This work finds that Arabidopsis KINETOCHORE NULL2 (KNL2) colocalizes with the centromere histone variant cenH3. Characterization of knl2 mutants showed reduction of cenH3 deposition at centromeres, abnormalities of mitosis and meiosis, seed abortion, and alterations in DNA methylation.
Abstract
The centromeric histone H3 variant cenH3 is an essential centromeric protein required for assembly, maintenance, and proper function of kinetochores during mitosis and meiosis. We identified a KINETOCHORE NULL2 (KNL2) homolog in Arabidopsis thaliana and uncovered features of its role in cenH3 loading at centromeres. We show that Arabidopsis KNL2 colocalizes with cenH3 and is associated with centromeres during all stages of the mitotic cell cycle, except from metaphase to mid-anaphase. KNL2 is regulated by the proteasome degradation pathway. The KNL2 promoter is mainly active in meristematic tissues, similar to the cenH3 promoter. A knockout mutant for KNL2 shows a reduced level of cenH3 expression and reduced amount of cenH3 protein at chromocenters of meristematic nuclei, anaphase bridges during mitosis, micronuclei in pollen tetrads, and 30% seed abortion. Moreover, knl2 mutant plants display reduced expression of suppressor of variegation 3-9 homologs2, 4, and 9 and reduced DNA methylation, suggesting an impact of KNL2 on the epigenetic environment for centromere maintenance.
INTRODUCTION
Centromeres are the chromosomal positions of assembly of the kinetochore proteins that are responsible for sister chromatid cohesion, chromosome movement, and cell cycle regulation (Allshire, 1997; Henikoff and Dalal, 2005). Proper kinetochore establishment depends on the centromeric histone variant cenH3 (Howman et al., 2000; Blower and Karpen, 2001), originally described as human CENP-A (Earnshaw and Rothfield, 1985). However, the primary signals specifying the position of cenH3 chromatin are still poorly understood.
In most eukaryotes, CenH3 localization is not specified by centromeric DNA sequences, but rather is determined by epigenetic marks such as DNA methylation (Woo et al., 2007; Zhang et al., 2008). CenH3 itself has been proposed to be the epigenetic mark triggering the assembly of functional kinetochores, since ectopically incorporated cenH3 can form de novo centromeres at positions free of centromeric repeats (Lo et al., 2001; Nasuda et al., 2005).
The cenH3 deposition pathway can be subdivided into three parts: initiation, deposition, and maintenance (De Rop et al., 2012). (1) Initiation generates the epigenetic context for correct cenH3 incorporation by means of the Mis18 complex (in mammals consisting of Mis18 binding protein1 (Mis18BP1) = KNL2, Mis18α, and Mis18β), which is located at centromeres before cenH3 assembly (De Rop et al., 2012). Its downregulation results in reduced amounts of cenH3 at centromeres. In Schizosaccharomyces pombe fission yeast, Mis18 remains associated with centromeres along the cell cycle, except from metaphase to mid-anaphase, recruits the cenH3 assembly machinery and generates the correct epigenetic context for cenH3 loading (Hayashi et al., 2004; Williams et al., 2009; Phansalkar et al., 2012). Also, human KNL2 (Hayashi et al., 2004; Fujita et al., 2007) and Caenorhabditis elegans KNL2 are involved in cenH3 loading (Maddox et al., 2007). C. elegans KNL2 and cenH3 colocalize at centromeres throughout the cell cycle and coordinate kinetochore assembly and chromosome segregation. Human KNL2 and other components of the human Mis18 complex are transiently present at centromeres after mitotic exit (Fujita et al., 2007; Maddox et al., 2007; Silva and Jansen, 2009). Knockout of murine Mis18α is embryo lethal (Kim et al., 2012). Cultured homozygous mutant embryos showed reduced DNA methylation, altered histone modifications, increased centromeric transcripts, misaligned chromosomes, anaphase bridges, and lagging chromosomes (Kim et al., 2012).
(2) Deposition of cenH3 to centromeres depends on chaperones. Two cenH3 chaperone types have been identified: in humans, the Holliday junction recognition protein (HJURP) and nuclear autoantigenic sperm protein and in yeast the respective homologs suppressor of chromosome missegregation protein3 and silencing in the middle of the centromere protein3. HJURP (yeast suppressor of chromosome missegregation protein3) is highly variable among different organisms, and no homologs have yet been identified for Drosophila melanogaster, C. elegans, or plants. In Drosophila, chromosome alignment defect1 (CAL1) protein plays the role of HJURP (Phansalkar et al., 2012). Barnhart et al. (2011) showed that recruitment of HJURP to endogenous centromeres is mediated by the Mis18 complex.
(3) For maintenance of newly incorporated cenH3, involvement of two subunits (remodeling and spacing factor1 and sucrose nonfermenting protein2 homolog H) of the ATP-depending chromatin remodeling and spacing factor in stabilization of incorporated cenH3 has been proposed (Obuse et al., 2004; Izuta et al., 2006).
Here, we examine KNL2 function and show that Arabidopsis KNL2 localizes at chromocenters during all stages of the mitotic cell cycle, except from metaphase to mid-anaphase, and its level is strictly regulated by the proteasome degradation pathway. Knockout of KNL2 via a T-DNA insertion resulted in a reduced amount of centromeric cenH3, mitotic and meiotic abnormalities, and reduced growth and fertility. Reduced expression of histone methyltransferases and reduced DNA methylation in the knl2 mutant suggest a role for KNL2 in determination of the epigenetic status of centromeres.
RESULTS
Identification and in Silico Analysis of the KNL2 Homolog of Arabidopsis thaliana
KNL2 homologs contain a conserved protein module of ∼90 amino acids designated as the SANTA domain (for SANT associated) due to its association with the SANT domain, a protein domain present in chromatin remodeling enzymes such as Swi3p, histone acetyltransferase Ada2p, deacetylase N-CoR, and transcription initiation factor TFIIIB (Aasland et al., 1996).
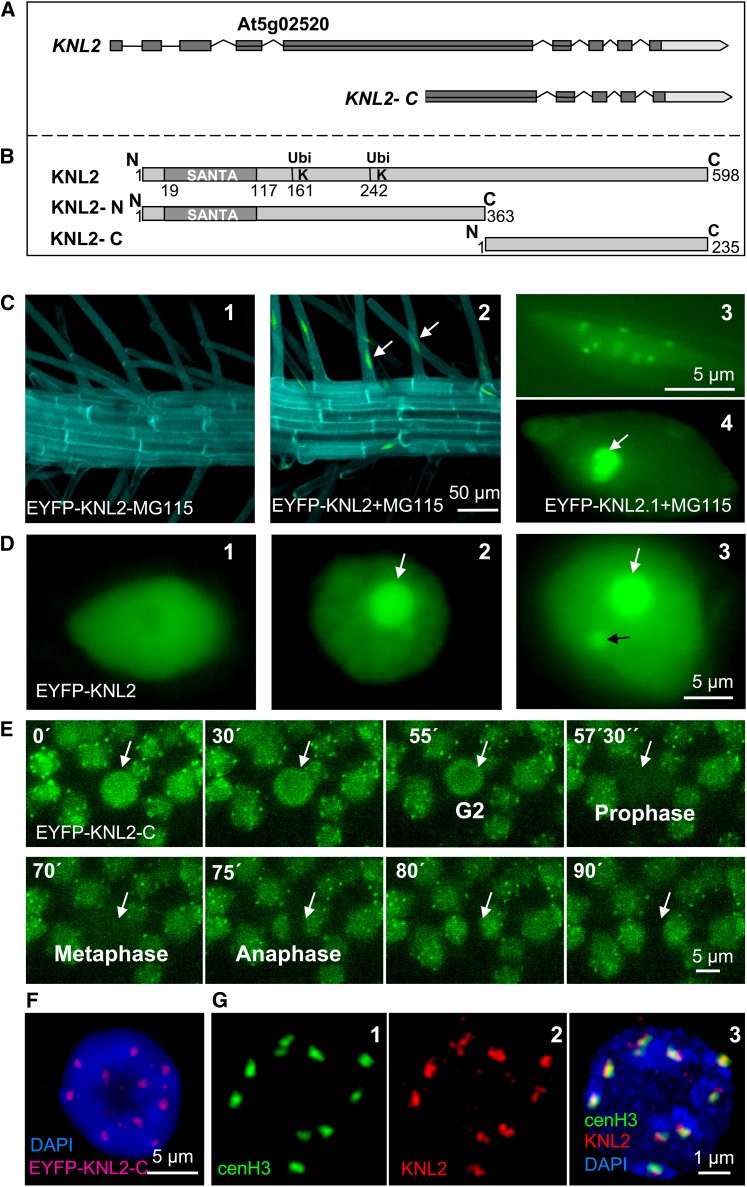
Metazoan genomes have only one gene encoding a protein with SANTA domain, but in representatives of higher plants (e.g., rice [Oryza sativa] and Arabidopsis), two genes were identified (D. Zhang et al., 2006). At5g02520 of Arabidopsis encodes a 598–amino acid protein containing a N-terminal SANTA domain at position 19 to 117 (Figure 1B). The second gene, At1g58210, encodes a 1246–amino acid protein, which has a SANTA domain, a kinase interacting protein 1 domain (KIP1), and a C-terminal structural maintenance of chromosomes, bacterial type domain (SMC_Prok_B). Among eukaryotes, KIP1 domain proteins are present exclusively in plants and are involved in signaling regulation through interaction with the kinase domain of PRK1, a receptor-like kinase in plant pollen (Skirpan et al., 2001). As At5g02520 encodes a protein with the most typical features of known KNL2-like proteins, we selected this gene for further analysis and named it KNL2. In silico analysis of the Arabidopsis KNL2 gene structure revealed nine exons (Figure 1A).
Figure 1.
KNL2 Gene and Protein Structures and Subcellular Localization of KNL2.
(A) Structure of KNL2 gene and the part of the gene encoding the C-terminal part of KNL2. Exons are shown as dark-gray boxes.
(B) Schematic of KNL2 protein structure (top fragment). The conserved protein domain SANTA (amino acids 19 to aa117) and putative ubiquitination sites at positions 161 and 242 are indicated. KNL2-N (middle fragment) and KNL2-C (bottom fragment) were separately fused with EYFP for localization studies. KNL2-C was used for expression of recombinant protein.
(C) Roots of EYFP-KNL2 transgenic plants without (1) and with (2) treatment with the proteasome inhibitor MG115. Expression of EYFP-KNL2 was detected only after treatment with MG115. KNL2 is detectable in nucleoplasm and at chromocenters (3) or in nucleoplasm and nucleolus (white arrow) and weak at chromocenters (4).
(D) Transient expression of the EYFP-KNL2 fusion construct in N. benthamiana. Three compartments are labeled: nucleoplasm (1), nucleoplasm and nucleoli (2), and nucleoplasm, nucleoli, and nuclear bodies (3).
(E) Live imaging of root tip cells of Arabidopsis transformed with the EYFP-KNL-C fusion construct. A cell undergoing mitosis is indicated by an arrow.
(F) Nucleus of an EYFP-KNL2-C transformed Arabidopsis plant immunostained with anti-GFP antibodies showed colocalization of EYFP-cenH3 immunosignals with bright DAPI-stained chromocenters.
(G) Double immunostaining of a meristematic nucleus of an EYFP-cenH3 transformant with anti-GFP for the EYFP-cenH3 (1) and anti-KNL2 antibodies (2). Colocalization of immunosignals for both proteins (3). In (F) and (G), (3) DNA is counterstained with DAPI.
To investigate the evolutionary relationship of Arabidopsis KNL2 and its putative homologs in other plant species, BLAST analysis of the conserved SANTA domain was performed. A multiple sequence alignment was generated using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/), visualized by the JalView program (see Supplemental Figure 1 online), and a phylogenetic tree for the conserved domains of KNL2 was constructed (see Supplemental Figure 2 and Supplemental Data Set 1 online) using the program MEGA4. SANTA domain containing proteins were found in all angiosperms with sequenced genomes. In most plant genomes analyzed so far (Brachypodium distachyon, sorghum [Sorghum bicolor], rice, grape [Vitis vinifera], A. thaliana, Arabidopsis lyrata, castor bean [Ricinus communis], Medicago truncatula, soybean [Glycine max], and Populus trichocarpa), except for barley (Hordeum vulgare) and maize (Zea mays), we found two genes encoding SANTA domain proteins.
Subcellular Localization and Dynamics of KNL2
To study the subcellular localization of Arabidopsis KNL2 in vivo, we constructed a fusion of KNL2 to enhanced yellow fluorescent protein (EYFP). To that end, the KNL2 cDNA was cloned into the pDONR221 vector and subcloned into pGWB41 (35Spro, C-EYFP) and pGWB42 (35Spro, N-EYFP) vectors, respectively. However, no EYFP-specific signal could be found in stably transformed Arabidopsis plants. Because the N-terminal part of KNL2 contains two putative ubiquitination sites according to the UbSite prediction program (http://protein.cau.edu.cn/cksaap_ubsite/) (Figure 1B), we assumed that the fusion protein is degraded via the proteasome degradation pathway. To test this assumption, we treated seedlings of transformants with the protease inhibitor MG115. Inhibition of the proteasome degradation pathway resulted in pronounced KNL2-specific signals at centromeric chromocenters and in nucleoplasm or in the nucleoli and nucleoplasm (Figure 1C).
To find out whether the putative ubiquitination sites at the N terminus of KNL2 are indeed responsible for the proteolytic degradation, cDNAs comprising the N-terminal part including the SANTA domain and the two putative ubiquitination sites or the C-terminal part without these regions were separately fused to EYFP (Figure 1B). Nicotiana benthamiana leaves were infiltrated with Agrobacterium tumefaciens transformed with KNL2-N-, KNL2-C-, and KNL2-EYFP fusion constructs. In all cases, fluorescence was detected in the nucleoplasm or in nucleoplasm, nucleoli, and nuclear bodies (Figure 1D). However, for the KNL2 and KNL2-N EYFP fusion constructs, the number of nuclei showing fluorescence was very low (approximately seven nuclei per 400 mm2). Treatment of N. benthamiana leaf discs with MG115 protease inhibitor increased the number of nuclei with fluorescence (32 per 400 mm2). In contrast with the KNL2 and KNL2-N constructs, expression of KNL2-C fused to EYFP resulted in a high number of nuclei showing fluorescence without application of protease inhibitor (300 per 400 mm2). These data indicate that the level of KNL2 is regulated by the proteasome degradation pathway via ubiquitination at its N-terminal part.
Since full-length KNL2 and its N- and C-terminal parts fused with EYFP showed the same localization pattern in transiently transformed N. benthamiana, but the C terminus appeared to be more stable than other variants, we generated Arabidopsis plants stably expressing EYFP-KNL2-C or KNL2-C-EYFP fusion constructs to study localization and dynamics of KNL2 in vivo. N-terminal as well as C-terminal fusion of KNL2-C with EYFP yielded no fluorescence during mitosis (Figure 1E). Interphase nuclei exhibited fluorescence signals at chromocenters and to a lesser degree in nucleoplasm, even without application of a proteasome inhibitor (Figures 1E and 1F). In differentiated root nuclei, EYFP-KNL2-C fluorescence was also present in the nucleolus. Despite the absence of the putative DNA binding domain, EYFP-KNL2-C was localized at chromocenters (Figure 1F), indicating that centromeric localization of Arabidopsis KNL2 does not require a SANTA domain.
To analyze the dynamics of EYFP-KNL2-C in root tips of Arabidopsis transformants, fluorescence loss in photobleaching (FLIP) and fluorescence recovery after photobleaching (FRAP) were applied. Bleaching of a selected area within the nucleoplasm of nuclei expressing EYFP-KNL2-C in 10-s intervals during 1 min (FLIP) resulted in disappearance of fluorescence also in nonbleached areas of nucleoplasm, chromocenters, and the nucleolus (see Supplemental Figure 3A online). FRAP experiments showed that bleached fluorescence of EYFP-KNL2-C at chromocenters recovers to ∼80% within 10 s (see Supplemental Figure 3B online). Both FLIP and FRAP experiments showed high turnover of KNL2. FLIP additionally revealed a high mobility of KNL2 between centromeres, nucleoplasm, and nucleoli.
To analyze the localization of endogenous KNL2 and to compare it with the localization of KNL2 fused to EYFP, antibodies against the C-terminal part of KNL2 (Figure 1B) were generated. Immunostaining of Arabidopsis root tip nuclei with anti-KNL2 antibodies revealed a similar localization pattern (Figure 1G, middle panel) as observed in meristematic nuclei of transformants stably expressing KNL2-C-EYFP (Figures 1E and 1F) or KNL2-EYFP, after treatment with protease inhibitor (Figure 1C, right panel, top part). No immunosignals were detected at centromeres during mitosis. Finally, double immunostaining experiments of Arabidopsis EYFP-cenH3 transformants with anti–green fluorescent protein (GFP) and anti-KNL2 antibodies (Figure 1G) confirmed the centromeric position of KNL2 in meristematic interphase nuclei (Figure 1G, right panel).
The Arabidopsis KNL2 Promoter Is Active in Dividing Tissues
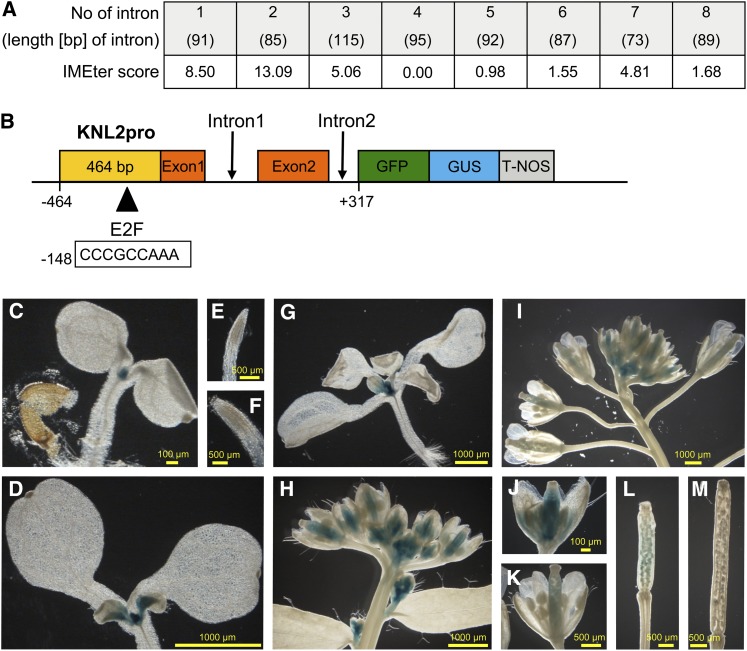
To test whether the Arabidopsis KNL2 promoter (KNL2pro) is active in diving tissues similar to cenH3pro (Heckmann et al., 2011), transgenic lines expressing a AtKNL2pro:GFP-GUS (for β-glucuronidase) reporter gene construct were generated. We performed an in silico analysis of all KNL2 introns using the IMEter online program (available through http://korflab.ucdavis.edu/cgi-bin/web-imeter.pl) that predicts intron mediated enhancement of transcription. The first and the second introns of KNL2 showed high IMEter scores (Figure 2A). As KNL2 is closely flanked by At5g02510 (its coding region terminates at –464 bp relative to KNL2 ATG), a genomic fragment ranging from –464 up to + 317 (up to the third exon) relative to the transcription initiation site of KNL2 was fused to the GFP-GUS reporter gene (Figure 2B).
Figure 2.
KNL2pro Activity in Arabidopsis during Development.
(A) IMEter scores, indicating predicted intron mediated enhancement of transcription, for all introns of KNL2 as determined by in silico analysis using the IMEter online program (Rose et al., 2008).
(B) Scheme of the KNL2pro:GFP-GUS reporter gene construct.
(C) to (M) Histochemical localization of GUS activity in Arabidopsis plants transgenic for AtKNL2pro:GFP-GUS: 4-d-old (C) and 7-d-old (D) seedling as well as their primary root tips ([E] and [F]). Fourteen-day-old seedling (G), inflorescences ([H] and [I]), mature flowers ([J] and [K]), and siliques ([L] and [M]) at different developmental stages.
Only half of the 30 T1 lines of Arabidopsis stably expressing the KNL2pro:GFP-GUS construct showed GUS staining. While GUS staining intensity varied, all positive lines showed a similar staining pattern. Five representative lines were chosen for detailed analysis.
In 4-d-old seedlings, the shoot apical meristem was stained (Figure 2C). In older plantlets (7 and 14 d old), KNL2pro activity was also detected in leaf primordia and in basal parts of emerging leaves (Figures 2D and 2G). Surprisingly, no GUS activity and no GFP fluorescence could be detected in root meristems (Figures 2E and 2F), while immunostaining with anti-KNL2 antibodies indicated the presence of KNL2 protein. Possibly, regulatory elements apart from the analyzed genomic fragment are required for KNL2 expression in root meristems. GUS activity was found in inflorescence meristems, young inflorescences, and developing flower buds (Figure 2H). Older inflorescences displayed GUS activity of variable intensity in developing sepals and pistils (excluding stigmata) (Figures 2H and 2J), while in mature flowers, GUS staining was limited to styles (Figures 2I and 2K). A negative relationship between GUS staining intensity and tissue age was found in leaves (Figures 2D and 2G), flowers (Figures 2H to 2K), and siliques (Figures 2L and 2M). For instance, GUS staining was detected in the basal part of young leaves containing actively dividing cells, but not in developed leaves.
GUS activity indicates that KNL2pro is active in dividing tissues similar to cenH3pro, except in roots meristems, suggesting that KNL2 expression might coincide with cenH3 expression as expected based on its putative involvement in cenH3 deposition to centromeres.
Expression of Arabidopsis cenH3 is regulated by the E2F transcription factor family (Heckmann et al., 2011). To see whether KNL2 is similarly regulated, the KNL2 promoter region was studied in silico using the NSITE program (available through www.softberry.com/berry.phtml?topic=promoter). A potential E2F binding site (CCCGCCAAA) was found at −148 bp upstream of ATG. Moreover, transgenic plants overexpressing E2Fa/DPa showed a 4.42-fold increase in KNL2 transcript level (De Veylder et al., 2002). Thus, KNL2 might be transcriptionally regulated in a similar way as cenH3.
Knockout of KNL2 Expression Results in Reduced cenH3 Expression and Deposition
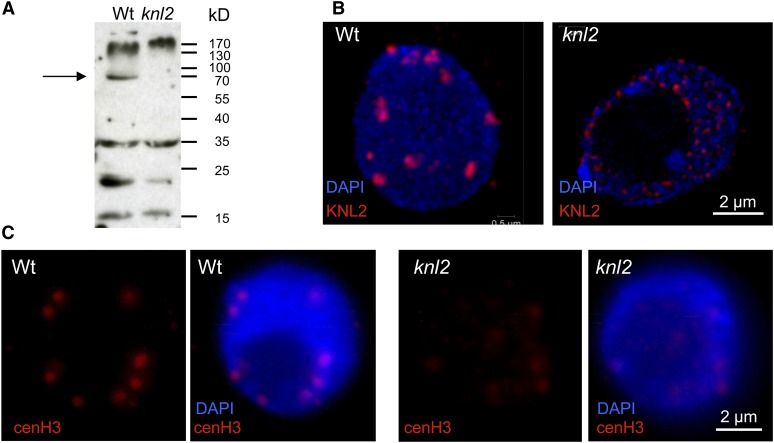
To test whether deregulation of KNL2 expression has an impact on the level of cenH3 at centromeres and subsequently on chromosome segregation, three T-DNA insertion mutants were identified for the KNL2 gene (see Supplemental Figure 4 online). The positions of T-DNA insertions were confirmed by sequencing of PCR products from the corresponding genomic positions. The T-DNA insertion lines SALK-051020 and SALK-050998 contain insertions within the third and fourth intron, respectively, and SALK-039432 within the fourth exon of At5g02520. RT-PCR analysis revealed for homozygous SALK-051020 and SALK-050998 mutants full-length KNL2 transcripts at the level of the wild type and no obvious phenotypic abnormalities. Homozygous SALK-039432 (see Supplemental Figure 4A online) yielded mainly transcripts from the region upstream the T-DNA insertion and a low amount of transcript from downstream the T-DNA (the latter likely mediated by the cauliflower mosaic virus 35S promoter within the T-DNA insertion; see Supplemental Figure 4B online). Immunoblot and indirect immunostaining confirmed the absence of KNL2 in SALK-039432. Only wild-type seedlings revealed the expected immunoblot signal of ∼66 kD (Figure 3A). Furthermore, in contrast with the wild type (Figure 3B, left panel), the knl2 mutant (SALK-039432) did not show KNL2 immunosignals at chromocenters (Figure 3B, right panel) and, most importantly, the cenH3 level at chromocenters of the knl2 mutant was strongly reduced compared with the wild type (Figure 3C). For both the knl2 mutant and Arabidopsis wild type, the immunostaining was repeated three times, and each time 10 root tips of the wild type and the knl2 mutants were analyzed. In all cases, cenH3 immunosignals at chromocenters of knl2 nuclei were weaker than in the wild type.
Figure 3.
KNL2 and cenH3 Protein Levels in Atknl2 Mutant Compared with the Wild Type.
(A) Immunoblot analysis with anti-KNL2 antibodies on protein extracts isolated from seedlings of Arabidopsis wild-type (Wt) and knl2 T-DNA insertion mutants. Arrow shows the position of KNL2 protein (∼66 kD) in the wild type and its absence in the knl2 mutant.
(B) Immunostaining of meristematic nuclei of Arabidopsis wild type and the knl2 mutant with anti-KNL2 antibodies.
(C) Immunostaining of meristematic nuclei of Arabidopsis wild type and knl2 mutant with anti-cenH3 antibodies. Immunostaining experiment was repeated three times, and each time 10 root tips of the wild type and knl2 mutant were analyzed. In (B) and (C), DNA is counterstained with DAPI.
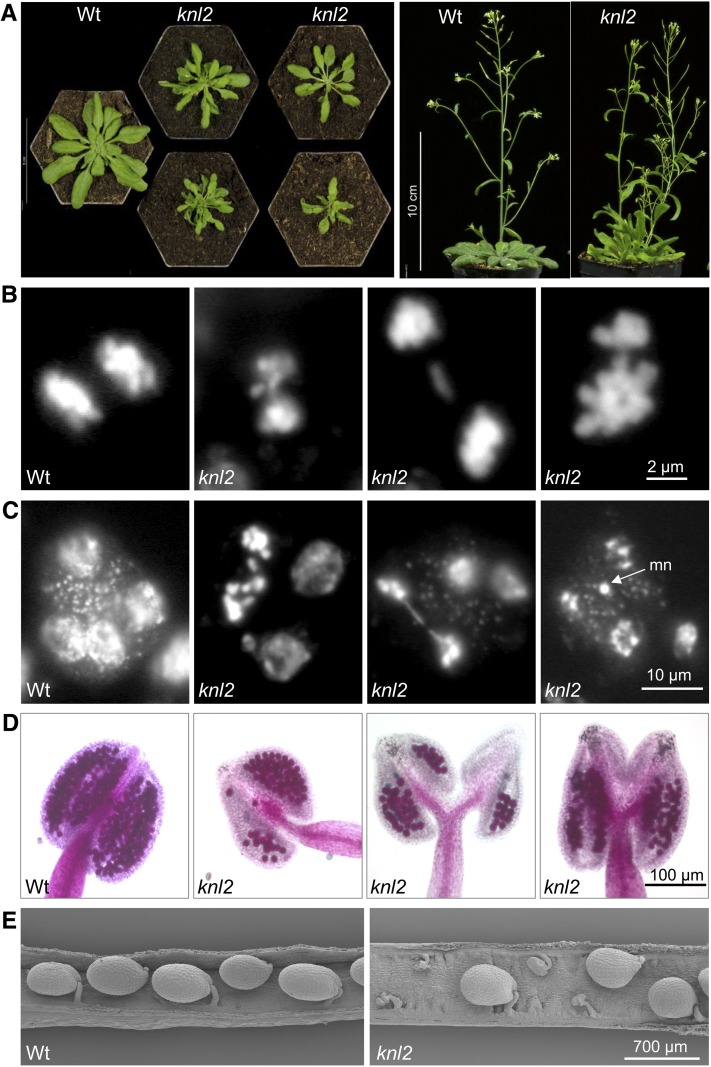
In a population of 60 homozygous knl2 mutant plants, the growth rate was highly variable (Figure 4A, left panel) and some plants showed an abnormal leaf shape. During flowering, the mutant plants formed multiple shoots with siliques of reduced size (Figure 4A, right panel). Mitosis in root tip meristems of knl2 mutants did not reveal obvious abnormalities, while somatic cell division in young flower buds (1 mm) of the knl2 mutant showed 10% of anaphases (out of 1500 cells) with bridges and/or lagging chromosomes (Figure 4B). Analysis of meiocytes of the knl2 mutant showed micronuclei within pollen tetrads (Figure 4C), a reduced number of vital pollen grains (Figure 4D) and seed abortion. In 50 siliques of the knl2 mutant, on average 30% of seeds were aborted, while only 7% aborted seeds were found in the wild type (Figure 4E).
Figure 4.
Phenotype of the knl2 Mutant.
(A) Comparison of the phenotype of knl2 mutant with that of the wild-type (Wt). Plants were grown for 4 weeks (left panel) or 6 weeks (right panel) on soil.
(B) Mitotic anaphases of the wild type and of a knl2 mutant plant with bridges and lagging chromosomes. Among 400 anaphases of the wild type and 1500 anaphases of the knl2 mutant, 0.5 and 10% showed bridges, respectively.
(C) Pollen tetrads of the wild type and of a knl2 mutant with bridges and micronuclei (mn), respectively.
(D) Anthers of the wild type and of a knl2 mutant after Alexander staining to indicate viable pollen.
(E) Scanning electron microscopy images of siliques of the wild type and a knl2 mutant.
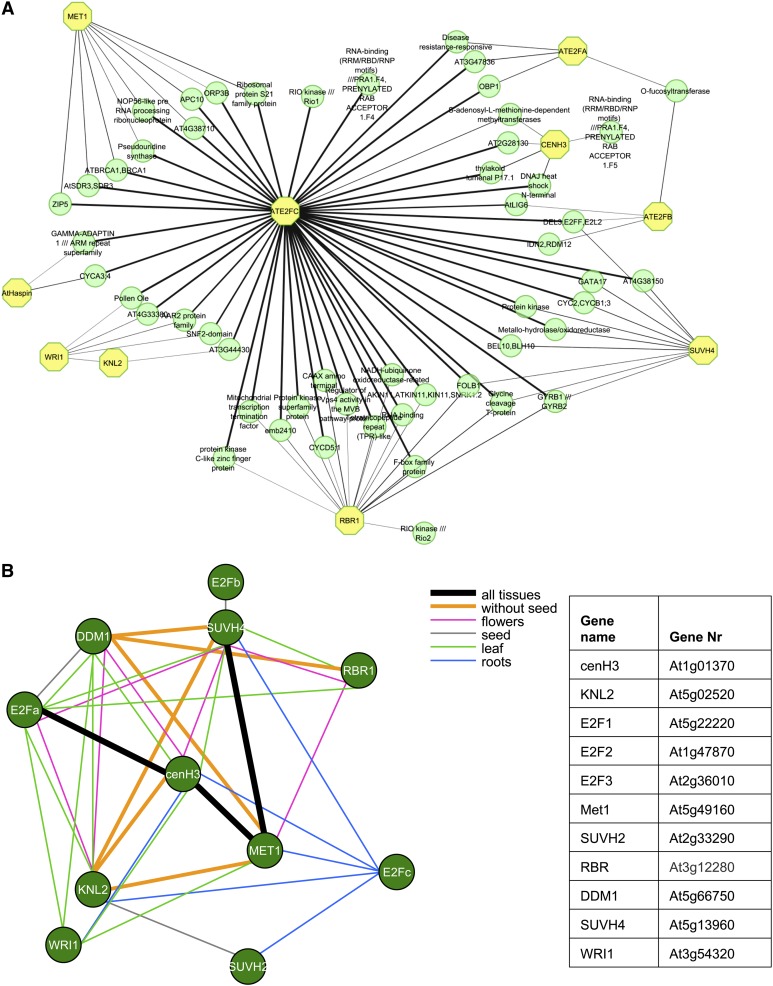
A Regulatory Network for cenH3 Assembly in Arabidopsis
Colocalization of KNL2 and cenH3 at Arabidopsis centromeres, expression activity of both promoters in meristematic tissues, and a reduction of cenH3 amount at centromeres of the knl2 mutant indicate an involvement of KNL2 in centromeric assembly of cenH3. Therefore, we addressed the question how KNL2 activity and cenH3 assembly are regulated. Since KNL2pro contains an E2F binding site and is active in dividing tissues, it might be regulated by E2F transcription factors in a cell cycle–dependent manner.
The promoter binding capacity of E2F transcription factors is regulated by the retinoblastoma-related (RBR) protein, which, when hypophosphorylated, binds E2Fs and inhibits their activity. Downregulation of RBR expression by RNA interference (RNAi) results in a 1.64-fold increase of KNL2 transcript (https://www.genevestigator.com/gv/). Thus, regulatory networks of cenH3, KNL2, E2Fa,b,c, and RBR genes were reconstructed based on gene-by-gene interactions. Functional relationships between genes can be reconstructed based on, for example, modified transcriptional activity, posttranscriptional modifications, protein–protein interactions of their products in different protein complexes, or their participation in several metabolic pathways.
Since histone modifications and DNA methylation of centromeric chromatin are important for assembly of cenH3 at centromeres (Gopalakrishnan et al., 2009; González-Barrios et al., 2012; Kim et al., 2012), genes encoding DNA (cytosine-5)-methyltransferase 1 (MET1), decreased DNA methylation1 (DDM1), and the histone methyltransferase SUVH4 were additionally included for analysis. The reconstituted regulatory network is composed of 113 genes, which are linked by a bar if the Mutual Information score was above 0.5 (Figure 5A). In a further step, genes encoding proteins with unknown function as well as proteins of metabolic pathways were manually omitted from the network. To categorize the remaining 61 genes, the gene list was processed with the Web-based AgriGO tool (Du et al., 2010), to perform an enrichment analysis for gene ontology terms. This analysis showed that 16 out of the 61 genes belong to the cell cycle and the cell cycle regulation and 25 to nucleic acid metabolism (see Supplemental Figure 5 online). A more comprehensive list of genes involved in these pathways is given in Supplemental Data Set 2 online.
Figure 5.
Gene Regulatory Network for cenH3 Deposition in Arabidopsis and Coexpression of KNL2 and cenH3 with Genes of the RBR-E2F Transcription Regulation Pathway and with DNA and H3 Methyltransferases.
(A) Key regulator genes are indicated by yellow octagons. The network is inferred using collected gene expression data from the National Center for Biotechnology Information Gene Expression Omnibus (see Methods). The edge width represents the interaction score calculated by MRNET algorithm (Meyer et al., 2008). KNL2, kinetochore null2; cenH3, centromeric histone H3; MET1, methyltransferase 1; E2Fa, E2Fb, E2Fc, Arabidopsis thaliana E2F transcription factors; RBR1, retinoblastoma related1; SUVH4, histomemethyltransferase; Haspin, Haspin kinase; WRI1, sequence-specific DNA binding transcription factor WRINKLED1.
(B) Tissue-specific coexpression patterns are derived from predefined microarray sets in the CORNET program. Different colors indicate the tissues that display a significant correlation of expression between the corresponding genes linked by a bar (Pearson coefficient at least 0.6 and P value < 0.05).
Additionally, the in silico analysis revealed a central role of the transcription factor E2Fc in this regulatory network and its direct or indirect connection to the expression of almost all genes of this network (Figure 5A).
Since this analysis represents a general pattern of gene-by-gene regulation, we inspected the tissue-specific expression pattern of selected Arabidopsis genes (Figure 5B) using the software CORNET and a set of microarrays predefined by the program (microarray-derived expression data from roots, leaves, flowers, and seeds). The results are summarized in Figure 5B. KNL2 expression showed a strong correlation to the expression of cenH3, MET1, and SUVH4 in roots, leaves, and flowers. Expression of cenH3 was correlated with that of E2Fa and MET1 in all analyzed tissues. Thus, microarray-derived data suggest that expression of KNL2 and of cenH3 is controlled by the E2F-RBR pathway and is tightly connected with the expression of DNA and histone methyltransferases.
To test whether the regulatory pathway of cenH3 assembly reconstituted for Arabidopsis is conserved among different organisms, we generated a cenH3 assembly pathway for human using the STRING tool (Search Tool for Retrieval of Interacting Genes/Proteins; http://string-db.org/newstring_cgi/show_input_page.pl?UserId=tliElpgeVlzFandsessionId=6_s6ILXTavzg).
Since we found that human components of the E2F-RBR pathway, DNA methyltransferases (DNMT1 and DNMT3B), and histone H3 lysine 9 (H3K9) methyltransferases (see Supplemental Table 1A online) have a strongly coordinated expression (see Supplemental Figure 6 online) and for some components of this regulatory network protein–protein interactions were demonstrated experimentally (see Supplemental Table 1B online), this network seems indeed to be highly conserved among eukaryotes.
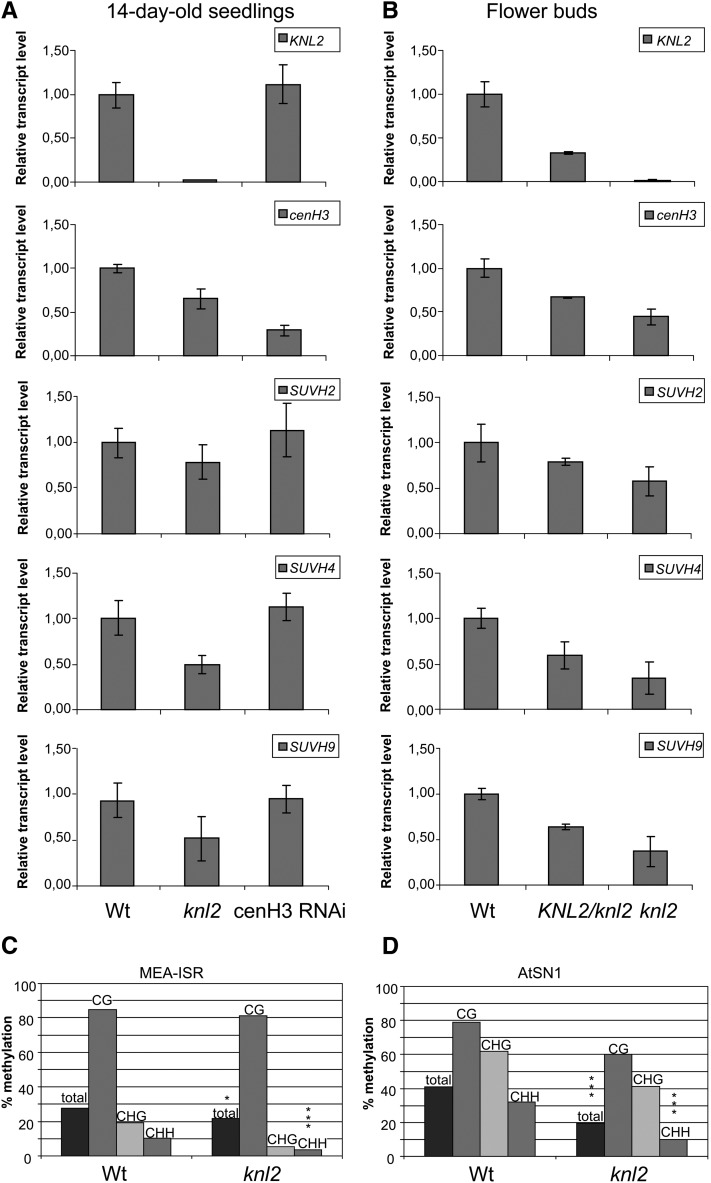
Expression of cenH3, SUVH2, SUVH4, and SUVH9 Genes and DNA Methylation Are Reduced in knl2 Mutant Plants
Real-time RT-PCR analysis showed the absence of KNL2 transcripts in seedlings and flower buds of the knl2 mutant and a strong transcript reduction in flower buds of hemizygous KNL2/knl2 plants (Figures 6A and 6B).
Figure 6.
Expression of KNL2, cenH3, SUVH2, SUVH4, and SUVH9 in the knl2 Mutant and cenH3 RNAi Plants and DNA Methylation at MEA-ISR and At-SN1 Loci in knl2 Mutants.
(A) and (B) Real-time quantitative RT-PCR analysis of KNL2, cenH3, SUVH2, SUVH4, and SUVH9 mRNA transcripts in 14-d-old seedlings of the knl2 mutant and cenH3 RNAi transformants compared with the wild type (A) and in flower buds of heterozygous KNL2/knl2 and homozygous knl2 mutants compared with the wild type (B). Amplification of ACTIN2 mRNA was used for data normalization.
(C) and (D) DNA methylation was determined by bisulfite sequencing of genomic DNA of 7-d-old seedlings. Individual clones were sequenced for the different genotypes. The bars mark the levels of DNA methylation in percentage of methylated cytosines relative to total cytosines. Wt, wild-type plants (Columbia-0).
(C) DNA methylation at MEA-ISR. Significant differences in total DNA methylation between the different genotypes were checked by χ2 tests. Wild type, n = 26; knl2, n = 19 (complete methylation: χ2, 5.54, *P < 0.05; asymmetric methylation: χ2, 18.24, ***P < 0.001).
(D) DNA methylation at At-SN1. Wild type, n = 12; At-knl2, n = 10 (χ2, 45.48, ***P < 0.001; asymmetric methylation: χ2, 51.30, ***P < 0.001).
Notably, in the knl2 mutant not only the amount of the cenH3 protein was reduced, but also the cenH3 transcript level in seedlings and flower buds of the knl2 mutant was reduced compared with that of the wild type (Figures 6A and 6B). The reduction was more pronounced in flower buds (45% of the wild-type level) than in seedlings (65%). Even in flower buds of hemizygous KNL2/knl2 mutants, the cenH3 transcript level was reduced to 67% of the wild type.
By contrast, we found that downregulation of cenH3 expression in cenH3 RNAi transformants does not influence the expression of KNL2, indicating an upstream function of KNL2 relative to cenH3 (Figures 6A and 6B).
As suggested by the predicted regulatory network (Figure 5A), the histone methyltransferase SUVH4 and members of the RNA-directed DNA methylation pathway (involved in de novo2, SUVH2, and DDM1) are candidates that are potentially influenced by reduced KNL2 expression. Therefore, we analyzed the expression of the histone methyltransferase encoding genes SUVH4, SUVH2, and SUVH9 in 14-d-old seedlings of Arabidopsis wild type, knl2 mutants, and cenH3 RNAi transformants (Lermontova et al., 2011a) by real-time RT-PCR (Figure 6A). We found that expression of SUVH4 and SUVH9 was reduced in the knl2 mutant to 50% of the wild-type level, but expression of SUVH2 was only slightly lower than in the wild type (Figure 6A). In cenH3 RNAi transformants, expression of all three genes remained at the level of the wild type (Figure 6A). Additionally, we analyzed expression of SUVH2, SUVH4, and SUVH9 in flower buds (tissues enriched in mitotic and meiotic divisions) of wild-type, hemizygous and homozygous knl2 mutant plants (Figure 6B). Already in hemizygous lines, all three histone methyltransferases analyzed showed a reduced transcript level. This effect was more pronounced in homozygous mutants (Figure 6B).
Since KNL2 expression was strongly correlated with that of MET1 and of histone methyltransferase SUVH4, we asked whether the level of DNA methylation mediated by RNA-dependent DNA methylation (RdDM) is also affected in knl2 mutants. Because it is difficult to analyze DNA methylation in centromeric regions, which cannot be distinguished from pericentromeric regions, we performed bisulfite conversion of the knl2 genome and sequenced the intergenic region at the MEDEA locus (MEA-ISR; Johnson et al., 2008) and the At-SN1 locus. Both loci have been used in several studies as marker regions for the effect of mutations on RdDM at endogenous sequences (Wierzbicki et al., 2008, 2009; Johnson et al., 2008). In knl2 seedlings, the DNA methylation in asymmetric context at MEA-ISR (Figure 6C) and at At-SN1 (Figure 6D) is significantly reduced compared with the wild-type seedlings. Such reduction of RdDM in the At-SN1 region was also described for suvh2 and suvh9 mutant plants (Johnson et al., 2008; Kuhlmann and Mette, 2012) and for other components of the RdDM pathway (Zilberman et al., 2003; He et al., 2009). Thus, we assume that KNL2 in Arabidopsis is involved in the RdDM pathway and might contribute to reduce histone H3 lysine 9 di-methylation (H3K9me2) via reduced cytosine methylation (Soppe et al., 2002).
DISCUSSION
KNL2 Is Characterized by a Conserved SANTA Domain
We identified an Arabidopsis KNL2 protein homolog. KNL2 homologs share a conserved SANTA domain, implying a putative function in regulating chromatin remodeling. Interestingly, KNL2 proteins of vertebrates contain SANTA and SANT domains together, but the KNL2 proteins of plants contain only the SANTA domain. D. Zhang et al. (2006), based on in silico analysis of SANTA domain–containing proteins, assumed that the highly conserved hydrophobic residues of SANTA domains might be responsible for a function of this domain in protein–protein interactions, but not for DNA binding. Nevertheless, the SANT domain is found in a number of chromatin remodeling proteins with multiple activities such as DNA binding, histone tail binding, and protein–protein interactions (Aasland et al., 1996; Wang et al., 2006).
The Subcellular Localization of Arabidopsis KNL2 Is Similar to That of Yeast, but Differs from That of Mammals
We showed that Arabidopsis KNL2 is associated with centromeres during all stages of the cell cycle, except from metaphase to mid-anaphase, similar to Mis18 of fission yeast (Hayashi et al., 2004; Williams et al., 2009), but different to the human Mis18 complex, which is only transiently present at centromeres after mitotic exit (Fujita et al., 2007; Maddox et al., 2007; Silva and Jansen, 2009).
We showed previously that in plants cenH3 loading to centromeres occurs during G2 and/or prophase (Lermontova et al., 2006, 2007), but in metazoans it is claimed to occur during anaphase-early G1 (Jansen et al., 2007; Schuh et al., 2007; Hemmerich et al., 2008). Likely this difference is predetermined by the different cell cycle periods during which KNL2 is located at centromeres. Localization of KNL2 at centromeres throughout the cell cycle (except M phase) might be also related to the at least limited cenH3 turnover throughout the cell cycle in Arabidopsis (Lermontova et al., 2011b), which was not observed for human cenH3 (Hemmerich et al., 2008).
Full-length KNL2 and its C-terminal fragment KNL2-C show similar nuclear localization patterns. Either KNL2-C possesses DNA binding capacity or it is located at centromeres as part of a protein complex. The second possibility gains support from the high turnover rate of KNL2-C (see Supplemental Figures 3A and 3B online) that is not typical for DNA-associated proteins, such as cenH3 (Lermontova et al., 2011b).
In differentiated nuclei of stable Arabidopsis transformants and in transiently transformed N. benthamiana leaves, all KNL2 variants fused with EYFP were localized in the nucleolus. A similar situation was found for assembly factors of nonplant species, such as the human HJURP (Kato et al., 2007; Dunleavy et al., 2009) and CAL1 of Drosophila (Schittenhelm et al., 2010). Chen et al. (2012) proposed a functional link between nucleolus and centromere assembly, but the detailed mechanism remains to be elucidated. The regulatory network suggested in Figure 5A is compatible with such a link because it includes genes encoding nucleolar proteins (see Supplemental Data Set 2 online).
Stable Arabidopsis transformants expressing full-length KNL2-EYFP under the control of the strong cauliflower mosaic virus 35S promoter displayed fluorescence signals at chromocenters, in nucleoplasm, and in nucleoli only after application of the proteasome inhibitor MG115 (Figure 1C). Thus, we uncovered proteolytic degradation as a mechanism regulating the level of KNL2. This degradation is most likely regulated by ubiquitination at Lys-161 and Lys-242 (Figure 1B). This assumption finds support by the increased number of nuclei expressing KNL2-N-EYFP containing Lys-161 and Lys-242 in N. benthamiana after application of proteasome inhibitor, while the C-terminal part (KNL2-C-EYFP) without Lys-161 and Lys-242 yielded many fluorescent interphase nuclei in N. benthamiana and stable transformants of Arabidopsis without application of the proteasome inhibitor.
Inactivation of KNL2 Affects cenH3 Loading and Mitotic and Meiotic Divisions
Knockout of KNL2 via insertion of T-DNA into the 4th exon caused a reduced level of cenH3 at centromeres as demonstrated by immunostaining experiment. The consequences were most pronounced during flowering stages, when 10% of mitotic anaphases showed anaphase bridges and lagging chromosomes in flower buds, micronuclei in pollen tetrads, and later aborted seeds, but the vegetative growth of the knl2 mutant was also reduced.
Depletion of KNL-2 in C. elegans yielded defects in mitotic and meiotic chromosome segregation, and depletion of human KNL-2 resulted in mitotic defects (Maddox et al., 2007). Similar to the knl2 mutant, formation of anaphase bridges was observed in the Mis18α mutant of mice (Kim et al., 2012). This might be due to disturbed localization, and therefore function, of spindle assembly checkpoint components. Indeed, it was shown that in KNL-2 depleted C. elegans, the BUB-1 spindle assembly checkpoint protein failed to localize to chromosomes (Maddox et al., 2007).
Thus, KNL2 plays a role in assembly of cenH3 at centromeres, and its knockout reduces, but does not abolish, centromeric localization of cenH3. Other proteins, such as the second SANTA domain–containing protein encoded by At1g58210, can possibly at least partially compensate for the loss of KNL2.
cenH3 Assembly Is Regulated at the Transcriptional Level
Besides reduced deposition of cenH3 at centromeres, inactivation of KNL2 also triggers a reduced cenH3 transcript level. Because seedlings of cenH3 RNAi plants displayed a KNL2 transcript level similar to that of the wild type, KNL2 apparently functions upstream of cenH3 expression and likely regulates cenH3 amount and deposition. Involvement of KNL2 in transcriptional regulation of genes is supported by the fact that human proteins harboring SANTA and SANT domains interact with the transcription factor Sp1 (Gunther et al., 2000) and participate in transcriptional regulation of gene expression (Ding et al., 2004). Also, depletion of CAL1, a cenH3 assembly factor of Drosophila, not only affects cenH3 localization, but also leads to a reduced cenH3 level (Pauleau and Erhardt, 2011).
We demonstrated, by means of an KNL2pro:GFP-GUS reporter construct, preferential expression of KNL2 in dividing tissues and showed that the KNL2 promoter contains an E2F binding site, similar to what was described for cenH3 (Heckmann et al., 2011). Joint expression regulation of both genes could be a prerequisite for the involvement of KNL2 in cenH3 deposition at centromeres in dividing tissues. Moreover, KNL2 transcripts are increased in E2Fa/DPa-overexpressing plants (De Veylder et al., 2002) and in plants (https://www.genevestigator.com/gv/) with a reduced RBR level, which enables E2Fs to activate E2F-responsive promoters (Lageix et al., 2007). Additionally, the proposed regulatory networks suggest a central role of the E2F/RBR regulatory pathway, particularly of E2Fc, in the regulation of cenH3 and KNL2 expression. Transcription of cenH3 and KNL2 in nondividing tissues might be repressed by (1) E2Fc, which generally inhibits E2Fc-responsive genes (del Pozo et al., 2002, 2006) and apparently occupies a central role in this regulatory network; (2) an interplay of typical and atypical E2Fs (Berckmans and De Veylder, 2009); and/or (3) E2F/RBR, recruiting enzymes with chromatin silencing/inactivating features to E2F-responsive promoters (Luo et al., 1998; van den Heuvel and Dyson, 2008), such as a DNA methyltransferase, which might methylate KNL2 and cenH3 promoters in nondividing tissues. The latter mechanism would explain the increased expression of cenH3 (3.3-fold) and KNL2 (3.5-fold) in met1-3 mutants with a reduced level of CG methylation (Mathieu et al., 2007). An increase in expression of cenH3 (approximately twofold) and of KNL2 (∼1.8-fold) was also observed after treatment of Arabidopsis seedlings with the DNA methylation inhibitor 5-aza-2′-deoxycytidine (https://www.genevestigator.com/gv/). Computational analysis showed a strong correlation of the expression of KNL2 and cenH3 with genes of the RBR-E2F regulatory pathway also in humans, indicating that this regulation might be highly conserved, as we suggested previously (Heckmann et al., 2011).
Arabidopsis KNL2 Is Involved in the Epigenetic Regulation of cenH3 Assembly in Addition to Regulation of cenH3 Transcript Level
We observed that in a population of knl2 mutant plants (∼60 plants), ∼30% of plants show reduced growth (Figure 4) and a phenotype similar to that of the cenH3 RNAi transformants (Lermontova et al., 2011a), while growth rate of another 70% of plants was only slightly reduced or did not differ from the wild type. Such phenotypic variation within a population of homozygous mutants was also observed for epigenetic mutants of Arabidopsis, such as met1-3 (Mathieu et al., 2007) and ddm1, the latter showing a gradual accumulation of developmental aberrations during inbreeding (Vongs et al., 1993; Kakutani et al., 1996). MET1 and the chromatin remodeling factor DDM1 (Soppe et al., 2002) together with the methyltransferases SUVH4, SUVH5, and SUVH6 (Ebbs et al., 2005; Ebbs and Bender, 2006) are needed to maintain DNA and H3K9 methylation at centromeric 180-bp repeats and to suppress transcription of centromeric sequences (X. Zhang et al., 2006). What is the function of KNL2 in this process? Downregulation of KNL2 expression results not only in reduced expression and centromeric localization of cenH3, but also in reduced expression of H3K9 methyltransferases (SUVH2, SUVH4, and SUVH9), in decreased H3K9me2 at centromeres, and in reduced DNA methylation of genomic regions subjected to RNA-directed DNA methylation. The regulatory network postulated for cenH3 assembly components of Arabidopsis indicates correlated gene expression of KNL2, cenH3, MET1, and SUVH4 (Figure 5). A correlation between H3K9 and DNA methylation and cenH3 assembly at centromeres was already shown for other organisms. Kim et al. (2012) tested epigenetic markers for silenced and open chromatin state of centromeres in mis18α mutant mouse embryos. They showed that H3K9me2 and H3K9me3 levels were decreased in homozygous compared with heterozygous mutants, whereas acetylation of H3 was increased, that Mis18α interacts with DNA methyltransferases DNMT3A/3B, and that this interaction is critical for maintaining DNA methylation, for regulating the epigenetic status of centromeric chromatin, and for licensing cenH3 loading.
In the fission yeast S. pombe, tethering of histone H3K9 methyltransferase Clr4 to euchromatic loci via specific DNA binding proteins promotes heterochromatin assembly and simultaneously de novo cenH3 incorporation, kinetochore assembly, and centromere function on plasmid-based minichromosomes (Folco et al., 2008; Kagansky et al., 2009). In mammalian cells, the kinetochore protein CENP-C recruits DNA methyltransferases to centromeric and pericentromeric satellite sequences (Gopalakrishnan et al., 2009). The loss of CENP-C or of DNA methylation leads to chromosome misalignment and mis-segregation during mitosis and to increased transcription of centromeric repeats. Because the presumed protein–protein interaction network for the human cenH3 assembly pathway also contains DNA and H3K9 methyltransferases (see Supplemental Figure 6 online), the regulation of cenH3 assembly seems to be conserved among eukaryotic phyla.
METHODS
Plasmid Construction and Plant Transformation
The entire open reading frames of KNL2 and KNL2-N and KNL2-C fragments were amplified by RT-PCR with RNA isolated from flower buds of Arabidopsis thaliana wild type and cloned into the pDONR221 vector (Invitrogen) via the Gateway BP reaction. From pDONR221 clones, they were recombined via Gateway LR reaction (Invitrogen) into the two attR recombination sites of the Gateway-compatible vectors pGWB41 and pGWB42 (http://shimane-u.org/nakagawa/gbv.htm), respectively, to study localization of both proteins in vivo. To study the activity of the KNL2 promoter in different tissues of Arabidopsis, a KNL2pro:GFP-GUS reporter construct was generated using a genomic fragment ranging from –464 up to + 317 relative to the transcriptional KNL2 start site and fused to the GFP-GUS reporter gene of the pKGWFS7.0 vector (http://gateway.psb.ugent.be/information).
To produce recombinant protein in Escherichia coli for the generation of antibodies, the KNL2-C fragment was subcloned into the Gateway-compatible pDEST17 vector (Invitrogen). Plants of Arabidopsis accession Columbia-0 were transformed according to the flower dip method (Clough and Bent, 1998). T1 transformants were selected on Murashige and Skoog medium (Murashige and Skoog, 1962) containing 50 mg/L of kanamycin and 50 mg/L hygromycin. Growth conditions in a cultivation room were 20°C 8 h light/18°C 16 h dark or 20°C 16 h light/18°C 8 h dark.
RNA Isolation and RT-PCR Analysis
Total RNA was isolated from 16-d-old seedlings and flower buds of control and knl2 mutant plants using the Qiagen RNeasy plant mini kit. To avoid contamination with genomic DNA, total RNA for RT-PCR analysis was treated with DNase. Reverse transcription was performed using a first-strand cDNA synthesis kit (Fermentas), oligo(dT)18 primer (Fermentas), and 2 µg of total RNA as a starting material. For quantitative real-time RT-PCR, an iCycler iQ (Bio-Rad) and an iQ SYBR Green Supermix (Bio-Rad) were used. Each transcript was quantified three times using three independent biological replicates. A fragment of ACTIN2 cDNA was amplified for data normalization. The cDNA equivalent to 10 or 100 ng of total RNA was used in a 25-μL PCR reaction to amplify ACTIN2 cDNA or KNL2, cenH3, SUVH2, SUVH4, and SUVH9 cDNAs, respectively. The following program was used to amplify all transcripts: initial denaturation, 5 min at 95°C; then 40 cycles with 15 s denaturation at 95°C, 30 s annealing at 62°C, and 30 s elongation at 72°C.
Histochemical GUS Enzyme Activity Assay
GUS activity was detected using 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (Jefferson et al., 1987) as described (Heckmann et al., 2011). A Nikon SMZ1500 stereomicroscope equipped with a Nikon Digital Sight DS-SMc camera was used for light microscopy, and images were processed via Nikon NIS-Elements AR 3.2x software.
Analysis of T-DNA Insertion Mutants
Seeds of T-DNA insertion lines were obtained from the European Arabidopsis Stock Center (http://Arabidopsis.info/). To confirm the presence of, and to identify heterozygous versus homozygous T-DNA insertions, we performed PCR with pairs of gene-specific primers flanking the putative positions of T-DNA (see Supplemental Table 2 online) and with a pair of gene-specific and T-DNA end-specific primers (LBb3.1; see Supplemental Table 2 online).
Expression and Purification of Recombinant Protein and Generation of Antibodies
The E. coli expression strain BL21 (Studier and Moffatt, 1986) was transformed with the expression vector pDEST17-KNL2-C. Recombinant protein synthesis was induced upon addition of isopropyl β-d-thiogalactopyranoside (1 mM final concentration) during exponential growth at OD600 = 0.4 for 4 h at 37°C. Recombinant protein was purified via His-tag using Ni2+-NTA beads (Qiagen) under denaturing conditions. The purified fusion proteins were refolded by dialysis with a gradient of decreasing concentration of urea using dialysis system from Scienova. Two rabbits were immunized with 500 µg of purified protein three times.
Immunoblot Analysis and Immunostaining
Protein extraction and immunoblot analysis were performed as described (Lermontova et al., 2008).
Immunostaining of nuclei/chromosomes was performed as described (Jasencakova et al., 2000). EYFP-KNL2-C and EYFP-cenH3 fusion proteins were detected with rabbit (1:500; Clontech) or mouse (1:500; Roche) polyclonal antisera against GFP and with goat anti-rabbit rhodamine (1:200; Jackson Immuno Research Laboratories) or with goat anti-mouse Alexa488 (1:200; Sigma-Aldrich). Endogenous KNL2 was detected using rabbit antibodies against the recombinant C-terminal part of KNL2 (1:1000) and goat anti-rabbit rhodamine (1:200; Jackson Immuno Research Laboratories).
Chromosome Preparation and Analysis of Mitosis and Meiosis
For mitosis analysis, flower buds or roots, and for meiosis analysis, flower buds were fixed in ethanol/acetic acid (3:1) overnight with several changes of fixation solution. The fixed material was washed 3× 10 min in 1× citric buffer (4 mM citric acid and 6 mM tri-sodium-citrate, pH 4.8) and digested in 2.5% PCP/1× citric buffer at 37°C for 1 h. The digested material was squashed in a drop of 45% acetic acid, and the slides were air-dried after removal of the cover slip. The slides were counterstained with 2 μg mL−1 of 4′,6-diamidino-2-phenylindole (DAPI) in Vectashield mounting medium (Vector Laboratories) and checked under an epifluorescence microscope (BX61, Olympus) equipped with a×100/1.30 plan apochromat objective.
Whole-Mount Preparation
Siliques of different developmental stages were fixed in ethanol-acetic acid (9:1) overnight at 4°C and dehydrated in 70 and 90% ethanol, for 1 h each. The preparation was then cleared in chloral hydrate (chloral hydrate:water:glycerol = 8:2:1) overnight at 4°C. Seeds in siliques were counted under a binocular (Carl Zeiss).
Alexander Staining
Flowers and flower buds were collected in 10% ethanol and incubated overnight at 10°C. Anthers were isolated and put on slides. Dissected anthers were incubated with Alexander stain (Alexander, 1969) under cover slips for 15 min at room temperature and evaluated using a light microscope Axiophot (Carl Zeiss).
Microscopy Analysis of Fluorescent Signals
For time-lapse microscopy, seedlings of transformants were grown in cover slip chambers (Nalge Nunc International) for 7 to 10 d and analyzed with a LSM 510 META confocal laser scanning microscope (Carl Zeiss). EYFP was excited with a 488-nm laser line and specific fluorescence recorded with a 505- to 550-nm band-pass filter. FLIP and FRAP were performed according to Lermontova et al. (2011b).
The proteasome inhibitor MG115 was used to prevent degradation and to increase accumulation of transgenic proteins in plants. One-week-old transgenic Arabidopsis seedlings were incubated in liquid Murashige and Skoog medium (Duchefa Biochemie) with either 50 μM MG115 (Serva) dissolved in DMSO or 50 μM DMSO solution (mock treatment) for 10 to 16 h. The fusion proteins were detected by microscopy.
Light Microscopy
Leaf samples were fixed with 1% glutaraldehyde and 4% formaldehyde in 50 mM phosphate buffer, pH 7.0. After dehydration and embedding in Spurr resin, semithin sections (1.0 µm) were cut on a Reichert-Jung Ultracut S (Leica) and stained with 2% crystal violet. Digital recordings were made on an Axiovert microscope equipped with an Axiocam (Carl Zeiss).
Scanning Electron Microscopy
Siliques were fixed with 1% glutaraldehyde and 4% formaldehyde in 50 mM phosphate buffer, pH 7.0, for 2 h. After washing and dehydration in an ethanol series, samples were critical point dried in a Bal-Tec critical point dryer. After gold coating in an Edwards S150B sputter coater (Edwards High Vacuum), probes were examined in a Hitachi S4100 scanning electron microscope (Hisco Europe) at 5 kV acceleration voltage. Digital recordings were made and saved as TIFF files.
Generation of Gene Regulatory Networks
To infer gene regulatory networks, the information flow between pairs of genes is mathematically transformed into a mutual information matrix instead of a correlation matrix. Mutual information can be considered as a generalization of correlation analysis by dealing with nonlinear, nonmonotonic dependencies among expression profiles. It is based on Shannon entropy and mutual information (MRNET, ARACNE, and CLR) (Meyer et al., 2008; Friedel et al., 2012). The gene expression data were taken from experiments with the following National Center for Biotechnology Information Gene Expression Omnibus accession numbers: GSE5529, GSE17496, GSE23950, and GSE25067 (sample numbers are GSM436179-GSM436182; GSM128749-GSM128756; GSM436174-GSM436178; and GSM615829-GSM615840) (http://www.ncbi.nlm.nih.gov/geo/).
DNA Methylation Analysis (Bisulfite Sequencing)
Genomic DNA for methylation analysis was extracted with a DNeasy plant maxi kit (Qiagen) from seedlings grown on GM medium for 1 week under long-day conditions.
Bisulfite-mediated chemical conversion of DNA was done using a Qiagen EpiTect bisulfite kit following the manufacturer’s instructions. Amplicons were transformed with the help of the StrataClone PCR cloning kit. Sequencing of single clonal colonies was done with the Cyclereader auto sequencing kit (Fermentas) and a Licor 4300 DNA analyzer. Statistical analysis was performed using the χ2 test comparing the number of methylation events out of all possible cytosine sites detected by bisulfite sequencing. Standard deviation is calculated by the QuickBasic program for Exact and Mid-P Confidence Intervals for a Binomial Proportion. Primers used for amplification of genomic regions are defined in the Supplemental Table 2 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: KNL2 (At5g02520), cenH3 (At1g01370), SUVH2 (At2g33290), SUVH4 (At5G13960), and SUVH9 (At4g13460).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Multiple Alignment of the N-Terminal Parts of Plant KNL2 Orthologs.
Supplemental Figure 2. Evolutionary Relationships of KNL2 Proteins of 25 Taxa.
Supplemental Figure 3. FLIP and FRAP on 10-d-Old Seedlings Expressing EYFP-KNL2-C.
Supplemental Figure 4. Schematic View of the KNL2 Gene with the Corresponding T-DNA Insertions and Analysis of KNL2 Transcript in the SALK-039432 Mutant.
Supplemental Figure 5. Gene Ontology Enrichment Analysis on Arabidopsis Genes Selected from Regulatory Network Using AgriGO Web Application.
Supplemental Figure 6. Protein–Protein Interaction Network of Selected Human Proteins in STRING.
Supplemental Table 1A. List of Selected Arabidopsis Genes Involved in Regulation of cenH3 Assembly and Their Homologs of H. sapiens.
Supplemental Table 1B. H. sapiens Proteins Involved in cenH3 Assembly and Their Interacting Partners.
Supplemental Table 2. Primers Used in This Study.
Supplemental Data Set 1. Alignment of the N-Terminal Parts of Plant KNL2 Orthologs of 25 Taxa Corresponding to Supplemental Figure 2.
Supplemental Data Set 2. List of Top Ranked Gene Interactions with Given Gene Annotation Taken from TAIR10 and MapMan in Arabidopsis.
Acknowledgments
We thank Andrea Kunze and Joachim Bruder for technical assistance, Udo Conrad and Ulrike Gresch for the generation of anti-KNL2 antibodies, Andreas Houben for critical reading of the article and helpful suggestions and discussions, and Karin Lipfert and Heike Ernst for help with preparation of figures. This work was supported by a grant from the Deutsche Forschungsgemeinschaft to I.L. (LE2299/1-1).
AUTHOR CONTRIBUTIONS
I.L. designed the research. I.L., M.K., T.R., S.H., M.S., D.D., and V.S. performed research. I.L., M.K., S.F., T.R., S.H., M.S., D.D., and V.S. analyzed data. I.L., M.K., S.F., and I.S. wrote the article.
Glossary
- EYFP
enhanced yellow fluorescent protein
- FLIP
fluorescence loss in photobleaching
- FRAP
fluorescence recovery after photobleaching
- GUS
β-glucuronidase
- RNAi
RNA interference
- RdDM
RNA-dependent DNA methylation
- DAPI
4′,6-diamidino-2-phenylindole
References
- Aasland R., Stewart A.F., Gibson T. (1996). The SANT domain: A putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 21: 87–88 [PubMed] [Google Scholar]
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Allshire R.C. (1997). Centromeres, checkpoints and chromatid cohesion. Curr. Opin. Genet. Dev. 7: 264–273 [DOI] [PubMed] [Google Scholar]
- Barnhart M.C., Kuich P.H., Stellfox M.E., Ward J.A., Bassett E.A., Black B.E., Foltz D.R. (2011). HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J. Cell Biol. 194: 229–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berckmans B., De Veylder L. (2009). Transcriptional control of the cell cycle. Curr. Opin. Plant Biol. 12: 599–605 [DOI] [PubMed] [Google Scholar]
- Blower M.D., Karpen G.H. (2001). The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3: 730–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.C., Greene E., Bowers S.R., Mellone B.G. (2012). A role for the CAL1-partner Modulo in centromere integrity and accurate chromosome segregation in Drosophila. PLoS ONE 7: e45094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- del Pozo J.C., Boniotti M.B., Gutierrez C. (2002). Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. Plant Cell 14: 3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo J.C., Diaz-Trivino S., Cisneros N., Gutierrez C. (2006). The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 18: 2224–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rop V., Padeganeh A., Maddox P.S. (2012). CENP-A: The key player behind centromere identity, propagation, and kinetochore assembly. Chromosoma 121: 527–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Beeckman T., Beemster G.T., de Almeida Engler J., Ormenese S., Maes S., Naudts M., Van Der Schueren E., Jacqmard A., Engler G., Inzé D. (2002). Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J. 21: 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Gillespie L.L., Mercer F.C., Paterno G.D. (2004). The SANT domain of human MI-ER1 interacts with Sp1 to interfere with GC box recognition and repress transcription from its own promoter. J. Biol. Chem. 279: 28009–28016 [DOI] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38 (Web Server issue): W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy E.M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., Almouzni-Pettinotti G. (2009). HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137: 485–497 [DOI] [PubMed] [Google Scholar]
- Earnshaw W.C., Rothfield N. (1985). Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91: 313–321 [DOI] [PubMed] [Google Scholar]
- Ebbs M.L., Bartee L., Bender J. (2005). H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol. Cell. Biol. 25: 10507–10515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs M.L., Bender J. (2006). Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18: 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco H.D., Pidoux A.L., Urano T., Allshire R.C. (2008). Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel S., Usadel B., von Wirén N., Sreenivasulu N. (2012). Reverse engineering: A key component of systems biology to unravel global abiotic stress cross-talk. Front. Plant Sci. 3: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., Yanagida M. (2007). Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell 12: 17–30 [DOI] [PubMed] [Google Scholar]
- González-Barrios R., Soto-Reyes E., Herrera L.A. (2012). Assembling pieces of the centromere epigenetics puzzle. Epigenetics 7: 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan S., Sullivan B.A., Trazzi S., Della Valle G., Robertson K.D. (2009). DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Hum. Mol. Genet. 18: 3178–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther M., Laithier M., Brison O. (2000). A set of proteins interacting with transcription factor Sp1 identified in a two-hybrid screening. Mol. Cell. Biochem. 210: 131–142 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M. (2004). Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118: 715–729 [DOI] [PubMed] [Google Scholar]
- He X.J., Hsu Y.F., Pontes O., Zhu J., Lu J., Bressan R.A., Pikaard C., Wang C.S., Zhu J.K. (2009). NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 23: 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann S., Lermontova I., Berckmans B., De Veylder L., Bäumlein H., Schubert I. (2011). The E2F transcription factor family regulates CENH3 expression in Arabidopsis thaliana. Plant J. 68: 646–656 [DOI] [PubMed] [Google Scholar]
- Hemmerich P., Weidtkamp-Peters S., Hoischen C., Schmiedeberg L., Erliandri I., Diekmann S. (2008). Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 180: 1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Dalal Y. (2005). Centromeric chromatin: What makes it unique? Curr. Opin. Genet. Dev. 15: 177–184 [DOI] [PubMed] [Google Scholar]
- Howman E.V., Fowler K.J., Newson A.J., Redward S., MacDonald A.C., Kalitsis P., Choo K.H. (2000). Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA 97: 1148–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izuta H., Ikeno M., Suzuki N., Tomonaga T., Nozaki N., Obuse C., Kisu Y., Goshima N., Nomura F., Nomura N., Yoda K. (2006). Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells 11: 673–684 [DOI] [PubMed] [Google Scholar]
- Jansen L.E., Black B.E., Foltz D.R., Cleveland D.W. (2007). Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasencakova Z., Meister A., Walter J., Turner B.M., Schubert I. (2000). Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell 12: 2087–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L.M., Law J.A., Khattar A., Henderson I.R., Jacobsen S.E. (2008). SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet. 4: e1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagansky A., Folco H.D., Almeida R., Pidoux A.L., Boukaba A., Simmer F., Urano T., Hamilton G.L., Allshire R.C. (2009). Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science 324: 1716–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T., Jeddeloh J.A., Flowers S.K., Munakata K., Richards E.J. (1996). Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl. Acad. Sci. USA 93: 12406–12411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Sato N., Hayama S., Yamabuki T., Ito T., Miyamoto M., Kondo S., Nakamura Y., Daigo Y. (2007). Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 67: 8544–8553 [DOI] [PubMed] [Google Scholar]
- Kim I.S., Lee M., Park K.C., Jeon Y., Park J.H., Hwang E.J., Jeon T.I., Ko S., Lee H., Baek S.H., Kim K.I. (2012). Roles of Mis18α in epigenetic regulation of centromeric chromatin and CENP-A loading. Mol. Cell 46: 260–273 [DOI] [PubMed] [Google Scholar]
- Kuhlmann M., Mette M.F. (2012). Developmentally non-redundant SET domain proteins SUVH2 and SUVH9 are required for transcriptional gene silencing in Arabidopsis thaliana. Plant Mol. Biol. 79: 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lageix S., Catrice O., Deragon J.M., Gronenborn B., Pélissier T., Ramírez B.C. (2007). The nanovirus-encoded Clink protein affects plant cell cycle regulation through interaction with the retinoblastoma-related protein. J. Virol. 81: 4177–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I., Fuchs J., Schubert I. (2008). The Arabidopsis checkpoint protein Bub3.1 is essential for gametophyte development. Front. Biosci. 13: 5202–5211 [DOI] [PubMed] [Google Scholar]
- Lermontova I., Fuchs J., Schubert V., Schubert I. (2007). Loading time of the centromeric histone H3 variant differs between plants and animals. Chromosoma 116: 507–510 [DOI] [PubMed] [Google Scholar]
- Lermontova I., Koroleva O., Rutten T., Fuchs J., Schubert V., Moraes I., Koszegi D., Schubert I. (2011a). Knockdown of CENH3 in Arabidopsis reduces mitotic divisions and causes sterility by disturbed meiotic chromosome segregation. Plant J. 68: 40–50 [DOI] [PubMed] [Google Scholar]
- Lermontova I., Rutten T., Schubert I. (2011b). Deposition, turnover, and release of CENH3 at Arabidopsis centromeres. Chromosoma 120: 633–640 [DOI] [PubMed] [Google Scholar]
- Lermontova I., Schubert V., Fuchs J., Klatte S., Macas J., Schubert I. (2006). Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell 18: 2443–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo A.W., Magliano D.J., Sibson M.C., Kalitsis P., Craig J.M., Choo K.H. (2001). A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res. 11: 448–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R.X., Postigo A.A., Dean D.C. (1998). Rb interacts with histone deacetylase to repress transcription. Cell 92: 463–473 [DOI] [PubMed] [Google Scholar]
- Maddox P.S., Hyndman F., Monen J., Oegema K., Desai A. (2007). Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 176: 757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O., Reinders J., Caikovski M., Smathajitt C., Paszkowski J. (2007). Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130: 851–862 [DOI] [PubMed] [Google Scholar]
- Meyer P.E., Lafitte F., Bontempi G. (2008). minet: A R/Bioconductor package for inferring large transcriptional networks using mutual information. BMC Bioinformatics 9: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Nasuda S., Hudakova S., Schubert I., Houben A., Endo T.R. (2005). Stable barley chromosomes without centromeric repeats. Proc. Natl. Acad. Sci. USA 102: 9842–9847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C., Yang H., Nozaki N., Goto S., Okazaki T., Yoda K. (2004). Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells 9: 105–120 [DOI] [PubMed] [Google Scholar]
- Pauleau A.L., Erhardt S. (2011). Centromere regulation: New players, new rules, new questions. Eur. J. Cell Biol. 90: 805–810 [DOI] [PubMed] [Google Scholar]
- Phansalkar R., Lapierre P., Mellone B.G. (2012). Evolutionary insights into the role of the essential centromere protein CAL1 in Drosophila. Chromosome Res. 20: 493–504 [DOI] [PubMed] [Google Scholar]
- Rose A.B., Elfersi T., Parra G., Korf I. (2008). Promoter-proximal introns in Arabidopsis thaliana are enriched in dispersed signals that elevate gene expression. Plant Cell 20: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm R.B., Althoff F., Heidmann S., Lehner C.F. (2010). Detrimental incorporation of excess Cenp-A/Cid and Cenp-C into Drosophila centromeres is prevented by limiting amounts of the bridging factor Cal1. J. Cell Sci. 123: 3768–3779 [DOI] [PubMed] [Google Scholar]
- Schuh M., Lehner C.F., Heidmann S. (2007). Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 17: 237–243 [DOI] [PubMed] [Google Scholar]
- Silva M.C., Jansen L.E. (2009). At the right place at the right time: Novel CENP-A binding proteins shed light on centromere assembly. Chromosoma 118: 567–574 [DOI] [PubMed] [Google Scholar]
- Skirpan A.L., McCubbin A.G., Ishimizu T., Wang X., Hu Y., Dowd P.E., Ma H., Kao T. (2001). Isolation and characterization of kinase interacting protein 1, a pollen protein that interacts with the kinase domain of PRK1, a receptor-like kinase of petunia. Plant Physiol. 126: 1480–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppe W.J., Jasencakova Z., Houben A., Kakutani T., Meister A., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F. (2002). DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21: 6549–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W., Moffatt B.A. (1986). Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189: 113–130 [DOI] [PubMed] [Google Scholar]
- van den Heuvel S., Dyson N.J. (2008). Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 9: 713–724 [DOI] [PubMed] [Google Scholar]
- Vongs A., Kakutani T., Martienssen R.A., Richards E.J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928 [DOI] [PubMed] [Google Scholar]
- Wang L., Rajan H., Pitman J.L., McKeown M., Tsai C.C. (2006). Histone deacetylase-associating Atrophin proteins are nuclear receptor corepressors. Genes Dev. 20: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A.T., Haag J.R., Pikaard C.S. (2008). Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki A.T., Ream T.S., Haag J.R., Pikaard C.S. (2009). RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 41: 630–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.S., Hayashi T., Yanagida M., Russell P. (2009). Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol. Cell 33: 287–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H.R., Pontes O., Pikaard C.S., Richards E.J. (2007). VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 21: 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Martyniuk C.J., Trudeau V.L. (2006). SANTA domain: A novel conserved protein module in Eukaryota with potential involvement in chromatin regulation. Bioinformatics 22: 2459–2462 [DOI] [PubMed] [Google Scholar]
- Zhang W., Lee H.R., Koo D.H., Jiang J. (2008). Epigenetic modification of centromeric chromatin: Hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize. Plant Cell 20: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Ecker J.R. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Jacobsen S.E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]