The soil salinity tolerance of an Arabidopsis mutant is shown to be caused by a mutation in the ETO1 gene that results in ethylene overproduction. Increased ethylene causes root stele reactive oxygen species (ROS)–dependent reductions in root Na influx and xylem loading and stelar ROS-independent enhancement of root K status, thus improving plant Na/K homeostasis and salinity tolerance.
Abstract
High soil Na concentrations damage plants by increasing cellular Na accumulation and K loss. Excess soil Na stimulates ethylene-induced soil-salinity tolerance, the mechanism of which we here define via characterization of an Arabidopsis thaliana mutant displaying transpiration-dependent soil-salinity tolerance. This phenotype is conferred by a loss-of-function allele of ETHYLENE OVERPRODUCER1 (ETO1; mutant alleles of which cause increased production of ethylene). We show that lack of ETO1 function confers soil-salinity tolerance through improved shoot Na/K homeostasis, effected via the ETHYLENE RESISTANT1–CONSTITUTIVE TRIPLE RESPONSE1 ethylene signaling pathway. Under transpiring conditions, lack of ETO1 function reduces root Na influx and both stelar and xylem sap Na concentrations, thereby restricting root-to-shoot delivery of Na. These effects are associated with increased accumulation of RESPIRATORY BURST OXIDASE HOMOLOG F (RBOHF)–dependent reactive oxygen species in the root stele. Additionally, lack of ETO1 function leads to significant enhancement of tissue K status by an RBOHF-independent mechanism associated with elevated HIGH-AFFINITY K+ TRANSPORTER5 transcript levels. We conclude that ethylene promotes soil-salinity tolerance via improved Na/K homeostasis mediated by RBOHF-dependent regulation of Na accumulation and RBOHF-independent regulation of K accumulation.
INTRODUCTION
Almost all important crop plants are sensitive to high soil salt concentrations (Frommer et al., 1999; Flowers, 2004). High soil salinity is therefore a severe problem in approximately one-third of the world’s irrigated agricultural land and is a major constraint on agricultural productivity (Greenway and Munns, 1980; Zhu, 2002; Munns and Tester, 2008). NaCl is the most soluble and widespread soil salt. High soil Na concentrations damage plants by causing increased tissue Na accumulation and K loss (Nassery, 1979; Greenway and Munns, 1980; Lynch and Läuchli, 1984; Zhu, 2002; Munns and Tester, 2008). Accordingly, plants have evolved a variety of salinity tolerance mechanisms, including mechanisms to restrict Na accumulation and K loss (Zhu, 2002; Smith et al., 2010). For example, in Arabidopsis thaliana, the product of the SALT OVERLY SENSITIVE1 (SOS1) gene, the plasma membrane Na+/H+ antiporter SOS1, transports Na+ accumulated in outer cell layers of the root back into soil solution, thereby reducing the net influx of Na+ into inner cell layers of the root and, in turn, root-to-shoot Na+ delivery (Shi et al., 2000, 2002; Zhu, 2002; Munns and Tester, 2008). Several other components (e.g., SOS2, SOS3, and the calcineurin B-like protein 10 [a putative calcium sensor]) (Liu and Zhu, 1998; J. Liu et al., 2000; Qiu et al., 2002; Quan et al., 2007) are also involved in the signaling pathways by which Na+ is eventually transported back out of the root, while the HIGH-AFFINITY K+ TRANSPORTER1 (HKT1) gene product (HKT1) can retrieve Na+ from the transpiration stream xylem sap before it reaches the shoot (Mäser et al., 2002; Sunarpi et al., 2005; Møller et al., 2009). High salinity also causes strong depolarization of the electrical potential difference across the plasma membrane, leading to tissue K+ loss via outward-rectifying K+ channels (Shabala and Cuin, 2008; Jayakannan et al., 2013), in turn impairing metabolic processes. Therefore, the ability of plants to retain tissue K concentration correlates with plant salinity tolerance (Maathuis and Amtmann, 1999; Chen et al., 2005; Cuin et al., 2008; Munns and Tester, 2008). Intriguingly, salinity tolerance can be achieved by manipulating the expression of both Na+ transporter genes (e.g., HKT1 and SOS1; Shi et al., 2003; Møller et al., 2009; Yang et al., 2009) and K+ transporter genes (e.g., ARABIDOPSIS K TRANSPORTER1 [AKT1]; Ardie et al., 2010), indicating that differential regulation of Na+ and K+ transporters, and thus of ionic homeostasis, can contribute significantly to salinity tolerance.
Soil-salinity stress causes in planta accumulation of reactive oxygen species (ROS), which can result in oxidative stress and cell damage (Miller et al., 2010; Smith et al., 2010). However, in addition to being a toxic agent in salinity stress, ROS also act as an important signaling mediator of plant salinity tolerance (Kaye et al., 2011; Jiang et al., 2012; Ma et al., 2011). For example, 5PTase7 regulates ROS production in saline conditions (likely through activation of NADPH oxidases), and Arabidopsis mutants lacking 5PTase7 activity are hypersensitive to salinity (Kaye et al., 2011). Our previous investigations indicated that RESPIRATORY BURST OXIDASE HOMOLOG F (RBOHF) encodes a specific isoform of NADPH oxidase (RBOHF) that plays a key role in soil-salinity tolerance (Jiang et al., 2012). Lack of RBOHF function causes a deficiency in salt-induced accumulation of ROS in the Arabidopsis root stele that results in increased Na concentrations both in root stelar cells and xylem sap in saline soil conditions, increased root-to-shoot Na delivery, increased shoot Na accumulation, and consequential soil-salinity hypersensitivity (Jiang et al., 2012). On the other hand, ROS (specifically the hydroxyl radical [OH∙]) may have adverse effects by activating root Na+-permeable nonselective cation channels (Demidchik et al., 2003; Foreman et al., 2003) and outward-rectifying K+ channels (Demidchik et al., 2010). These observations indicate that increased ROS levels play an important role in Na/K homeostasis and salinity tolerance. Since ROS production is influenced by many signals (e.g., biotic attack, abiotic stress, and hormonal signaling; Torres et al., 2002; Kwak et al., 2003; Marino et al., 2012), it is possible that additional factors regulate RBOHF in the mediation of soil-salinity tolerance. Here, we explore the possibility that the stress phytohormone ethylene is such a regulatory factor.
Phytohormones are known to play important roles in regulating ionic homeostasis and plant salt tolerance (e.g., Achard et al., 2006; Wu et al., 2008). For example, salt increases abscisic acid (ABA) levels and activates ABA-dependent signaling pathways (Zhu, 2002), in turn regulating transcriptome-level salt stress responses and physiological adaptation to salt stress (Xiong et al., 2001). Salicylic acid can prevent salinity-induced K loss, thus promoting Arabidopsis salinity tolerance (Jayakannan et al., 2013). Kinetin can inhibit K release into the xylem (Collins and Kerrigan, 1974; Hong and Sucoff, 1976), increase K+ channel-mediated root K uptake (Shabala et al., 2009), and thus alleviate the negative impact of salinity on plants (Tounekti et al., 2011). In addition, polyamines likely play important roles in salinity tolerance (Krishnamurthy and Bhagwat, 1989) and have been implicated in the regulation of ion channel activity (Brüggemann et al., 1998; K. Liu et al., 2000; Shabala et al., 2007; Pandolfi et al., 2010; Zepeda-Jazo et al., 2011). Finally, the stress phytohormone ethylene is also likely involved in the regulation of plant salt tolerance (Zhang et al., 2004; H. Wang et al., 2004; Cao et al., 2007). Ethylene binds to endoplasmic reticulum membrane–localized receptors (e.g., ETHYLENE RESISTANT1 [ETR1]; Chang et al., 1993), thus deactivating CONSTITUTIVE TRIPLE RESPONSE1 (CTR1; a putative mitogen-activated protein kinase kinase kinase; Kieber et al., 1993), in turn inhibiting the activity of a signal transduction cascade that targets the ETHYLENE INSENSITIVE3 (EIN3) transcription factor protein for destruction in the proteasome (Solano et al., 1998; Guo and Ecker, 2003; Potuschak et al., 2003; Yoo et al., 2008; Zhao and Guo, 2011). Accordingly, ethylene promotes the accumulation of EIN3 in the nucleus (Potuschak et al., 2003; Yoo et al., 2008) where EIN3 binds to the promoters of its target genes, thus triggering a diverse array of ethylene responses (Solano et al., 1998; Fujimoto et al., 2000). Amongst EIN3-regulated genes are a number encoding ETHYLENE RESPONSE FACTOR (ERF) transcription factors. Previous reports have shown that overexpression of selected ERF genes confers increased salt tolerance (H. Wang et al., 2004; Zhang et al., 2004; Zhang et al., 2012) and that mutants exhibiting constitutive ethylene responses are also relatively salt resistant (Achard et al., 2006). By contrast, mutants deficient for ethylene responses display salt hypersensitivity at different developmental stages (Achard et al., 2006; Lei et al., 2011). These findings suggest that ethylene-regulated signaling pathways play an important role in plant salinity stress adaptation. However, the precise causal molecular and physiological mechanisms underlying this adaptation are largely unknown.
The DELLA proteins, gibberellin-opposable growth-inhibitory components of the gibberellin-GID1-DELLA signaling pathway (Harberd et al., 2009), are also known to play critical roles in plant tolerance to salt (Achard et al., 2006). We previously found that salt-activated ABA and ethylene signaling pathways regulate plant growth and/or development via integration at the level of DELLA function (Achard et al., 2006), suggesting the importance of DELLAs in some aspects of plant growth responses to high salt environments. In this study, we aimed to identify DELLA-independent salinity tolerance mechanisms and hence conducted a genetic screen for the identification of salt-resistant mutants using a multiply mutant progenitor line devoid of all five of the DELLAs encoded by the Arabidopsis genome (the global DELLA-deficient line; see Methods). Identified in the absence of DELLAs, such mutants might be expected to define salt-tolerance pathways that operate in a DELLA-independent fashion. In addition, we conducted our genetic screen in soil-grown conditions resembling natural environments rather than in Petri dish conditions (Jiang et al., 2012), anticipating that this would enable identification of salt tolerance mechanisms important in natural and agricultural conditions.
Here, we describe the genetic, molecular, and physiological characterization of an Arabidopsis soil-salinity tolerant mutant, soil salinity tolerant1-1 (sst1-1). We first show that sst1-1 is a loss-of-function allele of the previously described ETHYLENE OVERPRODUCER1 (ETO1) locus, mutations in which cause increased production of ethylene (K.L.C. Wang et al., 2004). Our characterization of sst1-1 thus reveals an association between in vivo regulation of ethylene synthesis and salt stress tolerance. We next show that lack of ETO1 function promotes soil-salinity tolerance by reducing root-to-shoot Na delivery via an RBOHF-dependent mechanism. In addition, we show that ethylene promotion of soil-salinity tolerance is likely in part attributable to an increase in tissue K concentration, possibly via upregulation of the expression of the gene encoding the high-affinity K+ transporter HIGH-AFFINITY K+ TRANSPORTER5 (HAK5). Our observations provide a mechanistic understanding of how the stress phytohormone ethylene promotes plant tolerance of soil salinity, which has potential value for the development of salinity-tolerant crop plants.
RESULTS
Isolation and Genetic Characterization of sst1-1
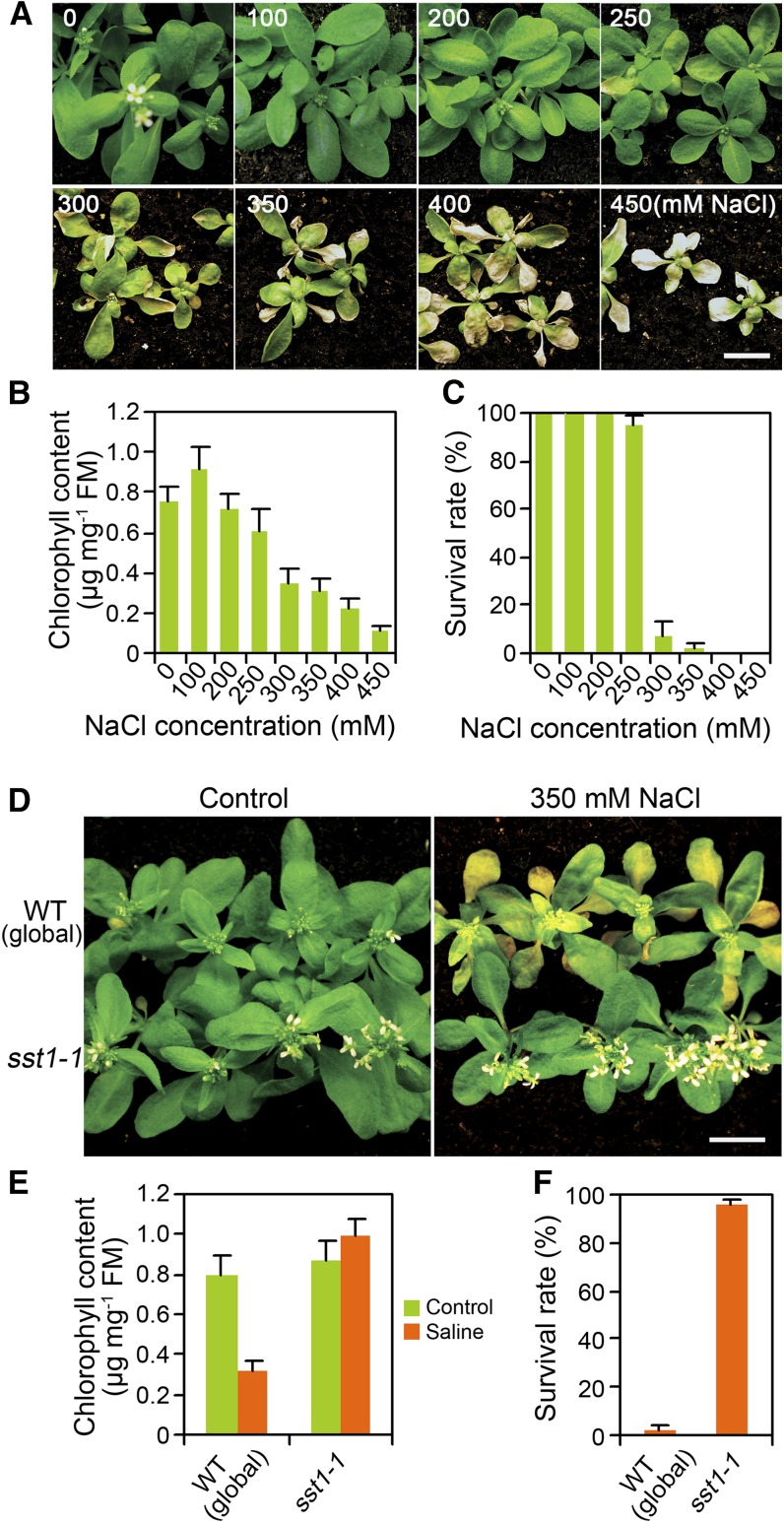
We aimed to identify naturally important salt tolerance mechanisms (using soil-based genetic screens; see Jiang et al., 2012) that may have been missed by previous studies using in vitro (Petri dish) conditions (e.g., J. Liu et al., 2000). This is particularly the case because transpirational flow is essentially blocked by lack of evaporation in enclosed in vitro conditions, and yet the transpiration stream is likely of paramount importance to delivery of Na to shoots in natural and agricultural conditions (Jiang et al., 2012). We therefore screened for mutants tolerant of soil salinity in conditions of active transpiration. Watering soil-grown Arabidopsis seedlings once with NaCl solution of increasing concentration (up to 450 mM) caused a progressive decrease in chlorophyll content (Figures 1A and 1B) and progressive increase in plant death (Figure 1C).
Figure 1.
Soil-Salinity Tolerance of sst1-1.
(A) Appearance of Arabidopsis thaliana wild-type (Landsberg erecta) plants following treatment with saline solutions of increasing NaCl concentration. Four-week-old soil-grown plants were watered (concentrations as indicated) once to soil capacity and then subsequently watered as required with nonsaline water. Pictures were taken 10 d after treatment.
(B) Chlorophyll content of plants shown in (A). Chlorophyll content measurements were as described in Methods. FM, fresh mass.
(C) Survival rate (%) of plants 3 weeks following salinity treatments.
(D) to (F) Appearance (D) and chlorophyll content (E) of global progenitor (wild type [WT]) and sst1-1 mutant plants 1 week following treatment with 350 mM NaCl. Survival rate (%) (F) 3 weeks following the salinity treatment.
Data shown in (B), (C), (E), and (F) are means ± se of three replicates. Bars = 1 cm in (A) and (D).
We performed a genetic screen for mutants exhibiting pronounced salt tolerance. Approximately 20,000 soil-grown fast-neutron-mutagenized DELLA-deficient (global) M2 seedlings (see Methods; Belfield et al., 2012; Jiang et al., 2012) were watered with 350 mM NaCl. Three weeks later, all surviving plants (as shown in Supplemental Figure 1 online) were selected and allowed to self-pollinate, and the heritability of salinity tolerance was determined in subsequent generations. Among ∼10 such mutants, the phenotype of one, sst1-1, was shown by backcrossing to be conferred by a single recessive mutant allele (a 3:1 ratio was obtained in the F2 generation of this cross).
sst1-1 conferred a clear soil-salinity tolerance phenotype (Figure 1D). Although there was no detectable difference between the shoots of the DELLA-deficient global progenitor and sst1-1 (also DELLA-deficient) plants grown in normal conditions (Figure 1D), progenitor plants were yellowing (decreased chlorophyll content) 1 week after being watered with 350 mM NaCl, while sst1-1 plants remained green after this treatment (Figures 1D and 1E). Whereas the majority of progenitor plants were dead 3 weeks after treatment, the majority of sst1-1 plants had survived (Figure 1F). By contrast, there were no detectable phenotypic differences between progenitor and sst1-1 plants following treatment with a sublethal mannitol concentration (see Supplemental Figure 2 online), suggesting that the NaCl tolerance of sst1-1 plants is distinct from any effect on osmotic tolerance. Notably, although the sst1-1 mutant was relatively tolerant of soil salinity (Figures 1D to 1F), there were no detectable differences between the salt sensitivities of progenitor and sst1-1 plants when grown in Petri dish conditions (see Supplemental Figures 3A and 3B online). Thus, sst1-1 identifies a gene that specifically affects plant sensitivity to soil salinity but not to in vitro salinity.
sst1-1 Is a Mutant ETO1 Allele
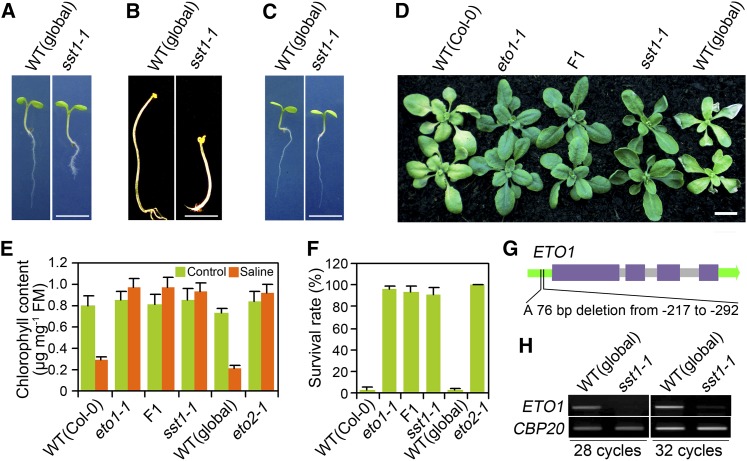
Although there was no detectable phenotypic difference between shoots of global and sst1-1 grown in normal soil (Figure 1D), we observed that sst1-1 seedling primary roots were shorter than those of global controls (Figure 2A). In addition, we found that dark-grown sst1-1 seedlings constitutively exhibited the triple response phenotype (shorter, radially swollen hypocotyl and exaggerated apical hook) characteristic of dark-grown seedlings treated with ethylene (Figure 2B; Guzmán and Ecker, 1990). These observations suggested that sst1-1 might confer changes in ethylene synthesis, signaling, or response. In further experiments, we found that growth of the sst1-1 primary root was restored by treatment with silver (Ag+) ions (Figure 2C). Silver ions block the binding of ethylene to endoplasmic reticulum membrane–localized ETR1-type ethylene receptors (Beyer, 1976), consequently suppressing the phenotype of ethylene overproducing mutants (e.g., eto1-1; K.L.C. Wang et al., 2004), but not suppressing the phenotype of constitutive ethylene response mutants (e.g., ctr1-1; Guzmán and Ecker, 1990). These latter observations suggested the possibility that sst1-1 might be a mutant ETO1 allele. ETO1 encodes a broad-complex/tramtrack/bric-a-brac protein that interacts with type-2 1-aminocyclopropane-1-carboxylate synthase (ACS) ethylene biosynthetic pathway enzymes (e.g., ACS5), thereby causing degradation of ACS proteins and reducing ethylene production (K.L.C. Wang et al., 2004). ETO1 loss-of-function alleles thus confer increased levels of ACS enzymes and of consequent ethylene production (K.L.C. Wang et al., 2004). Accordingly, we found that F1 heterozygote plants generated by crossing sst1-1 and eto1-1 displayed a similar level of salt tolerance to that of sst1-1 or eto1-1 homozygotes (Figures 2D to 2F), suggesting that sst1-1 is indeed a mutant ETO1 allele. We next determined the DNA sequence of the ETO1 gene in sst1-1, including 5′ and 3′ flanking regions, and identified a 76-bp deletion in the ETO1 promoter region (Figure 2G; see Supplemental Figure 4 online). Subsequent RT-PCR results indicated that this deletion causes a substantial decrease in the abundance of ETO1 transcripts (Figure 2H). In addition, the fact that the sst1-1/eto1-1 heterozygote (also heterozygous for the gai-t6 rga-t2 rgl1-1 rgl2-1 and rgl3-4 mutations that when homozygous confer the DELLA deficiency of the sst1-1 progenitor) exhibits tolerance of soil salinity demonstrates that the soil-salinity tolerance conferred by ETO1 loss-of-function alleles is not dependent on lack of DELLA function. Thus, we conclude that sst1-1 is a mutant ETO1 allele (hereafter named eto1-15) that results in reduced levels of ETO1 transcripts.
Figure 2.
sst1-1 Is a Mutant ETO1 Allele.
(A) to (C) Appearance of 6-d-old global progenitor (wild type [WT]) and sst1-1 mutant seedlings grown in light/dark cycles (A), constant darkness (B), or light/dark cycles in the presence of 10 µM Ag+ (C).
(D) Appearance of plants of various genotypes (as indicated) 10 d following treatment with 350 mM NaCl (plant growth and salinity treatment as described in Figure 1A). F1 is sst1-1/eto1-1 heterozygote. Col-0 is the wild-type control appropriate for eto1-1, global for sst1-1.
(E) Chlorophyll contents of plants shown in (D) (with additional eto2-1 mutant). Col-0 is the wild-type control appropriate for eto2-1. FM, fresh mass.
(F) Survival rate (%) of plants (genotypes as indicated) 3 weeks after treatment with 350 mM NaCl in soil-growth conditions.
(G) Representation of the ETO1 gene showing the location (promoter) of the deletion in the sst1-1 allele. Purple boxes represent coding sequences. Narrower pale boxes represent introns. Green boxes represent 5′ and 3′ untranslated regions.
(H) RT-PCR analysis of the abundance of ETO1 transcripts in wild-type progenitor and sst1-1 mutant. CBP20 transcripts provide control.
Bars = 0.5 cm in (A) to (C) and 1 cm in (D). Data shown in (E) and (F) are means ± se of three replicates.
We next determined if the soil-salinity tolerance of ETO1 loss-of-function alleles is a property peculiar to ETO1 or if other mutations conferring increased ethylene levels also lead to increased soil-salinity tolerance. We found that eto2-1, a gain-of-function ACS5 mutant allele of ETO2 that confers increased ethylene production (K.L.C. Wang et al., 2004), also displayed increased soil-salinity tolerance (see Supplemental Figure 5 online; Figures 2E and 2F). These observations indicated that the increased soil-salinity tolerance conferred by eto1-1, eto1-15, and eto2-1 is caused by increased ethylene level.
Lack of ETO1 Function Ameliorates Shoot Na Accumulation and K Depletion in Plants Grown on Saline Soils
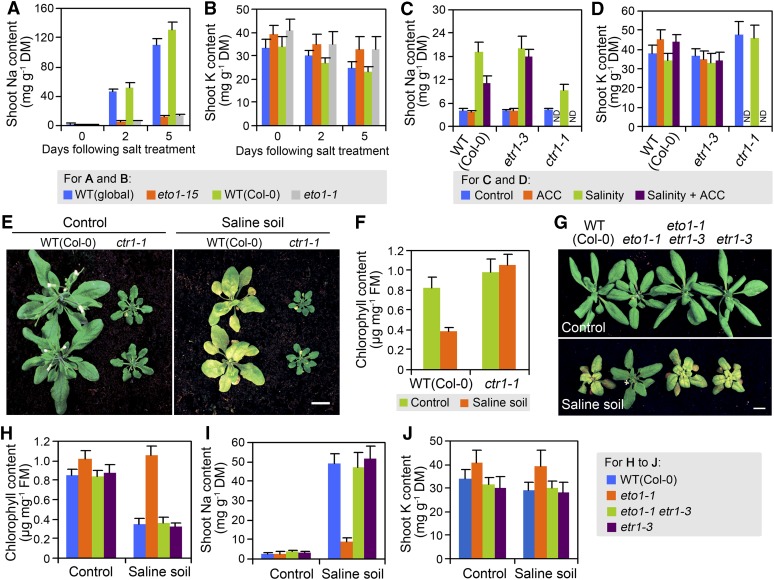
Growth in high-salinity environments causes in planta accumulation of Na, depletion of tissue K, and resultant cellular injury (Zhu, 2002; Munns and Tester, 2008). We therefore investigated whether the soil-salinity tolerance conferred by lack of ETO1 function might be due to improved Na/K homeostasis. When analyzed 2 or 5 d after watering with 350 mM NaCl solution, both the eto1-15 and eto1-1 mutants lacking ETO1 function had accumulated much less shoot Na than their respective wild-type controls (Figure 3A). Furthermore, we found that these genotypes also differed in their shoot K concentrations (Figure 3B): the eto1-15 and eto1-1 mutants had ∼10 to 15% higher (P < 0.05) shoot K concentrations (versus the wild type) in control soil conditions, and while shoot K concentrations declined significantly in the wild-type lines following salt treatment (day 0 compared with day 5, P < 0.05), the mutants lacking ETO1 function displayed no statistically significant decrease in K content (day 0 compared with day 5, P > 0.4; Figure 3B). These observations suggest that lack of ETO1 function confers salt tolerance by ameliorating shoot Na accumulation and salt-induced depletion of shoot K.
Figure 3.
Ethylene Modulates Shoot Na and K Homeostasis via ETR1-CTR1–Regulated Ethylene Signaling.
(A) and (B) Na (A) and K (B) contents of shoots (genotypes as indicated) 2 or 5 d following salt treatment (dry mass [DM]) in soil-grown conditions. Global provides the wild-type (WT) control for eto1-15 and Col-0 the wild-type control for eto1-1. Plant growth and salt treatment were as described in Figure 1A.
(C) and (D) Shoot Na (C) and K (D) contents of ACC-treated versus control plants (genotypes as indicated). ACC treatment was facilitated by use of hydroponically grown plants. Four-week-old plants (genotypes as indicated; see Methods) were treated with quarter-strength MS (control) or quarter-strength MS plus 100 mM NaCl. ACC treatment (50 μM) was applied 2 d prior to salinity treatment. Shoot samples were collected 2 d following onset of salinity treatment. ND, not determined.
(E) Wild-type (Col-0) and ctr1-1 mutant plants grown in control soil conditions (Control) or for 10 d following treatment with 350 mM NaCl (Saline soil). Plant growth and salinity treatment were as described in Figure 1A.
(F) Chlorophyll contents of plants shown in (E). FM, fresh mass.
(G) Plants (genotypes as indicated) grown in control conditions (Control) or for 10 d following treatment with 350 mM NaCl (Saline soil). Plant growth and salinity treatment were as described in Figure 1A.
(H) Chlorophyll contents of plants shown in (G).
(I) and (J) Shoot Na (I) and K (J) contents of plants (genotypes as indicated) grown in control conditions or for 2 d following salinity treatment (350 mM NaCl; Saline soil). Plant growth and salinity treatment were as described in Figure 1A.
Data shown in (A) to (D), (F), and (H) to (J) are means ± se of at least three replicates. Bars = 1 cm in (E) and (G).
Ethylene Regulates Shoot Na and K Content via the ETR1-CTR1–Regulated Ethylene Signaling Pathway
Since lack of ETO1 function causes an increase in ethylene production and ethylene activates downstream signaling pathways by binding to ethylene receptors (e.g., ETR1; Chang et al., 1993), we next determined if ethylene regulates shoot Na and K concentrations via ETR1-regulated ethylene signaling pathways. 1-Aminocyclopropane-1-carboxylic acid (ACC), the immediate precursor of ethylene, is a widely used chemical ethylene surrogate treatment (Wang et al., 2002). We found that ACC treatment of wild-type plants had no detectable effect on shoot Na concentration under control conditions but caused ∼50% reduction in shoot Na concentration 2 d following onset of salinity treatment (Figure 3C). In addition, ACC treatment of wild-type plants led to slightly but significantly (P < 0.05) elevated shoot K concentration, both under control conditions and 2 d following the onset of salinity treatment (Figure 3D). By contrast, ACC treatment of the ethylene-insensitive (ethylene receptor) mutant etr1-3 had no significant effect on shoot Na or K concentration in either control or salinity treatment conditions (Figures 3C and 3D), suggesting that ethylene regulates shoot Na and K concentrations via the ETR1-regulated ethylene signaling pathway. Consistent with this, ctr1-1 (a loss-of-function allele of CTR1, a gene encoding a downstream negative regulator of the ETR1-regulated ethylene signaling pathway; Kieber et al., 1993) displayed a reduced increase in shoot Na concentration compared with wild-type plants following salinity treatment (Figure 3C) and maintained a relatively high shoot K concentration in both control and salinity treatment conditions (Figure 3D). Accordingly, ctr1-1 displayed tolerance of soil salinity (Figures 3E and 3F). In further experiments with a double mutant etr1-3 eto1-1 line, we found that etr1-3 suppressed both the soil-salinity tolerance (Figures 3G and 3H) and the reduced Na and elevated K shoot concentrations (Figures 3I and 3J) characteristic of eto1-1. These observations indicate that the ethylene overproduction characteristic of loss of ETO1 function regulates shoot Na and K homeostasis via the ETR1-CTR1–regulated ethylene signaling pathway.
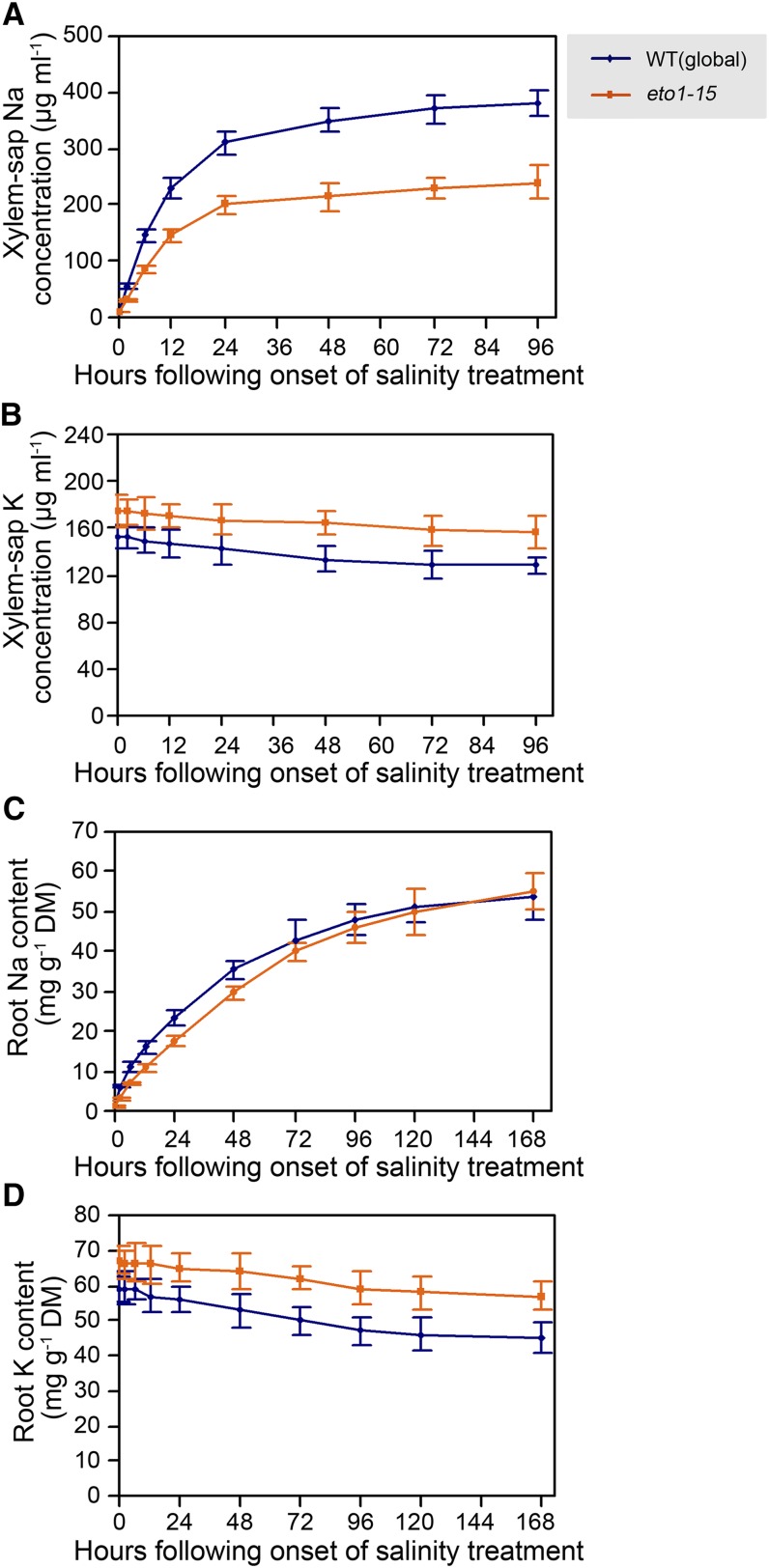
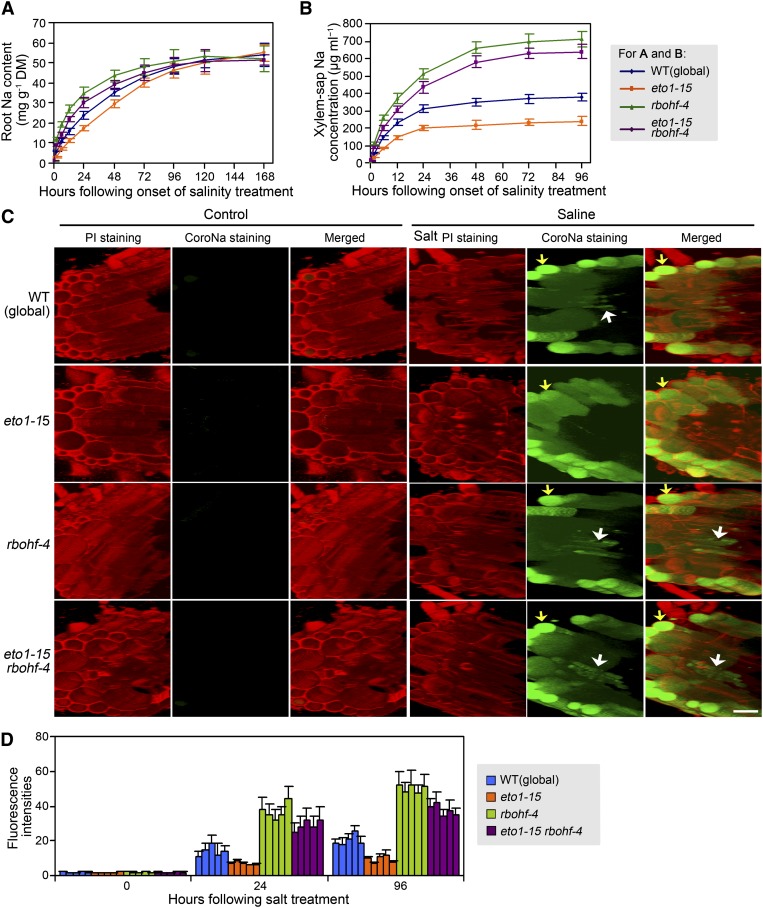
Lack of ETO1 Function Confers Decreased Xylem Sap Na Concentration and Increased Xylem Sap K Concentration
Growth of plants in high-salinity conditions causes excessive root-to-shoot Na delivery via the xylem in the transpiration stream, but mutants lacking ETO1 function were remarkably effective at restricting accumulation of Na in the shoot when exposed to salinity (Figure 3A). To determine if this effect was related to delivery of Na via the xylem, we compared ion concentrations in xylem sap obtained as root pressure exudate from detopped root systems of mutant plants lacking ETO1 function (eto1-15) versus wild-type plants at intervals following the onset of salinity treatment. While xylem sap Na concentrations of both wild-type and eto1-15 plants increased with a similar time course following the start of salinity treatment (Figure 4A), that of eto1-15 increased much less and averaged ∼60% of the wild type (Figure 4A). In addition, eto1-15 plants showed consistently higher xylem sap K concentrations (versus the wild type) in both control and salinity-treated plants (Figure 4B). The lower Na concentration and higher K concentration in the xylem sap of eto1-15 plants therefore contribute directly to the reduced shoot Na accumulation and enhanced shoot K accumulation characteristic of mutants lacking ETO1 function (Figures 3A and 3B).
Figure 4.
Lack of ETO1 Function Alters Root and Xylem Sap Na and K Homeostasis.
(A) and (B) Xylem sap Na (A) and K (B) concentrations at intervals following onset of salinity treatment. WT, the wild type.
(C) and (D) Root Na (C) and K (D) content at intervals following onset of salinity treatment. DM, dry mass.
To facilitate the collection of root samples and to compare the dynamics of changes in Na and K concentrations in xylem sap and root, hydroponically grown plants were used in these experiments. Four-week-old hydroponically grown wild-type (global) and eto1-15 plants were treated with quarter-strength MS (control) or quarter-strength MS plus 100 mM NaCl. Xylem sap (exudation rates comparable between the two genotypes) and root samples were collected as described in Methods. Data shown are means ± se of three replicates.
Lack of ETO1 Function Reduces Root Na Influx and Xylem Na Loading and Promotes Retention of Relatively High Root K
To determine whether ETO1 modulates xylem sap Na and K concentrations by regulating root Na and K uptake or xylem sap loading, or through a combination of both of these processes, we also examined the time course of changes in bulk root Na and K concentrations in wild-type and eto1-15 plants following exposure to salinity. We found that bulk root Na concentration was consistently lower in eto1-15 than in wild-type plants over the first 2 d of salinity treatment (Figure 4C), indicating that lack of ETO1 function reduced (net) Na influx into the root during this time. Estimates of the slopes of the curves at the start of salinity treatment indicated that the initial rate of Na uptake into roots of eto1-15 plants was ∼70% of that into wild-type plants. By the third day of exposure to salinity and thereafter, the bulk root Na concentrations of eto1-15 and wild-type plants were indistinguishable (Figure 4C), whereas eto1-15 plants maintained consistently lower xylem sap Na concentrations (versus the wild type) for at least 4 d following salinity treatment (Figure 4A), indicating that the rate of xylem loading of Na was lower in eto1-15 plants even at comparable bulk root Na content. Thus, it appears that reduced accumulation of shoot Na in eto1-15 plants is attributable both to lower influx of Na into the root and to reduced xylem Na loading. In addition, we found that eto1-15 bulk root K concentrations were consistently higher (P < 0.05) than those of the wild type in both salinity-treated and control plants (Figure 4D), an observation consistent with the high shoot and xylem sap K concentrations characteristic of eto1-15 (Figures 3B and 4B).
Enhanced Na Tolerance and Shoot Na Homeostasis of Mutants Lacking ETO1 Function Is Dependent upon RBOHF Function
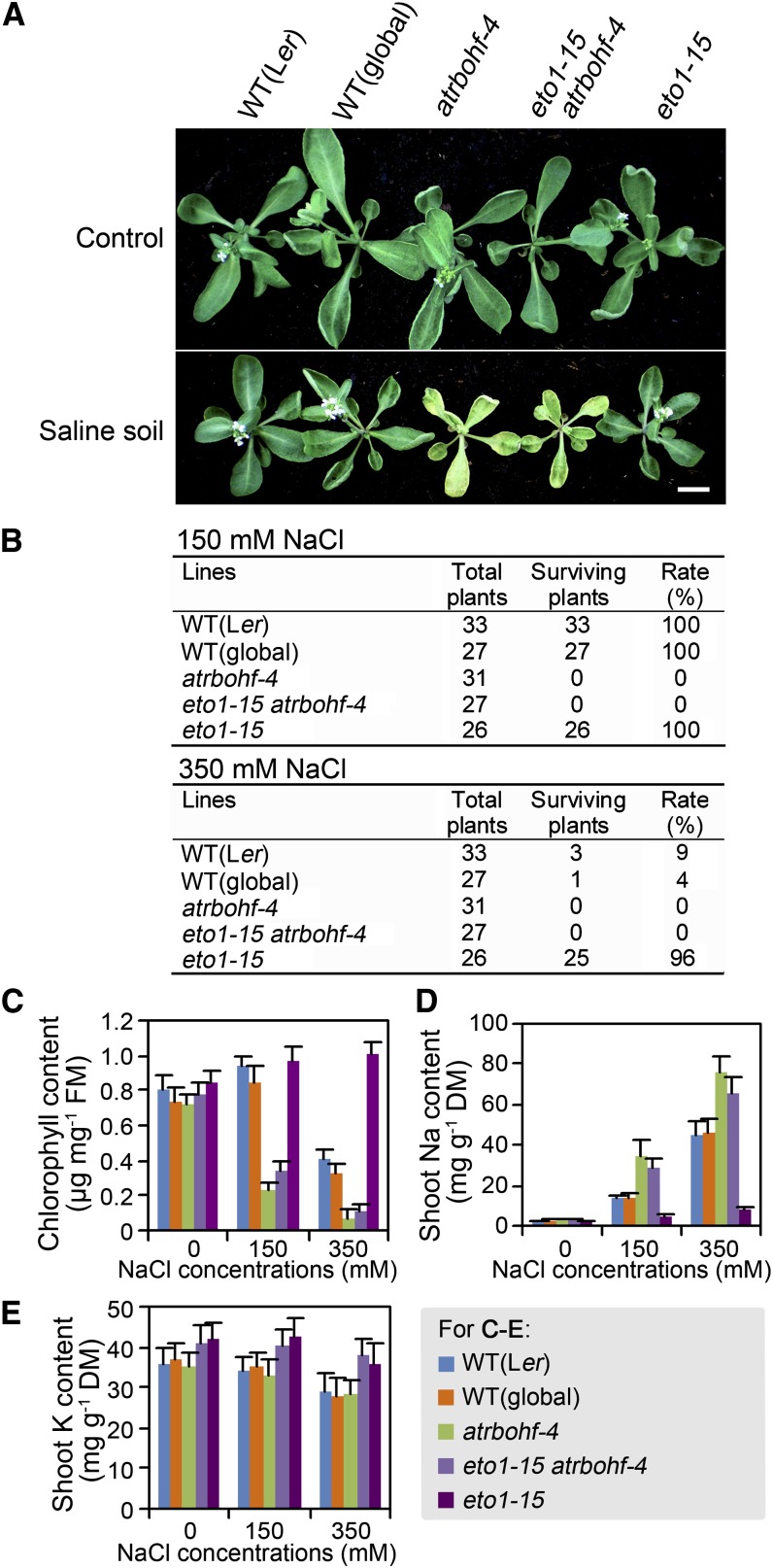
An earlier study suggested a linkage between ethylene and RBOHF in the induction of stomatal closure (Desikan et al., 2006). In addition, we previously showed that lack of RBOHF function causes overaccumulation of shoot Na and hypersensitivity to soil salinity (Jiang et al., 2012). We therefore next determined if the salinity tolerance conferred by lack of ETO1 function is dependent upon RBOHF function by making an eto1-15 rbohf-4 double mutant (rbohf-4 is an RBOHF loss-of-function allele [the same allele as sss1-1 described by Jiang et al., 2012]). eto1-15 rbohf-4 plants displayed the salinity hypersensitivity characteristic of rbohf-4 (Figures 5A to 5C), and, while eto1-15 survived treatment with 350 mM NaCl (Figures 1D to 1F, 5B, and 5C), eto1-15 rbohf-4 plants were bleached and killed by NaCl treatments at 150 mM and above (Figures 5A to 5C; see Supplemental Figure 6 online). These observations indicate that ethylene confers soil-salinity tolerance via mechanisms dependent upon the function of RBOHF.
Figure 5.
Ethylene Regulates Shoot Na Homeostasis via an RBOHF-Dependent Mechanism.
(A) Plants (genotypes as indicated) grown in control conditions (Control) or 7 d following treatment with 350 mM NaCl (Saline soil). Plant growth and salinity treatments were as described in Figure 1A. WT (Ler), wild-type Landsberg erecta. Bar = 1 cm.
(B) Plants (genotypes as indicated) surviving treatment with 150 or 350 mM NaCl (expressed as number of surviving plants and rate of survival [%] 3 weeks after treatment).
(C) Chlorophyll content (expressed as percentage of content of plants grown in control conditions) of shoots (genotypes as indicated) 7 d following treatment with 150 or 350 mM NaCl solutions. FM, fresh mass.
(D) and (E) Shoot Na (D) and shoot K (E) contents of plants (genotypes as indicated) 2 d following NaCl treatments (NaCl concentrations as indicated) in soil-growth conditions. DM, dry mass.
Results shown in (C) to (E) are means ± se of three independent replicates.
Analyzing the elemental content of plants treated with either 150 or 350 mM NaCl, we next found that, while eto1-15 as expected accumulated markedly less shoot Na than the wild type, the eto1-15 rbohf-4 double mutant exhibited shoot Na concentrations markedly higher than in wild-type plants and similar to those accumulated in the rbohf-4 single mutant (Figure 5D). These observations indicate that ethylene regulates shoot Na accumulation predominantly via an RBOHF-dependent mechanism. In contrast with the ethylene regulation of shoot Na concentration, we found that eto1-15 and eto1-15 rbohf-4 displayed similar shoot K concentrations under both control and salinity-treated conditions (Figure 5E), indicating that RBOHF function does not contribute to the ethylene regulation of shoot K concentration.
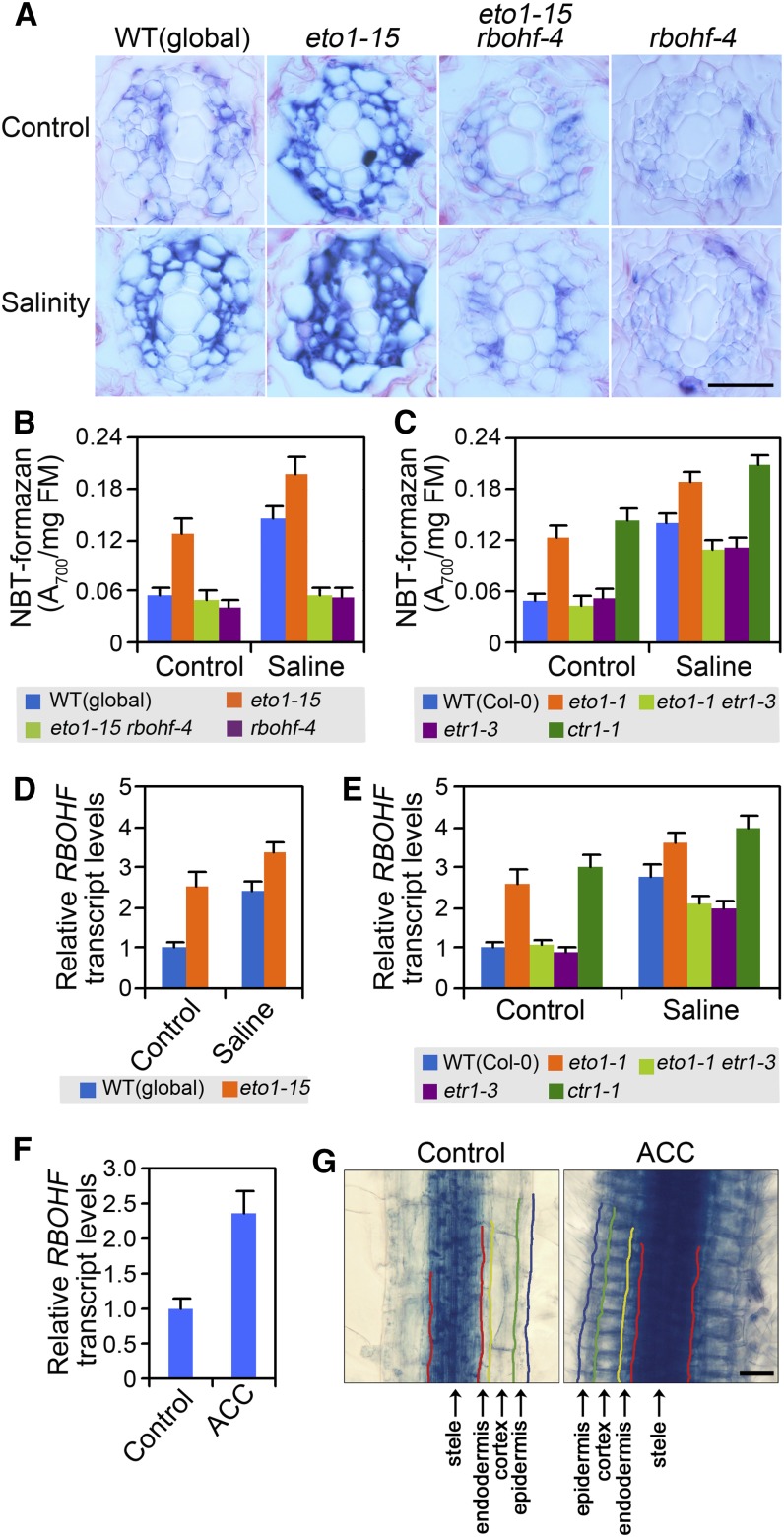
Lack of ETO1 Function Increases RBOHF-Dependent ROS Production in the Root Stele
Our previous work showed that RBOHF-dependent production of ROS in root stelar cells regulates root-to-shoot Na delivery (Jiang et al., 2012), while the observations above indicate that the effect of ethylene in ameliorating shoot Na accumulation is RBOHF dependent (Figure 5D). We therefore next determined if lack of ETO1 function reduces shoot Na concentrations by increasing RBOHF-regulated ROS production in the root stele. Intriguingly, we first found that eto1-15 displayed a constitutively elevated level of stelar cell ROS (as detected by nitroblue tetrazolium [NBT] staining; Figure 6A; see Supplemental Figure 7 online). Second, the eto1-15 rbohf-4 double mutant displayed an intensity of NBT staining similar to that of the rbohf-4 single mutant, indicating that the ethylene-induced stele ROS accumulation characteristic of eto1-15 is RBOHF dependent (Figure 6A). Thirdly, while salinity treatment increased the NBT signal in stelar cells of wild-type plants, it also further increased the already elevated NBT signal in eto1-15 plants (Figures 6A and 6B). The constitutively elevated levels of root stele ROS, together with the further increase induced by salinity treatment, could therefore explain the reduced root-to-shoot Na delivery conferred by eto1-15 (Figure 3A).
Figure 6.
Ethylene Regulates RBOHF-Mediated ROS Production in Root Vascular Tissue.
(A) The effect of salinity treatment on NBT-visualized ROS levels in the stele of wild-type (WT; global), eto1-15, eto1-15 rbohf-4, and rbohf-4 roots. Four-week-old hydroponically grown plants (genotypes as indicated) were treated with quarter-strength MS (control) or quarter-strength MS plus 100 mM NaCl. Root samples were collected for NBT staining (see Methods) 3 d following the treatments. Bar = 50 µm.
(B) and (C) Colorimetric quantification of NBT-formazan deposition in root tissues of plants (genotypes as indicated) under control conditions or 3 d following onset of salinity treatments (see Methods). FM, fresh mass. Data are means ± se of three replicates.
(D) to (G) Ethylene increases RBOHF transcript levels in root vasculature tissue. Real-time PCR analysis of RBOHF transcript levels in root tissues (genotypes and treatments as indicated) ([D] to [F]). Plant growth and treatments were as described in Figures 3C and 3D. Root tissues were collected 3 d following the onset of the salinity ([D] and [E]) or ACC (F) treatment. Data shown in (D) to (F) were expressed as fold increase of RBOHF transcript levels in experimental samples over that in untreated wild-type controls and are means ± se of three replicates. Effect of ACC treatment on GUS staining of mature pRBOHF:GUS roots (G). Colored lines have been added to demarcate the different tissues for ease of visualization. Bar = 50 µm.
We next determined if ethylene regulates stele ROS production through the ETR1-regulated ethylene signaling pathway and found that etr1-3 suppressed the high root stele NBT staining of eto1-1 in control conditions, while in these same conditions, ctr1-1 roots displayed constitutively high levels of NBT staining (Figure 6C). These observations suggest that stele ROS accumulation due to lack of ETO1 function is dependent on the ETR1-regulated ethylene signaling pathway. Interestingly, while etr1-3 suppressed the increased NBT staining of eto1-1, the etr1-3 mutant nevertheless displayed a salinity-induced increase in NBT staining, albeit of smaller magnitude than that of the wild-type control (P < 0.05; Figure 6C), thus suggesting that salinity-induced root stele ROS accumulation is partially but not fully dependent upon ETR1-regulated ethylene signaling.
Ethylene Increases Root Stele RBOHF Transcript Levels
Our previous study indicated that salinity causes root stele ROS accumulation at least partially via increased RBOHF transcript levels (Jiang et al., 2012). We next found that eto1-1, eto1-15, and ctr1-1 root tissues displayed elevated RBOHF transcript levels in control conditions (Figures 6D and 6E). Furthermore, while eto1-1 displayed elevated levels of RBOHF transcript, in the eto1-1 etr1-3 double mutant they were indistinguishable from wild-type levels (Figure 6E), suggesting that lack of ETO1 function increases RBOHF transcript levels via a mechanism dependent upon the ETR1-regulated ethylene signaling pathway. ACC treatment of wild-type plants also caused an increase in RBOHF transcript levels (Figure 6F), and ACC treatment of pRBOHF:GUS (for β-glucuronidase) plants caused an increase in root GUS staining, predominantly in stelar tissue (Figure 6G). These observations suggest that ethylene elevates ROS production in root stele at least partially by increasing RBOHF transcript levels.
Salinity treatment resulted in increased RBOHF transcript levels in wild-type plants and further increased the already elevated RBOHF transcript levels in eto1-1 and ctr1-1 plants (Figure 6E). The additive effects of salinity and ethylene/ethylene signaling on RBOHF transcript levels probably explains their additive effects on root stele ROS levels (Figures 6A to 6C). Salinity treatment also caused an increase in RBOHF transcript levels in the etr1-3 mutant, although the magnitude of this increase was smaller than in wild-type plants (P < 0.05; Figure 6E), suggesting that factors additional to the ETR1-regulated ethylene signaling pathway mediate salinity-induced increases in root stele RBOHF transcript levels.
Ethylene Regulation of Root Na Influx Is Dependent upon RBOHF Function
Our previous study indicated that RBOHF regulates root Na influx at relatively early stages of salinity treatments (Jiang et al., 2012). Furthermore, the observations described above show that lack of ETO1 function reduces net influx of Na into the root, at least during the first 2 d of salinity treatment (Figure 4C). We next determined if this effect of lack of ETO1 function is RBOHF dependent by comparing the temporal dynamics of root Na accumulation in eto1-15, rbohf-4, eto1-15 rbohf-4, and wild-type roots in response to salinity treatment. We observed that the rbohf-4 mutation conferred a significantly greater increase in bulk root Na content during the first 2 d of salinity treatment not only in the wild-type background but also in the eto1-15 mutant background (Figure 7A), indicating that ethylene regulation of root Na influx is largely RBOHF dependent. However, we also found that root Na concentration in eto1-15 rbohf-4 remained lower than that of rbohf-4 (at least for the first 2 d of salt treatment), suggesting that RBOHF-independent mechanisms also play subsidiary roles in ethylene regulation of root Na influx. After prolonged salinity treatment (≥4 d), all tested genotypes had accumulated similar bulk root Na concentrations (Figure 7A).
Figure 7.
ETO1 Regulates Root Na, Xylem Sap Na, and Root Vascular Tissue Na Concentration via an RBOHF-Dependent Mechanism.
(A) and (B) Root (A) and xylem sap (B) Na concentrations in control conditions or following salinity treatment (genotypes as indicated). Plant growth, salinity treatments, and sample collections were as described in Figure 4. Data are means ± se of three replicates. Data for global and eto1-15 are reproduced from Figure 4 to allow direct comparison of the effect of rbohf-4. DM, dry mass.
(C) Cellular localization of root Na+ (genotypes as indicated) in control conditions or following salinity treatment (Saline). Confocal images of roots stained with CoroNa Green (green) and propidium iodide (PI; red) are shown. Plant growth and salinity treatment was as described in Figure 4. CoroNa Green staining was as described in Methods. Arrows highlight the CoroNa Green signal in epidermal (yellow) or vascular (white) cells. Bar = 50 µm.
(D) Quantitative comparison of CoroNa Green fluorescence intensities in root pericycle cells 24 and 96 h following onset of salinity treatment. Five individual plants were measured for each genotype. Values are means ± se for 10 pericycle cells from each individual plant.
Ethylene Regulation of Xylem Sap and Root Stele Na Concentrations Are Dependent upon RBOHF Function
We next compared the temporal dynamics of xylem sap Na concentration in eto1-15, rbohf-4, eto1-15 rbohf-4, and wild-type plants in response to the onset of salinity treatment. We found that the rbohf-4 mutation conferred a significant increase in xylem sap Na concentration in both the wild-type background and the eto1-15 mutant background (Figure 7B), indicating that ethylene regulation of xylem Na loading is largely RBOHF dependent. However, the eto1-15 rbohf-4 xylem sap Na concentrations were consistently lower than those of rbohf-4, suggesting that RBOHF-independent mechanisms also play subsidiary roles in the ethylene regulation of xylem Na loading. Consistent with our previous observations that ethylene regulation of shoot K concentration is not dependent on RBOHF function (Figure 5E), ethylene regulation of bulk root and xylem sap K concentrations were unaffected by rbohf-4 (see Supplemental Figure 8 online).
Our previous studies indicated that RBOHF-catalyzed stelar ROS production decreases xylem sap Na concentrations mainly by reducing stelar Na concentrations (Jiang et al., 2012). In addition, our observations described above indicate that lack of ETO1 function causes an increase in RBOHF-dependent ROS accumulation in the root stele (Figures 6A and 6B) and a decrease in xylem sap Na concentration (Figure 4A). We next determined if the decrease in xylem sap Na concentration conferred by lack of ETO1 function is also attributable to decrease in root stelar Na concentration. We visualized Na accumulation in the stele of salinity-exposed roots using CoroNa Green dye (a green fluorescent indicator of Na+; Oh et al., 2009). Fluorescence was barely detectable in root cells of all tested genotypes under control conditions but was prominent in epidermal cells of roots following exposure to NaCl (Figure 7C). Fluorescence was faintly but clearly detectable in the stelar cells of salinity-treated wild-type roots (Figures 7C and 7D) but was not detected in equivalent cells of salinity-treated eto1-15 roots (Figures 7C and 7D). Thus, less Na+ accumulated in the root vasculature of salinity-treated eto1-15 versus wild-type plants. By contrast, fluorescence became clearly detectable in the vasculature tissue of salinity-treated eto1-15 rbohf-4 roots (Figures 7C and 7D), indicating that the reduced root vascular Na+ accumulation characteristic of eto1-15 is dependent upon RBOHF function.
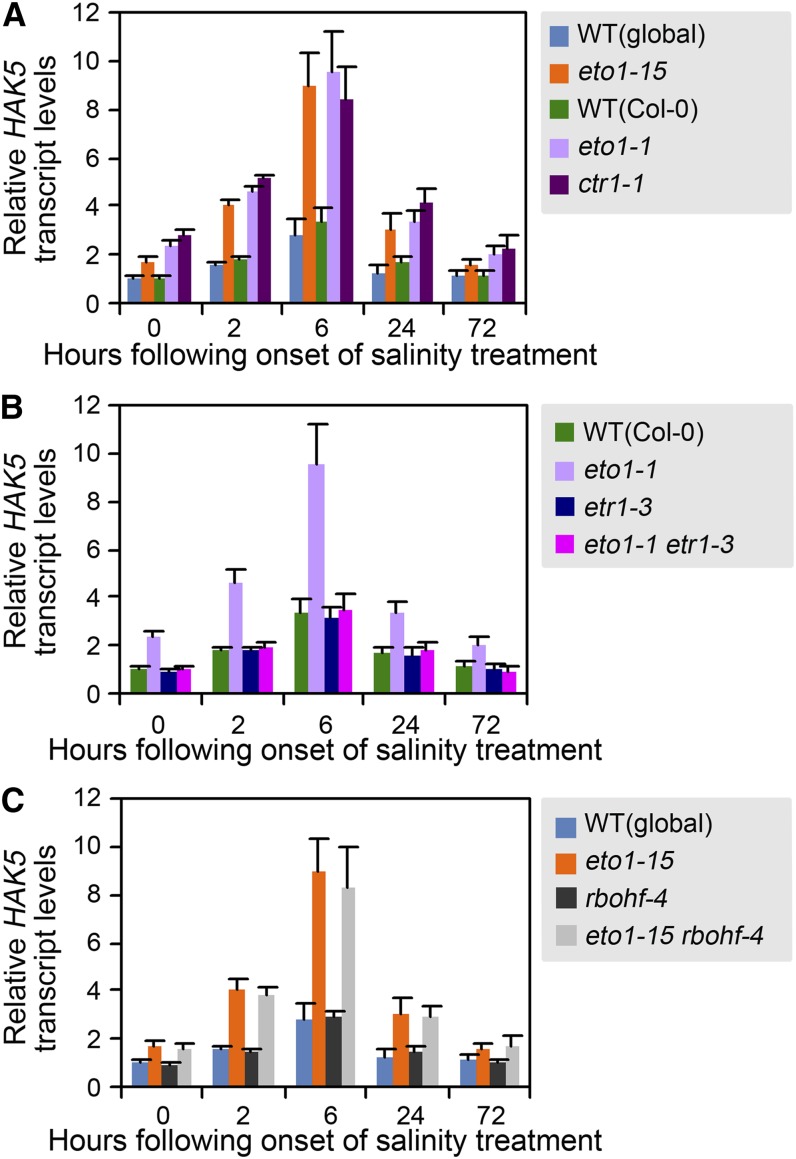
Salinity and Ethylene Increase the Abundance of Gene Transcripts Encoding the HAK5 K+ Transporter
The observations described above indicate that ETO1 confers tolerance of soil salinity by regulating RBOHF-dependent Na homeostasis and RBOHF-independent K homeostasis (Figures 3A, 3B, and 7; see Supplemental Figure 8 online). Previous studies identified possible roles for a number of different ion transporters in the regulation of Na and K homeostasis (reviewed in Demidchik and Maathuis, 2007; Munns and Tester, 2008; Shabala and Cuin, 2008; Kronzucker and Britto, 2011). Among the genes that have been functionally characterized, loss of function of genes encoding HKT1 or SOS1 Na+ transporters has been found to confer Na hypersensitivity (Rus et al., 2001; Shi et al., 2002), while ectopic overexpression of SOS1 or stele-specific expression of HKT1 increases salinity tolerance (Shi et al., 2003; Møller et al., 2009). In addition, AKT1 (a Shaker-type K+ channel) and HAK5 (a high-affinity K+ transporter belonging to the KUP/HAK/KT family) are the two K+ transporters that play a major role in K acquisition by the roots, particularly in the low concentration range (Hirsch et al., 1998; Gierth et al., 2005; Pyo et al., 2010). We therefore next determined if ethylene signaling modulates Na and K homeostasis by regulating the levels of gene transcripts encoding these transporters. We found that eto1-15, eto1-1, ctr1-1, and wild-type plants all displayed similar levels of expression of SOS1, HKT1, and AKT1 under control conditions and that transcript levels were generally unresponsive to salt treatment (except for a slight increase in SOS1 expression in all genotypes in response to salt; see Supplemental Figure 9 online). This suggests that ethylene does not regulate Na and K homeostasis via transcript-level regulation of the abundance of SOS1, HKT1, or AKT1. By contrast, we found that the levels of HAK5 transcripts in eto1-1, eto1-15, and ctr1-1 were consistently approximately twofold higher than in wild-type plants in control conditions (Figure 8A) and that HAK5 transcript levels displayed a transient three- to fivefold increase in response to salinity in all genotypes (peaking at 6 h following onset; Figure 8A). In addition, we found that etr1-3 suppressed the elevated HAK5 expression level conferred by eto1-1 (Figure 8B), indicating that lack of ETO1 function promotes the increase in HAK5 transcript level via the ETR1-CTR1–regulated ethylene signaling pathway. The elevated HAK5 transcript level in mutants lacking ETO1 function and in ctr1-1 potentially explains the relatively high K concentration characteristic of these mutants (Figures 3B, 3D, 4B, and 4D). Finally, consistent with the observation that lack of ETO1 function confers retention of high K concentrations in the shoot, xylem sap, and root via an RBOHF-independent mechanism (Figure 5E; see Supplemental Figure 8 online), we found that ethylene regulation of HAK5 transcript levels was RBOHF independent (Figure 8C).
Figure 8.
Salinity and Ethylene Regulate HAK5 Transcript Levels.
Effect of increasing duration of salinity treatment on HAK5 transcripts in root tissues of plants (genotypes as indicated). Plant growth and salinity treatments were as described in Figure 4A. Samples were collected at the time points indicated following onset of salinity treatments. Real-time PCR analysis was as described in Methods. Data were expressed as fold change of HAK5 transcript levels in experimental samples over that in untreated wild-type controls at the onset of salinity treatment. Values are means ± se of three biological replicates. Data for Col-0, eto1-1, global, and eto1-15 in (B) and (C) are reproduced from (A). WT, the wild type.
DISCUSSION
Increased in Vivo Ethylene Production Confers Soil-Salinity Tolerance
Ethylene synthesis is induced in plants by various biotic and abiotic environmental stresses, including pathogen attack, wounding, drought, flooding, salinity, low phosphorus, and low potassium (Borch et al., 1999; Wang et al., 2002; Achard et al., 2006; Broekaert, et al., 2006; Jung et al., 2009; Shi et al., 2012), and ethylene-mediated responses are often part of the crucial adaptive strategies that enable survival of those stresses. For example, the ethylene-inducible Sub1A gene (encoding an ERF-like protein) confers submergence tolerance in rice (Oryza sativa; Xu et al., 2006). In addition, ethylene signaling negatively regulates freezing tolerance (Shi et al., 2012) and positively regulates low potassium tolerance in Arabidopsis (Jung et al., 2009). Here, by employing a mutant screen using soil-grown plants in transpiring conditions, we discovered that a mutation causing enhanced ethylene production via loss of ETO1 function confers soil-salinity tolerance (Figures 1D to 1F). Our further demonstration that the eto2-1 mutant allele (which also causes enhanced ethylene production; K.L.C. Wang et al., 2004) similarly confers soil-salinity tolerance (Figures 2E and 2F; see Supplemental Figure 5 online) provides further genetic evidence that an increase in in vivo ethylene production confers plant soil-salinity tolerance.
While previous studies have indicated that enhanced ethylene signaling (as conferred by the ctr1-1 mutation or by overexpression of selected ERF factors) results in salinity tolerance in various conditions (H. Wang et al., 2004; Achard et al., 2006; Zhang et al., 2012), our discovery that increased ethylene production promotes soil-salinity tolerance is particularly notable since eto1-15 displays a survival rate comparable with that of wild-type controls under in vitro salinity conditions (see also Divi et al., 2010; see Supplemental Figure 3 online). This apparent discrepancy is likely to be due to the large differences in transpiration rate characteristic of the two experimental systems, which in turn may provide insight into the mechanisms underlying this conditional salinity tolerance phenotype. Since salinity stress is well known to increase the levels of transcripts specified by genes regulating ethylene biosynthesis (e.g., ACS2 and ACS7), and, hence, ethylene emission (Achard et al., 2006), our observations thus indicate that salinity-induced ethylene is an especially potent promoter of salt tolerance in conditions in which transpiration is active.
RBOHF Is an Important Mediator of Ethylene-Regulated Soil-Salinity Tolerance and Na Homeostasis
As mentioned in the Introduction, ROS production is both a consequence of the damage that stress causes and a signaling component that mediates adaptive stress responses. The results here indicate that ethylene promotes soil-salinity tolerance by increasing RBOHF-dependent ROS production (at least partially via increasing RBOHF transcript levels [Figure 6]) but also possibly via posttranslational activation of RBOHF (Sirichandra et al., 2009; Yun et al., 2011; Drerup et al., 2013). ROS are versatile molecules mediating a variety of biological responses in plants, including programmed cell death, development, hormone signaling, and environmental responses (Mittler et al., 2004, 2011; Torres et al., 2005; Kwak et al., 2006; Miller et al., 2009, 2010). The levels of in planta ROS are regulated by the interplay of ROS-generating and ROS-scavenging mechanisms (Mittler et al., 2004), and the NADPH oxidases, or respiratory burst oxidase homologs (RBOHs), are the most thoroughly studied plant ROS-generating enzymes (Marino et al., 2012). The Arabidopsis genome encodes 10 RBOH enzymes (encoded by genes RBOHA to RBOHJ; Torres et al., 2002), and expression of individual RBOH genes is commonly restricted to particular cells/tissues and regulates unique biological functions (e.g., RBOHC regulates the growth of root hair cells; Foreman et al., 2003). Our previous study indicated that ROS produced by RBOHF in the root stele confer soil-salinity tolerance (Jiang et al., 2012). Here, we show that the soil-salinity tolerance of eto1-15 is substantially dependent upon ethylene regulation of RBOHF-mediated ROS production in the root stele (Figures 5 to 7), indicating that RBOHF-produced ROS is an important mediator of ethylene signaling in soil-salinity tolerance. Our findings also need to be considered in light of a previous study indicating that ethylene causes stomatal closure via RBOHF-mediated ROS production (Desikan et al., 2006). Taken together, these observations indicate that RBOHF is likely to be an important mediator of ethylene signaling cascades and highlight the potential value of determining the function of the ethylene RBOHF regulon in response to other stresses and developmental cues.
As a key ROS-generating enzyme, RBOHF is an essential component of various biological processes, including induction of the phytohormone salicylic acid during the incompatible interaction (Chaouch et al., 2012), localized deposition of lignin (Lee et al., 2013), ABA and salicylic acid regulation of stomatal closing (Kwak et al., 2003; Khokon et al., 2011), jasmonic acid regulation of gene expression (Maruta et al., 2011), and regulation of sodium homeostasis in saline soil conditions (Jiang et al., 2012). Previous studies have demonstrated that the activity of RBOHF is regulated by many factors and at various levels. For example, ABA treatment and high salinity stress increase RBOHF transcript levels, thus increasing RBOHF-mediated ROS production (Kwak et al., 2003; Ma et al., 2011; Jiang et al., 2012). In addition, the activity of RBOHF may be positively regulated by phosphorylation (e.g., by OPEN STOMATA1; Sirichandra et al., 2009), by CBL1/9-CIPK26 complexes (Drerup et al., 2013), or by phosphatidic acid binding (Zhang et al., 2009), and alternatively may be negatively regulated by direct binding of CIPK26 (Kimura et al., 2013). Intriguingly, some of these actual or potential RBOHF regulatory factors are also involved in the regulation of salinity tolerance (e.g., ABA and phosphatidic acid; Yu et al., 2010).
When plants are grown in conditions of high soil salinity, a damaging excess of Na is delivered to the shoot via the xylem in the transpiration stream (Zhu, 2002; Munns and Tester, 2008). Our previous study indicated that RBOHF-mediated stele-specific ROS production plays an important role in regulation of root-to-shoot Na delivery and Na tolerance in transpiring plants exposed to soil salinity (Jiang et al., 2012). Here, we show that ethylene promotes soil-salinity tolerance by reducing root-to-shoot Na delivery via regulation of RBOHF-dependent ROS accumulation in root stele (Figures 5 to 7). Our observations indicate that, under high-salinity conditions, loss of ETO1 function increases RBOHF-dependent ROS accumulation in root stele (Figures 6A and 6B) and that this reduces stelar Na concentration (Figures 7A, 7C, and 7D), which in turn lowers the xylem sap Na concentration (Figure 7B), thus decreasing delivery of Na to the shoot, resulting in reduced shoot Na accumulation (Figure 3A) and enhanced salinity tolerance (Figure 1D). Lack of RBOHF function substantially suppresses the reduced shoot Na accumulation and soil-salinity tolerance characteristic of eto1-15 (Figure 5), indicating that ethylene regulation of salinity tolerance is largely dependent upon RBOHF-mediated regulation of tissue Na homeostasis. By contrast, a recent study has indicated that ethylene regulation of salt tolerance in Arabidopsis is not dependent upon a decrease in tissue Na content (Yang et al., 2013). This discrepancy is likely the consequence of the experimental system employed by Yang et al. (2013) being distinct from ours: under their experimental conditions, wild-type (Columbia-0 [Col-0]) plants treated with 200 mM NaCl exhibited a higher yield biomass than plants treated with 50 mM NaCl (Yang et al., 2013). However, our results are in accordance with this in showing that maintenance of high shoot K concentration is associated with ethylene-mediated salinity tolerance, although our genetic evidence indicates that this is independent of RBOHF function (Figures 3J and 5E; see Supplemental Figure 8 online).
Delivery of Na+ from the soil solution to shoot is a complex process that most probably involves the concerted action of several different transporters, but the key steps are believed to be (1) Na+ uptake into the root across the epidermis, (2) Na+ transport across the endodermis into cells of the central vascular cylinder (stele), (3) Na+ loading from stelar parenchyma cells into tracheary elements of the xylem, and (4) Na+ delivery to the shoot by mass flow in the transpiration stream (Amtmann and Sanders, 1999; Tester and Davenport, 2003; Munns and Tester, 2008; Kronzucker and Britto, 2011). Here, we show that, under saline conditions, the considerable reduction in shoot Na content caused by lack of ETO1 function (Figures 3A and 5D) is associated with reduced xylem sap Na concentration (Figure 4A). Interestingly, although the initial rate of root Na+ influx was lower in the eto1-15 mutant following onset of salinity treatment, xylem sap Na+ concentration was maintained at reduced levels compared with the wild type in the steady state even when bulkroot Na concentrations had reached identical values in the two lines (Figures 4A and 4C). The simplest explanation for this observation is that the reduced Na+ concentration in the root stelar tissue of the eto1-15 mutant (Figures 7C and 7D) leads directly to lower rates of xylem loading and hence to a lower xylem sap Na+ concentration. Other approaches, such as quantitative cryoanalytical electron microscopy, have also indicated the close relationship between Na+ concentration in stelar cells and xylem loading of Na+ for delivery to the shoot in the transpiration stream (Läuchli et al., 2008). In addition, our observations indicate that ethylene-regulated Na homeostasis is largely dependent on RBOHF-mediated ROS production in root stele (Figures 5 to 7), although the exact components of the signaling process that link salinity-induced ethylene production and enhanced levels of ROS remain to be defined.
Much interest surrounds the role of specific transport systems in the uptake and accumulation of mineral ions from the soil, so an important goal for future work will be identification of specific downstream targets of the signaling cascade initiated by salinity-induced ethylene production. Manipulation of expression of the Na+ transporters SOS1 and HKT1 has revealed their important roles in regulating root Na acquisition and xylem sap Na concentration (Shi et al., 2003; Møller et al., 2009), but our results show that RBOHF-produced ROS regulates Na homeostasis by a mechanism that is not dependent on changes in SOS1 or HKT1 transcript levels (see Supplemental Figure 9 online). However, the activity of such transporters can also be regulated by posttranscriptional mechanisms, as has been established for SOS1 (e.g., Qiu et al., 2002), so it remains possible that RBOHF-produced ROS could be involved in such posttranscriptional regulation. This is particularly possible because SOS1 interacts with the ROS signal regulators MAPK6 (Yu et al., 2010) and RCD1 (Katiyar-Agarwal et al., 2006) and because of indications that ROS may play a role in regulating Na+/K+ antiporter activity (Zhou et al., 2012). Root Na+ influx, and possibly a component of xylem Na+ loading, is also likely attributable to nonselective cation channels (Demidchik et al., 2002; Tester and Davenport, 2003; Demidchik and Maathuis, 2007; Munns and Tester, 2008). It will be interesting to determine if RBOHF-produced ROS is involved in regulation of the activity of these channels when the specific genes encoding them have been identified.
Ethylene Regulates HAK5 Transcript Levels and Tissue K Homeostasis
Plant salinity tolerance is achieved by some degree of Na/K homeostasis, which involves restricting tissue Na accumulation and maintaining adequate K concentrations, for example, by increasing the capacity for tissue K accumulation (Shi et al., 2003; Shabala and Cuin, 2008; Horie et al., 2009; Møller et al., 2009; Ardie et al., 2010; Kronzucker and Britto, 2011). We found that, compared with wild-type plants, mutants lacking ETO1 function show constitutively elevated tissue K concentrations under control conditions and less K depletion following salinity treatment (Figures 3B, 4B, and 4D). A link between the soil-salinity tolerance conferred by lack of ETO1 function and retention of higher tissue K concentrations may be provided by the constitutively elevated levels of transcripts encoding the high-affinity K+ transporter HAK5 observed in the eto1-1, eto1-15, and ctr1-1 mutants (Figure 8A). HAK5 is the only gene amongst those encoding KUP/HAK/KT family transporters whose transcript levels have consistently been found to be upregulated by K+ deficiency (Ahn et al., 2004; Armengaud et al., 2004; Gierth et al., 2005; Rubio et al., 2008), but there have been contradictory reports of the response of HAK5 transcript abundance to salinity. Long-term exposure of wild-type Arabidopsis to NaCl for 6 d (Ahn et al., 2004) or 16 d (Nieves-Cordones et al., 2010) was found to cause either no change or a reduction, respectively, in HAK5 transcript level. However, HAK5 transcript expression is less sensitive to downregulation by salt in the halophyte Thellungiella halophila than in Arabidopsis (Alemán et al., 2009), while transcript levels for four HAK genes actually increase in response to salt treatment in the halophilic plant Mesembryanthemum crystallinum (Su et al., 2002). Our results reveal that HAK5 expression increases in Arabidopsis in response to NaCl, but only transiently, with transcript levels peaking at 6 h and returning to close to basal levels within 24 h (Figure 8). Thus, both the elevated basal levels of HAK5 transcripts as well as the greater amplitude of the short-term response to salt treatment may contribute to the maintenance of tissue K concentrations and enhanced salinity tolerance conferred by lack of ETO1 function.
The ethylene-dependent increase in HAK5 transcripts in response to low K+ has been shown to be mediated by RAP2.11, an ethylene-inducible ERF transcription factor that binds to the HAK5 promoter (Kim et al., 2012). Although ethylene regulation of HAK5 expression is mediated via ROS production (Shin and Schachtman, 2004; Jung et al., 2009), our results show that this is independent of RBOHF function (Figure 8C). Interestingly, HAK5 is expressed mainly in the root hairs and epidermis (Ahn et al., 2004), in which Na accumulation is particularly pronounced (Figure 7C). The response of HAK5 transcripts to low K+ was found to be dependent upon the function of another member of the NADPH oxidase gene family, RBOHC (also known as ROOT HAIR DEFECTIVE2; Foreman et al., 2003), as HAK5 was not upregulated in response to K+ deprivation in rhd2 mutant plants (Shin and Schachtman, 2004). Thus, the elevated K status of roots (Figure 4) and improved salinity tolerance (Figures 1 to 3) associated with lack of ETO1 function may be attributable in part to enhanced uptake of K+ into the roots via the high-affinity transporter HAK5. ROSs have also been implicated in direct modulation of ion channel activity and could, for example, play a role in activating the K+ outward channels responsible for K+ loading from the stelar tissue into the xylem (Gaymard et al., 1998; Demidchik et al., 2010; Garcia-Mata et al., 2010). However, the increased xylem sap K+ concentration conferred by lack of ETO1 function is also independent of RBOHF-mediated ROS production (see Supplemental Figure 8B online), so the higher rate of xylem loading of K+ is probably a simple function of the elevated bulk root K+ concentration observed in the eto1-15 mutant plants (see Supplemental Figure 8A online).
The Mechanism of Ethylene Regulation of Soil-Salinity Tolerance
In summary, we have shown that ethylene regulates Arabidopsis soil-salinity tolerance via the canonical ETR1-CTR1 ethylene signaling pathway and that this regulation targets distinct aspects of organismal Na and K homeostasis. Ethylene production is increased in saline conditions, and here we show that increased ethylene reduces shoot exposure to soil Na by reducing bulk root Na influx and predominantly by reducing the Na content of the xylem sap, hence reducing delivery of Na to the shoot via the transpiration stream. The ethylene effect on xylem sap Na is dependent on the function of a specific respiratory burst oxidase, RBOHF, a function that we previously demonstrated to be necessary for soil-salinity tolerance and to be likely due to RBOHF-catalyzed ROS production (Jiang et al., 2012). In addition, we have shown that ethylene regulates K homeostasis via an RBOHF-independent mechanism that possibly involves upregulation of gene transcripts encoding the K+ transporter HAK5.
While our observations demonstrate that ethylene modulates root-to-shoot Na delivery via regulation of RBOHF-dependent ROS accumulation in root stelar tissues (Figures 5 to 7), a previous study indicated that engineering enhanced expression of a gene (HKT1;1) encoding the HKT1 Na+ transporter in root stelar cells of Arabidopsis can reduce root-to-shoot Na delivery, thus increasing salinity tolerance (Møller et al., 2009). These observations support the notion that regulation of Na concentration in root vasculature and xylem sap is crucial for regulating root-to-shoot Na delivery in saline conditions, suggesting the possibility that cell type–specific engineering of ethylene signaling or ROS production could increase the soil-salinity tolerance of plants. Since genes encoding components of the ethylene signaling pathway and RBOHF-related enzymes exist in the genomes of many crops (Xu et al., 2006; Wong et al., 2007; Lightfoot et al., 2008), it is possible that cell type–specific engineering of ethylene signaling or ROS production might provide a route towards increase in the soil-salinity tolerance of a wide range of crop species.
METHODS
Plant Materials
The global-DELLA (gai-t6 rga-t2 rgl1-1 rgl2-1 rgl3-4; global) Arabidopsis thaliana line and fast-neutron-mutagenized M2 seeds (global background) were as previously described (Belfield et al., 2012). eto1-1, eto2-1, and etr1-3 are in the Col-0 genetic background (Guzmán and Ecker, 1990; K.L.C. Wang et al., 2004). The pRBOHF-GUS transgenic line and rbohf-4 mutant (global background) were also as previously described (Jiang et al., 2012).
Soil and Hydroponic Plant Culture
Arabidopsis plants were grown on soil and in hydroponic conditions as previously described (Jiang et al., 2012).
Mutant Screen
To screen for soil-salinity–tolerant mutants, M2 seeds (global background, mutagenized via fast-neutron bombardment; Belfield et al., 2012) were first sown on soil (Erin multipurpose compost). Four-week-old seedlings were subsequently watered with 350 mM NaCl solution to soil capacity. After 3 weeks, all surviving plants were transferred to normal soil conditions and allowed to set seed. Heritability of the salt tolerance of candidate mutants was validated in subsequent generations.
Transcript Analyses
Total RNA was extracted using a Qiagen RNeasy kit. cDNA synthesis was performed using SuperScript III reverse transcriptase (Invitrogen). For quantitative RT-PCR, SYBR green PCR master mix (ABI) was used on an Applied Biosystems 7300 real-time PCR system. Expression levels of genes were determined from three independent biological replicates. For each biological replicate, we ran at least two technical replicates. The CBP20 (At5g44200) gene was used as an internal positive control. Primer pairs are as described in Supplemental Table 1 online.
Xylem Sap Collection
We developed an efficient method for collection of xylem sap from hydroponically grown Arabidopsis plants modified from Roosens et al. (2003). In brief, rosette leaves and inflorescence stems were excised at the top of the hypocotyl. The hypocotyls of detopped plants were then gently inserted into flexible plastic tubing (Tygon Laboratory Tubing) of suitable diameter to form a tight seal round the stem (see Supplemental Figure 10 online). Sap exuding from the detopped plants by root pressure was collected for up to 6 h (see Supplemental Figure 10 online). This method enabled the collection of up to 100 μL of xylem sap from a single detopped plant.
Determination of Elemental Concentrations
For determination of Na and K concentration in root or shoot tissue, samples were harvested, oven-dried for at least 24 h at 80°C, weighed, and then digested in concentrated (69%, v/v) HNO3 for at least 12 h for elemental extraction. Concentrations of Na and K were determined in appropriately diluted samples in an air-acetylene flame by atomic absorption spectrophotometry using a double-beam optical system with deuterium arc background correction (AAnalyst 100; Perkin-Elmer). Measurement of xylem sap Na and K concentration was performed as previously described (Jiang et al., 2012).
Determination of Chlorophyll Concentrations
Leaves from control and salt-treated plants were collected, weighed fresh, washed in distilled water, and extracted in 80% (v/v) acetone at room temperature in darkness (overnight), and the concentration of chlorophyll was determined according to Lichtenthaler (1987).
Determination of ROS Accumulation
We used NBT (Sigma-Aldrich) to detect ROS production. Four-week-old hydroponically grown plants were treated with quarter-strength Murashige and Skoog (MS; control) or quarter-strength MS plus 100 mM NaCl. Whole root systems were collected 3 d following treatment onset and stained with freshly made NBT solution (1.0 mg mL−1 NBT in 10 mM NaN3 and 10 mM phosphate buffer, pH 7.5) for 1 h. NBT-stained samples were then used to quantify the generation of formazan (Myouga et al., 2008) and to analyze the intensity of NBT staining in root vasculature. For histological sectioning of the NBT-stained samples, the samples were fixed in 4% paraformaldehyde in PBS at pH 7.0. After 24 h, samples were rinsed in PBS and dehydrated in an ethanol series of 30, 50, 70, and 85% (60 min each), then in 95% ethanol (0.1% eosin; overnight), and finally in 100% ethanol (0.1% eosin; 2 × 60 min). The samples were then processed in a Histoclear series of 25, 50, 75, and 100% (60 min each) and subsequently in Histoclear containing increasing volumes of wax chips (0.25, 0.5, and 0.75 volume) at 60°C for 2 h each. The samples were then placed in freshly melted wax overnight at 60°C. Finally, samples were placed in molds and sectioned with a Leica RM 2135 rotary microtome (10 μm). Sections were examined using a Leica DMRB light microscope, and photographs were obtained with a charge-coupled device digital camera (Micropublisher 5.0 RTV).
Visualization of Cellular Na+ Concentration
Four-week-old hydroponically grown seedlings were treated with 100 mM NaCl in liquid media, following which whole root systems were incubated for 8 h in a Petri dish in the same media supplemented with 2.5 μM CoroNa Green-AM (Invitrogen). Propidium iodide (2.5 μg mL−1; Invitrogen) was added as a counterstain for the plant cell wall prior to confocal microscopy. Z stacks of fluorescent images were recorded using a Zeiss LSM 510 confocal system with fixed settings of laser power, pinhole, and photomultiplier gain for all samples. Excitation and detection wavelengths were as previously described (Oh et al., 2010). The fluorescence intensity was calculated using Zeiss LSM 510 software.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: ETO1, At3g51770; ETR1, At1g66340; RBOHF, At1g64060; SOS1, At2g01980; HKT1, At4g10310; and HAK5, At4g13420.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Identification of a Soil-Salinity–Tolerant Mutant.
Supplemental Figure 2. The sst1-1 Mutation Has No Detectable Effect on Osmotic Stress Tolerance.
Supplemental Figure 3. sst1-1 Does Not Confer in vitro Salinity Tolerance.
Supplemental Figure 4. The sst1-1 Mutant Allele Carries a Mutant ETO1 Promoter.
Supplemental Figure 5. The eto2-1 Mutant Allele Confers Soil-salinity Tolerance.
Supplemental Figure 6. Lack of RBOHF Function Suppresses the Soil-Salinity Tolerance Conferred by eto1-15.
Supplemental Figure 7. The Effect of Salinity Treatment on NBT-Visualized ROS Levels in Mature Root Stele (Genotypes as Indicated).
Supplemental Figure 8. ETO1 Regulates Root and Xylem Sap K Concentration via an RBOHF-Independent Mechanism.
Supplemental Figure 9. The Effect of Salinity Treatment on At SOS1, At HKT1, and At AKT1 Transcript Levels.
Supplemental Figure 10. Xylem Sap Collection from a Detopped Hydroponically Grown Arabidopsis Plant.
Supplemental Table 1. List of Primers Used in RT-PCR Experiments (Figures 2H, 6D and 6E and 8; Supplemental Figure 9).
Acknowledgments
This article is based on work supported by Award KUK-I1-002-03, made by King Abdullah University of Science and Technology, and by Biotechnology and Biological Sciences Research Council Grant BB/F020759/1.
AUTHOR CONTRIBUTIONS
C.J., J.A.C.S., and N.P.H. designed the research. C.J., E.J.B., and J.A.C.S. performed the experiments. C.J., E.J.B., Y.C., J.A.C.S., and N.P.H. contributed to writing the article.
Glossary
- ROS
reactive oxygen species
- ABA
abscisic acid
- ACC
1-aminocyclopropane-1-carboxylic acid
- NBT
nitroblue tetrazolium
- Col-0
Columbia-0
- MS
Murashige and Skoog
References
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van Der Straeten D., Peng J., Harberd N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Ahn S.J., Shin R., Schachtman D.P. (2004). Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol. 134: 1135–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemán F., Nieves-Cordones M., Martínez V., Rubio F. (2009). Differential regulation of the HAK5 genes encoding the high-affinity K+ transporters of Thellungiella halophila and Arabidopsis thaliana. Env. Exp. Bot. 65: 263–269 [Google Scholar]
- Amtmann A., Sanders D. (1999). Mechanisms of Na+ uptake by plant cells. Adv. Bot. Res. 29: 75–112 [Google Scholar]
- Ardie S.W., Liu S., Takano T. (2010). Expression of the AKT1-type K+ channel gene from Puccinellia tenuiflora, PutAKT1, enhances salt tolerance in Arabidopsis. Plant Cell Rep. 29: 865–874 [DOI] [PubMed] [Google Scholar]
- Armengaud P., Breitling R., Amtmann A. (2004). The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol. 136: 2556–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfield E.J., et al. (2012). Genome-wide analysis of mutations in mutant lineages selected following fast-neutron irradiation mutagenesis of Arabidopsis thaliana. Genome Res. 22: 1306–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer E.M. (1976). A potent inhibitor of ethylene action in plants. Plant Physiol. 58: 268–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borch K., Bouma T.J., Lynch J.P., Brown K.M. (1999). Ethylene: A regulator of root architectural responses to soil phosphorus availability. Plant Cell Environ. 22: 425–431 [Google Scholar]
- Broekaert W.F., Delauré S.L., De Bolle M.F., Cammue B.P. (2006). The role of ethylene in host-pathogen interactions. Annu. Rev. Phytopathol. 44: 393–416 [DOI] [PubMed] [Google Scholar]
- Brüggemann L.I., Pottosin I.I., Schönknecht G. (1998). Cytoplasmic polyamines block the fast-activating vacuolar cation channel. Plant J. 16: 101–105 [Google Scholar]
- Cao W.H., Liu J., He X.J., Mu R.L., Zhou H.L., Chen S.Y., Zhang J.S. (2007). Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 143: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Kwok S.F., Bleecker A.B., Meyerowitz E.M. (1993). Arabidopsis ethylene-response gene ETR1: Similarity of product to two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Chaouch S., Queval G., Noctor G. (2012). AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J. 69: 613–627 [DOI] [PubMed] [Google Scholar]
- Chen Z., Newman I., Zhou M., Mendham N., Zhang G., Shabala S. (2005). Screening plants for salt tolerance by measuring K+ flux: A case study for barley. Plant Cell Environ. 28: 1230–1246 [Google Scholar]
- Collins J.C., Kerrigan A.P. (1974). The effect of kinetin and abscisic acid on water and ion transport in isolated maize roots. New Phytol. 73: 309–314 [Google Scholar]
- Cuin T.A., Betts S.A., Chalmandrier R., Shabala S. (2008). A root’s ability to retain K+ correlates with salt tolerance in wheat. J. Exp. Bot. 59: 2697–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V., Cuin T.A., Svistunenko D., Smith S.J., Miller A.J., Shabala S., Sokolik A., Yurin V. (2010). Arabidopsis root K+-efflux conductance activated by hydroxyl radicals: single-channel properties, genetic basis and involvement in stress-induced cell death. J. Cell Sci. 123: 1468–1479 [DOI] [PubMed] [Google Scholar]
- Demidchik V., Davenport R.J., Tester M. (2002). Nonselective cation channels in plants. Annu. Rev. Plant Biol. 53: 67–107 [DOI] [PubMed] [Google Scholar]
- Demidchik V., Maathuis F.J.M. (2007). Physiological roles of nonselective cation channels in plants: From salt stress to signalling and development. New Phytol. 175: 387–404 [DOI] [PubMed] [Google Scholar]
- Demidchik V., Shabala S.N., Coutts K.B., Tester M.A., Davies J.M. (2003). Free oxygen radicals regulate plasma membrane Ca2+- and K+-permeable channels in plant root cells. J. Cell Sci. 116: 81–88 [DOI] [PubMed] [Google Scholar]
- Desikan R., Last K., Harrett-Williams R., Tagliavia C., Harter K., Hooley R., Hancock J.T., Neill S.J. (2006). Ethylene-induced stomatal closure in Arabidopsis occurs via AtrbohF-mediated hydrogen peroxide synthesis. Plant J. 47: 907–916 [DOI] [PubMed] [Google Scholar]
- Divi U.K., Rahman T., Krishna P. (2010). Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol. 10: 151–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drerup M.M., Schlücking K., Hashimoto K., Manishankar P., Steinhorst L., Kuchitsu K., Kudla J. (2013). The Calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF. Mol. Plant 6: 559–569 [DOI] [PubMed] [Google Scholar]
- Flowers T.J. (2004). Improving crop salt tolerance. J. Exp. Bot. 55: 307–319 [DOI] [PubMed] [Google Scholar]
- Foreman J., Demidchik V., Bothwell J.H., Mylona P., Miedema H., Torres M.A., Linstead P., Costa S., Brownlee C., Jones J.D., Davies J.M., Dolan L. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Frommer W.B., Ludewig U., Rentsch D. (1999). Taking transgenic plants with a pinch of salt. Science 285: 1222–1223 [DOI] [PubMed] [Google Scholar]
- Fujimoto S.Y., Ohta M., Usui A., Shinshi H., Ohme-Takagi M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata C., Wang J., Gajdanowicz P., Gonzalez W., Hills A., Donald N., Riedelsberger J., Amtmann A., Dreyer I., Blatt M.R. (2010). A minimal cysteine motif required to activate the SKOR K+ channel of Arabidopsis by the reactive oxygen species H2O2. J. Biol. Chem. 285: 29286–29294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaymard F., Pilot G., Lacombe B., Bouchez D., Bruneau D., Boucherez J., Michaux-Ferrière N., Thibaud J.-B., Sentenac H. (1998). Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell 94: 647–655 [DOI] [PubMed] [Google Scholar]
- Gierth M., Mäser P., Schroeder J.I. (2005). The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol. 137: 1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway H., Munns R.A. (1980). Mechanisms of salt tolerance in non-halophytes. Annu. Rev. Plant Physiol. 31: 149 –190 [Google Scholar]
- Guo H., Ecker J.R. (2003). Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Guzmán P., Ecker J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd N.P., Belfield E., Yasumura Y. (2009). The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: How an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch R.E., Lewis B.D., Spalding E.P., Sussman M.R. (1998). A role for the AKT1 potassium channel in plant nutrition. Science 280: 918–921 [DOI] [PubMed] [Google Scholar]
- Hong S.G., Sucoff E. (1976). Effects of kinetin and root tip removal on exudation and potassium (rubidium) transport in roots of honey locust. Plant Physiol. 57: 230–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T., Hauser F., Schroeder J.I. (2009). HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 14: 660–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakannan M., Bose J., Babourina O., Rengel Z., Shabala S. (2013). Salicylic acid improves salinity tolerance in Arabidopsis by restoring membrane potential and preventing salt-induced K+ loss via a GORK channel. J. Exp. Bot. 64: 2255–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Belfield E.J., Mithani A., Visscher A., Ragoussis J., Mott R., Smith J.A.C., Harberd N.P. (2012). ROS-mediated vascular homeostatic control of root-to-shoot soil Na delivery in Arabidopsis. EMBO J. 31: 4359–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.-Y., Shin R., Schachtman D.P. (2009). Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21: 607–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S., Zhu J., Kim K., Agarwal M., Fu X., Huang A., Zhu J.-K. (2006). The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 18816–18821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye Y., Golani Y., Singer Y., Leshem Y., Cohen G., Ercetin M., Gillaspy G., Levine A. (2011). Inositol polyphosphate 5-phosphatase7 regulates the production of reactive oxygen species and salt tolerance in Arabidopsis. Plant Physiol. 157: 229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokon A.R., Okuma E., Hossain M.A., Munemasa S., Uraji M., Nakamura Y., Mori I.C., Murata Y. (2011). Involvement of extracellular oxidative burst in salicylic acid-induced stomatal closure in Arabidopsis. Plant Cell Environ. 34: 434–443 [DOI] [PubMed] [Google Scholar]
- Kieber J.J., Rothenberg M., Roman G., Feldmann K.A., Ecker J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Kim M.J., Ruzicka D., Shin R., Schachtman D.P. (2012). The Arabidopsis AP2/ERF transcription factor RAP2.11 modulates plant response to low-potassium conditions. Mol. Plant 5: 1042–1057 [DOI] [PubMed] [Google Scholar]
- Kimura S., Kawarazaki T., Nibori H., Michikawa M., Imai A., Kaya H., Kuchitsu K. (2013). The CBL-interacting protein kinase CIPK26 is a novel interactor of Arabidopsis NADPH oxidase AtRbohF that negatively modulates its ROS-producing activity in a heterologous expression system. J. Biochem. 153: 191–195 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy R., Bhagwat K.A. (1989). Polyamines as modulators of salt tolerance in rice cultivars. Plant Physiol. 91: 500–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronzucker H.J., Britto D.T. (2011). Sodium transport in plants: A critical review. New Phytol. 189: 54–81 [DOI] [PubMed] [Google Scholar]
- Kwak J.M., Mori I.C., Pei Z.M., Leonhardt N., Torres M.A., Dangl J.L., Bloom R.E., Bodde S., Jones J.D., Schroeder J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J.M., Nguyen V., Schroeder J.I. (2006). The role of reactive oxygen species in hormonal responses. Plant Physiol. 141: 323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Läuchli A., James R.A., Huang C.X., McCully M., Munns R. (2008). Cell-specific localization of Na+ in roots of durum wheat and possible control points for salt exclusion. Plant Cell Environ. 31: 1565–1574 [DOI] [PubMed] [Google Scholar]
- Lee Y., Rubio M.C., Alassimone J., Geldner N. (2013). A mechanism for localized lignin deposition in the endodermis. Cell 153: 402–412 [DOI] [PubMed] [Google Scholar]
- Lei G., Shen M., Li Z.G., Zhang B., Duan K.X., Wang N., Cao Y.R., Zhang W.K., Ma B., Ling H.Q., Chen S.Y., Zhang J.S. (2011). EIN2 regulates salt stress response and interacts with a MA3 domain-containing protein ECIP1 in Arabidopsis. Plant Cell Environ. 34: 1678–1692 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (1987). Chlorophylls and carotenoids - Pigments of photosynthetic biomembranes. Methods Enzymol. 148: 350–382 [Google Scholar]
- Lightfoot D.J., Boettcher A, Little A, Shirley N., Able A.J. (2008). Identification and characterisation of barley (Hordeum vulgare) respiratory burst oxidase homologue family members Funct. Plant Biol. 35: 347–359 [DOI] [PubMed] [Google Scholar]
- Liu J., Ishitani M., Halfter U., Kim C.S., Zhu J.K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 97: 3730–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhu J.K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280: 1943–1945 [DOI] [PubMed] [Google Scholar]
- Liu K., Fu H., Bei Q., Luan S. (2000). Inward potassium channel in guard cells as a target for polyamine regulation of stomatal movements. Plant Physiol. 124: 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J., Läuchli A. (1984). Potassium transport in salt-stressed barley roots. Planta 161: 295–301 [DOI] [PubMed] [Google Scholar]
- Ma L., Zhang H., Sun L., Jiao Y., Zhang G., Miao C., Hao F. (2011). NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na⁺/K⁺ homeostasis in Arabidopsis under salt stress. J. Exp. Bot. 63: 305–317 [DOI] [PubMed] [Google Scholar]
- Maathuis F.J.M., Amtmann A. (1999). K+ nutrition and Na+ toxicity: The basis of cellular K+/Na+ ratios. Ann Bot. 84: 123–133 [Google Scholar]
- Marino D., Dunand C., Puppo A., Pauly N. (2012). A burst of plant NADPH oxidases. Trends Plant Sci. 17: 9–15 [DOI] [PubMed] [Google Scholar]
- Maruta T., Inoue T., Tamoi M., Yabuta Y., Yoshimura K., Ishikawa T., Shigeoka S. (2011). Arabidopsis NADPH oxidases, AtrbohD and AtrbohF, are essential for jasmonic acid-induced expression of genes regulated by MYC2 transcription factor. Plant Sci. 180: 655–660 [DOI] [PubMed] [Google Scholar]
- Mäser P., et al. (2002). Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 531: 157–161 [DOI] [PubMed] [Google Scholar]
- Miller G., Schlauch K., Tam R., Cortes D., Torres M.A., Shulaev V., Dangl J.L., Mittler R. (2009). The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2: ra45. [DOI] [PubMed] [Google Scholar]
- Miller G., Suzuki N., Ciftci-Yilmaz S., Mittler R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33: 453–467 [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Suzuki N., Miller G., Tognetti V.B., Vandepoele K., Gollery M., Shulaev V., Van Breusegem F. (2011). ROS signaling: The new wave? Trends Plant Sci. 16: 300–309 [DOI] [PubMed] [Google Scholar]
- Møller I.S., Gilliham M., Jha D., Mayo G.M., Roy S.J., Coates J.C., Haseloff J., Tester M. (2009). Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21: 2163–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R., Tester M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Myouga F., Hosoda C., Umezawa T., Iizumi H., Kuromori T., Motohashi R., Shono Y., Nagata N., Ikeuchi M., Shinozaki K. (2008). A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell 20: 3148–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassery H. (1979). Salt-induced loss of potassium from plant roots. New Phytol. 83: 23–27 [Google Scholar]
- Nieves-Cordones M., Alemán F., Martínez V., Rubio F. (2010). The Arabidopsis thaliana HAK5 K+ transporter is required for plant growth and K+ acquisition from low K+ solutions under saline conditions. Mol. Plant 3: 326–333 [DOI] [PubMed] [Google Scholar]
- Oh D.H., Lee S.Y., Bressan R.A., Yun D.J., Bohnert H.J. (2010). Intracellular consequences of SOS1 deficiency during salt stress. J. Exp. Bot. 61: 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D.H., et al. (2009). Loss of halophytism by interference with SOS1 expression. Plant Physiol. 151: 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfi C., Pottosin I., Cuin T., Mancuso S., Shabala S. (2010). Specificity of polyamine effects on NaCl-induced ion flux kinetics and salt stress amelioration in plants. Plant Cell Physiol. 51: 422–434 [DOI] [PubMed] [Google Scholar]
- Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., Genschik P. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Pyo Y.J., Gierth M., Schroeder J.I., Cho M.H. (2010). High-affinity K+ transport in Arabidopsis: AtHAK5 and AKT1 are vital for seedling establishment and postgermination growth under low-potassium conditions. Plant Physiol. 153: 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q.S., Guo Y., Dietrich M.A., Schumaker K.S., Zhu J.K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 99: 8436–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan R., Lin H., Mendoza I., Zhang Y., Cao W., Yang Y., Shang M., Chen S., Pardo J.M., Guo Y. (2007). SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19: 1415–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosens N., Verbruggen N., Meerts P., Ximénez-Embún P., Smith J.A.C. (2003). Natural variation in cadmium tolerance and its relationship to metal hyperaccumulation for seven populations of Thlaspi caerulescens from western Europe. Plant Cell Environ. 26: 1657–1672 [Google Scholar]
- Rubio F., Nieves-Cordones M., Alemán F., Martínez V. (2008). Relative contribution of AtHAK5 and AtAKT1 to K+ uptake in the high-affinity range of concentrations. Physiol. Plant. 134: 598–608 [DOI] [PubMed] [Google Scholar]
- Rus A., Yokoi S., Sharkhuu A., Reddy M., Lee B.H., Matsumoto T.K., Koiwa H., Zhu J.K., Bressan R.A., Hasegawa P.M. (2001). AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc. Natl. Acad. Sci. USA 98: 14150–14155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S., Cuin T.A. (2008). Potassium transport and plant salt tolerance. Physiol. Plant. 133: 651–669 [DOI] [PubMed] [Google Scholar]
- Shabala S., Cuin T.A., Pottosin I. (2007). Polyamines prevent NaCl-induced K+ efflux from pea mesophyll by blocking non-selective cation channels. FEBS Lett. 581: 1993–1999 [DOI] [PubMed] [Google Scholar]
- Shabala S., Pang J., Zhou M., Shabala L., Cuin T.A., Nick P., Wegner L.H. (2009). Electrical signalling and cytokinins mediate effects of light and root cutting on ion uptake in intact plants. Plant Cell Environ. 32: 194–207 [DOI] [PubMed] [Google Scholar]
- Shi H., Ishitani M., Kim C., Zhu J.K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Lee B.H., Wu S.J., Zhu J.K. (2003). Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 21: 81–85 [DOI] [PubMed] [Google Scholar]
- Shi H., Quintero F.J., Pardo J.M., Zhu J.K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Tian S., Hou L., Huang X., Zhang X., Guo H., Yang S. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24: 2578–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R., Schachtman D.P. (2004). Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc. Natl. Acad. Sci. USA 101: 8827–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C., Gu D., Hu H.C., Davanture M., Lee S., Djaoui M., Valot B., Zivy M., Leung J., Merlot S., Kwak J.M. (2009). Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 583: 2982–2986 [DOI] [PubMed] [Google Scholar]
- Smith A.M., Coupland G., Dolan L., Harberd N., Jones J., Martin C., Sablowski R., Amey A. (2010). Plant Biology. (New York: Garland Science, Taylor & Francis Group; ). [Google Scholar]
- Solano R., Stepanova A., Chao Q., Ecker J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H., Golldack D., Zhao C., Bohnert H.J. (2002). The expression of HAK-type K+ transporters is regulated in response to salinity stress in common ice plant. Plant Physiol. 129: 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunarpi H., et al. (2005). Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J. 44: 928–938 [DOI] [PubMed] [Google Scholar]
- Tester M., Davenport R.J. (2003). Na+ tolerance and Na+ transport in higher plants. Ann. Bot. (Lond.) 91: 503–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M.A., Dangl J.L., Jones J.D. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M.A., Jones J.D., Dangl J.L. (2005). Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 37: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Tounekti T., Hernández I., Müller M., Khemira H., Munné-Bosch S. (2011). Kinetin applications alleviate salt stress and improve the antioxidant composition of leaf extracts in Salvia officinalis. Plant Physiol. Biochem. 49: 1165–1176 [DOI] [PubMed] [Google Scholar]
- Wang H., Huang Z., Chen Q., Zhang Z., Zhang H., Wu Y., Huang D., Huang R. (2004). Ectopic overexpression of tomato JERF3 in tobacco activates downstream gene expression and enhances salt tolerance. Plant Mol. Biol. 55: 183–192 [DOI] [PubMed] [Google Scholar]
- Wang K.L.C., Li H., Ecker J.R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14 (Suppl): S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.L.C., Yoshida H., Lurin C., Ecker J.R. (2004). Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428: 945–950 [DOI] [PubMed] [Google Scholar]
- Wong H.L., Pinontoan R., Hayashi K., Tabata R., Yaeno T., Hasegawa K., Kojima C., Yoshioka H., Iba K., Kawasaki T., Shimamoto K. (2007). Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 19: 4022–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Zhang Z., Zhang H., Wang X.C., Huang R. (2008). Transcriptional modulation of ethylene response factor protein JERF3 in the oxidative stress response enhances tolerance of tobacco seedlings to salt, drought, and freezing. Plant Physiol. 148: 1953–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Ishitani M., Lee H., Zhu J.K. (2001). The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13: 2063–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Xu X., Fukao T., Canlas P., Maghirang-Rodriguez R., Heuer S., Ismail A.M., Bailey-Serres J., Ronald P.C., Mackill D.J. (2006). Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442: 705–708 [DOI] [PubMed] [Google Scholar]
- Yang L., Zu Y.G., Tang Z.H. (2013). Ethylene improves Arabidopsis salt tolerance mainly via retaining K+ in shoots and roots rather than decreasing tissue Na+ content. Environ. Exp. Bot. 86: 60–69 [Google Scholar]
- Yang Q., Chen Z.Z., Zhou X.F., Yin H.B., Li X., Xin X.F., Hong X.H., Zhu J.K., Gong Z. (2009). Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol. Plant 2: 22–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Tena G., Xiong Y., Sheen J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Nie J., Cao C., Jin Y., Yan M., Wang F., Liu J., Xiao Y., Liang Y., Zhang W. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188: 762–773 [DOI] [PubMed] [Google Scholar]
- Yun B.-W., Feechan A., Yin M., Saidi N.B.B., Le Bihan T., Yu M., Moore J.W., Kang J.-G., Kwon E., Spoel S.H., Pallas J.A., Loake G.J. (2011). S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478: 264–268 [DOI] [PubMed] [Google Scholar]
- Zepeda-Jazo I., Velarde-Buendía A.M., Enríquez-Figueroa R., Bose J., Shabala S., Muñiz-Murguía J., Pottosin I.I. (2011). Polyamines interact with hydroxyl radicals in activating Ca2+ and K+ transport across the root epidermal plasma membranes. Plant Physiol. 157: 2167–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Huang Z., Xie B., Chen Q., Tian X., Zhang X., Zhang H., Lu X., Huang D., Huang R. (2004). The ethylene-, jasmonate-, abscisic acid- and NaCl-responsive tomato transcription factor JERF1 modulates expression of GCC box-containing genes and salt tolerance in tobacco. Planta 220: 262–270 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhu H., Zhang Q., Li M., Yan M., Wang R., Wang L., Welti R., Zhang W., Wang X. (2009). Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21: 2357–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wang J., Zhang R., Huang R. (2012). The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 71: 273–287 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Guo H.W. (2011). Paradigms and paradox in the ethylene signaling pathway and interaction network. Mol. Plant 4: 626–634 [DOI] [PubMed] [Google Scholar]
- Zhou H., Zhao J., Yang Y., Chen C., Liu Y., Jin X., Chen L., Li X., Deng X.W., Schumaker K.S., Guo Y. (2012). Ubiquitin-specific protease16 modulates salt tolerance in Arabidopsis by regulating Na+/H+ antiport activity and serine hydroxymethyltransferase stability. Plant Cell 24: 5106–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]