In the moss Physcomitrella patens, LHCSR and PSBS, which trigger nonphotochemical quenching (NPQ) in green algae and vascular plants, respectively, are both active. This work reports that zeaxanthin, an NPQ enhancer, is far more active on LHCSR-dependent NPQ than on the PSBS-dependent NPQ. Consistent with this, zeaxanthin binds LHCSR in excess light.
Abstract
Nonphotochemical quenching (NPQ) dissipates excess energy to protect the photosynthetic apparatus from excess light. The moss Physcomitrella patens exhibits strong NPQ by both algal-type light-harvesting complex stress-related (LHCSR)–dependent and plant-type S subunit of Photosystem II (PSBS)-dependent mechanisms. In this work, we studied the dependence of NPQ reactions on zeaxanthin, which is synthesized under light stress by violaxanthin deepoxidase (VDE) from preexisting violaxanthin. We produced vde knockout (KO) plants and showed they underwent a dramatic reduction in thermal dissipation ability and enhanced photoinhibition in excess light conditions. Multiple mutants (vde lhcsr KO and vde psbs KO) showed that zeaxanthin had a major influence on LHCSR-dependent NPQ, in contrast with previous reports in Chlamydomonas reinhardtii. The PSBS-dependent component of quenching was less dependent on zeaxanthin, despite the near-complete violaxanthin to zeaxanthin exchange in LHC proteins. Consistent with this, we provide biochemical evidence that native LHCSR protein binds zeaxanthin upon excess light stress. These findings suggest that zeaxanthin played an important role in the adaptation of modern plants to the enhanced levels of oxygen and excess light intensity of land environments.
INTRODUCTION
Light is essential for photosynthesis, but when its intensity exceeds the activity of the photosynthetic electron transport chain, light can damage the photosynthetic apparatus. Indeed, excess light increases the lifetime of chlorophyll singlet excited states (1Chl*) and, thus, the probability of triplet chlorophyll (3Chl*) formation. Triplet chlorophyll can react with molecular oxygen and produce singlet oxygen (1O2), leading to photodamage and photoinhibition (Barber and Andersson, 1992; Miller et al., 2008; Takahashi and Badger, 2011). Land environments have highly variable light intensities, temperatures, and water availability, which can cause rapid changes in the excitation pressure on photosystem II (PSII), leading to photooxidative damage. Plants have evolved multiple mechanisms of photoprotection, such as light avoidance through leaf and chloroplast movements, cyclic electron transport around photosystem I (PSI), and photorespiration (Takahashi and Badger, 2011).
Three additional photoprotection mechanisms involve carotenoids: (1) modulation of 3Chl* yield, (2) scavenging of reactive oxygen species (ROS), and (3) heat dissipation of energy from light absorbed in excess (nonphotochemical quenching [NPQ]). In vascular plants, these three functions are upregulated by the synthesis of zeaxanthin from violaxanthin in excess light conditions. This reaction is catalyzed by violaxanthin deepoxidase (VDE) and together with zeaxanthin reepoxidation by zeaxanthin epoxidase (ZEP) upon return to light-limiting conditions of photosynthesis constitutes the xanthophyll cycle. Previous work focused on the modulation of 3Chl* yield in leaves (Dall’Osto et al., 2012) and ROS scavenging in green algae (Baroli et al., 2003, 2004) and in higher plants (Havaux et al., 2007; Dall’Osto et al., 2010). npq1, the Arabidopsis thaliana mutant unable to synthesize zeaxanthin, shows NPQ reduction (Niyogi et al., 1998; Li et al., 2002), implying that zeaxanthin synthesis enhances the amplitude of both qE and qZ components. qZ has been directly attributed to zeaxanthin accumulation and in particular to its binding to light-harvesting complex (LHC) proteins, specifically LHCb5 (Dall’Osto et al., 2005). qE, the fastest quenching component, requires the PSII protein, S subunit of PSII (PSBS; Li et al., 2000), which transduces low lumenal pH signal into a quenching reaction by the reversible protonation of two lumen-exposed Glu residues (Li et al., 2004). Although PSBS is not a pigment binding protein (Dominici et al., 2002; Bonente et al., 2008a), its activity in triggering NPQ is enhanced by zeaxanthin, possibly by binding to LHCb proteins, where quenching occurs (Ruban et al., 1999; Caffarri et al., 2001; Holt et al., 2005; Ahn et al., 2008; Avenson et al., 2008; Betterle et al., 2009; Dall’Osto et al., 2012). In green algae, where PSBS is absent (Bonente et al., 2008b), the triggering of qE requires light-harvesting complex stress-related (LHCSR) (Peers et al., 2009), a chlorophyll binding protein sharing with PSBS the capacity of binding dicyclohexylcarbodiimide and transducing the low lumenal pH into a quenching reaction (Peers et al., 2009; Bonente et al., 2011a).
The moss Physcomitrella patens is particularly well suited for studying the evolution of photoprotection mechanisms and their modulation by zeaxanthin because of its position in the Viridiplantae clade, providing insights on the novel strategies evolved to face the stressful conditions upon emersion from water. Consistent with this, NPQ in P. patens depends on both PSBS and LHCSR (Alboresi et al., 2010; Gerotto et al., 2011) and presents an active xanthophyll cycle (Alboresi et al., 2008). The analysis of the P. patens genome (Koziol et al., 2007; Alboresi et al., 2008, 2011; Rensing et al., 2008) revealed that the composition of its antenna system is more similar to that of vascular plants than to unicellular green algae due to the presence of genes encoding LHCb3 and LHCb6, which are not found in Chlamydomonas reinhardtii (Elrad and Grossman, 2004). Both carotenoid accumulation and NPQ are enhanced by abiotic stress (Gerotto et al., 2011; Azzabi et al., 2012).
To study the role of zeaxanthin in P. patens, we produced a vde knockout (KO) mutant by homologous recombination in the wild type and in genotypes performing only LHCSR-dependent NPQ (psbs KO) or PSBS-dependent NPQ (lhcsr1 lhcsr2 KO) (hereafter indicated as lhcsr KO). vde KO plants completely lost their capacity to synthesize zeaxanthin under excess light conditions, implying the identified VDE gene encodes the only VDE enzyme in P. patens. We show that in P. patens, qE is highly dependent on zeaxanthin, particularly its LHCSR-dependent component. Biochemical evidence suggests that zeaxanthin binding to LHCSR is directly responsible for enhancing quenching activity.
RESULTS
Characterization of Genes Encoding the Carotenoid Biosynthetic Pathway in P. patens
According to previous reports, carotenoid content is enhanced by abiotic stress in the moss P. patens (Gerotto et al., 2011; Azzabi et al., 2012), suggesting that photoprotection is modulated by changes in carotenoid biosynthesis. The carotenoid biosynthetic enzymes are well conserved in Viridiplantae, and genes can be identified by similarity search (Coesel et al., 2008; Cazzonelli and Pogson, 2010). A summary of P. patens genes putatively encoding carotenoid biosynthesis enzymes is shown in Table 1. We observed a higher gene copy number for each carotenoid biosynthesis-related sequence than in other species, similar to the case of other metabolic pathways (Rensing et al., 2008). This feature is likely related to a recent genome duplication of P. patens followed by the retention of sequences encoding proteins with important functions for the adaptation to environment cues (Rensing et al., 2007). We identified two genes putatively encoding ZEP (Figure 1, Table 1) and single genes putatively encoding the ε-ring hydroxylase (LUT1) and VDE. The conversion of trans-violaxanthin to trans-neoxanthin in Arabidopsis requires the ABA4 gene, possibly encoding a neoxanthin synthase (NSY) (Dall’Osto et al., 2007; North et al., 2007). We identified two coding sequences sharing high identity with ABA4 in P. patens genome. The carotenoid biosynthesis genes of P. patens share high sequence identity with their pinophyta counterparts (Table 1).
Table 1. Genes of the P. patens Carotenoid Biosynthetic Pathway.
| Protein | Also Known | Length (aa) | Locus Name | Best Hit NCBI (Locus) | ID (%) |
|---|---|---|---|---|---|
| LCYE1 | LUT2 | 589 | Pp1s11_269V6 | Picea glauca (BT114509) | 65 |
| LCYE2 | LUT2 | 580 | Pp1s153_133V6 | P. glauca (BT114509) | 65 |
| CYP97C1 | LUT1 | 576 | Pp1s307_64V6 | P. glauca (BT111413) | 75 |
| CYP97A3 | LUT5 | 576 | Pp1s28_409V6 | P. glauca (BT113884) | 41 |
| LCYB1 | 559 | Pp1s55_202V6 | Taxodium distichum (AB161862) | 64 | |
| LCYB2 | 554 | Pp1s160_155V6 | Cryptomeria japonica (AB096553) | 65 | |
| CHYB1 | CHY2 | 346 | Pp1s373_23V6 | P. glauca (BT101326) | 80 |
| CHYB2 | CHY2 | 346 | Pp1s19_96V6 | Picea sitchensis (EF085473) | 67 |
| CHYB3 | CHY2 | 353 | Pp1s20_24V6 | P. sitchensis (EF085473) | 70 |
| ZEP1 | ABA1/NPQ2 | 685 | Pp1s91_16V6 | Selaginella moellendorffii (XM_002972191) | 68 |
| ZEP2 | ABA1/NPQ2 | 728 | Pp1s219_79V6 | S. moellendorffii (XM_002972191) | 63 |
| VDE | NPQ1 | 531 | Pp1s161_120V6 | Vitis vinifera (XM_002267116) | 60 |
| NSY1? | ABA4 | 283 | Pp1s108_75V6 | P. sitchensis (EF085729) | 67 |
| NSY2? | ABA4 | 283 | Pp1s96_157V6 | P. sitchensis (EF085729) | 59 |
Locus name identified by TBLASTN in Phytozome version 8 starting from Arabidopsis protein sequences. The best hit was identified by TBLASTN against the National Center for Biotechnology Information (NCBI) nucleotide collection. The identity between P. patens protein and its best hit is reported in the last column. aa, amino acids. Blank spaces indicate that there were no common names available.
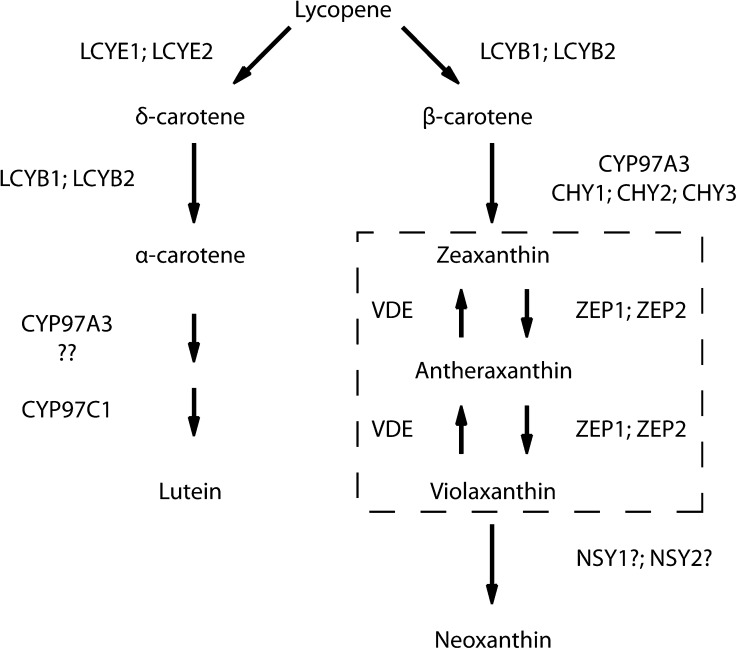
Figure 1.
Scheme of Carotenoid Biosynthetic Pathway in P. patens.
The genes identified in this study, lycopene b-cyclase (LCYB), lycopene ε-cyclase (LCYE), β-hydroxylase (CYP97A3. CHY), ε-hydroxylase (CYP97C1), ZEP, VDE, and the putative NSY, are indicated. The reaction of neoxanthin synthase is indicated by a question mark (NSY1?; NSY2?) because its catalytic activity has not yet been verified in vitro (North et al., 2007). Unknown genes could participate to β-hydroxylase activity and are here indicated by two question marks (??). The dashed box includes the xanthophyll cycle.
The VDE Pp1s161_120V6 Gene Fully Accounts for Zeaxanthin Accumulation upon Transition from Dark to Excess Light
In order to examine the photoprotective role of zeaxanthin, we proceeded to isolate vde null mutants in the moss P. patens, taking advantage of its capacity for efficient homologous recombination (see Supplemental Figure 1A online). We obtained targeted KO of VDE by using two different selection markers conferring either hygromycin B or zeocin resistance. Stably resistant plants were screened by PCR to verify that the expected event of homologous recombination took place (see Supplemental Figure 1B online), and immunoblotting analysis was performed on total protein extracts using a polyclonal antibody toward Arabidopsis VDE protein (see Supplemental Figures 1C and 2 online). Selected transformants were analyzed for their in vivo fluorescence behavior and NPQ kinetics using a fluorescence imaging system (see Supplemental Figures 1D and 1E online). Plants lacking VDE were also strongly affected in NPQ amplitude, irrespective of the hygromycin or zeocin resistance gene used as a marker for selection. Also, resistant lines in which gene targeting did not occur showed a wild-type NPQ kinetic (see Supplemental Figure 1E online). NPQ kinetics were calculated upon fluorometric measurements using a Dual-PAM 100 apparatus on plants grown on minimal medium (Figure 2A). NPQ of wild-type plants was 9.5-fold higher than in vde KO plants, and this phenotype was consistent with the effect measured on plants treated with the VDE inhibitor DTT (see Supplemental Table 1 and Supplemental Figure 3A online). Moreover, HPLC analysis performed on dark-adapted versus excess light–treated plants (Table 2) showed that vde KO plants were unable to convert violaxanthin into zeaxanthin. We conclude that the putative VDE gene (Pp1s161_120V6) identified in the P. patens genome is responsible for encoding the complete extent of VDE activity upon excess light treatment and that zeaxanthin synthesis is responsible for a very strong upregulation of NPQ activity.
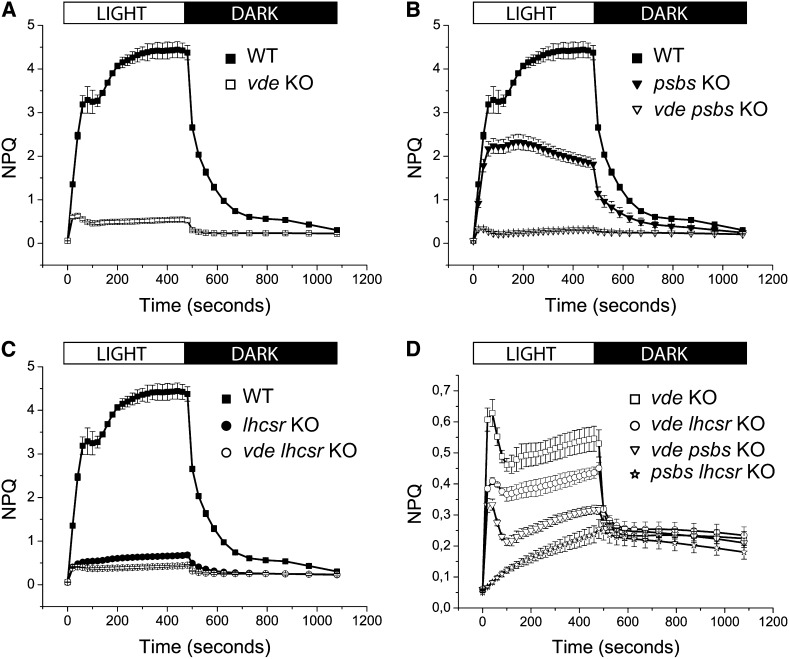
Figure 2.
NPQ Induction and Relaxation Kinetics of the vde KO Genotypes Described in Supplemental Figure 2 Online.
Chlorophyll fluorescence was measured after 45 min of dark adaptation using 830 μmol photons m−2 s−1 of actinic red light and 6000 μmol photons m−2 s−1 of saturating light. Exposure to actinic irradiance is indicated at the top of the figure. Data are mean values of plants grown on three independent plates ± sd.
(A) The wild type (WT; closed squares) is compared with the single vde KO (open squares).
(B) psbs KO (closed triangles) is compared with psbs vde KO (open triangles).
(C) lhcsr KO (closed circles) is compared with vde lhcsr KO (open circles). In (B) and (C), the wild type (closed squares) is shown as a reference.
(D) Comparison between psbs lhcsr KO (open stars) and the vde KO plants reported in (A) to (C).
Table 2. Xanthophyll Cycle Pigment Composition and Deepoxidation Index Determination of Wild-Type and vde KO P. patens Plants.
| Sample | Vio | Ant | Zea | V+A+Z | DEP |
|---|---|---|---|---|---|
| Wild type dark | 4.36 ± 0.27 | n.d. | n.d. | 4.36 ± 0.27 | n.d. |
| Wild type light | 1.15 ± 0.03 | 0.42 ± 0.04 | 3.88 ± 0.14 | 5.45 ± 0.15 | 0.75 ± 0.01 |
| vde dark | 5.16 ± 0.26 | n.d. | n.d. | 5.16 ± 0.26 | n.d. |
| vde light | 5.16 ± 0.36 | n.d. | n.d. | 5.16 ± 0.36 | n.d. |
| Wild-type thylakoids light | 1.08 ± 0.02 | 0.58 ± 0.00 | 3.81 ± 0.20 | 5.48 ± 0.18 | 0.75 ± 0.01 |
| psbs lhcsr KO thylakoids light | 1.06 ± 0.03 | 0.59 ± 0.10 | 3.80 ± 0.20 | 5.45 ± 0.17 | 0.75 ± 0.02 |
Plants were dark adapted for 45 min directly on minimum agar medium. Dark-adapted plants were then treated for 30 min with excess light (∼830 µmol photons m−2 s−1) and either used for total extract or for thylakoid preparation. Acetone extract of each sample was separated by HPLC to separate and quantify individual pigments. Carotenoid content (mol) is normalized to chlorophylls a+b (100 mol). Vio, violaxanthin; Ant, antheraxanthin; Zea, zeaxanthin. Deepoxidation index (DEP) is calculated as follows: (0.5·A+Z)/(A+Z+V). Data are expressed as means ± sd, n = 3. n.d., not detected.
Dependence of LHCSR- and PSBS-Dependent NPQ Quenching Components on Zeaxanthin
The strong inhibition of NPQ amplitude in the vde KO genotype raises the question whether both LHCSR- and PSBS-dependent NPQ mechanisms are upregulated by zeaxanthin in P. patens. In order to answer this question, we made double mutants by introducing the vde KO mutation in lhcsr KO and psbs KO plants and verifying that VDE enzyme did not accumulate in the resulting double mutants (see Supplemental Figure 2 online). We retained at least two independent lines for each double mutant, although a larger number shared the same phenotype (see Supplemental Figure 3B online). NPQ measurements showed that the vde KO mutation had a strong effect on the NPQ amplitude of both psbs KO and lhcsr KO lines (Figures 2B to 2D). However, we noticed that the effect of the vde KO mutation in decreasing NPQ amplitude is far stronger in psbs KO than in lhcsr KO double mutants (Figures 2B to 2D; see Supplemental Figure 3B online). Detailed analysis of NPQ kinetics showed that a residual VDE-independent qE activity is present in all vde KO mutants (Figure 2A), but it is absent in the triple lhcsr psbs KO mutant (Figure 2D). Thus, zeaxanthin needs either PSBS or LHCSR to exert any effect on the NPQ activation of P. patens cells.
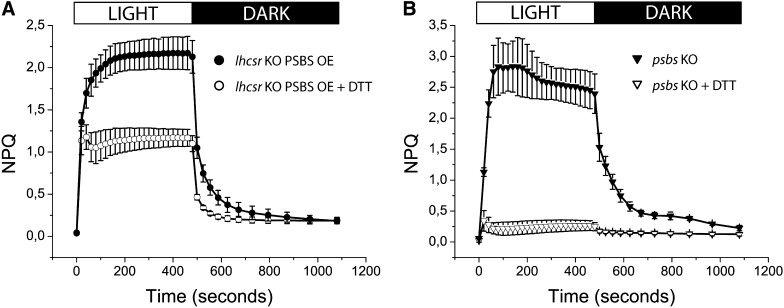
The weakness of the change of fluorescence yield undergone by lhcsr KO versus vde lhcsr KO casts some doubt on the capacity of zeaxanthin of upregulating PSBS-dependent qE (Figure 2C). In fact, a fraction of the quenching is likely to be of the qZ type (i.e., independent of the transmembrane pH gradient) (Dall’Osto et al., 2005; Kalituho et al., 2007; Nilkens et al., 2010). In order to verify this possibility, we used the PSBS overexpressor line on the lhcsr KO background (PSBS OE) (Gerotto et al., 2012), which exhibits similar NPQ amplitude as the psbs KO line but fully dependent on PSBS. When NPQ was measured upon infiltration of PSBS OE with the VDE inhibitor DTT, the amplitude was clearly lower than in the sample infiltrated with buffer, confirming that PSBS-dependent NPQ in P. patens indeed showed a zeaxanthin-dependent enhancement (Figure 3), although to a lower level than the LHCSR-dependent component (Figure 2B). To focus on the fast and pH dependent component of NPQ, we calculated qE from NPQ induction kinetics as the difference between NPQ at the end of the actinic light period and after 2.5 min of dark relaxation (see Supplemental Figure 4 online). On this basis, the zeaxanthin-dependent enhancement of qE was found to be higher in plants expressing LHCSR than in plants expressing PSBS (see Supplemental Figure 4 online). We conclude that both qE components are enhanced by zeaxanthin in P. patens and yet the effect is far larger on the LHCSR-dependent component.
Figure 3.
Effect of DTT on NPQ of lhcsr KO PSBS OE and psbs KO P. patens Plants Grown on Minimum Agar Medium for ∼10 d (24°C, 50 µmol Photons m−2 s−1).
Chlorophyll fluorescence was measured after 45 min of dark adaptation using 830 μmol photons m−2 s−1 of actinic red light and 6000 μmol photons m−2 s−1 of saturating light. Exposure to actinic irradiance is indicated at the top of the figure. Data are mean values of plants grown on three independent plates ± sd.
(A) lhcsr KO PSBS OE plants measured directly on the growth medium (closed circles) or infiltrated with 1 mM DTT (open circles).
(B) psbs KO plants measured directly on the growth medium (closed triangles) or infiltrated with 1 mM DTT (open triangles).
Effect of vde KO and lhcsr psbs KO Mutations on Resistance to Photoinhibitory Excess Light Treatment
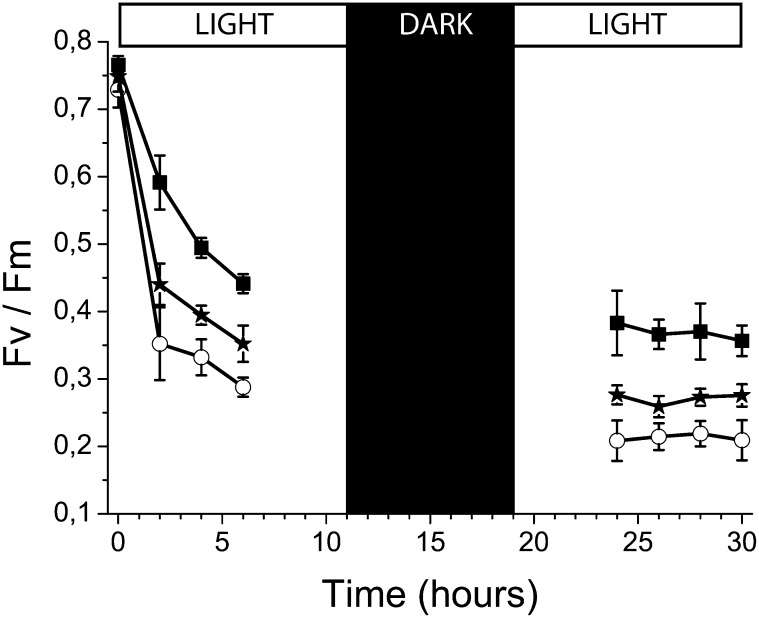
Zeaxanthin has been reported to increase resistance to photoinhibitory excess light treatment in plants and algae (Havaux and Niyogi, 1999; Baroli et al., 2004; Dall’Osto et al., 2005). To verify that this was also true in P. patens, 10-d-old plants grown in control light were exposed to 850 µmol photons m−2 s−1 for 6 h, and the treatment was then continued the following day after incubation overnight in the dark. The dark period during excess light treatment was maintained in order to avoid additional stress related to a sudden change in the circadian clock, in agreement with previous reports (Havaux and Niyogi, 1999). Determination of PSII quantum efficiency at different time points showed that the wild type underwent a decrease in Fv/Fm from 0.75 to 0.55 at the end of the first part of the treatment and to 0.45 at the end of the second (Figure 4). In the same conditions, the vde KO plants underwent a more rapid decrease (0.28 and then 0.20 at the end of the first and second period, respectively), clearly showing enhanced sensitivity to excess light. lhcsr psbs KO plants, completely unable to activate any NPQ but still able to synthesize zeaxanthin, showed an intermediate behavior (0.35 and then 0.28 at the end of the first and second period, respectively), implying that zeaxanthin had additional photoprotective actions besides its effect on NPQ.
Figure 4.
PSII Photoinhibition in P. patens Plants Monitored by Fv/Fm Measurements.
The wild type (black squares), vde KO (open circles), and lhcsr psbs KO (black stars). Ten-day-old plants were exposed to excess light stress (24°C, 850 µmol photons m−2 s−1) for 5 h. Photoinhibition was measured following the decrease of Fv/Fm calculated from chlorophyll fluorescence. Data are mean values of three to four independent plates, and eight spots per plate were analyzed after fluorescence imaging. Error bars indicate sd.
Step Solubilization of Thylakoid Membranes
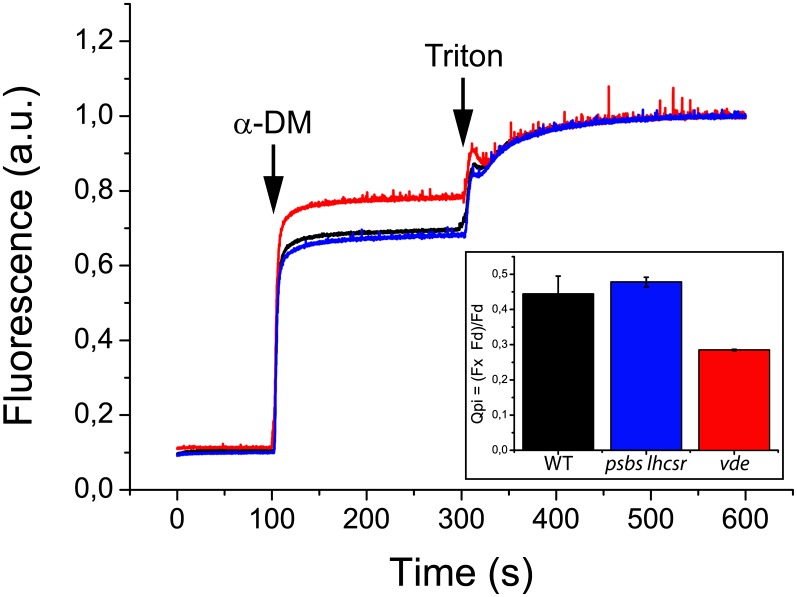
A direct quenching effect of zeaxanthin bound to LHC pigment-protein complexes, independent from ΔpH, has been shown to account for a slowly relaxing component in NPQ kinetics, called qZ (Dall’Osto et al., 2005; Nilkens et al., 2010), or qI in earlier reports (Farber et al., 1997; Horton et al., 2005). This effect can be measured by comparing the fluorescence yield of a thylakoid sample before and after the release of the pigment–protein interactions upon treatment with different detergents. In the original report, thylakoids isolated from genotypes either enriched in zeaxanthin or violaxanthin were shown to have different fluorescence yields on a chlorophyll basis, which was equalized upon disruption of pigment–protein interactions (Dall’Osto et al., 2005). In order to verify if such an effect was present in P. patens, we exposed the wild type, psbs lhcsr KO, and vde KO genotypes to excess light for 1 h (24°C, 850 µmol photons m−2 s−1), upon which the level of zeaxanthin accumulation was measured by HPLC analysis and thylakoid aliquots were submitted to step solubilization. The wild type and psbs lhcsr KO accumulated high levels of zeaxanthin but vde KO did not (Table 2). Fluorescence yield of a thylakoid suspension was measured under continuous stirring for 100 s. Membranes were first treated by the mild detergent n-dodecyl-alpha-d-maltoside (α-DM) at the final concentration of 0.075% (w/v), a treatment that releases the pigment-protein complexes from the thylakoid lipid bilayer without disrupting pigment–protein interactions (Dall’Osto et al., 2005). An increase in fluorescence yield was associated to membrane solubilization (Figure 5). In a second step, Triton X-100 was added to a final concentration of 1.8% (w/v), generating a further increase in fluorescence yield due to the release of chlorophyll, free from interactions with proteins and carotenoids in detergent micelles. Overall, the full solubilization of thylakoid membranes produced a ninefold increase in fluorescence yield (Qm+pi) with 63 to 78% due to the release of protein–protein interactions caused by the solubilization of the membrane (Qm) and 37 to 22% due to disruption of pigment–protein interactions (Qpi) depending on the genotype. A small difference in fluorescence yield was observed even before thylakoid solubilization, with the vde KO showing 6% higher fluorescence yield with respect to wild-type and psbs lhcsr KO samples. Upon the first mild solubilization step, the two samples showed a clear difference in fluorescence yield, implying that pigment-proteins containing zeaxanthin have a lower chlorophyll fluorescence yield. This was described by Qpi, the parameter that quantifies the quenching generated by pigment–protein interactions, scoring 0.29 in vde KO or in wild-type thylakoids from dark-adapted plants versus 0.44 in the sample from excess light–treated wild type (Figure 5, Table 3).
Figure 5.
Release of Chlorophyll Fluorescence Quenching by Step Solubilization of Thylakoid Membranes.
Thylakoids were purified from plants treated for 1 h by excess light (24°C, 850 µmol photons m−2 s−1). Fluorescence yield of wild-type (WT), npq4, and vde KO membranes was measured after solubilization with 0.075% α-DM and 1.8% Triton X-100. Chlorophyll concentration of isolated thylakoids was set to 0.1 µg/mL. The samples were measured at room temperature under continuous stirring. The inset reports the quenching of protein-pigment binding (Qpi). Data are expressed as means ± sd. a.u., arbitrary units.
Table 3. Fluorometric Analysis of the Effect of Step Solubilization on P. patens Thylakoids.
| Quenching Parameter | Wild Type | psbs lhcsr1 lhcsr2 KO | vde KO |
|---|---|---|---|
| Qm = (Fd − Ft)/Ft | 5.56 ± 0.07 | 5.82 ± 0.16 | 5.92 ± 0.17 |
| Qpi = (Fx − Fd)/Fd | 0.44 ± 0.05 | 0.48 ± 0.01 | 0.29 ± 0.00 |
| Qm+pi = (Fx − Ft)/Ft | 8.47 ± 0.34 | 9.08 ± 0.27 | 7.90 ± 0.23 |
Thylakoid membranes were purified from plants treated for 1 h by excess light (24°C, 850 µmol photons m−2 s−1). Fluorescence yield was measured after solubilization with 0.075% α-DM and 1.8% Triton X-100 as shown in Figure 4. The samples were measured at room temperature under continuous stirring. Data are expressed as means ± sd, n = 3. See Methods for details.
Zeaxanthin Binds to LHCSR in P. patens
The strong decrease in chlorophyll fluorescence quenching induced by the vde KO mutation on psbs lhcsr KO plants both in vitro (Figure 5) and in vivo upon excess light treatment (Figure 2) showed that NPQ activation strongly depends on zeaxanthin accumulation, especially for its LHCSR-dependent component (Figure 2B). Since zeaxanthin is not a fluorescent molecule, it must affect fluorescence yield through binding one or more chlorophyll binding proteins. In order to search for interactions of zeaxanthin and chlorophyll binding proteins, we proceeded to fractionation of thylakoid membranes, isolated from plants either harvested upon growth in control light or treated with excess light (lacking or accumulating zeaxanthin, respectively). Thylakoids were solubilized with the mild detergent α-DM and loaded onto a Suc density gradient, which was centrifuged at an average of 40,000g for 22 h. The separation pattern was very similar, irrespective of the light treatment (Figure 6). Each gradient was fractionated into 30 aliquots that were analyzed by spectrophotometry and SDS-PAGE to identify their pigment-protein content and polypeptide composition. By HPLC analysis of acetone-extracted pigments of each fraction, we identified protein-bound chromophores, particularly xanthophyll cycle pigments violaxanthin and zeaxanthin. In the lower part of the gradient (fractions 1 to 16), reaction center–containing green bands were found, namely, PSII-LHCII supercomplexes (fractions 2 to 7), PSI-LHCI (fractions 10 to 13), and PSII core complex (fractions 14 to 16). In the upper part of the gradient, there were three closely spaced green bands containing LHCb proteins in pentameric (LHCb4-LHCb6-LHCII trimer, fractions 17 and 18), trimeric (LHCII, fractions 19 to 21), and monomeric (fractions 22 to 24) aggregation states, and a faint yellow top band containing free pigments, enriched in carotenoids (fractions 25 to 27). Comparison of violaxanthin distribution among fractions in control sample versus violaxanthin plus zeaxanthin in excess light–treated samples (Figure 6) shows a very similar pattern, implying that newly synthesized zeaxanthin replaces preexisting violaxanthin within pigment-protein complexes. Zeaxanthin was present in all green bands containing LHC proteins, namely, PSII-LHCII and PSI-LHCI, LHCb pentamers, trimers, and monomers but not in the PSII core complexes, in agreement with their binding of β-carotene only (Bassi et al., 1993). The same level of deepoxidation upon exposure to excess light and zeaxanthin binding to pigment-proteins, as assessed by Suc gradient fractionation and HPLC analysis, was found in wild-type and lhcsr psbs KO plants, as shown by a deepoxidation index of 0.76 in both cases. We observed that the exchange of violaxanthin versus zeaxanthin is rapid and more complete on LHC binding sites with respect to previous reports on Arabidopsis, which has a deepoxidation index of 0.50 (Dall’Osto et al., 2010).
Figure 6.
Suc Density Gradient Fractionation of Wild-Type Solubilized Thylakoids, Western Blot Analysis, and Xanthophylls Distribution Along Gradients.
(A) Suc density gradient fractionation of solubilized thylakoids, isolated from wild-type plants grown under standard conditions. An equal volume of each fraction was used for immunoblotting analysis using antibody against LHCSR protein. LHCSR1 is indicated in red, LHCSR2 is indicated in green, and LHCII nonspecific signals are indicated in blue.
(B) Pictures on the left represent the Suc density gradient fractionation of wild-type solubilized thylakoids isolated from plants grown under control light conditions (CL) or treated for 30 min under ∼850 µmol photons m−2 s−1 (excess light [EL]). An equal volume of each fraction was used for the HPLC profiling of the xanthophyll cycle. Data are reported in the graphs on the right. Vio, violaxanthin; Zea, zeaxanthin. The content of each band has been attributed on the basis of spectroscopy analysis and earlier determinations in Arabidopsis (Betterle et al., 2009; Dall’Osto et al., 2010).
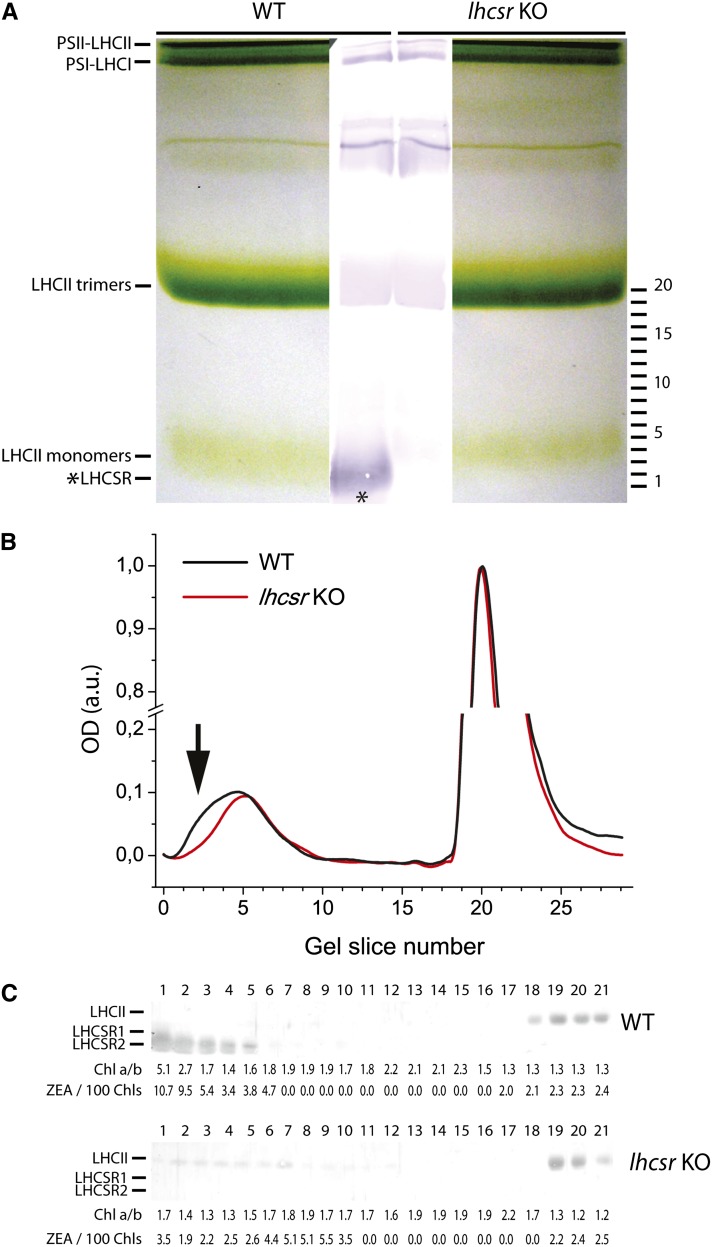
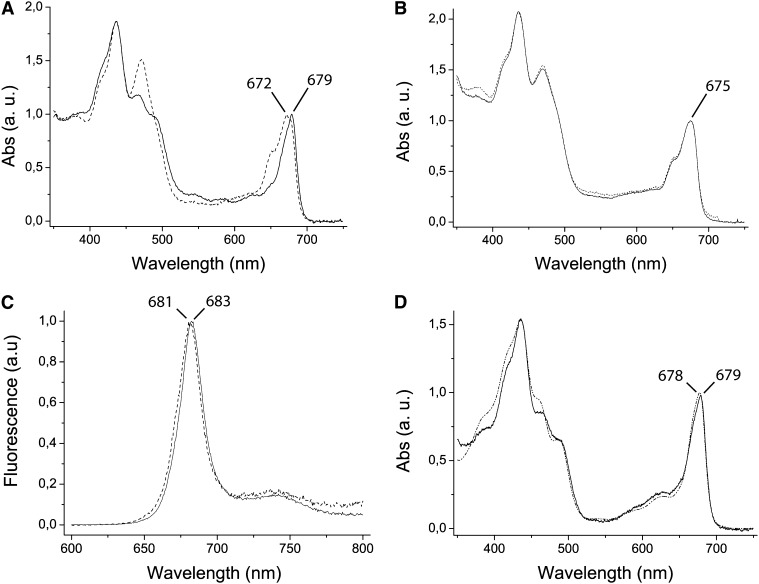
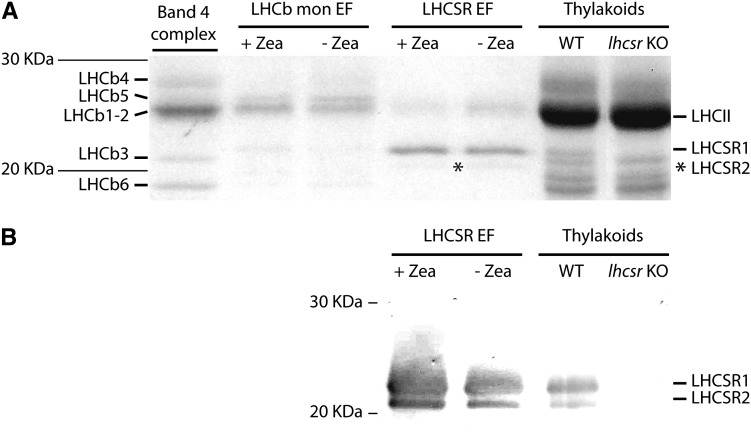
When comparing the zeaxanthin distribution pattern from wild-type excess light–treated samples, we observed an enrichment in fractions 23 to 27 of the Suc gradient (Figure 6B). Immunoblotting analysis of fractions using an antibody directed against C. reinhardtii-LHCSR protein (Bonente et al., 2011a) revealed that the same fractions were enriched in LHCSR proteins (Figure 6A), thus suggesting zeaxanthin might be a ligand for LHCSR. This hypothesis was verified by applying a fractionation method with high resolution in the low molecular mass range (i.e., nondenaturing gel electrophoresis). Upon separation of α-DM solubilized thylakoids from excess light–treated wild-type and lhcsr KO samples (Figure 7A), the middle part of the gel including both samples was blotted into a polyvinylidene fluoride (PVDF) membrane and the migration of the LHCSR protein was detected with an anti-Cr-LHCSR antibody. Faint reactions were detectable at different apparent molecular masses in both wild-type and lhcsr KO lanes, due to cross-reactions with members of LHC family as confirmed by their presence in both samples. The only differential reaction was observed at ∼25 kD, consistent with location of LHCSR in the upper fractions of the Suc gradient (Figure 6). The remaining part of the gel was used for detailed fractionation: To this aim, the 20- to 100-kD molecular mass range, including LHC monomers and trimers, was cut into thin slices that were eluted in a buffer solution containing 0.03% α-DM. The eluate from each slice was submitted to HPLC pigment analysis, absorption spectroscopy, and SDS-PAGE/immunoblotting. The monomeric LHCb green band extended toward lower molecular mass in the wild type compared with the lhcsr KO lane (Figure 7B). Densitometry of the Deriphat-PAGE showed that the monomeric band contained 20% more chlorophyll in the wild type versus lhcsr KO. Anti-LHCSR immunoreaction was clearly located in the same lower molecular mass range of the gel (Figure 7A), implying the LHCSR protein migrated at that position as a green band. HPLC analysis and absorption spectra of the fractions eluted from this gel region showed the presence of a complex with very low chlorophyll b content (Figure 7C) and characterized by a Qy absorption peak red-shifted to 679 nm versus 672 of the corresponding area of the gel in lhcsr KO lane and 675 nm in the bulk monomeric fractions from either the wild type of lhcsr KO (Figures 8A and 8B) and a small red shift in fluorescence emission spectra (Figure 8C). It is interesting to note that this spectrum closely matches that of the C. reinhardtii recombinant LHCSR reconstituted in vitro with pigments (Bonente et al., 2011a) (Figure 8D). When the individual bands eluted from the gel were run in denaturing SDS-PAGE and immunoblotted with an anti-CrLHCSR antibody, it clearly appeared that reactive bands were present in the same fractions from the wild type but not from lhcsr KO (Figure 7C) where, instead, only a faint reaction in the position of LHCII apoprotein, likely a cross-reaction with LHCSR, was observed. HPLC analysis showed that fraction 1 from the wild type has a high chlorophyll a/b ratio and is enriched in zeaxanthin (Figure 7C), together with lutein and violaxanthin. Neoxanthin, a major component of LHCb proteins, was absent. It is interesting to note that the zeaxanthin/chlorophyll ratio increased threefold in fractions with lower molecular mass within the monomeric LHCb band (fractions 1 to 3 versus 4 and 5) and that this effect was accompanied by an increase of the chlorophyll a/b ratio. Both these effects were absent in the corresponding fractions from lhcsr KO. The above results strongly suggest that native LHCSR from P. patens is a chlorophyll a/chlorophyll b/xanthophyll binding protein exchanging its bound violaxanthin with zeaxanthin upon exposure to excess light. Yet, we cannot fully exclude that components other than LHCSR might be involved. In order to verify this issue, we performed a complementary experiment: Thylakoids, either from control plants or loaded with zeaxanthin by incubating at low pH and ascorbate as reported in Methods, were fractionated by Deriphat-PAGE. The two moieties of the monomeric LHC band were excised from the gel using as a reference the migration pattern in lhcsr KO in order to separate a monomeric LHCb-enriched fraction from the LHCSR-enriched fraction. These were analyzed by SDS-PAGE for polypeptide composition, HPLC for pigment content, and immunoblotting for identification of LHCSR polypeptides (Figure 9; see Supplemental Table 2 online). Coomassie blue staining (Figure 9A) and the strong reaction of anti-LHCSR antibody (Figure 9B) showed that LHCSR1 and LHCSR2 polypeptides are the major components of the LHCSR-enriched fraction with only a small contamination from LHCII. The monomeric LHCb-enriched fractions still contains small amounts of LHCSR but are enriched in LHCb5, LHCb1-2, LHCb3, and LHCb6, as assessed by immunoblotting with specific antibodies (data not shown) and by comparing the comigration with band 4 complex, containing LHCII, LHCb4, and LHCb6 (Bassi and Dainese, 1992). Absorption spectra of LHCSR-enriched and LHCb-enriched fractions were as shown in Figure 8A. HPLC analysis showed low chlorophyll/carotenoid ratio (3.0 to 3.5) in LHCSR-enriched fractions versus monomeric LHCbs (5.0 to 5.3) consistent with a lower number of chlorophylls per polypeptide and/or a higher number of carotenoid ligands in the former samples. About half of violaxanthin was accessible to deepoxidation, consistent with the presence of two distinct violaxanthin binding sites in LHCSR (Bonente et al., 2011a). An additional feature of this protein can be inferred from the absence of any reaction in fractions of higher apparent molecular mass in the gel (6 to 21 in Figure 7), implying LHCSR is a monomer.
Figure 7.
LHCSR Localization among Pigment Binding Complexes of P. patens.
(A) Nondenaturing gel electrophoresis of pigment binding complexes from thylakoids of wild-type (WT) and lhcsr KO plants after solubilization with 0.8% α-DM. Plants were treated for 30 min with excess light (24°C, 850 µmol photons m−2 s−1) before thylakoid extraction. Chlorophylls (150 µg) were loaded for each sample. The stacking gel contained 3.5% (w/v) acrylamide, and the resolving gel contained 7% (w/v) acrylamide (48:1.5 acrylamide/bis-acrylamide). Different pigment binding complexes, identified from their mobility, are indicated on the left. The central part of the gel was transferred on a PVDF membrane for immunoblotting analysis using an antibody raised against LHCSR. The image of the membrane is superposed on the picture of the green gel. The position of gel slices excised from the nondenaturing gel is shown on the right side.
(B) Densitometric analysis of the non denaturing gel electrophoresis shown in (A). The wild type, black line; lhcsr KO, red line. Signals are normalized to sample number 20, which corresponds to the maximum intensity in the region of LHCII trimers. A black arrow indicates the region at low molecular mass in which LHCSR accumulates. a.u., arbitrary units.
(C) Immunoblotting and pigment profiles of the proteins eluted from the nondenaturing gel shown in (A) and separated in second dimension under denaturing conditions. The numbering of fractions reported on the top of the each filter corresponds to that of gel slices cut from the nondenaturing gel. LHCII and LHCSR position in denaturing gels is reported on the left side of each filter. The genotype is indicated on the right side. Chlorophyll a/b ratio and values of zeaxanthin/100 chlorophyll ratios are indicated underneath each filter.
Figure 8.
Spectroscopy Analysis of Native Proteins Purified from Deriphat-PAGE and Recombinant LHCSR.
(A) Absorption spectrum of fraction 1 eluted from the gel. Sample containing LHCSR protein from thylakoids of excess light–treated wild-type P. patens plants (continuous line) is compared with the corresponding fraction purified from lhcsr KO solubilized thylakoids (pointed line). The LHCSR-containing fractions from the wild type had absorption spectra strongly contrasting with those from lhcsr KO thylakoids. a.u., arbitrary units.
(B) Absorption spectrum of fraction 5, corresponding to LHC monomers of the wild type (continuous line) and lhcsr KO (pointed line).
(C) Fraction 1 fluorescence emission at 300K with excitation at 440 nm of the wild type (continuous line) and lhcsr KO (pointed line).
(D) Absorption spectrum of fraction 1 eluted from the gel and corrected for scattering. Sample containing LHCSR protein from thylakoids of wild-type excess light–treated P. patens plants (continuous line) is compared with the spectrum of recombinant LHCSR3 from C. reinhardtii (broken line; redrawn from Bonente et al., 2011a).
Figure 9.
Analysis of Fractions Isolated from Thylakoids upon Fractionation by Native PAGE or Suc Gradient Ultracentrifugation.
(A) Coomassie blue–stained SDS-PAGE gel of band 4 complex (LHCII-LHCb4-LHCb6 complex) isolated by Suc gradient as previously described (Betterle et al., 2009). LHCb monomer-enriched fraction (LHCb mon EF) and LHCSR-enriched fraction (LHCSR EF) were purified as described in Figure 7, either from thylakoids isolated under standard conditions at pH 7.8 (-Zea) or thylakoids treated at pH 5.1 to convert violaxanthin into zeaxanthin (+Zea). Thylakoids from the wild type (WT) and lhcsr KO were loaded as control for LHCSR mobility.
(B) Immunoblot analysis of moss preparations as in (A) probed with an anti-LHCSR antibody specific to P. patens LHCSR.
DISCUSSION
Zeaxanthin accumulates in excess light conditions in many photosynthetic organisms and has been shown to upregulate the efficiency of a number of photoprotective mechanisms, including NPQ (Demmig-Adams et al., 1989; Esteban et al., 2009), ROS scavenging (Dall’Osto et al., 2010), 3Chl* yield (Dall’Osto et al., 2012), and thylakoid membrane reorganization (Betterle et al., 2009), which together contribute to increase the resistance to excess light. The involvement of zeaxanthin in the different components of photoprotection does not appear to be the same in all organisms (Esteban et al., 2009), so that the role of master switch that zeaxanthin assumes in vascular plants (Morosinotto et al., 2003) is likely to result from the convergence of zeaxanthin dependence of distinct regulatory mechanisms. In this article, we provided three lines of evidence showing that zeaxanthin synthesis is crucial for photoprotection in P. patens. First, we showed that zeaxanthin accumulation is induced under exposure to abiotic stress, such as short excess light treatment, and that it plays an important role in the amplitude of NPQ (Table 2, Figure 2). Second, we generated vde KO mutants, which completely lose their capacity to convert violaxanthin into zeaxanthin, demonstrating that the product of a single VDE gene is essential for violaxanthin-to-zeaxanthin conversion under oxidative stress conditions. Finally, we showed that both the PSBS- and the LHCSR-dependent mechanisms activating NPQ in P. patens depend on zeaxanthin (Figure 2; see Supplemental Figure 4 online) and VDE mutants are impaired in their capacity to resist excess light treatments (Figures 5). The very high level of zeaxanthin dependence for NPQ activity is of particular interest in the light of the preeminent role of LHCSR versus PSBS in triggering this process in P. patens. Analysis of vde KO genotypes confirms preliminary indication of the importance of zeaxanthin for NPQ in P. patens from the effect of exposure to excess light (Table 2) (Alboresi et al., 2008; Gerotto et al., 2011). Moreover, it appears that both PSBS- and LHCSR-dependent NPQ mechanisms are dependent on zeaxanthin. In fact, P. patens is the only organism for which independent activity of PSBS and LHCSR has been fully demonstrated (Alboresi et al., 2010; Gerotto et al., 2012), and NPQ amplitude was strongly decreased in both psbs vde KO and lhcsr vde KO mutants (Figure 2), implying that both PSBS- and LHCSR-dependent quenching mechanisms need zeaxanthin for full activity. While this is a well-known property of PSBS-dependent NPQ, as supported by the phenotype of the npq1 Arabidopsis mutant (Niyogi et al., 1998; Bonente et al., 2008a), heat dissipation activity exhibits little (Niyogi et al., 1997) or no (Bonente et al., 2011a) dependence on zeaxanthin in C. reinhardtii, where the role of LHCSR in NPQ was first demonstrated (Peers et al., 2009). It should be noted that recombinant Cr-LHCSR can bind zeaxanthin and yet its fluorescence decay properties are only weakly affected by deepoxidation state of its xanthophyll cycle ligands (Bonente et al., 2011a) consistent with the absence of a bona fide VDE encoding gene, although C. reinhardtii is able to accumulate zeaxanthin in high light (Grossman et al., 2010). Here, we show that LHCSR quenching activity is highly dependent on zeaxanthin (Figure 2C). In P. patens, the upregulation by zeaxanthin of the LHCSR-dependent NPQ component is far stronger than the complementary PSBS-dependent component, leading to a strong control of zeaxanthin over NPQ to a level unprecedented in green algae and plants (Figures 2B and 2C). High zeaxanthin dependence of NPQ amplitude was reported in diatoms (Coesel et al., 2008; Nymark et al., 2009; Bailleul et al., 2010). While NPQ is decreased in a LHCSR knockdown genotype (lhcx1), clear evidence that LHCSR is the only agent for NPQ is lacking in these algae (Lavaud et al., 2012).
Molecular Basis for the Strong Dependence of qE on Zeaxanthin
NPQ is a composite phenomenon whose major component qE is rapidly reversible and depends on a transmembrane pH gradient. In vascular plants, zeaxanthin-dependent enhancement of qE has been proposed to be elicited by its binding to the L2 site of monomeric LHCb proteins. This allosteric binding induces conformational changes that lead to a new chromophore–chromophore interaction favoring the formation of intramolecular quenching sites (Ahn et al., 2008; Avenson et al., 2008; Ruban et al., 2007; Ballottari et al., 2010; Miloslavina et al., 2011) and the reorganization of PSII supercomplexes in thylakoid membranes (Betterle et al., 2009; Johnson et al., 2011). Analysis of the distribution of zeaxanthin in P. patens thylakoid membrane complexes from excess light–treated plants shows that most (75.6%) of the newly synthesized zeaxanthin is found as a complex with LHCb proteins, PSI and PSII supercomplexes (Figure 6). When compared with previous work on Arabidopsis, we observed on one hand that the extent of conversion of violaxanthin to zeaxanthin is higher in P. patens (de Bianchi et al., 2008) and, on the other hand, that LHCSR migrates as a monomer in the upper part of the Suc gradient (Figure 6A). This finding prompted us to isolate the LHCSR protein from excess light–treated plants by preparative Deriphat-PAGE. The LHCSR-enriched fraction has absorption spectrum closely resembling recombinant LHCSR from C. reinhardtii, including a red shift of the Qy transition peak (Bonente et al., 2011a; Figure 8A), a high chlorophyll a/b ratio of 5.1, and the binding of lutein, violaxanthin, antheraxanthin, and zeaxanthin but not neoxanthin (Bonente et al., 2011a; Figure 8B). Based on a chlorophyll complement of 7 to 8 per polypeptide (Bonente et al., 2011a), three xanthophyll binding sites one for lutein and two for violaxanthin/antheraxanthin/zeaxanthin are likely to be present in ppLHCSR. Deepoxidation index is 0.66 versus 0.58 in bulk monomeric LHCs, and the fraction of xanthophyll complement made by xanthophyll cycle pigments is 60% versus 23 to 29% in bulk monomeric LHCs. Thus, ppLHCSR appears to be highly enriched in xanthophyll cycle pigments, undergoing high levels of deepoxidation with respect to the overall LHCb antenna complement. Densitometric analysis of SDS-PAGE fractionation and immunoblotting showed that contamination of the LHCSR-enriched fraction by comigrating LHCII (Figure 9) was not more than 20%. Since LHCII has 4 times lower xanthophyll-cycle carotenoid content per chlorophyll due to a higher number of chlorophyll ligands per polypeptide and higher lutein content (Liu et al., 2004) with respect to LHCSR (Bonente et al., 2011a; see Supplemental Figure 2) the hypothesis of zeaxanthin binding to monomeric LHCbs rather than to LHCSR is inconsistent with the experimental data. In addition, the zeaxanthin binding properties of LHCSR is supported by the characteristics of fractions isolated from lhcsr KO thylakoids; these are far less abundant and bear contrasting features (i.e., high chlorophyll b content, blue-shifted absorption peak, and the presence of neoxanthin). It should be noted that zeaxanthin binding does not activate quenching sites in C. reinhardtii LHCSR either in vivo or in vitro (Bonente et al., 2011a), while both zeaxanthin and LHCSR are needed for qE activation in P. patens. This implies that C. reinhardtii and P. patens differ for the effect of zeaxanthin binding to LHCSR in enhancing quenching. Thus, photoprotection by scavenging ROS (Baroli et al., 2003) and by heat dissipation of excess energy (Peers et al., 2009) are independently activated in C. reinhardtii. Instead, mosses coordinately control these mechanisms through the binding of zeaxanthin to different binding sites. Zeaxanthin binding to monomeric Lhcb proteins was shown to downregulate 3Chl* quenching and ROS scavenging (Dall’Osto et al., 2012), and yet in the absence of PSBS, no quenching can be induced (Li et al., 2000). Similarly, qE in P. patens is fully dependent on LHCSR and/or PSBS (Alboresi et al., 2010; Figure 2). We conclude that during evolution from an aquatic unicellular free-living organism, C. reinhardtii, to a plant pioneering colonization to the subaerial environment, distinct photoprotection mechanisms became integrated through the involvement of zeaxanthin in activation of NPQ (Morosinotto et al., 2003).
Enhancement of LHCSR- versus PSBS-Dependent Components
It can be asked whether the quenching mechanism is the same for both the LHCSR- and PSBS-dependent qE components based on the lower zeaxanthin dependence shown by genotypes expressing PSBS versus those expressing LHCSR. Since PSBS, as isolated from thylakoid membranes, is not a pigment binding protein (Dominici et al., 2002; Bonente et al., 2008b), quenching in vascular plants was proposed to occur in LHCb proteins upon a conformational change elicited by the protonation of the interacting PSBS subunit (Bonente et al., 2008a), zeaxanthin dependence being conferred by its binding to LHCb proteins. In P. patens, the level of zeaxanthin binding to LHC proteins is higher than in plants (Figure 6, Table 2), thus suggesting that binding of zeaxanthin to LHCb proteins is not a limiting factor for either the amplitude of qE or its zeaxanthin-dependent enhancement (Gerotto et al., 2012; Figure 3). Thus, the low effect of zeaxanthin on PSBS-dependent NPQ component is consistent with its binding to LHCb proteins only, where it favors transition to a quenching conformation (Moya et al., 2001; Avenson et al., 2008; Ruban et al., 2012) while the LHCSR-dependent component, in addition, relies on binding to the LHCSR protein. At present we cannot state whether LHCSR acts uniquely through developing internal quenching sites (Bonente et al., 2011a) or if it can also induce quenching in neighbor LHCb proteins similar to PSBS. The low NPQ resulted upon depleting LHCbm1 (Elrad et al., 2002; Ferrante et al., 2012) is consistent with the second hypothesis. In this case, the effect of zeaxanthin binding to LHCSR and LHCb proteins (Figures 6 and 8) may yield a cooperative response and enhanced zeaxanthin dependence.
Other Photoprotective Activities of Zeaxanthin besides Quenching
Long-term light stress is more photoinhibiting on vde KO than on lhcsr psbs KO (Figure 4) despite the fact that the former has higher NPQ activity (Figure 2D), implying that zeaxanthin has additional photoprotective effects besides energy quenching. Previous work on C. reinhardtii (Baroli et al., 2003) and Arabidopsis (Havaux and Niyogi, 1999) showed an enhanced ROS scavenging activity of zeaxanthin with respect to preexisting violaxanthin both in the lipid phase (Havaux et al., 2007) and bound to pigment-proteins (Dall’Osto et al., 2010). Recent work has shown a photoprotective ROS-scavenging activity of zeaxanthin under salt and osmotic stress resistance in P. patens (Azzabi et al., 2012). On this basis, our results can be interpreted with the presence of zeaxanthin-dependent photoprotective ROS-scavenging activity, which confers increased resistance to photodamage to lhcsr psbs KO versus vde KO. This appears to be an early photoprotection mechanism of zeaxanthin conserved through the evolution up to higher plants.
Conclusions
We applied targeted insertion mutagenesis to attribute VDE activity to the product of the Pp1s161_120V6 gene in P. patens. The resulting KO mutant showed increased photosensitivity because of the lack of at least two components: first, an NPQ-independent activity, such as enhancement of ROS scavenging by zeaxanthin; and second, enhancement of multiple NPQ activities by zeaxanthin. The effect of zeaxanthin on PSBS-dependent NPQ is similar to that previously observed in plants (Niyogi et al., 1998). However, by contrast, the effect of zeaxanthin on LHCSR-dependent NPQ is far stronger. A search for the basis of the high zeaxanthin dependence of NPQ led to the identification of a new zeaxanthin binding site in the LHCSR protein upon its isolation in the form of native chlorophyll a/b–xanthophyll binding protein. Properties of native LHCSR closely fit those previously reported for recombinant C. reinhardtii-LHCSR whose activity, however, was zeaxanthin independent (Bonente et al., 2011a). In P. patens, the interplay of two NPQ mechanisms, triggered respectively by PSBS and LHCSR, with different levels of zeaxanthin dependence provides an extremely flexible NPQ, an essential feature for its response in the land environment characterized by highly fluctuating light.
METHODS
Plant Materials and Growth Conditions
Physcomitrella patens subsp patens was grown in controlled-environment chambers with 16 h of light (50 μmol photons m−2 s−1) and 8 h of dark at 24°C either on the minimal medium (PpNO3) or on the same medium supplemented with 0.5% Glc and 5 mM diammonium (+)-tartrate (rich medium, PpNH4) as previously described (Ashton et al., 1979). For excess light treatments, protonema grown on PpNO3 at 24°C and 50 μmol photons m−2 s−1 were transferred to 850 μmol photons m−2 s−1. The whole physiological characterization was performed on plants grown on PpNO3, while routine plant amplification was done on PpNH4.
In Vivo fluorescence measurements
Whenever a precise NPQ kinetic was needed, in vivo chlorophyll fluorescence was measured in P. patens plants at room temperature with a Dual PAM-100 fluorimeter (Heinz Walz), with a saturating light of 6000 μmol photons m−2 s−1 and an actinic red light of 830 μmol photons m−2 s−1. Actinic light intensity during the measurement was chosen to be just sufficient to saturate photosynthesis in control plants and ensure maximal NPQ amplitude. Plants were dark adapted for 40 min at room temperature before measurement. For DTT treatment, moss tissues were treated with 5 mM DTT in HEPES/sorbitol buffer during the 40 min of dark adaptation. The parameters Fv/Fm and NPQ were calculated as (Fm − Fo)/Fm and (Fm − Fm′)/Fm′ (Genty et al., 1989). Mutant screening and Fv/Fm under stress was monitored by chlorophyll fluorescence imaging with a Closed FluorCam FC 800MF (Photon Systems Instruments).
Chlorophyll and Carotenoid Analysis
Small pieces of 2 to 5 mm of diameter of protonema tissue were frozen in liquid nitrogen and grinded using pestles for 1.5-mL microcentrifuge tubes. Pigments were extracted in 0.5 mL of 85% acetone and analyzed by HPLC after two steps of centrifugation at maximum speed for 15 min at 4°C (Gilmore and Yamamoto, 1991).
Immunoblotting Analysis
SDS-PAGE analyses were performed as in by Laemmli (1970) with some modifications in order to separate LHCSR1 from LHCSR2. We used an acrylamide/bis-acrylamide ratio of 75: 1 and a total concentration of acrylamide of 4.5 and 15%, respectively, for stacking and running gel. Urea (6 M) was also added into the running gel. After SDS-PAGE, polypeptides were transferred onto an Immobilon PVDF membrane (Millipore) using a Mini Trans-Blot cell (Bio-Rad) and detected by specific antibodies generated in the lab.
Generation of Transgenic P. patens Plants
Genomic DNA was isolated from P. patens protonema (Allen et al., 2006) and used as a template for all molecular cloning. Two regions inside the VDE coding sequence (locus Pp1s161_120V6) were amplified by PCR and subcloned into pGEM-T Vector (catalog number A3600; Promega). The sequence of the primers used for molecular cloning is detailed in Supplemental Figure 1 online. These regions were cloned up- and downstream of the genes conferring either hygromycin B or zeocin resistance, respectively, into BHRf and BZRf plasmids (kindly provided by F. Nogue, Institut National de la Recherche Agronomique, Versailles, France). P. patens transformation was performed as described (Schaefer and Zrÿd, 1997) with minor modifications. Briefly, 5-d-old protonemal tissue was collected for protoplast generation and polyethylene glycol–mediated transformation. Primary resistant plants were selected on growth media containing 25 μg·L−1 hygromycin B (catalog number A2175; Applichem) or 50 μg·L−1 zeocin (Duchefa Biochemie). Stable transformants were successively selected for further molecular characterization as detailed in Supplemental Figure 1 online. Simple vde KO mutants were isolated by transforming wild-type plants, and multiple vde KO mutants were obtained by transforming psbs KO and lhcsr1 lhcsr2 KO (also called lhcsr KO).
Thylakoid Isolation and Pigment Binding Complexes Purification
Thylakoids were purified from protonemal tissue of P. patens plants following the same protocol used for seed plants with minor modifications (Bassi and Simpson, 1987). Tissues were harvested and freshly homogenized in cold extraction buffer (0.5% milk powder, 0.4 M NaCl, 0.02 M Tricine-KOH, pH 7.8, and 0.005M ε-aminocaproic acid, 0.001 phenylmethylsulfonyl fluoride and 0.001 benzamidine as protease inhibitors). After filtration, samples were precipitated by centrifugation at 1500g for 15 min at 4°C and then resuspended in hypotonic buffer (0.15 M NaCl, 0.005 M MgCl2, 0.02 M Tricine-KOH, pH 7.8, and protease inhibitor). After centrifugation for 10 min at 10,000g at 4°C, thylakoids were resuspended in a buffer containing 0.4 M sorbitol, 0.015 M NaCl, 0.005 M MgCl2, and 0.01 M HEPES-KOH, pH 7.5. Finally, thylakoids were either used directly or frozen in liquid nitrogen and stored at –80°C until use.
Thylakoid membranes corresponding to 500 µg of chlorophylls were washed with 5 mM EDTA and then resuspended in 0.5 mL of 10 mM solubilized HEPES, pH 7.5. Samples were then solubilized by adding 0.5 mL of 1.6% α-DM and 10 mM HEPES, pH 7.5, by vortexing for 1 min. The solubilized samples were kept 10 min in ice and then centrifuged at 15,000g for 10 min to eliminate unsolubilized material. The fractionation occurred by ultracentrifugation on a 0.1 to 1 M Suc gradient containing 0.03% α-DM and 10 mM HEPES, pH 7.5 (22 h at 40,000g at 4°C). The green bands of the Suc gradient were harvested continuously using a syringe needle connected to a peristaltic pump.
Violaxanthin Deepoxidation in Vitro
Thylakoids were diluted to 50 μg/mL of chlorophylls using a solution containing 330 mM sorbitol, 5 mM MgCl2, 10 mM NaCl, 40 mM MES/NaOH, pH 5.1, 20 mM ascorbate, 0.1% (w/v) BSA, and protease inhibitors. Thylakoids were stirred for 2 h at room temperature and then centrifuged at 13,000 rpm for 10 min at 4°C. The pellet was resuspended at 1 mg/mL of chlorophyll and solubilized with 0.8% α-DM before loaded in a nondenaturing Deriphat-PAGE.
Thylakoid Step Solubilization
Step solubilization of thylakoid membranes was performed by injecting the detergents into a cuvette in which the fluorescence was measured in the continuously stirred sample. The same procedure was already described (Dall’Osto et al., 2005), and we optimized the conditions for P. patens thylakoids. The excitation light at the sample level was of 16 µmol photons m−2 s−1 (excitation wavelength, 440 nm; emission wavelength, 680 nm). Unstacked thylakoids corresponding to a final chlorophyll concentration of 0.5 µg/mL were added into the cuvette, and two different detergents were added: first, α-DM at final 0.075% (w/v), then Triton X-100 at final 1.8% (w/v). Quenching and fluorescence parameters were defined as previously published (Dall’Osto et al., 2005). Ft corresponds to thylakoid fluorescence (unsolubilized membranes) at 1 min from the start of measurement, Fd is pigment-protein complexes fluorescence after 2.5 min of solubilization in α-DM, and Fx is free-pigment fluorescence measured after 5 min of solubilization in 1.8% Triton X-100. Quenching parameters are calculated as follows: Qm = (Fd − Ft)/Ft describes the quenching of chlorophyll fluorescence due to protein–protein and lipid–protein interactions in the membrane; Qpi = (Fx − Fd)/Fd is the quenching of free chlorophylls provided by their binding into chlorophyll-protein complexes; and Qm + pi = (Fx − Ft)/Ft is the total quenching of free pigment fluorescence.
Deriphat-PAGE
Nondenaturing Deriphat-PAGE was performed following the method previously developed (Peter et al., 1991) with the following modifications: The stacking gel contained 3.5% (w/v) acrylamide (38:1 acrylamide/bis-acrylamide) and the resolving gel contained 7% (w/v) acrylamide. Thylakoids concentrated at 1 mg/mL of chlorophyll were solubilized with 0.8% α-DM, and 120 μg of chlorophylls were loaded in each lane.
Absorption Spectroscopy
Steady state spectra were obtained directly on samples collected from the Suc gradients and diluted if needed with 10 mM HEPES, pH 7.5, 0.03% α-DM, and 0.5 M Suc. Native chlorophyll binding complexes were extracted from the acrylamide gel by grinding the slices in a buffer containing 10 mM HEPES, pH 7.5, and 0.03% α-DM. Absorption measurements were performed using an SLM-Aminco DW-2000 spectrophotometer at room temperature.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: DS544900 (Pp1s11_269V6, LCYE1), DS545042 (Pp1s153_133V6, LCYE2), DS545196 (Pp1s307_64V6, CYP97C1), DS544917 (Pp1s28_409V6, CYP97A3), DS544944 (Pp1s55_202V6, LCYB1), DS545049 (Pp1s160_155V6, LCYB2), DS545260 (Pp1s373_23V6, CHYB1), DS544908 (Pp1s19_96V6, CHYB2), DS544909 (Pp1s20_24V6, CHYB3), DS544980 (Pp1s91_16V6, ZEP1), DS545108 (Pp1s219_79V6, ZEP2), DS545050 (Pp1s161_120V6, VDE), DS544997 (Pp1s108_75V6, NSY1), DS544985 (Pp1s96_157V6, NSY2), DS545130 (Pp1s241_86V6, PSBS), DS545102 (Pp1s213_80V6, LHCSR1), and DS544988 (Pp1s99_95V6, LHCSR2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Representation of the VDE Gene and the Corresponding vde KO Mutant and Preliminary Analysis of the Isolated Mutants.
Supplemental Figure 2. Biochemical Characterization of Wild-Type, vde KO, psbs vde KO, and lhcsr vde KO Mutant Mosses Grown under Standard Conditions.
Supplemental Figure 3. NPQ Induction and Relaxation of Wild-Type, vde KO, psbs vde KO, and lhcsr vde KO Mutant Mosses Grown under Standard Conditions.
Supplemental Figure 4. Effect of vde KO Mutation and DTT on qE of Various P. patens Genotypes.
Supplemental Table 1. Xanthophyll Cycle Pigment Composition and Deepoxidation Index Determination of Wild-Type Physcomitrella patens Plants Treated with Excess Light and DTT.
Supplemental Table 2. Pigment Composition of Chlorophyll-Proteins Purified from Thylakoids of Wild-Type and lhcsr KO Physcomitrella patens Plants Treated with Excess Light.
Acknowledgments
This work was supported by European Commission EUFP7 SUNBIOPATH-GA-245070, the Cariverona foundation “Biomasse di Oggi e di Domani, and by project ACCLIPHOT PITN-GA-2012-316427.” We thank Ghazi Azzabi for his help during the early phase of plasmid preparation for the isolation of vde KO.
AUTHOR CONTRIBUTIONS
A.P. performed biochemical and physiological characterization of wild-type and vde KO mutants. C.G. performed NPQ measurements with PAM-100. L.D. was involved in the thylakoid step solubilization experiment, the photoinhibition experiment, data analysis, and critical review of the article. T.M. contributed to initial design of experiments and critical reviewing the article. R.B. coordinated the experiments and wrote the article. A.A. isolated and characterized the vde KO mutants, coordinated the experiments, and wrote the article.
Glossary
- PSII
photosystem II
- PSI
photosystem I
- ROS
reactive oxygen species
- LHC
light-harvesting complex
- KO
knockout
- α-DM
n-dodecyl-α-d-maltoside
- PVDF
polyvinylidene fluoride
References
- Ahn T.K., Avenson T.J., Ballottari M., Cheng Y.-C., Niyogi K.K., Bassi R., Fleming G.R. (2008). Architecture of a charge-transfer state regulating light harvesting in a plant antenna protein. Science 320: 794–797 [DOI] [PubMed] [Google Scholar]
- Alboresi A., Caffarri S., Nogue F., Bassi R., Morosinotto T. (2008). In silico and biochemical analysis of Physcomitrella patens photosynthetic antenna: Identification of subunits which evolved upon land adaptation. PLoS ONE 3: e2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alboresi A., Gerotto C., Cazzaniga S., Bassi R., Morosinotto T. (2011). A red-shifted antenna protein associated with photosystem II in Physcomitrella patens. J. Biol. Chem. 286: 28978–28987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alboresi A., Gerotto C., Giacometti G.M., Bassi R., Morosinotto T. (2010). Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proc. Natl. Acad. Sci. USA 107: 11128–11133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen G.C., Flores-Vergara M.A., Krasynanski S., Kumar S., Thompson W.F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1: 2320–2325 [DOI] [PubMed] [Google Scholar]
- Ashton N.W., Grimsley N.H., Cove D.J. (1979). Analysis of gametophytic development in the moss, Physcomitrella patens, using auxin and cytokinin resistant mutants. Planta 144: 427–435 [DOI] [PubMed] [Google Scholar]
- Avenson T.J., Ahn T.K., Zigmantas D., Niyogi K.K., Li Z., Ballottari M., Bassi R., Fleming G.R. (2008). Zeaxanthin radical cation formation in minor light-harvesting complexes of higher plant antenna. J. Biol. Chem. 283: 3550–3558 [DOI] [PubMed] [Google Scholar]
- Azzabi G., Pinnola A., Betterle N., Bassi R., Alboresi A. (2012). Enhancement of non-photochemical quenching in the Bryophyte Physcomitrella patens during acclimation to salt and osmotic stress. Plant Cell Physiol. 53: 1815–1825 [DOI] [PubMed] [Google Scholar]
- Bailleul B., Rogato A., de Martino A., Coesel S., Cardol P., Bowler C., Falciatore A., Finazzi G. (2010). An atypical member of the light-harvesting complex stress-related protein family modulates diatom responses to light. Proc. Natl. Acad. Sci. USA 107: 18214–18219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballottari M., Girardon J., Betterle N., Morosinotto T., Bassi R. (2010). Identification of the chromophores involved in aggregation-dependent energy quenching of the monomeric photosystem II antenna protein Lhcb5. J. Biol. Chem. 285: 28309–28321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber J., Andersson B. (1992). Too much of a good thing: Light can be bad for photosynthesis. Trends Biochem. Sci. 17: 61–66 [DOI] [PubMed] [Google Scholar]
- Baroli I., Do A.D., Yamane T., Niyogi K.K. (2003). Zeaxanthin accumulation in the absence of a functional xanthophyll cycle protects Chlamydomonas reinhardtii from photooxidative stress. Plant Cell 15: 992–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroli I., Gutman B.L., Ledford H.K., Shin J.W., Chin B.L., Havaux M., Niyogi K.K. (2004). Photo-oxidative stress in a xanthophyll-deficient mutant of Chlamydomonas. J. Biol. Chem. 279: 6337–6344 [DOI] [PubMed] [Google Scholar]
- Bassi, R. and Dainese, P. (1992). A supramolecular light-harvesting complex from chloroplast photosystem-II membranes. Eur. J. Biochem. 204: 317–26. [DOI] [PubMed]
- Bassi, R., Pineau, B., Dainese, P., and Marquardt, J. (1993). Carotenoid-binding proteins of photosystem II. Eur. J. Biochem. 212: 297–303. [DOI] [PubMed]
- Bassi, R. and Simpson, D. (1987). Chlorophyll-protein complexes of barley photosystem I. Eur. J. Biochem. 163: 221–230. [DOI] [PubMed]
- Betterle N., Ballottari M., Zorzan S., de Bianchi S., Cazzaniga S., Dall’osto L., Morosinotto T., Bassi R. (2009). Light-induced dissociation of an antenna hetero-oligomer is needed for non-photochemical quenching induction. J. Biol. Chem. 284: 15255–15266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonente G., Ballottari M., Truong T.B., Morosinotto T., Ahn T.K., Fleming G.R., Niyogi K.K., Bassi R. (2011a). Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biol. 9: e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonente G., Formighieri C., Mantelli M., Catalanotti C., Giuliano G., Morosinotto T., Bassi R. (2011b). Mutagenesis and phenotypic selection as a strategy toward domestication of Chlamydomonas reinhardtii strains for improved performance in photobioreactors. Photosynth. Res. 108: 107–120 [DOI] [PubMed] [Google Scholar]
- Bonente G., Howes B.D., Caffarri S., Smulevich G., Bassi R. (2008a). Interactions between the photosystem II subunit PsbS and xanthophylls studied in vivo and in vitro. J. Biol. Chem. 283: 8434–8445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonente G., Passarini F., Cazzaniga S., Mancone C., Buia M.C., Tripodi M., Bassi R., Caffarri S. (2008b). The occurrence of the psbS gene product in Chlamydomonas reinhardtii and in other photosynthetic organisms and its correlation with energy quenching. Photochem. Photobiol. 84: 1359–1370 [DOI] [PubMed] [Google Scholar]
- Caffarri S., Croce R., Breton J., Bassi R. (2001). The major antenna complex of photosystem II has a xanthophyll binding site not involved in light harvesting. J. Biol. Chem. 276: 35924–35933 [DOI] [PubMed] [Google Scholar]
- Cazzonelli C.I., Pogson B.J. (2010). Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 15: 266–274 [DOI] [PubMed] [Google Scholar]
- Coesel S., Oborník M., Varela J., Falciatore A., Bowler C. (2008). Evolutionary origins and functions of the carotenoid biosynthetic pathway in marine diatoms. PLoS ONE 3: e2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Osto L., Caffarri S., Bassi R. (2005). A mechanism of nonphotochemical energy dissipation, independent from PsbS, revealed by a conformational change in the antenna protein CP26. Plant Cell 17: 1217–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Osto L., Cazzaniga S., Havaux M., Bassi R. (2010). Enhanced photoprotection by protein-bound vs free xanthophyll pools: a comparative analysis of chlorophyll b and xanthophyll biosynthesis mutants. Mol. Plant 3: 576–593 [DOI] [PubMed] [Google Scholar]
- Dall’Osto L., Cazzaniga S., North H., Marion-Poll A., Bassi R. (2007). The Arabidopsis aba4-1 mutant reveals a specific function for neoxanthin in protection against photooxidative stress. Plant Cell 19: 1048–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Osto L., Holt N.E., Kaligotla S., Fuciman M., Cazzaniga S., Carbonera D., Frank H.A., Alric J., Bassi R. (2012). Zeaxanthin protects plant photosynthesis by modulating chlorophyll triplet yield in specific light-harvesting antenna subunits. J. Biol. Chem. 287: 41820–41834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bianchi S., Dall’Osto L., Tognon G., Morosinotto T., Bassi R. (2008). Minor antenna proteins CP24 and CP26 affect the interactions between photosystem II subunits and the electron transport rate in grana membranes of Arabidopsis. Plant Cell 20: 1012–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B., Winter K., Krüger A., Czygan F.C. (1989). Zeaxanthin synthesis, energy dissipation, and photoprotection of photosystem II at chilling temperatures. Plant Physiol. 90: 894–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici P., Caffarri S., Armenante F., Ceoldo S., Crimi M., Bassi R. (2002). Biochemical properties of the PsbS subunit of photosystem II either purified from chloroplast or recombinant. J. Biol. Chem. 277: 22750–22758 [DOI] [PubMed] [Google Scholar]
- Elrad D., Grossman A.R. (2004). A genome’s-eye view of the light-harvesting polypeptides of Chlamydomonas reinhardtii. Curr. Genet. 45: 61–75 [DOI] [PubMed] [Google Scholar]
- Elrad D., Niyogi K.K., Grossman A.R. (2002). A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell 14: 1801–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban R., Olano J.M., Castresana J., Fernández-Marín B., Hernández A., Becerril J.M., García-Plazaola J.I. (2009). Distribution and evolutionary trends of photoprotective isoprenoids (xanthophylls and tocopherols) within the plant kingdom. Physiol. Plant. 135: 379–389 [DOI] [PubMed] [Google Scholar]
- Farber A., Young A.J., Ruban A.V., Horton P., Jahns P. (1997). Dynamics of xanthophyll-cycle activity in different antenna subcomplexes in the photosynthetic membranes of higher plants (The relationship between zeaxanthin conversion and nonphotochemical fluorescence quenching). Plant Physiol. 115: 1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante P., Ballottari M., Bonente G., Giuliano G., Bassi R. (2012). LHCBM1 and LHCBM2/7 polypeptides, components of major LHCII complex, have distinct functional roles in photosynthetic antenna system of Chlamydomonas reinhardtii. J. Biol. Chem. 287: 16276–16288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genty B., Briantais J.-M., Baker N.R. (1989). The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 990: 87–92 [Google Scholar]
- Gerotto C., Alboresi A., Giacometti G.M., Bassi R., Morosinotto T. (2012). Coexistence of plant and algal energy dissipation mechanisms in the moss Physcomitrella patens. New Phytol. 196: 763–773 [DOI] [PubMed] [Google Scholar]
- Gerotto C., Alboresi A., Giacometti G.M., Bassi R., Morosinotto T. (2011). Role of PSBS and LHCSR in Physcomitrella patens acclimation to high light and low temperature. Plant Cell Environ. 34: 922–932 [DOI] [PubMed] [Google Scholar]
- Gilmore A.M., Yamamoto H.Y. (1991). Zeaxanthin formation and energy-dependent fluorescence quenching in pea chloroplasts under artificially mediated linear and cyclic electron transport. Plant Physiol. 96: 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A.R., Karpowicz S.J., Heinnickel M., Dewez D., Hamel B., Dent R., Niyogi K.K., Johnson X., Alric J., Wollman F.-A., Li H., Merchant S.S. (2010). Phylogenomic analysis of the Chlamydomonas genome unmasks proteins potentially involved in photosynthetic function and regulation. Photosynth. Res. 106: 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M., Dall’osto L., Bassi R. (2007). Zeaxanthin has enhanced antioxidant capacity with respect to all other xanthophylls in Arabidopsis leaves and functions independent of binding to PSII antennae. Plant Physiol. 145: 1506–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M., Niyogi K.K. (1999). The violaxanthin cycle protects plants from photooxidative damage by more than one mechanism. Proc. Natl. Acad. Sci. USA 96: 8762–8767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt N.E., Zigmantas D., Valkunas L., Li X.-P., Niyogi K.K., Fleming G.R. (2005). Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science 307: 433–436 [DOI] [PubMed] [Google Scholar]
- Horton P., Wentworth M., Ruban A. (2005). Control of the light harvesting function of chloroplast membranes: The LHCII-aggregation model for non-photochemical quenching. FEBS Lett. 579: 4201–4206 [DOI] [PubMed] [Google Scholar]
- Johnson M.P., Goral T.K., Duffy C.D.P., Brain A.P.R., Mullineaux C.W., Ruban A.V. (2011). Photoprotective energy dissipation involves the reorganization of photosystem II light-harvesting complexes in the grana membranes of spinach chloroplasts. Plant Cell 23: 1468–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalituho L., Beran K.C., Jahns P. (2007). The transiently generated nonphotochemical quenching of excitation energy in Arabidopsis leaves is modulated by zeaxanthin. Plant Physiol. 143: 1861–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol A.G., Borza T., Ishida K.-I., Keeling P., Lee R.W., Durnford D.G. (2007). Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms. Plant Physiol. 143: 1802–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lavaud J., Materna A.C., Sturm S., Vugrinec S., Kroth P.G. (2012). Silencing of the violaxanthin de-epoxidase gene in the diatom Phaeodactylum tricornutum reduces diatoxanthin synthesis and non-photochemical quenching. PLoS ONE 7: e36806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-P., Björkman O., Shih C., Grossman A.R., Rosenquist M., Jansson S., Niyogi K.K. (2000). A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Li X.-P., Gilmore A.M., Caffarri S., Bassi R., Golan T., Kramer D., Niyogi K.K. (2004). Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. J. Biol. Chem. 279: 22866–22874 [DOI] [PubMed] [Google Scholar]
- Li X.-P., Muller-Moule P., Gilmore A.M., Niyogi K.K. (2002). PsbS-dependent enhancement of feedback de-excitation protects photosystem II from photoinhibition. Proc. Natl. Acad. Sci. USA 99: 15222–15227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Yan H., Wang K., Kuang T., Zhang J., Gui L., An X., Chang W. (2004). Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428: 287–292 [DOI] [PubMed] [Google Scholar]
- Miller G., Shulaev V., Mittler R. (2008). Reactive oxygen signaling and abiotic stress. Physiol. Plant. 133: 481–489 [DOI] [PubMed] [Google Scholar]
- Miloslavina Y., de Bianchi S., Dall’Osto L., Bassi R., Holzwarth A.R. (2011). Quenching in Arabidopsis thaliana mutants lacking monomeric antenna proteins of photosystem II. J. Biol. Chem. 286: 36830–36840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morosinotto T., Caffarri S., Dall’Osto L., Bassi R. (2003). Mechanistic aspects of the xanthophyll dynamics in higher plant thylakoids. Physiol. Plant. 119: 347–354 [Google Scholar]
- Moya I., Silvestri M., Vallon O., Cinque G., Bassi R. (2001). Time-resolved fluorescence analysis of the photosystem II antenna proteins in detergent micelles and liposomes. Biochemistry 40: 12552–12561 [DOI] [PubMed] [Google Scholar]
- Nilkens M., Kress E., Lambrev P., Miloslavina Y., Müller M., Holzwarth A.R., Jahns P. (2010). Identification of a slowly inducible zeaxanthin-dependent component of non-photochemical quenching of chlorophyll fluorescence generated under steady-state conditions in Arabidopsis. Biochim. Biophys. Acta 1797: 466–475 [DOI] [PubMed] [Google Scholar]
- Niyogi K.K., Bjorkman O., Grossman A.R. (1997). Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell 9: 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi K.K., Grossman A.R., Björkman O. (1998). Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell 10: 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North H.M., De Almeida A., Boutin J.-P., Frey A., To A., Botran L., Sotta B., Marion-Poll A. (2007). The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J. 50: 810–824 [DOI] [PubMed] [Google Scholar]
- Nymark M., Valle K.C., Brembu T., Hancke K., Winge P., Andresen K., Johnsen G., Bones A.M. (2009). An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum. PLoS ONE 4: e7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peers G., Truong T.B., Ostendorf E., Busch A., Elrad D., Grossman A.R., Hippler M., Niyogi K.K. (2009). An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462: 518–521 [DOI] [PubMed] [Google Scholar]
- Peter G.F., Takeuchi T., Philip Thornber J. (1991). Solubilization and two-dimensional electrophoretic procedures for studying the organization and composition of photosynthetic membrane polypeptides. Methods 3: 115–124 [Google Scholar]
- Rensing S.A., Ick J., Fawcett J.A., Lang D., Zimmer A., Van de Peer Y., Reski R. (2007). An ancient genome duplication contributed to the abundance of metabolic genes in the moss Physcomitrella patens. BMC Evol. Biol. 7: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing S.A., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Ruban A.V., Berera R., Ilioaia C., van Stokkum I.H.M., Kennis J.T.M., Pascal A.A., van Amerongen H., Robert B., Horton P., van Grondelle R. (2007). Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature 450: 575–578 [DOI] [PubMed] [Google Scholar]
- Ruban A.V., Johnson M.P., Duffy C.D.P. (2012). The photoprotective molecular switch in the photosystem II antenna. Biochim. Biophys. Acta 1817: 167–181 [DOI] [PubMed] [Google Scholar]
- Ruban A.V., Lee P.J., Wentworth M., Young A.J., Horton P. (1999). Determination of the stoichiometry and strength of binding of xanthophylls to the photosystem II light harvesting complexes. J. Biol. Chem. 274: 10458–10465 [DOI] [PubMed] [Google Scholar]
- Schaefer D.G., Zrÿd J.P. (1997). Efficient gene targeting in the moss Physcomitrella patens. Plant J. 11: 1195–1206 [DOI] [PubMed] [Google Scholar]
- Takahashi S., Badger M.R. (2011). Photoprotection in plants: A new light on photosystem II damage. Trends Plant Sci. 16: 53–60 [DOI] [PubMed] [Google Scholar]