The Ubc13-type ubiquitin-conjugating enzyme catalyzes Lys-63–specific ubiquitination, which usually plays a nonproteolytic, regulatory role in the cell. In this study, Fni3, the tomato homolog of Ubc13, and its cofactor Suv were identified and shown to regulate Fen-mediated and other R protein–mediated, immunity-associated cell death, revealing an important mechanism for regulation of the plant immune system.
Abstract
The activation of an immune response in tomato (Solanum lycopersicum) against Pseudomonas syringae relies on the recognition of E3 ligase–deficient forms of AvrPtoB by the host protein kinase, Fen. To investigate the mechanisms by which Fen-mediated immunity is regulated, we characterize in this study a Fen-interacting protein, Fni3, and its cofactor, S. lycoperiscum Uev (Suv). Fni3 encodes a homolog of the Ubc13-type ubiquitin-conjugating enzyme that catalyzes exclusively Lys-63–linked ubiquitination, whereas Suv is a ubiquitin-conjugating enzyme variant. The C-terminal region of Fen was necessary for interaction with Fni3, and this interaction was required for cell death triggered by overexpression of Fen in Nicotiana benthamiana leaves. Fni3 was shown to be an active E2 enzyme, but Suv displayed no ubiquitin-conjugating activity; Fni3 and Suv together directed Lys-63–linked ubiquitination. Decreased expression of Fni3, another tomato Ubc13 homolog, Sl-Ubc13-2, or Suv in N. benthamiana leaves diminished cell death associated with Fen-mediated immunity and cell death elicited by several other resistance (R) proteins and their cognate effectors. We also discovered that coexpression of Fen and other R proteins/effectors with a Fni3 mutant that is compromised for ubiquitin-conjugating activity diminished the cell death. These results suggest that Fni3/Sl-Ubc13-2 and Suv regulate the immune response mediated by Fen and other R proteins through Lys-63–linked ubiquitination.
INTRODUCTION
An important component of induced plant defense against attempted pathogen infection involves recognition of pathogen effectors by plant resistance (R) proteins, which in turn activates downstream signaling pathways that culminate in the host response known as effector-triggered immunity (Hogenhout et al., 2009; Oh and Martin, 2011). Strain DC3000 of Pseudomonas syringae pv tomato (Pst) expresses and delivers ∼30 type III effectors into the host cell during infection of tomato (Solanum lycopersicum), causing bacterial speck disease on plants that lack genetic resistance (Pedley and Martin, 2003; Lindeberg et al., 2006). In tomato plants expressing Pto, a Ser/Thr protein kinase, and Prf, a coiled-coil–nucleotide binding site–leucine-rich repeat protein, either of two effector proteins, AvrPto or AvrPtoB, is recognized by Pto, leading to host resistance to Pst infection (Tang et al., 1996; Kim et al., 2002). The Pto gene belongs to a small gene family in which another member encodes the protein kinase Fen (Martin et al., 1994; Pedley and Martin, 2003). Fen does not recognize AvrPto or AvrPtoB but does detect certain truncated forms and natural variants of AvrPtoB that lack the C-terminal domain (Abramovitch et al., 2003; Lin et al., 2006; Rosebrock et al., 2007). Fen-mediated recognition of AvrPtoB variants activates a rapid, localized programmed cell death (PCD) referred to as the hypersensitive response on host plants (Heath, 2000; Abramovitch et al., 2003). This immunity-associated PCD eventually arrests the growth and development of the Pst pathogen at the sites of attempted infection.
AvrPtoB is a modular protein, and its C-terminal domain encodes a structural mimic of eukaryotic RING/U-box–like E3 ubiquitin ligases (Janjusevic et al., 2006). Host immunity mediated by Fen kinase was termed Rsb (for resistance suppressed by AvrPtoB C terminus) after the observation that it is undermined by AvrPtoB E3 ligase–mediated ubiquitination and consequent degradation of Fen (Abramovitch et al., 2003; Rosebrock et al., 2007). Rsb immunity exists in many cultivated and wild species of tomato and is believed to have evolved prior to the occurrence of Pto-mediated immunity (Rosebrock et al., 2007). Early work did not reveal a role for Fen in mediating immunity against Pst, and few investigations on Fen-mediated immunity have been reported (Martin et al., 1994). By contrast, nearly two dozen components involved in Pto-mediated immunity have been identified and characterized in tomato and/or Nicotiana benthamiana, a model plant closely related to tomato (Oh and Martin, 2011). Several of these components, including Prf, are also essential to Fen-mediated immunity, suggesting substantial overlap of Fen- and Pto-mediated signal transduction pathways (Rosebrock et al., 2007). Despite these findings, the mechanism by which Fen-mediated immunity is regulated remains largely unknown.
Ubiquitination has emerged in recent years as an important regulatory mechanism underlying plant immune signaling (Zeng et al., 2006; Trujillo and Shirasu, 2010). The ubiquitination process attaches ubiquitin, a highly conserved 76–amino acid protein, to a substrate through stepwise reactions mediated by members of three different classes of enzymes, a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3) (Ciechanover, 1998). The E3 ubiquitin ligases play a key role in governing substrate specificity and therefore have been the focus of many studies related to ubiquitination. By contrast, the E2 ubiquitin–conjugating enzyme is often considered as a “carrier of ubiquitin” with auxiliary roles and has seldom been investigated. However, increasing evidence suggests that E2 enzymes play an important role in determining the topology of the polyubiquitin chain and the fate of substrate proteins being modified by ubiquitination (Ye and Rape, 2009). Several E3 ligases have been shown to act as either positive or negative regulators of plant immunity and associated PCD (Zeng et al., 2004; González-Lamothe et al., 2006; Yang et al., 2006; Zeng et al., 2006; Trujillo et al., 2008; Spoel et al., 2009). Nevertheless, the role of E2 enzymes in plant immunity remains largely unexplored.
The E2 enzymes possess a characteristic catalytic core with a conserved tertiary structure in which a Cys residue is present at the active site. Eukaryotic genomes typically encode dozens of E2 proteins that can be classified into multiple subfamilies (Vierstra, 2003; Michelle et al., 2009). Among all E2 members, Ubc13 and its homologs are the only ones known to exclusively catalyze Lys-63–linked polyubiquitination, a form of ubiquitination that has been shown to serve as a nonproteolytic, regulatory signal in diverse cellular processes (Galan and Haguenauer-Tsapis, 1997; Hofmann and Pickart, 1999; Deng et al., 2000; Spence et al., 2000). Uniquely, Ubc13 requires cofactors with which it forms a heterodimer essential for its catalytic activity. These cofactors, termed ubiquitin-conjugating enzyme variants (Uev), are similar in structure and sequence to active E2 enzymes but lack the conserved active-site Cys residue and, thus, by themselves lack conjugating activity (VanDemark et al., 2001; Zhang et al., 2005; Eddins et al., 2006). Two types of Uev cofactors have been characterized in mammals to date (Andersen et al., 2005). The Methyl methanesulfonate sensitive2 (Mms2) type promotes error-free DNA replication during DNA repair (Xiao et al., 1998). The second type, Uev1a, differs from Mms2 by having an additional ∼30 amino acids at its N terminus (Andersen et al., 2005). Notably, Uev1a plays a critical role in the regulation of human and animal immunity (Bhoj and Chen, 2009).
Plant genomes have Ubc13- and Uev-like genes, although they are not well studied. Two Arabidopsis thaliana Ubc13-like genes were identified by sequence homology and were recently implicated in Arabidopsis epidermal cell differentiation and iron deficiency responses (Wen et al., 2006; Li and Schmidt, 2010). The Arabidopsis E2 variant Constitutive photomorphogenesis10 (COP10) negatively regulates photomorphogenesis and promotes activity of Ubc13/Uev1a and other E2 enzymes (Yanagawa et al., 2004). However, the activity of COP10 is clearly distinct from Mms2 or Uev1a, as it is not exclusively involved in the formation of Lys-63 ubiquitin linkages. In addition, four Arabidopsis Uev homologs were identified, and one of them, Uev1D, was shown to be involved in DNA damage response (Wen et al., 2008).
In order to investigate how Fen-mediated immunity is regulated, a screen for Fen-interacting (Fni) proteins was performed using a tomato yeast two-hybrid library (Halterman, 1999). The screen identified Fni3 (for Fen-interacting protein 3), a tomato Ubc13-type ubiquitin-conjugating enzyme, and subsequent experiments showed Fni3 does not interact with other members of the Pto family. In an independent screen using virus-induced gene silencing (VIGS), a tomato Uev homolog, Suv (for S. lycopersicum Uev), was identified (del Pozo et al., 2004) and found to interact with Fni3. Here, we report a role for Fni3/Suv in the regulation of plant immunity-associated PCD. Fni3 was found to encode an active E2, while Suv displayed no ubiquitin-conjugating activity. Fni3 acted with Suv to direct Lys-63–linked ubiquitination, and both were found to be required for Fen-mediated immunity in N. benthamiana. We revealed that Fni3 and its homologs, Ubc13-2 and Suv, are required for PCD associated with the immune response activated by several other plant R proteins. Together, our data suggest Ubc13/Uev-like ubiquitin-conjugating enzyme/E2 variant via Lys-63–specific ubiquitination plays an essential role in effector-triggered immunity.
RESULTS
A Tomato Ubc13-Type Ubiquitin-Conjugating Enzyme Interacts with Fen
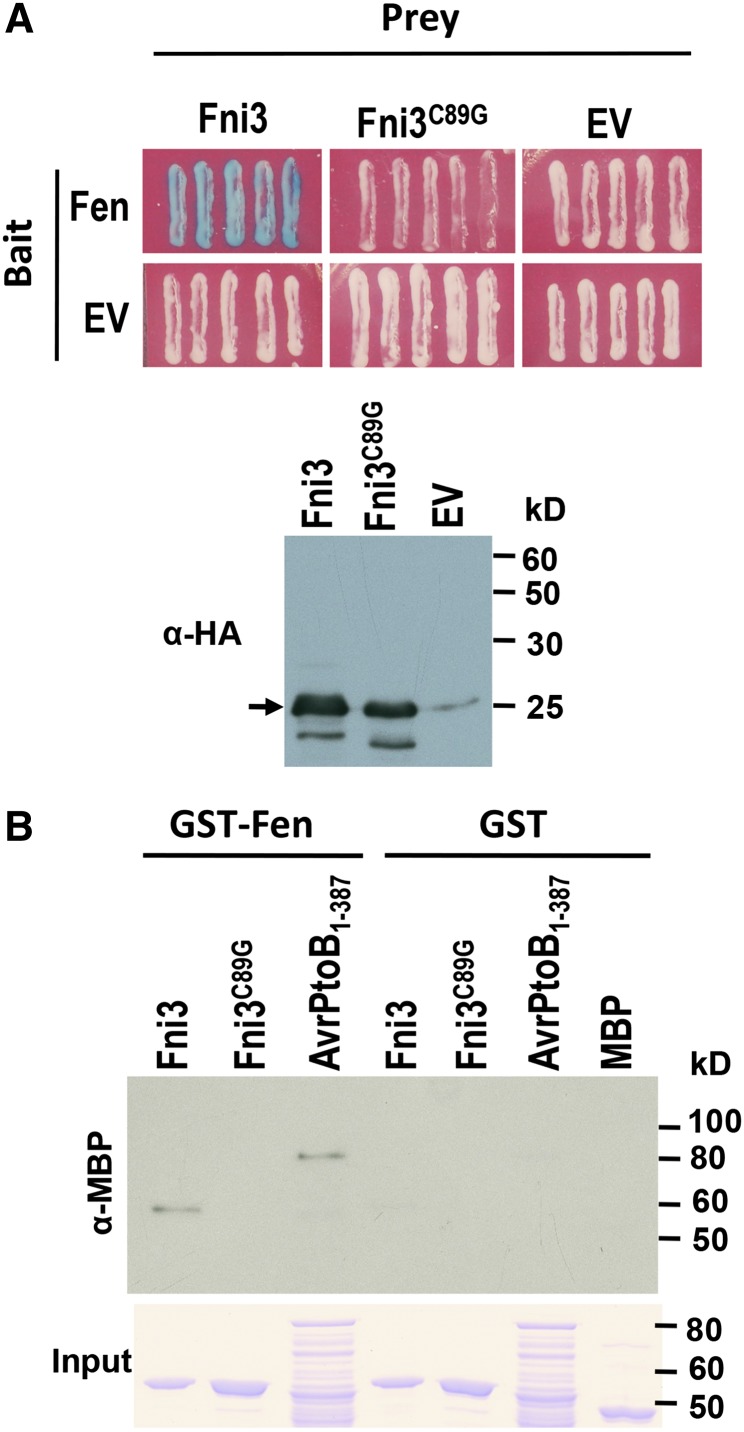
Host immunity mediated by the Fen kinase against the bacterial pathogen Pst occurs in many cultivars and wild relatives of tomato (Rosebrock et al., 2007). To understand how Fen-mediated immunity is regulated, a yeast two-hybrid screen for Fen-interacting (Fni) proteins was performed using a tomato cDNA library generated from leaves of Rio Grande-PtoR tomato plants challenged with Pst (Zhou et al., 1995; Halterman, 1999). From a screen of approximately two million initial transformants, the Fni3 was identified (Figure 1A). A bimolecular fluorescence complementation (BiFC) assay indicated that Fni3 associated with Fen in tomato cells as well (see Supplemental Figure 1A online). Alignment of Fni3 with two Ubc13 homologs from Arabidopsis and vertebrate Ubc13 proteins from frog (Xenopus tropicalis), human (Homo sapiens), or mouse (Mus musculus) indicated Fni3 shares 98 and 78% identity to Arabidopsis and vertebrate homologs, respectively (see Supplemental Figures 1B and 1C and Supplemental Data Set 1 online). As might be expected with this overall high level of amino acid sequence similarity between Fni3 and other Ubc13 homologs, Fni3 has all of the functionally important amino acids for biochemical activities of Ubc13, namely, Cys-89 located in the putative active site for ubiquitin thioester formation, Met-66, which is involved in the interaction with an E3 ligase, and three pocket residues (Glu-57, Phe-59, and Arg-72) that determine binding specificity for Uev protein (VanDemark et al., 2001; Wooff et al., 2004). Substitution of Cys-89 in the active site of Fni3 with Gly (Fni3C89G) essentially abolishes the Fen–Fni3 interaction (Figure 1A).
Figure 1.
A Tomato Ubc13-Like Protein, Fni3, Interacts with the Fen Protein.
(A) Fni3 interacts with Fen in a yeast two-hybrid assay. This was demonstrated by the activation of the lacZ reporter gene (blue patches). Photographs were taken 30 h after streaking onto plates containing X-Gal. Expression of Fni3 and Fni3C89G mutant proteins was examined by anti-HA immunoblotting. The arrow denotes the band in which Fni3 and Fni3C89G was detected.
(B) Fni3 interacts physically with Fen in a GST pull-down assay. Binding of GST-Fen to MBP-tagged fusion protein, MBP-Fni3, MBP-Fni3C89G, and MBP-AvrPtoB1-387, was examined by immunoblotting using anti-MBP monoclonal antibody. GST was used as a negative control. MBP-AvrPtoB1-387 was used as a positive control. The Coomassie blue–stained gel in the bottom panel (labeled as input) indicates equal amount of MBP fusion proteins were used for binding GST-Fen and GST in the assay. The numbers on the right show the molecular mass of marker proteins in kilodaltons.
Since Fen shares ∼80% amino acid identity with Pto and other members of the Pto family (Martin et al., 1994; Chang et al., 2002), we next evaluated the ability of Fni3 to interact with four Pto family members from the wild tomato species Solanum pimpinellifolium, homologs of Fen, Pto, and PtoD from the wild tomato species Solanum habrochaites, and three other kinases that are involved in tomato immunity against Pst, Adi3, MAPKKKα, and Pti1 (Zhou et al., 1995; del Pozo et al., 2004; Devarenne et al., 2006). Fni3 was found to interact with Fen and the S. habrochaites Fen homolog, but not with other Pto family members or with Adi3, Pti1, or MAPKKKα (see Supplemental Figures 2A and 2B online), indicating the interaction of Fen with Fni3 is specific.
Conventionally, ubiquitin-congugating enzymes (E2s) are thought to interact primarily with ubiquitin ligases (E3s). Nevertheless, E2 enzymes have been shown in several cases to interact with substrate proteins of ubiquitination (Laine et al., 2006; Shembade et al., 2007; Hong et al., 2008). Interaction of Fen and Fni3 in the yeast cell could be either direct or indirect, possibly involving an E3 ligase. To distinguish between these possibilities, we used a glutathione S-transferase (GST) pull-down assay to examine binding of Fni3 to Fen (Figure 1B). The result indicated that Fni3 interacts directly with Fen. Fen also interacted with the AvrPtoB1-387 protein (Rosebrock et al., 2007), but not with the Fni3 protein carrying the C89G substitution.
In addition to Fni3, the tomato genome also encodes another Ubc13-like protein we designated as Sl-Ubc13-2. Sl-Ubc13-2 is 99% identical to Fni3 in amino acid sequence and 91% identical in nucleotide sequence (see Supplemental Figures 3A and 3B online). Sl-Ubc13-2 physically interacted with Fen (see Supplemental Figure 2C online). However, it interacted only weakly with Fen in a yeast two-hybrid assay (see Supplemental Figure 2A online), possibly due to being modified in yeast (see Supplemental Figure 2A online, right panel). No interaction was detected between Sl-Ubc13-2 and other Pto family members.
The C-Terminal Region of Fen Is Necessary for Interaction with Fni3
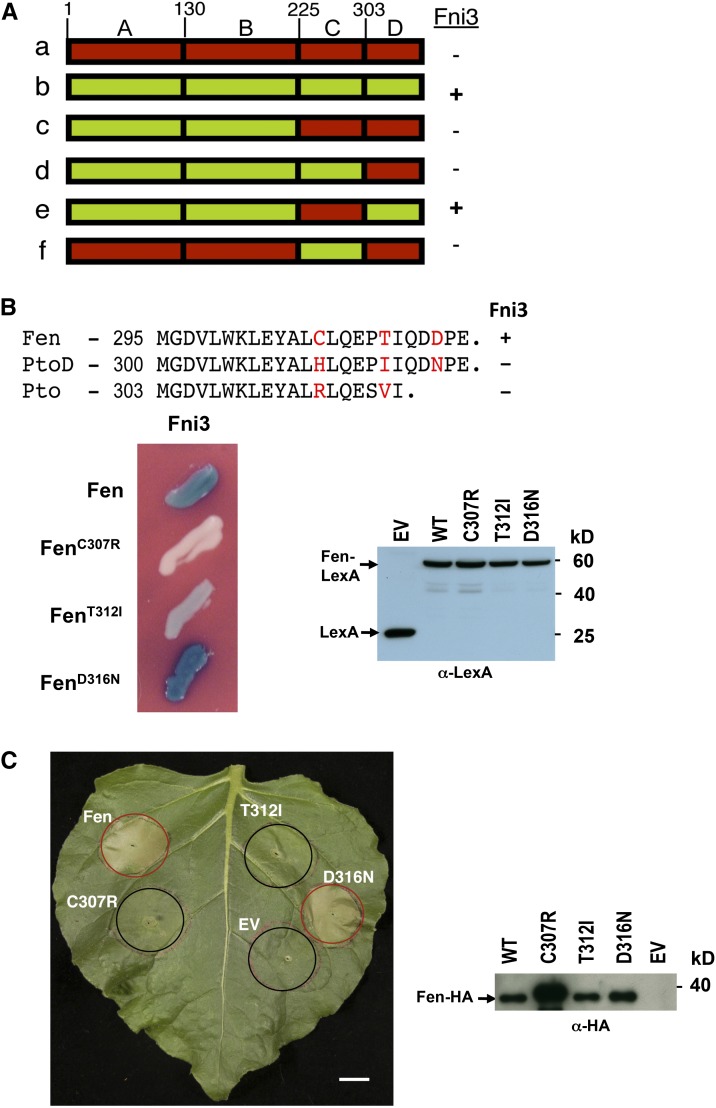
To determine which region of Fen is involved in the interaction with Fni3, we used a series of Fen/Pto chimeric proteins that were reported previously to be expressed in yeast (Tang et al., 1996) and have proven useful in the identification of Pto residues critical for AvrPto and AvrPtoB interaction (Frederick et al., 1998; Kim et al., 2002). While it is possible that the N-terminal part (A and B regions, Figure 2A) of Fen contributes to binding with Fni3, we found that only the chimeric protein that retains the C terminus of Fen (the D region) was able to interact with Fni3 (Figure 2A). Sequence alignment of the D regions of Fen and PtoD (the closest paralog of Fen that is unable to interact with Fni3) revealed differences at only three residues, Cys-307, Thr-312, and Asp-316, in the Fen protein (Figure 2B). Two of these residues, Cys-307 and Thr-312, also differ from Pto, although Pto also lacks the C-terminal five amino acids that are present in Fen and PtoD. To assess the possible involvement of these polymorphisms in interacting with Fni3, the three residues were individually substituted with the corresponding PtoD or Pto residue, and the variant Fen proteins were tested for their ability to interact with Fni3. The interaction of Fni3 with two variant Fen proteins, FenCys307Arg and FenThr312Ile, was compromised and significantly diminished, respectively (Figure 2B).
Figure 2.
Interaction of Fni3 with Fen Is Required for PCD Triggered by Overexpression of Fen.
(A) Identification of the region of Fen required for interaction with Fni3 using Pto/Fen chimeric proteins. Pto (in red)/Fen (in green) chimeric proteins composed of four regions, A to D, were tested for interaction with Fni3 in a yeast-two hybrid assay. + Denotes positive interaction, and − represents no interaction.
(B) Interaction of Fen and Fen variant proteins with Fni3. Sequence alignment of region D from Fen, PtoD, and Pto reveals polymorphism in three amino acid residues of Fen (Cys-307, Thr-312, and Asp-316). Fen mutant proteins containing substitution at these residues with corresponding amino acids from PtoD or Pto, FenCys307Arg (FenC307R or C307R), FenThr312Ile (FenT312I or T312I), and FenAsp316Asn (FenD316N or D316N) were tested for interaction with Fni3.The abundance of Fen and Fen variant proteins expressed in the yeast cells was examined by anti-LexA immunoblotting. Blue patches indicating activation of the lacZ reporter gene denote interaction occurs. Photographs were taken 24 h after streaking onto plates containing X-Gal.
(C) HA-tagged Fen and Fen D region variants were evaluated for their ability to elicit PCD in N. benthamiana. This experiment was repeated three times with similar results. The abundance of HA-tagged Fen and Fen mutant proteins was examined by anti-HA immunoblotting following expression in N. benthamiana via Agrobacterium-mediated transient expression. EV, pBTEX empty vector; WT, the wild type. Bar = 1 cm.
Interaction of Fen with Fni3 Is Required for PCD Triggered by Overexpression of Fen in N. benthamiana
Agrobacterium tumefaciens–mediated transient expression of Fen in N. benthamiana leaves causes constitutive signaling by Fen that leads to immunity-associated PCD in a Prf-dependent but effector-independent manner (Chang et al., 2002; Mucyn et al., 2009). We used this phenotype to evaluate the requirement of the Fen–Fni3 interaction for immunity-associated PCD initiation. The amino acid sequence of the N. benthamiana homolog of Fni3 (Nb-Fni3) is nearly identical to that of tomato Fni3 (see Supplemental Figure 3C online). All Fen variant proteins were expressed in N. benthamiana leaves, and an empty vector (EV) was used as a negative control (Figure 2C). The two proteins that interacted with Fni3, Fen and FenAsp316Asn, elicited a strong PCD response, whereas FenCys307Arg, FenThr312Ile, and the negative control failed to activate PCD (Figure 2C). These observations support the notion that interaction of Fen with Fni3 is important for induction of PCD. However, an alternative possibility is that the Cys307Arg or Thr312Ile substitutions grossly alter Fen structure and prevent it from interacting with other host proteins that may be necessary for Fen-mediated PCD. We therefore used FenThr312Ile as an example to assess the ability of the two mutants to interact with two additional proteins, Fni2 and AvrPtoB1-387. Fni2 is an uncharacterized Rcd-like kinase that was also found to interact with Fen (Halterman, 1999). FenThr312Ile interacted with both Fni2 and AvrPtoB1-387 in a manner similar to Fen but was not able to interact with Fni3 (see Supplemental Figure 4 online). These results suggest the interaction of Fen with Fni3 is required for PCD associated with Fen-mediated immunity.
Suv Encodes a Uev Homolog That Interacts with Fni3
A previously reported VIGS screen in N. benthamiana evaluated the role of ∼2400 random tomato cDNAs in Pto-elicited, plant immunity–associated PCD (del Pozo et al., 2004). Genes identified in this screen have been found to encode MAPKKKα, Cbl10, and Cipk6 (del Pozo et al., 2004; de la Torre et al., 2013). Of relevance here, another cDNA identified from the screen encodes a protein with striking similarity to Ubc13 cofactor Uev proteins, Mms2 and Uev1a. We named this gene Suv (for Solanum lycopersicum Uev). Phylogenetic analysis of Suv with two yeast Mms2 proteins from Schizosaccharomyces pombe and Saccharomyces cerevisiae, two variants of human Uev, a Uev protein from mouse, and Arabidopsis COP10 (as an outlier) revealed that Suv is more similar to the ancestor of Mms2 and Uev1a than to either protein individually (see Supplemental Figure 5A and Supplemental Data Set 2 online). As mentioned above, Ubc13-directed ubiquitination of substrate requires the formation of a dimeric complex with a Uev protein. Since Fni3 and Suv are each highly homologous to the corresponding component of the Ubc13/Uev heterodimeric complex, we hypothesized that Fni3 and Suv may interact in vivo. Indeed, strong interaction was detected between Fni3 and Suv, while no interaction between Fen and Suv was observed in yeast two-hybrid assay (see Supplemental Figure 5B online). Suv also showed strong interaction with Sl-Ubc13-2 (see Supplemental Figure 5C online).
Fni3 Is an Active Ubiquitin-Conjugating Enzyme, Whereas Suv Has No E2 Enzymatic Activity
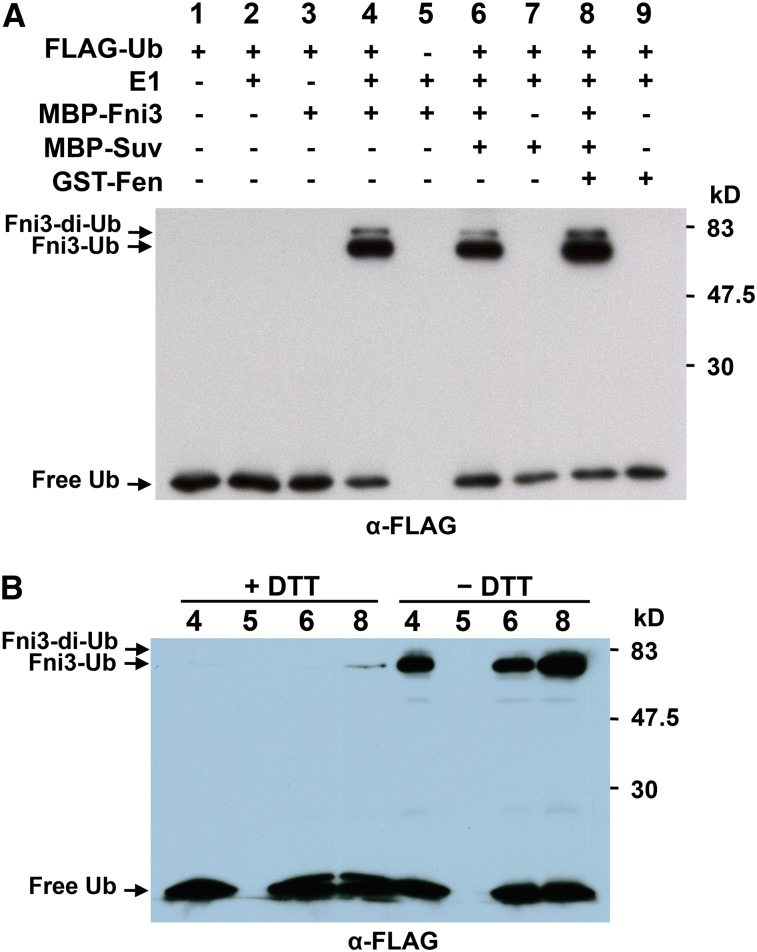
The E2 ubiquitin–conjugating enzymes mediate the transfer of an activated ubiquitin molecule from a ubiquitin-activating enzyme (E1) to its own active-site Cys through formation of a thioester bond. To determine whether Fni3 and Suv are active E2 enzymes, we used a thioester assay (Philip and Haystead, 2007) to assess the ability of Fni3 and Suv to accept an activated ubiquitin molecule from an E1. A recombinant E1 enzyme was first incubated with or without FLAG-tagged ubiquitin in the presence of ATP to allow for the formation of E1 ubiquitin thioester intermediates (charged E1). Fni3 and/or Suv were then added to the reaction. In the presence of the charged E1, Fni3 demonstrated conjugating activity, manifested by the formation of monoubiquitin- and diubiquitin-bound Fni3. As predicted, Suv did not exhibit conjugating activity, likely due to the lack of the active-site Cys required for conjugation to ubiquitin (Figure 3A).
Figure 3.
Fni3 Is an Active E2 Enzyme, Whereas Suv Has No Ubiquitin-Conjugating Activity.
(A) Anti-FLAG immunoblot following a thiolester assay with recombinant fusion proteins MBP-Fni3 and MBP-Suv, E1, and FLAG-ubiquitin (FLAG-Ub). The numbers on the top mark the lanes/reactions.
(B) Attachment of ubiquitin adducts to Fni3 in the thioester assay is DTT sensitive. The numbers on the top denote the corresponding reactions indicated in (A). The numbers on the right denote the molecular mass of marker proteins in kilodaltons.
[See online article for color version of this figure.]
We next examined the possibility that Fen might affect the ubiquitin-conjugating activity of Fni3 via phosphorylation or simply by binding to Fni3. First, we tested whether Fen could enhance or suppress Fni3 activity, not through phosphorylation, but simply through binding to Fni3 in the thioester assay (Figure 3A). The proper conformation of Fen is likely required for the ability to bind interacting proteins, and kinases like Fen often need to be phosphorylated to obtain this conformation. Therefore, we preincubated Fen with Fni3 under reaction conditions that allow Fen to be an active kinase to allow for autophosphorylation of Fen before the thioester assay. The formation of a ubiquitin-Fni3 linkage was similar to that displayed in the reaction without preincubation with Fen (Figure 3A, lanes 4, 6, and 8), indicating Fen does not affect the formation of Fni3-ubiquitin linkage in this assay. To ensure this observation is not affected by the epitope tag (maltose binding protein [MBP]) fused to Fni3/Suv, we also performed the assay using GST-tagged Fni3 and Suv, and a similar result was obtained (see Supplemental Figure 6A online).
The ubiquitin-Fni3 linkage formed in the above assay can be linked through a thioester bond to the catalytic Cys residue (Cys-89), which is sensitive to DTT treatment, or through the amino group of Fni3 (amide linkage, resistant to DTT treatment). To distinguish them, we split the reactions and treated the reaction separately either with DTT or without DTT. As shown in Figure 3B, essentially no ubiquitin-Fni3 linkage was retained after DTT treatment in the reactions in which Fni3 alone or Fni3/Suv was present, indicating that the linkage was via a thioester bond to Cys-89 of Fni3. However, after DTT treatment, a trace of the ubiquitin-Fni3 linkage was retained in the reaction in which Fen was present. Combined with data shown in Figure 1 that Cys-89 is important for interaction of Fni3 with Fen, we speculate that Fen in the reaction may affect (but not block) the access of ubiquitin molecules to the Cys-89 residue and hence increases the chance of ubiquitin linking to the amino group. Similar results were obtained when GST-tagged Fni3 and Suv were used (see Supplemental Figure 6B online).
Fni3 and Suv together Direct Lys-63–Specific Ubiquitination
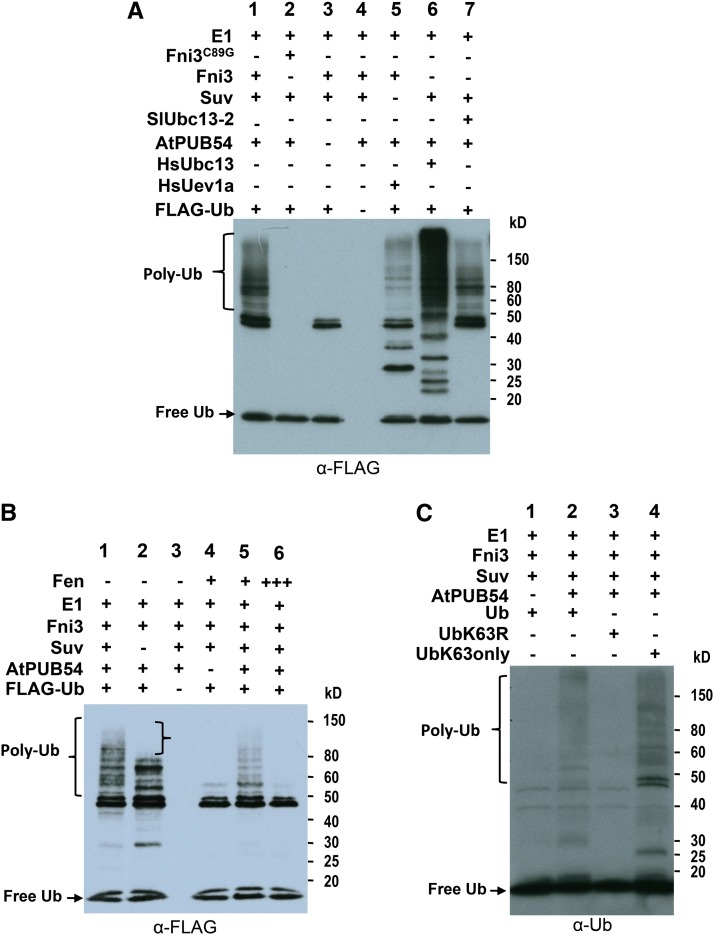
Ubc13 and its cofactor Uev are unique among E2 ubiquitin–conjugating enzymes in that they catalyze exclusively the formation of Lys-63–linked polyubiquitin chains (Lim and Lim, 2011). To examine if Fni3/Suv directs Lys-63–linked polyubiquitination, we first tested if they can direct polyubiquitination using in vitro ubiquitination assay (Zeng et al., 2004). Since the cognate tomato E3 ligase(s) that works with Fni3/Suv is unknown, we used purified recombinant protein of a U-box–type Arabidopsis E3 ligase, At-PUB54, for the assay (PUB stands for Plant U-box Protein). PUB54 was shown to work with Arabidopsis Ubc13 to direct formation of Lys-63–linked polyubiquitin chains (Wiborg et al., 2008). In the presence of E1, PUB54, ubiquitin, and other essential factors, Fni3/Suv acted with PUB54 to catalyze polyubiquitination (Figure 4A, lane 1). In the absence of Fni3, PUB54, or ubiquitin, no polyubiquitination was observed (Figure 4A, lanes 2 to 4). Fni3 and Suv also acted with human Uev1a and Ubc13, respectively, to direct polyubiquitination (Figure 4A, lanes 5 and 6), suggesting that this biochemical function of the plant and mammalian Ubc13-type E2 enzymes and their cofactors is conserved. Additionally, our in vitro ubiquitination assay indicated that Sl-Ubc13-2 also worked with Suv to catalyze polyubiquitination but not as efficiently as Fni3 (Figure 4A, lanes 1 and 7). Moreover, in the absence of Suv, no high molecular weight polyubiquitin chains were formed, supporting the role of Suv as a cofactor of Fni3 (Figure 4B, lane 2). In the presence of Fen, however, the formation of polyubiquitin chains was reduced and the reduction was dose dependent on the amount of Fen (Figure 4B, lanes 1, 5, and 6). This suggests Fen interferes with the activity of the E3 ligase PUB54 in the assay, likely because Fen interaction with Fni3 affects access to Fni3 by PUB54. As a control, GST alone did not affect the ubiquitination catalyzed by Fni3-PUB54 in the assay (see Supplemental Figure 6C online).
Figure 4.
Fni3 and Suv Catalyze Lys-63–Linked Ubiquitination.
(A) In vitro ubiquitination assay using GST-tagged Fni3, Suv, and At-PUB54 recombinant proteins. This experiment was repeated two times with similar results.
(B) Effect of Suv and Fen on Fni3-directed in vitro ubiquitination. One third and equal amount of Fen to AtPUB54 was added to the reactions of lane 5 and 6, respectively. This experiment was repeated three times with similar results.
(C) Fni3 and Suv catalyze Lys-63–specific ubiquitination. This experiment was repeated two times with similar results. The numbers on the right denote the molecular mass of marker proteins in kilodaltons.
[See online article for color version of this figure.]
We next examined whether Fni3/Suv direct Lys-63–linked ubiquitination using different ubiquitin mutants in the in vitro ubiquitination assay. Similar to results shown in Figure 4A, polyubiquitination was observed when intact ubiquitin was used in the reaction (Figure 4C, lane 2). However, the formation of polyubiquitin chains was compromised when the Lys-63 residue was substituted with Arg (Figure 4C, lane 3). When a ubiquitin variant in which all Lys residues were substituted with Arg except only Lys-63 remained intact was used, Fni3/Suv directed formation of polyubiquitin chains comparable to that of intact ubiquitin (Figure 4C, lane 4). Taken together, these results demonstrate that Fni3 and Suv together catalyze Lys-63–specific polyubiquitination.
Fni3/Ubc13-2 and Suv Are Required for Fen-Mediated Immune Signaling
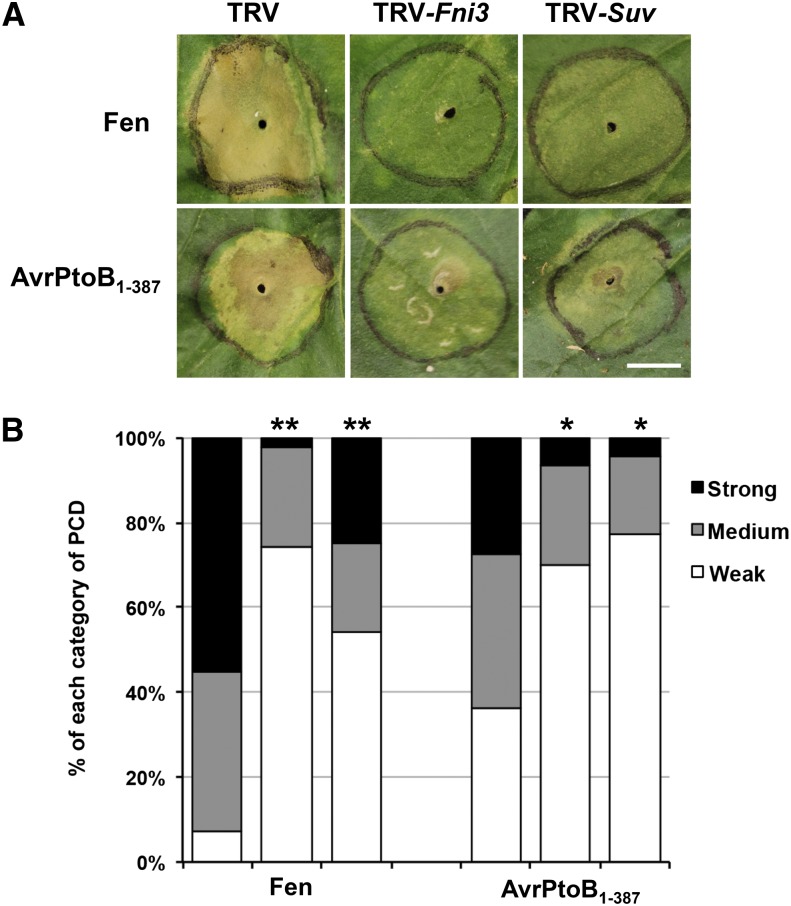
Our data showing that interaction of Fni3 with Fen is necessary for PCD triggered by overexpression of Fen in N. benthamiana suggests that Fni3 plays an important role in Fen-mediated immune signaling. To assess the contribution of Fni3 and Suv in Fen-mediated immunity, we relied on Rsb, which is activated by Fen or Fen homologs in response to AvrPtoB lacking E3 ligase activity and is typically manifested as immunity-associated PCD on N. benthamiana plants (Abramovitch et al., 2003, 2006). We used N. benthamiana instead of tomato because it gives higher efficiency of VIGS and better transient expression of PCD elicitors. We evaluated the activation of Rsb-associated PCD as well as PCD induced by overexpression of Fen under circumstances in which the expression of Fni3 and Suv in N. benthamiana plants either is normal or is reduced by VIGS using a tobacco rattle virus (TRV) vector (del Pozo et al., 2004). For the latter experiment, we infected individual N. benthamiana plants with a TRV vector carrying the Fni3 or Suv gene (TRV-Fni3 and TRV-Suv, respectively) or the empty TRV vector (as control). The N. benthamiana homologs of Fni3, Sl-Ubc13-2, and Suv share high identity in nucleotide sequence to their counterparts in tomato (see Supplemental Figures 7A and 7B online) and the TRV-Fni3– and TRV-Suv–infected plants had greatly reduced expression of N. benthamiana Fni3 (Nb-Fni3), Sl-Ubc13-2 (Nb-Ubc13-2), and Suv (Nb-Suv), respectively, compared with TRV-only infected plants (see Supplemental Figure 8 online). Plants infected with TRV-Fni3 or TRV-Suv grew normally except that the leaves of TRV-Fni3 plants were slightly curved (see Supplemental Figure 9 online). Four weeks after induction of VIGS, leaves of TRV- and TRV-Fni3– or TRV-Suv–infected plants were agroinfiltrated with transgenes expressing Fen or AvrPto B1-387. As shown in Figures 5A and 5B, TRV-Fni3– or TRV-Suv–infected plants exhibited greatly reduced PCD in response to Fen or the Rsb inducer AvrPtoB1-387, whereas control plants inoculated with TRV vector showed a strong PCD in response to both of these proteins. The extent of reduction of cell death in TRV-Suv–infected plants was less than that of TRV-Fni3–infected plants. These results indicate that Nb-Fni3 and Nb-Suv are required for Fen-mediated immune signaling in N. benthamiana.
Figure 5.
Fni3 and Suv Positively Regulate PCD Associated with Fen-Mediated Immunity.
(A) VIGS of Fni3 or Suv gene significantly diminished PCD initiated by transient expression of Fen or the Rsb elicitor, AvrPtoB1-387, in N. benthamiana. The TRV vector was used to knock down expression of Fni3 (TRV-Fni3) or Suv (TRV-Suv). TRV vector only was used as a control. The PCD was initiated by syringe-infiltrating Agrobacterium carrying T-DNA with the genes Fen or AvrPtoB1-387 into the N. benthamiana leaves. The infiltrated area of the leaf is outlined by a black circle. Photographs were taken 3 d after infiltration. Bar = 1 cm.
(B) The degree of reduction in PCD elicited by transient expression of Fen and AvrPtoB1-387 in N. benthamiana. The degree of PCD induced by Fen or AvrPtoB1-387 as shown on the y axis was monitored visually and classified into three categories: strong (75 to 100%), partial (25 to 75%), or weak (<25%). The degree of PCD of each treatment was statistically analyzed by Fisher’s exact test using the number of spots in each category (*P < 0.05 and **P < 0.01). Asterisks indicate significant reduction of PCD by silencing either Fni3 (second lane of each PCD elicitor) or Suv (third lane of each PCD elicitor) when compared with TRV-only plants (first lane of each PCD elicitor). The results are the sum of four independent experiments in which at least 15 infiltrations for each PCD elicitor were performed on TRV-only, TRV-Fni3, and TRV-Suv plants, respectively.
In TRV-Fni3–infected N. benthamiana plants, the expression of both Nb-Fni3 and Nb-Ubc13-2 was greatly reduced (see Supplemental Figure 8 online), suggesting knockdown of either Fni3 alone or both genes accounted for the decrease in Fen-mediated immunity. To distinguish these possibilities, we generated a VIGS construct, TRV-Fni3-3′ , in which the TRV vector carried only the 3′ end of the Fni3 gene (see Supplemental Figure 10A online). The sequence identity of Fni3 and Sl-Ubc13-2 in the 3′ end of the gene is low (see Supplemental Figure 10B online); thus, TRV-Fni3-3′ was used to silence Fni3 only. In contrast with TRV-Fni3–infected plants, in which cell death caused by overexpression of Fen and AvrPtoB1-387 was significantly diminished, the TRV-Fni3-3′–infected plants developed slightly reduced but nearly comparable cell death to that of the TRV vector–infected plants (see Supplemental Figure 10C online), suggesting that both Fni3 and Ubc13-2 are involved in Fen-mediated immunity and associated cell death.
Fni3 and Suv Are Involved in Immunity-Associated PCD Elicited by Other R Proteins
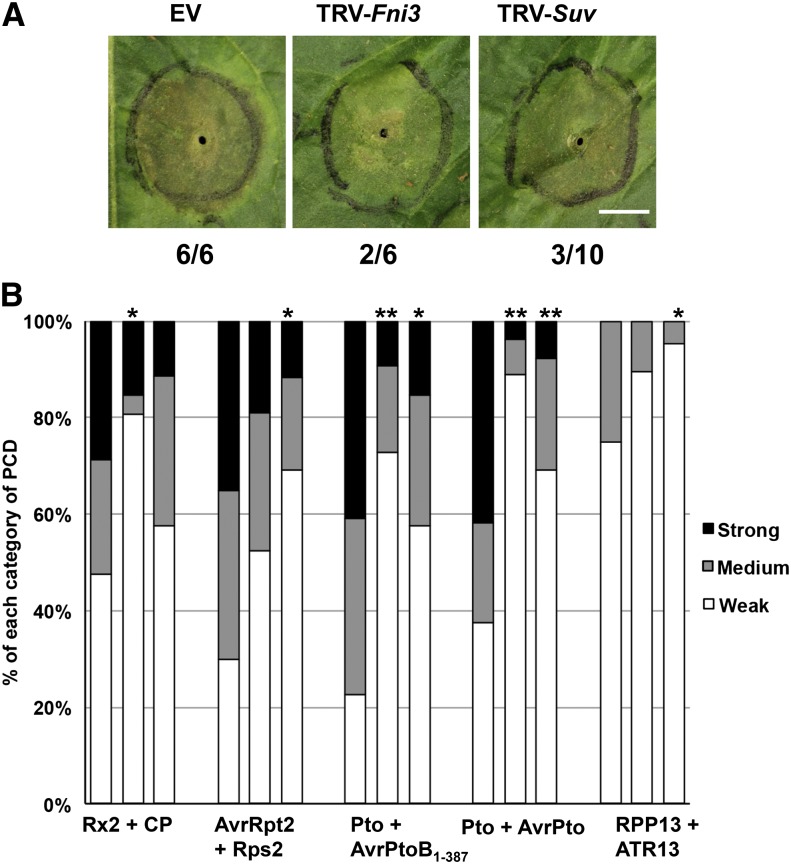
To assess whether Fni3 and Suv affect nonhost immunity, a type of plant immunity against microbes that are not natural pathogen of the host, we examined the response of N. benthamiana to the tomato bacterial pathogen Pst DC3000. This bacterial strain expresses an effector protein, HopQ1-1, that is recognized by a yet unknown R protein in N. benthamiana, leading to immunity-associated PCD (Wei et al., 2007). Leaves of N. benthamiana plants in which expression of Fni3 or Suv had been knocked down by VIGS were infiltrated with 5 × 106 colony-forming units/mL of DC3000, and the occurrence of PCD was scored 4 d after infiltration. As shown in Figure 6A, PCD was greatly reduced in both TRV-Fni3– and TRV-Suv–infected plants compared with TRV EV-infected plants, supporting a role for Fni3/Ubc13-2 and Suv as positive regulators of immunity-associated PCD in this nonhost plant–pathogen interaction.
Figure 6.
Silencing of Fni3 or Suv Reduces Immunity-Associated PCD Induced by Other R Protein/Effector Pairs.
(A) Immunity-associated PCD triggered by the nonhost bacterial pathogen Pst DC3000 in N. benthamiana. A suspension of 5 × 106 colony-forming units/mL of Pst strain DC3000 was infiltrated into the outlined area of N. benthamiana leaves where either Fni3 or Suv has been silenced. The photographs were taken 3 d after inoculation. The numbers below the images denote number of infiltrated areas that showed strong PCD and the total number of infiltrated areas. This experiment was repeated two times with similar results. Bar = 1 cm.
(B) Effect of silencing Fni3 on PCD caused by Agrobacterium-mediated transient expression of other R protein/effector. The degree of PCD induced by other R protein/effector interactions on Fni3- or Suv-silenced N. benthamiana plants was visually measured and statistically analyzed using Fisher’s exact test as described in Figure 5B. Asterisks (*P < 0.05 and **P < 0.01) indicate significant reduction of PCD by silencing either Fni3 (second lane of each PCD elicitor) or Suv (third lane of each PCD elicitor) compared with PCD in TRV-only plants (first lane of each PCD elicitor). The results are the sum of at least five independent experiments in which at least 20 infiltrations for each PCD elicitor were conducted on TRV-only, TRV-Fni3, and TRV-Suv plants, respectively.
The requirement of Fni3/Ubc13-2 and Suv for both Fen-mediated and HopQ1-1–elicited immunity-associated PCD prompted us to investigate whether they might play a broader role in PCD induction mediated by other plant immunity-associated elicitors. We examined four additional PCD-inducing R protein/effector pairs originating from different host/pathogen systems: potato (Solanum tuberosum) Rx2/PVX coat protein (CP) (Bendahmane et al., 2000), Arabidopsis RPS2/AvrRPt2 from P. syringae (Day et al., 2005), tomato Pto/AvrPto from P. syringae, and Arabidopsis RPP13/ATR13Emco5Δ41aa from the oomycete Hyaloperonospora parasitica (Rentel et al., 2008). We also included Pto/AvrPtoB1-387 in the experiment (Abramovitch et al., 2003). PCD was monitored for 4 to 5 d after agroinfiltration of the PCD-eliciting gene pairs into leaves of TRV-Fni3– or TRV-Suv–infected plants or TRV-only plants (Figure 6B). With the exception of Rx2/CP, PCD elicited by other four R protein/effector pairs was reduced in Suv-silenced plants by a statistically significant level. PCD elicited by Rx2/CP, Pto/AvrPtoB1-387, and Pto/AvrPto was also reduced in TRV-Fni3–infected plants. The decrease in cell death initiated by Pto/AvrPto was unexpected considering that we have not observed an interaction in the yeast two-hybrid system of Pto with Fni3 and Sl-Ubc13-2. These data suggest Nb-Fni3, Nb-Ubc13-2, and Nb-Suv play a broad role in diverse immunity-associated PCD in N. benthamiana.
The Ubiquitin-Conjugating Activity of Fni3 and Sl-Ubc13-2 Is Necessary for the Development of Cell Death Associated with Effector-Triggered Immunity
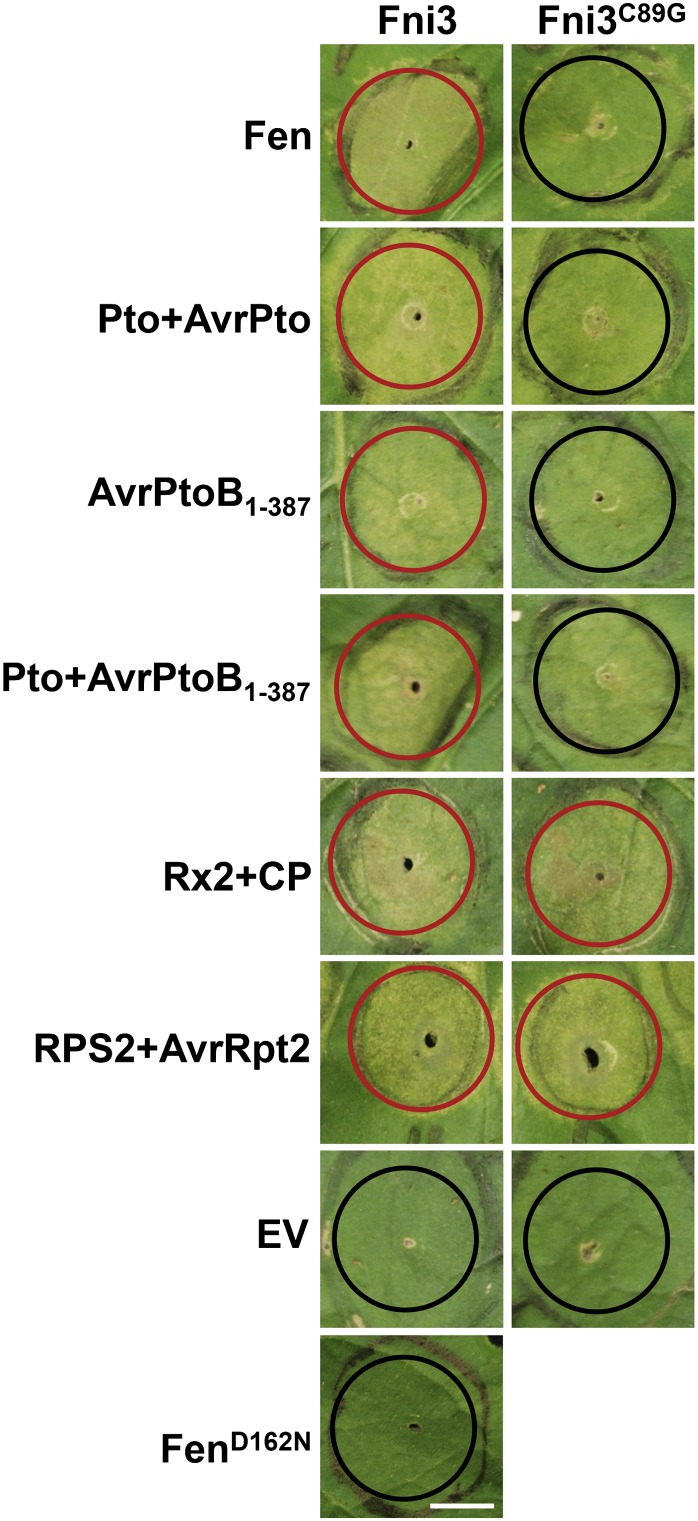
To test whether the ubiquitin-conjugating activity of Fni3 is involved in immunity-associated PCD initiated by cell death triggers, we next coexpressed the R protein (and its cognate effector) with Fni3 or the Fni3 mutant, Fni3Cys89Gly, in N. benthamiana. The highly conserved Cys residue located in the putative active site of Fni3 is substituted with Gly in Fni3Cys89Gly, and the mutation results in loss of Fni3 E2 activity (Figure 4A, lane 2; see Supplemental Figure 11 online). We therefore hypothesize that Fni3Cys89Gly might act as a dominant-negative form of Fni3/Sl-Ubc13-2 when transiently expressed in N. benthamiana, similar to what is observed in the human and animal Ubc13 mutant Ubc13C87A (Trompouki et al., 2003; Slotman et al., 2012). Indeed, compared with coexpression with Fni3, which led to strong cell death in N. benthamiana, cell death induced by transient expression of Fen, Pto/AvrPto, AvrPtoB1-387, and Pto/AvrPtoB1-387 was significantly reduced when Fni3C89G was coexpressed (Figure 7). By contrast, the cell death induced by AvrRpt2/Rps2 was not affected when Fni3C89G was coexpressed. The cell death induced by Rx2/CP was only slightly reduced when coexpressed with Fni3C89G compared with coexpression with Fni3. Agrobacterium-mediated transient expression of Fni3 or Fni3C89G with the EV did not cause cell death in N. benthamiana, indicating overexpression of Fni3 does not result in cell death. Additionally, no cell death was observed when Fni3 was coexpressed with FenD162N encoding a kinase-deficient Fen mutant protein (Figure 7), consistent with the previous finding that cell death caused by overexpression of Fen is kinase activity dependent (Mucyn et al., 2009). Together, the result suggests immunity-associated PCD triggered by Fen and other R protein/effectors indeed requires the ubiquitin-conjugating activity of Fni3/Sl-Ubc13-2.
Figure 7.
The Ubiquitin-Conjugating Activity of Fni3 Is Required for PCD Associated with Effector-Triggered Immunity.
Coexpression of an E2 activity mutant of Fni3, Fni3C89G, with Fen and other PCD triggers diminished cell death that was caused by expression of corresponding R protein/cognate effector. Agrobacterium-mediated transient coexpression of Fen, Pto/AvrPto, Rx/CP, Pto/AvrPtoB1-387, AvrPtoB1-387, RPS2/AvrRpt2, and Fni3 or Fni3C89G in N. benthamiana was performed through infiltration of Agrobacterium carrying T-DNA with the corresponding gene or the EV pBTEX in the area indicated by red (denoting strong cell death) or black (denoting no or reduced cell death) circles in fully expanded leaves of 4- to 5-week-old plants. At least five spots of infiltration were performed for each cell death trigger/Fni3 (or Fni3C89G) combination on five different plants, and a typical result is shown here. Photographs were taken 3.5 d after infiltration. The experiment was repeated three times with similar results. Bar = 1 cm.
Fen Does Not Phosphorylate Fni3 in Vitro
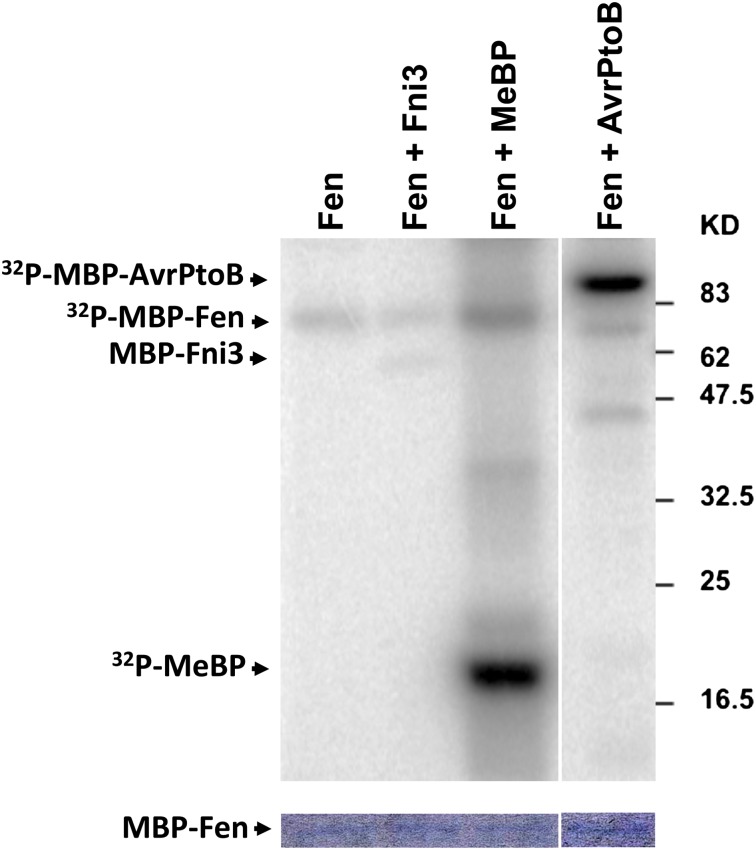
We predicted that there were two possible outcomes from the interaction between Fen and Fni3. First, Fen might regulate the activity of Fni3 by phosphorylating the Fni3 protein. Indeed, phosphorylation of E2 enzymes has been reported as a mechanism to either promote or suppress their activity (Sarcevic et al., 2002; Semplici et al., 2002; Oh et al., 2006; Philip and Haystead, 2007). Second, Fni3 might direct ubiquitination of Fen to alter its functions in the cell. To test these possibilities, we used a standard in vitro kinase activity assay to test whether Fen can phosphorylate Fni3. We found that Fen weakly autophosphorylated and it very strongly phosphorylated both the commonly used substrate for kinase activity assay, myelin basic protein (MeBP), and AvrPtoB (Figure 8). However, Fen could not phosphorylate Fni3, perhaps instead regulating Fen functions by Fni3/Suv-directed Lys-63–linked ubiquitination.
Figure 8.
Fni3 Is Not Phosphorylated by Fen.
In vitro kinase activity assay of Fen using MBP-Fni3 and MBP-AvrPtoB fusion protein and MeBP as substrates. The Coomassie blue–stained gel at the bottom shows equal (but low) abundance of Fen protein in each reaction. This experiment was repeated two times with a similar result. The numbers on the right denote the molecular mass of marker proteins in kilodaltons.
[See online article for color version of this figure.]
Fni3 Does Not Change the Stability of Fen in Vivo
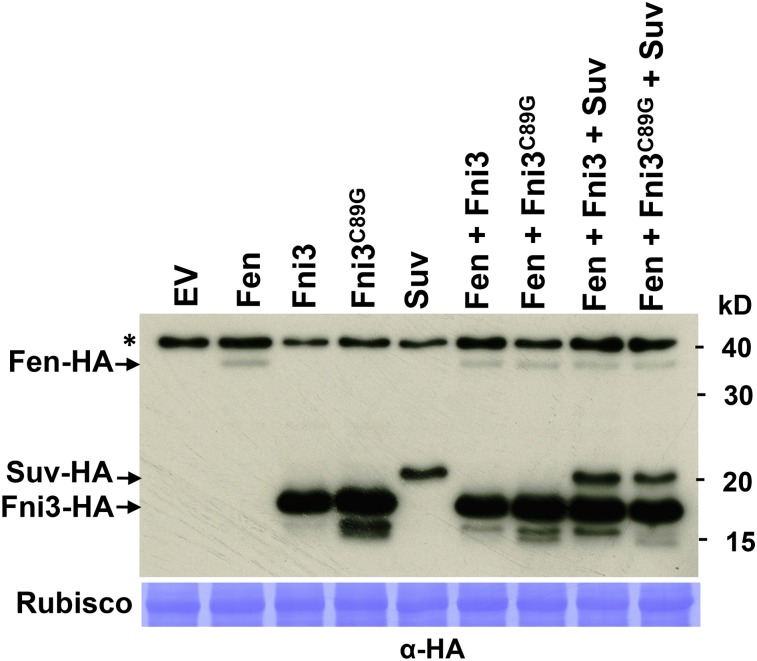
The result that Fen does not phosphorylate Fni3 prompted us to examine the possible effect of Fni3 on Fen. We coexpressed via Agrobacterium-mediated transient expression Fen and Fni3, Fen and Fni3C89G, Fen, Fni3 and Suv, and Fen, Fni3C89G, and Suv on tomato RG-pto11 plants where the Pto gene is nonfunctional (Pedley and Martin, 2003). Expression of the EV and Fen by itself was used as control. Unlike on N. benthamiana plants, transient expression of Fen on leaves of RG-pto11 plants does not cause cell death. The expression level of Fen protein was examined 2 d after the agroinfiltration. Compared with that coexpressed with Fni3, the level of Fen protein was not altered when it was coexpressed with Fni3C89G (Figure 9). A similar result was obtained when FLAG-tagged Fen was used (see Supplemental Figure 12 online). Therefore, Fni3 does not appear to affect the stability of Fen in plant cells.
Figure 9.
Fni3 Does Not Affect the Stability of Fen in Vivo.
Agrobacterium-mediated transient expression of HA-tagged Fni3, Fni3C89G, Suv, and Fen alone or coexpressed in the combinations as specified were performed on tomato RG-pto11 plants. The abundance of Fen, Fni3/Fni3C89G, and Suv were examined by immunoblotting using anti-HA antibody. Staining of ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) subunits by Coomassie blue demonstrated equal loading. Asterisk denotes nonspecific hybridization band in the immunoblot. The numbers on the right denote the molecular mass of marker proteins in kilodaltons. This experiment was repeated two times with a similar result.
[See online article for color version of this figure.]
DISCUSSION
As part of our investigation to understand how Fen-mediated immunity is regulated, we characterized Fni3, a Ubc13-type ubiquitin carrier protein that interacts with Fen and its cofactor Suv. We also identified another Ubc13-type gene, Sl-Ubc13-2, from the tomato genome that encodes a protein nearly identical to Fni3, raising the possibility that they may have some functional redundancy. Fni3 was shown to be an active E2 enzyme, whereas Suv displayed no ubiquitin-conjugating activity. Both Fni3 and Sl-Ubc13-2 acted with Suv to catalyze polyubiquitination. Fni3 and Suv together were shown to direct Lys-63–linked ubiquitination. Knocking down the expression of both Fni3 and Ubc13-2 by VIGS significantly diminished PCD associated with Fen-mediated immunity. In addition, knocked-down expression of Suv reduced PCD associated with Fen and several other R proteins. The fact that we observed diminished PCD upon coexpression of Fen or other R proteins with a Fni3 mutant that has no E2 enzymatic activity further suggests Lys-63–linked ubiquitination is involved in the plant immune response. Taken together, data presented in this study supports the hypothesis that Ubc13/Suv modulates effector-triggered immunity likely through Lys-63–linked ubiquitination.
Unlike conventional Lys-48–linked ubiquitination that mainly serves as a signal for 26S proteasome–mediated degradation of substrate proteins, Lys-63–linked polyubiquitination has been shown to play nonproteolytic, regulatory roles in various cellular processes (Fisk and Yaffe, 1999; Duncan et al., 2006; Bhoj and Chen, 2009; Lauwers et al., 2009; Martinez-Forero et al., 2009). In human and animals, Lys-63–linked ubiquitination directed by Ubc13 plays a crucial role in regulating both innate and adaptive immunity (Bhoj and Chen, 2009). Consequently, the Ubc13 protein has become a target of pathogens in suppressing host immunity (Sanada et al., 2012). The identification and characterization of a role for Fni3/Suv in affecting the immunity-related protein kinase, Fen, and likely other R protein–mediated cell death therefore suggests a mechanism in the regulation of plant immune signaling. Noteworthy, the Fen-mediated cell death appears to be due to its misregulation in N. benthamiana and is dependent on the endogenous R protein, Prf (Chang et al., 2002; Mucyn et al., 2009). Interestingly, it was shown recently that the type III effectors PthA2 and PthA3, which are important for pathogenesis of the bacterial pathogen Xanthomonas axonopodis pv citri in the development of citrus canker disease, interacted with citrus Ubc13 and Uev proteins and inhibited Ubc13/Uev-mediated Lys-63–linked ubiquitination (Domingues et al., 2010). This supports the notion that Ubc13/Uev-mediated Lys-63–linked ubiquitination plays an essential role in plant immunity as well and has become a target of manipulation by plant pathogens.
In the ubiquitination system, the E2 enzyme usually interacts with the E3 ligase and the E3 associates with the substrate. Nonetheless, we discovered in this study the ubiquitin-conjugating enzyme Fni3 interacts with Fen, a protein kinase. In addition to the effector proteins of pathogens mentioned above, approximately a dozen other non-E3 ligase host proteins have so far been reported to interact with E2 proteins, including a few kinases (Sarcevic et al., 2002; Semplici et al., 2002; Oh et al., 2006; Philip and Haystead, 2007; Jean and Moss, 2008). Either these kinases regulate the enzymatic activity of the E2 by phosphorylating it or their own stability/activity is affected by the E2 enzyme. Similarly, the outcome of Fen–Fni3 interaction can be either that Fni3 affects Fen function(s) presumably through Lys-63–linked ubiquitination of Fen and/or its interactor(s) or that Fen exerts its impact on Fni3 via its kinase activity, or both. Our thioester assay indicated that Fen does not affect the conjugating activity of Fni3, and in vitro kinase activity assays showed that Fen does not phosphorylate Fni3, suggesting Fen unlikely affects the enzymatic activity of Fni3 by phosphorylation. However, we found Fen reduces the ubiquitination catalyzed in vitro by PUB54 and Fni3 in a dose-dependent manner probably due to the interaction of Fen with Fni3 blocks the access to Fni3 by PUB54. This finding is reminiscent of Ubc13 attenuating the Lys-48–linked polyubiquitination of p53 by the E3 ligase Hdm2 (Laine et al., 2006). Because the cognate tomato E3 ligase of Fni3 is unknown, we used PUB54 in the assay. Therefore, whether Fen affects the ubiquitination catalyzed by Fni3 and its cognate tomato E3 ligase in vivo and, if yes, whether the effect is biologically relevant to the positive role of Fni3/Suv in regulating Fen (which is Prf-dependent) and other R protein–mediated, immunity-associated cell death warrants further investigation. Additionally, a recent study of Ubc13 and p53 indicated that Lys-63–linked ubiquitination of p53 at the polysomes by Ubc13 resulting in monomerization of p53 accounts for the regulation of p53 localization and activity by Ubc13 (Topisirovic et al., 2009). Likewise, Lys-63–linked ubiquitination of Fen and/or its interactor(s) by Fni3 may be responsible for the regulation of Fen-mediated cell death by Fni3.
In the case of Ubc13 and p53, the Ubc13 protein affects the localization and, hence, stability of p53 (Laine et al., 2006). However, our transient coexpression experiment indicated that Fni3 does not affect the stability of Fen in plant cells. It is unknown at present how Fni3/Suv might affect the cellular functions of Fen and downstream signaling. In the regulation of human and animal innate immunity, which is triggered upon recognition of pathogen-associated molecular patterns by pattern recognition receptors, such as Toll-like receptors and NOD-like receptors, Lys-63–linked polyubiquitination is intimately involved in promoting the formation and activation of protein kinase complexes, such as those involving the IκB kinase (Jiang and Chen, 2012). These kinases in turn switch on downstream transcription factors, such as nuclear factor-κB, which orchestrate gene expression programs to protect cells and organisms from pathogen infection. Therefore, one possibility is that Fni3/Suv-mediated Lys-63 polyubiquitination affects the kinase activity of Fen, which in turn affects the phosphorylation of downstream substrate(s) by Fen. Alternatively, Fni3 might regulate the cellular functions of Fen through affecting interaction of Fen with other protein(s) or its subcellular localization in the cell. Indeed, Lys-63–linked ubiquitination has been shown to regulate subcellular localization of protein kinases (Yang et al., 2009) and protein–protein interactions (Zhang et al., 2012) in animal cells. The importance of subcellular localization to Fen’s function is supported by the finding that Fen-mediated cell death in N. benthamiana is N-myristoylation dependent and the N-myristoylation is believed to affect the subcellular localization of Fen (Rommens et al., 1995; Mucyn et al., 2009). Further experiments to distinguish these possibilities will facilitate our understanding of the mechanism by which Lys-63–linked ubiquitination acts on Fen-mediated immune signaling.
The Lys-63–linked ubiquitination in human and animal innate and adaptive immune signaling is often catalyzed by RING domain–containing E3 ligases, such as Tumor Necrosis Factor receptor-associated factors (such as TRAF6) and Tripartite Motif-containing protein (such as TRIM25) together with E2 enzyme Ubc13 and its cofactor Uev1a (also known as UBE2V1) (Jiang and Chen, 2012), which are close homologs of Fni3 and Suv, respectively. It therefore seems plausible that one or more tomato E3 ligase(s) works together with Fni3/Suv to catalyze Lys-63–linked ubiquitination that is related to Fen-mediated immunity. The unknown E3 ligase, together with Fni3/Suv, might either act to ubiquitinate Fen or its interactor(s) by Lys-63–linked polyubiquitin chains, or, similar to what occurs in human and animals, the E3 ligase might autoubiquitinate with the Lys-63–linked polyubiquitin chains serving as the signal and scaffold to promote formation and activation of a downstream kinase complex. However, our observation that Fni3 interacts physically with Fen argues for the first possibility, analogous to what was observed in the case of regulating p53 localization and activity by Ubc13 (Laine et al., 2006; Topisirovic et al., 2009). Identification and characterization of the tomato E3 ligase therefore will be a key next step in elucidating the regulation of Fen-mediated immunity by Fni3.
Fni3 was found to interact specifically with Fen among members of the Pto family. In particular, Fni3 (and its close homolog Sl-Ubc13-2) did not interact with Pto despite the fact it has 80% amino acid identity with Fen and is actively involved in tomato immunity against Pst (Pedley and Martin, 2003), and the downstream signaling of Pto-mediated immunity overlaps with that of Fen-mediated immune signaling (Rosebrock et al., 2007). Nevertheless, we observed that silencing of either Fni3 or Suv diminished PCD elicited by Pto/AvrPto in leaves of N. benthamiana. This is consistent with the fact that Suv was identified in a VIGS screen that tested ∼2400 random tomato cDNAs for effects on plant immunity-associated PCD elicited by Pto (del Pozo et al., 2004). One explanation for these observations is that Lys-63–linked ubiquitination acts at and therefore is essential to the regulation of multiple points of plant immune signaling pathway and Fni3/Suv-directed Lys-63–linked ubiquitination might, unlike in the case of Fen, act somewhere other than at Pto itself in Pto-mediated signaling. Ubc13-directed Lys-63 ubiquitination has indeed been found to act at several positions in human and animal immune signaling pathways (Bhoj and Chen, 2009). In the future, it will be important to find out why Pto and Fen potentially employ different mechanisms of Lys-63 ubiquitination in regulating immune signaling mediated by them.
METHODS
Growth of Bacteria and Plant Materials
Agrobacterium tumefaciens strains GV2260 and GV3101 and Pseudomonas syringae pv tomato DC3000 were grown at 28°C in Luria-Bertani and King’s B medium, respectively, with appropriate antibiotics. Nicotiana benthamiana seeds were germinated and plants were grown on autoclaved soil in a growth chamber with 16 h light (∼215 μmol/m2/s at the leaf surface of the plants) and 25°C/23°C day/night temperature. For VIGS, plants were transferred to a growth chamber with 16 h of light and 24°C/22°C day/night temperature.
DNA Manipulations and Plasmid Constructions
All DNA manipulations were performed according to standard techniques (Sambrook and Russell, 2001). Plasmids in the pTRV2 vector for VIGS were constructed by Gateway cloning using the pENTR/SD/D-TOPO entry vector according to protocols provided by the manufacturer (Invitrogen; now Life Technologies). The FenCys307Arg, FenThr312Ile, FenAsp316Asn, FenAsp162Asn, and Fni3Cys89Gly mutations were generated using the QuickChange site-directed mutagenesis kit II according to the protocol provided by the manufacturer (Stratagene; now Agilent). Primers used for cloning of Fni3, Suv, and Sl-Ubc13-2 and N. benthamiana homologs of Fni3 (Nb-Fni3), Suv (Nb-Suv), and Sl-Ubc13-2 (Nb-Ubc13-2) are listed in Supplemental Table 1 online.
Yeast Two-Hybrid Assay
The LexA-based yeast two-hybrid system and procedures used for screening for Fen-interacting proteins was as described (Golemis et al., 2008). For testing the interaction of two proteins using the yeast two-hybrid assay, plasmids of the bait and prey constructs were transformed into yeast cells using the lithium acetate/polyethylene glycol method as described in the Yeast Protocols Handbook (Clontech). Yeast transformants were selected on plates containing dropout base (Sunrise Science Product) plus Glc but without uracil, His, and Trp. Successful transformants were then patched on dropout base with Gal/raffinose and 5-Bromo-4-chloro-3-indolyl β-d-galactopyranoside, but lacking uracil, His, and Trp. Blue color of the patch indicates interaction of the two proteins in yeast cells. Expression of fusion proteins was verified by immunoblot using either mouse anti-LexA (Dualsystems Biotech) or rat anti-hemagglutinin (HA; 3F10) horseradish peroxidase–conjugated (Roche) monoclonal antibodies.
Sequence Alignment and Phylogenetic Analysis
For sequence alignment, sequences of interest in the FASTA format were input into the ClustalX 2.0 program and aligned using the ClustalX algorithm (Larkin et al., 2007). The aligned sequences were then used for phylogenetic analysis using the MEGA5 program (Tamura et al., 2011). To build an unrooted phylogenetic tree using MEGA5, the evolutionary history was inferred using the neighbor-joining method. The bootstrap consensus tree inferred from 2000 replicates was taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in <50% bootstrap replicates were collapsed. The evolutionary distances were computed using the Jones-Thornton-Taylor matrix-based method (Jones et al., 1992), with units representing the number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1).
Agrobacterium-Mediated Transient Expression
Fen and its derivatives (FenCys307Arg, FenThr312Ile, and FenAsp316Asn) were cloned into a pBTEX 35S cauliflower mosaic virus promoter expression cassette (provided by R. Bressan at Purdue University, West Lafayette, IN) with HA tag at the C terminus. Each construct was then transformed into Agrobacterium strain GV2260. AvrPtoB1-387, Pto/AvrPto, RPS2/AvrRpt2 (Kunkel et al., 1993), RPP13/ATR13Emco5Δ41aa (Rentel et al., 2008), and Rx2/CP (Bendahmane et al., 2000) under the control of the 35S promoter were expressed transiently in N. benthamiana using Agrobacterium-mediated infiltration as previously described (Abramovitch et al., 2003). For coexpression of PCD triggers and Fni3 (or Fni3C89G), the final OD600 of Agrobacterium harboring pBTEX-Fni3 (or -Fni3C89G) for infiltration was 0.4. In testing the effect of Fni3/Suv on the stability of Fen by transient coexpression, equal amounts (OD600 = 0.4) of Agrobacterium strains harboring each gene (pBTEX-Fni3, Fni3C89G, Suv, or Fen) were used. The total OD600 of each inoculum for infiltration was brought to 1.2 using cells harboring the pBTEX EV when necessary.
VIGS
Gene silencing was induced using TRV vectors (Liu et al., 2002) delivered by Agrobacterium strain GV2260 or GV3101, as previously described (del Pozo et al., 2004). The Fni3 and Suv genes were cloned into the pTRV2 vector. Agrobacterium (OD600 = 0.5) containing appropriate pTRV plasmids was induced with acetosyringone and used to infiltrate 3-week-old N. benthamiana seedlings. VIGS-treated N. benthamiana plants were maintained for 4 to 5 weeks at 24°C /22°C, 16/8 h day/night condition to allow silencing to occur.
Effectiveness of VIGS of Nb-Fni3, Nb-Ubc13-2, and Nb-Suv
To determine the effectiveness of silencing Nb-Fni3, Nb-Ubc13-2, and Nb-Suv by VIGS in N. benthamiana, the expression of these genes was determined by RT-PCR at low PCR cycles. In brief, three leaf discs of ∼2.5-cm2 area were collected separately from three different leaves of plants for transient assays, as shown in Figures 5A, 5B, 6A, and 6B. Total RNA was isolated from the leaf tissue using RNeasy Plant Mini Kit with in-column DNase treatment (Qiagen) by following the protocol provided by the manufacturer. This was followed by synthesis of the first-strand cDNA using the Superscript III reverse transcriptase (Invitrogen) using the protocol from the manufacturer. PCR amplification (25 cycles) of Nb-Fni3, Nb-Ubc13-2, Nb-Suv, and Nb-EF1α using the first-strand cDNA as template and primers specified in the Supplemental Table 1 and separation and staining of the PCR product were performed as described (Sambrook and Russell, 2001). The separated RT-PCR products were photographed using the Gel Doc SR system (Bio-Rad) and quantified using the software Image Lab (Bio-Rad) by following instructions from the manufacturer. Nb-EF1α was used as an internal reference to determine the amount of cDNA template to be used. The relative transcript level of Nb-Fni3, Nb-Ubc13-2, and Nb-Suv was normalized by the abundance of the Nb-EF1α gene. For comparison of gene expression in VIGS plants to TRV vector-only control plants, the expression level of the Nb-Fni3, Nb-Ubc13-2, and Nb-Suv genes in the control plants was set as 100%. Three biological replicates were used for the experiment.
Extraction and Purification of Proteins
MBP or MBP fused to Fni3, Fni3C89G, or AvrPtoB1-387 was expressed in Escherichia coli strain BL21(DE3) and purified using amylose resin (New England Biolabs) according to the manufacturer’s instructions. GST or GST-tagged Fen, PUB54, Fni3, Suv, and Sl-Ubc13-2 were expressed in E. coli strain BL21(DE3) and purified with Glutathione Sepharose 4 Fast Flow beads (GE Healthcare) by following the protocol provided by the manufacturer. Yeast protein extractions were performed according to previously described methods (Munkvold et al., 2008). Total proteins from N. benthamiana leaf tissues were prepared by grinding ∼3 cm2 of leaf discs in liquid nitrogen and homogenizing the tissue in 400 μL of protein extraction buffer (1.5% N-lauryl sarcosine, 2 M thiourea, 20 mM DTT, 50 mM Tris-HCl (Tris-hydroxymethyl-aminomethane-hydrogen chloride), pH 7.5, and plant protease inhibitors [Sigma Aldrich]). Cell debris was pelleted by centrifugation at 15,000g at 4°C for 15 min. The concentration of total protein in supernatants was determined using protein assay agent (Bio-Rad). Approximately 30 μg of total proteins of each sample was resolved by SDS-PAGE and analyzed by immunoblotting. To examine the effect of Fni3 on the stability of Fen (Figure 9; see Supplemental Figure 12 online), the whole tomato (Solanum lycopersicum) leaf infiltrated with Agrobacterium was harvested, ground in liquid nitrogen, and homogenized using 2.5 mL of protein sample loading buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 0.01% bromophenol blue, 10% glycerol, 5% freshly added β-mercaptoethanol, and a plant protease inhibitors cocktail [Sigma-Aldrich]) (Laemmli, 1970) followed by heating at 100°C for 10 min before proceeding to the centrifugation step. Rat monoclonal anti-HA (3F10) horseradish peroxidase–conjugated antibody (Roche) was used for immunoblotting.
Kinase Activity Assay
In vitro kinase activity assays were performed as described previously (Zeng et al., 2012). The reaction was performed in a 30-μL reaction containing 200 ng of MBP-Fen, 7.5 μg of myelin basic protein (MeBP) or MBP-Fni3, 5 μCi of [γ-32P]ATP, 20 μL of kinase buffer (25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 10 mM NaCl, 1 mM DTT, 50 μM ATP, and 1 mM EDTA) at room temperature for 30 min. Reactions were stopped by adding SDS loading buffer and heated at 100°C for 5 min before electrophoresis on 12.5% SDS polyacrylamide gels. Gels were dried and radioactivity visualized using a phosphor imager (Molecular Dynamics).
GST Pull-Down Assay
The in vitro binding assays were performed as described with modifications (Wilson et al., 2003). For the assays, 0.04 nM of MBP or MBP-fused protein was mixed with 0.03 nM GST or GST-Fen in 370-μL reactions containing PBS/0.5% Triton X-100 at 4°C for 45 min using a nutating mixer prior to addition of 30 μL of Glutathione Sepharose 4 Fast Flow beads. The reactions were mixed at 4°C for another 15 min and were then loaded onto 0.8-mL Pierce centrifuge columns (Thermo Scientific) equilibrated with PBS/0.5% Triton X-100. The columns were spun at 500g for 2 min, and the flow-through was discarded. The columns containing the Glutathione Sepharose 4 beads were washed with 600 μL PBS/0.5% Triton X-100 six times. The columns were spun 2 min at 850g before proteins bound to the beads were eluted with 50 μL of SDS sample buffer, resolved by SDS-PAGE, and analyzed by immunoblotting. An anti-MBP monoclonal antibody (New England Biolabs) was used as primary antibody for immunoblotting.
Thioester Assay of E2 Ubiquitin–Conjugating Activity
The ubiquitin-conjugating activity assays of Fni3 and Suv were performed as described with modifications (Kraft et al., 2005; Philip and Haystead, 2007). Briefly, in a 15-μL reaction, 80 ng (50 nM) of Yeast E1 (Boston Biochem) was preincubated with 3 μg of FLAG-ubiquitin in 20 mM Tris HCl, pH 7.5, 10 mM MgCl2, and 1 mM ATP for 10 min. To the reaction was added 100 ng of MBP-Fni3 (or GST-Fni3) and continued for 20 min. The reaction was then stopped with SDS sample loading buffer and was resolved by 10% SDS-PAGE. To examine the E2 ubiquitin linkage in the assay (Figure 3B), the reaction volume was scaled up to 30 μL. The reactions were split and terminated by addition of either SDS sample loading buffer with 100 mM DTT or 8 M urea sample buffer (-DTT). The proteins were transferred to polyvinyl difluoride membrane and probed with mouse monoclonal ANTI-FLAG M2-peroxidase-conjugated (horseradish peroxidase) antibody (Sigma-Aldrich) before being detected using ECL kit (GE Healthcare). To test whether Fen affects the ubiquitin-conjugating activity of Fni3, 500 ng of MBP-Fni3 (or GST-Fni3) and 500 ng GST-Fen were incubated in a 10-μL total volume mixture consisting of 20 mM HEPES, pH 7.5, 10 mM MgCl2, and 1 mM ATP for 30 min at 30°C. Two microliters of this solution containing MBP-Fni3 (or GST-Fni3) (100 ng) was then used for thioester assay reactions.
In Vitro Ubiquitination Assay
The in vitro ubiquitination assay was performed as described (Rosebrock et al., 2007). The reactions were resolved by SDS-PAGE and analyzed by immunoblotting using mouse monoclonal ANTI-FLAG M2-peroxidase-conjugated (horseradish peroxidase) antibody (Sigma-Aldrich). For the examination of Lys-63 specificity of Fni3-directed ubiquitination, 12 μg of ubiquitin or ubiquitin mutants was used for each reaction, and rabbit antiubiquitin antibody (Boston Biochem) was used as the primary antibody for immunoblotting. For testing the effect of Fen on Fni3 in the in vitro ubiquitination assay, 500 ng (∼8 pmol) or 1.5 μg (∼25 pmol) of purified GST-Fen was added to the reactions, respectively.
Tomato Protoplast Preparation and BiFC Assay
Tomato RG-pto11 plants were grown in a growth chamber with 16 h of light (∼215 μmol/m2/s at the leaf surface of the plants) and 24°C/22°C day/night temperature. Protoplasts were prepared from the young fully expanded leaves of 3- to 4-week-old plants as described (Yoo et al., 2007) with modifications. Briefly, leaves were gently rubbed with carborundum and sliced to remove veins. Leaf strips were submerged upside down in a Petri dish containing the digestion solution (1% Cellulase R10, 0.2% Macerozyme R10, 0.4 M mannitol, 20 mM KCl, 20 mM MES, pH 5.7, 10 mM CaCl2, and 0.1% BSA) and vacuum infiltrated for 7 min followed by digestion at 25◦C with shaking at 60 rpm for 3 h. The digestion solution with released protoplasts was poured into a 40-mL round-bottom test tube. The protoplasts were collected by centrifugation at 60g for 3 min, washed twice with W5 solution (154 mM sodium chloride, 125 mM calcium chloride, 5 mM potassium chloride, and 2 mM MES, pH 5.7), and incubated with W5 on ice for 30 min for recovery. The pellet of protoplasts was gently resuspended in MMG solution (0.4 M mannitol, 15 mM MgCl2, and 4 mM MES, pH 5.7) and maintained on ice. For the BiFC assay, the split-Yellow Fluorescent Protein (YFP) constructs pA7-N-terminal of YFP and pA7-C-terminal of YFP were used as described (Chen et al., 2006). The tomato protoplasts were transferred to a round-bottom 2-mL Eppendorf tube containing 10 µg of plasmid DNA of each construct or EV as indicated in Supplemental Figure 1A online and mixed gently, followed by transfection using 40% Fluka polyethylene glycol for 7 min at 26◦C. The transfection was stopped with the addition of and subsequent washing with W5 solution. The transfected protoplast samples were incubated in W5 solution with 5 mM Glc overnight at 24°C in the dark followed by examination of fluorescence using a Nikon 90i fluorescence microscope. Images were captured using NIS Elements software provided by the manufacturer.
Accession Numbers
Sequence data that were used for phylogenetic analysis in this article can be found in the GenBank data library under the following accession numbers: Hs-Ubc13 (P61088), Mm-Ubc13 (AAI38212), Xt-Ubc13 (CAJ82488.1, At-Ubc13A (Q94A97), At-Ubc13B (Q9FZ48), At-Uev1a (NP_565834.1), At-Uev1b (NP_564994.1), At-Uev1c (NP_850259.1), At-Uev1d (NP_566968.1), Hs-Uev1a (NP_068823.2), Hs-Mms2 (NP_003341.1), Sp-Mms2 (O74983.1), Mm-Mms2 (NP_076074.2), Mm-Uev1 (NP_075719.1), Sc-Mms2 (NP_011428.1), and At-COP10 (AAK57749.1). Sequence data from this article can be found in GenBank under the following accession numbers: Fni3 (KF496880), Sl-Ubc13-2 (KF496882), Suv (KF496884), Nb-Fni3 (KF496881), Nb-Ubc13-2 (KF496883), and Nb-Suv (KF496885).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Fen Interacts with Fni3 in Vivo, and Fni3 Encodes a Protein Homologous to Ubc13-Type E2 Ubiquitin–Conjugating Enzyme.
Supplemental Figure 2. Fni3 Interacts Specifically with Fen.
Supplemental Figure 3. Alignment of the Nucleotide and Amino Acid Sequence of Fni3 and Sl-Ubc13-2 and the Amino Acid Sequence of Fni3 and Nb-Fni3.
Supplemental Figure 4. Fen and FenThr312Ile (FenT312I) Were Evaluated for Their Ability to Interact with Two Additional Proteins, Fni2 and AvrPtoB1-387.
Supplemental Figure 5. Identification of Suv (Solanum lycoperiscum Uev) as a Uev Homolog That Interacts with Fni3 and Sl-Ubc13-2.
Supplemental Figure 6. Thioester Assay of GST-Fused Fni3 and Suv and the Effect of GST on Ubiquitination Catalyzed by Fni3 and At-PUB54.
Supplemental Figure 7. Homolog of Fni3 (Nb-Fni3), Ubc13-2 (Nb-Ubc13-2), and Suv (Nb-Suv) from N. benthamiana Share High Nucleotide Sequence Identity to That of Tomato.
Supplemental Figure 8. N. benthamiana Homolog of Fni3, Sl-Ubc13-2, and Suv Was Silenced in TRV-Fni3 and TRV-Suv Plants, Respectively.
Supplemental Figure 9. Morphological Phenotype of N. benthamiana Plants in Which Fni3 or Suv Is Silenced.
Supplemental Figure 10. Both Fni3 and Sl-Ubc13-2 Are Involved in Fen-Mediated Immune Signaling.
Supplemental Figure 11. Substitution of the Cys-89 of Fni3 with Gly Compromises Its Ubiquitin-Conjugating Activity.
Supplemental Figure 12. The Effect of Fni3 on the Stability of Fen in Tomato Cells.
Supplemental Table 1. Primer Sequences Used for This Study.
Supplemental Data Set 1. Sequences and Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 1C.
Supplemental Data Set 2. Sequences and Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 5A.
Acknowledgments
We thank Olga del Pozo (Martin Universidad de Sevilla, Spain) for sharing data from her VIGS screen. We also thank Stephen Grace, Kleve Maurice, William Baltosser, and Hongli Wang of the Biology Department at University of Arkansas at Little Rock for allowing use of their laboratory equipment. The pBTEX 35S cauliflower mosaic virus promoter expression cassette was provided by R. Bressan (Purdue University, West Lafayette, IN). This research was supported, in part, by start-up funds from University of Arkansas at Little Rock to L.Z. and by grants from the National Institutes of Health (R01-GM078021) to G.B.M. and the National Science Foundation (IOS-1052495) to L.Z.
AUTHOR CONTRIBUTIONS
R.V.M., Y.L., L.Z., T.R.R., S.H., and J.J.B. performed the experiments. L.Z., Y.L., R.V.M., T.R.R., G.B.M., and R.A.C. analyzed data. G.B.M. designed some of the experiments and edited the article. L.Z. designed the majority of experiments and wrote and edited the article.
Glossary
- Pst
Pseudomonas syringae pv tomato
- PCD
programmed cell death
- BiFC
bimolecular fluorescence complementation
- GST
glutathione S-transferase
- EV
empty vector
- VIGS
virus-induced gene silencing
- TRV
tobacco rattle virus
- HA
hemagglutinin
References
- Abramovitch R.B., Janjusevic R., Stebbins C.E., Martin G.B. (2006). Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc. Natl. Acad. Sci. USA 103: 2851–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramovitch R.B., Kim Y.-J., Chen S., Dickman M.B., Martin G.B. (2003). Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 22: 60–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen P.L., Zhou H., Pastushok L., Moraes T., McKenna S., Ziola B., Ellison M.J., Dixit V.M., Xiao W. (2005). Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A. J. Cell Biol. 170: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A., Querci M., Kanyuka K., Baulcombe D.C. (2000). Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: Application to the Rx2 locus in potato. Plant J. 21: 73–81 [DOI] [PubMed] [Google Scholar]
- Bhoj V.G., Chen Z.J. (2009). Ubiquitylation in innate and adaptive immunity. Nature 458: 430–437 [DOI] [PubMed] [Google Scholar]
- Chang J.H., Tai Y.-S., Bernal A.J., Lavelle D.T., Staskawicz B.J., Michelmore R.W. (2002). Functional analyses of the Pto resistance gene family in tomato and the identification of a minor resistance determinant in a susceptible haplotype. Mol. Plant Microbe Interact. 15: 281–291 [DOI] [PubMed] [Google Scholar]
- Chen S., Tao L., Zeng L., Vega-Sanchez M.E., Umemura K., Wang G.L. (2006). A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol. Plant Pathol. 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Ciechanover A. (1998). The ubiquitin-proteasome pathway: On protein death and cell life. EMBO J. 17: 7151–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day B., Dahlbeck D., Huang J., Chisholm S.T., Li D., Staskawicz B.J. (2005). Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell 17: 1292–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre, F., Gutiérrez-Beltrán, E., Pareja-Jaime, Y., Chakravarthy, S., Martin, G.B., and del Pozo, O. (2013). The tomato calcium sensor Cbl10 and its interacting protein kinase Cipk6 define a signaling pathway in plant immunity. Plant Cell 25: 2748–2764. [DOI] [PMC free article] [PubMed]
- del Pozo O., Pedley K.F., Martin G.B. (2004). MAPKKKalpha is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 23: 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z.J. (2000). Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103: 351–361 [DOI] [PubMed] [Google Scholar]
- Devarenne T.P., Ekengren S.K., Pedley K.F., Martin G.B. (2006). Adi3 is a Pdk1-interacting AGC kinase that negatively regulates plant cell death. EMBO J. 25: 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues M.N., De Souza T.A., Cernadas R.A., de Oliveira M.L., Docena C., Farah C.S., Benedetti C.E. (2010). The Xanthomonas citri effector protein PthA interacts with citrus proteins involved in nuclear transport, protein folding and ubiquitination associated with DNA repair. Mol. Plant Pathol. 11: 663–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan L.M., Piper S., Dodd R.B., Saville M.K., Sanderson C.M., Luzio J.P., Lehner P.J. (2006). Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 25: 1635–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddins M.J., Carlile C.M., Gomez K.M., Pickart C.M., Wolberger C. (2006). Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 13: 915–920 [DOI] [PubMed] [Google Scholar]
- Fisk H.A., Yaffe M.P. (1999). A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J. Cell Biol. 145: 1199–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick R.D., Thilmony R.L., Sessa G., Martin G.B. (1998). Recognition specificity for the bacterial avirulence protein AvrPto is determined by Thr-204 in the activation loop of the tomato Pto kinase. Mol. Cell 2: 241–245 [DOI] [PubMed] [Google Scholar]
- Galan J.M., Haguenauer-Tsapis R. (1997). Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 16: 5847–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis, E.A., Serebriiskii, I., Finley, R.L., Jr., Kolonin, M.G., Gyuris, J., and Brent, R. (2008). Interaction trap/two-hybrid system to identify interacting proteins. In Current Protocols in Molecular Biology, F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, eds (New York: John Wiley), pp. 20.21.21–20.21.35. [Google Scholar]
- González-Lamothe R., Tsitsigiannis D.I., Ludwig A.A., Panicot M., Shirasu K., Jones J.D.G. (2006). The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18: 1067–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halterman, D. (1999). Characterization of the Fenthion Response in Tomato and the Identification of Genes That Encode Fen-Interacting Proteins. PhD dissertation (Lafayette, IN: Purdue University). [Google Scholar]
- Heath M.C. (2000). Hypersensitive response-related death. Plant Mol. Biol. 44: 321–334 [DOI] [PubMed] [Google Scholar]
- Hofmann R.M., Pickart C.M. (1999). Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96: 645–653 [DOI] [PubMed] [Google Scholar]
- Hogenhout S.A., Van der Hoorn R.A.L., Terauchi R., Kamoun S. (2009). Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact. 22: 115–122 [DOI] [PubMed] [Google Scholar]
- Hong S., Lee S., Cho S.-G., Kang S. (2008). UbcH6 interacts with and ubiquitinates the SCA1 gene product ataxin-1. Biochem. Biophys. Res. Commun. 371: 256–260 [DOI] [PubMed] [Google Scholar]
- Janjusevic R., Abramovitch R.B., Martin G.B., Stebbins C.E. (2006). A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 311: 222–226 [DOI] [PubMed] [Google Scholar]
- Jean S., Moss T. (2008). A ubiquitin-conjugating enzyme, ube2d3.2, regulates xMLK2 and pronephros formation in Xenopus. Differentiation 76: 431–441 [DOI] [PubMed] [Google Scholar]
- Jiang X., Chen Z.J. (2012). The role of ubiquitylation in immune defence and pathogen evasion. Nat. Rev. Immunol. 12: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Kim Y.J., Lin N.C., Martin G.B. (2002). Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109: 589–598 [DOI] [PubMed] [Google Scholar]
- Kraft E., Stone S.L., Ma L., Su N., Gao Y., Lau O.-S., Deng X.-W., Callis J. (2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel B.N., Bent A.F., Dahlbeck D., Innes R.W., Staskawicz B.J. (1993). RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell 5: 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Laine A., Topisirovic I., Zhai D., Reed J.C., Borden K.L.B., Ronai Z.e. (2006). Regulation of p53 localization and activity by Ubc13. Mol. Cell. Biol. 26: 8901–8913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lauwers E., Jacob C., André B. (2009). K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 185: 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Schmidt W. (2010). A lysine-63-linked ubiquitin chain-forming conjugase, UBC13, promotes the developmental responses to iron deficiency in Arabidopsis roots. Plant J. 62: 330–343 [DOI] [PubMed] [Google Scholar]
- Lim, K.-L., and Lim, G.G.Y. (2011). K63-linked ubiquitination and neurodegeneration. Neurobiol. Dis. 43: 9–16. [DOI] [PubMed]
- Lin N.-C., Abramovitch R.B., Kim Y.J., Martin G.B. (2006). Diverse AvrPtoB homologs from several Pseudomonas syringae pathovars elicit Pto-dependent resistance and have similar virulence activities. Appl. Environ. Microbiol. 72: 702–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg M., Cartinhour S., Myers C.R., Schechter L.M., Schneider D.J., Collmer A. (2006). Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol. Plant Microbe Interact. 19: 1151–1158 [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S.P. (2002). Virus-induced gene silencing in tomato. Plant J. 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Martin G.B., Frary A., Wu T., Brommonschenkel S., Chunwongse J., Earle E.D., Tanksley S.D. (1994). A member of the tomato Pto gene family confers sensitivity to fenthion resulting in rapid cell death. Plant Cell 6: 1543–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Forero I., Rouzaut A., Palazon A., Dubrot J., Melero I. (2009). Lysine 63 polyubiquitination in immunotherapy and in cancer-promoting inflammation. Clin. Cancer Res. 15: 6751–6757 [DOI] [PubMed] [Google Scholar]
- Michelle C., Vourc’h P., Mignon L., Andres C.R. (2009). What was the set of ubiquitin and ubiquitin-like conjugating enzymes in the eukaryote common ancestor? J. Mol. Evol. 68: 616–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucyn T.S., Wu A.-J., Balmuth A.L., Arasteh J.M., Rathjen J.P. (2009). Regulation of tomato Prf by Pto-like protein kinases. Mol. Plant Microbe Interact. 22: 391–401 [DOI] [PubMed] [Google Scholar]
- Munkvold K.R., Martin M.E., Bronstein P.A., Collmer A. (2008). A survey of the Pseudomonas syringae pv. tomato DC3000 type III secretion system effector repertoire reveals several effectors that are deleterious when expressed in Saccharomyces cerevisiae. Mol. Plant Microbe Interact. 21: 490–502 [DOI] [PubMed] [Google Scholar]
- Oh C.-S., Martin G.B. (2011). Effector-triggered immunity mediated by the Pto kinase. Trends Plant Sci. 16: 132–140 [DOI] [PubMed] [Google Scholar]
- Oh R.S., Bai X., Rommens J.M. (2006). Human homologs of Ubc6p ubiquitin-conjugating enzyme and phosphorylation of HsUbc6e in response to endoplasmic reticulum stress. J. Biol. Chem. 281: 21480–21490 [DOI] [PubMed] [Google Scholar]
- Pedley K.F., Martin G.B. (2003). Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu. Rev. Phytopathol. 41: 215–243 [DOI] [PubMed] [Google Scholar]
- Philip N., Haystead T.A. (2007). Characterization of a UBC13 kinase in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 104: 7845–7850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel M.C., Leonelli L., Dahlbeck D., Zhao B., Staskawicz B.J. (2008). Recognition of the Hyaloperonospora parasitica effector ATR13 triggers resistance against oomycete, bacterial, and viral pathogens. Proc. Natl. Acad. Sci. USA 105: 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommens C.M., Salmeron J.M., Baulcombe D.C., Staskawicz B.J. (1995). Use of a gene expression system based on potato virus X to rapidly identify and characterize a tomato Pto homolog that controls fenthion sensitivity. Plant Cell 7: 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosebrock T.R., Zeng L., Brady J.J., Abramovitch R.B., Xiao F., Martin G.B. (2007). A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448: 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Sanada T., et al. (2012). The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature 483: 623–626 [DOI] [PubMed] [Google Scholar]
- Sarcevic B., Mawson A., Baker R.T., Sutherland R.L. (2002). Regulation of the ubiquitin-conjugating enzyme hHR6A by CDK-mediated phosphorylation. EMBO J. 21: 2009–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semplici F., Meggio F., Pinna L.A., Oliviero S. (2002). CK2-dependent phosphorylation of the E2 ubiquitin conjugating enzyme UBC3B induces its interaction with beta-TrCP and enhances beta-catenin degradation. Oncogene 21: 3978–3987 [DOI] [PubMed] [Google Scholar]
- Shembade N., Harhaj N.S., Yamamoto M., Akira S., Harhaj E.W. (2007). The human T-cell leukemia virus type 1 Tax oncoprotein requires the ubiquitin-conjugating enzyme Ubc13 for NF-kappaB activation. J. Virol. 81: 13735–13742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman J.A., da Silva Almeida A.C., Hassink G.C., van de Ven R.H.A., van Kerkhof P., Kuiken H.J., Strous G.J. (2012). Ubc13 and COOH terminus of Hsp70-interacting protein (CHIP) are required for growth hormone receptor endocytosis. J. Biol. Chem. 287: 15533–15543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J., Gali R.R., Dittmar G., Sherman F., Karin M., Finley D. (2000). Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 102: 67–76 [DOI] [PubMed] [Google Scholar]
- Spoel S.H., Mou Z., Tada Y., Spivey N.W., Genschik P., Dong X. (2009). Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Frederick R.D., Zhou J., Halterman D.A., Jia Y., Martin G.B. (1996). Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274: 2060–2063 [DOI] [PubMed] [Google Scholar]
- Topisirovic I., Gutierrez G.J., Chen M., Appella E., Borden K.L.B., Ronai Z.A. (2009). Control of p53 multimerization by Ubc13 is JNK-regulated. Proc. Natl. Acad. Sci. USA 106: 12676–12681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E., Hatzivassiliou E., Tsichritzis T., Farmer H., Ashworth A., Mosialos G. (2003). CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature 424: 793–796 [DOI] [PubMed] [Google Scholar]
- Trujillo M., Ichimura K., Casais C., Shirasu K. (2008). Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 18: 1396–1401 [DOI] [PubMed] [Google Scholar]
- Trujillo M., Shirasu K. (2010). Ubiquitination in plant immunity. Curr. Opin. Plant Biol. 13: 402–408 [DOI] [PubMed] [Google Scholar]
- VanDemark A.P., Hofmann R.M., Tsui C., Pickart C.M., Wolberger C. (2001). Molecular insights into polyubiquitin chain assembly: Crystal structure of the Mms2/Ubc13 heterodimer. Cell 105: 711–720 [DOI] [PubMed] [Google Scholar]
- Vierstra R.D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8: 135–142 [DOI] [PubMed] [Google Scholar]
- Wei C.-F., Kvitko B.H., Shimizu R., Crabill E., Alfano J.R., Lin N.-C., Martin G.B., Huang H.-C., Collmer A. (2007). A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. 51: 32–46 [DOI] [PubMed] [Google Scholar]
- Wen R., Newton L., Li G., Wang H., Xiao W. (2006). Arabidopsis thaliana UBC13: Implication of error-free DNA damage tolerance and Lys63-linked polyubiquitylation in plants. Plant Mol. Biol. 61: 241–253 [DOI] [PubMed] [Google Scholar]
- Wen R., Torres-Acosta J.A., Pastushok L., Lai X., Pelzer L., Wang H., Xiao W. (2008). Arabidopsis UEV1D promotes Lysine-63-linked polyubiquitination and is involved in DNA damage response. Plant Cell 20: 213–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiborg J., O’Shea C., Skriver K. (2008). Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem. J. 413: 447–457 [DOI] [PubMed] [Google Scholar]
- Wilson M.I., Gill D.J., Perisic O., Quinn M.T., Williams R.L. (2003). PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol. Cell 12: 39–50 [DOI] [PubMed] [Google Scholar]
- Wooff J., Pastushok L., Hanna M., Fu Y., Xiao W. (2004). The TRAF6 RING finger domain mediates physical interaction with Ubc13. FEBS Lett. 566: 229–233 [DOI] [PubMed] [Google Scholar]
- Xiao W., Lin S.L., Broomfield S., Chow B.L., Wei Y.F. (1998). The products of the yeast MMS2 and two human homologs (hMMS2 and CROC-1) define a structurally and functionally conserved Ubc-like protein family. Nucleic Acids Res. 26: 3908–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa Y., Sullivan J.A., Komatsu S., Gusmaroli G., Suzuki G., Yin J., Ishibashi T., Saijo Y., Rubio V., Kimura S., Wang J., Deng X.W. (2004). Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 18: 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.-W., González-Lamothe R., Ewan R.A., Rowland O., Yoshioka H., Shenton M., Ye H., O’Donnell E., Jones J.D.G., Sadanandom A. (2006). The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18: 1084–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.-L., Wang J., Chan C.-H., Lee S.-W., Campos A.D., Lamothe B., Hur L., Grabiner B.C., Lin X., Darnay B.G., Lin H.-K. (2009). The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 325: 1134–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Rape M. (2009). Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10: 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zeng L., Velásquez A.C., Munkvold K.R., Zhang J., Martin G.B. (2012). A tomato LysM receptor-like kinase promotes immunity and its kinase activity is inhibited by AvrPtoB. Plant J. 69: 92–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L.-R., Qu S., Bordeos A., Yang C., Baraoidan M., Yan H., Xie Q., Nahm B.H., Leung H., Wang G.-L. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L.-R., Vega-Sánchez M.E., Zhu T., Wang G.-L. (2006). Ubiquitination-mediated protein degradation and modification: An emerging theme in plant-microbe interactions. Cell Res. 16: 413–426 [DOI] [PubMed] [Google Scholar]
- Zhang J., Hu M.M., Wang Y.Y., Shu H.B. (2012). TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J. Biol. Chem. 287: 28646–28655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Windheim M., Roe S.M., Peggie M., Cohen P., Prodromou C., Pearl L.H. (2005). Chaperoned ubiquitylation—Crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 20: 525–538 [DOI] [PubMed] [Google Scholar]
- Zhou J.-M., Loh Y.-T., Bressan R.A., Martin G.B. (1995). The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell 83: 925–935 [DOI] [PubMed] [Google Scholar]