Brassinosteroids are essential regulators of plant growth and development. While BRI1 signaling regulates cellular growth in the majority of plant tissues, our study uncovers the composition of the BRL3 receptor complex in Arabidopsis and addresses cell type–specific roles for BRL3 signaling in root development.
Abstract
Brassinosteroid (BR) hormones are primarily perceived at the cell surface by the leucine-rich repeat receptor-like kinase BRASSINOSTEROID INSENSITIVE1 (BRI1). In Arabidopsis thaliana, BRI1 has two close homologs, BRI1-LIKE1 (BRL1) and BRL3, respectively, which are expressed in the vascular tissues and regulate shoot vascular development. Here, we identify novel components of the BRL3 receptor complex in planta by immunoprecipitation and mass spectrometry analysis. Whereas BRI1 ASSOCIATED KINASE1 (BAK1) and several other known BRI1 interactors coimmunoprecipitated with BRL3, no evidence was found of a direct interaction between BRI1 and BRL3. In addition, we confirmed that BAK1 interacts with the BRL1 receptor by coimmunoprecipitation and fluorescence microscopy analysis. Importantly, genetic analysis of brl1 brl3 bak1-3 triple mutants revealed that BAK1, BRL1, and BRL3 signaling modulate root growth and development by contributing to the cellular activities of provascular and quiescent center cells. This provides functional relevance to the observed protein–protein interactions of the BRL3 signalosome. Overall, our study demonstrates that cell-specific BR receptor complexes can be assembled to perform different cellular activities during plant root growth, while highlighting that immunoprecipitation of leucine-rich repeat receptor kinases in plants is a powerful approach for unveiling signaling mechanisms with cellular resolution in plant development.
INTRODUCTION
Plant steroid hormones brassinosteroids (BRs) are perceived by the plasma membrane–localized BRASSINOSTEROID INSENSITIVE1 (BRI1; Li and Chory, 1997). BRI1 is one of the best-characterized leucine-rich repeat (LRR) receptor-like kinase (RLK) proteins in plants. Brassinolide (BL) binding occurs at the BRI1 extracellular domain. This domain consists of 25 LRRs interrupted by a 70–amino acid island domain between the 21st and 22nd LRR, which creates a surface pocket for ligand binding (Wang et al., 2001; Kinoshita et al., 2005; Hothorn et al., 2011; She et al., 2011). Ligand-mediated BRI1 receptor activation results in mutual transphosphorylation events with one or several of the SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK) coreceptors, one of which is SERK3/BRI1 ASSOCIATED KINASE1 (BAK1) (Li et al., 2002; Russinova et al., 2004; Wang et al., 2005; Karlova et al., 2006; Wang et al., 2008). Recent evidence suggests that SERK coreceptors are essential for BRI1-mediated signaling (Gou et al., 2012). Downstream of BRI1 and SERKs, members of the BRASSINOSTEROID SIGNALING KINASE (BSK) cytoplasmic kinase family are subject to BRI1-mediated phosphorylation (Tang et al., 2008), and subsequently the signal is transmitted to BRI1-EMS SUPPRESSOR1 (BES1) and BRASSINAZOLE-RESISTANT1 (BZR1) transcription factors (Wang et al., 2002; Yin et al., 2005).
Many of the components of the BRI1 pathway have been identified using forward or reverse genetic approaches (Wang et al., 2012), while structural studies of the extracellular domain of BRI1 confirmed the behavior of BRI1 mutant alleles (Hothorn et al., 2011; She et al., 2011). As an alternative approach, we previously employed immunoprecipitation (IP) of the green fluorescent protein (GFP)–tagged SERK1 coreceptor (Karlova et al., 2006; Smaczniak et al., 2012). This resulted in the identification of BRI1 as well as BAK1, suggesting that at least in part, protein–protein interactions mirror the genetic evidence.
In Arabidopsis thaliana, there are two closely related members of the small BRI1-like family, BRASSINOSTEROID RECEPTOR-LIKE1 (BRL1) and BRL3, respectively, that share the overall structure of BRI1, including the ligand binding island domain, and can bind to BL with higher (BRL1) or similar (BRL3) binding affinity as the main BRI1 receptor (Caño-Delgado et al., 2004; Kinoshita et al., 2005). While BRI1 is expressed in most if not all cells (Friedrichsen et al., 2000), the expression of BRL1 and BRL3 is enriched in the vascular tissues. The analysis of the bri1 brl1 brl3 mutant in the inflorescence stem suggested a redundant role with BRI1 for these two receptors in regulating cell proliferation during vascular bundle patterning (Caño-Delgado et al., 2004). Since then, the discrete localization of BRLs together with the dramatic phenotype of triple BR receptor mutants has hampered the identification of novel specific roles for BRL receptors in plant growth and development. In this study, it was of interest to unravel the composition of the BRL3 receptor complex and its contribution to plant growth and development.
Here, we report the identification of proteins associated with BRL3 receptors using IP and liquid chromatography–tandem mass spectrometry (LC/MS/MS) techniques. We found that the BRL3 complex contains the BAK1 coreceptor and several other previously described BR-signaling components. BRs regulate the normal cell cycle progression of root meristematic cells, including that of the rarely dividing quiescent center (QC) cells, during root growth (González-García et al., 2011), yet a role for BRLs in the root had not hitherto been reported. The genetic analysis of brl1 brl3 mutants in combination with bak1-3 confirms a novel cell-specific role for the BRL3 complex in regulating QC cell renewal in response to BRs. Our study unveils the functional relevance of BR receptor complexes in regulating BR-mediated responses with cellular resolution in plant development.
RESULTS
Protein Expression of BRI1-Like Family Members
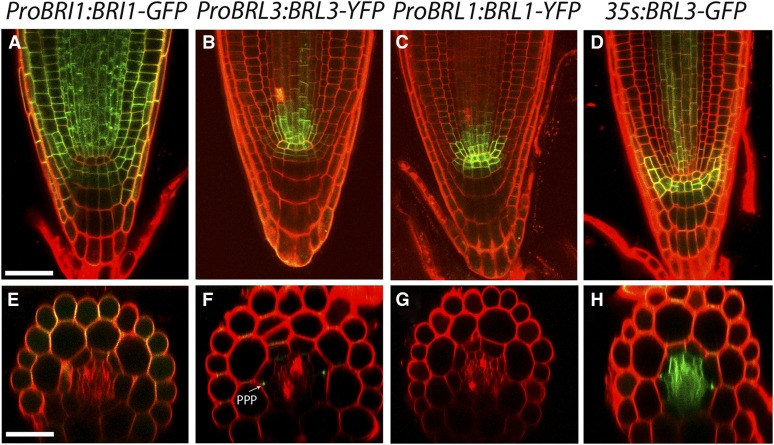
In Arabidopsis, expression of BRL1 and BRL3 is enriched in the vascular tissues, whereas that of BRI1 appears in most plant cells (Caño-Delgado et al., 2004). To reveal the localization of BRL1 and BRL3 receptors, the full-length genomic sequences of BRL1 and BRL3 were fused to yellow fluorescent protein (YFP) under the control of the native promoters consisting of a region 2 kb upstream of the start codon for BRL1 and BRL3 (ProBRL1:BRL1-YFP and ProBRL3:BRL3-YFP, respectively). Localization of these receptor fusions in stable T4 homozygous plants was compared with that of ProBRI1:BRI1-GFP plants, a construct previously shown to complement bri1 null mutants (Geldner et al., 2007). Root analysis of 6-d-old plants revealed the presence of BRI1-GFP in all cell files of the root apical meristem (Figure 1A), similar to that reported (Friedrichsen et al., 2000; Geldner et al., 2007; Wilma van Esse et al., 2011). Further up in the meristem, confocal microscopy of a transverse section showed a predominant BRI1 localization at the outer cell files (epidermis/cortex) at the differentiation zone (Figure 1E). In contrast with BRI1, the localization pattern for BRL1 and BRL3 was specific to a few cell files. At the root apex BRL1 and BRL3 are similarly localized at the QC, columella stem cells, and a group of provascular cells, including vascular initials cells located right above the QC (Figures 1B and 1C). Transverse view at the differentiation zone of the root showed the presence of BRL3 at the phloem-pole pericycle cells (Figure 1F), whereas BRL1 was absent from these cells (Figure 1G).
Figure 1.
Spatial Localization of BRI1-Like Family Receptors in Arabidopsis.
Six-day-old seedlings expressing ProBRI1:BRI1-GFP ([A] and [E]), ProBRL3:BRL3-YFP ([B] and [F]), ProBRL1:BRL1-YFP ([C] and [G]), and 35S:BRL3-GFP ([D] and [H]) in the meristematic zone of the primary root. Bars = 50 µm.
(A) Ubiquitous expression of ProBRI1:BRI1-GFP in the meristematic zone.
(B) ProBRL3:BRL3-YFP expression in the QC, columella stem cell, and vascular initials of the meristematic zone.
(C) ProBRL1:BRLI1-YFP plants show provascular expression overlapping with ProBRL3:BRL3-YFP in the meristematic zone.
(D) 35S:BRL3-GFP plants show a similar localization pattern to plants expressing ProBRL3:BRL3-YFP.
(E) Confocal radial sections of ProBRI1:BRI1-GFP expression in the epidermis and cortex at the differentiation zone of the root.
(F) ProBRL3:BRL3-YFP localization in the phloem-pole pericycle (F) compared with ProBRL1:BRLI1-GFP (G), which is absent at the differentiation zone of the root. PPP, phloem-pole pericycle.
(H) A confocal radial view of 35S:BRL3-GFP denotes BRL3 localization in the stele of the differentiation zone of the root.
Since the expression of BRL1 and BRL3 receptors under their native promoters was much lower than that of ProBRI1:BRI1-GFP lines, additional 35S:BRL3-GFP–overexpressing plants were established in order to increase the amount of BRL3 protein in the plant (Figures 1D and 1H). The analysis of 35S:BRL3-GFP plants revealed a localization pattern similar to the ProBRL3:BRL3-YFP native lines (Figures 1B and 1D), suggesting an additional spatial control at the protein level for BRL3. At the elongation and differentiation zone of the root, a stele-specific localization was found in 35S:BRL3-GFP plants (Figure 1H).
In mature plant organs, BRL1 and BRL3 were predominantly localized at leaf veins and associated with phloem tissues in the vascular bundles of the shoot inflorescence stem (see Supplemental Figure 1 online). While 35S:BRL3-GFP plants showed a strong localization in the vascular tissues, GFP was also present at the epidermis where BRI1 is predominantly localized (see Supplemental Figure 2 online).
Identification of Proteins That Coimmunoprecipitate with BRL3
Previously identified BRI1 interactors have been essential for understanding BR signal transduction in the plant. Here, we use an alternative approach, to identify a native GFP-tagged plant receptor complex from young seedlings directly by IP using anti-GFP antibodies immobilized on beads (see Methods). In preliminary experiments, wild-type Columbia-0 (Col-0) seedlings were used as a control to detect proteins that bind nonspecifically to anti-GFP beads. Three independent biological replicates were performed for each tagged receptor and wild-type complex purified in pairs. The resulting peptides were analyzed by LC/MS/MS as previously described (Smaczniak et al., 2012). First, the ProBRL3:BRL3-YFP line was used. In this line, BRL3-YFP was undetectable in total protein extracts analyzed directly by immunoblot, but successfully enriched after IP (see Supplemental Figure 3A online). Peptide measurement by LC/MS/MS of native BRL3 IPs only yielded a few proteins besides the BRL3 bait, including BRL1, DET3, and clathrin binding protein At4g18060 (see Supplemental Table 1 online). To increase the sensitivity of our approach, 35S:BRL3-GFP lines were subsequently used for IP (Figures 1C, 1D, 1G, and 1H; see Supplemental Figures 1A and 4 online and Supplemental References 1 online). Furthermore, a revised procedure (see Methods) that employs protein cross-linking, microsomal membrane isolation, and complete solubilization after ultracentrifugation, in addition to removal of putative GFP-only coimmunoprecipitated proteins by comparison with a 35S:GFP-expressing control rather than the wild type, was employed to minimize false positives. Using this protocol and LC/MS/MS analysis combined with stringent peptide and protein scoring parameters, a total of 128 BRL3 coimmunoprecipitated proteins were significantly enriched (see Supplemental Data Set 1 online).
Previously described components of BR signaling, such as BSK1 and BSK3 (Kim et al., 2011), were found among the proteins identified in BRL3 IPs (Table 1). Indeed, the coreceptor BAK1 (Li et al., 2002; Nam and Li, 2002) was also purified as a highly significant interactor of BRL3. Interestingly, BRI1 itself was not detected among the proteins that coimmunoprecipitate with BRL3 using our experimental conditions.
Table 1. Proteins That Coimmunoprecipitate with BRL3 and Are Implicated in BRI1-Mediated BR Signaling.
| Gene Locus | Protein Name | Localization | Type | BZR1/BES1 Targeta | Fold Change |
|---|---|---|---|---|---|
| BRs | |||||
| At3g13380 | BRL3 | PMb | LRR-RLK | Yes/yes | Infinite |
| At4g00710 | BSK3 | PM | Kinase | Yes/low | 118 |
| At4g35230 | BSK1 | PM | Kinase | Yes | 1037 |
| At4g33430 | BAK1 | PM, endosome | LRR-RLK | Yes/yes | Infinite |
| At3g51550 | FERONIA | PM | RLK | Yes (low) | Infinite |
| Signaling | |||||
| At4g23650 | CDPK6, | PM, cytosol | Kinase | Yes (low) | 205 |
| At1g63500 | Protein kinase protein with tetratricopeptide repeat domain | PM | Kinase | Yes (low) | 160 |
| At3g57530 | CDPKW and calcium-dependent protein kinase 32 (CDPK32) | Cytosol | Kinase | Yes | Infinite |
| Transport | |||||
| At2g21410 | VHA-A2 proton pump | Vacuole, tonoplast | ATPase | 110 | |
| At4g30190 | AHA2 ATPase 2 | Vacuole | ATPase | Yes | 254 |
| Trafficking | |||||
| At3g60190 | Dynamin-related protein 1E (DRP1E) | PM, cell plate | GTPase | Yes | 208 |
A summary of selected proteins that coimmunoprecipitate with BRL3. Only proteins that passed the cutoff threshold described in Methods and have been implicated or known to be involved in BR signaling are shown. For the complete list of proteins that coimmunoprecipitate with BRL3, see Supplemental Data Set 1 online. The last column lists the fold change when compared to the GFP-only control. Infinite means that peptides were not detected in the control sample. Representative product ion spectra of peptides identified for each of these proteins are shown in Supplemental Figure 8 online.
Data from Sun et al. (2010) and Yu et al. (2011).
PM, plasma membrane.
The VHA-A2 (Dettmer et al., 2006) and AHA2 ATPases (related to the AHA1 plasma membrane ATPase; Witthöft et al., 2011) were enriched in BRL3 IPs as well as the RLK FERONIA (Guo et al., 2009). A number of other cytoplasmic kinases, including CPK3 CALCIUM-DEPENDENT PROTEIN KINASE6 (CDPK6), were also detected. Notably absent were the PP2AA1/RCN1 phosphatase (Wu et al., 2011) and the inhibitor protein BKI1 (Wang and Chory, 2006). Taken together, our results show that proteins that coimmunoprecipitate with BRL3 under our conditions include known components of the BR signaling pathway, but lack the main receptor BRI1.
BRL3 Interacts with BAK1 in Vivo
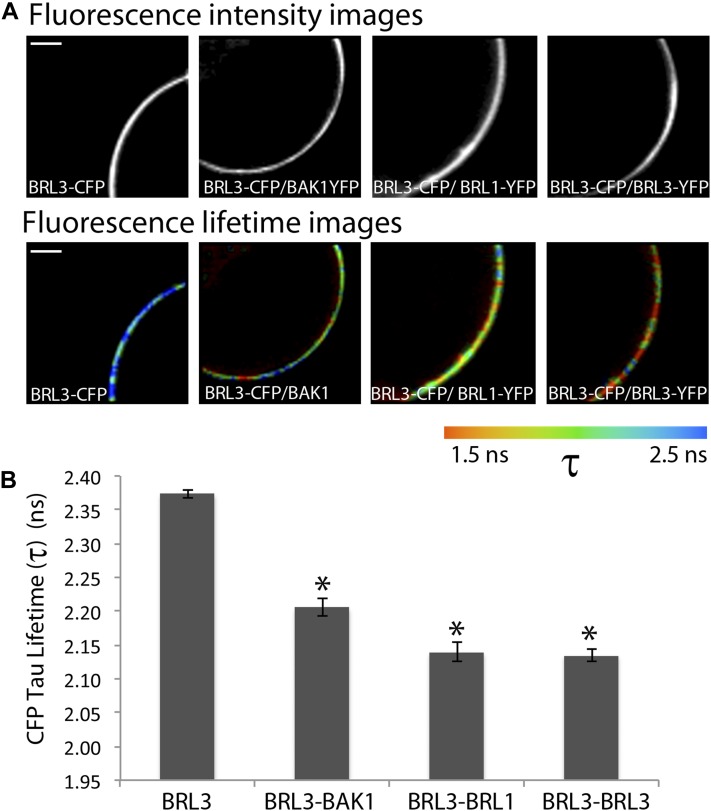
To confirm the newly identified putative interactions with the BRL3 complex, we transiently expressed selected cyan fluorescent protein (CFP)- and YFP-tagged receptors in Arabidopsis protoplasts and subjected these protoplasts to fluorescence resonance energy transfer (FRET) combined with fluorescence lifetime imaging (FLIM). Positive colocalization of CFP-tagged BRL3 was observed in protoplasts expressing BAK1, BRL1, and BRL3 fused to YFP. FLIM microscopy allowed the selection of certain membrane fluorescence areas where CFP lifetime was imaged (Figure 2A). The average lifetimes of the BRL3-CFP fluorescence of all double-transfected protoplast combinations were measured and compared with the control BRL3-CFP protoplasts. Significantly reduced CFP lifetimes were found between the BRL3-CFP and BAK1-YFP receptor pairs (Figure 2B), confirming their in vivo interaction properties. A positive interaction was also detected for BRL3-CFP/BRL3-YFP homodimers, but also between BRL3-CFP/BRL1-YFP receptors (Figure 2B). Overall, these results confirm BAK1 as a BRL3 interactor in live protoplasts.
Figure 2.
FRET-FLIM Validation Analysis in Arabidopsis Protoplasts.
(A) and (B) FRET combined with FLIM in Arabidopsis protoplasts transfected with BRL3-CFP alone or coexpressing BRL3-CFP with BAK1-YFP, BRL3-YFP, and BRL1-YFP.
(A) Top panel: Fluorescence intensity images show expression along the entire membrane. Bottom panel: Fluorescence lifetime images show τ lifetime for CFP represented by a colored scale. Bars = 10 µm.
(B) Graphical representation for significantly reduced CFP τ lifetimes between the BRL3-CFP and BAK1-YFP receptor pairs confirming their in vivo interaction. Reduced lifetime also significant for protoplasts coexpressing BRL3 with BRL3 and BRL1 membrane proteins. Results are the average of three independent replicate experiments ± se (n = 45). Student’s t test indicates that differences are statistically significant between BRL3-CFP and BAK1-YFP as well as between BRL3-CFP homodimers and BRL1-YFP heterodimers (*P value <0.01).
[See online article for color version of this figure.]
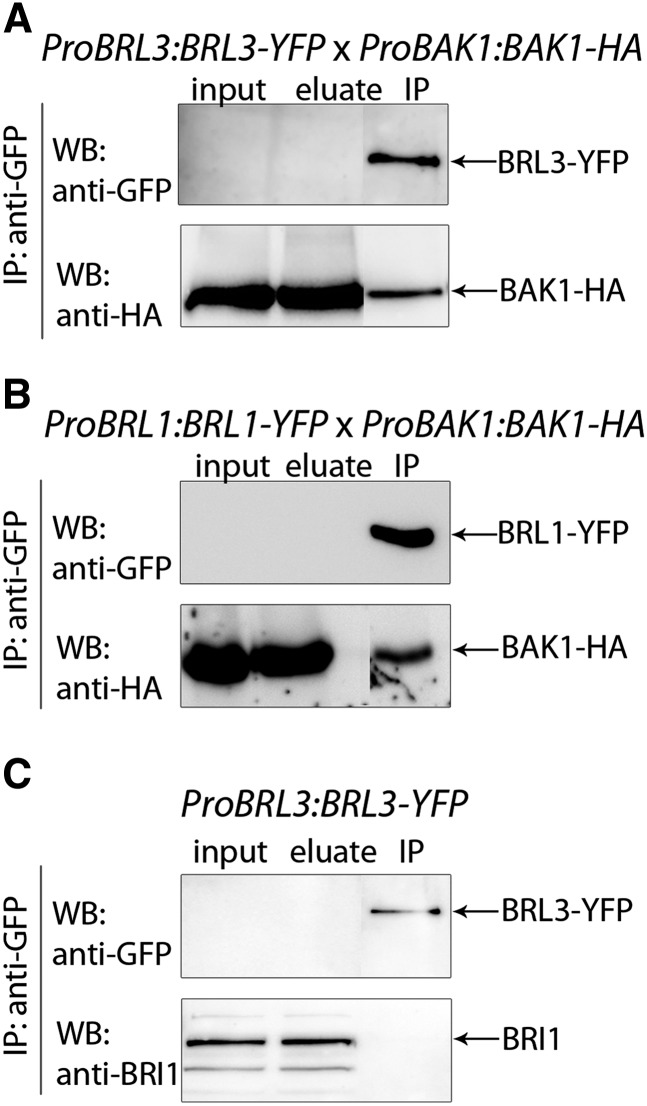
To confirm the observed interactions, in vivo coimmunoprecipitation was done in double tagged Arabidopsis plants coexpressing ProBAK1:BAK1-HA and ProBRL3:BRL3-YFP constructs. BAK1 was successfully detected in an immunoprecipitated BRL3-YFP sample, demonstrating the interaction between BRL3 and BAK1 proteins in planta (Figure 3A). Similar results were obtained for BRL1-BAK1 (Figure 3B). By contrast, the BRI1 receptor was not detected in immunoprecipitates from ProBRL3:BRL3-GFP plants (Figure 3C). Nonetheless, BRI1/BRL3 heterodimerization was only detected by FRET-FLIM analysis (see Supplemental Figure 5 online).
Figure 3.
BRL3 Forms Stable Hetero-Oligomers with BAK1 but Not BRI1 in Vivo.
(A) Top panel: Coimmunoprecipitation using double tagged transgenic plants coexpressing ProBAK1:BAK1-HA and ProBRL3:BRL3-YFP. In these plants, the amount of BRL3-YFP is almost undetectable in total protein extracts by direct immunoblot (input), yet successfully enriched after anti-GFP IP. Bottom panel: BAK1 is successfully detected using anti-HA antibodies in both the direct immunoblot (input) and immunoprecipitated samples, demonstrating that the two receptors heterodimerize in planta.
(B) Top panel: In the ProBRL1:BRL1-YFP line, BRL1-YFP is undetectable in total protein extracts analyzed directly by immunoblot (input), yet successfully enriched after anti-GFP IP. Bottom panel: BAK1 is successfully detected using anti-HA antibodies in the BRL1-YFP protein immunoprecipitated fraction.
(C) Top panel: In the ProBRL3:BRL3-YFP line, BRL3-YFP is undetectable in total protein extracts analyzed directly by immunoblot (input), yet successfully enriched after anti-GFP IP. Bottom panel: Immunoblot with anti-BRI1 native antibodies does not detect BRI1 receptor in the ProBRL3:BRL3-GFP immunoprecipitated fraction.
IP, immunoprecipitation; WB, Western Blot.
In conclusion, our data show that BRL3 receptors interact with BAK1 in planta, while a direct interaction of BRI1/BRL3 could not be confirmed in vivo.
The BRL1/BRL3/BAK1 Receptor Complex Accounts for Root Growth and QC Organization
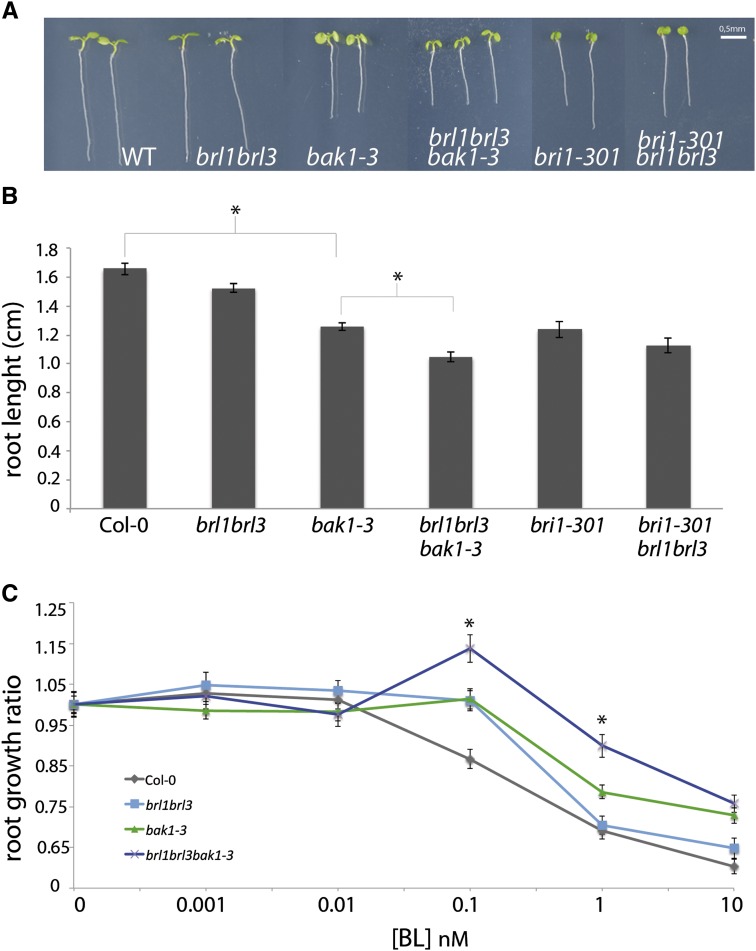
BRI1-mediated signaling regulates normal cell cycle progression of root meristematic cells, including the rarely divided QC cells during root growth (González-García et al., 2011), yet a role for BRLs in the root has not hitherto been reported. To address the biological relevance of the observed BRL1 and BRL3 interactions with BAK1, we performed a genetic analysis using multiple combinations of BRI1-like receptors and the BAK1 coreceptor. Root length analysis of 6-d-old seedlings showed that bak1-3 roots are significantly shorter than Col-0 wild-type plants (Figures 4A and 4B), in agreement with previous reports (Nam and Li, 2002; Albrecht et al., 2008), whereas brl1 brl3 double mutant roots were of similar length as wild-type ones. Strikingly, brl1 brl3 bak1-3 triple mutants enhanced the bak1-3 short root phenotype (Figures 4A and 4B), supporting the notion that biochemical interaction between BRL1/BRL3 and BAK1 is required for BR-mediated root growth. By contrast, the roots of bri1-301 brl1 brl3 (Figures 4A and 4B) and bri1-116 brl1 brl3 (see Supplemental Figure 6 online) were of similar length as those of their respective bri1 parents.
Figure 4.
The Root and BR Insensitivity Phenotypes of the brl1 brl3 bak1-3 Triple Mutant Are More Severe Than Those of bak1-3.
(A) Phenotype of 6-d-old wild-type (WT), bak1-3, brl1 brl3, brl1 brl3 bak1-3, bri1-301, and bri1-301 brl1 brl3 mutants. Bar = 0.5 mm.
(B) Root length assay of 6-d-old wild-type, bak1-3, brl1 brl3, brl1 brl3 bak1-3, bri1-301, and bri1-301 brl1 brl3 seedlings. Results are average ± se (n = 90). Student’s t test indicates that root length differences are statistically significant between Col-0 wild type and bak1-3 as well as between bak1-3 and brl1 brl3 bak1-3 (*P value < 0.01).
(C) Dose–response curve of exogenous BL treatments (0.001, 0.01, 0.1, 1, and 10 nM) in 6-d-old seedlings. Ratios of root shortening were calculated for each BL concentration and represented in the graph. Results are the average of three independent replicate experiments ± se (n = 85). Student’s t test indicates that brl1 brl3 bak1-3 is statistically less sensitive to BL when compared with Col-0 wild type at 0.1 and 1 nM of BL (*P value < 0.01).
[See online article for color version of this figure.]
BR sensitivity of brl1 brl3 bak1-3 mutants was analyzed in a dose–response curve. Increasing BL concentrations significantly reduced the root length of wild-type plants (Figure 4C; in agreement with González-García et al., 2011). At 0.1 nM BL, a 20% reduction of root growth was observed in the wild-type plants that was not observed in brl1 brl3, bak1-3, and brl1 brl3 bak1-3 (Figure 4C). In response to 1 nM BL, the root length of brl1 brl3 and bak1-3 was similar to that of the wild type, whereas in the same conditions, brl1 brl3 bak1-3 showed a significantly reduced sensitivity to BR-mediated root shortening (Figure 4C).
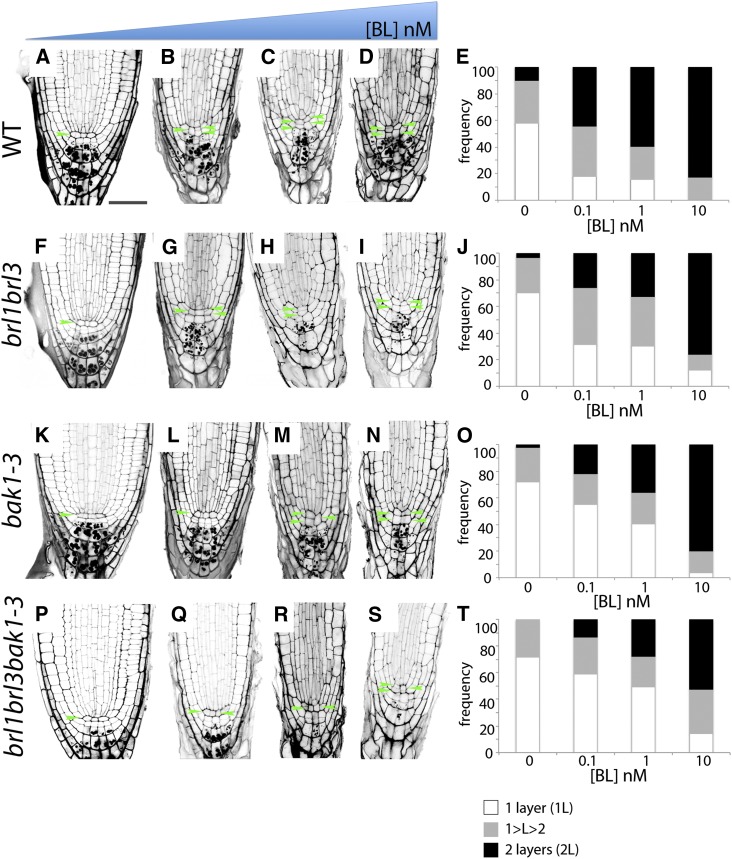
A previous mutant analysis showed that BRs are required to maintain quiescence at the root stem cell niche (González-García et al., 2011). Since BRL receptors localized to these cells (Figures 1B and 1C), we asked whether the BRL3 complex is necessary to preserve the competence of QC cells to divide at the root apex. Confocal microscopy and quantitative analysis of QC cells in a wild-type population showed that 58% of the plants had one layer of QC cells (1L), whereas 10% had two layers (2L; Figures 5A and 5E). The remaining 32% of the roots analyzed exhibited an intermediary phenotype, where only some of the QC cells appeared divided (Figures 5A and 5E). This transition state was referred to as (1<L<2). Analysis of QC organization was studied in different mutant combinations. Compared with wild-type plants (Figures 5A and 5E), a reduction in the frequency of QC division was observed in brl1 brl3 (Figures 5F and 5J), bak1-3 (Figures 5K and 5O), and brl1 brl3 bak1-3 (Figures 5P and 5T) mutants, showing 70% of the plants with one layer of QC cells. Strikingly, treatment with 0.1 nM BL enhanced the observed insensibility to BL in brl1 brl3 double mutants (25% 2L; Figures 5G and 5J) and bak1-3 mutant (20% 2L; Figures 5L and 5O) compared with the wild type (45% 2L; Figures 5B and 5E) in the QC cells. This phenotype was even stronger in the brl1 brl3 bak1-3 triple mutants (13% 2L; Figures 5Q and 5T), in agreement with our previous results in the BL dose–response curve.
Figure 5.
The BRL3/BRL1/BAK1 Complex Promotes Activity of QC Cells in the Primary Root.
The effect of exogenously applied BL on modified PseudoSchiff-PI–stained root tips of the wild type (WT; [A] to [E]), brl1 brl3 ([F] to [J]), bak1-3 ([K] to [O]), and brl1 brl3 bak1-3 ([P] to [T]) in response to a BL dose curve. Images illustrate longitudinal median confocal images of 6-d-old primary Col-0 roots treated with the indicated amounts of BL. Wild-type root tip control (A), treated with 0.1 nM BL (B), 1 nM BL (C), and 10 nM BL continuous treatment. brl1 brl3 control (F) and treated with 0.1 nM BL (G), 1 nM BL (H), and 10 nM BL continuous treatment (I). bak1-3 root tip without (K) and after 0.1 nM BL (L), 1 nM BL (M), and 10 nM BL continuous treatment (N). brl1 brl3 bak1-3 without (P) and with 0.1 nM BL (Q), 1 nM BL (R), and 10 nM BL continuous treatment (S). Quantitative analysis of the effects of exogenous BL treatment at different concentrations (from left to right: 0, 0.1, 1, and 10 nM of BL) in QC division for the wild type, brl1 brl3, bak1-3, and brl1 brl3 bak1-3 triple mutant, respectively ([E], [J], [O], and [T]). Frequency distribution of the number of QC layers: one layer (1L; white), 1<layer<2 (1<L2; gray) and two layers (2L; black) of QC cells for the different BL treatments. Green arrowheads indicate the QC cell layer. Bar = 50 µm.
[See online article for color version of this figure.]
Similar to our results obtained for the BL dose–response curve in roots (Figure 4C), this phenotype was dose specific and 1 nM BL promoted QC division in wild-type plants (60% 2L; Figures 5C and 5E), whereas the QC cells of brl1 brl3 and bak1-3 plants retained some insensitivity to this hormone concentration (35% 2L; Figures 5H, 5J, 5M, and 5O). This insensitivity was stronger in brl1 brl3 bak1-3 mutants treated with 1 nM BL (28% 2L; Figures 5R and 5T) and was confirmed when these plants were treated with 10 nM BL (Figures 5S and 5T). In agreement, brl1 brl3 bak1-3 mutants showed an increased number of plants with one QC layer compared with the remaining genotypes for all BL concentrations analyzed (Figures 5E, 5J, 5O, and 5T). The complete quantitative analysis for three independent biological replicates is shown in Supplemental Supplemental Table 2 online. Furthermore, we observed that the brl1 brl3 bak1-3 mutants showed hypersensitivity to BR in the stele when compared with bak1 or brl1 brl3 mutants (see Supplemental Figure 7 online). These results unveil a concerted action of these BR receptors in the QC and provascular cells and provide biological significance for the observed biochemical interactions. In conclusion, our analysis shows that BRLs regulate BR-mediated responses of a specific cellular environment at the innermost located tissues of the plant.
DISCUSSION
Our study provides biochemical and genetic evidence for the interaction of the BR receptors BRL3 and BRL1 and the coreceptor BAK1 in vivo, while demonstrating that the BRL3 signalosome complex is required for normal root growth and development in Arabidopsis. Several lines of evidence indicate that using IP linked to mass spectrometry analysis is a powerful tool to identify true interactors of the BRL3 receptor complex. First, we purified the BRL3 receptor complex in Arabidopsis and identified a number of interactors of BRI1, which have been previously characterized as BR signaling components (Li et al., 2002; Deng et al., 2007). In addition, our analysis revealed a number of potential interactors that coimmunoprecipitated with the BRL3 receptor protein. Second, our validation analysis by FRET-FLIM and coimmunoprecipitation assays demonstrates that BRL3 and BAK1 interact in native conditions in the plant. Third, the genetic analysis of brl1 brl3 bak1-3 triple mutants addresses the functional relevance of the observed biochemical interactions in Arabidopsis root development. Further evidence supporting that the BRL3 complex is required for BR-mediated root development comes from our data showing the localization of BRL1 and BRL3 receptors in the stem cell niche/provascular cells and the deficient QC renewal activity and stele defects revealed in our analysis of brl1 brl3 bak1-3.
Despite numerous examples describing the connection between SERK proteins and BR signaling (Albrecht et al., 2008; Gou et al., 2012), only SERK1, BAK1, and BKK1 have been shown to interact with BRI1 in vivo (Karlova et al., 2006; He et al., 2007). Our analysis reporting the functional interaction of BAK1 with the BRL3 receptor in vivo reveals the importance of the BRL3 signalosome complex in BR-mediated root growth and development. However, whether BRL3 triggers common and/or independent signaling outputs with BRI1 in the regulation of plant growth and development remains to be elucidated. On the one hand, several lines of evidence support a concerted action of BRI1 and BRL3 receptors in Arabidopsis root development. The root phenotypes of brl1 brl3 bak1-3 mutants cannot rule out that BRI1 is involved in the developmental phenotypes and pathways initiated by the BRL3 receptor. Our data showing that brl1 brl3 bak1-3 mutant roots enhance the bak1-3 mutant short root phenotype argue in favor of the contribution of BRL receptors to root growth jointly with BRI1. Supporting this, a role for BRI1 signaling in BR-mediated root growth and QC division has been established (González-García et al., 2011), showing that bri1 mutants have impaired QC divisions in response to BRs, similar to the phenotype of brl1 brl3 bak1-3 mutants. The functional redundancy of the BRL3 receptor has been previously proposed in Arabidopsis, where the shoot of bri1 brl1 brl3 triple mutants displayed enhanced defects of bri1 mutant vascular phenotypes (Caño-Delgado et al., 2004). Further supporting this, bri1 phenotypes in the shoot appeared to be rescued when BRL1 or BRL3 were expressed under the BRI1 promoter (Caño-Delgado et al., 2004), although the root phenotypes remain to be investigated. Altogether, these results support the hypothesis that BRI1 and BRL3 receptors signal together in BR-mediated root growth and QC division dynamics.
On the other hand, our study also suggests a spatial regulation of BR signaling that begins with the formation of different BR receptor complexes among the different root cell types. First, our data show a reduction of BRI1 receptor levels in the QC and provascular cells at the primary root in agreement with previous findings (Wilma van Esse et al., 2011), whereas the highest levels of BRL1 and BRL3 proteins were detected in these specific cells. Second, the localization of native BRL3 receptors in the innermost located tissues of the plant and the overexpression of BRL3 in 35S:BRL3-GFP lines suggest that the existence of posttranscriptional modifications might exclude BRL3 receptors at the outer root cell layers. As BRI1 signals from the epidermis to regulate root growth (Hacham et al., 2011), it is plausible that BRLs regulate the BR-mediated responses of a specific cellular environment at the innermost located tissues of the plant. Further supporting this idea, brl1 brl3 bak1-3 mutants showed hypersensitivity to BRs in the stele when compared with bak1 and/or brl1 brl3 mutants.
Finally, the lack of BRI1-specific peptides in BRL3 coimmunoprecipitates subjected to mass spectrometry analysis together with the inability to detect native BRI1 receptors in coimmunoprecipitation of ProBRL3:BRL3-YFP lines are consistent with the formation of distinct BR receptor complexes, although negative mass spectrometry and coimmunoprecipitation results cannot be considered conclusive. Considering the complementary localization of BRI1 an BRL3 receptors in the root, good candidates for BRL3-specific interactors might be proteins that coimmunoprecipitate with BRL3 that appeared enriched in the stem cell niche, including BAK1, PIN FORMED7, ATP-binding cassette-36, and an uncharacterized LRR-RLK (At1g53440), as well as proteins enriched in provascular/stele tissues, such as an uncharacterized LRR-RLK (At2g37050) and an ATP-binding cassette-2 transporter. Future identification and comparison of proteins that coimmunoprecipitate with BRI1 and BRL3, along with independent confirmation of their interaction, will be key to further investigations of the common and specific functions of both receptors in the plant.
To date, the majority of BRI1 signaling components identified are broadly expressed in most plant tissues and result in pleiotropic growth defects when mutated or overexpressed. Beyond the overall plant dwarfism, detailed phenotypic analysis has permitted the characterization of tissue-specific defects in plant vasculature (Caño-Delgado et al., 2004; Ibañes et al., 2009; Fàbregas et al., 2010), pollen development (Ye et al., 2010), root hair patterning (Kuppusamy et al., 2009), and QC activity (González-García et al., 2011). Signal amplification from the BRI1 receptor implies the transcriptional regulation of thousands of genes that, as direct targets of BES1 and BRZ1, may act in different cell types regulating different responses (Sun et al., 2010; Yu et al., 2011). Alternatively, the analysis of stomata patterning defects in BR-deficient mutants has set a role for mitogen-activated protein kinases as cell type–specific regulators of BR-mediated responses independently of BES1/BRZ1 via BIN2 (Gudesblat et al., 2012 ; Kim et al., 2012). Another example is the contribution of BRs to the lateral organ boundary at the plant shoot by reducing BR levels in the boundary domain, while repressing BZR transcription of CUP-SHAPED COTYLEDON genes outside the boundary (Bell et al., 2012; Gendron et al., 2012). Future studies tracing back the control of cell type–specific targets to specific BR receptor complexes will help delineate the different BR signaling pathways that control plant development.
METHODS
Plant Materials
Transgenic 35S:(BRI1-like3)BRL3-GFP lines in the Col-0 wild-type background were generated using the DNA clone reported by Caño-Delgado et al. (2004). Pro(BRI1-like1)BRL1-YFP and ProBRL1:BRL1-YFP constructs were cloned using a recombination Gateway Multisite Cloning system. DNA sequences were amplified from respective BAC clones (MIRP15 and F20N20). The purified gene PCR product was placed into the Gateway pDONR221 donor vector by the BP reaction. The same procedure was done for the 2-kb promoter PCR product of both receptors, placed into the Gateway P4P1R vector. For the tagged YFP, a P2RP3 donor vector was used. A recombination LR reaction was performed using the three sequenced pENTRY vectors in a three-component pDEST vector, pB7m34GW, and transferred to plants by floral dipping (Clough, and Bent, 1998). Arabidopsis thaliana Col-0 ecotype plants expressing BRI1 fused to GFP were described previously (Pro[Brassinosteroid Insensitive 1]BRI1:BRI1-GFP; Geldner et al., 2007). Wild-type Col-0 seedlings were used as a control.
Sterilized seeds were vernalized for 48 h at 4°C and grown for either 6 or 10 d in agar or liquid Murashige and Skoog (MS) medium containing 10 g/L Suc and 5 mM [2-(N-Morpholino) ethanesulfonic acid]/potassium hydroxide, pH 5.7, under normal conditions (16 h of light/8 h of dark; 20 to 23°C).
Six-day-old T4 homozygous plants of ProBRL1:BRL1-YFP and ProBRL3:BRL3:YFP grown on MS agar were used for the confocal microscopy expression analysis. Ten-day-old T4 homozygous seedlings expressing ProBRI1:BRI1-GFP, ProBRL3:BRL3-YFP, and 35S:BRL3-GFP were used for the IP experiments.
Root Length and BL Sensitivity Assays
The root length of 6-d-old seedlings grown vertically on half-strength MS agar without Suc was measured and the data were analyzed with ImageJ software (http://rsb.info.nih.gov/ij/). All experiments were repeated at least three times. A Student’s t test was used to show statistical differences of root lengths among Col-0, bak1-3, and brl1 brl3 bak1-3 mutants. For BL dose–response curve assays, the average root length for each genotype was calculated by dividing the average root length for a certain BL concentration by the average root length of the preceding BL concentration. The se associated with each ratio was calculated with the corresponding formula for se divisions. The mean ratios between the three replicates were calculated and plotted as well as their corresponding se. A one-way analysis of variance test was done to calculate the BL curve replicates significance (*P value < 0.01).
IP and Immunoblot Analysis
Initial pull-down experiments using ProBRL3:BRL3-expressing seedlings were performed using the protocols described by Smaczniak et al. (2012). Coimmunoprecipitation experiments were done in the same conditions as the IPs described above, but instead of 10 g of starting material, 5 g was used for homozygous lines coexpressing ProBRL3:BRL3-YFP and ProBAK1:BAK1-HA.
Total Membrane Protein Isolation and IP
Seedlings expressing 35S:BRL3-GFP and 35S:GFP (control line) in the Col-0 background were grown in shaking liquid culture. Approximately 100 mg of seeds per treatment were surface-sterilized, placed at 4°C for 48 h, and placed in a 1-liter flask containing 75 mL of Gamborg's B-5 salts (Gamborg et al., 1968), pH 5.8, supplemented with 2% Suc. Flasks were shaken at 80 rpm at room temperature in constant light for 11 d. In vivo protein cross-linking was performed on 40 g of whole Arabidopsis seedlings as previously described (Rohila et al., 2004) followed by flash freezing in liquid nitrogen.
Total membrane protein was isolated by grinding 40 g of frozen Arabidopsis tissue in 80 mL of cold lysis buffer (20 mM (hydroxymethyl)aminomethane hydrochloride (Tris-HCl), pH 8.8, 150 mM NaCl, 1 mM EDTA, 20% glycerol, 1 mM phenylmethylsulfonyl fluoride, 20 mM NaF, 50 nM microcystin, and protease inhibitor cocktail tablets; Roche Diagnostics). The extract was centrifuged at 6000g for 20 min (4°C), and the resultant supernatant was further centrifuged at 100,000g for 2 h (4°C) to pellet the microsomal fraction. The microsomal pellet was resuspended in 4 mL of resuspension buffer (10 mM Tris-HCl, pH 7.3, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 20 mM NaF, 500 nM microcystin, and half a tablet of protease inhibitor cocktail; Roche Diagnostics) and sonicated using a probe sonicator. The sample was cleared of any residual insoluble material by centrifugation at 21,000g for 30 min, and total solubilized membrane protein in the supernatant was quantified using the Bradford Assay. Total membrane protein extracts were equalized to 40 mg protein in 4 mL of resuspension buffer containing 1% Triton X-100 (above). Fifty microliters of anti-GFP magnetic beads (Miltenyi Biotec) was added to the protein extracts, which were then rotated for 1 h at 4°C. Magnetic beads with attached proteins were immobilized on a magnetic separator (Miltenyi Biotec). The beads were washed with 1 mL of resuspension buffer (above), 500 μL of wash buffer 1 (150 mM NaCl, 1% Igepal CA-630, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris HCl, pH 8.0; Miltenyi Biotec), and 250 μL of wash buffer 2 (20 mM Tris-HCl pH 7.5; Miltenyi Biotec). Proteins were eluted from the beads with 120 μL of elution buffer (50 mM Tris-HCl, pH 6.8, 50 mM DTT, 1% SDS, 1 mM EDTA, 0.005% bromophenol blue, and 10% glycerol; Miltenyi Biotec). Three independent biological replicates were prepared for the BRL3 and GFP control IPs. While our protocol and controls eliminated many false positives, we cannot completely rule out that detergent-insoluble membrane components may still be present in our IP samples. Therefore, we refer to the proteins identified by LC/MS/MS after IP as proteins coimmunoprecipitating with BRL3, rather than as BRL3-interacting proteins.
SDS-PAGE and LC/MS/MS Analysis
After IP, 100 μL of eluted proteins was separated on a 4 to 20% NuPage gradient gel (Invitrogen). After staining to ensure equal protein loading, each lane was segmented into bands and subjected to in-gel tryptic digestion, and peptides were extracted as previously described (Mitra et al., 2012). Peptides were analyzed by LC/MS/MS with an Easy-nanoLC-1000 coupled to an Orbitrap Elite mass spectrometer (Thermo Scientific). Peptides were initially preconcentrated and desalted online using an Acclaim PepMap100 C18 5-µm trapping column (100-µm i.d.; length of 20 mm; Thermo Scientific). The nano-liquid chromatography separation was performed using a 30-cm reversed-phase column comprised of a 360 µm o.d. × 75 µm i.d. PicoFrit Capillary (New Objective) that was slurry packed in-house with a 3-µm Magic C18 stationary phase (Bruker-Michrom). The mobile phase consisted of (A) 0.1% formic acid in water and (B) 0.1% formic acid in acetonitrile. After loading essentially the entire peptide sample from each band onto the reversed-phase column, the mobile phase was held at 5% B for 5 min and the peptides were then separated using a linear gradient from 5 to 40% B over 60 min followed by 15 min of isocratic separation at 95% B at a flow rate of 300 nL/min. The data were acquired using data-dependent acquisition, where the 10 most intense precursor ions were selected from the mass spectrometry scan (resolving power of 60,000 at mass-to-charge ratio of 400) for collision-induced dissociation fragmentation in the ion trap using a 35% normalized collision energy setting. Monoisotopic precursor selection was enabled, and precursor ions with unassigned charge or a charge state of +1 were excluded. Fragmented precursor ion masses were excluded from further selection for 60 s. For internal mass calibration, the ion at mass-to-charge ratio of 445.120025 was used as the lock mass.
For LC/MS/MS analysis, water was distilled and purified using a High-Q 103S water purification system. Acetonitrile (Chromasolv for HPLC, gradient grade, ≥99.9%) and formic acid (98 to 100%, American Chemical Society reagent grade) were obtained from Sigma-Aldrich. All other materials were purchased from Sigma-Aldrich unless otherwise noted.
Database Searching and Protein Quantification of LC/MS/MS Data
A workflow within Proteome Discoverer 1.4 (PD) (Thermo Scientific) was used to process, conduct a database search, and determine peak areas for the identified peptides. The database searches were parsed to a Mascot server (Matrix Science) to search against the TAIR10 database (www.Arabidopsis.org) using the following parameters: a precursor ion tolerance of 10 ppm, a product ion mass tolerance of 0.80 D, carbamidomethylation of Cys specified as a fixed modification, and oxidation of Met and phosphorylation of Ser, Thr, and Tyr specified as variable modifications. The Precursor Ions Area Detector node within PD was used in the PD workflow to report peak areas for each Mascot identified peptide.
Next, individual msf result files were loaded into Scaffold 3.6.5 (Proteome Software) for data visualization and cross-sample comparison. Implementing the Peptide Prophet (Keller et al., 2002) and Protein Prophet (Nesvizhskii et al., 2003) algorithms within Scaffold, statistical validation was performed to classify identifications as having at least two unique peptides with a protein identification probability of at least 95% and a peptide identification probability of at least 95%. Individual protein quantification was accomplished using the Total Precursor Intensity option within Scaffold. Fold changes (BRL3-GFP over GFP control) were also calculated and reported within Scaffold. Identified proteins were considered enriched in the BRL3 data set if (1) the protein was identified in at least two out of three BRL3 replicates, and (2) when the average abundance of the protein in the BRL3 replicates was quantified to be at least 100-fold greater than the average abundance in the GFP control replicates.
Confocal Microscopy
To analyze the GFP localization in ProBRL1:BRL1-YFP, ProBRL3:BRL3-YFP, 35S:BRL3-GFP, and ProBRI1:BRI1-GFP lines, 6-d-old roots were stained in 10 mg/mL propidium iodide (PI) and visualized after excitation by a Kr/Ar 488-nm laser line. PI and GFP were detected with a 570- to 670-nm and 500- to 545-nm band-pass filter, respectively. For YFP, the excitation wavelength was 488 nm, and fluorescence was collected in the range of 493 to 536 nm (rendered in green). An FV 1000 confocal microscope (Olympus) was used. Different Z stacks and transversal optical sections were processed using Olympus FV software and assembled with Photoshop CS (Adobe Systems). Starch granules in columella cells were visualized using a modified PseudoSchiff-PI staining method (Truernit et al., 2008).
Arabidopsis Protoplast FRET-FLIM Analysis
For determining heterodimerization among receptors, PMON999YFP/CFP vectors containing 35S:BRL3-YFP/CFP, 35S:BRL1-YFP, 35S:BRI1-YFP or 35S:BAK1-YFP were generated by J. Russinova and F. Breukelen in the laboratory of S. de Vries, Wageningen University. Maxiprep DNA extraction was done to obtain 1 µg/mL plasmid DNA for all of the protoplast experiments. Protoplast isolation from rosette leaves of 4-week-old Col-0 wild-type plants was done as described (Wu et al., 2009). Protoplast transfection by adding polyethylene glycol/ Ca2+ was performed as described (Russinova et al., 2004). For FLIM measurements, a Hamamatsu R3809U multichannel plate photomultiplier tube was used, which has a time resolution of 50 ps. FRET between CFP and YFP was detected by monitoring donor emission using a 470- to 500-nm band-pass filter. Donor FLIM lifetimes (CFP) were analyzed with SPCImage 3.10 software (Becker and Hickl) using a two-component decay model. Several cells (n > 20) were analyzed for each experiment. Statistical significance of differences between samples was determined using a two-tailed Student’s t test (*P value<0.01).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: BRL3 (At3g13380), BRl1 (At4g39400), BRL1 (At1g55610), and BAK1 (At4g33430).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Vascular Expression Pattern of the BRI1-Like Receptor Family in Other Plant Organs.
Supplemental Figure 2. ProBRI1:BRI1-GFP and 35S:BRL3-GFP Show Different and Overlapping Localization Patterns in the Leaves.
Supplemental Figure 3. Immunoprecipitation of the BRL3 Receptor Complex in ProBRL3:BRL3-YFP Plants.
Supplemental Figure 4. The Localization of BRL3 in 35S:BRL3-GFP Lines Is Not Altered in MS Liquid under Agitation Conditions.
Supplemental Figure 5. BRL3 and BRI1 Have the Capacity to Heterodimerize in Overexpression but Not in Native Conditions.
Supplemental Figure 6. bri1-116 brl1 brl3 Shows the Same Mutant Root Phenotype as bri1.
Supplemental Figure 7. The brl1 brl3 bak1-3 Mutant Is Hypersensitive to BR in the Stele.
Supplemental Figure 8. Product Ion Spectra of Proteins That Coimmunoprecipitate with BRL3 and Are Listed in Table 1.
Supplemental Table 1. Table of Specific and Unique Peptides Identified for BRL1, DET3, and Clathrin Binding Protein by Immunoprecipitation of the ProBRL3:BRL3-YFP Line Coupled to LC/MS/MS Analysis Using Procedures Previously Described (Smaczniak et al., 2012).
Supplemental Table 2. Quantification of mPS-PI–Stained Root Tips of the Wild Type, brl1 brl3, bak1-3, and brl1 brl3 bak1-3 in Response to a BL Dose Curve.
Supplemental References 1. References for Supplemental Figures 1-8.
Supplemental Data Set 1. Complete List of Proteins That Coimmunoprecipitate with BRL3, Identified by LC/MS/MS Analysis of BRL3-GFP versus GFP-Only Control.
Acknowledgments
We thank Mary-Paz González-García, Santiago Mora-García, Jenny Russinova, and Josep Vilarrasa for insightful comments on the article, Emma Sudrià and Eloi Franco-Trepat for technical support, Juan Vegas for statistical analysis, Joanne Chory for anti-BRI1 antibodies, Jan Willem Borst for FRET-FLIM experiments, and Kevin Blackburn for assistance with the mass spectrometry analysis. S.D.C., T.E.N., and M.B.G. are funded by grants from the U.S. National Science Foundation (MCB-1021363 and DBI-1126244). N.F. is funded by an Formació de personal Investigador PhD fellowship from the Generalitat de Catalunya. A.I.C.-D. and S.d.V. are recipients of a Marie-Curie Initial Training Network “BRAVISSIMO” (Grant PITN-GA-2008-215118). S.d.V. was a sabbatical professor funded by AGAUR (Generalitat de Catalunya) in A.I.C.-D.’s lab. A.I.C.-D. is funded by a grant from the Spanish Ministry of Education and Science (BIO2010/00505).
AUTHOR CONTRIBUTIONS
N.F., S.d.V., and A.I.C.-D. conceived the project, designed experiments, and wrote the article. N.F. performed biochemical and microscopy experiments, DNA manipulation, and segregation and analysis of brl1 brl3 bak1 and bri1 brl1 brl3 mutants. N.F., N.L., S.B., T.E.N., M.B.G., and S.D.C. performed mass spectrometry analysis of proteins coimmunoprecipitated with BRL3.
Glossary
- BR
brassinosteroids
- LRR
leucine-rich repeat
- RLK
receptor-like kinase
- BL
brassinolide
- GFP
green fluorescent protein
- QC
quiescent center
- YFP
yellow fluorescent protein
- LC/MS/MS
liquid chromatography–tandem mass spectrometry
- IP
immunoprecipitation
- Col-0
Columbia-0
- CFP
cyan fluorescent protein
- FRET
fluorescence resonance energy transfer
- FLIM
fluorescence lifetime imaging
- MS
Murashige and Skoog
- PD
Proteome Discoverer 1.4
- PI
propidium iodide
References
- Albrecht C., Russinova E., Kemmerling B., Kwaaitaal M., de Vries S.C. (2008). Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE proteins serve brassinosteroid-dependent and -independent signaling pathways. Plant Physiol. 148: 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, E.M., Lin, W.C., Husbands, A.Y., Yu, L., Jaganatha, V., Jablonska, B., Mangeon, A., Neff, M.M., Girke, T., and Springer, P.S. (2012). Arabidopsis LATERAL ORGAN BOUNDARIES negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl. Acad. Sci. USA 109: 21146–21151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caño-Delgado A., Yin Y., Yu C., Vafeados D., Mora-García S., Cheng J.C., Nam K.H., Li J., Chory J. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131: 5341–5351 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Deng Z., Zhang X., Tang W., Oses-Prieto J.A., Suzuki N., Gendron J.M., Chen H., Guan S., Chalkley R.J., Peterman T.K., Burlingame A.L., Wang Z.Y. (2007). A proteomics study of brassinosteroid response in Arabidopsis. Mol. Cell. Proteomics 6: 2058–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fàbregas N., Ibañes M., Caño-Delgado A.I. (2010). A systems biology approach to dissect the contribution of brassinosteroid and auxin hormones to vascular patterning in the shoot of Arabidopsis thaliana. Plant Signal. Behav. 5: 903–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen D.M., Joazeiro C.A., Li J., Hunter T., Chory J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamborg O.L., Miller R.A., Ojima K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Geldner N., Hyman D.L., Wang X., Schumacher K., Chory J. (2007). Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 21: 1598–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron, J.M., Liu, J.S., Fan, M., Bai, M.Y., Wenkel, S., Springer, P.S., Barton, M.K., and Wang, Z.Y. (2012). Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 21152–21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García M.P., Vilarrasa-Blasi J., Zhiponova M., Divol F., Mora-García S., Russinova E., Caño-Delgado A.I. (2011). Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138: 849–859 [DOI] [PubMed] [Google Scholar]
- Gou X., Yin H., He K., Du J., Yi J., Xu S., Lin H., Clouse S.D., Li J. (2012). Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 8: e1002452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudesblat G.E., Schneider-Pizoń J., Betti C., Mayerhofer J., Vanhoutte I., van Dongen W., Boeren S., Zhiponova M., de Vries S., Jonak C., Russinova E. (2012). SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 14: 548–554 [DOI] [PubMed] [Google Scholar]
- Guo H., Li L., Ye H., Yu X., Algreen A., Yin Y. (2009). Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 106: 7648–7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y., Holland N., Butterfield C., Ubeda-Tomas S., Bennett M.J., Chory J., Savaldi-Goldstein S. (2011). Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, K., Gou, X., Yuan, T., Lin, H., Asami, T., Yoshida, S., Russell, S.D., and Li, J. (2007). BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr. Biol. 17: 1109–1115. [DOI] [PubMed]
- Hothorn M., Belkhadir Y., Dreux M., Dabi T., Noel J.P., Wilson I.A., Chory J. (2011). Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibañes M., Fàbregas N., Chory J., Caño-Delgado A.I. (2009). Brassinosteroid signaling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proc. Natl. Acad. Sci. USA 106: 13630–13635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R., Boeren S., Russinova E., Aker J., Vervoort J., de Vries S. (2006). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18: 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A.I., Kolker E., Aebersold R. (2002). Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- Kim T.W., Guan S., Burlingame A.L., Wang Z.Y. (2011). The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 43: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Michniewicz M., Bergmann D.C., Wang Z.Y. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Caño-Delgado A., Seto H., Hiranuma S., Fujioka S., Yoshida S., Chory J. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Kuppusamy K.T., Chen A.Y., Nemhauser J.L. (2009). Steroids are required for epidermal cell fate establishment in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 106: 8073–8076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K.A., Doke J.T., Tax F.E., Walker J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Mitra S.K., Goshe M.B., Clouse S.D. (2012). Experimental analysis of receptor kinase phosphorylation. Methods Mol. Biol. 876: 1–15 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A.I., Keller A., Kolker E., Aebersold R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Rohila, J.S., Chen, M., Cerny, R., and Fromm, M.E. (2004). Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J. 38: 172–181. [DOI] [PubMed]
- Russinova E., Borst J.W., Kwaaitaal M., Caño-Delgado A., Yin Y., Chory J., de Vries S.C. (2004). Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She J., Han Z., Kim T.W., Wang J., Cheng W., Chang J., Shi S., Wang J., Yang M., Wang Z.Y., Chai J. (2011). Structural insight into brassinosteroid perception by BRI1. Nature 474: 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C., Li N., Boeren S., America T., van Dongen W., Goerdayal S.S., de Vries S., Angenent G.C., Kaufmann K. (2012). Proteomics-based identification of low-abundance signaling and regulatory protein complexes in native plant tissues. Nat. Protoc. 7: 2144–2158 [DOI] [PubMed] [Google Scholar]
- Sun Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Kim T.W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., Barthélémy J., Palauqui J.C. (2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chory J. (2006). Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313: 1118–1122 [DOI] [PubMed] [Google Scholar]
- Wang X., Kota U., He K., Blackburn K., Li J., Goshe M.B., Huber S.C., Clouse S.D. (2008). Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 15: 220–235 [DOI] [PubMed] [Google Scholar]
- Wang X., Goshe M.B., Soderblom E.J., Phinney B.S., Kuchar J.A., Li J., Asami T., Yoshida S., Huber S.C., Clouse S.D. (2005). Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Bai M.Y., Oh E., Zhu J.Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46: 701–724 [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Seto H., Fujioka S., Yoshida S., Chory J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wilma van Esse G., Westphal A.H., Surendran R.P., Albrecht C., van Veen B., Borst J.W., de Vries S.C. (2011). Quantification of the brassinosteroid insensitive1 receptor in planta. Plant Physiol. 156: 1691–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witthöft J., Caesar K., Elgass K., Huppenberger P., Kilian J., Schleifenbaum F., Oecking C., Harter K. (2011). The activation of the Arabidopsis P-ATPase 1 by the brassinosteroid receptor BRI1 is independent of threonine 948 phosphorylation. Plant Signal. Behav. 6: 1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.H., Shen S.C., Lee L.Y., Lee S.H., Chan M.T., Lin C.S. (2009). Tape-Arabidopsis Sandwich - A simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Wang X., Li X., Kamiya Y., Otegui M.S., Chory J. (2011). Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci. Signal. 4: ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Zhu W., Li L., Zhang S., Yin Y., Ma H., Wang X. (2010). Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc. Natl. Acad. Sci. USA 107: 6100–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yu, X., Li, L., Zola, J., Aluru, M., Ye, H., Foudree, A., Guo, H., Anderson, S., Aluru, S., Liu, P., Rodermel, S., and Yin, Y. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana Plant J. 65: 634–646. [DOI] [PubMed]