This work identifies a major role for the cytosolic structural protein of plant-infecting bunyaviruses in the concentration of viral glycoprotein cargo at endoplasmic reticulum export sites before their traffic to the Golgi complex where they further mature.
Abstract
In contrast with animal-infecting viruses, few known plant viruses contain a lipid envelope, and the processes leading to their membrane envelopment remain largely unknown. Plant viruses with lipid envelopes include viruses of the Bunyaviridae, which obtain their envelope from the Golgi complex. The envelopment process is predominantly dictated by two viral glycoproteins (Gn and Gc) and the viral nucleoprotein (N). During maturation of the plant-infecting bunyavirus Tomato spotted wilt, Gc localizes at endoplasmic reticulum (ER) membranes and becomes ER export competent only upon coexpression with Gn. In the presence of cytosolic N, Gc remains arrested in the ER but changes its distribution from reticular into punctate spots. Here, we show that these areas correspond to ER export sites (ERESs), distinct ER domains where glycoprotein cargo concentrates prior to coat protein II vesicle–mediated transport to the Golgi. Gc concentration at ERES is mediated by an interaction between its cytoplasmic tail (CT) and N. Interestingly, an ER-resident calnexin provided with Gc-CT was similarly recruited to ERES when coexpressed with N. Furthermore, disruption of actin filaments caused the appearance of a larger amount of smaller ERES loaded with N-Gc complexes, suggesting that glycoprotein cargo concentration acts as a trigger for de novo synthesis of ERES.
INTRODUCTION
Plant viruses are pathogens known to hijack the host cell in various ways for their own benefit (e.g., by using its translational machinery for viral protein synthesis to support ongoing replication and assembly of progeny virus). During these processes, viral proteins traffic along elements of the cytoskeleton to either reach the site of virus replication or move toward the so-called viral factories (i.e., sites where structural proteins and progeny DNA/RNA genomes concentrate to enable particle assembly). In many cases, plant RNA viruses replicating in the cytoplasm do this in association with membranes (e.g., endoplasmic reticulum [ER], chloroplast, and vacuoles) that often become rearranged or induced to proliferate (reviewed in Ritzenthaler and Elamawi, 2006). Once progeny virus is synthesized, particles move to neighboring cells to spread the viral infection. To this end, plant viruses encode the so-called movement proteins that upon synthesis are delivered to plasmodesmata, subsequently anchor on a docking complex (Thomas et al., 2008), and induce a modification and dilation in these channels to allow virus particles, or nucleoprotein complexes, to pass to neighboring cells (Lee et al., 2005; Ashby et al., 2006; Benitez-Alfonso et al., 2010; Verchot-Lubicz et al., 2010; Harries and Ding, 2011; Niehl and Heinlein, 2011; Schoelz et al., 2011). Also during this process a role for the cytoskeleton has been implicated (Lee et al., 2005; Wright et al., 2007; Harries et al., 2010).
Whereas most plant viruses are surrounded by a capsid shell, only two virus families, Rhabdoviridae and Bunyaviridae, include plant-infecting viruses that are surrounded by a lipid membrane containing virally encoded glycoproteins. The requirement of these plant viruses for a lipid membrane is likely to assist in receptor-mediated endocytosis to enter their insect vector, a property inherited from an ancestor shared with their animal-infecting (rhabdo-/bunyaviral) counterparts (Kormelink et al., 2011).
By default, (viral) glycoproteins generally are synthesized on rough endoplasmic reticulum membranes; after translocation, they are transported to the Golgi complex to ultimately be secreted from cells. The presence of a specific transmembrane domain (TMD), an ER-retention signal (KDEL/HDEL), or other cell sorting signals causes protein trafficking to deviate from the default setting. Instead, glycoproteins may thereby end up in ER, Golgi, or plasma membrane, etc., where they retain and/or exert their function. During passage through these cellular compartments, glycoproteins are posttranslationally modified (Vitale and Denecke, 1999; Neumann et al., 2003).
For many viruses surrounded by a membrane, the various steps within the process of particle assembly remain an enigma. These viruses usually obtain their lipid membranes by budding into one of the host cellular membranes. Viruses of the large family of arthropod-borne Bunyaviridae, whose members are primarily restricted to animal-infecting viruses (Elliott, 1990; Kormelink et al., 2011; Plyusnin and Elliott, 2011), obtain their lipid envelop from the Golgi complex, and the virus particles are secreted from the cell by transport from the Golgi to the plasma membrane (Griffiths and Rottier, 1992; Elliott, 1996; Peterson and Melin, 1996). During particle assembly of the plant-infecting bunyavirus Tomato spotted wilt virus (TSWV), ribonucleoproteins (RNPs) are enwrapped by Golgi cisternae containing the two viral glycoproteins (Gn and Gc), leading to the formation of doubly enveloped virus particles (Kikkert et al., 1999). These fuse with each other and ER-derived membranes and give rise to large vesicles containing an accumulation of mature singly enveloped virus particles, where they remain until uptake by their insect vector for further transmission (Kikkert et al., 1997, 1999, 2001). In insect cells, enwrapment of RNPs has never been observed, but mature virus particles are secreted from the salivary gland cells (the primary site of virus replication) into the salivary gland ducts. Hence, TSWV particle assembly in insect cells resembles that of the animal-infecting bunyaviruses, where secretion of mature virus particles from the cell is observed (Griffiths and Rottier, 1992; Elliott, 1996; Peterson and Melin, 1996).
Since the viral glycoproteins are the key mediators in the process of particle assembly and release, they have been the focus of intense studies for different animal-infecting bunyaviruses. These studies revealed that (for most bunyaviruses) Gc is arrested in the ER but upon coexpression with Gn becomes redirected to the Golgi, likely due to heterodimerization (Matsuoka et al., 1988; Chen et al., 1991; Lappin et al., 1994). Their accumulation and retention in the Golgi is due to the presence of a Golgi retention signal in Gn (Andersson et al., 1997a; Gerrard and Nichol, 2002; Shi et al., 2004), and budding of RNPs into the lumen of the Golgi is assumed to be triggered by a specific interaction between the cytoplasmic tail (CT) of one of the glycoproteins (Andersson et al., 1997b; Strandin et al., 2013) and the major structural component of the RNPs (i.e., the nucleocapsid [N] protein). Prior to this, RNPs have to concentrate at glycoprotein-containing foci in the Golgi complex.
The TSWV glycoproteins exhibit a similar trafficking behavior in mammalian cells (Kikkert et al., 2001), likely due to their close ancestral relation and similarity to members of the orthobunyavirus genus, and a Golgi retention signal was initially mapped at the CT and TMD of Gn (Snippe et al., 2007a). More recent studies have shown that the TMD of Gn is sufficient to enable ER exit and Golgi targeting of Gc (Ribeiro et al., 2009b). An interaction between N and Gc was suggested to trigger Golgi membrane envelopment of RNPs based on the occurrence of an interaction measured in vivo by Förster resonance energy transfer (FRET) and fluorescence lifetime imaging microscopy (FLIM) (Snippe et al., 2007b). Furthermore, in mammalian cells, transiently expressed TSWV N has been shown to accumulate as large aggregates in a non-Golgi perinuclear region, in a microtubule- and actin filament–dependent manner (Snippe et al., 2005a). Upon coexpression of N and Gc, both proteins similarly accumulate in a perinuclear region, likely also in a microtubules/actin dependent way (Snippe et al., 2007b).
In plant cells, transiently expressed TSWV glycoprotein Gc similarly remains in the ER and upon coexpression with Gn becomes competent for ER-to-Golgi transport (Ribeiro et al., 2008). Furthermore, and interestingly, both Gc and Gn are able to individually induce ER and Golgi membrane curving, respectively, as observed with the formation of (pseudo-) circular and pleomorphic membrane structures (Ribeiro et al., 2008). Whether this points to an interaction with or reflects a feature shared with reticulons is not yet known (Yang and Strittmatter, 2007; Diaz and Ahlquist, 2012), although recently the glycoproteins have been proposed to act as matrix protein (Strandin et al., 2013). Opposed to its perinuclear accumulation in mammalian cells, the N protein in plant cells localizes as large inclusion bodies distributed throughout the cytoplasm (Ribeiro et al., 2009a), and FRET/FLIM analysis has demonstrated interactions between N and both glycoproteins at the interface of ER and Golgi (Ribeiro et al., 2009a). Remarkably, the interaction between N and Gc does not release the glycoprotein from its ER arrest but renders its distribution within this organelle from reticular to punctate spots (Ribeiro et al., 2009a).
Here, we specifically localized these N-Gc complexes and analyzed these interactions using FRET/FLIM while studying the role of the plant cell cytoskeleton in cytosolic N trafficking. N-Gc complexes localize at ER export sites (ERESs), and inhibitors of actin and microtubule polymerization do not prevent N protein from oligomerization or interaction with the glycoproteins at ER or Golgi. An increase in number of N-Gc (ERES) foci in the presence of cytoskeleton inhibitors was observed and suggested a de novo synthesis of ERESs.
RESULTS
Distribution of N Protein within the Cytoplasm Is Actin Dependent but Microtubule Independent
During (plant) bunyavirus replication in the cytoplasm and prior to virus particle assembly, newly synthesized viral RNPs ultimately concentrate at the interface of Golgi at foci containing viral mature glycoproteins. It is not known whether this localization involves movement of the N protein, the major structural protein of RNPs, along elements of the cytoskeleton. In order to investigate a possible dependence of TSWV N protein trafficking and behavior on cytoskeleton elements, tobacco (Nicotiana tabacum) protoplasts were cotransfected with yellow fluorescent protein (YFP)–fused N (N-YFP; Ribeiro et al., 2009a; Figure 1) and green fluorescent protein (GFP)–α-tubulin (a marker of the microtubule network) (Kumagai et al., 2001).
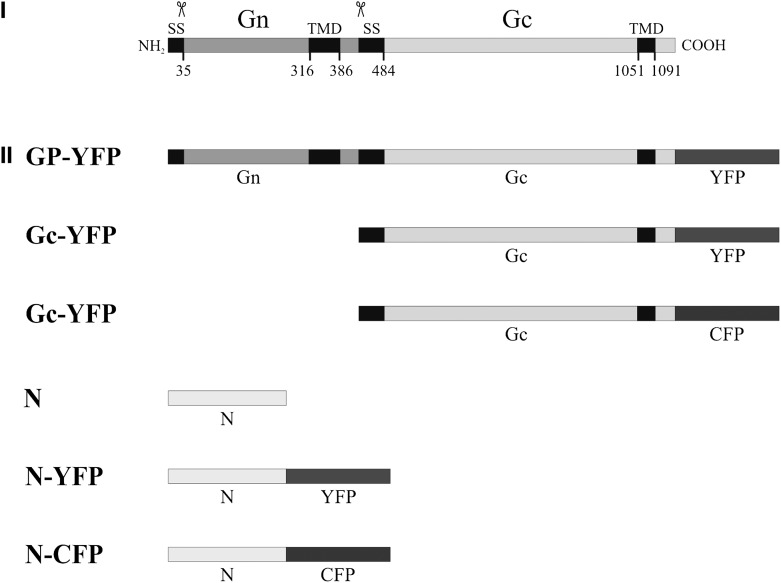
Figure 1.
Schematic Presentation of Constructs Used in This Study.
(I) Schematic representation of the topology of the TSWV glycoprotein precursor. (II) Viral protein constructs used for expression analysis (the glycoprotein constructs are aligned below the precursor). Predicted cleavage sites (scissor symbols), hydrophobic areas (black boxes), and amino acid positions are indicated (SS stands for signal sequence). In all constructs, the YFP or CFP fluorophores were fused in frame at the position of the stop codon.
The cells were incubated either in the presence of colchicine, an inhibitor of microtubule polymerization (Morejohn and Fosket, 1991; Guha and Bhattacharyya, 1997), or cytochalasin D, an inhibitor of actin polymerization (Goddette and Frieden, 1986; Laporte et al., 2003). Several control experiments were performed in order to determine the optimal drug concentrations to be used while maintaining the viability of the cells (as verified by, for example, the presence of mobile Golgi units analyzed in at least two independent experiments sets of minimally 20 cells per condition). These ended up in the range determined earlier as optimal while maintaining cell viability (Laporte et al., 2003). Furthermore, time-course experiments on cells expressing GFP-α-tubulin under the influence of the drugs were performed in order to determine the earliest time point where an effect could be observed (as show in Supplemental Figure 1 online for three different time points). Combining these data with immunolocalization experiments using anti-α-tubulin antibodies, the microtubule network was observed already disrupted at 3 h after the application of either drug (see Supplemental Figures 1C and 1F online). In our experiments, Cytochalasin D also affected the microtubule network (see Supplemental Figures 1E and 1F online), likely due to the tight association of the actin filaments with the microtubules, causing a general cytoskeleton disruption. Hence, in the following experiments, depolymerization of microtubules was used as an indirect marker to monitor the effect of the actin polymerization inhibitor.
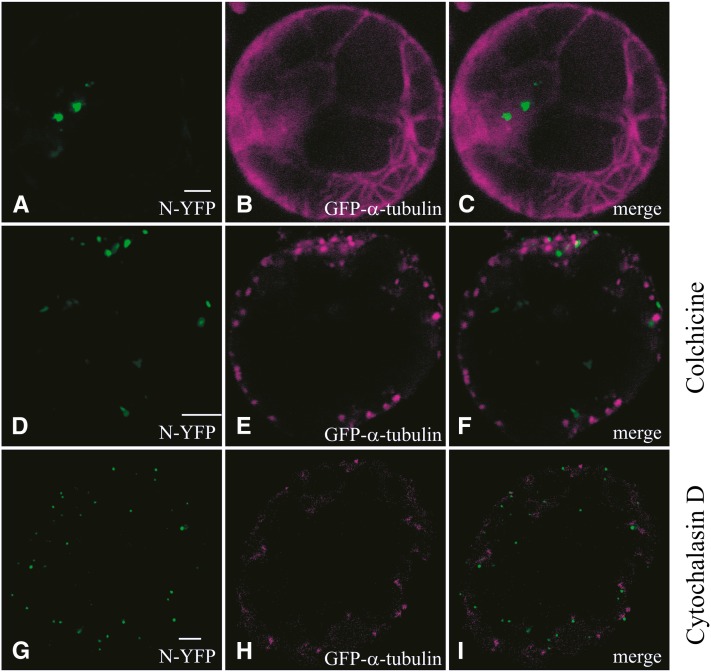
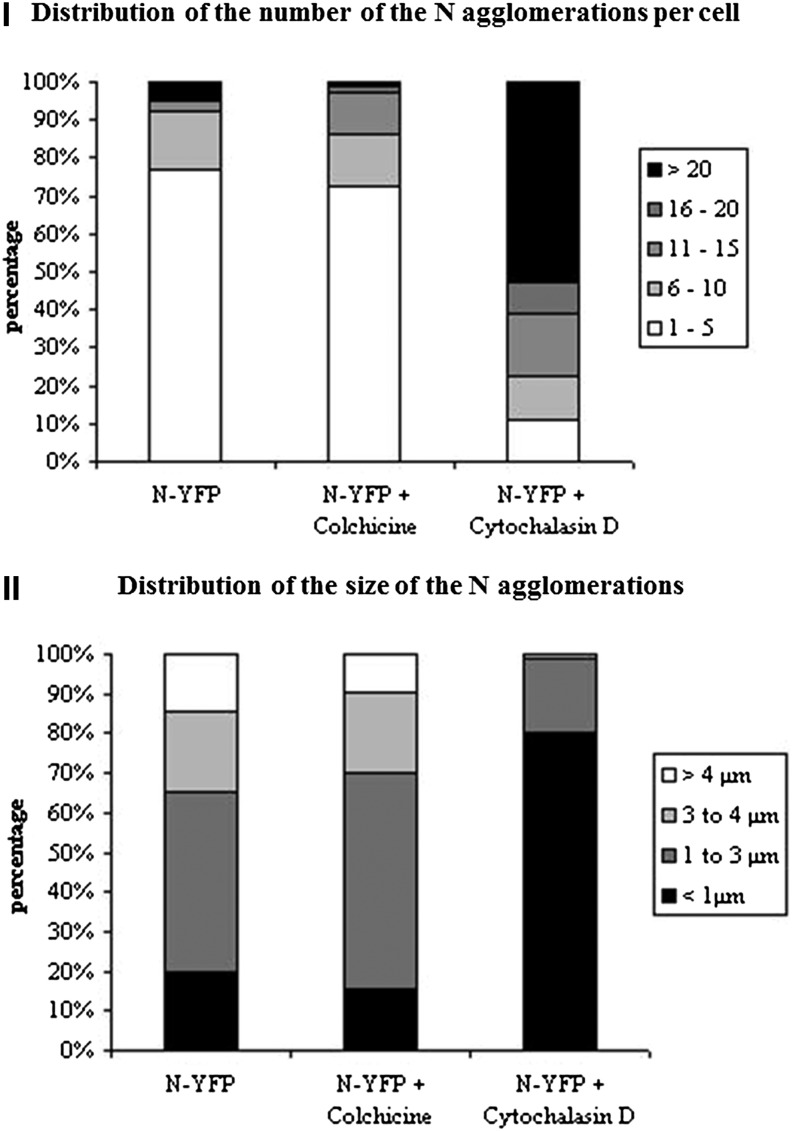
Having set the conditions for depolymerization of microtubules and actin filaments, the effects on the behavior of the N protein were analyzed by confocal microscopy 24 to 48 h posttransfection (p.t.). When N was expressed and analyzed in nontreated cells, no colocalization was observed between the N agglomerations and microtubules (Figures 2A to 2C). Upon colchicine treatment, the microtubule network was already disturbed after 24 h (Figure 2E), but no change could be observed in the distribution of N agglomerations; N agglomerates were formed in similar number (Mann-Whitney U test: Unumber of agglomerations = 2329.0; n = 70; Significance = 0.613) and size (Mann-Whitney U test: Usize of agglomerations = 153647.5; n = 70; Significance = 0.990) compared with nontreated cells (Figure 2D). By contrast, 24 h after cytochalasin D treatment, the number of agglomerations increased (Mann-Whitney U test: Unumber of agglomerations = 567.5; n = 70; Significance = 0.000), and their size became significantly smaller (Mann-Whitney U test: Usize of agglomerations = 92,599.5; n = 70; Significance = 0.000) (Figure 2G). During both treatments, no colocalization was observed between N and depolymerized α-tubulin (Figures 2D to 2I). The observed differences have been graphically presented (Figure 3) and altogether indicate that in plant cells, the formation of N protein agglomerations is actin dependent and microtubule independent.
Figure 2.
Cellular Distribution of Transiently Expressed N Protein under the Influence of Cytoskeleton Inhibitors.
(A) to (C) Protoplasts transfected with N-YFP and GFP-α-tubulin 24 h p.t. N-YFP (A), GFP-α-tubulin (B), and merge image of (A) and (B) in (C).
(D) to (F) Protoplasts transfected with N-YFP and GFP-α-tubulin and treated with colchicine 24 h p.t. N-YFP (D), GFP-α-tubulin (E), and merge image of (D) and (E) in (F).
(G) to (I) Protoplasts transfected with N-YFP and GFP-α-tubulin and treated with cytochalasin D 24 h p.t. N-YFP (G), GFP-α-tubulin (H), and merge image of (G) and (H) in (I).
The images were taken 24 h p.t. Bars = 5 µm.
Figure 3.
Graphical Representations of the Influence of Colchicine and Cytochalasin D on the Number and Size of Cytoplasmic N Aggregations.
(I) Distribution of the number of N agglomerations per cell. (II) Distribution of the size of N agglomerations.
Interactions among the Structural TSWV Proteins Are Not Affected by Changes in the Cytoskeleton
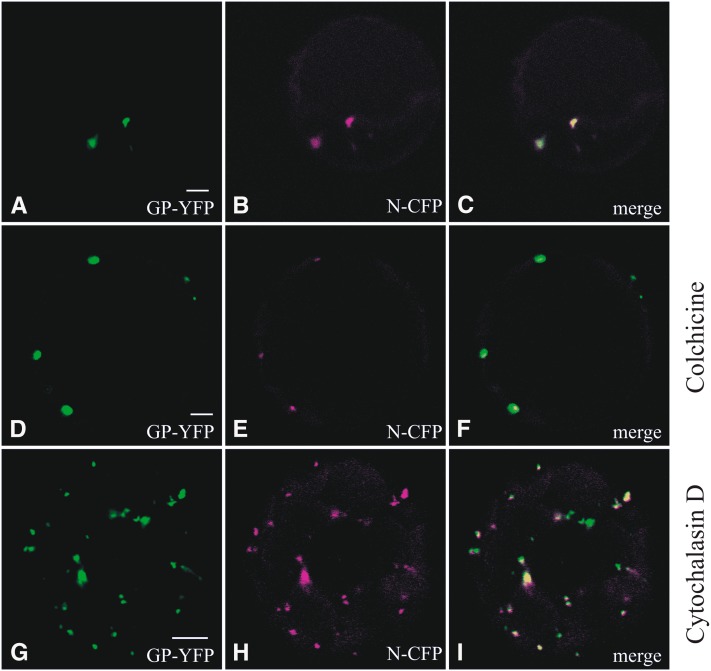
Previous studies have shown a colocalization and interaction between N and both viral glycoproteins at ER and Golgi (Ribeiro et al., 2009a). To investigate the requirement of actin filaments and/or microtubules for the migration of cytoplasmic N to these sites, protoplasts were cotransfected with a fluorophore-fused glycoprotein precursor construct GP-YFP, which allowed the monitoring of Gc-YFP in the presence of Gn (Ribeiro et al., 2008), and N-CFP (for cyan fluorescent protein) (Ribeiro et al., 2009a; Figure 1). As expected, and similarly to what was observed in nontreated cells (Figures 4A to 4C), 24 h after colchicine application, the N protein still colocalized with the glycoproteins (Figures 4D to 4F). Surprisingly, similar results were obtained upon treatment with cytochalasin D (Figures 4G to 4I). However, the number of sites where the glycoproteins and N colocalized increased in number and decreased in size (Figures 4A and 4G), in analogy to what was previously observed for N-YFP in cells treated with the same drug. These findings indicated that the spatial distribution observed for the colocalizing glycoproteins and N highly depended on actin-dependent formation of cytoplasmic N agglomerations, but the ability of these N agglomerations to reach the sites of colocalization with the viral glycoproteins did not seem to require actin filaments.
Figure 4.
Cellular Distribution of N and the Viral Glycoproteins under the Influence of Cytoskeleton Inhibitors.
(A) to (C) Protoplasts transfected with GP-YFP and N-CFP 24 h p.t. GP-YFP (Gc-YFP) (A), N-CFP (B), and merge image of (A) and (B) in (C).
(D) to (F) Protoplasts transfected with GP-YFP and N-CFP and treated with colchicine 24 h p.t. GP-YFP (Gc-YFP) (D), N-CFP (E), and merge image of (D) and (E) in (F).
(G) to (I) Protoplasts transfected with GP-YFP and N-CFP and treated with cytochalasin D 24 h p.t. GP-YFP (Gc-YFP) (G), N-CFP (H), and merge image of (G) and (H) in (I).
Bars = 5 µm.
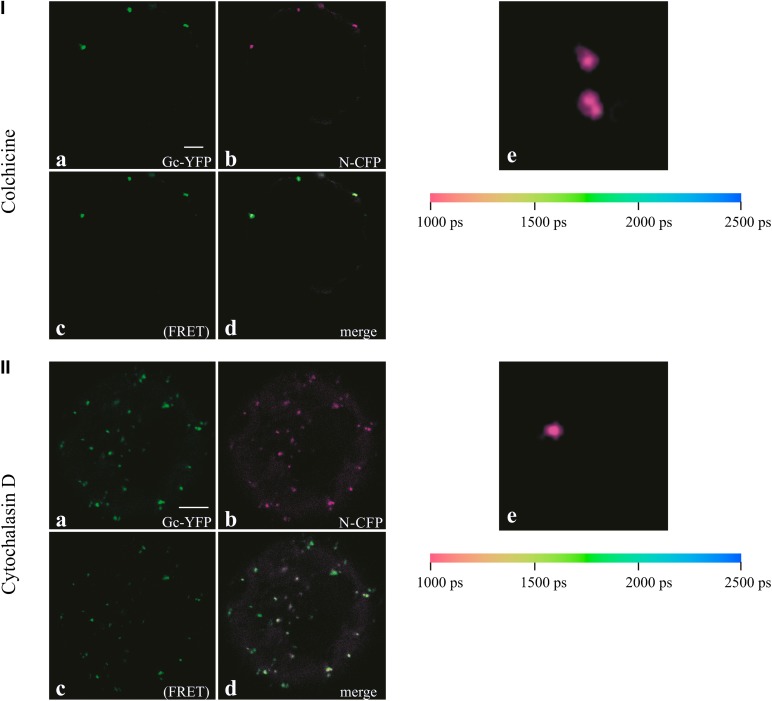
To substantiate the findings that disruption of microtubules and actin filaments did not abrogate their colocalization, nor affect their interaction, FRET/FLIM analyses were performed on protoplasts coexpressing GP-YFP and N-CFP and treated with either colchicine or cytochalasin D (Figure 5). Confocal imaging (Figures 5Ia to 5Id and 5IIa to 5IId) showed a weaker N-CFP fluorescence in the presence of GP-YFP in cells treated with colchicine (Figure 5Ib) as well as in cells treated with cytochalasin D (Figure 5IIb) compared with cells that only expressed N-CFP. A high YFP fluorescence was detected upon excitation of CFP (Figures 5Ic and 5IIc) and suggested the occurrence of FRET. To exclude CFP crosstalk and/or direct YFP excitation, FLIM was performed, which revealed a strong decay in the lifetime of the donor fluorophore, even after cells were treated with either colchicine (Figure 5Ie) or cytochalasin D (Figure 5IIe). In previous studies (Ribeiro et al., 2009a), the interaction between Gc and N in the presence of Gn revealed a decrease of 30 to 50% (with an average of 38%) in the lifetime of the donor fluorophore. Here, in cells treated with colchicine (where a total of 12 cells were analyzed, with different regions of interest within the cells), a 33 to 58% decrease in CFP lifetime was still observed with an average of ∼45%. The example in Figure 5Ie shows an average lifetime decrease from 2380 to 1271 ps, corresponding to 46.5%. Similar results were obtained for the cells treated with cytochalasin D (for a total of 10 cells with different regions of interest within the cells), where a 39 to 62% decrease was observed, with an average of 47%. The example in Figure 5IIe shows an average lifetime decrease from 2380 to 1424 ps, corresponding to 40%. Altogether these results indicated that, even after microtubules and/or actin filaments were disrupted, the viral N protein was still able to translocate to the sites where the glycoproteins are concentrated (ER and Golgi) and to interact with them in similar conformations as in nontreated cells.
Figure 5.
FRET and FLIM Analysis of N–Gc Interactions in the Presence of Gn and Cytoskeleton Inhibitors.
(I) Protoplasts transfected with N-CFP and GP-YFP treated with colchicine 24 h p.t. GP-YFP (Gc-YFP) (a), N-CFP (b), YFP fluorescence upon CFP excitation (indication of FRET) (c), and merge image of (a) to (c) in (d). (e) Color-coded image of CFP lifetime (different protoplast).
(II) Protoplasts transfected with N-CFP and GP-YFP treated with cytochalasin D 24 h p.t. GP-YFP (Gc-YFP) (a), N-CFP (b), YFP fluorescence upon CFP excitation (indication of FRET) (c), and merge image of (a) to (c) in (d). (e) Color-coded image of CFP lifetime (different protoplast).
The color legends indicate the CFP lifetime in picoseconds. The CFP lifetime measured upon single N-CFP expression is 2380 ps. Bars = 5 µm.
Glycoprotein Gc Accumulates at ERESs upon Coexpression with N
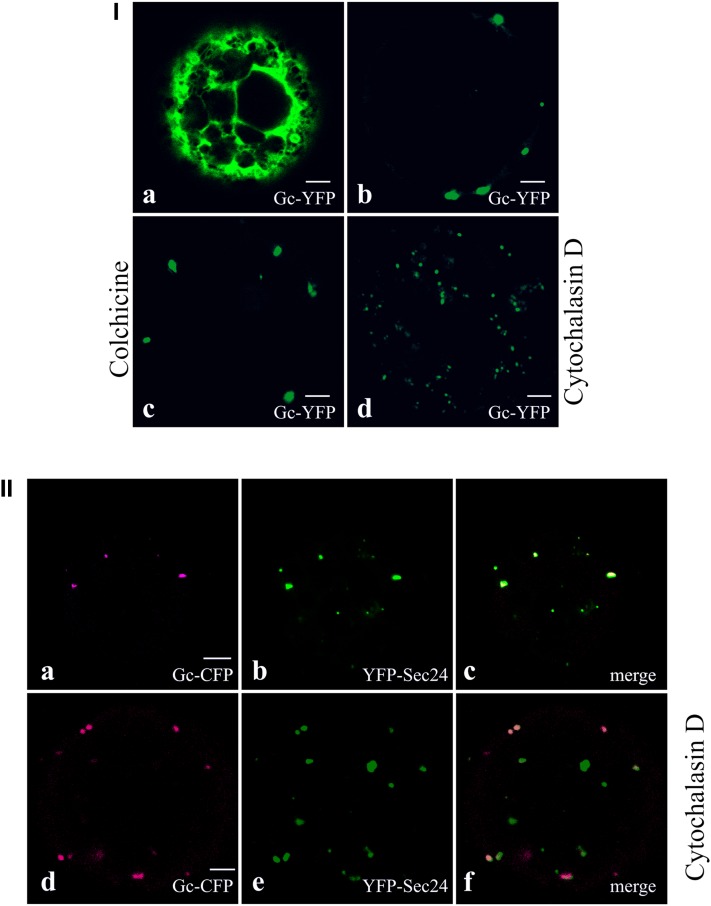
Previous studies have shown that upon coexpression, the TSWV Gc glycoprotein interacts with Gn and becomes ER export competent. During this process, most heterodimers rapidly end up at the Golgi complex, though small amounts can be detected at ERESs, which suggested that the glycoproteins most likely employ coat protein II (COPII)-coated membranes/vesicles to reach the Golgi (Ribeiro et al., 2008). When Gc was coexpressed with the cytoplasmic N protein in the absence of Gn, Gc also interacted with N but did not become ER export competent. Instead, a dramatic change from its reticular ER distribution into accumulating punctuate spots was discerned (Figures 6Ia and 6Ib) (Ribeiro et al., 2009a). As anticipated, the additional presence of colchicine did not have an effect, but cytochalasin D again increased the number and size of these spots (Figures 6Ib to 6Id).
Figure 6.
Localization of TSWV Gc/N Complexes within the ER.
(Ia) Protoplast transfected with Gc-YFP 24 h p.t.
(Ib) protoplast transfected with Gc-YFP and N 24 h p.t.
(Ic) protoplast transfected with Gc-YFP and N treated with colchicine 24 h p.t.
(Id) protoplast transfected with Gc-YFP and N treated with cytochalasin D 24 h p.t.
(IIa) to (IIc) Protoplasts transfected with Gc-CFP, N, and YFP-Sec24 at 24 h p.t. Gc-CFP (a), YFP-Sec24 (b), and merge of (a) and (b) in (c).
(IId) to (IIf) Protoplasts transfected with Gc-CFP, N, and YFP-Sec24 and incubated in the presence of cytochalasin D, 24 h p.t. Gc-CFP (d), YFP-Sec24 (e), and merge image of (d) and (e) in (f).
The presence of N in the protoplasts presented in (Ib) to (Id) and (IIa) to (IIf) is demonstrated by the punctate localization of Gc-CFP. Bars = 5 µm.
Due to the earlier observations, it was tempting to assume that N, although cytosolic, was also able to recruit and relocate Gc to ERESs. With this in mind, tobacco protoplasts were transfected with Gc-CFP, N, and ERES marker YFP-Sec24 (Stefano et al., 2006). The results showed that, indeed, N was able to interact with Gc and redirect it specifically to ERESs (Figures 6IIa to 6IIc). Even in the presence of cytochalasin D, the agglomerations of Gc-N still colocalized with the ERES marker (Figures 6IId to 6IIf). To exclude the possibility that the increased number in ERESs was triggered by depolymerization of the actin filaments, protoplasts were transfected with YFP-Sec24 ERES marker only and subjected to the same concentration of cytochalasin D. Samples were taken and analyzed at the same time point, and during repeated experiments in which 40 cells were observed (20 of which treated with cytochalasin D), the number of ERESs presented by the marker did not reveal any significant difference in the presence or absence of the drug. The increase in the number of ERES during coexpression of Gc and N in the presence of cytochalasin D thus likely involved a de novo formation of ERES.
Gc Is Recruited to ERES after Interaction of Its CT with the N Protein
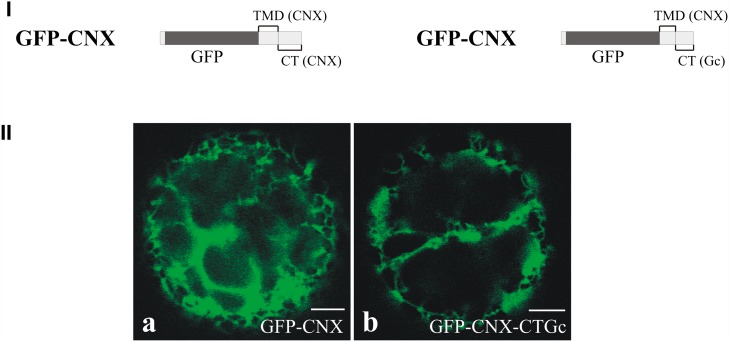
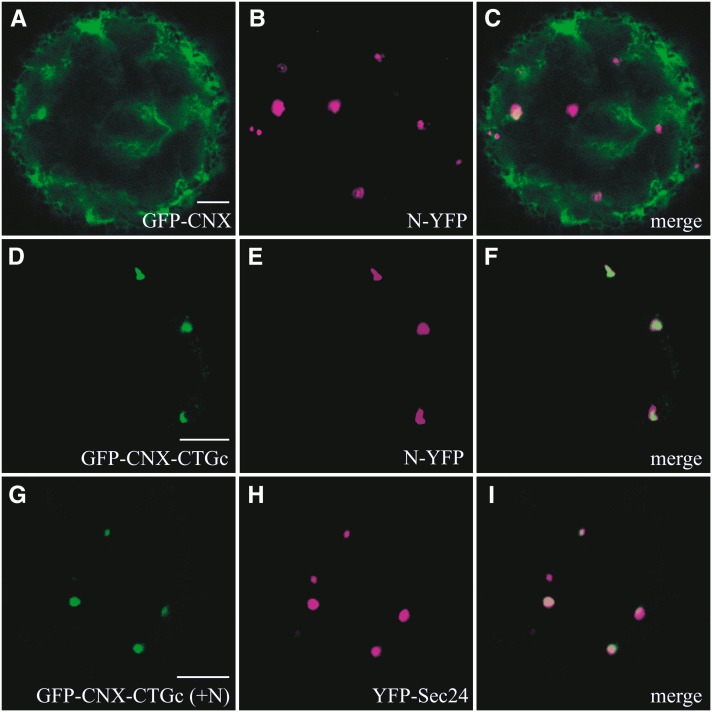
The recruitment of Gc to ERES was likely due to an interaction between N and Gc, most probably involving the cytoplasmic tail (CT) of Gc. To test this hypothesis, a calnexin mutant (denoted GFP-CNX-CTGc; Figure 7I) was constructed containing the CT of Gc (CTGc). This mutant originated from a calnexin derivative (named GFP-CNX; Figures 7I and 7IIa) in which the luminal domain was exchanged by GFP and, like calnexin, still localized in a reticular pattern (daSilva et al., 2005, 2006). Upon transfection of protoplasts with GFP-CNX-CTGc, similarly to GFP-CNX, a typical ER localization was observed (Figure 7IIb), which demonstrated that the CT of Gc did not alter the localization of this ER-resident protein. However, upon coexpression of GFP-CNX-CTGc with N-YFP, the reticular pattern disappeared and both proteins were observed to concentrate and colocalize at punctate spots (Figures 8D to 8F), similarly to what had previously been observed upon coexpression of Gc and N. As a control, GFP-CNX was coexpressed with N-YFP, but no change in the reticular localization of GFP-CNX nor any colocalization with N was observed (Figures 8A to 8C). To exclude the possibility that the observed change in localization was due to a deletion of the calnexin CT, another mutant was constructed, equivalent to GFP-CNX but lacking its CT (denoted GFP-CNXΔCT; daSilva et al., 2006). Expression of this protein resulted in a clear reticular ER localization (data not shown). Coexpression of GFP-CNX-CTGc, N, and YFP-Sec24 (Figures 8G to 8I) demonstrated that these punctate spots corresponded to ERESs and showed that N was also able to redirect this chimeric calnexin within the ER to ERES.
Figure 7.
Localization of Calnexin Mutants.
(I) Schematic representation of the calnexin mutants used in this study. TMDCNX and CTCNX refer to calnexin’s TMD and CT, respectively. CTGc refers to the CT of Gc. (II) Localization analysis of the two calnexin mutants. (a) Protoplast transfected with GFP-CNX 24 h p.t. (b) Protoplast transfected with GFP-CNX-CTGc 24 h p.t. Bars = 5 μm.
[See online article for color version of this figure.]
Figure 8.
Influence of N Protein on the Localization of a Calnexin Mutant Containing the CT of Gc.
(A) to (C) Protoplasts transfected with GFP-CNX and N-YFP at 24 h p.t. GFP-CNX (A), N-YFP (B), and merge of (A) and (B) in (C).
(D) to (F) Protoplasts transfected with GFP-CNX-CTGc and N-YFP at 24 h p.t. GFP-CNX-CTGc (D), N-YFP (E), and merge of (D) and (E) in (F).
(G) to (I) Protoplasts transfected with GFP-CNX-CTGc, N, and YFP-Sec24 at 24 h p.t. GFP-CNX-CTGc (G), YFP-Sec24 (H), merge of (G) and (H) in (I). The presence of N in the protoplasts presented in (G) to (I) is demonstrated by the punctate localization of GFP-CNX-CTGc.
Bars = 5 μm.
DISCUSSION
The plant-infecting bunyaviruses are among the very few plant viruses that are surrounded by a lipid membrane, obtained from the Golgi complex and spiked with two viral glycoproteins. So far, only limited information is available on the maturation mechanisms of these glycoproteins and their interplay with various elements of the cellular endomembrane system and cytoskeleton, ultimately leading to particle assembly at the Golgi interface. Unraveling this process might be of interest for development of future antiviral drugs that target glycoprotein maturation and functioning of the animal-infecting members of this family (of which viruses like Rift Valley Fever and Crimean Congo heamorraghic fever are listed as biological threats) and also might contribute novel insights into the functioning of the endomembrane system and how viruses manipulate this to their own benefit.
The glycoproteins from the plant-infecting bunyavirus TSWV have previously been studied in mammalian cells, and their trafficking and localization behavior were shown to be similar to the glycoproteins from their animal-infecting counterparts, which might be explained by their ancestral relationship (Snippe et al., 2005b; Kormelink et al., 2011; Plyusnin and Elliott, 2011). Animal and plant cells have a different organization of their endomembrane systems and cytoskeleton (Brandizzi et al., 2002). However, considering the functional and structural similarities between the plant- and animal-infecting bunyaviruses (Kikkert et al., 2001; Cortez et al., 2002; Snippe et al., 2005a), these viruses are likely to hijack the endomembrane system in a similar way.
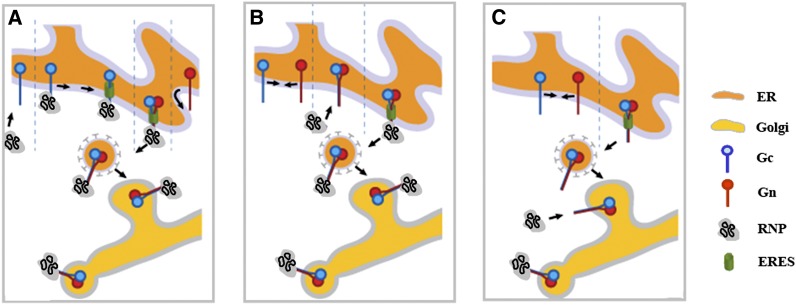
The data presented in this study, combined with earlier studies on the TSWV glycoproteins (Ribeiro et al., 2008, 2009a, 2009b), provide some novel insights into the very first (sequential) steps of viral glycoprotein maturation and their exit from ER that may be representative for all bunyaviruses, irrespective of plant or animal infection. Upon coexpression, both glycoproteins are ER export competent and exit via ERESs involving COPII vesicles as demonstrated by inhibition of ER-Golgi trafficking in the presence of COPII vesicle transport inhibitor Sec12p (Ribeiro et al., 2009b). Here, it is shown that, interestingly, Gc is also able to concentrate at ERES when in the presence of the cytosolic N protein, although further transport from there is halted. These results show that Gc localization does not entirely depend on its interaction with Gn. In light of the abundance of cytosolic N protein during the natural infection cycle and Gc being processed from the precursor following Gn, it becomes tempting to postulate a maturation pathway for the viral structural proteins in which the interaction between N and Gc may present a first and efficient step to concentrate Gc glycoprotein cargo and recruit host factors to ERESs prior to COPII vesicle formation and their subsequent scission from the ER (Figure 9). The fact that Gc in the presence of N is not able to escape from the ER points to a major role for Gn in the final stages of COPII vesicle formation/release. A previous study had already shown that a chimeric Gc provided with the longer TMD of Gn altered Gc into an ER export–competent glycoprotein (Ribeiro et al., 2009b) in agreement with and supporting other studies that indicated that the TMD length of type 1 membrane proteins has a major influence on their final destination within the endomembrane system (Brandizzi et al., 2002). Whether Golgi localization for all bunyavirus Gn glycoproteins relies on its TMD size, considering the absence of a clear (conserved) Golgi retention signal, remains to be investigated. Recently, the CTs of bunyavirus Gn glycoproteins have been postulated to act as matrix protein surrogates (Strandin et al., 2013), proteins that normally are able to alter membrane curvature and induce the formation of virus-like particles aided by cellular protein complexes from the endosomal sorting complexes required for transport (ESCRT). Although for bunyaviruses the involvement of ESCRT proteins in particle assembly has not yet been demonstrated, some of their Gn proteins contain amino acid sequences in their TMD-CT part that show similarity to Pro-rich late domains motifs from matrix proteins and are known to be involved in the recruitment of the cellular budding machinery (Strandin et al., 2013). The TSWV Gn glycoprotein contains a large TMD (amino acids 316 to 381; Kormelink et al., 1992; Cortez et al., 2002), and a closer look reveals a YPVL sequence (amino acids 351 to 354) in an internal hydrophilic core that shows a clear sequence similarity to the late domains of matrix proteins. No such similarity has been found with sequences within the CT of TSWV Gc. In plant cells, transient expression of Gn also induces the formation of pleomorphic, mostly circular, membrane structures somewhat reflecting a process of virus-like particle formation (Ribeiro et al., 2008). The observation that chimeric Gc containing the TMD of Gn become ER export competent (Ribeiro et al., 2009b) together makes it tempting to believe that Gn, in specific amino acids 316 to 381, enables recruitment of the cellular budding machinery to finalize ERES maturation and subsequent scission of COPII vesicles for anterograde transport to the Golgi. The various sequential protein interactions that may finally support TSWV glycoprotein Gn and Gc escape from ER and trafficking to Golgi are summarized in Figure 9. During this process, Gn plays the most important role, as only this viral protein supports the exit from ER of viral structural protein complexes, although speculative, possibly to support maturation and scission of COPII vesicles from ERES.
Figure 9.
Model Describing the Three Variants of Sequential Protein Interactions Leading to Gc Recruitment to ERES and Subsequent Exit via COPII Vesicles to Golgi, Where Bunyavirus Particle Assembly Primarily Takes Place.
In (A), Gc first becomes engaged in an interaction with N/RNPs prior to the interaction with Gn and subsequent ER exit via ERES; alternatively, Gc and Gn first heterodimerize. Next, although already ER exit competent, they immediately interact with N/RNPs to exit ER as a large structural viral protein complex (B) or only interact with RNPs at the Golgi interface while having transported there first as a Gn-Gc heterodimer (C).
[See online article for color version of this figure.]
Although for the animal-infecting bunyaviruses anterograde transport from ER to Golgi has not yet been shown to involve COPII vesicles, it is tempting to postulate that the maturation of the glycoproteins and their escape from ER to Golgi follows a similar pathway as described above for TSWV. In agreement with this model are studies on animal-infecting bunyaviruses in which particle assembly is also observed to occur at the ER-Golgi intermediate compartment (Jäntti et al., 1997; Snippe et al., 2005b). After all, this requires that all structural proteins (Gn, Gc, and N) already engage in an interaction at the ER to allow the envelopment of RNPs at the very first next organelle where COPII vesicles deliver their cargo, which in animal cells happens to be the ER-Golgi intermediate compartment.
Transport of individual (or small clusters of) N molecules through the cytoplasm to form larger inclusion bodies in plant cells seems to involve the actin network, but not microtubules. Although depolymerization of actin filaments by cytochalasin D led to an increase in number and decrease in size of N agglomerations, it did not prevent N from targeting to, colocalizing, and interacting with both glycoproteins at the ER and Golgi complex, as demonstrated by FRET/FLIM. Nevertheless, here, cytochalasin D did affect the number and size of the protein complexes, most likely as a direct consequence of a primary effect on the number and size of N inclusion bodies, prior to their targeting to the membranes where the viral glycoproteins localize. Similarly, the relocation of Gc by N from a reticular ER distribution to ERES did not require microtubules or actin filaments. During several control experiments, in the absence as well presence of viral proteins, ERESs surprisingly colocalized with depolymerized α-tubulin (see Supplemental Figure 2 online), while ERESs themselves were not affected by the inhibitors. In mammalian cells, microtubules are involved in the transport of proteins from ER to the Golgi (Murshid and Presley, 2004; Watson et al., 2005). Although in plants the actin network is involved in the maintenance of the organization of the Golgi apparatus and movement of the ER network (Boevink et al., 1998), cytoskeleton elements in general appear not to be required for protein transport from ER to the Golgi (Boevink et al., 1998; Brandizzi et al., 2002; Saint-Jore et al., 2002). The colocalization of ERESs and α-tubulin point to a correlation between ERES and microtubules. Whether this indicates a possible role of microtubules in the transport of cytoplasmic proteins to ERESs or in the formation of the latter remains to be investigated.
The earlier observation that Gn-Gc traffic to the Golgi system by COPII vesicles via ERESs (Ribeiro et al., 2009b) and the results from this study showing the concentration and arrest of Gc-N complexes at ERESs indicate the importance of ERES in the route of TSWV glycoproteins anterograde transport. ERESs are discrete domains of the ER where the machinery responsible for the formation of COPII vesicles is assembled (reviewed in Marti et al., 2010). In mammalian and yeast cells, the COPII coat components (the GTPase Sar1 and two heterodimeric complexes Sec23/24 and Sec13/31) are thought to be sequentially recruited at ERESs (Aridor et al., 2001; Matsuoka et al., 2001). However, the mechanism and regulation of ERES formation and cargo recruitment in plant cells are less well understood (Marti et al., 2010). It is possible that the cargo is recruited to defined ERES, as was observed to occur in mammalian cells (Aridor et al., 2001), but, alternatively, the cargo molecules themselves may recruit the COPII components to ERES (daSilva et al., 2004). Another study has shown that cargo transport is not only mediated by recruitment of COPII components to existing ERES, but also by de novo formation of ERES (Hanton et al., 2007). Our results seem to support the idea of de novo formation of ERES since we have shown that upon actin filaments disruption the increased number of Gc-N spots in the ER (though smaller in size) still correspond to ERES. The presented data also suggest that recruitment to ERES is not sufficient to grant ER export competence to the protein cargo and for Gc, as argued above, does rely on properties of Gn.
The role of the N protein in the accumulation of Gc at ERES is quite surprising. It is subject to different interpretations and raises several questions. First, the ability of N to colocalize with Gc at ER in a cytoskeleton-independent manner suggests that N possibly moves through the cytosol by cytoplasmic flow and, once in vicinity of the ER surface, interacts with the CT of Gc to catalyze (ERES) complex formation. The latter might involve a redirecting to existing ERES or de novo formation of ERES with recruitment of all the required COPII components. Whether the cytosolic N protein contains a sorting signal sequence to concentrate at existing or prestages of ERES is speculative. So far, experiments using organelle-specific marker proteins have never provided convincing indications for a colocalization of N with the endomembrane system, and during repeated coexpression experiments of N with the ERES marker Sec24, they never colocalized. While Gc-N has earlier been demonstrated to not colocalize with Golgi marker ST-GFP (Ribeiro et al., 2009a), it has been reported that, at least in leaf epidermal cells, Sec24 and Golgi markers are closely associated (Stefano et al., 2006). Thus, one would expect to see Gc+N colocalize with ST-GFP as well. However, coexpression of ST-GPF and Sec24 in tobacco protoplasts only showed partial colocalization (see Supplemental Figure 3 online). The absence of Gc+N colocalization with ST-GFP thus clearly marks the Gc+N colocalization at ERES as distinct.
The observation that a calnexin chimera provided with the CT of Gc was recruited to ERES upon coexpression with the N protein not only demonstrates that the interaction of Gc with N involves Gc-CT, but also supports the idea that generation of glycoprotein cargo complexes via protein interactions (e.g., with cytosolic N oligomers) likely presents an important trigger for de novo synthesis of ERES. The TSWV N and Gc proteins or N and Gc-CT–tailed chimeric proteins, upon coexpression, thereby not only present a novel bona fide ERES marker, but also form an interesting tool for the study of ERES biogenesis and dynamics.
METHODS
Plasmids and Organelle Markers
For the transient expression experiments, we made use of previously described constructs: pMON N, pMON N-YFP, and pMON N-CFP (Ribeiro et al., 2009a), as well as pMON Gc-YFP, pMON Gc-CFP, and pMON GP-YFP (Ribeiro et al., 2008).
The fluorescent proteins used in this study were the pH-insensitive forms of EYFP, ECFP, and mGFP5 (Haseloff et al., 1997). The spectral properties of mGFP5 allow efficient spectral separation from YFP (Brandizzi et al., 2002; Ribeiro et al., 2008, 2009a, 2009b).
In these experiments, we used GFP-α-tubulin, a marker of the microtubule network (Kumagai et al., 2001) and YFP-Sec24, a marker of ERESs (Stefano et al., 2006).
Plasmids encoding the GFP-fused TMD of Arabidopsis thaliana calnexin and CT of Gc (GFP-CNX-CTGc) mutant were obtained by assembly PCR. A PCR fragment containing the cauliflower mosaic virus 35S promoter, GFP, and the TMD of calnexin, flanked on its 3′-end with 24 nucleotides that overlap with the first 24 nucleotides at the 5′-terminal end of the CT of Gc, was amplified from a GFP-CNX construct (daSilva et al., 2006) (with primers 35SFwd, 5′-GGGCACTATCCTTCGCAAGACC-3′, and TMDCNXGcRvs, 5′-TTATAAGATTCATTCTTTACATATCGCCTTTTTGCCACCAAAGAT-3′). Another PCR fragment containing the CT of Gc, flanked on its 3′-end with a SpeI restriction site, was amplified from a construct that encodes Gc (pMONGc; prepared from the previously established pSFVGc; Kikkert et al., 2001), subjected to restriction enzyme digestion with BamHI, which resulted in the isolation of a cloning cassette that was inserted in the previously digested and dephosphorylated pMON999 vector, through the BamHI multiple cloning site, with the primers CTGcFwd, 5′-ATATGTAAAGAATGAATCTTATTA-3′, and CTGcRvs, 5′-GGGTCAGACAA GGTGAGAGAAATCCAT-3′. Both PCR fragments were fused by assembly PCR, and the product was digested with NcoI and SpeI and ligated to a predigested (with NcoI and XbaI) GFP-CNX construct (daSilva et al., 2006), leading to the final product. The integrity of all constructs was verified by sequence analysis.
Plant Material, Transient Expression, and Treatments
Tobacco plants (Nicotiana tabacum cv Petit Havana) were grown as described (Ribeiro et al., 2008) in controlled sterile conditions at 25°C with 16 h of light per day. Tobacco leaf protoplast preparation and transfection were performed as previously described (Ribeiro et al., 2008). When applicable, colchicine (Duchefa) or cytochalasin D (Sigma-Aldrich) were added immediately after recovery of the protoplasts from the electroporation procedure. The colchicine and cytochalasin D were prepared according to the manufacturer’s instructions. In brief, colchicine was dissolved in water, and cytochalasin D was initially dissolved in a stock solution of 1000× DMSO and afterwards further dissolved depending on the experiment (caution was taken not to exceed the maximum of 0.1% DMSO advised by the manufacturer to maintain good cell culture conditions). Several concentrations of colchicine and cytochalasin D were applied, and time-course analyses were performed to establish an optimal concentration for this specific type of cells and conditions. In the experiments presented in this study, the final concentrations of 100 μM colchicine and 20 μM cytochalasin D were used.
Sampling and Imaging
Confocal section images of living protoplasts (isolated at 24 to 48 h p.t.) were obtained using an inverted Zeiss 510 laser scanning microscope and a ×40 oil or ×63 oil and water immersion objective.
When imaging single expression of YFP-fused viral proteins, excitation lines of an argon ion laser of 488 nm were used with a 505/530-nm band-pass filter in the single-track facility of the microscope. For the imaging of the coexpressing YFP-fused and CFP-fused (as well as YFP-fused and GFP-fused) proteins, excitation lines of an argon ion laser of 458 nm for CFP (and GFP) and 514 nm for YFP were alternately used with line switching using the multitrack facility of the microscope. Fluorescence was detected using a 458/514-nm dichroic beam splitter and a 470/500-nm band-pass filter for CFP (and GFP) and a 535/590-nm band-pass filter for YFP. Appropriate controls were performed to exclude possible crosstalk and energy transfer between fluorochromes (Ribeiro et al., 2008, 2009a, 2009b).
Quantitative and Statistical Analysis
The quantitative analysis of the number and size of N agglomerations present in protoplasts upon treatment with the inhibitor and in nontreated cells was performed on a total of 70 cells per treatment. The sizes of the agglomerations were divided into four categories: 0 to 1 µm, 1 to 3 µm, 3 to 4 µm, and >4 µm. The number of N-YFP agglomerations per protoplast was counted and divided into five categories: 1 to 5, 6 to 10, 11 to 15, 16 to 20, and >20. The Mann-Whitney U test was applied, and the significance level was set at 0.05 (Ott and Longnecker, 2001).
FRET/FLIM Analyses
The FRET and FLIM analyses were performed with the same methodology and equipment as previously described (Ribeiro et al., 2009a).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: S48091 (TSWV GP precursor to the glycoproteins Gn and Gc) and AT3G07100 (ERES marker Sec24).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Time-Course Experiment for the Study of the Effect of Colchicine and Cytochalasin D on Microtubules.
Supplemental Figure 2. Localization Analysis of Gc N Complexes, ERES, and Depolymerized α-Tubulin.
Supplemental Figure 3. Localization and Behavior of Gc in the Presence of N Relative to ERES and Golgi Complex.
Acknowledgments
We thank Toshio Sano for kindly providing the GFP-α-tubulin construct, Federica Brandizzi for kindly providing the YFP-Sec24 marker construct, Gerard van der Krogt for the CFP and the pH-insensitive form of YFP, Jürgen Denecke for the GFP-CNX and GFP-CNXΔCT constructs as well as for valuable advice, and Maria João Cardoso for help with image processing. This work was financially supported by European Union-Research Training Network Grant HPRN-CT-2002-00262 and the Netherlands Organization for Scientific Research, section Earth and Life Sciences.
AUTHOR CONTRIBUTIONS
D.R., J.W.B., M.J., and S.M. performed the experiments. D.R., R.G., and R.K. designed the research. D.R. and R.K. wrote the article.
Glossary
- ER
endoplasmic reticulum
- TMD
transmembrane domain
- TSWV
Tomato spotted wilt virus
- RNP
ribonucleoprotein
- FRET
Förster resonance energy transfer
- FLIM
fluorescence lifetime imaging microscopy
- ERES
ER export site
- GFP
green fluorescent protein
- YFP
yellow fluorescent protein
- p.t.
posttransfection
- CFP
cyan fluorescent protein
- CT
cytoplasmic tail
References
- Andersson A.M., Melin L., Bean A., Pettersson R.F. (1997a). A retention signal necessary and sufficient for Golgi localization maps to the cytoplasmic tail of a Bunyaviridae (Uukuniemi virus) membrane glycoprotein. J. Virol. 71: 4717–4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson A.M., Melin L., Persson R., Raschperger E., Wikström L., Pettersson R.F. (1997b). Processing and membrane topology of the spike proteins G1 and G2 of Uukuniemi virus. J. Virol. 71: 218–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridor M., Fish K.N., Bannykh S., Weissman J., Roberts T.H., Lippincott-Schwartz J., Balch W.E. (2001). The Sar1 GTPase coordinates biosynthetic cargo selection with endoplasmic reticulum export site assembly. J. Cell Biol. 152: 213–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashby J., Boutant E., Seemanpillai M., Groner A., Sambade A., Ritzenthaler C., Heinlein M. (2006). Tobacco mosaic virus movement protein functions as a structural microtubule-associated protein. J. Virol. 80: 8329–8344 Erratum. J. Virol. 80: 12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y., Faulkner C., Ritzenthaler C., Maule A.J. (2010). Plasmodesmata: Gateways to local and systemic virus infection. Mol. Plant Microbe Interact. 23: 1403–1412 [DOI] [PubMed] [Google Scholar]
- Boevink P., Oparka K., Santa Cruz S., Martin B., Betteridge A., Hawes C. (1998). Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 15: 441–447 [DOI] [PubMed] [Google Scholar]
- Brandizzi F., Snapp E.L., Roberts A.G., Lippincott-Schwartz J., Hawes C. (2002). Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: Evidence from selective photobleaching. Plant Cell 14: 1293–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.Y., Matsuoka Y., Compans R.W. (1991). Assembly and polarized release of Punta Toro virus and effects of brefeldin A. J. Virol. 65: 1427–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez I., Aires A., Pereira A.M., Goldbach R., Peters D., Kormelink R. (2002). Genetic organisation of Iris yellow spot virus M RNA: Indications for functional homology between the G(C) glycoproteins of tospoviruses and animal-infecting bunyaviruses. Arch. Virol. 147: 2313–2325 [DOI] [PubMed] [Google Scholar]
- daSilva L.L.P., Foresti O., Denecke J. (2006). Targeting of the plant vacuolar sorting receptor BP80 is dependent on multiple sorting signals in the cytosolic tail. Plant Cell 18: 1477–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- daSilva L.L.P., Snapp E.L., Denecke J., Lippincott-Schwartz J., Hawes C., Brandizzi F. (2004). Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell 16: 1753–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- daSilva L.L.P., Taylor J.P., Hadlington J.L., Hanton S.L., Snowden C.J., Fox S.J., Foresti O., Brandizzi F., Denecke J. (2005). Receptor salvage from the prevacuolar compartment is essential for efficient vacuolar protein targeting. Plant Cell 17: 132–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A., Ahlquist P. (2012). Role of host reticulon proteins in rearranging membranes for positive-strand RNA virus replication. Curr. Opin. Microbiol. 15: 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott R.M. (1990). Molecular biology of the Bunyaviridae. J. Gen. Virol. 71: 501–522 [DOI] [PubMed] [Google Scholar]
- Elliott, R.M. (1996). The Bunyaviridae. (New York: Plenum Press). [Google Scholar]
- Gerrard S.R., Nichol S.T. (2002). Characterization of the Golgi retention motif of Rift Valley fever virus G(N) glycoprotein. J. Virol. 76: 12200–12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddette D.W., Frieden C. (1986). Actin polymerization. The mechanism of action of cytochalasin D. J. Biol. Chem. 261: 15974–15980 [PubMed] [Google Scholar]
- Griffiths G., Rottier P. (1992). Cell biology of viruses that assemble along the biosynthetic pathway. Semin. Cell Biol. 3: 367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha S., Bhattacharyya B. (1997). The colchicine-tubulin interaction: A review. Curr. Sci. 73: 351–358 [Google Scholar]
- Hanton S.L., Chatre L., Renna L., Matheson L.A., Brandizzi F. (2007). De novo formation of plant endoplasmic reticulum export sites is membrane cargo induced and signal mediated. Plant Physiol. 143: 1640–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries P., Ding B. (2011). Cellular factors in plant virus movement: At the leading edge of macromolecular trafficking in plants. Virology 411: 237–243 [DOI] [PubMed] [Google Scholar]
- Harries P.A., Schoelz J.E., Nelson R.S. (2010). Intracellular transport of viruses and their components: Utilizing the cytoskeleton and membrane highways. Mol. Plant Microbe Interact. 23: 1381–1393 [DOI] [PubMed] [Google Scholar]
- Haseloff J., Siemering K.R., Prasher D.C., Hodge S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäntti J., Hildén P., Rönkä H., Mäkiranta V., Keränen S., Kuismanen E. (1997). Immunocytochemical analysis of Uukuniemi virus budding compartments: Role of the intermediate compartment and the Golgi stack in virus maturation. J. Virol. 71: 1162–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkert M., Van Lent J., Storms M., Bodegom P., Kormelink R., Goldbach R. (1999). Tomato spotted wilt virus particle morphogenesis in plant cells. J. Virol. 73: 2288–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkert M., van Poelwijk F., Storms M., Kassies W., Bloksma H., van Lent J., Kormelink R., Goldbach R. (1997). A protoplast system for studying tomato spotted wilt virus infection. J. Gen. Virol. 78: 1755–1763 [DOI] [PubMed] [Google Scholar]
- Kikkert M., Verschoor A.D., Kormelink R., Rottier P., Goldbach R. (2001). Tomato spotted wilt virus glycoproteins exhibit trafficking and localization signals that are functional in mammalian cells. J. Virol. 75: 1004–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormelink, R., De Haan, P., Meurs, C., Peters, D., and Goldbach, R.(1992). The nucleotide sequence of the M RNA segment of tomato spotted wilt virus, a bunyavirus with two ambisense RNA segments. J. Gen. Virol. 73: 2795–804. [DOI] [PubMed]
- Kormelink R., Garcia M.L., Goodin M., Sasaya T., Haenni A.-L. (2011). Negative-strand RNA viruses: The plant-infecting counterparts. Virus Res. 162: 184–202 [DOI] [PubMed] [Google Scholar]
- Kumagai F., Yoneda A., Tomida T., Sano T., Nagata T., Hasezawa S. (2001). Fate of nascent microtubules organized at the M/G1 interface, as visualized by synchronized tobacco BY-2 cells stably expressing GFP-tubulin: Time-sequence observations of the reorganization of cortical microtubules in living plant cells. Plant Cell Physiol. 42: 723–732 [DOI] [PubMed] [Google Scholar]
- Laporte C., Vetter G., Loudes A.M., Robinson D.G., Hillmer S., Stussi-Garaud C., Ritzenthaler C. (2003). Involvement of the secretory pathway and the cytoskeleton in intracellular targeting and tubule assembly of Grapevine fanleaf virus movement protein in tobacco BY-2 cells. Plant Cell 15: 2058–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappin D.F., Nakitare G.W., Palfreyman J.W., Elliott R.M. (1994). Localization of Bunyamwera bunyavirus G1 glycoprotein to the Golgi requires association with G2 but not with NSm. J. Gen. Virol. 75: 3441–3451 [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Taoka K., Yoo B.C., Ben-Nissan G., Kim D.J., Lucas W.J. (2005). Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins. Plant Cell 17: 2817–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti L., Fornaciari S., Renna L., Stefano G., Brandizzi F. (2010). COPII-mediated traffic in plants. Trends Plant Sci. 15: 522–528 [DOI] [PubMed] [Google Scholar]
- Matsuoka K., Schekman R., Orci L., Heuser J.E. (2001). Surface structure of the COPII-coated vesicle. Proc. Natl. Acad. Sci. USA 98: 13705–13709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y., Ihara T., Bishop D.H., Compans R.W. (1988). Intracellular accumulation of Punta Toro virus glycoproteins expressed from cloned cDNA. Virology 167: 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morejohn L.C., Fosket D.E. (1991). The biochemistry of compounds with anti-microtubule activity in plant cells. Pharmacol. Ther. 51: 217–230 [DOI] [PubMed] [Google Scholar]
- Murshid, A., and Presley, J.F.(2004). ER-to-Golgi transport and cytoskeletal interactions in animal cells. Cell. Mol. Life Sci. 61: 133–145. [DOI] [PMC free article] [PubMed]
- Neumann U., Brandizzi F., Hawes C. (2003). Protein transport in plant cells: In and out of the Golgi. Ann. Bot. (Lond.) 92: 167–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehl A., Heinlein M. (2011). Cellular pathways for viral transport through plasmodesmata. Protoplasma 248: 75–99 [DOI] [PubMed] [Google Scholar]
- Ott, R., and Longnecker, M.(2001). An Introduction to Statistical Methods and Data Analysis, 5th ed. (Pacific Grove, CA: Duxbury). [Google Scholar]
- Peterson, R., and Melin, L.(1996). Synthesis, assembly, and intracellular transport of Bunyaviridae membrane proteins. In The Bunyaviridae, R.M. Elliott, ed (New York: Plenum Press), pp. 159–188. [Google Scholar]
- Plyusnin, A., and Elliott, R.(2011). Bunyaviridae: Molecular and cellular biology, A. Plyusnin and R. Elliott, eds (Norfolk, UK: Caister Academic Press), pp. 193–206. [Google Scholar]
- Ribeiro D., Borst J.W., Goldbach R., Kormelink R. (2009a). Tomato spotted wilt virus nucleocapsid protein interacts with both viral glycoproteins Gn and Gc in planta. Virology 383: 121–130 [DOI] [PubMed] [Google Scholar]
- Ribeiro D., Foresti O., Denecke J., Wellink J., Goldbach R., Kormelink R.J.M. (2008). Tomato spotted wilt virus glycoproteins induce the formation of endoplasmic reticulum- and Golgi-derived pleomorphic membrane structures in plant cells. J. Gen. Virol. 89: 1811–1818 [DOI] [PubMed] [Google Scholar]
- Ribeiro D., Goldbach R., Kormelink R. (2009b). Requirements for ER-arrest and sequential exit to the Golgi of Tomato spotted wilt virus glycoproteins. Traffic 10: 664–672 [DOI] [PubMed] [Google Scholar]
- Ritzenthaler, C. and Elamawi, R. (2006). The ER in replication of positive-strand RNA viruses. In Plant Cell Monographs, Vol. 4, The Plant Endoplasmic Reticulum, D.G. Robinson, ed. (Berlin: Springer Berlin Heidelberg), pp. 309–330. [Google Scholar]
- Saint-Jore C.M., Evins J., Batoko H., Brandizzi F., Moore I., Hawes C. (2002). Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 29: 661–678 [DOI] [PubMed] [Google Scholar]
- Schoelz J.E., Harries P.A., Nelson R.S. (2011). Intracellular transport of plant viruses: Finding the door out of the cell. Mol. Plant 4: 813–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Lappin D.F., Elliott R.M. (2004). Mapping the Golgi targeting and retention signal of Bunyamwera virus glycoproteins. J. Virol. 78: 10793–10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippe M., Borst J.W., Goldbach R., Kormelink R. (2005a). The use of fluorescence microscopy to visualise homotypic interactions of tomato spotted wilt virus nucleocapsid protein in living cells. J. Virol. Methods 125: 15–22 [DOI] [PubMed] [Google Scholar]
- Snippe M., Goldbach R., Kormelink R. (2005b). Tomato spotted wilt virus particle assembly and the prospects of fluorescence microscopy to study protein-protein interactions involved. Adv. Virus Res. 65: 63–120 [DOI] [PubMed] [Google Scholar]
- Snippe M., Smeenk L., Goldbach R., Kormelink R. (2007a). The cytoplasmic domain of tomato spotted wilt virus Gn glycoprotein is required for Golgi localisation and interaction with Gc. Virology 363: 272–279 [DOI] [PubMed] [Google Scholar]
- Snippe M., Willem Borst J., Goldbach R., Kormelink R. (2007b). Tomato spotted wilt virus Gc and N proteins interact in vivo. Virology 357: 115–123 [DOI] [PubMed] [Google Scholar]
- Stefano G., Renna L., Chatre L., Hanton S.L., Moreau P., Hawes C., Brandizzi F. (2006). In tobacco leaf epidermal cells, the integrity of protein export from the endoplasmic reticulum and of ER export sites depends on active COPI machinery. Plant J. 46: 95–110 [DOI] [PubMed] [Google Scholar]
- Strandin T., Hepojoki J., Vaheri A. (2013). Cytoplasmic tails of bunyavirus Gn glycoproteins-Could they act as matrix protein surrogates? Virology 437: 73–80 [DOI] [PubMed] [Google Scholar]
- Thomas C.L., Bayer E.M., Ritzenthaler C., Fernandez-Calvino L., Maule A.J. (2008). Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 6: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot-Lubicz J., Torrance L., Solovyev A.G., Morozov S.Y., Jackson A.O., Gilmer D. (2010). Varied movement strategies employed by triple gene block-encoding viruses. Mol. Plant Microbe Interact. 23: 1231–1247 [DOI] [PubMed] [Google Scholar]
- Vitale, A., and Denecke, J.(1999). The endoplasmic reticulum-gateway of the secretory pathway. Plant Cell 11: 615–628. [DOI] [PMC free article] [PubMed]
- Watson P., Forster R., Palmer K.J., Pepperkok R., Stephens D.J. (2005). Coupling of ER exit to microtubules through direct interaction of COPII with dynactin. Nat. Cell Biol. 7: 48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright K.M., Wood N.T., Roberts A.G., Chapman S., Boevink P., Mackenzie K.M., Oparka K.J. (2007). Targeting of TMV movement protein to plasmodesmata requires the actin/ER network: Evidence from FRAP. Traffic 8: 21–31 [DOI] [PubMed] [Google Scholar]
- Yang Y.S., Strittmatter S.M. (2007). The reticulons: A family of proteins with diverse functions. Genome Biol. 8: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]