This work reports the discovery of a cis-regulatory motif in the promoters of genes that are subject to mitochondrial retrograde regulation and the identification of transcription factors that bind to this element, thereby steering mitochondrial retrograde-induced gene expression.
Abstract
Upon disturbance of their function by stress, mitochondria can signal to the nucleus to steer the expression of responsive genes. This mitochondria-to-nucleus communication is often referred to as mitochondrial retrograde regulation (MRR). Although reactive oxygen species and calcium are likely candidate signaling molecules for MRR, the protein signaling components in plants remain largely unknown. Through meta-analysis of transcriptome data, we detected a set of genes that are common and robust targets of MRR and used them as a bait to identify its transcriptional regulators. In the upstream regions of these mitochondrial dysfunction stimulon (MDS) genes, we found a cis-regulatory element, the mitochondrial dysfunction motif (MDM), which is necessary and sufficient for gene expression under various mitochondrial perturbation conditions. Yeast one-hybrid analysis and electrophoretic mobility shift assays revealed that the transmembrane domain–containing NO APICAL MERISTEM/ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR/CUP-SHAPED COTYLEDON transcription factors (ANAC013, ANAC016, ANAC017, ANAC053, and ANAC078) bound to the MDM cis-regulatory element. We demonstrate that ANAC013 mediates MRR-induced expression of the MDS genes by direct interaction with the MDM cis-regulatory element and triggers increased oxidative stress tolerance. In conclusion, we characterized ANAC013 as a regulator of MRR upon stress in Arabidopsis thaliana.
INTRODUCTION
Plants are regularly exposed to adverse environmental conditions, such as drought, extreme temperatures, lack of nutrients, and pathogen assaults. These factors negatively affect plant growth and development and are responsible for major yield losses in agriculture (Bray et al., 2000). At the cellular level, the functioning of organelles such as chloroplasts and mitochondria is perturbed by these environmental stresses (Huner et al., 1998; Taylor et al., 2002); in response, feedback mechanisms are triggered that steer changes in nuclear gene expression to sustain and/or restore the organellar and at large, cellular functions (Rhoads and Subbaiah, 2007; Piñas Fernández and Strand, 2008). Mitochondria-to-nucleus signaling is referred to as mitochondrial retrograde regulation (MRR) that is a key event in eukaryotic cells during various stress situations (Butow and Avadhani, 2004).
Among the mitochondrial retrograde pathways, the retrograde (RTG) pathway in yeast (Saccharomyces cerevisiae) is the best studied, mainly regarding its role in metabolic compensation of mitochondrial dysfunction in ageing and nutrient depletion scenarios. Mitochondrial dysfunction provokes a cascade of cytosolic events that activates heterodimeric RTG transcription factors through their nuclear translocation (Liao and Butow, 1993; Liu and Butow, 2006). A related pathway, involving the central stress-mediating transcription factor NF-κB, has been identified in mammals (Biswas et al., 1999; Srinivasan et al., 2010). Similarly to the RTG pathway, the NF-κB RTG system stimulates the glycolysis-derived ATP production under impaired mitochondrial respiration (Jazwinski and Kriete, 2012), but it is much more complex, with alternative and specialized functions during innate immune responses (Hayden et al., 2006), reflecting the higher metabolic intricacy. Other (RTG/NF-κB independent) RTG pathways have been reported in yeast and mammals that trigger various responses to specific mitochondrial defects (Epstein et al., 2001; Jones et al., 2012).
Research on MRR in plants has been centered principally on the induction of components of the alternative respiratory chain in response to mitochondrial perturbation, namely, the ALTERNATIVE OXIDASE (AOX) gene (Dojcinovic et al., 2005; Zarkovic et al., 2005; Ho et al., 2008). AOX is induced by various treatments with chemical inhibitors and mutations that disrupt mitochondrial function at the respiratory chain level (mitochondrial electron transport chain [mtETC]) or the tricarboxylic acid (TCA) cycle. Therefore, AOX is used as a marker for the MRR response in plants and ALTERNATIVE OXIDASE1a (AOX1a) is used specifically in Arabidopsis thaliana (Vanlerberghe and McIntosh, 1996; Clifton et al., 2005; Rhoads and Subbaiah, 2007). AOX1a is also induced by various external stress treatments that might indirectly target mitochondrial function (Van Aken et al., 2009). A common component of various stresses that might result from mtETC inhibition is an increase in mitochondrial reactive oxygen species (mtROS) (Prasad et al., 1994; Maxwell et al., 1999). Induction of AOX serves to lower reactive oxygen species (ROS) formation from the impaired respiratory chain (Maxwell et al., 1999; Cvetkovska and Vanlerberghe, 2012). In addition to their damaging effects, mtROS probably act as MRR-triggering signaling molecules (Vanlerberghe et al., 2002; Rhoads et al., 2006).Calcium originating from the mitochondria might also be an MRR signal (Subbaiah et al., 1998), as seen in mammalian cells (Butow and Avadhani, 2004), as well as changes in mitochondrial redox status and metabolites (Schwarzländer and Finkemeier, 2013).
To date, three proteins have been shown to play a role in MRR in plants. ABSCISIC ACID INSENSITIVE4 (ABI4) represses the basal expression of the Arabidopsis AOX1a gene, thereby allowing derepression by MRR (Giraud et al., 2009), and is also a common component of multiple chloroplast RTG pathways (Koussevitzky et al., 2007). In addition, CYCLIN-DEPENDENT KINASE E;1 and WRKY40 have been identified as positive and negative regulators of MRR induction of AOX1a in Arabidopsis, respectively (Ng et al., 2013a; Van Aken et al., 2013). Although the effect of mitochondrial dysfunction on nuclear gene expression has been elucidated (Schwarzländer et al., 2012), the current understanding of the underlying regulatory mechanisms in plants is still limited.
Here, we followed a bottom-up approach to discover unknown regulatory components of MRR. Among the Arabidopsis gene promoters that are common and robust responsive targets of mitochondrial dysfunction, a cis-regulatory element was identified. This promoter element was necessary and sufficient to drive MRR-mediated gene expression and revealed the regulatory role of a transcription factor of the NO APICAL MERISTEM/ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR/CUP-SHAPED COTYLEDON (NAC) family in the plant MRR.
RESULTS
Identification of a cis-Regulatory Motif in the Promoters of MRR-Regulated Genes
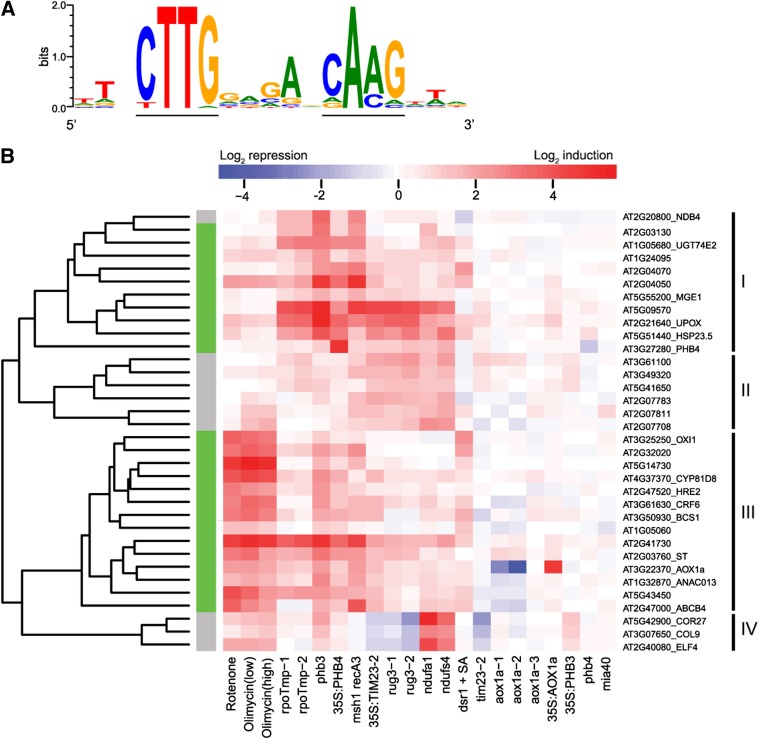
To identify transcriptional regulators of MRR, we looked for shared cis-regulatory elements in the promoters of MRR-regulated genes. To select these genes, we assembled a compendium of 12 publicly available and microarray-derived transcriptome data sets that encompassed 22 perturbation experiments in which mitochondrial function was impaired by short-term treatments with respiratory inhibitors or by genetic mutation of mitochondrial proteins: oligomycin and rotenone (Clifton et al., 2005); AOX1a-overexpressing and knockout mutants (35S:AOX1a and aox1a) (National Center for Biotechnology Information Gene Expression Omnibus database; Edgar et al., 2002; accession number GSE4113); 35S:PROHIBITIN3 (PHB3), 35S:PHB4; phb3 and phb4 (Van Aken et al., 2007); aox1a (Giraud et al., 2008); mutants defective in NADH dehydrogenase (ubiquinone) fragment S subunit 4 and fragment A subunit 1 (Meyer et al., 2009); RNA polymerase of the T3/T7 type dual-targeted to mitochondria and plastids mutants (Kühn et al., 2009); MutS Homolog1 and RECA homolog3 double mutant (Shedge et al., 2010); mitochondrial intermembrane space assembly machinery40 mutant (Carrie et al., 2010); disrupted in stress responses1 mutant (Gleason et al., 2011); RCC1/UVR8/GEF-like3 mutant (Kühn et al., 2011); and translocase inner membrane subunit23-2–overexpressing and knockout (35S:TIM23-2 and tim23-2) mutants (Wang et al., 2012). For experimental details of the individual microarray studies, see Supplemental Table 1 online. In this meta-analysis, we defined transcripts upregulated significantly (P value < 0.01 and log2-fold > 1) in five or more of the 22 conditions as robust general mitochondrial stress-responsive genes (see Supplemental Table 2 online). In the resulting 34 genes, we assessed the presence of shared sequence elements within the first 1-kb upstream intergenic region. Three de novo motif discovery algorithms (Bailey and Elkan 1994; Linhart et al., 2008; Thomas-Chollier et al., 2011) commonly identified a motif with the consensus sequence CTTGNNNNNCA[AC]G and the corresponding position weight matrix shown in Figure 1A. This cis-regulatory motif occurred at least once in 24 out of the 34 genes (see Supplemental Table 3 online), and the consensus was significantly enriched in this gene set compared with the genome (16.6-fold, hypergeometric P value 1.31E-18). As we hypothesized that this motif is a cis-regulatory element for transcriptional regulation in response to mitochondrial dysfunction, we designated it the mitochondrial dysfunction motif (MDM). Hereafter, these 24 MDM-containing genes are referred to as the MITOCHONDRIAL DYSFUNCTION STIMULON (MDS) genes (see Supplemental Table 4 online).
Figure 1.
Identification of the MDM cis-Regulatory Element in MRR-Upregulated Genes.
(A) Position weight matrix of MDM representing the occurrence in the 24 MDS promoters, showing the probability of (a) nucleotide(s) at each position. The MDM consensus (CTTGNNNNNCA[AC]G) is underlined.
(B) Hierarchical clustering of expression profiles of the 34 MRR-upregulated genes (P < 0.01, log2-fold change > 1, in five or more mitochondrial dysfunction conditions). Color codes represent the actual log2-fold changes in transgenic or treated plants compared with wild-type or untreated plants, respectively. The MDS genes containing the cis-regulatory MDM in their 1-kb upstream sequence are indicated with a green bar.
The 24 MDM-containing MDS genes were among the most responsive and upregulated genes with respect to the number of perturbation experiments and the response magnitude, implying they are common and robust MRR targets (Figure 1B). Hierarchical clustering of the 34 significantly upregulated genes upon mitochondrial dysfunction revealed that the 24 MDS genes clustered in two groups with distinct transcriptional profiles: one that comprises genes affected by treatments and in mutants (cluster III) and one containing genes mainly affected in mutants and, to a much lesser extent, by treatments (cluster I). As evolutionary conservation is suggestive of functional importance (Freeling and Subramaniam, 2009), we investigated the biological relevance of our candidate cis-regulatory element by examining its evolutionary conservation in six related dicot species (see Supplemental Methods 1 and Supplemental Table 5 online). Of the 24 MDS genes, nine shared a significant evolutionarily conserved MDM motif with orthologous genes in at least two other species (see Supplemental Table 3 online), suggesting a functional role of the motif in MRR.
MDM Is a cis-Regulatory Element Required for MRR-Induced Gene Expression
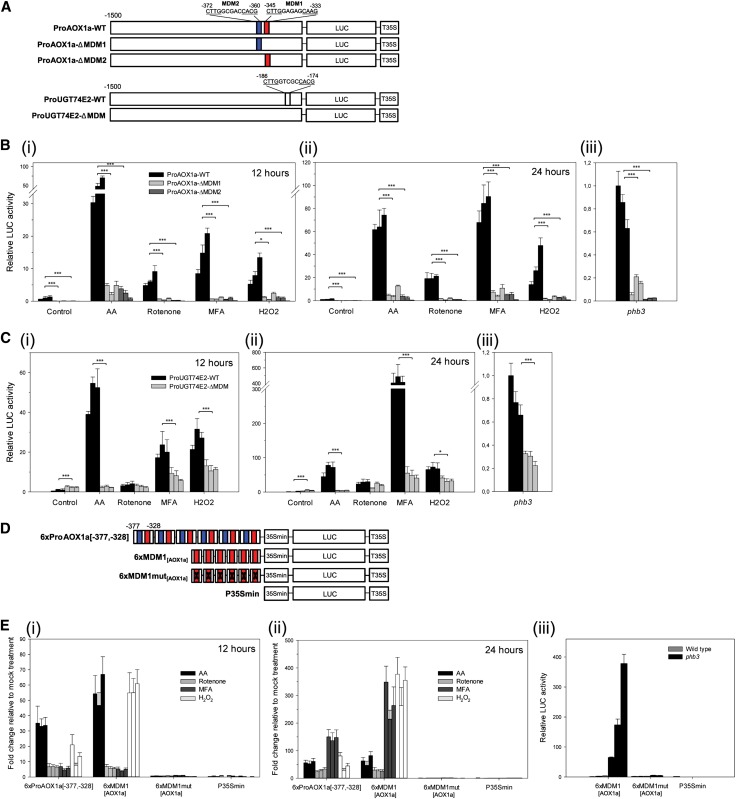
To assess the functionality of this cis-regulatory element in mediating MRR-induced gene expression, we studied the effect of its deletion on the promoter activity of AOX1a, which is a model gene for MRR studies (Rhoads and Subbaiah, 2007) and contains the MDM1[AOX1a] and MDM2[AOX1a] sequences (Figure 2A). MDM deletions in the 1.5-kb AOX1a promoter were generated and fused to the luciferase (LUC) reporter gene. Three independent transgenic ProAOX1a-WT:LUC, ProAOX1a-ΔMDM1:LUC, and ProAOX1a-ΔMDM2:LUC Arabidopsis lines were treated with antimycin A (AA), rotenone, monofluoroacetate (MFA), or hydrogen peroxide (H2O2) for 12 and 24 h, prior to LUC activity measurements. The mtETC is inhibited by AA and rotenone and the TCA cycle by MFA. H2O2 was included because components of the mtETC and TCA cycle are sensitive to oxidative stress (Sweetlove et al., 2002) and because it has been proposed as a signal within MRR (Vanlerberghe et al., 2002; Rhoads and Subbaiah, 2007). The absolute LUC signal in ProAOX1a-ΔMDM1:LUC and ProAOX1a-ΔMDM2:LUC was significantly lower than that of the ProAOX1a-WT-LUC plants after treatment with AA, rotenone, MFA, or H2O2 as well as the basal expression levels of the deletion constructs under nonstressed conditions (Figures 2Bi and 2Bii).
Figure 2.
The MDM Is Necessary and Sufficient for MRR-Mediated Promoter Activation.
(A) Schematic overview of AOX1a and UGT74E2 promoter deletion constructs. MDM deletions were generated in the 1.5-kb promoters and fused in frame to the LUC reporter gene.
(B) Regulatory characteristics of the MDM elements from the AOX1a promoter tested by comparison of the LUC expression driven by the AOX1a promoter (ProAOX1a-WT) to the same promoter construct with deletion of either MDM1[AOX1a] (ProAOX1a-ΔMDM1) or MDM2[AOX1a] (ProAOX1a-ΔMDM2) in transgenic Arabidopsis plants. Promoter activities were analyzed after mock (Control), AA, rotenone, MFA, or H2O2 treatment for 12 h (i) or 24 h (ii) and in phb3 mutants (iii). Bars indicate average relative LUC activities from eight biological replicates ± se. Per construct, data of three independent transgenic lines are shown. Asterisk indicates significant differences from ProAOX1a-WT (Student’s t test; *P < 0.05, **P < 0.01, and ***P < 0.001).
(C) Regulatory characteristics of the MDM element from the UGT74E2 promoter tested by comparing the LUC expression driven by the UGT74E2 promoter (ProUGT74E2-WT) to the same promoter construct with deletion of MDM[UGT74E2] (ProUGT74E2-ΔMDM) under the same conditions as in (B).
(D) Schematic overview of gain-of-function promoter constructs containing hexamers of the AOX1a promoter regions cloned upstream of the minimal CaMV 35S promoter (P35Smin) that drives the LUC gene transcription.
(E) Regulatory activity of the synthetic sequence containing six consecutive repeats of the 50-bp AOX1a promoter fragment, including two MDM elements (6xProAOX1a[-377,-328]) and one of the MDM sequences alone (6xMDM1[AOX1a]) in transgenic Arabidopsis plants. Constructs mutated in the MDM sequence (6xMDM1mut[AOX1a]) or without promoter fragment (P35Smin) were included as negative control. Average fold changes of LUC activity after 12 h (i) or 24 h (ii) of AA, rotenone, MFA, or H2O2 treatment relative to mock treatment are shown for three independent transgenic lines (±se; n = 8 biological replicates). (iii) Average relative LUC activity of the synthetic sequences in phb3 mutants (±se; n = 8 biological replicates).
[See online article for color version of this figure.]
Additionally, we assessed the effect of the MDM deletions on the AOX1a promoter activity in phb3 mutants of Arabidopsis. Prohibitins play an important role in mitochondrial biogenesis and activity in plants (Ahn et al., 2006), and the Arabidopsis phb3 mutant has altered mitochondrial morphology and strongly induced MDS genes (Figure 1B) (Van Aken et al., 2007). Similarly to the chemical inhibition results, the activity of ProAOX1a-ΔMDM1 and ProAOX1a-ΔMDM2 was significantly lower than that of ProAOX1a-WT in phb3 mutants (Figure 2Biii). These results imply that proficient ProAOX1a activity requires the contribution of both MDM1[AOX1a] and MDM2[AOX1a], under either control or mitochondrial stress conditions.
The effect of MDM deletion was analyzed on the promoter activity of the UDP-GLYCOSYL TRANSFERASE 74E2 (UGT74E2) gene that is responsive to mitochondrial dysfunction (Figure 1B), is one of the most strongly H2O2-responsive genes, and is induced by various abiotic stresses (Tognetti et al., 2010). The UGT74E2 promoter contains only one MDM consensus sequence (MDM[UGT74E2]; Figure 2A), thereby avoiding potential redundancy issues. Under nonstressed conditions, the activity of ProUGT74E2-ΔMDM was slightly higher than that of ProUGT74E2-WT, but, after AA and MFA treatments, the absolute LUC activity of the deletion construct was significantly lower than that of the wild-type construct (Figures 2Ci and 2Cii). After H2O2 treatment and in the phb3 mutant, but not after rotenone treatment, the MDM deletion resulted in significantly reduced UGT74E2 promoter activity, although to a lesser extent than after AA and MFA treatments (Figures 2Ci, 2Cii, and 2Ciii). These results indicate that MDM[UGT74E2] is necessary for UGT74E2 promoter activation under mitochondrial stress conditions, especially during mitochondrial perturbation mediated by AA and MFA.
MDM Is Sufficient for MRR-Mediated Gene Activation
To evaluate to what extent the MDM is sufficient to direct MRR-induced gene expression, we assessed the activity of the 50-bp region at −377 to −328 from the AOX1a promoter containing both the MDM1[AOX1a] and MDM2[AOX1a] elements by gain-of-function experiments. Therefore, reporter plasmids with a six-tandem repeat of this ProAOX1a[-377, -328] fused to the minimal cauliflower mosaic virus (CaMV) 35S promoter (P35Smin) located upstream of the LUC reporter gene were stably transformed in Arabidopsis plants (Figure 2D). Three independent transgenic 6xProAOX1a[-377, -328]-P35Smin:LUC lines were treated with AA, rotenone, MFA, or H2O2 for 12 and 24 h, prior to the LUC activity measurements. All treatments activated 6xProAOX1a[-377, -328] (Figures 2Ei and 2Eii). After 12 h, the induction of the LUC gene was the highest by AA and H2O2 and by MFA after 24 h. To determine whether the MDM alone was sufficient to trigger the gene expression after mitochondrial perturbations, we built a similar reporter construct with a hexamer of the 23-bp AOX1a promoter sequence containing the 13 bp of MDM1[AOX1a] with neighboring 5 bp at each end (5′-TCCATCTTGGAGAGCAAGAAAAA-3′), hereafter designated 6xMDM1[AOX1a]. The MDM1[AOX1a] was chosen because its similarity was the highest to the position weight matrix representation of the MDM consensus motif (Figure 1A). Like 6xProAOX1a[-377, -328], the P35Smin:LUC reporter-driving 6xMDM1[AOX1a] construct was strongly activated by AA, rotenone, MFA, and H2O2. Similar constructs with base substitutions in the MDM1[AOX1a] sequence (5′-TCCATAAAAAAAGGGGGGAAAAA-3′ and designated 6xMDM1mut[AOX1a]) or lacking the MDM promoter fragment (P35Smin) did not display any increase in LUC expression after mitochondrial perturbations. Furthermore, the responsiveness of MDM1[AOX1a] to mitochondrial dysfunction was confirmed in the phb3 mutant (Figure 2Eiii). Together, these data indicate that the MDM is regulated by mitochondrial perturbations and is a regulatory unit sufficient to confer MRR-mediated gene expression.
NAC Transcription Factors Specifically Bind to the MDM of Several MDS Promoters
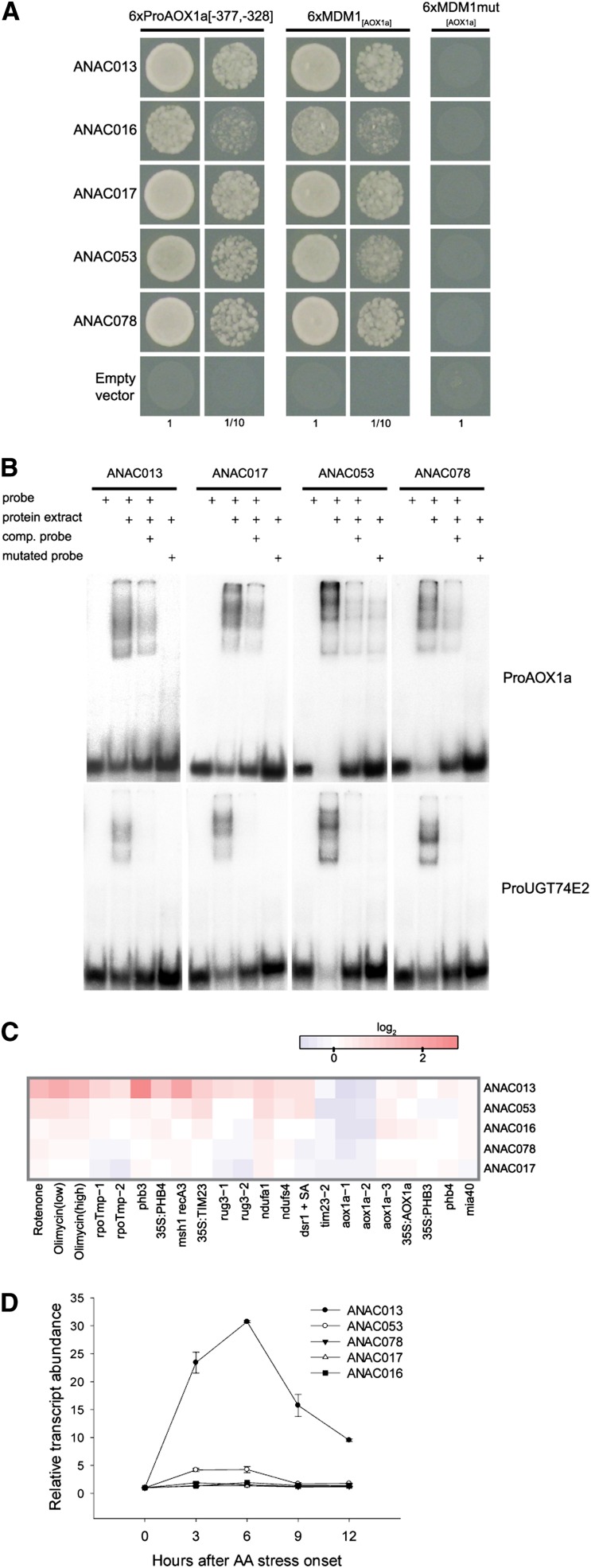
To identify transcriptional regulators that interact with the MDM, yeast one-hybrid (Y1H) screening was performed with the 6xProAOX1a[-377, -328] promoter fragment as bait against a cDNA expression library enriched for stress-responsive genes (Jaspers et al., 2009). In this effort, five NAC family transcription factors (Ooka et al., 2003; Olsen et al., 2005a) were found that bound to the AOX1a promoter fragment: ANAC013, ANAC016, ANAC017, ANAC053, and ANAC078 (Figure 3A). In addition, these NAC transcription factors also interacted with a promoter fragment containing only the MDM element with 5-bp flanking sequences at each end (6xMDM1[AOX1a]). This interaction was completely abolished when base substitutions were introduced into the MDM sequence (6xMDM1mut[AOX1a]). Thus, the five NAC proteins specifically interact with MDM1[AOX1a] in the Y1H system and not with its 5′- or 3′-flanking ends.
Figure 3.
Binding of NAC Transcription Factors to the MDM.
(A) Interaction of NAC transcription factors with the MDM in yeast as shown by Y1H assays. The promoter sequences of interest were fused to histidinol-phosphate aminotransferase imidazole acetol phosphate transaminase (HIS3). The interaction was positive upon growth on 20 mM 3-amino-1,2,4-triazole (3-AT), a competitive inhibitor of HIS3, and was observed with synthetic sequences containing six consecutive repeats of the 50-bp AOX1a promoter fragment, including two MDM elements (6xProAOX1a[-377,-328]) and one of the MDM sequences alone (6xMDM1[AOX1a]), but was abolished when the MDM sequence was mutated (6xMDM1mut[AOX1a]).
(B) Binding of NAC transcription factors with the MDM in vitro as shown by electrophoretic mobility shift assays. Purified NAC-GST proteins interact with radioactively labeled probes of the MDM element–containing AOX1a and UGT74E2 promoter regions. Interactions were abolished in the presence of excess unlabeled competitor probes or when the MDM sequence was mutated.
(C) and (D) Expression pattern of the isolated NAC transcription factors under mitochondrial dysfunction conditions. Expression data were obtained from publicly available microarray data (C) or from own quantitative RT-PCR analyses of AA time series (±se; n = 2 biological replicates) (D).
To confirm the binding of the NAC transcription factors to the MDM, we performed electrophoretic mobility shift assays with five different MDS promoters. Radioactively labeled DNA probes were synthesized that contained 30- to 50-bp MDM-surrounding regions of the AOX1a, UGT74E2, UPREGULATED BY OXIDATIVE STRESS (UPOX), At5g09570, and At2g04050 promoters (see Supplemental Table 6 online). Recombinant NAC-glutathione S-transferase (GST) fusion proteins for ANAC013, ANAC017, ANAC053, and ANAC078 were successfully produced and purified in Escherichia coli. For the four tested NAC proteins, specific shifts with the AOX1a, UGT74E2, UPOX, At5g09570, and At2g04050 probes were observed that could be abolished with a nonlabeled competitor against these probes, whereas the labeled MDM-mutated probes did not give retardation complexes (Figure 3B; see Supplemental Figure 1 online). Hence, ANAC013, ANAC017, ANAC053, and ANAC078 bind specifically to promoter fragments carrying the MDM element from several MDS genes.
The five MDM-binding NAC transcription factors are all putative membrane-associated NAC proteins, containing a C-terminal transmembrane (TM) motif and designated NAC WITH TRANSMEMBRANE MOTIF1-LIKE (NTL) (Kim et al., 2007, 2010b). NTL transcription factors are proteolytically cleaved at the membrane to release an active transcription factor that can enter the nucleus (Kim et al., 2006, 2007). As ANAC013 itself belongs to the MDS stimulon, it displayed expression characteristics similar to those of other genes that contain the MDM element in their promoters, including AOX1a (Pearson correlation coefficient 0.7; Figure 1B). This coexpression occurred not only under mitochondrial, but also under various environmental stress conditions (see Supplemental Figure 2 online). Of the five isolated NTL genes, ANAC013 and, to a lesser extent, ANAC053, were transcriptionally regulated under mitochondrial dysfunction conditions (Figure 3C), as also evidenced by their induction after AA treatments (Figure 3D), whereas ANAC016, ANAC078, and ANAC017 showed only minor transcriptional changes.
ANAC013 Binds and Transactivates MDM in Planta
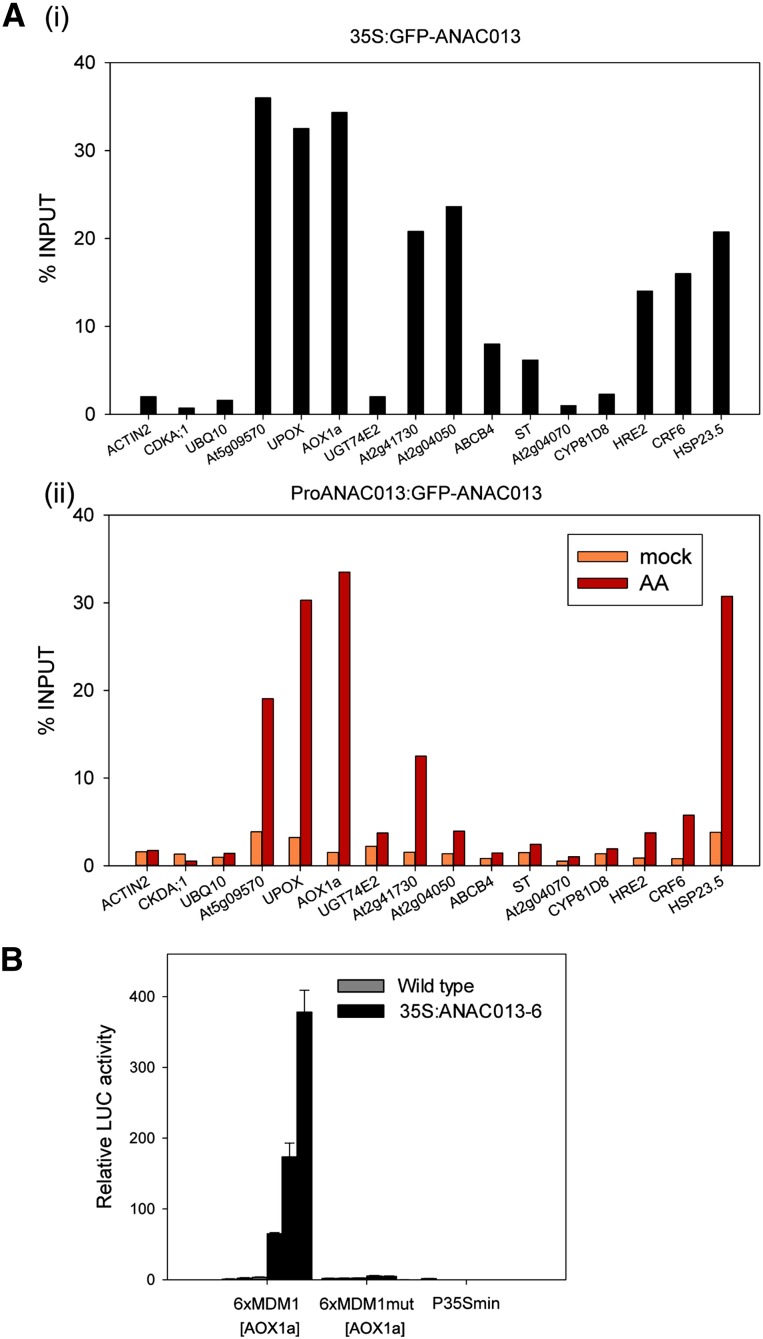
For a more detailed in planta functional analysis, we focused on ANAC013. By means of chromatin immunoprecipitation (ChIP), we examined whether ANAC013 was able to bind in Arabidopsis to 13 MDS promoters that had been selected from the 24 MDS genes based on their responsiveness under multiple mitochondrial dysfunction conditions and occurrence of an MDM sequence with strong similarity to its position weight matrix representation (Figures 1A and 1B; see Supplemental Figure 3 online). ChIP experiments were done on transgenic Arabidopsis seedlings containing a green fluorescent protein (GFP)–tagged version of ANAC013 under the control of the CaMV 35S promoter (35S:GFP-ANAC013) and of its own promoter (ProANAC013:GFP-ANAC013). In 35S:GFP-ANAC013 seedlings, the MDM-containing promoter regions of AOX1a, UPOX, At5g09570, At2g04050, At2g41730, HEAT SHOCK PROTEIN23.5 (HSP23.5), CYTOKININ RESPONSE FACTOR6 (CRF6) relative to ACTIN2, CYCLIN-DEPENDENT KINASE A;1 (CDKA;1), and UBIQUITIN10 (UBQ10) were more than 10-fold enriched after precipitation with an anti-GFP antibody, with the strongest enrichment for AOX1a, UPOX, and At5g09570. The enrichment was smaller (∼9.5-fold, 5.5-fold, and fourfold, respectively) for HYPOXIA RESPONSIVE ETHYLENE RESPONSE FACTOR2 (HRE2), ATP BINDING CASSETTE B4, and SULFOTRANSFERASE (Figure 4Ai; see Supplemental Figure 4 online). The binding of ANAC013 to MDM-containing promoter regions was also assessed under mitochondrial stress conditions. ProANAC013:GFP-ANAC013 seedlings were either mock or AA treated. Whereas in mock-treated plants binding of ANAC013 was not observed, after AA treatment, enrichment for most MDS promoters was evidenced (Figure 4Aii; see Supplemental Figure 4 online), validating the in vivo binding of ANAC013 to MDS promoter regions.
Figure 4.
Interaction of ANAC013 with the MDM from Several MDS Genes in Arabidopsis.
(A) Interaction of ANAC013 with MDS promoters in planta as shown by ChIP. Enrichment of promoter fragments surrounding the MDM after ChIP on control-grown 35S:GFP-ANAC013 (i) and mock- and AA-treated ProANAC013:GFP-ANAC013 (ii) seedlings with anti-GFP antibodies. ACTIN2, CDKA;1, and UBQ10 fragments were used as negative controls. Bars represent fold enrichment relative to the total genomic DNA from one biological sample (% INPUT). Similar data were obtained in at least one other biological repeat experiment and with different independent transgenic lines.
(B) Transactivation of the MDM from the AOX1a promoter by ANAC013 in Arabidopsis. ANAC013 activated the 6xMDM1[AOX1a]-driven LUC reporter gene in ANAC013-overexpressing plants (35S:ANAC013-6) when compared with wild-type plants. The induction was abolished when the MDM was mutated (6xMDM1mut[AOX1a]) or in the absence of the promoter fragment (P35Smin). Bars indicate average relative LUC activities ± se (n = 8 biological replicates).
[See online article for color version of this figure.]
To determine whether ANAC013 could transactivate a MDM-containing promoter fragment in planta, the 6xMDM1[AOX1a]-P35Smin:LUC, 6xMDM1mut[AOX1a]-P35Smin:LUC, and P35Smin:LUC reporter constructs were transformed into Arabidopsis plants overexpressing ANAC013 (35S:ANAC013-6). On average, the 6xMDM1[AOX1a]-containing reporter constructs were 100-fold more active in 35S:ANAC013-6 plants than in the wild-type background, whereas the mutated and control reporter constructs were not activated (Figure 4B).
ANAC013 Mediates MRR-Induced Gene Expression
To assess the role of ANAC013 in MRR-mediated gene expression, we determined the level of MDS transcripts in ANAC013 gain-of-function plants (35S:ANAC013-6). For the above-mentioned 13 MDS genes, including AOX1a, the transcript levels were induced in 35S:ANAC013-6 and most strongly for At5g09570, At2g04050, and UPOX (see Supplemental Figure 5 online). Next, the AA-mediated induction of the MDS genes in ANAC013 loss-of-function plants was tested. As no true loss-of-function T-DNA knockout mutants were available for ANAC013, transgenic lines were generated containing artificial ANAC013-targeting microRNA (miR) constructs. In these ANAC013-miR plants, the ANAC013 transcript levels were 67% lower than those of the wild type. ANAC013-miR and wild-type plants were either mock treated or treated with AA for 3 or 6 h. Analysis of the expression pattern of the 13 MDS genes revealed that under nonstressed conditions (mock treatment), the MDS expression levels were very low and did not significantly differ between ANAC013-miR and wild-type plants (see Supplemental Figure 5 online). After 3 h of AA treatment, upregulation of the MDS transcripts in ANAC013-miR plants was not significantly lower than in the wild-type plants, but significantly reduced after 6 h of AA stress, except for AOX1a, CRF6, and CYP81D8, with the most dramatic effect on the At5g09570 and UPOX genes. Together, these data indicate that ANAC013 is a positive MRR regulator necessary for AA-mediated induction of the MDS.
To verify that the transcriptional enhancement of AOX1a and UPOX in 35S:ANAC013-6 plants also resulted in increased protein production, their protein levels were examined by protein gel blot analysis on isolated mitochondria. Wild-type, 35S:ANAC013-6, and ANAC013-miR plants were either mock treated or treated with AA for 6 h. Under nonstressed conditions, the AOX1a and UPOX protein levels were very low in wild-type plants, but highly induced by ANAC013 overexpression, accumulating to levels higher than those detected in AA-treated wild-type plants (see Supplemental Figure 6 online). After treatment with AA, the AOX1a and UPOX protein levels were induced in wild-type plants and remained higher in 35S:ANAC013-6 than in wild-type plants. In ANAC013-miR lines, the AOX1a and UPOX protein levels did not significantly differ from those of wild-type plants both under nonstressed and AA treatment conditions.
ANAC013 Autoregulates Its Promoter Activity
As the expression profile of ANAC013 was similar to that of its target MDS genes (Figure 1B; see Supplemental Figure 2 online), we hypothesized that it could autoregulate its own expression. The ANAC013 promoter also contained the MDM cis-regulatory element (CTTGGAGAAGAAG; see Supplemental Table 3 online) that overlapped with two ANAC013 promoter elements shown previously to be required for UV-B induction of ANAC013 (Safrany et al., 2008). To examine the effect of ANAC013 on its own promoter activity, ProANAC013 fused to β-glucuronidase (GUS) constructs (ProANAC013:GUS) were transformed in 35S:ANAC013-6 plants. Whereas in wild-type ProANAC013:GUS plants, GUS staining was visible only in the shoot apical meristem (Skirycz et al., 2010) and hydathodes of 2-week-old seedlings, the ANAC013 promoter activity was strongly induced throughout the whole plant in the 35S:ANAC013-6 background (see Supplemental Figure 7 online). Electrophoretic mobility shift assays confirmed binding to an MDM-containing ANAC013 promoter fragment for ANAC013, as well as for the ANAC017, ANAC053, and ANAC078 proteins (see Supplemental Figure 7 and Supplemental Table 6 online). ChIP experiments on 35S:GFP-ANAC013 plants also revealed an enrichment of an MDM-surrounding ANAC013 promoter fragment (see Supplemental Figures 3 and 4 online). Moreover, although ANAC013 did not bind on ProANAC013 in ProANAC013:GFP-ANAC013 under nonstressed conditions, enrichment of an ANAC013 promoter fragment was evident under AA-mediated mitochondrial stress conditions. Hence, these results indicate that ANAC013 positively regulates its own promoter-mediated production through binding with the MDM.
ANAC013 Colocalizes with the Endoplasmic Reticulum
Overexpression of ANAC013 translationally fused to GFP had been shown previously to have a nucleocytosolic subcellular localization in transiently transformed Nicotiana benthamiana cells (Inzé et al., 2012). To analyze the subcellular localization of ANAC013, stable transgenic 35S:GFP-ANAC013 Arabidopsis lines were used, but none of them showed any detectable GFP fluorescence. Immunoelectron microscopy studies on these stable GFP fusion lines were also unsuccessful, possibly due to the low GFP-ANAC013 protein abundance and consistent with the low protein stability potentially due to proteasomal degradation observed for other NTL transcription factors (Kim et al., 2006; Seo et al., 2008). Protein gel blot analysis of these stable transgenic 35S:GFP-ANAC013 Arabidopsis lines with an anti-GFP antibody revealed that the overproduced GFP-ANAC013 was partially processed (see Supplemental Figure 8 online). Based on their molecular mass, the cross-reactive protein bands reflected most probably the full-length GFP-ANAC013 and the processed version lacking the C-terminal TM motif (Kim et al., 2010a), in agreement with the nucleocytosolic localization reported (Inzé et al., 2012).
We analyzed the subcellular localization pattern of GFP-ANAC013 in more detail by Agrobacterium tumefaciens–mediated transient transformation of 35S:GFP-ANAC013 constructs in N. benthamiana, hinting at a localization in the endoplasmic reticulum (ER), in addition to that in the nucleus. Comparison of the GFP-ANAC013 localization pattern to that of an ER marker indicated a putative ER localization for ANAC013 (see Supplemental Figure 8 online), without overlap with the mCherry-labeled mitochondrial marker. In another approach, epidermal cells of onion (Allium cepa) were transformed biolistically with C-terminal GFP fusions to the full-length ANAC013 (ANAC013-GFP), the full-length ANAC013 minus the TM domain (ANAC013ΔTM-GFP), and the isolated TM domain (TMANAC013-GFP) under the control of the 35S promoter. For the full-length and TM-deleted constructs, no GFP signal could be detected, whereas TMANAC013-GFP colocalized with the ER marker (see Supplemental Figure 8 online), but not with mitochondrial and peroxisomal marker proteins. Thus, these data indicate that ANAC013 is potentially targeted to the ER.
ANAC013-Overexpressing Plants Exhibit Increased Tolerance to Stress Induced by Methyl Viologen and Rotenone
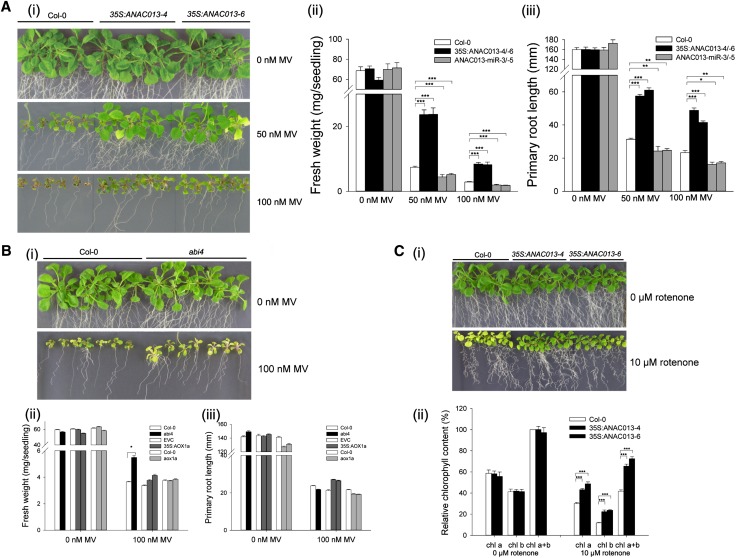
We assessed growth performance of ANAC013-overexpressing (35S:ANAC013-4 and 35S:ANAC013-6) and miR (ANAC013-miR-3 and ANAC013-miR-5) plants under oxidative (methyl viologen [MV], H2O2, 3-AT) and mitochondrial (AA, rotenone, and MFA) stress conditions. Wild-type and transgenic plants were germinated and grown on half-strength Murashige and Skoog (1/2MS) medium supplemented with 50 or 100 nM MV; 1, 2, 4, or 8 mM H2O2; 1, 2, 4, or 8 μM 3-AT; 50 or 100 μM AA; 10 or 20 μM rotenone; or 0.1 or 1 mM MFA.
During growth in the presence of MV, the lines showed significant growth differences. Postgermination growth inhibition by MV was significantly lower for 35S:ANAC013-4 and 35S:ANAC013-6 than for wild-type seedlings (Figure 5Ai). When grown on 100 nM MV, development of wild-type plants was severely retarded, whereas 35S:ANAC013 seedlings developed with milder symptoms. When grown on 50 nM MV, the rosette area of the overexpressing plants was visibly larger than that of wild-type plants. Fresh biomass of 35S:ANAC013 seedlings grown for 3 weeks on 50 or 100 nM MV was approximately threefold higher than that of wild-type plants (Figure 5Aii). Furthermore, root growth was improved in 35S:ANAC013 seedlings at both MV concentrations tested (Figure 5Aiii). The fresh weight and root length of ANAC013-miR seedlings were significantly lower than that of wild-type plants in the presence of MV. In addition, the performance of the AOX1a gain- and loss-of-function (Giraud et al., 2008) and abi4 mutant lines (Kerchev et al., 2011) was benchmarked in the presence of 100 nM MV. Whereas phenotypic differences were not as prominent as those observed for the ANAC013 transgenic lines, the growth performance of abi4 mutants was higher than that of wild-type plants (Figure 5B), but no major phenotypes were observed for AOX1a-overexpressing (35S:AOX1a) and knockout (aox1a) lines under these conditions (Figure 5B).
Figure 5.
Increased MV and Rotenone Tolerance of ANAC013-Overexpressing Plants.
(A) Three-week-old wild-type (Col-0), 35S:ANAC013-4, and 35S:ANAC013-6 seedlings germinated and grown on 1/2MS medium supplemented with 0, 50, or 100 nM MV (i). Fresh weight (ii) and primary root length (iii) of 3-week-old wild-type (Col-0), 35S:ANAC013-4, 35S:ANAC013-6, ANAC013-miR-3, and ANAC013-miR-5 seedlings germinated and grown in the presence of 0, 50, or 100 nM MV. Data represent average ± se (n = 20 to 25 plants). Asterisk indicates significant differences from Col-0 (Student’s t test; *P < 0.05, **P < 0.01, and ***P < 0.001).
(B) Three-week-old wild-type (Col-0) and abi4 mutant seedlings (i) germinated and grown on 1/2MS medium supplemented with 0 or 100 nM MV. Fresh weight (ii) and primary root length (iii) of 3-week-old wild-type (Col-0), abi4 mutant, empty-vector control (EVC), AOX1a-overexpressing (35S:AOX1a-1 and 35S:AOX1a-2), and AOX1a knockout (aox1a-1 and aox1a-2) seedlings germinated and grown in the presence of 0 or 100 nM MV. Data represent average ± se (n = 20 to 25 plants). Asterisk indicates significant differences from Col-0 (Student’s t test; *P < 0.05, **P < 0.01, and ***P < 0.001).
(C) Three-week-old wild-type (Col-0) and 35S:ANAC013 seedlings germinated and grown on 1/2MS medium supplemented with 0 or 10 μM rotenone (i) and relative chlorophyll content (ii). Chlorophyll content was measured and normalized per gram fresh weight of green tissue (ii). The total chlorophyll (Chl a+b) content in the wild type was set to 100%, and the relative chlorophyll contents were calculated accordingly. Data represent average ± se (n = 5 biological repeat samples). Chl a, chlorophyll a; Chl b, chlorophyll b. Asterisk indicates significant differences from Col-0 (Student’s t test; *P < 0.05, **P < 0.01, and ***P < 0.001).
In contrast with AOX1a transgenic lines, perturbed ANAC013 and ABI4 levels did not affect plant growth performance under AA stress conditions (see Supplemental Figure 9 online). Also under MFA, H2O2, or 3-AT stress, no phenotype was observed for ANAC013 transgenic lines (see Supplemental Figure 9 online). In the presence of 10 μM rotenone, wild-type and ANAC013-miR seedlings were bleached, but the chlorophyll a and b contents were significantly higher in 35S:ANAC013 lines than those of the control, indicating an increased tolerance under these conditions (Figure 5C).
As the phenotype was the most pronounced under MV conditions, we assessed whether the MV resistance of ANAC013-overexpressing plants during postgermination and early development occurred also when plants were treated at a later developmental stage. Two-week-old seedlings grown on a nylon mesh on 1/2MS medium were transferred to 2-μM MV-containing medium, and MV stress tolerance was monitored by quantification of the photosystem II (PSII) maximum efficiency (Fv’/Fm’) (Baker, 2008). Young leaves were examined for decrease in Fv’/Fm’ as an indication of stress sensitivity because they were more affected by MV treatment than the older leaves. The PSII maximum efficiency was similar in transgenic and wild-type plants before transfer to MV, but after 7 d and later, the decrease in Fv’/Fm’ of the overexpressing plants was significantly lower, implying an increase in MV tolerance (see Supplemental Figure 10 online). Furthermore, prolonged exposure to MV (6 weeks) was fatal to wild-type plants, whereas overexpressing plants still remained partially green. These results show that ANAC013 positively affects tolerance to oxidative stress induced by MV, and to a lesser extent by rotenone, and that this phenotype is the most pronounced during the early developmental stages.
DISCUSSION
When eukaryotic cells are exposed to environmental stresses, mitochondrial function can be disturbed and transmit the stress-induced signals to the nucleus to activate a transcriptional defense response with the aim to restore cellular function, a process often designated MRR. In plants, MRR has been shown to be involved in signal transduction events during biotic and various abiotic stresses, such as cold, oxygen deprivation, heat, and heavy metals (Lee et al., 2002; Bailey-Serres and Chang, 2005; Rhoads et al., 2005; Keunen et al., 2011). In contrast with yeast and mammalian systems, signal transducing proteins and transcriptional regulators of MRR in plants remain largely unknown. Here, we report on the discovery of a cis-regulatory motif in the promoters of genes that are under MRR control and on the identification of transcription factors that bind to this element, hereby steering mitochondrial RTG-induced gene expression.
The MDM cis-Regulatory Element Steers Mitochondrial RTG-Induced Gene Expression
To identify cellular components of the mitochondrial RTG signal transduction pathways in plants, we compiled a set of genes induced in response to perturbations of mitochondrial function, the MDS. By selecting genes that were significantly modified under multiple MRR-related conditions, we wanted to avoid potential off-target effects of chemical inhibitors and acclimation responses in stable mutants. The MDS encompasses, in addition to well-established general mitochondrial stress markers, such as AOX1a and UPOX (Ho et al., 2008; Schwarzländer et al., 2012; Van Aken and Whelan, 2012), several other mitochondrial proteins, TM transporters, UDP-glycosyltransferase, N-acetyltransferase, cytochrome P450, steroid sulfotransferase, and three transcription factors (see Supplemental Table 4 online). Based on the MDS coexpression under mitochondrial dysfunction conditions, we aimed at identifying transcription factor binding sites involved in MRR via the presence of common sequence motifs in the MDS promoters and discovered an unknown DNA motif. This cis-regulatory motif is necessary and sufficient for mitochondrial stress-responsive gene expression during inhibition of the cytochrome respiratory pathway and TCA cycle, during oxidative stress, and in the mitochondria-defective phb3 mutant, hence, its designation as the MDM. Accordingly, the MDM element is present in the ∼100-bp promoter region of AOX1a that, upon deletion and mutation, impairs induction by AA and MFA (Dojcinovic et al., 2005). However, further experiments will be needed to elucidate whether MDM exclusively regulates responses to mitochondrial stress conditions or whether it also mediates transcriptional responses triggered by stresses that do not involve mitochondrial events, but rather signals originating from other subcellular compartments. The responsiveness of MDM to various pharmacological and genetic perturbations that act on different mitochondrial components is similar to that demonstrated for other promoter elements responsive to mitochondrial stress provoked by rotenone and H2O2 application (Ho et al., 2008). Different mitochondrial perturbations act via distinct, but overlapping, pathways (Zarkovic et al., 2005; see accompanying article, Ng et al., 2013b), indicating a convergence of different MRR-transducing events at the promoter level or, more plausibly, more upstream by provoking the production and/or release of a common signal shortly after triggering mitochondrial dysfunction. Perturbation in mtROS homeostasis is a common component of stress conditions as a consequence of mitochondrial damage (Prasad et al., 1994; Maxwell et al., 1999). As ROS can act as signaling molecules in plant stress responses, they are potential candidates for the common signals produced upon mitochondrial dysfunction (Gechev et al., 2006; Rhoads et al., 2006). However, mtROS are probably not required for all plant MRR (Gray et al., 2004). Although the strong MDM responsiveness to H2O2 supports ROS as early MRR signals, we cannot exclude that external application of H2O2 in our experiments caused mitochondrial damage and dysfunction, such as inhibition of the cytochrome respiratory pathway and TCA cycle components (Sweetlove et al., 2002), thereby initiating MRR through another unidentified mechanism.
TM Domain–Containing NAC Transcription Factors Bind the MDM
We identified five closely related NAC transcription factors that interact with the MDM cis-regulatory element. Several NAC proteins have been reported to bind the so-called NAC binding site (NACBS) CGT[GA] or its reverse complement [TC]ACG (Olsen et al., 2005b; Jensen et al., 2010; Lee et al., 2012). NAC transcription factors bind DNA as dimers with two palindromically oriented NACBS repeats as preferred binding site (Olsen et al., 2005b), but with some degree of flexibility because a NAC dimer can bind to a high-affinity NACBS with one monomer and to a lower-affinity (suboptimal) binding site in a nonpalindromic sequence with a second monomer (Welner et al., 2012). The MDM contains the imperfect inverted-repeat structure CTTGN5CA[AC]G, and the flexibility in dimer binding might explain the degeneracy of the identified consensus. Furthermore, our data are in agreement with the previously determined consensus recognition sequence of ANAC078, sharing the MDM core sequence CA[AC]G (Yabuta et al., 2010), but slightly diverging from a reported binding site for ANAC053 (TACG[AC]CA) in the respiratory burst oxidase homolog C (RBOHC) and RBOHE gene promoters (Lee et al., 2012).
The MDM-binding NAC proteins belong to the subclass of putative membrane-bound NTL transcription factors (Kim et al., 2007, 2010b) and represent a complete phylogenetic subgroup in this NTL family. Moreover, phylogenetic analysis of all Arabidopsis NAC domains revealed that these proteins belong to the same subgroup (NAC2) (Ooka et al., 2003; Kim et al., 2010b), reflecting their common DNA-binding capacities to the MDM. This NAC2 subgroup comprises four additional (non-TM) NAC proteins (ANAC050, ANAC051/52, ANAC082, and ANAC103). By using the Y1H system, we could show binding of ANAC050 and ANAC051/52 to the MDM, but not of ANAC082 and ANAC103 (see Supplemental Figure 11 online). Additional experiments will be needed to elucidate the biological relevance of these interactions in planta.
ANAC013 Regulates MRR
Previous functional studies indicated that ANAC078 regulates light stress–dependent flavonoid biosynthesis and proteasome levels (Morishita et al., 2009; Yabuta et al., 2011). In transgenic Arabidopsis plants overexpressing ANAC053, leaf senescence was promoted during drought stress (Lee et al., 2012). Individual overexpression of ANAC013, ANAC053, and ANAC078 (see Supplemental Figures 5 and 12 online), or expression of a truncated ANAC017 form devoid of the TM domain (Ng et al., 2013b), constitutively activates the MDS genes, regardless of stress conditions, hinting at functional redundancy in the MDS regulation. However, the distinct spatiotemporal expression patterns of the five NTL proteins (Figures 3C and 3D; Kim et al., 2007) most probably suggest specialized subfunctionalization. From the five MDM-binding NTL proteins identified, ANAC013 is strongly regulated by MRR at the transcript level, and its transcript profile highly resembles the expression characteristics of the MDS genes under mitochondrial perturbation as well as under environmental stress conditions (Figure 1B; see Supplemental Figure 2 online). In the ANAC013 knockdown transgenic plants, the MDS induction is reduced in response to the mitochondrial complex-III inhibitor AA. However, to some degree, these plants could still transduce the MRR signal for MDS induction, possibly due to the residual ANAC013 transcript masking the knockdown readout and/or partial redundancy with respect to other (MDM-binding) transcription factors, such as ANAC017 (Ng et al., 2013b). Taken together, our results indicate that ANAC013 positively regulates MDS gene expression upon mitochondrial dysfunction by direct interaction with the MDM cis-regulatory element and, hence, can be considered a positive signal-transducing component of MRR (see Supplemental Figure 13 online).
TM transcription factors are anchored to intracellular membranes, and in response to various stresses, they are released via proteolysis and subsequently translocated to the nucleus, allowing prompt activation of the downstream transcriptional responses (Kim et al., 2007; Seo et al., 2008). Transient expression of GFP-tagged ANAC013 suggests that ANAC013 is localized in the ER under nonstressed conditions (see Supplemental Figure 8 online). The ER represents a dynamic network throughout the cell that is continuously rearranged and physically associates at some points with the mitochondria, allowing local communication between the two organelles (Hayashi et al., 2009). This physical interaction could facilitate proteolytic activation of ANAC013 by mitochondrial signals, such as ROS or calcium, which have previously been implicated in MRR (Rhoads and Subbaiah, 2007). Moreover, ANAC013 is seemingly strongly regulated at the gene transcription level by mitochondrial perturbations, as observed by its early and strong responsiveness and its very low expression levels in the absence of stress. In addition, DNA-binding and gain-of-function studies (see Supplemental Figure 7 online) reveal that a potential posttranslational ANAC013 activation can initiate a positive feedback loop, autoamplifying its own transcription and driving the expression of its target genes. Moreover, ANAC017 and ANAC053 also contain the MDM cis-regulatory element in their promoter, indicative of regulatory interactions with other NTL proteins involved in MRR regulation as well (Ng et al., 2013b).
In accordance with the MRR function in stress signal transduction, overexpression of ANAC013 causes a stress-tolerant phenotype in transgenic Arabidopsis plants under oxidative stress conditions induced by MV or rotenone (Figure 5). The tolerance of ANAC013-overexpressing lines is enhanced, most probably due to the constitutive production of MDS proteins, including AOX1a. AOX1a function is best studied with respect to mitochondria-initiated oxidative stress responses (Maxwell et al., 1999; Umbach et al., 2005) but is not limited to mitochondrial stress responses as evidenced by its importance in chloroplast protection during high-light stress (Yoshida et al., 2007; Giraud et al., 2008). Furthermore, the MDS is enriched for members of multigene families associated with detoxification pathways (cytochrome P450 monooxygenase, N-acetyltransferase, glycosyltransferase, ATP binding cassette, and multi antimicrobial extrusion protein transporters) (Manabe et al., 2007), together with two mitochondrial proteins (HSP23.5 and MITOCHONDRIAL GroP-like gene E1 [MGE1]) that might have chaperone-like activities (Visioli et al., 1997; Hu et al., 2012). Other Arabidopsis genes that contain evolutionarily conserved MDM instances are also enriched for components of the alternative respiratory chain, proteins involved in multidrug/xenobiotic transport, mitochondrial HSPs, and subunits of the 20S proteasome (see Supplemental Tables 5 and 7 online). The potential involvement of the MDM in proteasome regulation is consistent with the reported regulation of 20S and 26S proteasome levels by ANAC078, resulting in increased high-light stress tolerance (Yabuta et al., 2011). However, the MDS gene expression is not altered in ANAC078-overexpressing plants under high-light stress conditions (Morishita et al., 2009). Thus, from these data and previous observations, we suggest that ANAC013 and the MDS integrate oxidative stress responses, xenobiotic stress resistance, and protein quality control. This coordinated response presumably allows cells to prevent ROS formation, inactivate and eliminate the offending agent or toxic byproducts, and rapidly monitor and repair the damage under adverse conditions.
In yeast, a highly interconnected transcriptional network links oxidative stress, multidrug resistance, protein degradation, and protein folding responses through the coordinated action of several transcription factors (Salin et al., 2008; Teixeira et al., 2008). Interestingly, one of them (the zinc finger transcription factor Pleiotropic Drug Resistance Protein3) is involved in an MRR response in which mitochondrial dysfunction triggers the expression of an ABC transporter involved in multidrug resistance (Hallstrom and Moye-Rowley, 2000; Traven et al., 2001; Devaux et al., 2002). Similarly to yeast, multidrug resistance transporter, mitochondrial chaperone, and oxidative stress genes are induced by mitochondrial dysfunction in mammalian cells (Martinus et al., 1996; Park et al., 2004; Ferraresi et al., 2008), indicating that this process is conserved in eukaryotes. Furthermore, the basic leucine zipper YEAST ADAPTOR PROTEIN-1–like transcription factor (Yap1), which is a central node in the above-mentioned stress transcriptional network in yeast, is redox activated by the GLUTATHIONE PEROXIDASE3 (GPX3) in response to elevated H2O2 levels and xenobiotics (Delaunay et al., 2002; Azevedo et al., 2003). Although plants lack orthologs of Yap1-like transcription factors, a previously reported ANAC013-interacting protein, RADICAL-INDUCED CELL DEATH1 (RCD1), was hypothesized to be the plant equivalent of Yap1 (Miao et al., 2006; Jaspers et al., 2009). RCD1 complemented the oxidative stress–sensitive phenotype of the Yap1-deficient yeast strain and interacted with the Arabidopsis GPX3 protein that functions in H2O2 sensing and signal transduction (Belles-Boix et al., 2000; Miao et al., 2006). A similar redox-sensing regulatory system in mammals contains the basic leucine zipper transcription factor Nuclear factor erythroid2-Related Factor2 (Nrf2) that mediates oxidative and xenobiotic stress responses (Kobayashi and Yamamoto, 2006). Unlike Yap1, Nrf2 does not directly sense the stress but is activated by redox regulation of its interaction partner Kelch-like epichlorohydrin-associated protein1 (Keap1) (Itoh et al., 1999). Under unstressed conditions, Keap1 negatively regulates Nrf2 by proteosomal degradation and cytoplasmic sequestration, but, upon stimulation by oxidative or chemical stresses, Keap1 releases Nrf2 to escape proteosomal degradation and translocate to the nucleus (Kang et al., 2004; Kobayashi et al., 2004). Nrf2 induction and nuclear translocation have also been shown in response to an intramitochondrial inhibitor of specific mitochondrial proteins (such as HSPs, and aconitase and α-ketoglutarate dehydrogenase of the TCA cycle), although this activation probably does not involve increased ROS levels, but rather ER stress signaling induced by dysfunctional mitochondria (Ho et al., 2005). Taking the above described similarities in yeast, plants, and mammals into account, we can assume that the Arabidopsis MDS might be the functional equivalent of these yeast and mammalian oxidative and xenobiotic stress responses.
Besides its association with RCD1, ANAC013 also interacts with another member of the SIMILAR TO RCD-ONE (SRO) family, SRO5 (Jaspers et al., 2009). Interestingly, SRO5 has been suggested to function in mitochondria-nucleus communication, based on its reported localization in mitochondria or nucleus (Borsani et al., 2005; Jaspers et al., 2010). The interaction of RCD1 and SRO5 with transcription factors through the conserved RCD-SRO-TATA box-binding protein–associated factor 4 domain implies a role in the regulation of the transcription factor activity (Jaspers et al., 2009, 2010). As RCD1 is localized in the nucleus under nonstressed conditions and rcd1 mutants exhibit an elevated expression of ANAC013 target genes, RCD1 might negatively affect the ANAC013 function in the nucleus in the absence of stress (Jaspers et al., 2009). Similarly, RCD1 negatively regulates the stability of the DEHYDRATION-RESPONSE ELEMENT BINDING PROTEIN2A (DREB2A) transcription factor but is rapidly degraded upon stress, promoting the proper DREB2A function under these conditions (Vainonen et al., 2012). Furthermore, it might be necessary for correct meristem function, presumably by influencing the redox/ROS balance (Teotia and Lamb, 2011). Like ANAC013-overexpressing plants, rcd1 mutants display increased MV tolerance (Fujibe et al., 2004). ANAC013 and MDS genes are predominantly expressed in meristematic tissues (Skirycz et al., 2010) and the stress phenotype of ANAC013-overexpressing plants is the most pronounced when plants are stressed during early seedling development (Figure 5), suggesting that ANAC013 is important for the stress response in young tissues. Accordingly, maintenance of optimal mitochondrial function has been postulated to be required for appropriate meristem development under stress conditions (Skirycz et al., 2010; Van Aken et al., 2010) because dividing cells need energy and ROS can damage the mitochondrial function.
METHODS
Microarray Analysis, Motif Detection, and Evolutionary Conservation Analysis
For the meta-analysis of the transcriptome data of the MRR compendium, raw Affymetrix Cell Intensity files (http://www.ncbi.nlm.nih.gov/geo/) were preprocessed per data set in Bioconductor (http://www.bioconductor.org/), comprising normalization by Robust Multi-array Average with a custom Computer Document Format based on The Arabidopsis Information Resource (TAIR10) and provided by Brainarray (TAIR10 genes v14; http://brainarray.mbni.med.umich.edu). Differential gene expression was analyzed by the limma package (Smyth, 2004), P values were adjusted for multiple hypothesis testing by the Benjamini-Hochberg false discovery rate, and the significance cutoff was stringently set at a corrected P value < 0.01 and a log2-fold change > 1. Genes that were upregulated in five or more conditions were hierarchically clustered after gene centering and scaling of the log2 expression ratios, with Euclidian distance as distance measure and average linkage. For the meta-analysis of the biotic and abiotic stress compendium, the raw Affymetrix Cell Intensity files were obtained from the stress data set in CORNET (De Bodt et al., 2012) and preprocessed together in Bioconductor and clustered as described above.
For the de novo motif discovery, Multiple Expectation-Maximization for Motif Elicitation (MEME) version 4.8.1 (Bailey and Elkan 1994), Amadeus (Linhart et al., 2008), and the Regulatory Sequence Analysis Tools (RSAT)–spaced dyad tool (Thomas-Chollier et al., 2011) were used with default parameter settings (motif length 6 to 50 for MEME and 12 for Amadeus; monad length 3 for RSAT-spaced dyad tool). Application of different motif lengths confirmed the identified MDM motif. The position weight matrix of all MDM matches in the MDS promoters (see Supplemental Table 2 online) was visualized by generating a sequence logo by means of WebLogo 3 (http://weblogo.berkeley.edu/) (Crooks et al., 2004).
To evaluate the evolutionary conservation of an individual motif instance, for each Arabidopsis thaliana gene the orthologous genes from six other dicot species, Arabidopsis lyrata, papaya (Carica papaya), soybean (Glycine max), apple tree (Malus domestica), black cottonwood (Populus trichocarpa), and common grape vine (Vitis vinifera), were retrieved with the PLAZA 2.0 Integrative Orthology method that combines orthology information from phylogenetic trees, OrthoMCL families, and Best-Hits-and-Inparalogs families (Van Bel et al., 2012). Integrative orthologous genes supported by at least one orthology prediction method were retained for the conservation analysis. Based on the 1-kb orthologous upstream intergenic sequences, a test motif was mapped with DNA pattern (Thomas-Chollier et al., 2011) and the number of conserved motif matches was determined for each Arabidopsis gene (and its orthologs). Finally, the significance of the observed motif conservation per Arabidopsis gene was tested by random sampling of 1000 nonorthologous gene sets, maintaining the gene and species composition as observed in the real orthologous data set, and scoring the number of random gene sets with a similar or improved motif conservation level. As A. lyrata is closely related to A. thaliana, only genes with conserved motif matches in at least two other species (not including A. lyrata) and conservation P value < 0.05 were defined as significantly evolutionarily conserved. Gene Ontology enrichment was analyzed with the PLAZA Workbench (Van Bel et al., 2012).
Plant Growth Conditions and Stress Treatments
Arabidopsis (accession Columbia-0 [Col-0]) plants were grown on 1/2MS medium (Duchefa) supplemented with 1% (w/v) Suc, 0.75% (w/v) agar, and B5 vitamins, pH 5.7, at 21°C and 100 μE m−2 s−1 light intensity in a 16-h/8-h light/dark photoperiod. Unless stated otherwise, seedlings were grown for 2 weeks until stage 1.04 (Boyes et al., 2001). For the AA induction experiments, seedlings were sprayed with 0.01% (v/v) polysorbate 20–containing 0.1% (v/v) DMSO supplemented with 50 μM AA (Sigma-Aldrich) or without (mock). Plants were germinated and grown on 1/2MS medium supplemented with 50 or 100 μM AA for the AA stress assays, with 10 or 20 μM rotenone (Sigma-Aldrich), or with 0.1 or 1 mM MFA (Sigma-Aldrich) for the other mitochondrial stress assays, and with 1, 2, 4, or 8 mM H2O2 (Merck); with 1, 2, 4, or 8 μM 3-AT (Acros Organics); or with 50 or 100 nM MV (Acros Organics) for the oxidative stress assays. The PSII maximum efficiency (Fv’/Fm’) (Baker, 2008) was determined with a PAM-2000 chlorophyll fluorometer and ImagingWin software application (Walz) on light-adapted plants. Two or more independent experiments were performed for all stress assays.
Generation of Transgenic Arabidopsis Plants
Overexpressing plants were generated by cloning the open reading frame (ORF) of ANAC013, ANAC053, and ANAC078 into pK7WG2D (see Supplemental Table 8 online) (Karimi et al., 2002). To generate artificial microRNA plants, ANAC013-specific sequences were identified with the Web MicroRNA Designer (www.weigelworld.org). The microRNA precursors were constructed according to Schwab et al. (2006) and cloned into pK7WG2D. 35S:GFP-ANAC013 plants were generated by cloning the ANAC013 ORF into pK7WGF2 (Karimi et al., 2002). The 1.5-kb upstream region of the translational start site of ANAC013 (ProANAC013) was amplified by PCR from Arabidopsis Col-0 genomic DNA with primers (see Supplemental Table 8 online) and cloned into pBGWFS7 (Karimi et al., 2002), generating an in-frame GFP-GUS fusion. ProANAC013:GFP-ANAC013 constructs were created by recombining pEN-L4-ProANAC013-R1, pEN-L1-GFP-L2, and pEN-R2-ANAC013(ORF)-L3 into the MultiSite destination vector pK7m34GW (Karimi et al., 2005). Constructs were transformed into Arabidopsis Col-0 by Agrobacterium tumefaciens–mediated floral dipping (Clough and Bent, 1998). AOX1a-overexpressing lines (35S:AOX1a-1 [N6591], 35S:AOX1a-2 [N6593], and the empty control vector [N6590]) were obtained from the Nottingham Arabidopsis Stock Centre.
Promoter-LUC Constructs
The 1.5-kb upstream region from the translational start site of AOX1a and UGT74E2 was cloned into the pDONRP4-P1r vector (Invitrogen). The MDM sequence was deleted according to the PCR fusion/Gateway cloning procedure (Atanassov et al., 2009) (see Supplemental Table 8 online). Promoter-LUC fusion constructs were created by recombining the above promoter plasmids with pEN-L1-LUC+-L2 into the destination vector pB7m24GW by means of the MultiSite Gateway technology (Invitrogen) (Karimi et al., 2005).
The artificial promoter constructs were synthesized by DNA2.0, provided in a pJ244 vector backbone (6xP-AOX1a[-377,-328] and 6xMDM1[AOX1a]) or as an oligonucleotide (6xMDM1mut[AOX1a]) (Invitrogen), annealed by heating followed by gradual cooling, and subsequently cloned into pDONRP4-P1r (see Supplemental Table 9 online). The −46-bp CaMV 35S minimal promoter (P35Smin) was synthesized as an oligonucleotide (Invitrogen) and subsequently cloned into pDONR221 and pDONRP4-P2. Promoter-LUC constructs were created by recombining the synthetic promoter plasmids pEN-L4-promoter-R1, pEN-L1-P35Smin-L2, and pEN-R2-LUC+-L3 into the MultiSite destination vector pB7m34GW. The P35Smin:LUC reporter construct was obtained by recombining pEN-L4-P35Smin-L2 and pEN-R2-LUC+-L3 into pB7m34GW (Karimi et al., 2005).
LUC Assay
Plants were grown for 10 d in a 96-well white CulturPlate-96 (Perkin-Elmer) as sample holder. Five seedlings were grown per well containing 150 μL 1/2MS medium with 0.5% (w/v) Suc. The promoter activity was detected in the T2 generation of promoter-LUC mutants. As stress treatments, AA (Sigma-Aldrich) was added at a final concentration of 50 μM (0.1% [v/v] DMSO), rotenone at 100 μM (0.1% [v/v] DMSO), MFA at 25 mM, or H2O2 at 10 mM. Control plants were mock treated with 0.1% (v/v) DMSO. After 12 or 24 h of treatment, 100 μL luciferin (One Glo; Promega) was added to each well, followed by a 10-min dark incubation. Luminescence was measured with a LUMIstar Galaxy luminometer (BMG Labtechnologies).
Y1H Assays
Yeast strain YM4271 and destination vector pMW#2 were obtained from M. Walhout (University of Massachusetts Medical School, Worcester, MA). Design of the yeast reporter strains and cDNA library screening were done as described (Deplancke et al., 2006). Y1H screening was performed against a cDNA expression library enriched for stress-responsive genes (Jaspers et al., 2009). From the ∼6 × 106 screened transformants obtained from three independent cDNA library transformation experiments, 106 potential positives were selected to confirm their growth on 20 mM 3-AT. From the positive yeast clones, plasmids were isolated and retransformed in the reporter yeast strain for growth confirmation on 20 mM 3-AT. The pYESTrp2 empty vector containing only the transactivation domain was used as a negative control.
The ORF of ANAC082 was cloned into pDEST22 as a transactivation domain fusion (see Supplemental Table 8 online) (Invitrogen). ANAC050, ANAC051/52, and ANAC103 ORF clones in pDEST22 were obtained from the Regulatory Gene Initiative in Arabidopsis (REGIA) transcription factor collection (Paz-Ares, 2002). Yeast reporter strains were transformed with the pDEST22 prey constructs and growth was tested at different concentrations (5, 10, 20, 40, 60, and 80 mM) of 3-AT to test interactions. The pDEST22 empty vector containing only the transactivation domain was used as a negative control.
Electrophoretic Mobility Shift Assays
Oligonucleotide probes of 30 to 50 bp (see Supplemental Table 6 online) with wild-type or mutated MDM promoter sequences were annealed by heating at 99°C, followed by gradual cooling. Annealed probes were radiolabeled with [γ-32P]ATP (Perkin-Elmer) and polynucleotide kinase (Roche) and purified with Sephadex G-25–radiolabeled DNA Quick Spin columns (Roche). ANAC013, ANAC016, ANAC017, ANAC053, and ANAC078 were cloned into the GST-tag expression vector pDEST15 (Invitrogen) and transformed into Escherichia coli Rosetta 2 (DE3) pLysS-competent expression cells. Proteins were produced in 500 mL of culture overnight with 0.2 mM isopropyl-β-d-thio-galactoside at 18°C and shaking at 250 rpm. Cells were harvested, resuspended in extraction buffer (5× extraction buffer: 250 mM Tris-HCl, pH 8.5, 500 mM NaCl, 5 mM EDTA, and 1 mM DTT), and lysed by digestion with 1 mg/mL lysozyme and sonication. Lysates were clarified by centrifugation at 16,000g for 20 min. The filtered lysates were incubated with glutathione-agarose beads for 1 h at 4°C (Thermo Fisher Scientific) and washed with 10 volumes of extraction buffer containing the Complete EDTA-free protease inhibitor cocktail (Roche). Purified proteins were eluted with the extraction buffer containing 10 mM reduced glutathione. For gel shift assays, 20-μL reactions were set up with 4 μL 5× binding buffer (100 mM HEPES, pH 7.8, 0.5 M KCl, 5 mM MgCl2, 2.5 mM DTT, 5 mM EDTA, 0.25 mg/mL poly dI-dC, and 50% [v/v] glycerol), 1 fmol radiolabeled probe, 500 fmol unlabeled probe for competitor reactions, and 1.5 μg purified protein extract. Reactions were incubated for 20 min and separated on polyacrylamide gels (0.5× Tris/Borate/EDTA, 2.5% [v/v] glycerol, and 6% [w/v] acrylamide) for 2 h at 200 V on a 16 × 20-cm2 Protean II gel system (Bio-Rad). Gels were dried on Whatman paper in a gel dryer, exposed overnight (or longer), and visualized with PhosphorImager detection plates.
Quantitative RT-PCR
Total RNA and first-strand cDNA were prepared with TRIzol reagent (Invitrogen) and the iScript cDNA synthesis kit (Bio-Rad), respectively, according to the manufacturer’s instructions. As a template in the subsequent PCR, 5 μL of a 1:8 diluted first-strand cDNA was run on the iCycler iQ (Bio-Rad) with the SYBR Green I Master kit (Roche Diagnostics), according to the manufacturer’s instructions. All individual reactions were done in triplicate. Primers were designed with the Universal ProbeLibrary Assay Design center ProbeFinder software (Roche; http://www.roche-applied-science.com/; see Supplemental Table 8 online). For the expression analysis, values were normalized against ACTIN-RELATED PROTEIN7 (ARP7), whereas two reference genes, ARP7 and At2g28390 (Czechowski et al., 2005), were used to normalize data from the ANAC013-miR experiments. The Δ cycle threshold method (Livak and Schmittgen, 2001) was applied for relative transcript quantification.
ChIP
The ChIP experiments were performed as described (Bowler et al., 2004; Berckmans et al., 2011) with minor modifications. 35S:GFP-ANAC013, ProANAC013:GFP-ANAC013, 35S:ANAC013-6, and wild-type plants were grown for 8 d. ProANAC013:GFP-ANAC013 and wild-type plants were either mock treated (0.1% [v/v] DMSO) or treated with 50 μM AA for 24 h. Of whole seedlings, 1.5 g was harvested for cross-linking in 1% (v/v) formaldehyde under vacuum for 10 min. Gly was added to a final concentration of 0.125 M and vacuum was applied for 5 min. Chromatin was isolated and fragmented by sonication with a Bioruptor sonicator (Diagenode), resulting in fragments of 200 to 600 bp. The chromatin was precleared with 80 μL Protein A Agarose/Salmon Sperm DNA (Millipore). Ten microliters was used as INPUT, whereas the remainder was split into two samples, of which one was treated with 30 μL of anti-GFP antibody coupled to agarose beads (GFP-Trap_A; Chromotek) and the other without antibody. The samples were incubated overnight at 4°C and subsequently eluted from the beads. All centrifugation steps with bead-containing samples were done at 100g. Proteins were de-cross-linked and DNA was extracted by phenol/chloroform/isoamyl alcohol extraction followed by purification with the MinElute PCR purification kit (Qiagen). Quantitative PCRs were analyzed on a LightCycler 480 apparatus (Roche Diagnostics) with the SYBR Green I Master kit (Roche Diagnostics), and each reaction was done in triplicate. ACTIN2, CDKA;1, and UBQ10 were used as negative controls. ChIP–quantitative PCR data were normalized against the amount of chromatin used in the ChIP (INPUT) and were represented as % INPUT. For the quantitative PCR analysis, specific primers were designed for the MDM-containing promoter regions by means of primer-BLAST at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) (Rozen and Skaletsky, 2000) and tested for amplification specificity by melting curve analysis before use (see Supplemental Table 8 online). Data were obtained from single experiments, but similar data were obtained in independent runs and with independent 35S:GFP-ANAC013 and ProANAC013:GFP-ANAC013 lines.
Promoter-GUS Analysis
GUS assays were performed as described (Beeckman and Engler, 1994). Samples were photographed with a stereomicroscope (Stemi SV11; Zeiss) or with a Nomarski differential interference contrast microscope (Olympus BX51; Leica).
Chlorophyll Determinations
Chlorophyll was measured on 100 mg of fresh green tissue from 3-week-old plants and extracted by addition of 1 mL of 80% (v/v) acetone to the ground tissue and incubation for 30 min in the dark with shaking. The debris was pelleted by centrifugation at 1500g for 5 min. The optical densities of the supernatant at 646 and 663 nm were determined and used to calculate chlorophyll a and b and total chlorophyll concentrations according to Lichtenthaler and Wellburn (1985).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ANAC013, At1g32870; ANAC016, At1g34180; ANAC017, At1g34190; ANAC053, At3g10500; ANAC078, At5g04410; PHB3, At5g40770; ABCB4, At2g47000; AOX1a, At3g22370; CRF6, At3g61630; CYP81D8, At4g37370; HRE2, At2g47520; HSP23.5, At5g51440; ST, At1g32870; UGT74E2, At1g05680; UPOX, At2g21640; MGE1, At5g55200; ANAC050, At3g10480; ANAC051/52, At3g10490; ANAC082, At5g09330; ANAC103, At5g64060; ACT2, At3g18780; CDKA;1, At3g48750; UBQ10, At4g05320; TOM40, At3g2000; GPX3, At2g43350; RCD1, At1g32230; SRO5, At5g62520; and DREB2A, AT5G05410.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Binding of NAC Transcription Factors with the MDM of the At5g09570, UPOX, and At2g04050 Promoter in Vitro as Shown by Electrophoretic Mobility Shift Assays.
Supplemental Figure 2. Hierarchical Clustering of the MRR Upregulated Genes in the CORNET Stress Data Set, a Compilation of Biotic and Abiotic Stress Conditions.
Supplemental Figure 3. Schematic Overview of the Amplicons Used in ChIP-qPCR Analyses.
Supplemental Figure 4. Interaction of ANAC013 with MDS Promoters in Planta as Shown by ChIP.
Supplemental Figure 5. MRR-Mediated Gene Expression of the MDS Regulated by ANAC013 in Arabidopsis.
Supplemental Figure 6. Increase in AOX1a and UPOX Protein Abundance upon ANAC013 Overexpression.
Supplemental Figure 7. Autoregulation of ANAC013 Expression.
Supplemental Figure 8. ANAC013 Localization to the ER and the Nucleus.
Supplemental Figure 9. Phenotypes of ANAC013-Overexpressing and miRNA Lines under Mitochondrial and Oxidative Stress Conditions.
Supplemental Figure 10. Increased Photosynthetic Performance of ANAC013-Overexpressing Plants after Exposure to MV-Mediated Oxidative Stress.
Supplemental Figure 11. Y1H Assays Showing the Interaction with the MDM of Nontransmembrane NAC Transcription Factors of the NAC2 Phylogenetic Subgroup (Ooka et al., 2003).
Supplemental Figure 12. MDS Gene Expression Regulated by ANAC053 and ANAC078 in Arabidopsis.
Supplemental Figure 13. Diagram Summarizing the Role of ANAC013 in MRR.
Supplemental Table 1. Overview of MRR Microarray Data Sets Used in the Meta-Analysis.
Supplemental Table 2. Expression Values of the 34 MRR-Upregulated Genes under Five or More Mitochondrial Dysfunction Conditions.
Supplemental Table 3. Presence of the MDM in the MDS Gene Set and Its Conservation in Orthologous Genes of Related Dicot Species.
Supplemental Table 4. Functional Annotations of the MDS Genes.
Supplemental Table 5. Arabidopsis Genes Containing Conserved MDM Motif Instances.
Supplemental Table 6. Probes Used for Electrophoretic Mobility Shift Assays.
Supplemental Table 7. Overrepresented Gene Ontology Terms in the MDS Gene Set and in the Gene Set Containing All Arabidopsis Genes with Conserved MDM Motif Instances.
Supplemental Table 8. PCR Primers Used.
Supplemental Table 9. Probes Used for Generation of Artificial Promoter Constructs.
Supplemental Methods 1. Nanostring nCounter Assays, UPOX Antibody Production, Protein Gel Blots, Determination of Subcellular Localization, and Statistical Analyses.
Acknowledgments
We thank our colleagues Sandy Vanderauwera and Frank Hoeberichts for providing the ANAC013-miRNA lines and overexpression lines, Véronique Storme for help with the statistical analysis, Mária Šimášková, Bert Waelkens, and Aurine Verkest for help with the ChIP and Y1H experiments, Riet De Rycke for help with immunoelectron microscopy, Daniel Van Damme for help with the subcellular localization analysis, and Martine De Cock for help in preparing the article. This work was supported by grants from Ghent University Multidisciplinary Research Partnership (“Ghent BioEconomy” [Project 01MRB510W] and “Bioinformatics: from nucleotides to networks” [Project 01MR0310W]), the Interuniversity Attraction Poles Programme (IAP P7/29), initiated by the Belgian Science Policy Office, and the Australian Research Council Centre of Excellence Grant CEO561495. I.D.C. is indebted to the Agency for Innovation by Science and Technology for a predoctoral fellowship.
AUTHOR CONTRIBUTIONS
I.D.C. and F.V.B designed the research. I.D.C., O.V.A., M.W.M., S.R.L., A.Inzé, D.R., and B.v.d.C. performed the experiments. I.D.C, V.V., and K.V. designed and performed computational analyses. I.D.C, V.V., O.V.A, K.V., M.W.M, S.R.L, A.Ivanova, S.N., A.Inzé, P.J., Y.V.d.P., J.K., J.W., and F.V.B. analyzed the data and interpreted the results. I.D.C. wrote the article with the help of V.V., O.V.A., K.V., M.R.M., S.W.L., J.W., and F.V.B.
Glossary
- MRR
mitochondrial retrograde regulation
- RTG
retrograde
- mtETC
mitochondrial electron transport chain
- TCA
tricarboxylic acid
- mtROS
mitochondrial reactive oxygen species
- ROS
reactive oxygen species
- MDM
mitochondrial dysfunction motif
- AA
antimycin A
- MFA
monofluoroacetate
- H2O2
hydrogen peroxide
- CaMV
cauliflower mosaic virus
- Y1H
yeast one-hybrid
- TM
transmembrane
- ChIP
chromatin immunoprecipitation
- GUS
β-glucuronidase
- ER
endoplasmic reticulum
- MV
methyl viologen
- 3-AT
3-amino-1,2,4-triazole
- 1/2MS
half-strength Murashige and Skoog
- PSII
photosystem II
- NACBS
NAC binding site
- Col-0
Columbia-0
- ORF
open reading frame
References
- Ahn C.S., Lee J.H., Reum Hwang A., Kim W.T., Pai H.-S. (2006). Prohibitin is involved in mitochondrial biogenesis in plants. Plant J. 46: 658–667 [DOI] [PubMed] [Google Scholar]
- Atanassov I.I., Atanassov I.I., Etchells J.P., Turner S.R. (2009). A simple, flexible and efficient PCR-fusion/Gateway cloning procedure for gene fusion, site-directed mutagenesis, short sequence insertion and domain deletions and swaps. Plant Methods 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo D., Tacnet F., Delaunay A., Rodrigues-Pousada C., Toledano M.B. (2003). Two redox centers within Yap1 for H2O2 and thiol-reactive chemicals signaling. Free Radic. Biol. Med. 35: 889–900 [DOI] [PubMed] [Google Scholar]
- Bailey, T.L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, R. Altman, D. Brutlag, P. Karp, R. Lathrop, and D. Searls, eds (Menlo Park, CA: AAAI Press), pp. 28–36. [PubMed] [Google Scholar]
- Bailey-Serres J., Chang R. (2005). Sensing and signalling in response to oxygen deprivation in plants and other organisms. Ann. Bot. (Lond.) 96: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N.R. (2008). Chlorophyll fluorescence: A probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59: 89–113 [DOI] [PubMed] [Google Scholar]
- Beeckman T., Engler G. (1994). An easy technique for the clearing of histochemically stained plant tissue. Plant Mol. Biol. Rep. 12: 37–42 [Google Scholar]
- Belles-Boix E., Babiychuk E., Van Montagu M., Inzé D., Kushnir S. (2000). CEO1, a new protein from Arabidopsis thaliana, protects yeast against oxidative damage. FEBS Lett. 482: 19–24 [DOI] [PubMed] [Google Scholar]
- Berckmans B., et al. (2011). Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell 23: 3671–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G., Adebanjo O.A., Freedman B.D., Anandatheerthavarada H.K., Vijayasarathy C., Zaidi M., Kotlikoff M., Avadhani N.G. (1999). Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: A novel mode of inter-organelle crosstalk. EMBO J. 18: 522–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O., Zhu J., Verslues P.E., Sunkar R., Zhu J.-K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C., Benvenuto G., Laflamme P., Molino D., Probst A.V., Tariq M., Paszkowski J. (2004). Chromatin techniques for plant cells. Plant J. 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Boyes D.C., Zayed A.M., Ascenzi R., McCaskill A.J., Hoffman N.E., Davis K.R., Görlach J. (2001). Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, E.A., Bailey-Serres, J., and Weretilnyk, E. (2000). Responses to abiotic stress. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 1158–1203. [Google Scholar]
- Butow R.A., Avadhani N.G. (2004). Mitochondrial signaling: The retrograde response. Mol. Cell 14: 1–15 [DOI] [PubMed] [Google Scholar]
- Carrie C., Giraud E., Duncan O., Xu L., Wang Y., Huang S., Clifton R., Murcha M., Filipovska A., Rackham O., Vrielink A., Whelan J. (2010). Conserved and novel functions for Arabidopsis thaliana MIA40 in assembly of proteins in mitochondria and peroxisomes. J. Biol. Chem. 285: 36138–36148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton R., Lister R., Parker K.L., Sappl P.G., Elhafez D., Millar A.H., Day D.A., Whelan J. (2005). Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol. Biol. 58: 193–212 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E. (2004). WebLogo: a sequence logo generator. Genome Res. 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovska M., Vanlerberghe G.C. (2012). Alternative oxidase modulates leaf mitochondrial concentrations of superoxide and nitric oxide. New Phytol. 195: 32–39 [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.-R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S., Hollunder J., Nelissen H., Meulemeester N., Inzé D. (2012). CORNET 2.0: Integrating plant coexpression, protein–protein interactions, regulatory interactions, gene associations and functional annotations. New Phytol. 195: 707–720 [DOI] [PubMed] [Google Scholar]
- Delaunay A., Pflieger D., Barrault M.-B., Vinh J., Toledano M.B. (2002). A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111: 471–481 [DOI] [PubMed] [Google Scholar]
- Deplancke, B., Vermeirssen, V., Arda, H.E., Martinez, N.J., and Walhout, A.J.M.(2006). Gateway-compatible yeast one-hybrid screens. CSH Protoc. 2006:10.1101/pdb.prot4590. [DOI] [PubMed]
- Devaux F., Carvajal E., Moye-Rowley S., Jacq C. (2002). Genome-wide studies on the nuclear PDR3-controlled response to mitochondrial dysfunction in yeast. FEBS Lett. 515: 25–28 [DOI] [PubMed] [Google Scholar]
- Dojcinovic D., Krosting J., Harris A.J., Wagner D.J., Rhoads D.M. (2005). Identification of a region of the Arabidopsis AtAOX1a promoter necessary for mitochondrial retrograde regulation of expression. Plant Mol. Biol. 58: 159–175 [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein C.B., Waddle J.A., Hale W., IV, Davé V., Thornton J., Macatee T.L., Garner H.R., Butow R.A. (2001). Genome-wide responses to mitochondrial dysfunction. Mol. Biol. Cell 12: 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraresi R., Troiano L., Pinti M., Roat E., Lugli E., Quaglino D., Taverna D., Bellizzi D., Passarino G., Cossarizza A. (2008). Resistance of mtDNA-depleted cells to apoptosis. Cytometry A 73: 528–537 [DOI] [PubMed] [Google Scholar]
- Freeling M., Subramaniam S. (2009). Conserved noncoding sequences (CNSs) in higher plants. Curr. Opin. Plant Biol. 12: 126–132 [DOI] [PubMed] [Google Scholar]
- Fujibe T., Saji H., Arakawa K., Yabe N., Takeuchi Y., Yamamoto K.T. (2004). A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive rcd1, is tolerant to supplemental ultraviolet-B irradiation. Plant Physiol. 134: 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev T.S., Van Breusegem F., Stone J.M., Denev I., Laloi C. (2006). Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. BioEssays 28: 1091–1101 [DOI] [PubMed] [Google Scholar]
- Giraud E., Ho L.H.M., Clifton R., Carroll A., Estavillo G., Tan Y.-F., Howell K.A., Ivanova A., Pogson B.J., Millar A.H., Whelan J. (2008). The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol. 147: 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E., Van Aken O., Ho L.H.M., Whelan J. (2009). The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 150: 1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason C., Huang S., Thatcher L.F., Foley R.C., Anderson C.R., Carroll A.J., Millar A.H., Singh K.B. (2011). Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense. Proc. Natl. Acad. Sci. USA 108: 10768–10773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray G.R., Maxwell D.P., Villarimo A.R., McIntosh L. (2004). Mitochondria/nuclear signaling of alternative oxidase gene expression occurs through distinct pathways involving organic acids and reactive oxygen species. Plant Cell Rep. 23: 497–503 [DOI] [PubMed] [Google Scholar]
- Hallstrom T.C., Moye-Rowley W.S. (2000). Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275: 37347–37356 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Rizzuto R., Hajnoczky G., Su T.-P. (2009). MAM: More than just a housekeeper. Trends Cell Biol. 19: 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden M.S., West A.P., Ghosh S. (2006). NF-κB and the immune response. Oncogene 25: 6758–6780 [DOI] [PubMed] [Google Scholar]
- Ho H.K., White C.C., Fernandez C., Fausto N., Kavanagh T.J., Nelson S.D., Bruschi S.A. (2005). Nrf2 activation involves an oxidative-stress independent pathway in tetrafluoroethylcysteine-induced cytotoxicity. Toxicol. Sci. 86: 354–364 [DOI] [PubMed] [Google Scholar]
- Ho L.H.M., Giraud E., Uggalla V., Lister R., Clifton R., Glen A., Thirkettle-Watts D., Van Aken O., Whelan J. (2008). Identification of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in Arabidopsis. Plant Physiol. 147: 1858–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C., Lin S-j., Chi W-t., Charng Y-y. (2012). Recent gene duplication and subfunctionalization produced a mitochondrial GrpE, the nucleotide exchange factor of the Hsp70 complex, specialized in thermotolerance to chronic heat stress in Arabidopsis. Plant Physiol. 158: 747–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huner N.P.A., Öquist G., Sarhan F. (1998). Energy balance and acclimation to light and cold. Trends Plant Sci. 3: 224–230 [Google Scholar]
- Inzé A., Vanderauwera S., Hoeberichts F.A., Vandorpe M., Van Gaever T., Van Breusegem F. (2012). A subcellular localization compendium of hydrogen peroxide-induced proteins. Plant Cell Environ. 35: 308–320 [DOI] [PubMed] [Google Scholar]
- Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J.D., Yamamoto M. (1999). Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13: 76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers P., Blomster T., Brosché M., Salojärvi J., Ahlfors R., Vainonen J.P., Reddy R.A., Immink R., Angenent G., Turck F., Overmyer K., Kangasjärvi J. (2009). Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J. 60: 268–279 [DOI] [PubMed] [Google Scholar]
- Jaspers P., Overmyer K., Wrzaczek M., Vainonen J.P., Blomster T., Salojärvi J., Reddy R.A., Kangasjärvi J. (2010). The RST and PARP-like domain containing SRO protein family: Analysis of protein structure, function and conservation in land plants. BMC Genomics 11: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski S.M., Kriete A. (2012). The yeast retrograde response as a model of intracellular signaling of mitochondrial dysfunction. Front. Physiol. 3: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M.K., Kjaersgaard T., Nielsen M.M., Galberg P., Petersen K., O’Shea C., Skriver K. (2010). The Arabidopsis thaliana NAC transcription factor family: Structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426: 183–196 [DOI] [PubMed] [Google Scholar]
- Jones A.W.E., Yao Z., Vicencio J.M., Karkucinska-Wieckowska A., Szabadkai G. (2012). PGC-1 family coactivators and cell fate: Roles in cancer, neurodegeneration, cardiovascular disease and retrograde mitochondria-nucleus signalling. Mitochondrion 12: 86–99 [DOI] [PubMed] [Google Scholar]
- Kang M.-I., Kobayashi A., Wakabayashi N., Kim S.-G., Yamamoto M. (2004). Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc. Natl. Acad. Sci. USA 101: 2046–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., De Meyer B., Hilson P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Keunen E., Remans T., Bohler S., Vangronsveld J., Cuypers A. (2011). Metal-induced oxidative stress and plant mitochondria. Int. J. Mol. Sci. 12: 6894–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchev P.I., Pellny T.K., Diaz Vivancos P., Kiddle G., Hedden P., Driscoll S., Vanacker H., Verrier P., Hancock R.D., Foyer C.H. (2011). The transcription factor ABI4 Is required for the ascorbic acid-dependent regulation of growth and regulation of jasmonate-dependent defense signaling pathways in Arabidopsis. Plant Cell 23: 3319–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-G., Lee S., Ryu J., Park C.-M. (2010a). Probing protein structural requirements for activation of membrane-bound NAC transcription factors in Arabidopsis and rice. Plant Sci. 178: 239–244 [Google Scholar]
- Kim S.-G., Lee S., Seo P.J., Kim S.-K., Kim J.-K., Park C.-M. (2010b). Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics 95: 56–65 [DOI] [PubMed] [Google Scholar]
- Kim S.-Y., Kim S.-G., Kim Y.-S., Seo P.J., Bae M., Yoon H.-K., Park C.-M. (2007). Exploring membrane-associated NAC transcription factors in Arabidopsis: Implications for membrane biology in genome regulation. Nucleic Acids Res. 35: 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-S., Kim S.-G., Park J.-E., Park H.-Y., Lim M.-H., Chua N.-H., Park C.-M. (2006). A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18: 3132–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A., Kang M.-I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. (2004). Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24: 7130–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Yamamoto M. (2006). Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv. Enzyme Regul. 46: 113–140 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S., Nott A., Mockler T.C., Hong F., Sachetto-Martins G., Surpin M., Lim J., Mittler R., Chory J. (2007). Multiple signals from damaged chloroplasts converge on a common pathway to regulate nuclear gene expression. Science 316: 715–719 Erratum. Science 316, 1698. [PubMed] [Google Scholar]
- Kühn K., Carrie C., Giraud E., Wang Y., Meyer E.H., Narsai R., Colas des Francs-Small C., Zhang B., Murcha M.W., Whelan J. (2011). The RCC1 family protein RUG3 is required for splicing of nad2 and complex I biogenesis in mitochondria of Arabidopsis thaliana. Plant J. 67: 1067–1080 [DOI] [PubMed] [Google Scholar]
- Kühn K., Richter U., Meyer E.H., Delannoy E., Falcon de Longevialle A., O’Toole N., Börner T., Millar A.H., Small I.D., Whelan J. (2009). Phage-type RNA polymerase RPOTmp performs gene-specific transcription in mitochondria of Arabidopsis thaliana. Plant Cell 21: 2762–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.H., Lee H., Xiong L., Zhu J.-K. (2002). A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 14: 1235–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Seo P.J., Lee H.-J., Park C.-M. (2012). A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 70: 831–844 [DOI] [PubMed] [Google Scholar]
- Liao X., Butow R.A. (1993). RTG1 and RTG2: Two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell 72: 61–71 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H.K., Wellburn A.R. (1985). Determination of total carotenoids and chlorophylls a and b of leaf in different solvents. Biochem. Soc. Trans. 11: 591–592 [Google Scholar]
- Linhart C., Halperin Y., Shamir R. (2008). Transcription factor and microRNA motif discovery: The Amadeus platform and a compendium of metazoan target sets. Genome Res. 18: 1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Butow R.A. (2006). Mitochondrial retrograde signaling. Annu. Rev. Genet. 40: 159–185 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Manabe Y., Tinker N., Colville A., Miki B. (2007). CSR1, the sole target of imidazolinone herbicide in Arabidopsis thaliana. Plant Cell Physiol. 48: 1340–1358 [DOI] [PubMed] [Google Scholar]
- Martinus R.D., Garth G.P., Webster T.L., Cartwright P., Naylor D.J., Høj P.B., Hoogenraad N.J. (1996). Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. Eur. J. Biochem. 240: 98–103 [DOI] [PubMed] [Google Scholar]
- Maxwell D.P., Wang Y., McIntosh L. (1999). The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. USA 96: 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E.H., Tomaz T., Carroll A.J., Estavillo G., Delannoy E., Tanz S.K., Small I.D., Pogson B.J., Millar A.H. (2009). Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol. 151: 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Lv D., Wang P., Wang X.-C., Chen J., Miao C., Song C.-P. (2006). An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 18: 2749–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita T., Kojima Y., Maruta T., Nishizawa-Yokoi A., Yabuta Y., Shigeoka S. (2009). Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol. 50: 2210–2222 [DOI] [PubMed] [Google Scholar]
- Ng S., et al. (2013a). Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. J. Biol. Chem. 288: 3449–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S., et al. (2013b). A membrane-bound NAC transcription factor, ANAC017, controls mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25: 3450–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A.N., Ernst H.A., Lo Leggio L., Skriver K. (2005a). NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Olsen A.N., Ernst H.A., Lo Leggio L., Skriver K. (2005b). DNA-binding specificity and molecular functions of NAC transcription factors. Plant Sci. 169: 785–797 [Google Scholar]
- Ooka H., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10: 239–247 [DOI] [PubMed] [Google Scholar]
- Park S.Y., Chang I., Kim J.-Y., Kang S.W., Park S.-H., Singh K., Lee M.-S. (2004). Resistance of mitochondrial DNA-depleted cells against cell death: Role of mitochondrial superoxide dismutase. J. Biol. Chem. 279: 7512–7520 [DOI] [PubMed] [Google Scholar]
- Paz-Ares J., the Regia Consortium (2002). REGIA, an EU project on functional genomics of transcription factors from Arabidopsis thaliana. Comp. Funct. Genomics 3: 102–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñas Fernández A., Strand Å. (2008). Retrograde signaling and plant stress: Plastid signals initiate cellular stress responses. Curr. Opin. Plant Biol. 11: 509–513 [DOI] [PubMed] [Google Scholar]
- Prasad T.K., Anderson M.D., Stewart C.R. (1994). Acclimation, hydrogen peroxide, and abscisic acid protect mitochondria against irreversible chilling injury in maize seedlings. Plant Physiol. 105: 619–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads D.M., Subbaiah C.C. (2007). Mitochondrial retrograde regulation in plants. Mitochondrion 7: 177–194 [DOI] [PubMed] [Google Scholar]
- Rhoads D.M., Umbach A.L., Subbaiah C.C., Siedow J.N. (2006). Mitochondrial reactive oxygen species. Contribution to oxidative stress and interorganellar signaling. Plant Physiol. 141: 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads D.M., White S.J., Zou Y., Muralidharan M., Elthon T.E. (2005). Altered gene expression in plants with constitutive expression of a mitochondrial small heat shock protein suggests the involvement of retrograde regulation in the heat stress response. Physiol. Plant. 123: 435–444 [Google Scholar]
- Rozen, S., and Skaletsky, H. (2000). Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols, Methods in Molecular Biology, Vol. 132, S. Misener and S.A. Krawetz, eds (Totowa, NJ: Humana Press), pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Safrany J., Haasz V., Mate Z., Ciolfi A., Feher B., Oravecz A., Stec A., Dallmann G., Morelli G., Ulm R., Nagy F. (2008). Identification of a novel cis-regulatory element for UV-B-induced transcription in Arabidopsis. Plant J. 54: 402–414 [DOI] [PubMed] [Google Scholar]
- Salin H., Fardeau V., Piccini E., Lelandais G., Tanty V., Lemoine S., Jacq C., Devaux F. (2008). Structure and properties of transcriptional networks driving selenite stress response in yeasts. BMC Genomics 9: 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzländer M., Finkemeier I. (2013). Mitochondrial energy and redox signaling in plants. Antioxid. Redox Signal. 18: 2122–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzländer M., König A.-C., Sweetlove L.J., Finkemeier I. (2012). The impact of impaired mitochondrial function on retrograde signalling: A meta-analysis of transcriptomic responses. J. Exp. Bot. 63: 1735–1750 [DOI] [PubMed] [Google Scholar]
- Seo P.J., Kim S.-G., Park C.-M. (2008). Membrane-bound transcription factors in plants. Trends Plant Sci. 13: 550–556 [DOI] [PubMed] [Google Scholar]
- Shedge V., Davila J., Arrieta-Montiel M.P., Mohammed S., Mackenzie S.A. (2010). Extensive rearrangement of the Arabidopsis mitochondrial genome elicits cellular conditions for thermotolerance. Plant Physiol. 152: 1960–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A., De Bodt S., Obata T., De Clercq I., Claeys H., De Rycke R., Andriankaja M., Van Aken O., Van Breusegem F., Fernie A.R., Inzé D. (2010). Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol. 152: 226–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: article 3. [DOI] [PubMed] [Google Scholar]
- Srinivasan V., Kriete A., Sacan A., Jazwinski S.M. (2010). Comparing the yeast retrograde response and NF-κB stress responses: Implications for aging. Aging Cell 9: 933–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah C.C., Bush D.S., Sachs M.M. (1998). Mitochondrial contribution to the anoxic Ca2+ signal in maize suspension-cultured cells. Plant Physiol. 118: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove L.J., Heazlewood J.L., Herald V., Holtzapffel R., Day D.A., Leaver C.J., Millar A.H. (2002). The impact of oxidative stress on Arabidopsis mitochondria. Plant J. 32: 891–904 [DOI] [PubMed] [Google Scholar]
- Taylor N.L., Day D.A., Millar A.H. (2002). Environmental stress causes oxidative damage to plant mitochondria leading to inhibition of glycine decarboxylase. J. Biol. Chem. 277: 42663–42668 [DOI] [PubMed] [Google Scholar]
- Teixeira M.C., Dias P.J., Simões T., Sá-Correia I. (2008). Yeast adaptation to mancozeb involves the up-regulation of FLR1 under the coordinate control of Yap1, Rpn4, Pdr3, and Yrr1. Biochem. Biophys. Res. Commun. 367: 249–255 [DOI] [PubMed] [Google Scholar]
- Teotia S., Lamb R.S. (2011). RCD1 and SRO1 are necessary to maintain meristematic fate in Arabidopsis thaliana. J. Exp. Bot. 62: 1271–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M., Defrance M., Medina-Rivera A., Sand O., Herrmann C., Thieffry D., van Helden J. (2011). RSAT 2011: Regulatory sequence analysis tools. Nucleic Acids Res. 39 (Web Server issue): W86–W91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti V.B., et al. (2010). Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22: 2660–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven A., Wong J.M.S., Xu D., Sopta M., Ingles C.J. (2001). Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial DNA mutant. J. Biol. Chem. 276: 4020–4027 [DOI] [PubMed] [Google Scholar]
- Umbach A.L., Fiorani F., Siedow J.N. (2005). Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol. 139: 1806–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainonen J.P., Jaspers P., Wrzaczek M., Lamminmäki A., Reddy R.A., Vaahtera L., Brosché M., Kangasjärvi J. (2012). RCD1-DREB2A interaction in leaf senescence and stress responses in Arabidopsis thaliana. Biochem. J. 442: 573–581 [DOI] [PubMed] [Google Scholar]
- Van Aken O., Giraud E., Clifton R., Whelan J. (2009). Alternative oxidase: A target and regulator of stress responses. Physiol. Plant. 137: 354–361 [DOI] [PubMed] [Google Scholar]
- Van Aken O., Pečenková T., van de Cotte B., De Rycke R., Eeckhout D., Fromm H., De Jaeger G., Witters E., Beemster G.T.S., Inzé D., Van Breusegem F. (2007). Mitochondrial type-I prohibitins of Arabidopsis thaliana are required for supporting proficient meristem development. Plant J. 52: 850–864 [DOI] [PubMed] [Google Scholar]
- Van Aken O., Whelan J. (2012). Comparison of transcriptional changes to chloroplast and mitochondrial perturbations reveals common and specific responses in Arabidopsis. Front. Plant Sci. 3: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O., Whelan J., Van Breusegem F. (2010). Prohibitins: Mitochondrial partners in development and stress response. Trends Plant Sci. 15: 275–282 [DOI] [PubMed] [Google Scholar]
- Van Aken O., Zhang B., Law S., Narsai R., Whelan J. (2013). AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol. 162: 254–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel M., Proost S., Wischnitzki E., Movahedi S., Scheerlinck C., Van de Peer Y., Vandepoele K. (2012). Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 158: 590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe G.C., McIntosh L. (1996). Signals regulating the expression of the nuclear gene encoding alternative oxidase of plant mitochondria. Plant Physiol. 111: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe G.C., Robson C.A., Yip J.Y.H. (2002). Induction of mitochondrial alternative oxidase in response to a cell signal pathway down-regulating the cytochrome pathway prevents programmed cell death. Plant Physiol. 129: 1829–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visioli G., Maestri E., Marmiroli N. (1997). Differential display-mediated isolation of a genomic sequence for a putative mitochondrial LMW HSP specifically expressed in condition of induced thermotolerance in Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 34: 517–527 [DOI] [PubMed] [Google Scholar]
- Wang Y., Carrie C., Giraud E., Elhafez D., Narsai R., Duncan O., Whelan J., Murcha M.W. (2012). Dual location of the mitochondrial preprotein transporters B14.7 and Tim23-2 in complex I and the TIM17:23 complex in Arabidopsis links mitochondrial activity and biogenesis. Plant Cell 24: 2675–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welner D.H., Lindemose S., Grossmann J.G., Møllegaard N.E., Olsen A.N., Helgstrand C., Skriver K., Lo Leggio L. (2012). DNA binding by the plant-specific NAC transcription factors in crystal and solution: A firm link to WRKY and GCM transcription factors. Biochem. J. 444: 395–404 [DOI] [PubMed] [Google Scholar]
- Yabuta Y., Morishita T., Kojima Y., Maruta T., Nishizawa-Yokoi A., Shigeoka S. (2010). Identification of recognition sequence of ANAC078 protein by the cyclic amplification and selection of targets technique. Plant Signal. Behav. 5: 695–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta Y., Osada R., Morishita T., Nishizawa-Yokoi A., Tamoi M., Maruta T., Shigeoka S. (2011). Involvement of Arabidopsis NAC transcription factor in the regulation of 20S and 26S proteasomes. Plant Sci. 181: 421–427 [DOI] [PubMed] [Google Scholar]
- Yoshida K., Terashima I., Noguchi K. (2007). Up-regulation of mitochondrial alternative oxidase concomitant with chloroplast over-reduction by excess light. Plant Cell Physiol. 48: 606–614 [DOI] [PubMed] [Google Scholar]
- Zarkovic J., Anderson S.L., Rhoads D.M. (2005). A reporter gene system used to study developmental expression of alternative oxidase and isolate mitochondrial retrograde regulation mutants in Arabidopsis. Plant Mol. Biol. 57: 871–888 [DOI] [PubMed] [Google Scholar]