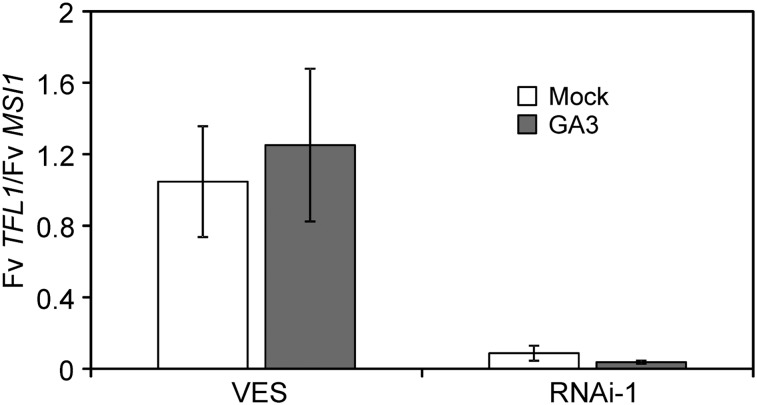
Our results indicated that Fv SOC1 promotes the expression of both Fv TFL1 and several genes of the GA biosynthetic pathway. This raised the question of whether Fv SOC1 upregulates Fv TFL1 through the GA pathway. In such a scenario, GA3 treatment should restore the normal Fv TFL1 expression level in Fv SOC1-RNAi plants in the SD strawberry background. Therefore, we treated SD strawberry and Fv SOC1-RNAi plants with GA3 and analyzed the expression of Fv TFL1 in the shoot apex samples. GA3 had no clear effect on the expression of Fv TFL1 in either RNAi or nontransgenic control plants (Figure 8; see Supplemental Figure 16 online), although it affected runner formation in RNAi plants (Figure 7B). Therefore, Fv SOC1 likely regulates vegetative and floral development in strawberry through GA-dependent and -independent pathways, respectively.
Figure 8.
GA Does Not Activate Fv TFL1.
Effect of GA3 on the relative expression of Fv TFL1 in SD strawberry (VES) and Fv SOC1-RNAi line #1. Shoot apex samples of plants grown under LDs for 5 weeks were collected 7 d after 50 ppm GA3 or mock treatment. Mean fold change was calculated relative to SD strawberry. Error bars represent se; n = 2.
DISCUSSION
Fv SOC1 Activates Fv TFL1 to Repress Flowering under LDs
SOC1 and SOC1-like genes encode MADS box transcription factors that are reported to function as floral activators in annual LD and SD plants (Menzel et al., 1996; Lee et al., 2000; Ferrario et al., 2004; Lee et al., 2004) and in a perennial species Cardamine flexuosa (Zhou et al., 2013). Here, we show that in strawberry, which is a seasonal flowering perennial SD plant (SD strawberry), the overexpression of the strawberry ortholog of SOC1 suppresses photoperiodic flowering, whereas Fv SOC1-RNAi plants flower continuously without inductive SD treatment, similarly to perpetual flowering mutants, which lack the functional floral repressor Fv TFL1 (Figures 3A to 3D; Iwata et al., 2012; Koskela et al., 2012). Our result that Fv SOC1 does not repress flowering under LDs in the perpetual flowering H4 genotype (Figure 5A) suggests that Fv SOC1 may repress flowering through Fv TFL1. Consistent with this idea, Fv TFL1 was upregulated in Fv SOC1-OX lines and downregulated in RNAi lines in the SD strawberry background, whereas the floral meristem identity gene Fv AP1 was oppositely regulated, correlating with flowering time. These results suggest that Fv SOC1 may activate Fv TFL1 to repress Fv AP1 and flowering under LDs in SD strawberry. Under SDs, however, both Fv SOC1 and Fv TFL1 are downregulated and flower induction occurs (Figures 2B to 2D; Koskela et al., 2012).
Guttridge and Thompson (1964) showed that exogenous GA application delays flowering in strawberry. Since we observed changes in the expression of several GA biosynthetic genes and GA-dependent vegetative phenotypes in Fv SOC1 transgenic lines in the SD strawberry background (see below), we tested if Fv SOC1 controls Fv TFL1 through GA. Although a recent study suggested that GA may activate the rose TFL1 homolog KOUSHIN (KSN) through GA responsive cis-elements, which are also present in the promoter of Fv TFL1 (Randoux et al., 2012), GA3 did not activate the expression of Fv TFL1 in our Fv SOC1-RNAi lines or in SD strawberry. These results suggest that GA is not involved in the regulation of Fv TFL1 mRNA expression. However, further studies are needed to explore whether changes in endogenous GAs have an effect on flowering time in Fv SOC1 RNAi and overexpression lines or in nontransgenic strawberry.
In Arabidopsis, TFL1 regulates both flowering time and inflorescence architecture (Bradley et al., 1997). TFL1 is highly expressed in the center of the inflorescence meristem in order to maintain indeterminacy of the meristem. However, AP1 and LFY downregulate TFL1 in the flanks of the inflorescence meristem to specify floral meristems (Liljegren et al., 1999; Ratcliffe et al., 1999). AP1 binds to the MADS box element downstream of the TFL1 coding sequence (Kaufmann et al., 2010), and a recent report showed that SOC1 is involved in the AP1-dependent regulation of TFL1 homologs in both Arabidopsis and rice (Liu et al., 2013). Our time-course gene expression analysis suggests that at least initial downregulation of Fv TFL1 in the shoot apex of SD strawberry does not depend on the activation of floral meristem identity genes. In SD strawberry, Fv TFL1 is downregulated after only 2 weeks of SD in parallel with Fv SOC1, but both Fv AP1 and Fv FUL1 are unaffected by SDs at this time point (Figure 2D; Koskela et al., 2012). However, Fv TFL1 mRNA levels still decrease during the following two weeks of SDs, when Fv AP1 and flowering, but not Fv FUL1, are induced (Heide and Sønsteby, 2007; Koskela et al., 2012). Therefore, the downregulation of Fv SOC1 by SDs may cause an initial decrease of Fv TFL1 mRNA levels independently of Fv AP1, but the role of Fv AP1 in the later stages cannot be excluded. Strawberry produces a cymose inflorescence, which does not have an indeterminate inflorescence meristem (Jahn and Dana, 1970), suggesting that the possible role of Fv AP1 in the regulation of Fv TFL1 is at least spatially different from that of the homologs in Arabidopsis. In addition, Fv LFY may play a role in later floral development, since its mRNA expression follows Fv AP1 with a delay in the shoot apex (Mouhu et al., 2009).
The promoter region of Fv TFL1 contains two predicted binding sites of MADS domain transcription factors called CArG boxes 2178 bp (TT[ACTTTTTAGT]C) and 1159 bp (TT[TCTTTTATGG]CAAA) upstream of Fv TFL1 start codon, supporting the hypothesis that Fv SOC1 may directly bind to the Fv TFL1 promoter. Furthermore, the latter putative CArG box is almost identical with the SOC1 binding site in the AGL24 promoter (Liu et al., 2008) and has an adjacent triple AAA, which is probably required for MADS box protein binding (Deng et al., 2011; Tao et al., 2012). A detailed study of these promoter elements should be performed to reveal their potential role in the regulation of Fv TFL1 mRNA expression.
The Function of Fv SOC1 Depends on Fv TFL1
In contrast with SD strawberry, perpetual flowering accessions containing nonfunctional Fv TFL1 flower earlier under LDs than under SDs (Sønsteby and Heide, 2008; Mouhu et al., 2009). We previously reported that Fv FT1 promotes flowering in H4 under LDs and proposed that in SD strawberry, this LD promotion pathway is masked by the repressor Fv TFL1 (Koskela et al., 2012). Our results for Fv SOC1-RNAi lines in the H4 background further support the presence of a LD-promoting pathway. In H4, opposite to SD strawberry, RNAi silencing of Fv SOC1 caused reduced Fv AP1 mRNA levels and delayed flowering specifically under LD conditions. However, the overexpression of Fv SOC1 had no effect on flowering time under LDs, suggesting that Fv SOC1 mRNA levels higher than a specific threshold have no additional effect on flowering time. We also found that the RNAi silencing of Fv FT1 in the H4 background downregulated Fv SOC1 and Fv AP1 (Figure 2E; Koskela et al., 2012), indicating that Fv FT1 may function upstream of Fv SOC1 to promote flowering in H4, similarly to FT and SOC1 in Arabidopsis (Samach et al., 2000; Moon et al., 2005; Yoo et al., 2005). Given that Fv FT1 activates Fv SOC1 in H4, it may also upregulate Fv TFL1 through Fv SOC1 in SD strawberry. Therefore, the activation of Fv TFL1 by Fv SOC1 may account for the divergence in the photoperiodic pathways between the LD plant Arabidopsis and the SD plant strawberry. In this scenario, H4, which lacks functional Fv TFL1, uses the default pathway. This default pathway may be present also in SD strawberry, but it is probably masked by the activation of Fv TFL1 by Fv SOC1. Also in Arabidopsis, the upregulation of TFL1 by FT was recently predicted by modeling, and a clear positive correlation was found in the expression of these genes (Jaeger et al., 2013), but the functional significance of this interaction remains to be shown.
How Fv SOC1 regulates flowering in H4 is an open question. In Arabidopsis, SOC1 directly activates the expression of LFY and AGL24 by binding to their promoters (Moon et al., 2005; Lee et al., 2008; Liu et al. 2008), but we did not find clear evidence that Fv SOC1 would regulate strawberry LFY and AGL24 homologs. Similarly, in mustard (Sinapis alba) and evergreen azalea (Rhododendron × pulchrum), SOC1 and LFY homologs are expressed independently of each other (D’Aloia et al., 2009; Cheon et al., 2012). One possibility is that Fv SOC1 directly activates Fv AP1. However, Fv SOC1 is likely to have several roles in the floral regulatory pathways in strawberry, since SOC1 interacts with several other MADS box proteins and binds to the promoters of multiple floral regulators in Arabidopsis (de Folter et al., 2005; van Dijk et al., 2010; Immink et al., 2012; Tao et al., 2012).
Fv SOC1 Regulates Vegetative Development by Activating GA Biosynthesis
In strawberries, flower initiation is tightly connected to changes in vegetative development within yearly growth cycles (Hytönen and Elomaa, 2011). Under LDs, strawberries continuously produce runners from axillary buds, whereas under flower inductive SDs, axillary buds differentiate into branch crowns (Konsin et al., 2001; Hytönen et al., 2004, 2009). We show here that in addition to its role in the regulation of flowering, Fv SOC1 plays a role in the photoperiodic regulation of vegetative development. While runner formation in SD strawberry ceased after a few weeks of SDs, Fv SOC1-OX plants in the SD strawberry background continued to produce runners under these conditions. By contrast, Fv SOC1-RNAi lines were dwarfed, with several branch crowns. Young RNAi plants formed a few runners before they completely stopped runner development in both LDs and SDs. Since both Fv SOC1-OX and RNAi lines lost the photoperiodic response of vegetative development, we reason that normal photoperiodic control of Fv SOC1 may be crucial for vegetative responses. However, we showed earlier that Fv TFL1 has no effect on the photoperiodic regulation of axillary bud differentiation to runners or branch crowns (Koskela et al., 2012); therefore, Fv SOC1 may regulate this process through other genes. RUNNERING LOCUS, which is an unknown but dominant regulator of runner formation (Brown and Wareing, 1965), is one candidate for such a gene.
Vegetative phenotypes of Fv SOC1 transgenic lines suggest that Fv SOC1 may affect the activity of the GA biosynthetic pathway (Guttridge and Thompson, 1964; Wiseman and Turnbull, 1999; Hytönen et al., 2009). This hypothesis is supported by the findings that prohexadione-calcium, the inhibitor of GA biosynthesis, stopped runner formation in Fv SOC1-OX plants, and GA3 induced runner development in Fv SOC1-RNAi lines in the SD strawberry background. We also revealed the role of Fv SOC1 as the putative regulator of GA biosynthetic genes. In Fv SOC1-OX plants in the SD strawberry background, several strawberry homologs of GA20ox and GA3ox genes were activated and an opposite trend was found in Fv SOC1-RNAi plants. Since GA-20 oxidation is a rate-limiting step in GA biosynthesis (Appleford et al., 2006; Middleton et al., 2012), twofold to severalfold changes in the expression levels of Fv GA20ox genes in our transgenic lines is likely to affect GA levels and, consequently, growth. In Arabidopsis, SOC1 is known to mediate the effect of GA on flowering (Moon et al., 2003; Searle et al., 2006), but its role in the regulation of GA biosynthesis is not clear. In concordance with our findings, the expression of GA20ox1 is slightly downregulated in the Arabidopsis soc1 mutant (Dorca-Fornell et al., 2011), but the effect of this change on growth is unknown. SOC1 may also regulate the GA biosynthetic pathway by repressing TEMPRANILLO1 (Tao et al., 2012), which downregulates GA3ox1 and GA3ox2 by binding to their promoters (Osnato et al., 2012). Although our results strongly suggest that Fv SOC1 controls the GA biosynthetic pathway in strawberry, genetic experiments combining silencing of GA biosynthetic genes with Fv SOC1 overexpression or vice versa would be necessary to confirm our findings.
Seasonal Regulation of Vegetative and Generative Development in Strawberry
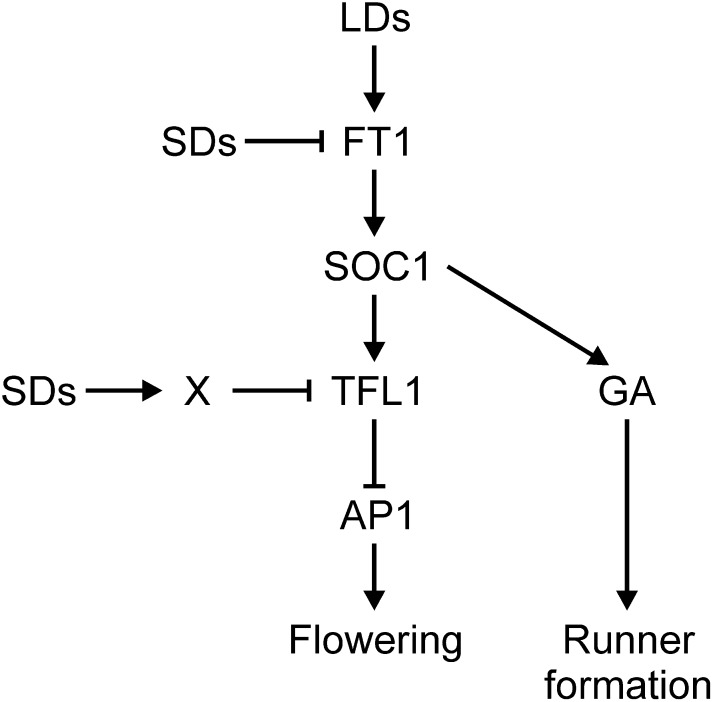
We recently presented a model suggesting that the seasonal regulation of Fv TFL1 mRNA expression regulates yearly flowering cycles in SD strawberry (Koskela et al., 2012). Here, we extend this genetic model and show evidence that branching of the flowering pathway may explain the seasonal regulation of vegetative development (Figure 9). We found that both Fv SOC1 and Fv TFL1 are highly expressed in the shoot apex under LDs, whereas under SDs, their gradual downregulation is followed by the induction of Fv AP1 and flowering. Under subsequent LDs, the expression of both Fv SOC1 and Fv TFL1 is reactivated in the newly emerged vegetative axillary shoots (Figure 2D; Koskela et al., 2012). Given that Fv SOC1 likely activates Fv TFL1 mRNA expression (Figures 3E, 3G, and 4B) and that Fv SOC1 is downregulated in Fv FT1 RNAi lines (Figure 2E), we propose that Fv FT1 may mediate the photoperiodic regulation of Fv SOC1, which regulates Fv TFL1 expression according to seasonal changes in photoperiod; therefore, flower initiation only occurs during short photoperiods in autumn, whereby plants flower the next spring. However, additional regulators are likely involved in the photoperiodic regulation of Fv TFL1 (indicated by x in Figure 9), or the activity of Fv SOC1 is modulated by light, since the overexpression of Fv SOC1 did not fully nullify the photoperiodic regulation of Fv TFL1. Our data indicate that the photoperiod sensing and transmission pathway is shared between Arabidopsis and strawberry. However, the divergence of the response pathway downstream of SOC1 orthologs may cause opposite photoperiodic flowering responses in Arabidopsis and SD strawberry.
Figure 9.
Model Showing the Photoperiodic Regulation of Flowering and Runner Formation in Strawberry.
Arrows indicate activation, and bars indicate repression.
Independently of its effect on flowering, Fv SOC1 may also promote GA biosynthesis (Figures 7 and 9), which is involved in the photoperiodic regulation of axillary bud differentiation to runners or branch crowns (Hytönen et al., 2009). The LD activation of Fv SOC1 could increase GA levels in summer to promote runner development, whereas the downregulation of Fv SOC1 and the GA biosynthetic pathway in autumn could cause the differentiation of axillary buds into branch crowns. In conclusion, we revealed the role of Fv SOC1 as a central signaling hub that regulates photoperiodic development in strawberry. A detailed analysis of Fv SOC1/GA and Fv SOC1/Fv TFL1 pathways may open new possibilities to control vegetative and generative development in strawberries and other rosaceous crops.
METHODS
Plant Material
Strawberry (Fragaria vesca) experiments were performed with seasonal flowering SD strawberry and perpetual flowering accession Hawaii-4 (H4). SD strawberry (PI551792) was obtained from the National Clonal Germplasm Repository, Corvallis, OR, and H4 (National Clonal Germplasm Repository accession number PI551572) was provided by Kevin Folta. Either seedlings or plants clonally propagated from runner cuttings were used for the experiments as indicated in the text and figure legends. For Arabidopsis thaliana experiments, Columbia-0 ecotype was used.
Growth Conditions and Phenotyping
Plants were grown in a greenhouse at the University of Helsinki, Finland. The greenhouse temperature was 18 ± 2°C, and plants were illuminated by high-pressure sodium lamps (HPS; Airam 400 W) for 18 h daily with a light intensity of ∼150 µmol m−2 s−1 (standard LD growing conditions). Photoperiodic experiments were conducted in greenhouse rooms during the winter season (October to March) when the natural light irradiance is low in Finland. In the photoperiodic treatments, strawberry plants were subjected to 12-h SD and 18-h LD photoperiods. In both photoperiods, plants were illuminated with 12 h of HPS (∼150 µmol m−2 s−1) light. In the LD treatment, 12 h of HPS illumination was extended with low-intensity incandescent light (∼8 µmol m−2 s−1) for 6 h in the evening. Darkening curtains were used to exclude any additional light during daylength extension treatment and to ensure darkness during the night. Temperature in photoperiodic experiments was constant 18 ± 1°C. Flowering time was measured either as the number of leaves initiated in the main crown below the terminal inflorescence or as the number of days to the first open flower. For Arabidopsis flowering observations, plants were grown under 8-h SD or 18-h LD at 20 ± 1°C.
Growth Regulator Treatments.
For prohexadione-calcium (BAS125; BASF) treatment, 100 mg L−1 solution was prepared in milli-Q water. GA3 (Duchefa) was first dissolved in ethanol at a concentration of 5 µg µL−1, and a 50 mg L−1 dilution was made in milli-Q water (Millipore). Growth regulators were sprayed on clonally propagated plants until drip-off. Prohexadione-calcium treatment was performed once and GA3 treatment twice with a 14-d interval. Mock treatments without growth regulators were performed for control plants.
Expression Analysis
Leaf and shoot apex samples were frozen in liquid nitrogen and stored at −70°C before total RNA was extracted using a modification of the pine tree method (Monte and Somerville, 2002). cDNAs were synthesized from 1 µg of total RNA using Superscript III reverse transcriptase (Invitrogen). SYBR Green Master (Roche) was used for real-time PCR reactions, which were performed in the Light Cycler 480 (Roche) instrument as described previously (Mouhu et al., 2009). Real-time PCR reactions were performed with three technical replicates and two to four biological replicates as mentioned in the figure legends. Relative expression levels were calculated by the ΔΔCt (cycle threshold) method with Fv MSI1 as the normalization gene as described previously (Mouhu et al., 2009). Primers used in the real-time PCR are listed in Supplemental Table 2 online. Primer efficiencies were almost equal for all primer pairs.
In Situ Hybridization
In situ hybridization was performed on longitudinal sections of the apex of the main shoot as described previously (Kurokura et al., 2006). To avoid cross-hybridization, the probe was designed in the 3′-region of the gene, which is less conserved among MADS box transcription factor encoding genes. Fv SOC1 probe template fragment was amplified by RT-PCR using forward and reverse primers 5′-GAAGGCACAGGTTTTCAAGG-3′ and 5′-CAGCCTTGGCTTGGATAGAG-3′, respectively, and cloned into the pDrive cloning vector (Qiagen). Antisense probe was synthesized using the SP6 promoter (Qiagen). Fv TFL1 sense probe described earlier was used as a control (Koskela et al., 2012).
Plasmid Constructs
Plasmid constructs for overexpression and RNAi silencing lines were created according to Gateway technology with Clonase II (Invitrogen). For Fv SOC1 overexpression and RNAi constructs, cDNA from F. vesca var semperflorens 'Baron Solemacher' was amplified with primer pairs 5′-AAAAAGCAGGCTGGTTGCGCTCATAATCTTCTCT-3′ (attB1) and 5′-AGAAAGCTGGGTTGTTCACACTCCTCTCCAACTG-3′ (attB2), and 5′-AAAAAGCAGGCTTCTCAAGAACTTGCTGGGTTCA-3′ (attB1) and 5′-AGAAAGCTGGGTGCTAGTGCTTCGATCTCCTTTCTG-3′ (attB2), respectively. The construct reported by Koskela et al. (2012) was used to overexpress mutated Fv TFL1 in SD strawberry. The destination vectors were p7WG2D for overexpression and PK7GWIWG2(II) for RNAi silencing (Karimi et al., 2002). Both vectors contain green fluorescent protein as a positive selection marker.
Transformation
Vectors carrying overexpression and RNAi constructs were electroporated into Agrobacterium tumefaciens strain GV3101 and transformed into SD strawberry and H4 as described previously (Oosumi et al., 2006). Several transgenic lines were generated for both constructs in each genetic background. Transgenic lines were selected for the experiments based on their phenotypes and Fv SOC1 expression levels. Arabidopsis Columbia-0 ecotype was transformed using the floral dip method, and transgenic T1 seedlings were selected based on green fluorescent protein fluorescence (Zhang et al., 2006).
Statistical Analyses
When appropriate, averages were subjected to analysis of variance using the general linear model procedure and differences between means were tested using contrasts in the SAS/STAT software (version 9.2 of the SAS System for Windows; SAS Institute).
Phylogenetic Analysis
Maximum likelihood phylogenetic analysis was performed to gain insight into the relationships among SOC1-like genes. Amino acid sequences of Fv SOC1 and other selected eudicots were aligned using Muscle (Edgar, 2004). A single most-optimal tree was computed using the randomized axelerated maximum likelihood BlackBox Web server (Stamatakis et al., 2008). Default settings were used with a gamma distribution and the Whelan and Goldman model of molecular evolution. One hundred bootstrap samples were generated to assess support for the inferred relationships.
Accession Numbers
Sequence data from this article can be found in the GenBank/National Center for Biotechnology Information data library under the following accession numbers: Fv SOC1 (FJ531999), Fv TFL1 (JN172097), Fv FT1 (JN172098), and Fv LFY (FJ532000). Predicted gene models (Hybrid V2) can be found in the Genome Database for Rosaceae (http://www.rosaceae.org): Fv AP1 (gene04564), Fv AGL24 (gene30741), Fv MSI1 (gene03001), Fv GA20ox2 (gene19438), Fv GA20ox4 (gene09034), Fv GA3ox1 (gene06004), Fv GA3ox2 (gene01056), Fv GA3ox3 (gene01058), Fv GA3ox4 (gene01059), and Fv GA3ox6 (gene11192).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Maximum Likelihood Tree of Amino Acid Sequences Showing Phylogenetic Relationships of SOC1-Like Proteins in Eudicots.
Supplemental Figure 2. Syntenic Analysis of Fv SOC1 and SOC1 from Arabidopsis.
Supplemental Figure 3. The Alignment of SOC1-Like Proteins.
Supplemental Figure 4. Overexpression of Fv SOC1 Advances Flowering in Arabidopsis.
Supplemental Figure 5. Diurnal Rhythm of Fv SOC1 mRNA Expression.
Supplemental Figure 6. Localization of Fv SOC1 mRNA in the Shoot Apex.
Supplemental Figure 7. The Expression of Fv SOC1 in Fv TFL1 Transgenic Lines.
Supplemental Figure 8. Phenotypes of Strong Fv SOC1-OX and RNAi Lines.
Supplemental Figure 9. The Expression of Fv AP1 in Young Fv SOC1-RNAi Plants.
Supplemental Figure 10. The Expression of Fv AGL24 and Fv LFY in Fv SOC1-RNAi Plants.
Supplemental Figure 11. The Expression of Fv AGL24 and Fv LFY in Fv SOC1 Transgenic Lines in the Hawaii-4 Background.
Supplemental Figure 12. Overexpression of Mutated Fv TFL1 Does Not Affect Flowering Time in Fragaria vesca.
Supplemental Figure 13. Fv SOC1 Regulates Vegetative Growth in Fragaria vesca.
Supplemental Figure 14. Overexpression of Fv SOC1 Suppresses the Formation of Branch Crowns in Fragaria vesca.
Supplemental Figure 15. The Effect of Fv SOC1 on the Expression of Gibberellin Pathway Genes in Fragaria vesca.
Supplemental Figure 16. The Effect of GA3 Treatment on the Expression of Fv TFL1.
Supplemental Table 1. Relative Expression of Fv SOC1 in Leaves of Fv SOC1-RNAi and OX Lines.
Supplemental Table 2. List of Primers Used in Real-Time PCR Reactions.
Supplemental Data Set 1. Alignments Used to Generate the Phylogeny in Supplemental Figure 1 Online.
Acknowledgments
We thank Lilia Sarelainen for technical assistance in genetic transformation and Teemu Teeri for help in generation of the sequence alignments for the phylogenetic analysis. The work was funded by the Academy of Finland (Grant 137439) and the University of Helsinki (Grant DW-4881545211) to T.H. K.M. belongs to Finnish Doctoral Program in Plant Science and E.A.K. to the Viikki Doctoral Program in Molecular Biosciences.
AUTHOR CONTRIBUTIONS
K.M., T.K., P.E., and T.H. designed the experiments. K.M., T.K., E.A.K., V.A.A., and T.H. performed the research. K.M. and T.H. wrote the article with input from all the authors.
Glossary
- GA
gibberellin
- SD
short-day
- LD
long-day
- RNAi
RNA interference
- HPS
high-pressure sodium
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Albani M.C., Battey N.H., Wilkinson M.J. (2004). The development of ISSR-derived SCAR markers around the SEASONAL FLOWERING LOCUS (SFL) in Fragaria vesca. Theor. Appl. Genet. 109: 571–579 [DOI] [PubMed] [Google Scholar]
- An H., Roussot C., Suárez-López P., Corbesier L., Vincent C., Piñeiro M., Hepworth S., Mouradov A., Justin S., Turnbull C., Coupland G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Appleford N.E.J., Evans D.J., Lenton J.R., Gaskin P., Croker S.J., Devos K.M., Phillips A.L., Hedden P. (2006). Function and transcript analysis of gibberellin-biosynthetic enzymes in wheat. Planta 223: 568–582 [DOI] [PubMed] [Google Scholar]
- Böhlenius H., Huang T., Charbonnel-Campaa L., Brunner A.M., Jansson S., Strauss S.H., Nilsson O. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312: 1040–1043 [DOI] [PubMed] [Google Scholar]
- Bradley D., Ratcliffe O., Vincent C., Carpenter R., Coen E. (1997). Inflorescence commitment and architecture in Arabidopsis. Science 275: 80–83 [DOI] [PubMed] [Google Scholar]
- Brown T., Wareing P.F. (1965). Genetical control of everbearing habit and 3 other characters in varieties of Fragaria vesca. Euphytica 14: 97–112 [Google Scholar]
- Cheon K., Nakatsuka A., Tasaki K., Kobayashi N. (2012). Seasonal changes in the expression pattern of flowering-related genes in evergreen azalea ‘Oomurasaki’ (Rhododendron × pulchrum). Sci. Hortic. (Amsterdam) 134: 176–184 [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- D’Aloia M., Tamseddak K., Bonhomme D., Bonhomme F., Bernier G., Périlleux C. (2009). Gene activation cascade triggered by a single photoperiodic cycle inducing flowering in Sinapis alba. Plant J. 59: 962–973 [DOI] [PubMed] [Google Scholar]
- de Folter S., Immink R.G.H., Kieffer M., Parenicová L., Henz S.R., Weigel D., Busscher M., Kooiker M., Colombo L., Kater M.M., Davies B., Angenent G.C. (2005). Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Ying H., Helliwell C.A., Taylor J.M., Peacock W.J., Dennis E.S. (2011). FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 6680–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorca-Fornell C., Gregis V., Grandi V., Coupland G., Colombo L., Kater M.M. (2011). The Arabidopsis SOC1-like genes AGL42, AGL71 and AGL72 promote flowering in the shoot apical and axillary meristems. Plant J. 67: 1006–1017 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J.R., Evans R.R., Regusci C.L., Rademacher W. (1999). Mode of action, metabolism, and uptake of BAS 125W, prohexadione-calcium. HortScience 34: 1200–1201 [Google Scholar]
- Ferrario S., Busscher J., Franken J., Gerats T., Vandenbussche M., Angenent G.C., Immink R.G.H. (2004). Ectopic expression of the petunia MADS box gene UNSHAVEN accelerates flowering and confers leaf-like characteristics to floral organs in a dominant-negative manner. Plant Cell 16: 1490–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttridge C.G., Thompson P.A. (1964). Effect of gibberellins on growth and flowering of Fragaria and Duchesnea. J. Exp. Bot. 15: 631–646 [Google Scholar]
- Hanano S., Goto K. (2011). Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23: 3172–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd N.P., Belfield E., Yasumura Y. (2009). The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: How an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R., Yokoi S., Tamaki S., Yano M., Shimamoto K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722 [DOI] [PubMed] [Google Scholar]
- Hecht V., Foucher F., Ferrándiz C., Macknight R., Navarro C., Morin J., Vardy M.E., Ellis N., Beltrán J.P., Rameau C., Weller J.L. (2005). Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 137: 1420–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V., Laurie R.E., Vander Schoor J.K., Ridge S., Knowles C.L., Liew L.C., Sussmilch F.C., Murfet I.C., Macknight R.C., Weller J.L. (2011). The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell 23: 147–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P., Thomas S.G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444: 11–25 [DOI] [PubMed] [Google Scholar]
- Heide O.M. (1977). Photoperiod and temperature interactions in growth and flowering of strawberry. Physiol. Plant. 40: 21–26 [Google Scholar]
- Heide O.M., Sønsteby A. (2007). Interactions of temperature and photoperiod in the control of flowering of latitudinal and altitudinal populations of wild strawberry (Fragaria vesca). Physiol. Plant. 130: 280–286 [Google Scholar]
- Hsu C.Y., et al. (2011). FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl. Acad. Sci. USA 108: 10756–10761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hytönen T., Elomaa P. (2011). Genetic and environmental regulation of flowering and runnering in strawberries. Genes Genomes Genomics 5: 56–64 [Google Scholar]
- Hytönen T., Elomaa P., Moritz T., Junttila O. (2009). Gibberellin mediates daylength-controlled differentiation of vegetative meristems in strawberry (Fragaria x ananassa Duch). BMC Plant Biol. 9: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hytönen T., Palonen P., Mouhu K., Junttila O. (2004). Crown branching and cropping potential in strawberry (Fragaria x ananassa Duch.) can be enhanced by day-length treatments. J. Hortic. Sci. Biotechnol. 79: 466–471 [Google Scholar]
- Immink R.G.H., Posé D., Ferrario S., Ott F., Kaufmann K., Valentim F.L., de Folter S., van der Wal F., van Dijk A.D.J., Schmid M., Angenent G.C. (2012). Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol. 160: 433–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata H., Gaston A., Remay A., Thouroude T., Jeauffre J., Kawamura K., Oyant L.H.-S., Araki T., Denoyes B., Foucher F. (2012). The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J. 69: 116–125 [DOI] [PubMed] [Google Scholar]
- Jaeger K.E., Pullen N., Lamzin S., Morris R.J., Wigge P.A. (2013). Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell 25: 820–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn O.L., Dana M.N. (1970). Crown and inflorescence development in the strawberry, Fragaria ananassa. Am. J. Bot. 57: 607–612 [Google Scholar]
- Jung J.H., Ju Y., Seo P.J., Lee J.H., Park C.M. (2012). The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 69: 577–588 [DOI] [PubMed] [Google Scholar]
- Kang C., Darwish O., Geretz A., Shahan R., Alkharouf N., Liu Z. (2013). Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell 25: 1960–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Wellmer F., Muiño J.M., Ferrier T., Wuest S.E., Kumar V., Serrano-Mislata A., Madueño F., Krajewski P., Meyerowitz E.M., Angenent G.C., Riechmann J.L. (2010). Orchestration of floral initiation by APETALA1. Science 328: 85–89 [DOI] [PubMed] [Google Scholar]
- Kim D.-H., Doyle M.R., Sung S., Amasino R.M. (2009). Vernalization: Winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 25: 277–299 [DOI] [PubMed] [Google Scholar]
- Konsin M., Voipio I., Palonen P. (2001). Influence of photoperiod and duration of short-day treatment on vegetative growth and flowering of strawberry (Fragaria X ananassa duch). J. Hortic. Sci. Biotechnol. 76: 77–82 [Google Scholar]
- Koskela E.A., Mouhu K., Albani M.C., Kurokura T., Rantanen M., Sargent D.J., Battey N.H., Coupland G., Elomaa P., Hytönen T. (2012). Mutation in TERMINAL FLOWER1 reverses the photoperiodic requirement for flowering in the wild strawberry Fragaria vesca. Plant Physiol. 159: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokura T., Inaba Y., Sugiyama N. (2006). Histone H4 gene expression and morphological changes on shoot apices of strawberry (Fragaria × ananassa Duch.) during floral induction. Sci. Hortic. (Amsterdam) 110: 192–197 [Google Scholar]
- Lee H., Suh S.S., Park E., Cho E., Ahn J.H., Kim S.G., Lee J.S., Kwon Y.M., Lee I. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Oh M., Park H., Lee I. (2008). SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy. Plant J. 55: 832–843 [DOI] [PubMed] [Google Scholar]
- Lee S., Kim J., Han J.J., Han M.J., An G. (2004). Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J. 38: 754–764 [DOI] [PubMed] [Google Scholar]
- Li D., Liu C., Shen L., Wu Y., Chen H., Robertson M., Helliwell C.A., Ito T., Meyerowitz E., Yu H. (2008). A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Liljegren S.J., Gustafson-Brown C., Pinyopich A., Ditta G.S., Yanofsky M.F. (1999). Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell 11: 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Chen H., Er H.L., Soo H.M., Kumar P.P., Han J.H., Liou Y.C., Yu H. (2008). Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135: 1481–1491 [DOI] [PubMed] [Google Scholar]
- Liu C., Teo Z.W.N., Bi Y., Song S., Xi W., Yang X., Yin Z., Yu H. (2013). A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Dev. Cell 24: 612–622 [DOI] [PubMed] [Google Scholar]
- Lyons E., Freeling M. (2008). How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 53: 661–673 [DOI] [PubMed] [Google Scholar]
- Menzel G., Apel K., Melzer S. (1996). Identification of two MADS box genes that are expressed in the apical meristem of the long-day plant Sinapis alba in transition to flowering. Plant J. 9: 399–408 [DOI] [PubMed] [Google Scholar]
- Middleton A.M., Úbeda-Tomás S., Griffiths J., Holman T., Hedden P., Thomas S.G., Phillips A.L., Holdsworth M.J., Bennett M.J., King J.R., Owen M.R. (2012). Mathematical modeling elucidates the role of transcriptional feedback in gibberellin signaling. Proc. Natl. Acad. Sci. USA 109: 7571–7576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte, D., and Somerville, S. (2002). Pine tree method for isolation of plant RNA. In DNA Microarrays: A Molecular Cloning Manual, D. Bowtell and J. Sambrook, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 124–126. [Google Scholar]
- Moon J., Lee H., Kim M., Lee I. (2005). Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol. 46: 292–299 [DOI] [PubMed] [Google Scholar]
- Moon J., Suh S.S., Lee H., Choi K.R., Hong C.B., Paek N.C., Kim S.G., Lee I. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Mouhu K., Hytönen T., Folta K., Rantanen M., Paulin L., Auvinen P., Elomaa P. (2009). Identification of flowering genes in strawberry, a perennial SD plant. BMC Plant Biol. 9: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutasa-Göttgens E., Hedden P. (2009). Gibberellin as a factor in floral regulatory networks. J. Exp. Bot. 60: 1979–1989 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Song I.-J., Fukuda T., Yokoyama J., Maki M., Ochiai T., Kameya T., Kanno A. (2005). Characterization of TrcMADS1 gene of Trillium camtschatcense (Trilliaceae) reveals functional evolution of the SOC1/TM3-like gene family. J. Plant Res. 118: 229–234 [DOI] [PubMed] [Google Scholar]
- Oosumi T., Gruszewski H.A., Blischak L.A., Baxter A.J., Wadl P.A., Shuman J.L., Veilleux R.E., Shulaev V. (2006). High-efficiency transformation of the diploid strawberry (Fragaria vesca) for functional genomics. Planta 223: 1219–1230 [DOI] [PubMed] [Google Scholar]
- Osnato M., Castillejo C., Matías-Hernández L., Pelaz S. (2012). TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat. Commun. 3: 808. [DOI] [PubMed] [Google Scholar]
- Peng J., Carol P., Richards D.E., King K.E., Cowling R.J., Murphy G.P., Harberd N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin P.A., Nilsson O. (2012). The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 35: 1742–1755 [DOI] [PubMed] [Google Scholar]
- Potter D., Eriksson T., Evans R.C., Oh S., Smedmark J.E.E., Morgan D.R., Kerr M., Robertson K.R., Arsenault M., Dickinson T.A., Campbell C.S. (2007). Phylogeny and classification of Rosaceae. Plant Syst. Evol. 266: 5–43 [Google Scholar]
- Randoux M., Jeauffre J., Thouroude T., Vasseur F., Hamama L., Juchaux M., Sakr S., Foucher F. (2012). Gibberellins regulate the transcription of the continuous flowering regulator, RoKSN, a rose TFL1 homologue. J. Exp. Bot. 63: 6543–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe O.J., Bradley D.J., Coen E.S. (1999). Separation of shoot and floral identity in Arabidopsis. Development 126: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Riechmann J.L., Meyerowitz E.M. (1997). MADS domain proteins in plant development. Biol. Chem. 378: 1079–1101 [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Kröber S., Amasino R.A., Coupland G. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo E., Lee H., Jeon J., Park H., Kim J., Noh Y.-S., Lee I. (2009). Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell 21: 3185–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V., et al. (2011). The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 43: 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G.G. (2004). The autonomous pathway: Epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr. Opin. Plant Biol. 7: 570–574 [DOI] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J.A. (2008). A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57: 758–771 [DOI] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Sønsteby A., Heide O. (2008). Long-day rather than autonomous control of flowering in the diploid everbearing strawberry Fragaria vesca ssp. semperflorens. J. Hortic. Sci. Biotechnol. 83: 360–366 [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Tao Z., Shen L., Liu C., Liu L., Yan Y., Yu H. (2012). Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J. 70: 549–561 [DOI] [PubMed] [Google Scholar]
- Taoka K., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–335 [DOI] [PubMed] [Google Scholar]
- Torti S., Fornara F., Vincent C., Andrés F., Nordström K., Göbel U., Knoll D., Schoof H., Coupland G. (2012). Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 24: 444–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainin T., Bar-Ya’akov I., Holland D. (2013). ParSOC1, a MADS-box gene closely related to Arabidopsis AGL20/SOC1, is expressed in apricot leaves in a diurnal manner and is linked with chilling requirements for dormancy break. Tree Genet. Genomes 9: 753–766 [Google Scholar]
- Tsuji H., Taoka K., Shimamoto K. (2011). Regulation of flowering in rice: Two florigen genes, a complex gene network, and natural variation. Curr. Opin. Plant Biol. 14: 45–52 [DOI] [PubMed] [Google Scholar]
- Turck F., Fornara F., Coupland G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59: 573–594 [DOI] [PubMed] [Google Scholar]
- Turnbull C. (2011). Long-distance regulation of flowering time. J. Exp. Bot. 62: 4399–4413 [DOI] [PubMed] [Google Scholar]
- Vandenbussche M., Theissen G., Van de Peer Y., Gerats T. (2003). Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frameshift mutations. Nucleic Acids Res. 31: 4401–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk A.D.J., Morabito G., Fiers M., van Ham R.C.H.J., Angenent G.C., Immink R.G.H. (2010). Sequence motifs in MADS transcription factors responsible for specificity and diversification of protein-protein interaction. PLoS Comput. Biol. 6: e1001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Farrona S., Vincent C., Joecker A., Schoof H., Turck F., Alonso-Blanco C., Coupland G., Albani M.C. (2009). PEP1 regulates perennial flowering in Arabis alpina. Nature 459: 423–427 [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Wiseman N.J., Turnbull C.G.N. (1999). Effects of photoperiod and paclobutrazol on growth dynamics of petioles in strawberry (Fragaria × ananassa). Aust. J. Plant Physiol. 26: 353–358 [Google Scholar]
- Yoo S.K., Chung K.S., Kim J., Lee J.H., Hong S.M., Yoo S.J., Yoo S.Y., Lee J.S., Ahn J.H. (2005). CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 139: 770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.R., Henriques R., Lin S.S., Niu Q.W., Chua N.H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646 [DOI] [PubMed] [Google Scholar]
- Zhou C.-M., Zhang T.-Q., Wang X., Yu S., Lian H., Tang H., Feng Z.-Y., Zozomova-Lihová J., Wang J.-W. (2013). Molecular basis of age-dependent vernalization in Cardamine flexuosa. Science 340: 1097–1100 [DOI] [PubMed] [Google Scholar]