This work finds that Arabidopsis PHOSPHOLIPID:DIACYLGLYCEROL ACYLTRANSFERASE1 (PDAT1) is a critical enzyme in triacylglycerol assembly in vegetative tissues. Overexpression of PDAT1 enhances both fatty acid and triacylglycerol synthesis in leaves. The results suggest genetic engineering strategies to increase oil accumulation in biomass crops used for feed and for biofuel production.
Abstract
There is growing interest in engineering green biomass to expand the production of plant oils as feed and biofuels. Here, we show that PHOSPHOLIPID:DIACYLGLYCEROL ACYLTRANSFERASE1 (PDAT1) is a critical enzyme involved in triacylglycerol (TAG) synthesis in leaves. Overexpression of PDAT1 increases leaf TAG accumulation, leading to oil droplet overexpansion through fusion. Ectopic expression of oleosin promotes the clustering of small oil droplets. Coexpression of PDAT1 with oleosin boosts leaf TAG content by up to 6.4% of the dry weight without affecting membrane lipid composition and plant growth. PDAT1 overexpression stimulates fatty acid synthesis (FAS) and increases fatty acid flux toward the prokaryotic glycerolipid pathway. In the trigalactosyldiacylglycerol1-1 mutant, which is defective in eukaryotic thylakoid lipid synthesis, the combined overexpression of PDAT1 with oleosin increases leaf TAG content to 8.6% of the dry weight and total leaf lipid by fourfold. In the plastidic glycerol-3-phosphate acyltransferase1 mutant, which is defective in the prokaryotic glycerolipid pathway, PDAT1 overexpression enhances TAG content at the expense of thylakoid membrane lipids, leading to defects in chloroplast division and thylakoid biogenesis. Collectively, these results reveal a dual role for PDAT1 in enhancing fatty acid and TAG synthesis in leaves and suggest that increasing FAS is the key to engineering high levels of TAG accumulation in green biomass.
INTRODUCTION
In plants, fatty acids (FAs), the building blocks for membrane lipids and storage triacylglycerol (TAG), are almost exclusively synthesized in the plastid (Ohlrogge and Browse, 1995). They are incorporated into glycerolipids through two parallel pathways, with the prokaryotic pathway confined to plastids and the eukaryotic pathway involving the endoplasmic reticulum (ER) and plastids (Roughan and Slack, 1982; Ohlrogge and Browse, 1995). Both pathways begin with the stepwise acylation of glycerol-3-phosphate (G3P), leading to the generation of phosphatidic acid (PA). Dephosphorylation of PA by PA phosphohydrolase gives rise to diacylglycerol (DAG), which in the plastid serves almost exclusively as a precursor for assembly of photosynthetic membrane lipids, through the prokaryotic pathway. DAG synthesized de novo in the ER through G3P acylation is mostly used to synthesize extraplastidic membrane phospholipids, such as phosphatidylcholine (PC) and phosphatidylethanolamine (PE) in growing leaves (Bates et al., 2007) and in developing seeds (Bates et al., 2009).
PC plays a central role in glycerolipid metabolism. It is the precursor for the synthesis of glycolipids, the predominant lipid species found in photosynthetic membranes. This synthesis occurs through the eukaryotic pathway (Roughan and Slack, 1982; Ohlrogge and Browse, 1995; Somerville and Browse, 1996), although the extent to which the eukaryotic pathway contributes to thylakoid lipid assembly varies depending on plant species and tissue within the plant. For example, in pea (Pisum sativum) plants (Mongrand et al., 1998) and in developing seeds of Arabidopsis thaliana (Xu et al., 2005), the eukaryotic pathway of thylakoid lipid synthesis predominates, whereas in leaves of Arabidopsis and spinach (Spinacia oleracea; Browse et al., 1986), about half of the photosynthetic membrane lipids are derived from the eukaryotic pathway and half from the prokaryotic pathway. Because the substrate specificity of acyltransferases in the plastid and ER differs, lipids synthesized via the prokaryotic pathway are characterized by the almost exclusive presence of a 16-carbon (C16) FA at the sn-2 position of the glycerol backbone, whereas those made via the eukaryotic pathway have a C18 at the same position (Frentzen et al., 1983; Heinz and Roughan, 1983; Frentzen, 1998). The Arabidopsis plastidic glycerol-3-phosphate acyltransferase1 (act1) mutants are defective in the first step of G-3-P acylation in the plastid (Kunst et al., 1988; Xu et al., 2006). As a consequence, these mutants lack prokaryotic thylakoid glycolipids and instead synthesize their thylakoid lipids almost exclusively via the eukaryotic pathway. On the other hand, the trigalactosyldiacylglycerol1 (tgd1-1) mutant is deficient in eukaryotic thylakoid lipid synthesis, and most of the photosynthetic membrane lipids in this mutant are assembled via the prokaryotic pathway (Xu et al., 2003, 2005).
PC is the dominant entry point for acyl groups exported from the plastid into glycerolipids through acyl editing (Bates et al., 2007, 2009; Tjellström et al., 2012). It is the major site of the eukaryotic pathway of FA desaturation (Sperling and Heinz, 1993; Shanklin and Cahoon, 1998), an important precursor for the synthesis of eukaryotic thylakoid lipids (Ohlrogge and Browse, 1995), and a potential carrier of newly synthesized FAs from plastid envelope membranes to the ER (Tjellström et al., 2012). In developing seeds, PC is the predominant source of DAG for TAG synthesis via the acyl-CoA–dependent and –independent pathway catalyzed by diacylglycerol:acyl-CoA acyltransferase (DGAT) and phospholipid:diacylglycerol acyltransferase (PDAT), respectively (Chapman and Ohlrogge, 2012). TAG is packaged in a dynamic subcellular organelle termed oil droplets (ODs) or oil bodies, which are composed of a central matrix of TAG surrounded by a monolayer of phospholipids with a subset of specific embedded proteins (Huang, 1996). The most abundant proteins coating seed ODs are oleosins, and genetic studies in Arabidopsis mutants have suggested a key role for oleosins in preventing ODs from coalescing, thereby maintaining ODs as small discrete entities to facilitate TAG mobilization and confer freezing tolerance during postgerminative seedling growth (Siloto et al., 2006; Shimada et al., 2008).
In the model plant Arabidopsis, DGAT1 and PDAT1 play overlapping roles in TAG synthesis in developing seeds and pollen (Zhang et al., 2009). Both DGAT1 and PDAT1 genes are expressed in leaves, roots, and stems, in addition to developing seeds and flowers (Lu et al., 2003; Ståhl et al., 2004). DGAT1 has been implicated in TAG biosynthesis in senescent leaves (Kaup et al., 2002; Slocombe et al., 2009), but the functional role of PDAT in leaf tissue remains largely unknown, although knockout of PDAT1 has recently been shown to have a small negative effect on TAG accumulation in roots of sdp1 mutants defective in SUGAR-DEPENDENT1 TAG lipase (Kelly et al., 2013). Here, we provide evidence that PDAT1 plays a critical role in TAG synthesis in Arabidopsis leaves. Overexpression of PDAT1 enhances both FA and TAG synthesis in leaves. The possible functional role of PDAT1 in membrane lipid turnover is also discussed.
RESULTS
The Relative Contribution of PDAT1 and DGAT1 to TAG Synthesis in Leaves
The Arabidopsis dgat1-1 and pdat1-2 mutants harbor an ethyl methanesulfonate–induced lesion in the DGAT1 locus (Zou et al., 1999) and a T-DNA insertion in the PDAT1 gene (Zhang et al., 2009), respectively. As the first step toward understanding the role of PDAT1 and DGAT1 in TAG synthesis in vegetative tissues, we quantified TAG levels in developing and senescing leaves of wild-type, dgat1-1, and pdat1-2 mutant plants. On a dry weight (DW) basis, the TAG levels measured as fatty acid methyl esters (FAMEs) were 0.04 and 0.15% in developing and senescing leaves of wild-type plants, respectively (Figure 1). Compared with the wild type, the pdat1-2 mutant displayed a 57% decrease in TAG content in developing leaves and 39% in senescing leaves. By contrast, the dgat1-1 mutant showed a less pronounced decrease in TAG level in developing leaves (31%) but a more severe drop in TAG content in senescing leaves (63%). These results suggest that PDAT1 has a more important role in TAG synthesis in growing leaves than DGAT1, whereas the opposite is true in senescing leaves. Consistent with this notion, analyzing the database of lipid-related gene expression during leaf senescence (Troncoso-Ponce et al., 2013) revealed that PDAT1 is expressed at much higher levels than DGAT1 during early leaf growth and development (see Supplemental Figure 1 online). In addition, the PDAT1 transcript level decreases with age in all three microarray studies compiled in the database, whereas DGAT1 expression tends to increase as the leaf senesces. The involvement of DGAT1 in TAG synthesis during leaf senescence is consistent with previous reports (Kaup et al., 2002; Slocombe et al., 2009).
Figure 1.
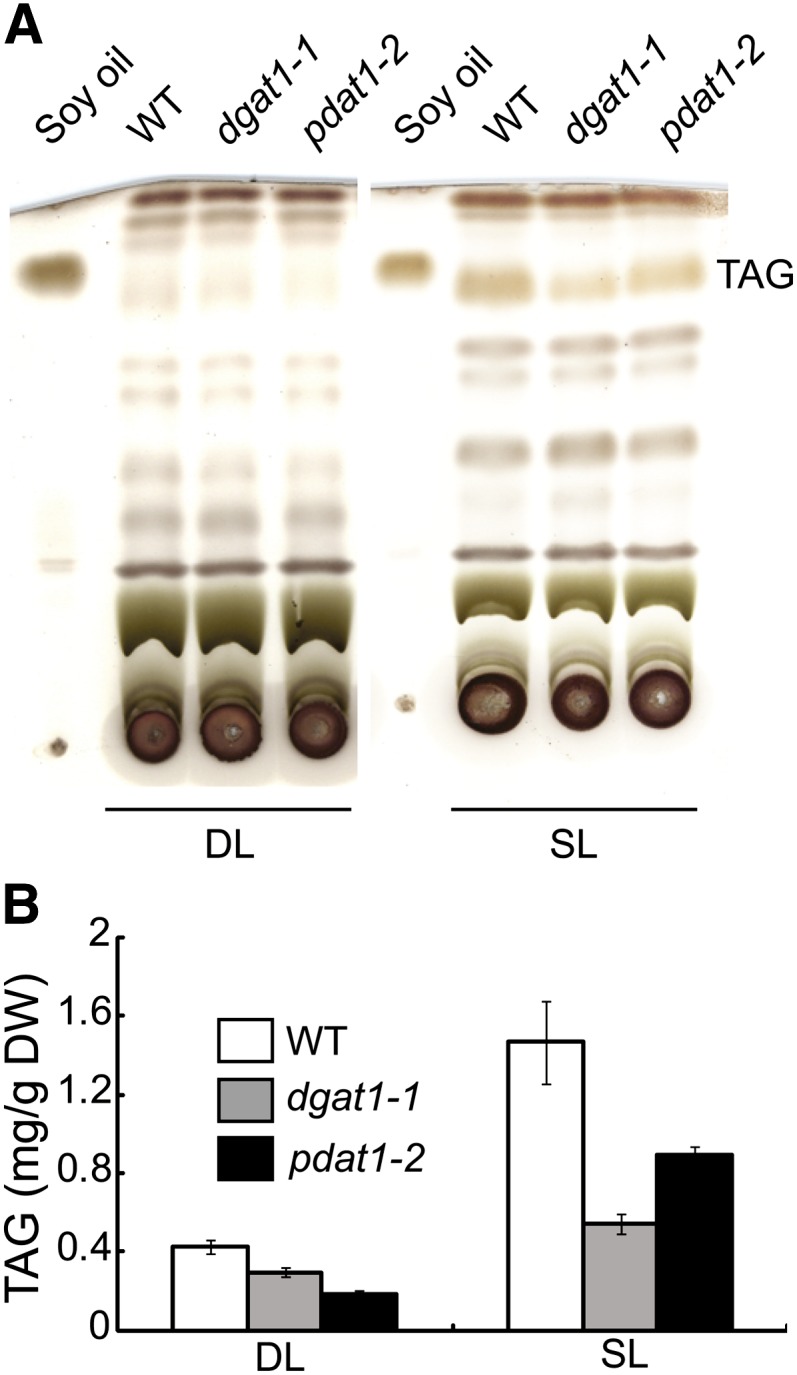
Roles of DGAT1 and PDAT1 in TAG Synthesis during Leaf Development.
Lipids were extracted from developing leaves (DL; the thirteenth leaf of 5-week-old plants) and senescing leaves (SL; the fifth leaf of 7-week-old plants) of the wild type (WT), dgat1-1, and pdat1-2. Values are means and sd of three to five biological replicates.
[See online article for color version of this figure.]
Overexpression of PDAT1 Enhances TAG Accumulation, Leading to OD Overexpansion in Leaves
A previous study showed that overexpressing PDAT1 driven by the constitutive 35S cauliflower mosaic virus promoter did not affect TAG levels or membrane lipid content and FA composition in the young seedlings of three independent transgenic Arabidopsis lines (Ståhl et al., 2004). We extended this study by examining TAG content in the leaves of a large number of adult transgenic plants grown in soil. On average, there was a sevenfold increase in TAG content in 22 randomly chosen primary transgenic plants compared with the wild type (see Supplemental Figure 2A online). By comparison, overexpressing DGAT1 driven by the same promoter resulted in only a marginal, if any, increase in the average leaf TAG levels in 21 randomly selected primary transformants tested. These results are in accordance with the absence of significant increases in total leaf FA content in Arabidopsis transgenic plants constitutively overexpressing DGAT1 as reported recently by others (Winichayakul et al., 2013). Together, these results indicate that PDAT1 exerts a much higher impact than DGAT1 on TAG synthesis in Arabidopsis leaves.
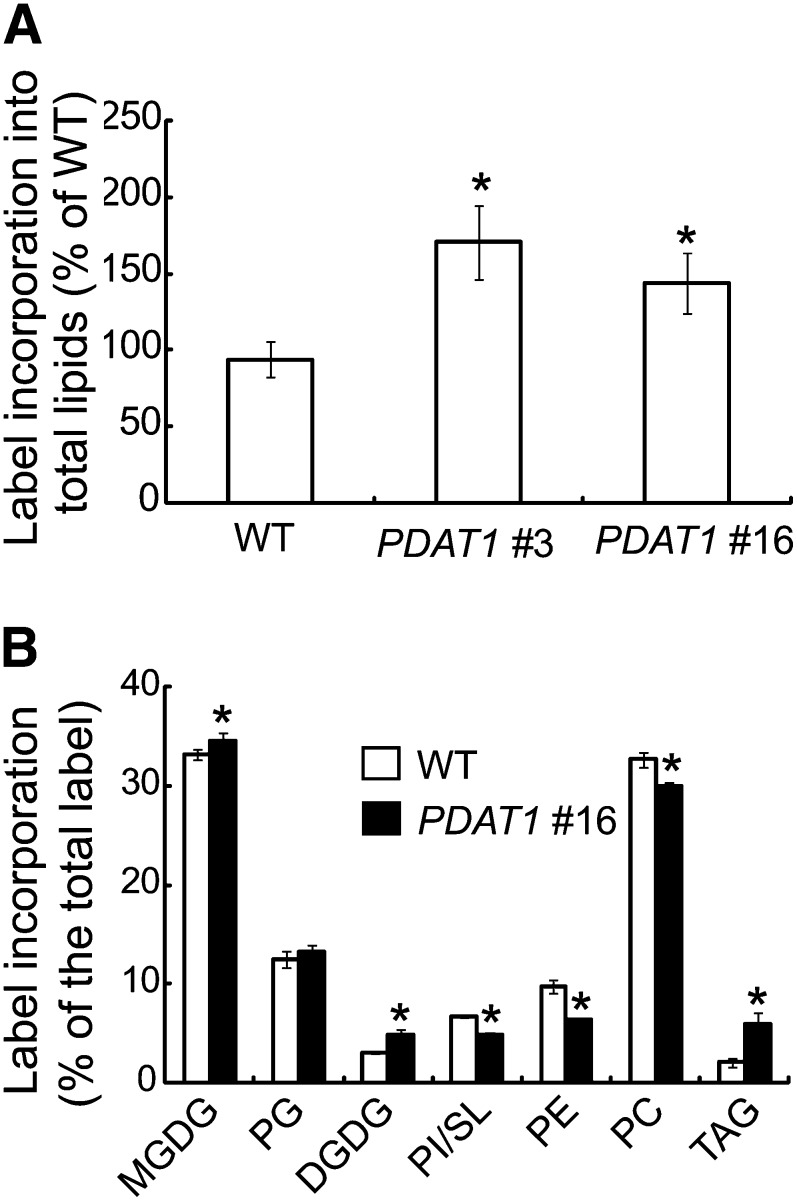
We subsequently obtained three homozygous transgenic lines overexpressing PDAT1 for detailed characterization. On a DW basis, the TAG level in mature rosette leaves of 7-week-old PDAT1 overexpressors grown in soil was 2.6%, a 28-fold increase compared with the wild type (Figure 2A). TAG compositional analysis showed that the predominant acyl chains in TAG derived from the leaves of PDAT1 overexpressors were polyunsaturated FAs with 18:2 and 18:3 accounting for over 75% of the total acyl chains. By comparison, the FAs recovered from wild-type plants by analysis of the TAG band from thin layer chromatography (TLC) plates were largely saturated acyl chains (Figure 2B). In contrast with TAGs isolated from Arabidopsis seedlings ectopically overproducing a Chlamydomonas reinhardtii DGAT type-two enzyme (Sanjaya et al., 2013) or transcription factors involved in storage product accumulation, such as LEAF COTYLEDON2 (LEC2) (Santos Mendoza et al., 2005), TAGs derived from leaves of PDAT1 overexpressors contained very limited amounts of very-long-chain fatty acids (Figure 2B).
Figure 2.
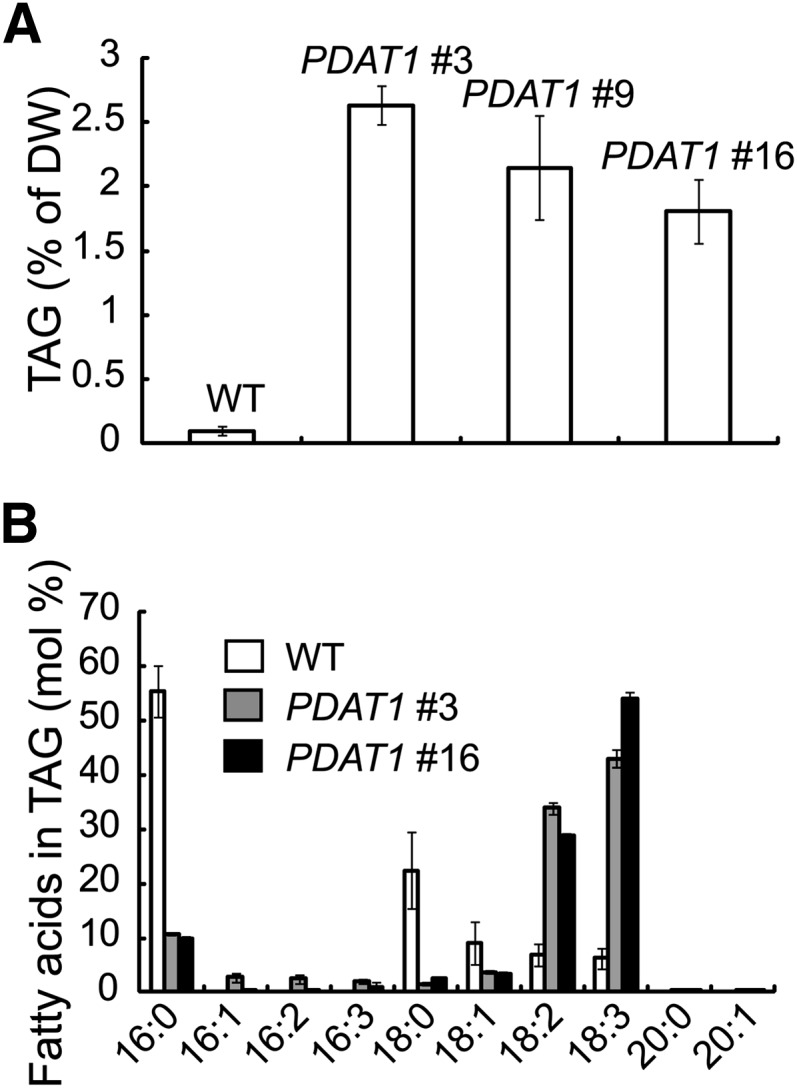
TAG Accumulation in Leaves of Transgenic Plants Overexpressing PDAT1.
(A) Leaf TAG content in 7-week-old wild type (WT) and three independent transgenic lines.
(B) FA composition of TAGs in the wild type and two independent transgenic lines. Values are means and sd of three biological replicates.
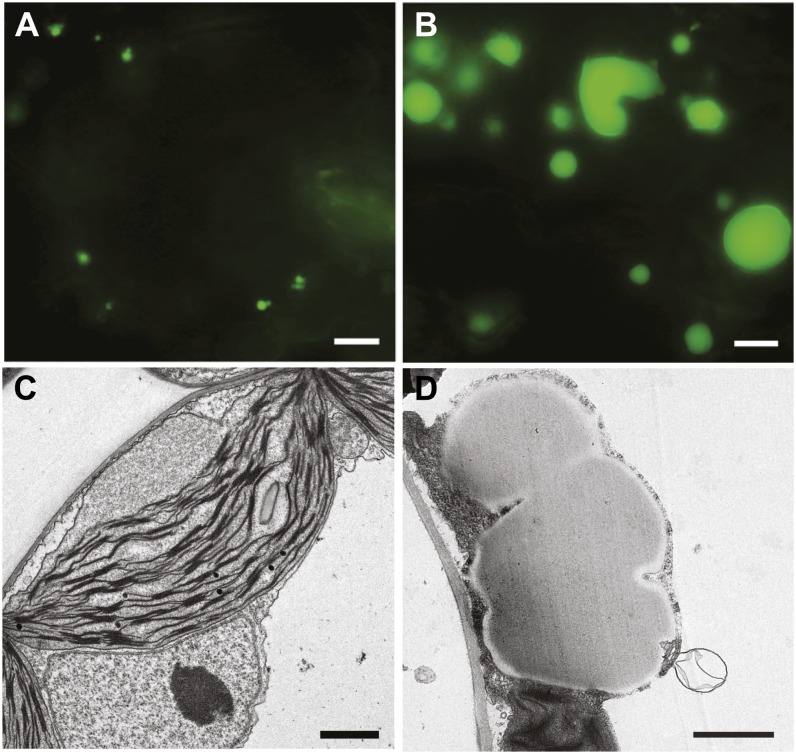
Consistent with the marked increase in TAG content, microscopy examination following Nile Red staining showed that, whereas only a few or no OD-like structures were seen in the leaves of wild-type plants under our growth conditions (Figures 3A and 3C), overexpression of PDAT1 led to OD accumulation in leaves (Figure 3B). Those ODs were often irregular in shape and frequently appeared to coalesce. Ultrastructural examination using transmission electron microscopy (TEM) confirmed that overexpression of PDAT1 led to the formation of large, irregularly shaped ODs in leaves (Figure 3D).
Figure 3.
OD Accumulation in Leaves of Transgenic Plants Overexpressing PDAT1.
(A) and (B) Images of ODs in leaves of 7-week-old wild type (A) and PDAT1-overexpressing line 16 (B) stained with Nile red. Bars = 5 µm.
(C) and (D) TEM images of leaf cells from 7-week-old wild type (C) and PDAT1-overexpressing line 16 (D). Bars = 1 µm.
Ectopic Expression of Oleosin Promotes the Clustering of Small ODs and Boosts Oil Accumulation in PDAT1 Transgenic Plants
Oleosins are known to play a key role in preventing ODs from coalescing in oilseeds (Siloto et al., 2006; Shimada et al., 2008). In addition, in yeast and mammalian model systems, the ectopic expression of OD-associated proteins often leads to increased TAG storage (Brasaemle et al., 2000; Froissard et al., 2009). To test the functional role of oleosins in TAG accumulation in vegetative tissues, OELOSIN1 (OLE1), the most abundant seed OD-specific protein of Arabidopsis (Huang, 1996; Shimada et al., 2008), was C-terminally tagged with green fluorescent protein (GFP), and this fusion gene was expressed in Arabidopsis wild-type plants under the control of the 35S cauliflower mosaic virus promoter. Three independent transgenic lines exhibiting high levels of green fluorescence signal in young seedlings were obtained. Analyzing leaf lipid extracts from 3-week-old transgenic plants revealed an up to sevenfold increase in TAG content compared with the wild type (see Supplemental Figure 2B online). TAGs isolated from OLE1 overexpressors were mainly composed of 16:0, 18:2, and 18:3 FAs, whereas very-long-chain fatty acids, which typically accumulate in Arabidopsis seeds, were present at very low levels (see Supplemental Figure 2C online), suggesting that TAG accumulation in 3-week-old seedlings is not a result of reduced hydrolysis of seed oil due to the ectopic overexpression of OLE1.
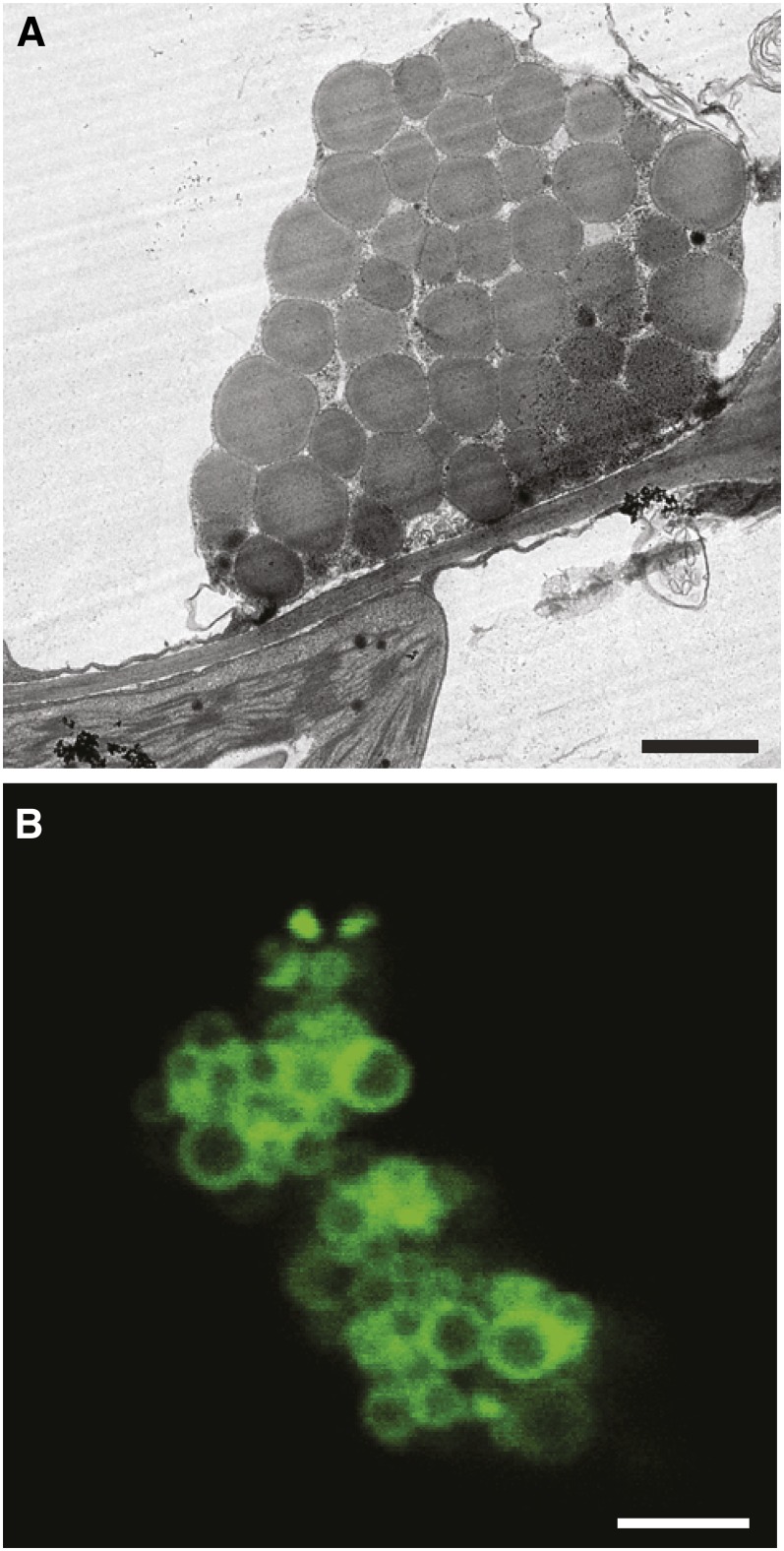
TEM analysis showed that ectopic expression of OLE1 induced the formation of clusters of small spherical ODs (Figure 4A), unlike the large aberrant ODs observed in transgenic plants overexpressing PDAT1 (Figure 3). Using confocal microscopy, we found that the OLE1-GFP fusion protein was exclusively associated with OD clusters and appeared as a ring-like pattern surrounding the perimeters of ODs (Figure 4B).
Figure 4.
Ectopic Expression of OLE1 Promotes the Clustering of Small ODs.
(A) TEM imaging of ODs in leaves of the OLE1-GFP–overexpressing line 1. Bar = 1 µm.
(B) Confocal microscopy of OD clusters in leaves of the OLE1-GFP–overexpressing line 1. Bar = 2 µm.
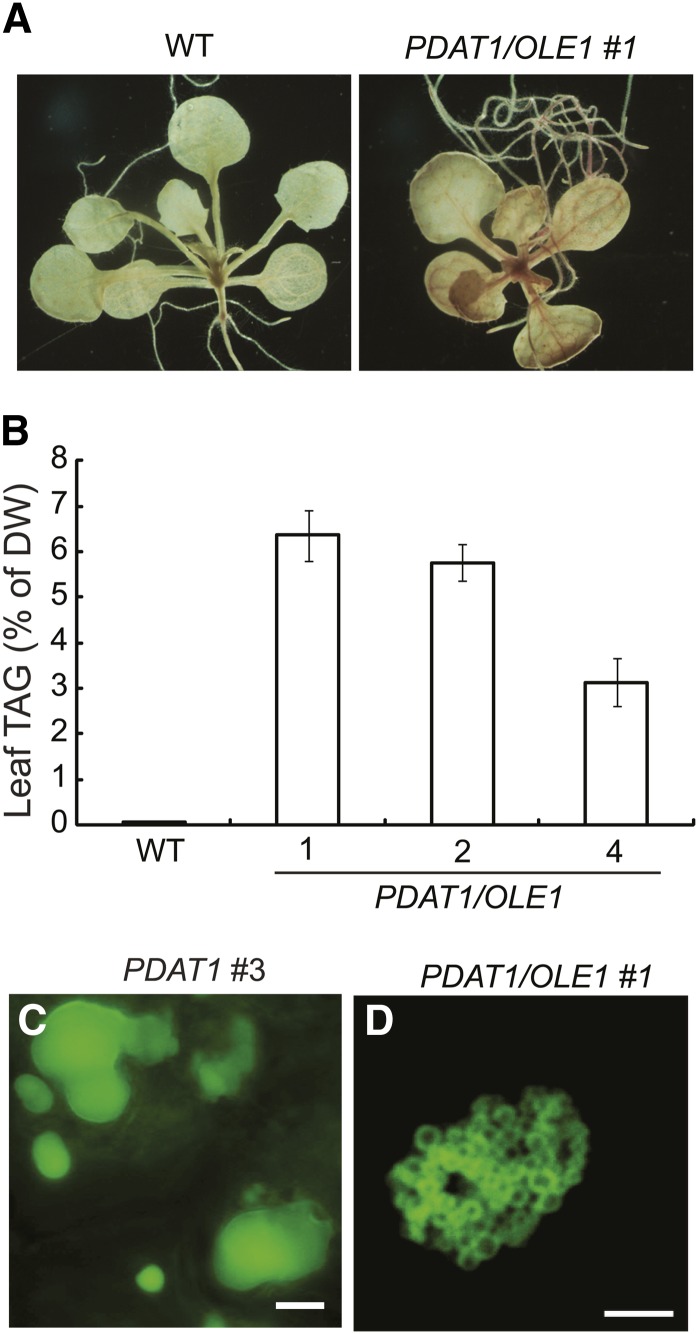
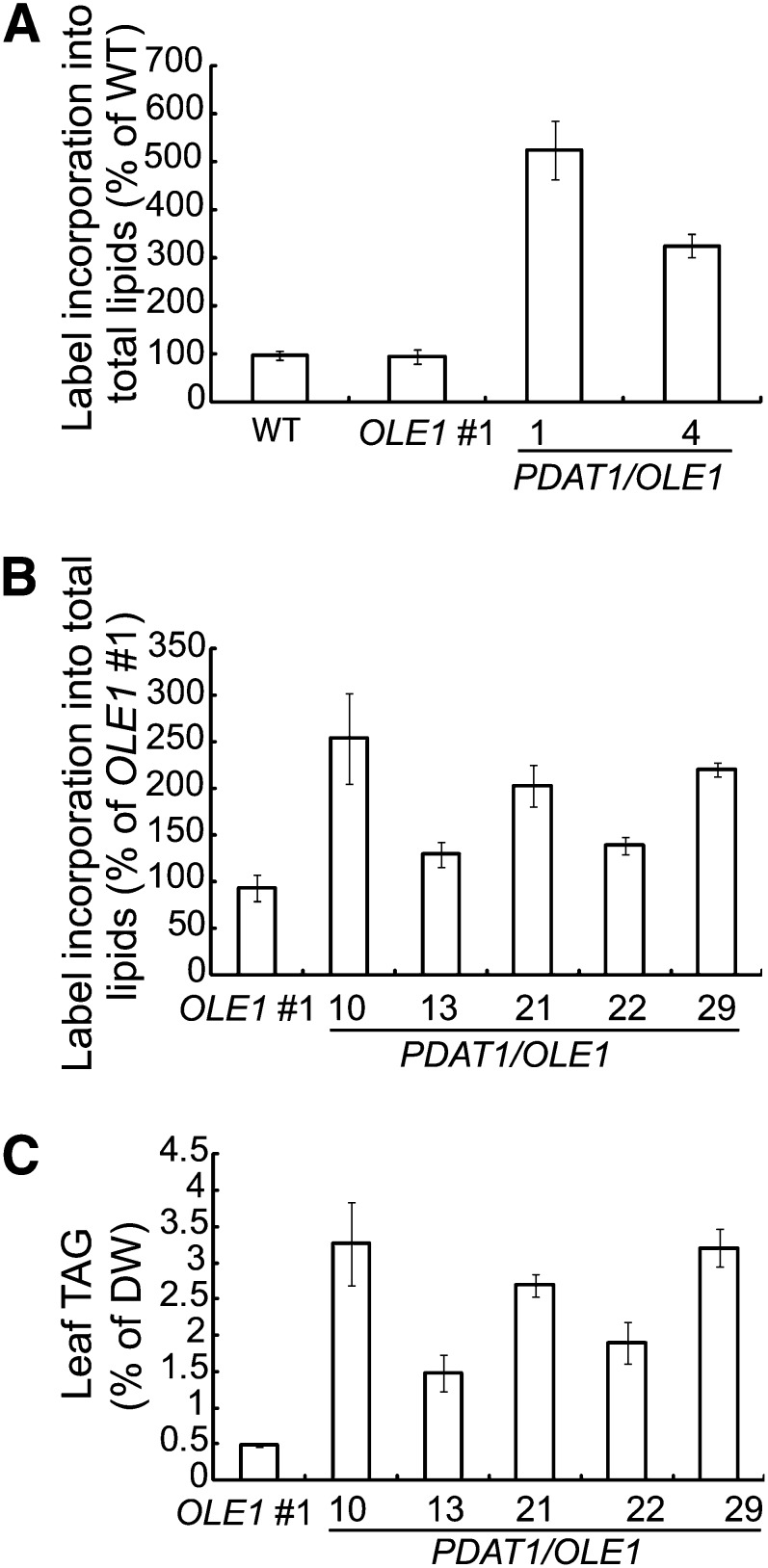
We next transformed the homozygous OLE1 overexpressor line 1 with the PDAT1 construct. Three independent PDAT1/OLE1 double transgenic lines were propagated to the T3 generation and used for detailed analysis. Using Fat Red 7B, a nonfluorescent dye that specifically stains neutral lipids (Brundrett et al., 1991), we found that the double transgenic seedlings were stained red in both roots and leaves (Figure 5A), whereas the wild-type seedlings did not stain. On soil, the PDAT1/OLE1 double transgenic lines displayed no obvious growth or developmental abnormalities (see Supplemental Figure 3A online). Quantification of TAG levels in the rosette leaves of 7-week-old soil-grown double transgenic plants showed an up to 74-fold increase compared with the wild type. On a DW basis, the TAG content in wild-type leaves was 0.05%, while in the three PDAT1/OLE1 double transgenic lines, it was between 5.7 and 6.4% (Figure 5B). There was also an up to twofold increase in the total leaf FA content in 7-week-old transgenic plants coexpressing PDAT1 and OLE1 compared with the wild type (see Supplemental Figure 3B online). In addition, TAG accumulated up to 1.5% per DW in the stems of the double transgenic plants, a 30-fold increase compared with the wild type (see Supplemental Figure 3C online). Microscopy analysis showed that, unlike large and often irregularly shaped ODs in plants carrying the PDAT1 transgene only (Figure 5C), the double transgenic plants contained clusters of spherical ODs in leaves (Figure 5D), similar to those seen in plants ectopically expressing OLE1 alone (Figure 4), thus confirming the role of OLE1 in promoting the clustering of ODs.
Figure 5.
TAG Accumulation in Transgenic Lines Coexpressing PDAT1 and OLE1.
(A) Fat Red 7B staining of 3-week-old seedlings of the wild type (WT), PDAT1 and OLE1-GFP double transgenic line 1 (PDAT1/OLE1 #1).
(B) TAG content in leaves of 7-week-old plants of the wild type and three transgenic lines coexpressing PDAT1 and OLE1-GFP (PDAT1/OLE1). Values are means and sd of three biological replicates.
(C) Images of ODs in leaves of 7-week-old PDAT1-overexpressing line 3 (PDAT1 #3) stained with Nile red. Bar = 5 µm.
(D) Confocal imaging of ODs in leaves of 7-week-old soil-grown transgenic line 1 coexpressing PDAT1 and OLE1-GFP (PDAT1/OLE1 #1). Bar = 5 µm.
The FA Composition of Membrane Lipids Is Altered in PDAT1 Transgenic Plants
To investigate the potential impact of PDAT1 overexpression on membrane lipids, we first surveyed the FA profiles of total polar lipids in leaves of three independent transgenic lines overexpressing PDAT1. The relative proportions of 16:3, 18:1, and 18:2 increased, with a concomitant decrease in the proportion of 18:3 (see Supplemental Figure 4A online). We next separated individual polar lipids by TLC. Analysis by gas chromatography of the FAMEs revealed no discernible differences in levels of the individual classes of major polar lipids between PDAT1 overexpressor 16 and the wild type (see Supplemental Figure 4B online). However, there was a substantial decrease in 18:3 with a corresponding increase in 18:1 and 18:2 in both extraplastidic (PE and PC) and thylakoid (digalactosyldiacylglycerol [DGDG] and monogalactosyldiacylglycerol [MGDG]) lipids (see Supplemental Figures 4C to 4F online). The marked decrease in 18:3 in PC and PE may reflect the substrate preference of PDAT1 as demonstrated by in vitro studies by Ståhl et al. (2004). This is supported by the particular enrichment of 18:3 in TAG accumulated in leaves of PDAT1 transgenic plants (Figure 2B). On the other hand, the decrease in the proportion of 18:3 in galactolipids, particularly in DGDG, may reflect the fact that PC is the precursor of galactolipids synthesized by the eukaryotic pathway. In addition to changes in C18 FAs, the levels of C16 acyl chains tended to decrease in PC but increase in DGDG and MGDG when compared with the wild type (see Supplemental Figures 4C to 4F online).
Overexpression of PDAT1 Alters the Positional Distribution of FAs in Galactolipids
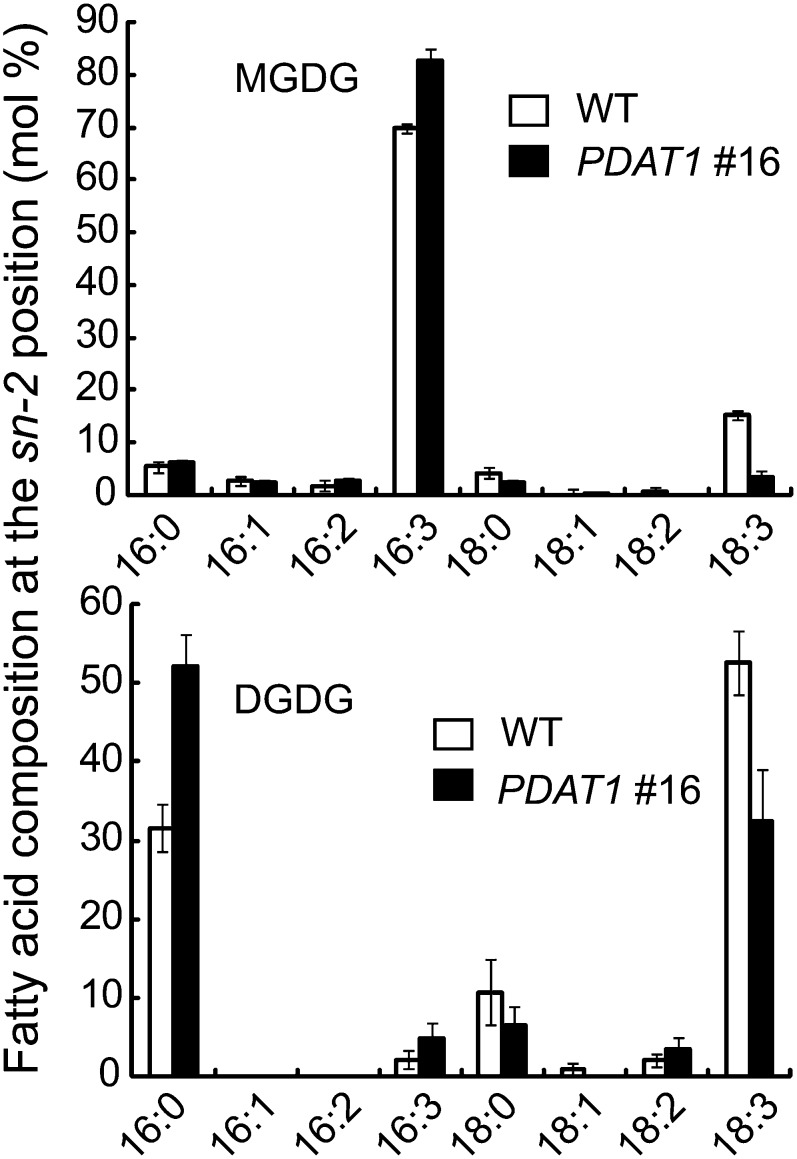
The increase in C16 FA content in galactolipids may suggest that the prokaryotic pathway of thylakoid lipid synthesis is enhanced in PDAT1 overexpressors. To test this possibility, the positional distribution of FAs in galactolipids was analyzed following Rhizopus lipase digestion. As shown in Figure 6, the relative proportion of C16 FAs at the sn-2 position of MGDG increased from 80% in the wild type to 94% in PDAT1 transgenic line 16. In the case of DGDG, the increase was from 34 to 57%. The finding that sn-2 C16 FA content increased to a greater degree in DGDG than in MGDG reflects the fact that the prokaryotic pathway makes a much smaller contribution to the synthesis of DGDG than to MGDG in the leaves of wild-type plants (Browse et al., 1986); consequently, DGDG is more affected than MGDG when prokaryotic thylakoid lipid synthesis is enhanced.
Figure 6.
FA Composition Exclusively at the sn-2 Position of the Glycerol Backbone of Galactolipids Isolated from Leaves of 7-Week-Old Wild-Type (WT) and PDAT1-Overexpressing Line 16 (PDAT1 #16).
Values are means and sd of three biological replicates.
Rate of FA Synthesis Is Enhanced in PDAT1 Transgenic Plants
The accumulation of large amounts of TAG without a concomitant decrease in membrane lipid content in PDAT1 transgenic plants suggests an increase in fatty acid synthesis (FAS). To test this possibility, we conducted 14C-acetate feeding experiments with leaf strips from expanding leaves for a period of 1 h, which was within the linear range of label incorporation (see Supplemental Figure 5 online). Because acetate incorporation into FAs can be influenced by levels of endogenous metabolites (Cronan et al., 1975; Nunn et al., 1977), the labeling experiments were performed in the presence of 1 mM unlabeled acetate in an attempt to eliminate potential variations in endogenous substrate pools between the transgenics and the wild type. We detected 72 and 44% increases in radiolabeled lipids in the PDAT1 overexpressor 9 and 16, respectively, compared with the wild type (Figure 7A), suggesting an increase in the rate of FAS due to PDAT1 overexpression. Analysis of label distribution showed that overexpression of PDAT1 significantly increased the label incorporation into galactolipids, phosphatidylglycerol (PG), and TAG, whereas the proportions of label in PC, PE, and sulfoquinovosyldiacylglycerol (SL)/phosphatidylinositol (PI) decreased (Figure 7B). These results are consistent with an increase in acyl fluxes toward galactolipids and PDAT1-mediated TAG synthesis in response to PDAT1 overproduction.
Figure 7.
Overexpression of PDAT1 Enhances the Rate of FAS in Leaves.
(A) Initial rates of FAS in growing leaves of 7-week-old wild-type (WT), PDAT1-overexpressing line 3 (PDAT1 #3), and PDAT1-overexpressing line 16 (PDAT1 #16) measured by 14C-acetate labeling.
(B) The distribution of label into TAG and polar lipids after labeling of detached leaves with 14C-acetate for 60 min.
Values in (A) and (B) are means and sd of three biological replicates. Asterisks indicate statistically significant differences from the wild type based on Student’s t test (P < 0.05).
Rate of FAS Strongly Correlates with Leaf TAG Content in Transgenic Plants
The leaf TAG levels in double transgenic plants coexpressing PDAT1 and OLE1 (Figure 5B) were much higher than in transgenic plants carrying PDAT1 alone (Figure 2A). To begin to dissect the biochemical basis for the increased TAG accumulation in double transgenic lines, growing leaves of PDAT1 single and PDAT1/OLE1 double transgenic plants were labeled with 14C-acetate. Compared with the transgenic plants overexpressing PDAT1 alone (Figure 7A), the initial rate of acetate incorporation into total lipids was much higher for the PDAT1/OLE1 double transgenic lines (Figure 8A). In addition, in five additional independent PDAT1/OLE1 double transgenic lines tested, there was a strong correlation between the rate of FAS (Figure 8B) and TAG content (Figure 8C). These results suggest that increased TAG accumulation in leaves of PDAT1/OLE1 double transgenic lines may be due to enhanced FAS. On the other hand, no apparent difference in acetate incorporation into total lipids was found between the wild type and transgenic plants overexpressing the OLE1 single transgene (Figure 8A), suggesting that the increased TAG accumulation in transgenic plants carrying OLE1 may be attributed to decreased TAG turnover, likely because of the shielding of ODs by OLE1 from the access of cytosolic lipases, in a manner similar to that observed for mammalian OD-associated protein perilipins (Brasaemle et al., 2000).
Figure 8.
TAG Accumulation Positively Correlates with the Rate of FAS.
Initial rates of FAS in growing leaves of 5-week-old wild-type (WT), OLE1-GFP–overexpressing line 1 (OLE1 #1), and PDAT1 and OLE1-GFP double transgenic line 1 (PDAT1/OLE1 #1) and 4 (PDAT1/OLE1 #4) measured by 14C-acetate labeling is shown in (A). Initial rates of FAS in growing leaves of 5-week-old OLE1 #1 and five double transgenic lines coexpressing PDAT1 and OLE1-GFP (PDAT1/OLE1) measured by 14C-acetate labeling is presented in (B). TAG content in leaves of 7-week-old OLE1 #1 and five double transgenic lines coexpressing PDAT1 and OLE1-GFP (PDAT1/OLE1) is shown in (C). Values are means and sd of three biological replicates.
Disruption of the Eukaryotic Thylakoid Lipid Pathway Enhances TAG Accumulation in Double Transgenic Plants Coexpressing PDAT1 and OLE1
In Arabidopsis, the prokaryotic and eukaryotic glycerolipid pathway each consumes about half of acyl chains synthesized de novo in the plastid (Browse et al., 1986). Disruption of either glycerolipid pathway may therefore be expected to increase acyl chains for TAG synthesis. To test this possibility, the tgd1-1 mutant, which is defective in the eukaryotic pathway of thylakoid lipid synthesis (Xu et al., 2003), was crossed with the PDAT1/OLE1 double transgenic line 1. Analysis of leaf lipid extracts from mature leaves of 7-week-old soil-grown plants showed that, on a DW basis, TAG content in the PDAT1/OLE1 double transgenic line in the tgd1-1 background was increased to 8.6% and total lipid content to 16.2%, a 97-fold and fourfold increase, respectively, compared with tgd1-1 (Figure 9A). As expected, coexpression of PDAT1 with OLE1 in tgd1-1 induced the formation of OD clusters in leaves (Figure 9B). In addition to leaves, TAG also accumulated in stems of tgd1-1/PDAT1/OLE1 transgenic plants to 3.0% ± 0.18% per DW (n = 3, ±sd), a 60-fold increase relative to tgd1-1 (0.05% ± 0.01% per DW, n = 3, ±sd). Remarkably, despite a large increase in TAG and total lipid levels, the growth and development of the double transgenic plants were only slightly affected (see Supplemental Figure 6A online). Furthermore, no apparent difference in total polar lipid content was found between tgd1-1/PDAT1/OLE1 and tgd1-1 (see Supplemental Figure 6B online).
Figure 9.
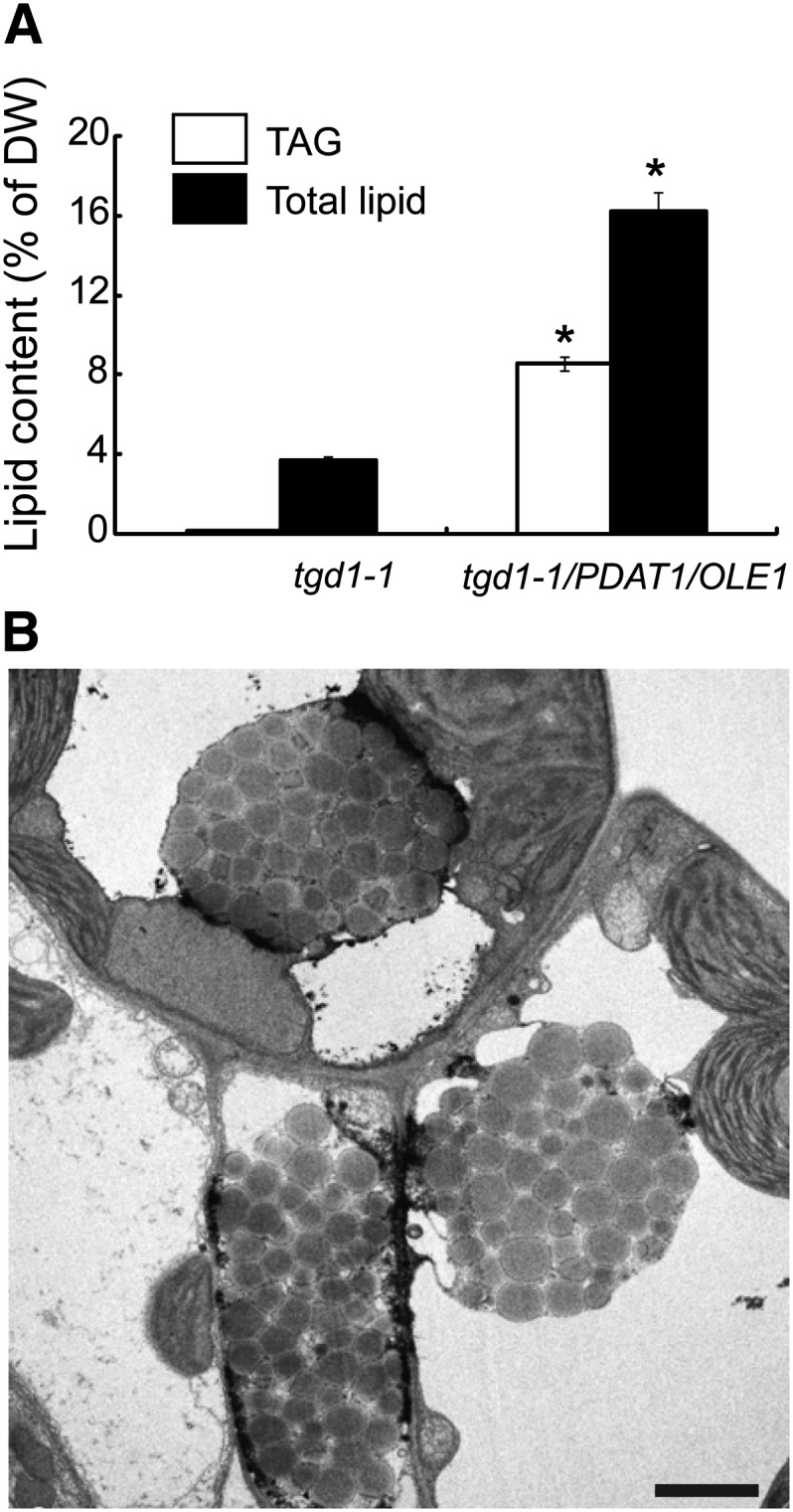
Disruption of the Eukaryotic Thylakoid Lipid Pathway Enhances TAG Accumulation in Transgenic Plants Coexpressing PDAT1 and OLE1.
(A) TAG and total lipid levels in leaves of 7-week-old tgd1-1 and transgenic plants coexpressing PDAT1 and OLE1-GFP in the tgd1-1 background (tgd1-1/PDAT1/OLE1). Values are means and sd of three biological replicates. Asterisks indicate statistically significant differences from tgd1-1 based on Student’s t test (P < 0.05).
(B) TEM imaging of ODs in leaves of 7-week-old tgd1-1/PDAT1/OLE1 plants. Bar = 2 µm.
Disruption of PDAT1 Partially Alleviates the Phenotype of the act1 Mutant
We next constructed a double mutant between pdat1-2 and act1 defective in the prokaryotic thylakoid lipid pathway. When grown in soil, the act1 pdat1-2 double mutant seedlings were visibly darker in color compared with the act1 mutant (Figure 10A). Consistent with the dark-green phenotype, the total chlorophyll content was increased by 9.8%, from 0.90 ± 0.02 in act1 to 0.99 ± 0.04 mg/g fresh weight (FW) (n = 3, ±sd) in the double mutant. Lipid analysis showed that disruption of PDAT1 in act1 resulted in small but significant increases in the levels of thylakoid lipids MGDG and PG (Figure 10B). In addition, the combined amount of SL and PI was slightly increased, whereas the level of PC, PE, and DGDG stayed largely unaltered in the act1 pdat1-2 double mutant relative to act1. The FA composition of all polar lipid classes examined did not show noticeable deviations from those in act1, with the exception of PC, which exhibited a significant increase in relative proportions of 18:2 and 18:3 with a corresponding decrease in 18:1 (Figure 10C). The mutation in act1 has been shown to result in decreases in the relative levels of chloroplast lipids PG, SL, and MGDG and increases in the relative amounts of 18:1 in extrachloroplast lipids, particularly in PC (Kunst et al., 1988), and a slight reduction in the amount of chlorophyll on a FW basis (Kunst et al., 1989). Thus, our lipid data, together with the increased chlorophyll content in act1 pdat1-2 plants, suggest that disruption of PDAT1 partially alleviates the phenotypes of the act1 mutant, presumably as a consequence of the increased diversion of FAS from TAG to eukaryotic thylakoid lipids in the double mutant.
Figure 10.
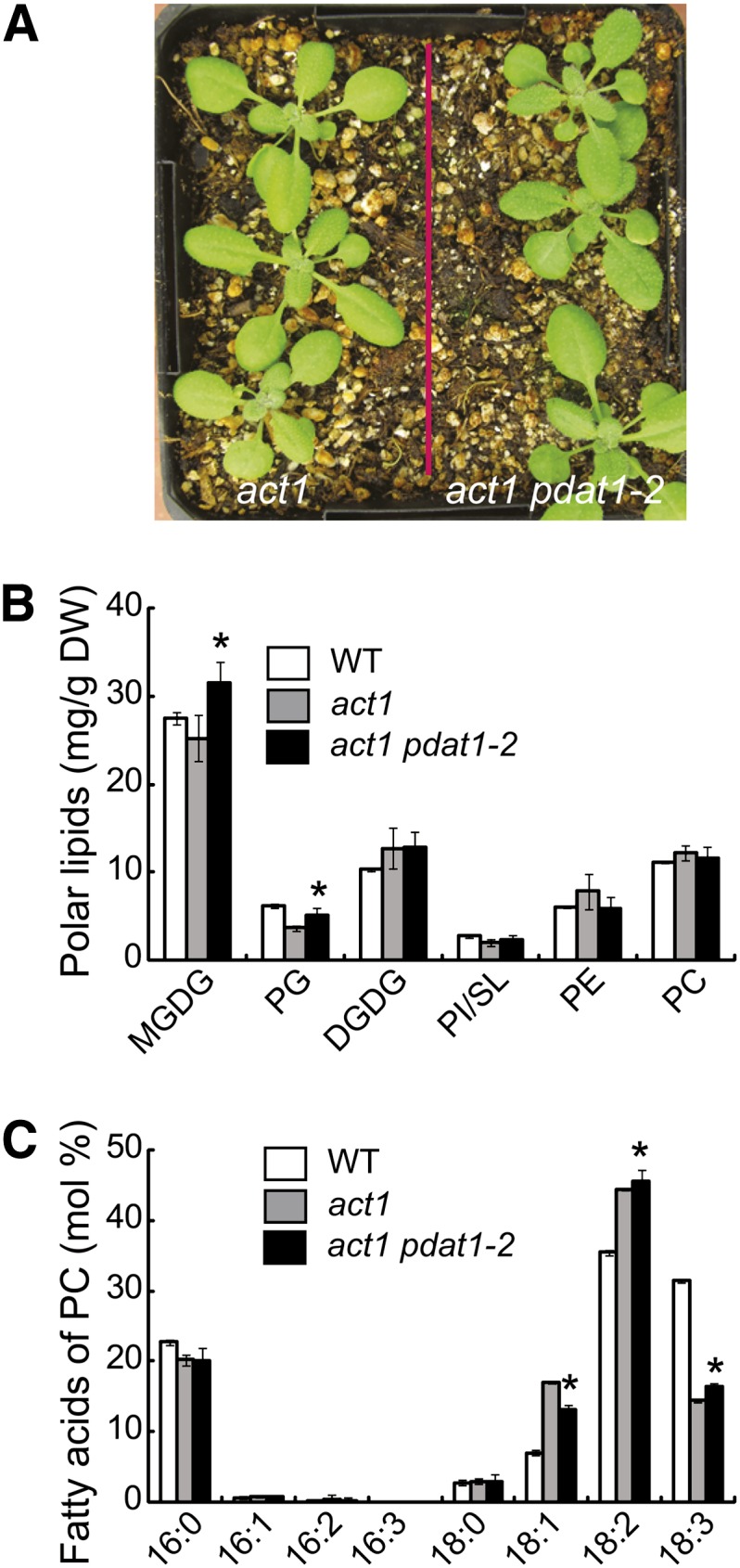
Disruption of PDAT1 Partially Complements the Phenotype of the act1 Mutant.
Images of 4-week-old act1 and act1 pdat1-2 plants are shown in (A). Polar lipid content in leaves of act1 and act1 pdat1-2 plants is shown in (B). FA composition of PC isolated from leaves of 4-week-old wild-type (WT) act1 and act1 pdat1-2 plants grown on agar plates is shown in (C). Values are means and sd of three biological replicates. Asterisks indicate a statistically significant difference from act1 based on Student’s t test (P < 0.05).
[See online article for color version of this figure.]
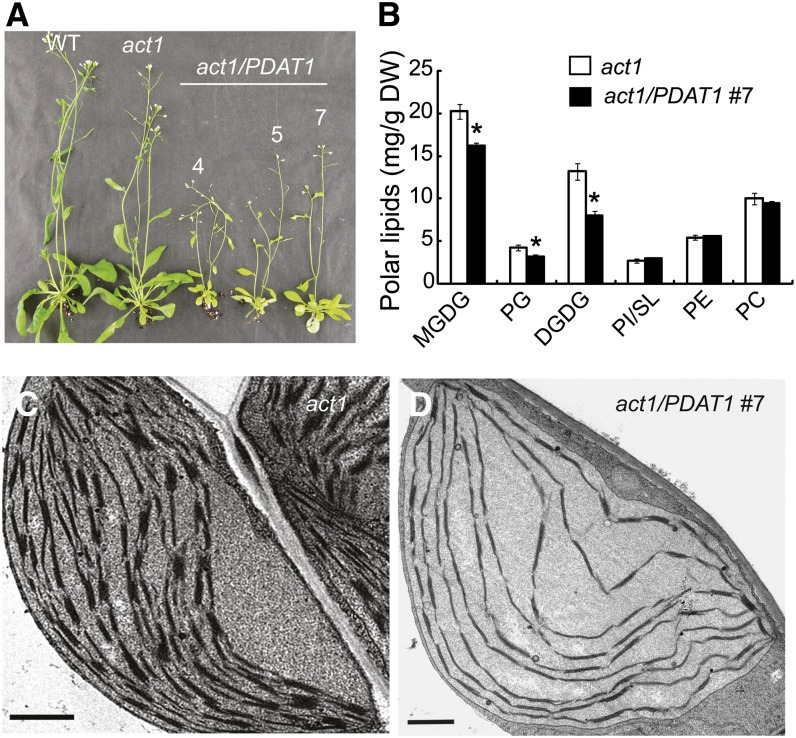
Overexpression of PDAT1 in act1 Enhances TAG Accumulation at the Expense of Thylakoid Lipids
Next, the PDAT1 construct was introduced into act1 by Agrobacterium tumefaciens–mediated transformation. When grown in soil, transgenic plants overexpressing PDAT1 in act1 (act1/PDAT1) were frequently found to be pale in color and stunted in growth (Figure 11A). Consistent with the pale phenotype, the amount of the major thylakoid lipids MGDG, DGDG, and PG was reduced by 25, 46, and 31%, respectively, in act1/PDAT1 line 7 compared with act1, whereas the levels of PC, PE, and SL/PI remained largely unchanged (Figure 11B). On a FW basis, the sum of MGDG, DGDG, and PG decreased from 2826 in act1 to 2065 µg/g FW in act1/PDAT1 line 7, resulting in a net loss of 761 µg/g FW, while the amount of TAG increased from 57.3 ± 9.8 to 760.9 ± 164.6 µg/g FW (n = 3, ±sd), a net gain of 704 µg/g FW. These results suggest that TAG accumulation in act1 PDAT1 overexpressing lines is largely accounted for by the increased diversion of acyl groups from thylakoid membrane lipid synthesis to TAG. Consistent with this, the total leaf FA content was comparable in act1/PDAT1 line 7 (5.61% ± 0.61% per DW, n = 3, ±sd) and act1 (5.91% ± 0.84% per DW, n = 4, ±sd).
Figure 11.
Overexpression of PDAT1 in act1 Promotes TAG Accumulation at the Expense of Thylakoid Lipids.
(A) Images of 6-week-old wild type (WT), act1, and three PDAT1-overexpressing lines in the act1 background (act1/PDAT1).
(B) Polar lipid levels in leaves of act1 and the act1/PDAT1 #7. Values are means and sd of three to four biological replicates. Asterisks indicate statistically significant differences from act1 based on Student’s t test (P < 0.05).
(C) and (D) TEM analysis of chloroplasts in leaves of act1 (C) and the act1/PDAT1 #7 (D). Bars = 1 µm.
[See online article for color version of this figure.]
To further investigate the nature of the pale-green phenotype in act1 overexpressing PDAT1, microscopy examination of leaf cross sections was performed. Compared with act1, the transgenic act1 plants displayed increased leaf thickness and increased cell size, but decreased cell number (see Supplemental Figure 7 online). The chloroplasts of the transgenic line appeared to be larger. The number of chloroplasts per cell cross section was reduced from 6.63 ± 0.05 in act1 to 3.33 ± 0.05 (n = 4, ± SD) in the act1/PDAT1 line 7. At the ultrastructural level, the amount of thylakoid membranes and thylakoid membrane stacking per chloroplast in mature leaves of act1 plants overexpressing PDAT1 were greatly reduced when compared with chloroplasts from act1 leaves at the same developmental stage (Figures 11C and 11D).
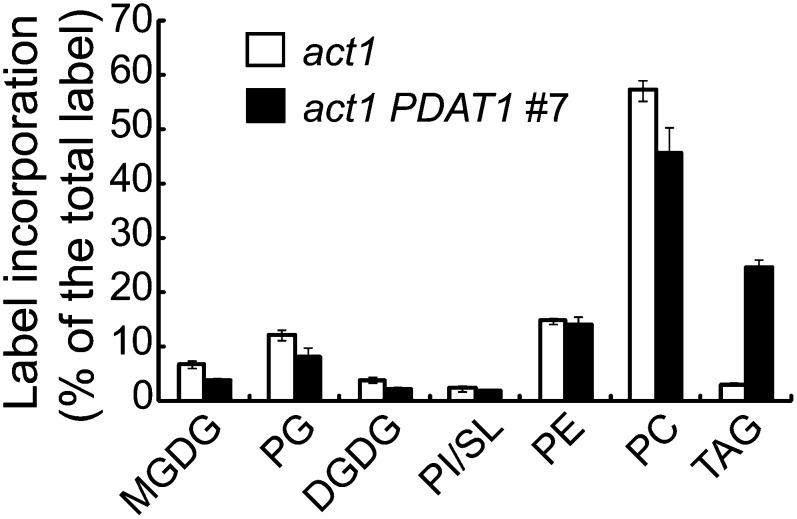
To gain more information on the metabolic changes leading to TAG accumulation due to PDAT1 overproduction in act1, we performed 14C-acetate pulse-chase labeling experiments using detached leaves. During the 1-h pulse, the rates of 14C-acetate incorporation were approximately threefold higher in the act1/PDAT1 line 7 (232.0 ± 18.9 disintegrations per minute/mg FW/min, n = 3, ±sd) compared with act1 (636.6 ± 127.7 disintegrations per minute/mg FW/min, n = 3, ±sd), suggesting an increase in FAS due to PDAT1 overexpression in the act1 background. Because the total leaf lipid content remained largely unchanged in act1/PDAT1 line 7 relative to act1, the rate of FA degradation must be increased concurrently such that a constant lipid level is maintained. Indeed, the average label decay rate was increased by threefold in act1/PDAT1 line 7 (22% per day) compared with act1 (7% per day) (see Supplemental Figure 8A online). Analysis of label distribution showed that PC was the most radioactive lipid immediately following the pulse, followed by PG and PE. Galactolipids MGDG and DGDG, despite accounting for over 70% of the total membrane lipid mass, contained <10% of total initial label in act1 (Figure 12). Overexpression of PDAT1 led to a 19% drop in initial label in PC, no change in PE, but a 25% decline in SL/PI, 42% in DGDG, 35% in PG, and 44% in MGDG, whereas TAG label increased from 3% in act1 to 25% in the act1/PDAT1 line 7, thus representing a nearly fourfold increase in the initial label partitioning into TAG (Figure 12). During the chase, the PC label declined, whereas labeled MGDG and DGDG increased in both act1 and act1 transgenic plants, reflecting a well-documented precursor-product relationship between PC and galactolipids, but the extent of increases in label in MGDG and DGDG was smaller in act1 transgenic plants (see Supplemental Figure 8C online) than in act1 (see Supplemental Figure 8B online). In both act1 and the act1 PDAT1 overexpressor, the label in TAG slightly increased during the initial 24 h of chase. This was followed by substantial decreases in labeled TAG during the remainder of the chase, reflecting turnover of TAG in both act1 and act1 transgenic plants.
Figure 12.
The Distribution of Radioactivity in Polar Lipids and TAG after Labeling of Detached Leaves with 14C-Acetate for 60 min in act1 and act1 PDAT1-Overexpressing Line #7 (act1/PDAT1 #7).
Values are means and sd of three biological replicates.
DISCUSSION
PDAT Is Functionally Conserved among Yeast, Microalgae, and Plants
In plants as well as in microalgae, mammals, and fungi, the terminal and committed step in the pathway of TAG biosynthesis is catalyzed by multiple DAG acyltransferases encoded by distinct gene families. Although the basic enzymes involved in the DAG esterification reaction have been extensively characterized at the biochemical and molecular level in several model systems, fundamental questions remain regarding the roles of the individual DAG acyltransferases and the functional significance of highly redundant activities. At least three classes of DAG acyltransferases consisting of acyl-CoA–dependent DGAT1 and DGAT2 and acyl-CoA–independent PDAT, contribute to TAG synthesis in yeasts (Kohlwein, 2010), microalgae (Liu and Benning, 2013), and plants (Chapman and Ohlrogge, 2012). Studies in both yeast (Oelkers et al., 2000, 2002) and the green alga C. reinhardtii (Yoon et al., 2012) have indicated that PDAT plays a major role in TAG synthesis during phases of active cell growth and division, while DGATs appear to be more important in cells entering the stationary phase of growth (Oelkers et al., 2002; Sandager et al., 2002). In Arabidopsis, most evidence suggests that DGAT1 is a major contributor to TAG synthesis in developing seeds (Katavic et al., 1995; Routaboul et al., 1999; Jako et al., 2001) and in leaves during senescence (Kaup et al., 2002; Slocombe et al., 2009). On the other hand, although PDAT1 is known to have an overlapping role in TAG synthesis in seeds and pollen (Zhang et al., 2009), neither disruption (Ståhl et al., 2004; Mhaske et al., 2005) nor overexpression of PDAT1 (Ståhl et al., 2004) appears to affect lipid content and FA composition in seeds and in young seedlings. Through extensive analyses of PDAT1 knockout mutants and overexpressing lines, particularly in the act1 mutant background, this work provides insight into the biological functions of PDAT1. We show that PDAT1 plays an important role in TAG synthesis in leaves; overexpression of PDAT1, but not DGAT1, increased leaf TAG content, whereas disruption of PDAT1 substantially decreased TAG levels with the most pronounced decrease occurring in rapidly growing leaves and the least in senescing leaves. Collectively, these results point to evolutionary conservation of PDAT functions associated with rapid cell growth and membrane proliferation in yeast, microalgae, and plants.
According to current knowledge about glycerolipid metabolism in plants, most of the acyl chains exported outside the plastid are first incorporated into PC, the substrate of PDAT, through acyl editing in growing leaf cells (Bates et al., 2007, 2009; Tjellström et al., 2012). Because lipid metabolism in growing leaf cells is directed primarily toward membrane lipid synthesis, the acyl chains released as a result of acyl editing of PC are primarily channeled into G3P acylation reactions to generate DAG for de novo PC synthesis (Bates et al., 2007) and therefore may be limiting for DGAT1-mediated TAG synthesis. As leaf tissues age, the rate of FAS declines (Bao et al., 2000; Hellgren and Sandelius, 2001) and fewer nascent FAs are fluxed through PC and more are used directly in the chloroplast to sustain thylakoid lipid turnover (Hellgren and Sandelius, 2001). This may explain why the PDAT1-mediated acyl-CoA–independent route is more involved in TAG synthesis in growing but not in senescing leaves of Arabidopsis than the DGAT1-catalyzed acyl-CoA–dependent reaction.
PDAT Plays a Role in Membrane Lipid Turnover
In vitro biochemical assays showed that Arabidopsis PDAT1 displays the highest activity toward oxygenated acyl groups, such as hydroxyl acyl chains (Ståhl et al., 2004), much as does the enzyme from castor (Ricinus communis) oil seeds that accumulates high levels of hydroxyl FAs (Dahlqvist et al., 2000). It has thus been hypothesized that Arabidopsis PDAT1, like PDAT in castor, may play a role in cell membrane repair by removing oxygenated FAs from membrane phospholipids (Ståhl et al., 2004). However, direct evidence in support of this hypothesis is still lacking.
Recent studies in other model organisms have indicated that membrane lipid and TAG metabolism are tightly linked processes (Rajakumari et al., 2010; Horvath et al., 2011; Yoon et al., 2012). In yeast acyl-CoA–deficient mutants, disruption of PDAT blocked phospholipid deacylation, resulting in an increase in levels of PC and PE (Mora et al., 2012). Likewise, knockdown of PDAT in C. reinhardtii has been shown to alter the molecular species composition of major thylakoid membrane lipids (Yoon et al., 2012). In this study, we showed that disruption of PDAT1 in the act1 mutant increased the proportions of polyunsaturated FAs in PC with a corresponding decrease in 18:1, whereas the reverse trend was found for PDAT1-overexpressing plants. Because PC is the site of FA desaturation, the opposite effects of PDAT1 knockout and overproduction on levels of FA unsaturation are in line with PDAT1’s function in the removal of acyl groups from PC. In addition to changes in the FA composition of PC, knockout of PDAT1 increased, whereas overexpression of this gene decreased levels of thylakoid lipid content in the act1 mutant. These results provide direct in vivo evidence that Arabidopsis PDAT1, like its homologs in yeast and C. reinhardtii, is also involved in the turnover of membrane lipids in leaves.
In the pdat1-2 single mutant, no appreciable change in membrane lipid content and FA composition was observed, despite the marked reduction in TAG content compared with the wild type. The lack of detectable phenotype in pdat1-2 may reflect the operation of lipid homeostatic mechanisms that fully compensate for the changes in membrane compositions resulting from the loss of PDAT1 function in the wild type. In this context, in PDAT1-overexpressing lines in the wild-type background, the increased diversion of acyl groups from membrane lipid synthesis to TAG was entirely compensated for by enhanced FAS and increased thylakoid lipid assembly via the prokaryotic pathway, thus resulting in little net change in the membrane lipid content and composition. Although the factors responsible for such metabolic adjustments are unknown, it is tempting to speculate that biochemical feedback regulation of FAS plays a key role. In this scenario, PDAT1-mediated TAG accumulation consumes, in the cytosol, acyl moieties destined for eukaryotic thylakoid lipid synthesis. This could (1) provide a driving force for FA export outside the chloroplast through vectorial acylation and (2) increase the demand for FAs needed for the prokaryotic thylakoid lipid synthesis. Both of these metabolic responses could lead to a depletion of 18:1-acyl carrier protein in the plastid, thereby releasing its feedback inhibition on the plastidic acetyl-CoA carboxylase (Andre et al., 2012). This could in turn lead to an increase in the rate of FAS and an increase in flow of FAs toward the prokaryotic pathway to compensate for the loss of lipid precursors normally shuttled from the ER to the chloroplast for eukaryotic thylakoid lipid synthesis.
In contrast with PDAT1 overexpressors in the wild-type background, overexpression of PDAT1 in act1 resulted in substantial reductions in levels of thylakoid lipids, leading to defects in chloroplast biogenesis, despite marked increases in the rates of FAS. These results indicate that the prokaryotic pathway plays a critical role in maintaining membrane lipid homeostasis when the eukaryotic pathway is severely compromised. In this regard, we showed previously that the genetic defects in the eukaryotic pathway of thylakoid lipid synthesis in tgd1 mutants can be almost fully compensated for by the augmented prokaryotic pathway without major growth and developmental consequences (Xu et al., 2005).
Biotechnological Implications for PDAT
FAs are the predominant component of TAG. Therefore, increasing FAS is a prerequisite for attaining high oil yield in rational genetic engineering studies aimed at enhancing oil accumulation in vegetative tissues of plants. To date, much attention has been focused on overexpression of transcription factors involved in seed storage product accumulation and seed maturation, such as WRINKLED1 (WRI1) and LEC2 (Slocombe et al., 2009; Andrianov et al., 2010; Kelly et al., 2013; Vanhercke et al., 2013). Aside from the growth and developmental defects associated with the ectopic overexpression of such seed-specific master regulators (Stone et al., 2001; Cernac and Benning, 2004; Baud et al., 2007), recent genetic evidence suggests that the WRI class of transcription factors are not involved in the regulation of FAS in vegetative tissues of Arabidopsis (To et al., 2012). In addition, it has been reported that WRI1-mediated TAG accumulation is dependent on the addition of soluble sugar in growth media (Cernac and Benning, 2004), and ectopic overexpression of WRI1 alone only results in marginal increases in TAG accumulation in leaves of soil-grown plants (Sanjaya et al., 2011). Foliar tissues constitute a major portion of the harvestable biomass of dedicated bioenergy crops such as switchgrass (Panicum virgatum). So far, however, only modest levels of leaf TAG have been achieved (Troncoso-Ponce et al., 2013), with the highest level among genetically modified Arabidopsis plants being 5% per DW found in the sdp1 TAG lipase mutant coexpressing WRI1 and DGAT1 (Kelly et al., 2013). Our study shows that PDAT1 has a dual role in enhancing FAS and directing FAs from membrane lipids to TAG in Arabidopsis leaves. We show that the combined expression of PDAT1 and OLE1 increases leaf TAG to 6.4% per DW in the wild type and 8.6% per DW in tgd1 without major negative growth consequences. Given the growing recognition of the potential benefits of maximizing TAG content in vegetative tissues of crops (Durrett et al., 2008; Ohlrogge et al., 2009; Chapman et al., 2013; Troncoso-Ponce et al., 2013), this clarification of the role of PDAT1 in plants may enable new strategies for future genetic engineering efforts aimed at enhancing oil accumulation in biomass crops used for biofuel production.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana plants used in this study were of the Columbia ecotype. The tgd1 mutant was previously described (Xu et al., 2003), as were dgat1-1 and pdat1-2 (Zhang et al., 2009). For growth on plates, surface-sterilized seeds of Arabidopsis were germinated on 0.6% (w/v) agar-solidified half-strength Murashige and Skoog (MS) (Murashige and Skoog, 1962) medium supplemented with 1% (w/v) Suc in an incubator with a photon flux density of 80 to 120 µmol m–2 s–1, a light period of 16 h (22°C), and a dark period of 8 h (18°C). For growth in soil, plants were first grown on MS medium for 10 d and then transferred to soil and grown under a photosynthetic photon flux density of 150 to 200 μmol m−2 s−1 at 22/18°C (day/night) with a 16-h-light/8-h-dark period.
Generation of Plant Expression Vectors and Plant Transformation
The full-length coding regions of PDAT1 and DGAT1 were amplified by RT-PCR using the primers 5′-GCGTGGTACCATGCCCCTTATTCATCGGA-3′ and 5′-ACGTCTGCAGTCACAGCTTCAGGTCAATACGCTC-3′ for PDAT1 and 5′-ACCTGGAGCTCATGGCGATTTTGGATTCTGC-3′ and 5′-CCGAGGTACCTCATGACATCGATCCTTTTCGGT-3′ for DGAT1. The resulting PDAT1 and DGAT1 PCR products were digested with KpnI-PstI or SacI-KpnI, respectively, and inserted into the respective sites of a binary vector derived from pPZP212 (Hajdukiewicz et al., 1994). To generate the OLE1-GFP fusion construct, the entire genomic DNA encoding the Arabidopsis OLE1 was amplified using the primers 5′-ATGGCGGATACAGCTAGAG-3′ and 5′-AGTAGTGTGCTGGCCACC-3′ and ligated into pCR8 topo-cloning entry vector (Invitrogen). The gene was then fused in frame with GFP at the C terminus through the LR reaction to the destination vector pGKPGWG (Zhong et al., 2008). After confirming the integrity of the constructs by sequencing, plant stable transformation was performed according to Clough and Bent (1998). Transgenic plants were selected in the presence of the respective antibiotics for the vectors on MS medium lacking Suc.
Lipid and FA Analyses
Plant tissues were frozen in liquid nitrogen, and total lipids were extracted by homogenization in chloroform/methanol/formic acid (1:1:0.1, by volume) and 1 M potassium chloride-0.2 M phosphoric acid according to Dörmann et al. (1995). Neutral and total polar lipids were separated on silica plates (Si250 with a preadsorbant layer; Mallinckrodt Baker) by TLC using a solvent system of hexane/diethyl ether/acetic acid (70:30:1, by volume). Polar lipids were separated using a solvent system consisting of acetone/toluene/water (91:30:7, by volume). Lipids were visualized by spraying 5% H2SO4 followed by charring. For quantitative analysis, lipids were visualized by brief exposure to iodine vapor and identified by cochromatography with lipid standards. Individual lipids were scraped from the plate and used to prepare FAMEs. Separation and identification of the FAMEs was performed on an HP5975 gas chromatography–mass spectrometer (Hewlett-Packard) fitted with 60 m × 250-µm SP-2340 capillary column (Supelco) with helium as a carrier gas. The methyl esters were quantified using heptadecanoic acid as the internal standard as described by Fan et al. (2011). The TAG content was calculated as described previously (Li et al., 2006). The FA composition at the sn-2 position of the glycerol backbone was determined by Rhizopus arrhizus lipase digestion as described by Härtel et al. (2000). Pigments were quantified according to Lichtenthaler (1987).
In Vivo Acetate Labeling
In vivo labeling experiments with 14C-acetate were done according to Koo et al. (2005). Briefly, rapidly growing leaves of 7-week-old plants were cut in strips and then incubated in the light (60 µmol m−2 s−1) at 22°C with shaking in 10 mL of medium containing 1 mM unlabeled acetate, 20 mM MES, pH 5.5, one-tenth strength of MS salts, and 0.01% Tween 20. The assay was started by the addition of 0.1 mCi of 14C-acetate (106 mCi/mmol; American Radiolabeled Chemicals). At the end of the incubation, leaf strips were washed three times with water and blotted onto filter paper. For the chase period, leaf tissue was incubated in the same medium lacking 14C-acetate under the same conditions as used for the pulse. Total lipids were extracted and separated as described above and radioactivity associated with total lipids or different lipid classes was determined by liquid scintillation counting or phosphorimaging, respectively.
Staining with Fat Red 7B
Seedlings were stained with a 0.1% (w/v) solution of Fat Red 7B (Sigma-Aldrich) overnight essentially as described by Brundrett et al. (1991). Following brief rinsing with distilled water, samples were visualized with a Wild Heerbrugg dissecting microscope.
Microscopy
For OD imaging, leaf tissues were stained with a neutral lipid-specific fluorescent dye, Nile red (Sigma-Aldrich), at a final concentration of 10 µg/mL and observed under a Zeiss epifluorescence microscope (Carl Zeiss Axiovert 200M) with a GFP filter. For TEM, leaf tissues were fixed with 2.5% (v/v) glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, for 2 h and then postfixed with 1% osmium tetroxide in the same buffer for 2 h at room temperature. After dehydration in a graded series of ethanol, the tissues were embedded in EPON812 resin (Electron Microscopy Sciences) and sectioned and stained with 2% uranyl acetate and lead citrate before viewing under a JEOL JEM-1400 LaB6 120KeV transmission electron microscope. For light microscopy observation, the sections were stained with 1% toluidine blue and examined using a Zeiss epifluorescence microscope as described above.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: DGAT1, At2g19450; OLE1, At4g25140; and PDAT1, At5g13460.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Changes in Levels of DGAT1 and PDAT1 Transcript during Leaf Development.
Supplemental Figure 2. TAG Levels in Transgenic Plants Overexpressing DGAT1, PDAT1, or OEL1.
Supplemental Figure 3. Growth and Lipid Phenotypes of the Transgenic Lines Coexpressing PDAT1 and OLE1.
Supplemental Figure 4. FA Composition of Individual Membrane Lipids from Leaves of PDAT1 Overexpressors.
Supplemental Figure 5. Time Course of 14C-Acetate Incorporation into Total Lipids of Leaf Strips from 7-Week-Old Wild-Type Plants Grown in Soil.
Supplemental Figure 6. Growth Phenotype and Total Leaf Polar Lipid Content of Transgenic Plants Coexpressing PDAT1 and GFP-OLE1 in the tgd1-1 Background.
Supplemental Figure 7. Overexpression of PDAT1 in act1 Leads to Defects in Chloroplast Division and Thylakoid Biogenesis.
Supplemental Figure 8. Pulse-Chase Acetate Labeling of FAs Associated with Individual Lipids of the act1 Mutant and act1 PDAT1-Overexpressing Line.
Acknowledgments
We thank John Ohlrogge for providing pdat1-2 mutant seeds. We also thank John Shanklin, John Ohlrogge, and Jitao Zou for critical reading of the article. This work was supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy through Grant DEAC0298CH10886 (BO-163) to C.X. Use of the transmission electron microscope and confocal microscope at the Center of Functional Nanomaterials was supported by the Office of Basic Energy Sciences, U.S. Department of Energy, under Contract DEAC02-98CH10886.
AUTHOR CONTRIBUTIONS
C.X. and J.F. designed the experiments. C.X., J.F., C.Y., and X.Z. performed the research. C.X. and J.F. analyzed the data and wrote the article.
Glossary
- FA
fatty acid
- TAG
triacylglycerol
- ER
endoplasmic reticulum
- G3P
glycerol-3-phosphate
- PA
phosphatidic acid
- DAG
diacylglycerol
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- OD
oil droplet
- DW
dry weight
- FAME
fatty acid methyl ester
- TLC
thin layer chromatography
- TEM
transmission electron microscopy
- GFP
green fluorescent protein
- DGDG
digalactosyldiacylglycerol
- MGDG
monogalactosyldiacylglycerol
- FAS
fatty acid synthesis
- PG
phosphatidylglycerol
- PI
phosphatidylinositol
- SQDG
sulfoquinovosyldiacylglycerol
- FW
fresh weight
- MS
Murashige and Skoog
References
- Andre C., Haslam R.P., Shanklin J. (2012). Feedback regulation of plastidic acetyl-CoA carboxylase by 18:1-acyl carrier protein in Brassica napus. Proc. Natl. Acad. Sci. USA 109: 10107–10112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianov V., Borisjuk N., Pogrebnyak N., Brinker A., Dixon J., Spitsin S., Flynn J., Matyszczuk P., Andryszak K., Laurelli M., Golovkin M., Koprowski H. (2010). Tobacco as a production platform for biofuel: Overexpression of Arabidopsis DGAT and LEC2 genes increases accumulation and shifts the composition of lipids in green biomass. Plant Biotechnol. J. 8: 277–287 [DOI] [PubMed] [Google Scholar]
- Bao X., Focke M., Pollard M., Ohlrogge J. (2000). Understanding in vivo carbon precursor supply for fatty acid synthesis in leaf tissue. Plant J. 22: 39–50 [DOI] [PubMed] [Google Scholar]
- Bates P.D., Durrett T.P., Ohlrogge J.B., Pollard M. (2009). Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol. 150: 55–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates P.D., Ohlrogge J.B., Pollard M. (2007). Incorporation of newly synthesized fatty acids into cytosolic glycerolipids in pea leaves occurs via acyl editing. J. Biol. Chem. 282: 31206–31216 [DOI] [PubMed] [Google Scholar]
- Baud S., Mendoza M.S., To A., Harscoët E., Lepiniec L., Dubreucq B. (2007). WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 50: 825–838 [DOI] [PubMed] [Google Scholar]
- Brasaemle D.L., Rubin B., Harten I.A., Gruia-Gray J., Kimmel A.R., Londos C. (2000). Perilipin A increases triacylglycerol storage by decreasing the rate of triacylglycerol hydrolysis. J. Biol. Chem. 275: 38486–38493 [DOI] [PubMed] [Google Scholar]
- Browse J., Warwick N., Somerville C.R., Slack C.R. (1986). Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the ‘16:3’ plant Arabidopsis thaliana. Biochem. J. 235: 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundrett M.C., Kendrick B., Peterson C.A. (1991). Efficient lipid staining in plant material with sudan red 7B or fluorol [correction of fluoral] yellow 088 in polyethylene glycol-glycerol. Biotech. Histochem. 66: 111–116 [DOI] [PubMed] [Google Scholar]
- Cernac A., Benning C. (2004). WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Chapman K.D., Dyer J.M., Mullen R.T. (2013). Commentary: Why don’t plant leaves get fat? Plant Sci. 207: 128–134 [DOI] [PubMed] [Google Scholar]
- Chapman K.D., Ohlrogge J.B. (2012). Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 287: 2288–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cronan J.E., Jr, Weisberg L.J., Allen R.G. (1975). Regulation of membrane lipid synthesis in Escherichia coli. Accumulation of free fatty acids of abnormal length during inhibition of phospholipid synthesis. J. Biol. Chem. 250: 5835–5840 [PubMed] [Google Scholar]
- Dahlqvist A., Ståhl U., Lenman M., Banas A., Lee M., Sandager L., Ronne H., Stymne S. (2000). Phospholipid:diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 97: 6487–6492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann P., Hoffmann-Benning S., Balbo I., Benning C. (1995). Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7: 1801–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett T.P., Benning C., Ohlrogge J. (2008). Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 54: 593–607 [DOI] [PubMed] [Google Scholar]
- Fan J., Andre C., Xu C. (2011). A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett. 585: 1985–1991 [DOI] [PubMed] [Google Scholar]
- Frentzen M. (1998). Acyltransferases from basic science to modified seed oils. Eur. J. Lipid Sci. Technol. 100: 161–166 [Google Scholar]
- Frentzen M., Heinz E., McKeon T.A., Stumpf P.K. (1983). Specificities and selectivities of glycerol-3-phosphate acyltransferase and monoacylglycerol-3-phosphate acyltransferase from pea and spinach chloroplasts. Eur. J. Biochem. 129: 629–636 [DOI] [PubMed] [Google Scholar]
- Froissard M., D’andréa S., Boulard C., Chardot T. (2009). Heterologous expression of AtClo1, a plant oil body protein, induces lipid accumulation in yeast. FEMS Yeast Res. 9: 428–438 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Härtel H., Dörmann P., Benning C. (2000). DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc. Natl. Acad. Sci. USA 97: 10649–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz E., Roughan P.G. (1983). Similarities and differences in lipid metabolism of chloroplasts isolated from 18:3 and 16:3 plants. Plant Physiol. 72: 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellgren L.I., Sandelius A.S. (2001). Age-dependent variation in membrane lipid synthesis in leaves of garden pea (Pisum sativum L.). J. Exp. Bot. 52: 2275–2282 [DOI] [PubMed] [Google Scholar]
- Horvath S.E., Wagner A., Steyrer E., Daum G. (2011). Metabolic link between phosphatidylethanolamine and triacylglycerol metabolism in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1811: 1030–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A.H.C. (1996). Oleosins and oil bodies in seeds and other organs. Plant Physiol. 110: 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jako C., Kumar A., Wei Y.D., Zou J.T., Barton D.L., Giblin E.M., Covello P.S., Taylor D.C. (2001). Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol. 126: 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katavic V., Reed D.W., Taylor D.C., Giblin E.M., Barton D.L., Zou J.T., Mackenzie S.L., Covello P.S., Kunst L. (1995). Alteration of seed fatty acid composition by an ethyl methanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol. 108: 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaup M.T., Froese C.D., Thompson J.E. (2002). A role for diacylglycerol acyltransferase during leaf senescence. Plant Physiol. 129: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.A., van Erp H., Quettier A.L., Shaw E., Menard G., Kurup S., Eastmond P.J. (2013). The sugar-dependent1 lipase limits triacylglycerol accumulation in vegetative tissues of Arabidopsis. Plant Physiol. 162: 1282–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlwein S.D. (2010). Triacylglycerol homeostasis: Insights from yeast. J. Biol. Chem. 285: 15663–15667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo A.J.K., Fulda M., Browse J., Ohlrogge J.B. (2005). Identification of a plastid acyl-acyl carrier protein synthetase in Arabidopsis and its role in the activation and elongation of exogenous fatty acids. Plant J. 44: 620–632 [DOI] [PubMed] [Google Scholar]
- Kunst L., Browse J., Somerville C. (1988). Altered regulation of lipid biosynthesis in a mutant of Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc. Natl. Acad. Sci. USA 85: 4143–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L., Browse J., Somerville C. (1989). Altered chloroplast structure and function in a mutant of Arabidopsis deficient in plastid glycerol-3-phosphate acyltransferase activity. Plant Physiol. 90: 846–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Beisson F., Pollard M., Ohlrogge J. (2006). Oil content of Arabidopsis seeds: The influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (1987). Chlorophylls and carotenoids: Pigments of photosynthetic membranes. Methods Enzymol. 148: 350–382 [Google Scholar]
- Liu B., Benning C. (2013). Lipid metabolism in microalgae distinguishes itself. Curr. Opin. Biotechnol. 24: 300–309 [DOI] [PubMed] [Google Scholar]
- Lu C.L., de Noyer S.B., Hobbs D.H., Kang J.L., Wen Y.C., Krachtus D., Hills M.J. (2003). Expression pattern of diacylglycerol acyltransferase-1, an enzyme involved in triacylglycerol biosynthesis, in Arabidopsis thaliana. Plant Mol. Biol. 52: 31–41 [DOI] [PubMed] [Google Scholar]
- Mhaske V., Beldjilali K., Ohlrogge J., Pollard M. (2005). Isolation and characterization of an Arabidopsis thaliana knockout line for phospholipid:diacylglycerol transacylase gene (At5g13640). Plant Physiol. Biochem. 43: 413–417 [DOI] [PubMed] [Google Scholar]
- Mongrand S., Bessoule J.J., Cabantous F., Cassagne C. (1998). The C-16:3/C-18:3 fatty acid balance in photosynthetic tissues from 468 plant species. Phytochemistry 49: 1049–1064 [Google Scholar]
- Mora G., Scharnewski M., Fulda M. (2012). Neutral lipid metabolism influences phospholipid synthesis and deacylation in Saccharomyces cerevisiae. PLoS ONE 7: e49269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Nunn W.D., Kelly D.L., Stumfall M.Y. (1977). Regulation of fatty acid synthesis during the cessation of phospholipid biosynthesis in Escherichia coli. J. Bacteriol. 132: 526–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelkers P., Cromley D., Padamsee M., Billheimer J.T., Sturley S.L. (2002). The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J. Biol. Chem. 277: 8877–8881 [DOI] [PubMed] [Google Scholar]
- Oelkers P., Tinkelenberg A., Erdeniz N., Cromley D., Billheimer J.T., Sturley S.L. (2000). A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J. Biol. Chem. 275: 15609–15612 [DOI] [PubMed] [Google Scholar]
- Ohlrogge J., Allen D., Berguson B., Dellapenna D., Shachar-Hill Y., Stymne S. (2009). Energy. Driving on biomass. Science 324: 1019–1020 [DOI] [PubMed] [Google Scholar]
- Ohlrogge J., Browse J. (1995). Lipid biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S., Rajasekharan R., Daum G. (2010). Triacylglycerol lipolysis is linked to sphingolipid and phospholipid metabolism of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1801: 1314–1322 [DOI] [PubMed] [Google Scholar]
- Roughan P.G., Slack C.R. (1982). Cellular organization of glycerolipid metabolism. Annu. Rev. Plant Physiol. 33: 97–132 [Google Scholar]
- Routaboul J.M., Benning C., Bechtold N., Caboche M., Lepiniec L. (1999). The TAG1 locus of Arabidopsis encodes for a diacylglycerol acyltransferase. Plant Physiol. Biochem. 37: 831–840 [DOI] [PubMed] [Google Scholar]
- Sandager L., Gustavsson M.H., Ståhl U., Dahlqvist A., Wiberg E., Banas A., Lenman M., Ronne H., Stymne S. (2002). Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 277: 6478–6482 [DOI] [PubMed] [Google Scholar]
- Sanjaya D., Durrett T.P., Weise S.E., Benning C. (2011). Increasing the energy density of vegetative tissues by diverting carbon from starch to oil biosynthesis in transgenic Arabidopsis. Plant Biotechnol. J. 9: 874–883 [DOI] [PubMed] [Google Scholar]
- Sanjaya M., Miller R., Durrett T.P., Kosma D.K., Lydic T.A., Muthan B., Koo A.J., Bukhman Y.V., Reid G.E., Howe G.A., Ohlrogge J., Benning C. (2013). Altered lipid composition and enhanced nutritional value of Arabidopsis leaves following introduction of an algal diacylglycerol acyltransferase 2. Plant Cell 25: 677–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos Mendoza M., Dubreucq B., Miquel M., Caboche M., Lepiniec L. (2005). LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett. 579: 4666–4670 [DOI] [PubMed] [Google Scholar]
- Shanklin J., Cahoon E.B. (1998). Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 611–641 [DOI] [PubMed] [Google Scholar]
- Shimada T.L., Shimada T., Takahashi H., Fukao Y., Hara-Nishimura I. (2008). A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J. 55: 798–809 [DOI] [PubMed] [Google Scholar]
- Siloto R.M.P., Findlay K., Lopez-Villalobos A., Yeung E.C., Nykiforuk C.L., Moloney M.M. (2006). The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocombe S.P., Cornah J., Pinfield-Wells H., Soady K., Zhang Q., Gilday A., Dyer J.M., Graham I.A. (2009). Oil accumulation in leaves directed by modification of fatty acid breakdown and lipid synthesis pathways. Plant Biotechnol. J. 7: 694–703 [DOI] [PubMed] [Google Scholar]
- Somerville C., Browse J. (1996). Dissecting desaturation: Plants prove advantageous. Trends Cell Biol. 6: 148–153 [DOI] [PubMed] [Google Scholar]
- Sperling P., Heinz E. (1993). Isomeric sn-1-octadecenyl and sn-2-octadecenyl analogues of lysophosphatidylcholine as substrates for acylation and desaturation by plant microsomal membranes. Eur. J. Biochem. 213: 965–971 [DOI] [PubMed] [Google Scholar]
- Ståhl U., Carlsson A.S., Lenman M., Dahlqvist A., Huang B., Banas W., Banas A., Stymne S. (2004). Cloning and functional characterization of a phospholipid:diacylglycerol acyltransferase from Arabidopsis. Plant Physiol. 135: 1324–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Kwong L.W., Yee K.M., Pelletier J., Lepiniec L., Fischer R.L., Goldberg R.B., Harada J.J. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjellström H., Yang Z., Allen D.K., Ohlrogge J.B. (2012). Rapid kinetic labeling of Arabidopsis cell suspension cultures: Implications for models of lipid export from plastids. Plant Physiol. 158: 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A., Joubès J., Barthole G., Lécureuil A., Scagnelli A., Jasinski S., Lepiniec L., Baud S. (2012). WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 24: 5007–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troncoso-Ponce M.A., Cao X., Yang Z., Ohlrogge J.B. (2013). Lipid turnover during senescence. Plant Sci. 205-206: 13–19 [DOI] [PubMed] [Google Scholar]
- Vanhercke T., El Tahchy A., Shrestha P., Zhou X.R., Singh S.P., Petrie J.R. (2013). Synergistic effect of WRI1 and DGAT1 coexpression on triacylglycerol biosynthesis in plants. FEBS Lett. 587: 364–369 [DOI] [PubMed] [Google Scholar]
- Winichayakul S., Scott R.W., Roldan M., Hatier J.H., Livingston S., Cookson R., Curran A.C., Roberts N.J. (2013). In vivo packaging of triacylglycerols enhances Arabidopsis leaf biomass and energy density. Plant Physiol. 162: 626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Fan J., Froehlich J.E., Awai K., Benning C. (2005). Mutation of the TGD1 chloroplast envelope protein affects phosphatidate metabolism in Arabidopsis. Plant Cell 17: 3094–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Fan J., Riekhof W., Froehlich J.E., Benning C. (2003). A permease-like protein involved in ER to thylakoid lipid transfer in Arabidopsis. EMBO J. 22: 2370–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Yu B., Cornish A.J., Froehlich J.E., Benning C. (2006). Phosphatidylglycerol biosynthesis in chloroplasts of Arabidopsis mutants deficient in acyl-ACP glycerol-3- phosphate acyltransferase. Plant J. 47: 296–309 [DOI] [PubMed] [Google Scholar]
- Yoon K., Han D., Li Y., Sommerfeld M., Hu Q. (2012). Phospholipid:diacylglycerol acyltransferase is a multifunctional enzyme involved in membrane lipid turnover and degradation while synthesizing triacylglycerol in the unicellular green microalga Chlamydomonas reinhardtii. Plant Cell 24: 3708–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Fan J., Taylor D.C., Ohlrogge J.B. (2009). DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21: 3885–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S.L., Lin Z.F., Fray R.G., Grierson D. (2008). Improved plant transformation vectors for fluorescent protein tagging. Transgenic Res. 17: 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Wei Y., Jako C., Kumar A., Selvaraj G., Taylor D.C. (1999). The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J. 19: 645–653 [DOI] [PubMed] [Google Scholar]