ABSTRACT
CTnDOT is a 65-kb conjugative transposon that is found in Bacteroides spp., which are one of the more abundant members within the lower human gastrointestinal tract. CTnDOT encodes resistance to the antibiotics erythromycin and tetracycline (Tc). An interesting feature of CTnDOT is that exposure to low levels of Tc induces a cascade of events that ultimately results in CTnDOT conjugative transfer. However, Tc is apparently not a switch that activates transfer but rather a signal that appears to override a series of negative regulators that inhibit premature excision and transfer of CTnDOT. In this minireview, we summarize over 20 years of research that focused on elucidating the highly coordinated regulation of excision, mobilization, and transfer of CTnDOT.
IMPORTANCE
Bacteroides spp. are abundant commensals in the human colon, but they are also considered opportunistic pathogens, as they can cause life-threatening infections if they should escape the colon. Bacteroides spp. are the most common cause of anaerobic infections and are rather difficult to treat due to the prevalence of antibiotic resistance within this genus. Today over 80% of Bacteroides are resistant to tetracycline (Tc), and a study looking at both clinical and community isolates demonstrated that this resistance was specifically due to the conjugative transposon CTnDOT.
Introduction
Bacteroides has recently garnered much recognition due to their abundance in the complex assemblage of microbes that inhabit the colon. This community within the gut may contain up to 1,500 species, present at a density of approximately 3 × 1011 bacterial cells per gram of feces (1). Although the microbiota is quite diverse, Bacteroidetes and Firmicutes are the two most numerically abundant phyla within the human colon. Early cultivation-based studies had identified Bacteroides as abundant and suggested that they comprise roughly 25 to 30% of the colonic microbiota (2–4). However, more-recent sequence-based studies have demonstrated that the members of the Bacteroidetes phylum represent upwards of 40% of the human gut microbiota (5, 6).
ROLES OF BACTEROIDES SPP. BENEFICIAL TO HUMAN HEALTH
The abundance of Bacteroides within the gut community appears to be vital to human health, as a decrease in the proportion of Bacteroidetes within the colon has been implicated in diseases such as obesity (7–9) and type 1 diabetes (10). Bacteroides bacteria are important for the acquisition of nutrients by the host by virtue of their role in the biosynthesis of vitamins and breakdown of complex polysaccharides that would otherwise go undigested (11, 12). This capacity for carbohydrate utilization is due to over 200 encoded gene products involved in the hydrolysis of glycosidic bonds, resulting in Bacteroides having one of the largest glycobiomes among any of the bacteria sequenced (13, 14). The short-chain fatty acids produced by Bacteroides as a result of this carbohydrate metabolism are also a significant source of caloric energy for the host (15). The role of Bacteroides with respect to nutrient acquisition is so paramount that malnutrition can result if there is a deficit of Bacteroides in the colon (16). In addition to providing important vitamins, sugars, and fatty acids to the host, Bacteroides spp. also protect the host from colonization by pathogens such as Clostridium difficile and Helicobacter hepaticus (15, 17).
Detrimental effects of Bacteroides on human health.
Despite the health benefits conferred by Bacteroides in the lower gastrointestinal tract, Bacteroides can have significant deleterious effects on human health if they escape the confines of the colon through disease or most commonly, when the lining of the colon has been compromised due to surgical trauma (15). When Bacteroides escapes the colon, opportunistic infections result and some can be life-threatening. The most common problem is intra-abdominal sepsis, but other complications include appendicitis and necrotizing soft tissue infections. Bacteroides spp. are the most commonly isolated organism from anaerobic infections. Although rare, Bacteroides can also cause endocarditis, meningitis, and septic arthritis (15). Bacteroides infections are becoming increasingly difficult to treat due to the prevalence of antibiotic resistance among members of this genus, which makes Bacteroides infections a serious public health threat (18). The problem of increasing antibiotic resistance in this genus is illustrated by tetracycline (Tc), which used to be a frontline antibiotic used for the treatment of Bacteroides infections due to its broad spectrum, ease of use, low incidence of side effects, and cost (19). After the debut of Tc in the 1950s, few isolates were resistant. By the 1970s, 20 to 30% of clinical and community isolates were Tc resistant, and by the 1990s, over 80% of Bacteroides isolates from clinical and community sources were resistant to Tc (20–23). Resistance to Tc has become so commonplace that Bacteroides strains are often not tested for susceptibility and are assumed to be resistant (24, 25). In many cases, Tc-resistant strains carried a conjugative transposon, also referred to as an integrated conjugative element, and most of those strains harbored the conjugative transposon CTnDOT (22, 26, 27).
Conjugative transposons.
Conjugative transposons (CTns) are mobile elements that normally reside in a single copy on the bacterial chromosome. These elements account for the majority of antibiotic resistance transfer within Bacteroides (28). CTns range in size from 18 to 500 kb with the majority of CTns averaging roughly 50 to 80 kb. The large size of most CTns is due to the need to encode the entire suite of gene products necessary to facilitate conjugative transfer, such as relaxases, coupling proteins, and mating bridge proteins (29, 30). Interestingly, these elements contain genes reminiscent of transposons, plasmids, and bacteriophages and thus share characteristics with these other mobile DNA elements. CTns possess the transposon-like property of maintenance within the chromosome, yet they can excise from the chromosome and integrate elsewhere. However, molecular mechanisms of excision and integration more closely resemble that of bacteriophage rather than transposition. The CTnDOT integrase and excision proteins themselves are quite similar to those from bacteriophage. Last, conjugative transposons are plasmid-like in their ability to form a covalently closed circular intermediate, while their lack of autonomous replication distinguishes CTns from plasmids (26).
Many conjugative transposons are able to mobilize other elements. For example, many coresident plasmids are mobilized by a conjugative transposon in trans. This occurs when a plasmid containing an oriT utilizes the CTn-provided mating pore proteins for transfer to a recipient cell. The Bacteroides CTns have also been shown to mobilize elements when in cis, a feature that is not typical for CTns. For example, if CTnDOT excises from the chromosome and integrates on a plasmid, it can provide the mating pore, an oriT, and the mobilization (relaxase/coupling) proteins, allowing it to transfer the entire plasmid by acting “in cis.” This ability to use both trans and cis mechanisms of mobilization is unusual and suggests that the Bacteroides CTns have a greater capacity to mobilize other elements (31).
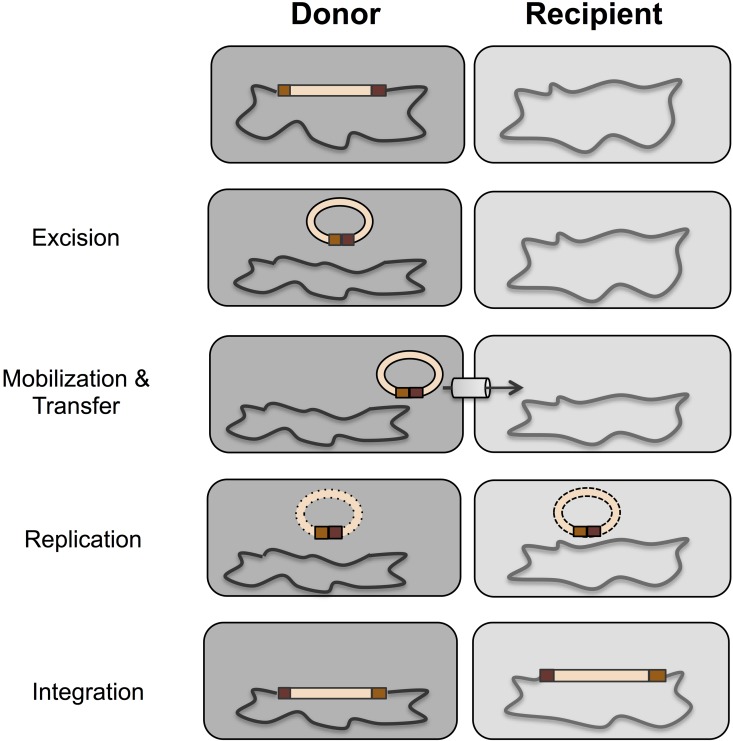
In order for transfer of the conjugative transposon to occur, there are three main steps that must take place (Fig. 1). The first step is excision from the chromosome to form a covalently closed circular intermediate. Second, a single-stranded copy is then transferred through the mating pore to a recipient cell, after which the copy becomes double stranded. Third, the intact double-stranded CTn integrates into the chromosome of the recipient (30). Conjugative transposition is replicative, as a copy of the CTn is retained in the donor cell. Because the element resides within the chromosome, it is also transferred vertically to progeny cells. This is important because when antibiotic resistance determinants are present on CTns, they are not only transferred readily within the population, but they are also very stably maintained from generation to generation. Further, it is believed that Bacteroides may serve as a reservoir of antibiotic resistance determinants which disseminates these genes to other organisms outside the Bacteroides genus, possibly even transferring these elements to organisms that are transiently passing through the gut (28, 32, 33). The vast spread of these conjugative transposons may thus pose a serious threat to the future ability to treat infections with antibiotics. The remainder of this minireview will focus on the Bacteroides conjugative transposon CTnDOT, which is a model CTn and was the first CTn identified in Gram-negative bacteria (34).
FIG 1 .
Overview of the CTnDOT life cycle. CTnDOT is a 65-kb conjugative transposon present in Bacteroides spp. that encodes resistance to the antibiotics erythromycin and tetracycline. The conjugative transfer of CTnDOT is stimulated by tetracycline induction. The first step in conjugative transfer is that CTnDOT must first excise from the host chromosome to form a circular intermediate. CTnDOT is then nicked at the oriT after which CTnDOT is then replicated and made double-stranded before integrating into the chromosomes of both the donor and recipient.
CTnDOT OVERVIEW
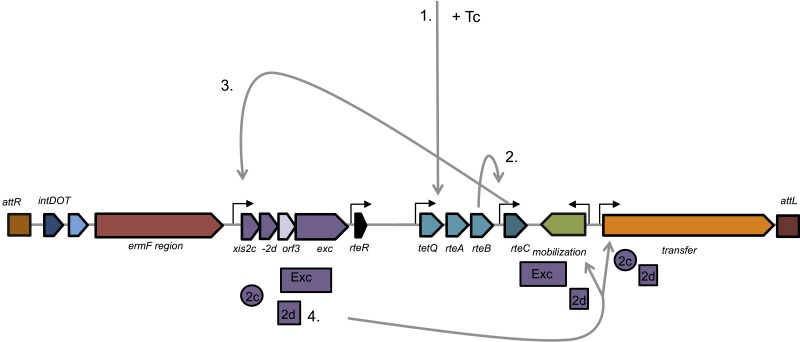
CTnDOT is a 65-kb conjugative transposon that carries genes encoding resistance to the antibiotics erythromycin (ermF) and tetracycline (tetQ). Other elements within the CTnDOT-like family include CTnERL and CTn341, although these elements differ from CTnDOT, as they lack the 13-kb ermF-encoding region (35–37). A notable feature of CTnDOT is that exposure to low levels of Tc stimulates its excision and conjugative transfer (38). Upon Tc induction (Fig. 2), a regulatory cascade is initiated, leading to translation of the tetQ-rteA-rteB operon. RteB then activates the transcription of rteC, and RteC in turn activates transcription of the excision operon, xis2c-xis2d-orf3-exc (xis2c and xis2d were referred to as orf2c and orf2d, respectively, in publications prior to 2012). As the names of genes in this operon imply, the encoded proteins (with the exception of Orf3) are needed to excise CTnDOT from the chromosome. Deletions of orf3 reveal no phenotype (38). Even more interesting is that these proteins have a regulatory function, as Xis2c, Xis2d, and Exc also collectively activate transcription of the tra and mob operons, which encode other products required for transfer and mobilization, and ultimately allow CTnDOT conjugative transfer (Fig. 2). When Tc is absent, there is no detectable excision or transfer of CTnDOT (38–41). This is due largely to a repressor of conjugative transfer, the small RNA RteR. In addition, we recently showed that the CTnDOT-encoded integrase, IntDOT, may also inhibit conjugative transfer (42, 43). An overview of the CTnDOT transcriptional cascade is shown in Fig. 2, and each of the gene products and their regulatory mechanism(s) are described in further detail in the following sections. Table 1 provides a summary of each of the key regulators described, in addition to their functional roles.
FIG 2 .
Positive regulation of CTnDOT upon exposure to tetracycline. In step 1, the tetQ-rteA-rteB operon is regulated by a translational attenuation mechanism where after exposure to tetracycline (+ Tc), translation of this region resumes; translation of this region ultimately allows for RteB to activate the transcription of RteC (step 2). RteC then activates transcription of the excision operon (step 3). Proteins encoded by the excision operon not only excise CTnDOT from the chromosome but are also involved in transcriptional regulation (step 4). Xis2c and Xis2d are involved in regulation of the tra region, whereas Xis2d and Exc are involved in enhancement of mob transcription. We have yet to identify any functional role for Orf3.
TABLE 1 .
Summary of CTnDOT regulatory proteins and RNAs
| Regulator | Function(s) | Reference(s) |
|---|---|---|
| RteB | Activates transcription of rteC | 40 |
| RteC | Activates transcription of the excision operon (xis2c, xis2d, orf3, and exc) | 42, 54 |
| Xis2c | Required for excision. Activates transcription of the tra operon. | 62, 71 |
| Xis2d | Required for excision. Activates transcription of both the tra and mob operons. | 44, 72 |
| Exc | Required for excision in vivo and enhances in vitro excision. Required for transcriptional activation of the mob operon. | 44, 55 |
| IntDOT | Required for integration and excision. Putative negative regulator. | 44, 55, 71 |
| RteR | Small RNA that negatively regulates the tra operon | 43 |
The tetQ-rteA-rteB operon.
Upon tetracycline (Tc) induction, the first step in the CTnDOT regulatory cascade occurs through an operon containing tetQ, rteA, and rteB. This operon is regulated via translational attenuation. Although the operon is transcribed constitutively, it is only upon exposure to Tc that translation occurs (19, 44–46). Similar mechanisms of inducible antibiotic resistance have also been reported for Tc induction in the Enterococcus Tn916 and related elements, as well as vancomycin induction of the Gram-positive Tn1546 (47).
CTnDOT tetQ encodes resistance to Tc via ribosomal protection, which appears to be one of the most common forms of Tc resistance, along with tetracycline efflux, in comparison to either modification or other rarer mechanisms (48). The two most heavily investigated genes encoding ribosomal protection in Gram-negative and Gram-positive bacteria are tetO and tetM, respectively (49).
Sequence homology suggests that RteA and RteB comprise a two-component regulator where RteA recognizes a signal and RteB is a response regulator. It is not yet clear what signal RteA is sensing, but findings suggest that it is not Tc (26). Earlier studies of RteA and RteB demonstrated that these two proteins were quite important; insertional disruptions of rteA-rteB eliminated conjugative transfer of CTnDOT. Additionally, mobilization of coresident plasmids and the Bacteroides nonreplicating Bacteroides units (NBUs) was also abolished (50, 51). Later studies from that laboratory demonstrated that the primary role for RteB is transcriptional activation of the downstream-encoded rteC. RteA and RteB are not involved in the regulation of the tetQ-rteA-rteB operon itself.
Although activating transcription within CTnDOT appears to be the primary function of RteA and RteB, these two proteins may have a more widespread role within the cell. RteB has demonstrated an ability to also regulate the expression of genes carried on chromosomes (52). The first class of affected genes was not unexpected; a microarray-based approach revealed four cryptic CTns that had not yet been identified in the Bacteroides thetaiotaomicron chromosome. More surprising was finding that chromosomal genes unlinked to such elements were also affected. For example, an ompA homologue, a polysaccharide export protein, an aminotransferase, and the chaperone protein groE were all upregulated by at least 7-fold. Although most of the genes that were downregulated were identified as hypothetical proteins of unknown function, an ATP-dependent helicase, glycosyltransferase, and a type 1 restriction enzyme were included in these regulated proteins (52). Although most genes identified in this study were regulated by RteA and RteB, two genes were regulated by RteC, the regulatory protein that is encoded downstream of rteB. Many questions remain as to why genes not involved with conjugative transfer were affected. Nonetheless, these observations further demonstrate the complexity of CTnDOT.
The regulatory protein, RteC.
RteC is a regulatory protein that is expressed only after Tc induction, due to transcriptional activation from the upstream-encoded RteB. The primary function of RteC is to activate transcription of the operon containing xis2c, xis2d, and exc, which are required for excision from the chromosome as well as activation of the transfer region (the excision operon is discussed in further detail below). A structural homologue search revealed that RteC contains a winged-helix motif, which is common among DNA-binding proteins (53). Later studies demonstrated that putting rteC under the control of a heterologous promoter, and thus bypassing the need for RteB, resulted in transcriptional activation of the excision operon (41). This further confirmed that the primary roles of the tetQ-rteA-rteB operon are resistance to Tc and activation of rteC transcription for downstream gene regulation (39, 41). RteC-mediated transcriptional activation of the excision operon occurs by the binding of RteC to the region located 50 to 70 nucleotides (nt) upstream of the excision promoter region, which was defined using both site-directed mutagenesis and electrophoretic mobility shift assay (EMSA) analysis (53).
The excision operon.
The excision operon is located downstream of the ermF region of CTnDOT and contains xis2c, xis2d, orf3, and exc. As the name implies, proteins encoded within this region are involved in excising CTnDOT from the chromosome in order to form the circular transfer intermediate. We have not yet identified any structural or regulatory function for Orf3, as orf3 deletions yield no phenotype, as excision and conjugative transfer are unaffected (38, 40). In addition to Xis2c, Xis2d, and Exc, the chromosomally encoded host factor Bhfa and the CTnDOT-encoded integrase, IntDOT, are also required for excision (54).
Xis2c and Xis2d, formerly known as Orf2c and Orf2d, respectively, are both small basic proteins that contain helix-turn-helix DNA binding domains. Xis2c shares similarities with lambda Xis, and Xis2d is homologous to the Escherichia coli excisionase TorI and the Tn916 excisionase (54–57). Exc contains a topoisomerase III domain and is capable of relaxing DNA in vitro (58). Not surprisingly, a mutation in Exc that changes the catalytic tyrosine residue to phenylalanine in the topoisomerase domain can no longer relax supercoiled DNA and can no longer catalyze the in vitro excision reaction. However, there is no noticeable defect in the in vivo excision reaction (58). These observations suggest that although the topoisomerase activity is relevant for the Exc function, it is likely not critical.
Xis2c, Xis2d, and Exc play an additional role, as they are required for positively regulating two very important regions of CTnDOT upon Tc induction, the transfer and mobilization regions.
The transfer operon.
The transfer (tra) genes are assembled in a 13-kb operon containing 17 protein-encoding genes, traA through traQ (59). These Tra proteins are required to assemble the mating bridge, a type IV secretion-like apparatus that is required for conjugative transfer. The tra genes have a much higher GC content (48 to 60%) than the Bacteroides chromosome (approximately 42%), which suggests that the tra genes originated from another genus of bacteria (60).
Evidence for a regulatory role of the CTnDOT excision operon was provided by studies in which this operon was sufficient to increase expression of a plasmid, pLYL72, that contains an 18-kb region of CTnDOT that is sufficient for self-transfer. Due to the limitations of Bacteroides genetics, we have extensively used pLYL72 to study regulation of CTnDOT conjugative transfer. While TraG, TraN, and TraP were translated from pLYL72 with or without Tc induction, if CTnDOT was integrated in the chromosome, no Tra gene products were detected without Tc and elevated levels of Tra protein were observed in the presence of Tc. Not only were levels of the Tra proteins affected, but transfer of pLYL72 was affected in parallel (38). A plasmid containing the excision operon and RteR was sufficient to mediate this effect. In this section, we will focus on positive regulation by the proteins of the excision operon; RteR-mediated repression is discussed later in this minireview.
Subcloning analysis suggested that the excision operon (xis2c-xis2d-orf3-exc) was mediating regulation of the transfer operon. The excision operon was placed under the control of a heterologous inducible promoter (PsusA, which is induced with maltose) to liberate expression of this operon from the requirement for RteC. Jeters et al. demonstrated that expressing the excision genes under control of the heterologous promoter activated a traA::uidA fusion, similar to that seen when the excision genes were dependent on RteC. This observation confirmed that the excision proteins are mediating activation of the tra operon and that rteC is required in this context only for activation of the excision operon (59).
A recent study confirmed more specifically that the excision proteins Xis2c and Xis2d are necessary and sufficient for transcriptional activation of the transfer operon. A deletion of either orf3 or exc, contained in this same operon, revealed no observable defect in activation of the tra operon. Xis2c and Xis2d appear to mediate transcriptional activation by binding upstream of the tra promoter (Fig. 2) as indicated by EMSA analysis, demonstrating that Xis2c and Xis2d DNA binding is sequence specific. Mutational analysis within the upstream tra promoter region showed a loss of Xis2d binding, further supporting the idea that binding upstream of the tra promoter results in regulation of the transfer region (61).
Although no studies have been performed on the CTnDOT Tra proteins to confirm specific roles of each protein in conjugative transfer, some work has defined which of the Tra proteins are required for conjugative transfer. Insertion deletion analysis was performed in CTnERL, a Bacteroides conjugative transposon that for the most part is similar to CTnDOT but lacks the ermF region, thereby allowing the use of erythromycin as a selectable marker (35).
Insertion deletions in traG, traI, traJ, and traM were sufficient to abolish CTnERL transfer, which suggested that these proteins were essential for conjugative transfer. Mutations in traH and traN resulted in a 100-fold decrease in CTnERL transfer, measured as transconjugants per recipient. A surprising finding from this study was that some insertions actually resulted in increased conjugative transfer. Disruption of traO resulted in a 10-fold increase in transfer, while insertions in traP and traQ resulted in a 100-fold increase. No insertions were generated in traABCD because this region was too poorly conserved between CTnERL and CTnDOT to make reliable interpretations. No insertions were made in traF or traL, because the open reading frame was too small to construct a reliable insertion, and Bonheyo et al. were unsuccessful in the attempt to construct an insertion in traK (62).
Thus far, we have reported that the tra promoter is independently always in the “on” state (i.e., when detected from pLYL72), but when the excision operon is present, Xis2c and Xis2d bind upstream to further activate transcription through this operon. While the focus has been on transcription initiation, there appears to be post- initiation effects. A deletion of the 5′ tra leader region results in a 10-fold activation of a tra::uidA fusion (Rob Jeters, unpublished results). An earlier hypothesis was that this region contained the RteR binding site, but we later ruled this out, as similar levels of traA are detected whether or not rteR is present. This observation warrants further investigation into the regulation of the tra leader region.
The mobilization operon.
The mobilization region, which is divergently transcribed from the transfer region, is an approximately 4-kb operon containing mobA, mobB, and mobC. Recently, a study of a CTnDOT-like element, CTn341, revealed that MobA and MobB function as the relaxase proteins that nick the oriT to initiate transfer. MobC is the coupling protein that shuttles the circular transfer intermediate to the mating pore. A null mutation made in either mobA or mobB abolished transfer of CTn341, yet coresident elements were still able to transfer to recipient cells. An unforeseen finding was that a null mutation of mobC still resulted in transfer of CTn341. This was unexpected because in other systems, the coupling protein is absolutely required for mobilization. Further insight revealed the presence of four mobC homologues in the chromosome, so it is likely then that at least one of these chromosomally encoded proteins could compensate for the deletion of mobC which ultimately resulted in wild-type transfer of CTn341 (63).
We have recently shown that the mob operon is actually transcribed constitutively with respect to Tc induction when detected from the self-transmissible plasmid pLYL72, which contains an 18-kb region of CTnDOT containing the mobilization and transfer regions as well as an oriT (64). However, when the mob genes are detected from a chromosomal copy of CTnDOT or CTnERL, a transcript is detectable only upon Tc induction.
When the intact excision operon is provided in trans to pLYL72, an approximately 10-fold increase in mob transcript is detected relative to the constitutive level of transcription. Sequential deletion analysis of the excision operon demonstrates that an in-frame deletion of either xis2d or exc is sufficient to diminish mob transcription to the constitutive levels, while a deletion of either xis2c or orf3 has no effect (43). While both the mob operon and tra operon share a requirement for xis2d to enhance transcription, they differ in that the mob operon also requires exc, and the tra operon requires xis2c in addition to xis2d (Fig. 2).
Similar to regulation of the transfer region, transcription of the mob promoter is constitutive from pLYL72, yet detectable only from CTnDOT upon Tc induction. This observation suggests that a negative mob regulator is present on CTnDOT. At this time, however, we do not know what is preventing mob transcription in the absence of Tc, but we have ruled out RteR.
NEGATIVE REGULATION OF CTnDOT
Thus far, we have described a multilayered regulatory cascade that initiates upon exposure of cells to Tc, but it is important to clarify that regulation of transfer is not this simple. Tc does not simply act like a switch, triggering a domino-like effect resulting in CTnDOT transfer. Instead, this induction is overriding a series of negative regulators that serve to prevent premature transfer of CTnDOT.
The small RNA, RteR.
The most extensively characterized of the CTnDOT negative regulators is the regulatory noncoding RNA, RteR. This small RNA was first identified in an attempt to identify the positive regulators of CTnDOT transfer and is the first small RNA to be characterized in the Bacteroidetes phylum. When an approximately 7-kb region containing the excision operon and rteR was provided in trans to pLYL72, this was sufficient to not only enhance transfer upon Tc induction 100- to 1,000-fold but also entirely prevent pLYL72 transfer in the absence of Tc. A 500-bp region downstream of exc was sufficient for inhibition of pLYL72 conjugative transfer whether or not Tc was present in the growth medium, suggesting that the CTnDOT negative regulator was localized to this fragment (38).
This 500-bp region encodes the small RNA, RteR, which is a 90-nt transcript that is transcribed from an independent promoter that is constitutive with respect to Tc induction. Previous studies demonstrated that no TraG protein (using TraG-specific antibodies) or traG mRNA was detected from pLYL72 when a fragment containing RteR was in trans (38, 59). We later demonstrated that similar levels of traA are detected when rteR is in trans to pLYL72, yet the downstream tra genes are barely detectable. Further, the tra half-life is the same whether or not rteR was present. Taken together, these observations suggested that RteR may be initiating premature transcription termination within the tra operon (42).
IntDOT.
The CTnDOT-encoded integrase IntDOT is a member of the tyrosine recombinase family, similar to the integrase on the conjugative transposon Tn916. IntDOT is required for integration of CTnDOT into the chromosome, a reaction that also requires the Bacteroides host factor Bhfa (65, 66). Although structurally similar, the Tn916 integrase and IntDOT differ with respect to integration. Tn916 integration appears to be random, whereas CTnDOT integrates into specific sites, although there are multiple sites within the Bacteroides chromosome (67). Another interesting feature of IntDOT is that while most other tyrosine recombinases require perfect homology in the overlap sequences they bind in order for recombination to occur, IntDOT can still catalyze integration even with a large mismatch of this 7-bp sequence. Further, the frequency of integration is the same whether the overlap sequence is homologous or whether mismatches are present, as long as the 2 bp adjacent to the catalytic site are homologous (68, 69). IntDOT is also required for the excision of CTnDOT from the chromosome, and interestingly, the homology of the overlap sites matters for excision. The excision reaction is more efficient when there is perfect homology compared to a mismatched overlap sequence (70, 71).
Although the CTnDOT integrase IntDOT has been well studied, we recently made the serendipitous discovery that when intDOT is provided in trans to the self-transmissible plasmid pLYL72, there is no detectable transfer, with or without Tc induction. A possible role for IntDOT as a negative regulator of CTnDOT transfer was first investigated when trying to elucidate the negative regulator of the mob operon. The first suspect was RteR, which was ruled out as the mob genes are transcribed constitutively with or without rteR in trans (42). The integrase of Tn916 can bind the Tn916 oriT region to prevent premature conjugative transfer, which prompted us to investigate whether the CTnDOT integrase could perform a similar function (72). Although the mob genes were transcribed whether or not IntDOT was in trans, pLYL72 transfer was not detectable from this same strain. This additional role of IntDOT remains to be further explored.
CONCLUSIONS
CTnDOT has established itself as a fascinating element, as the regulation of self-transfer is highly coordinated so as to promote conjugative transfer promptly upon ideal conditions—in this case exposure to the antibiotic Tc. Further, this prevents the unnecessary transfer until it is vital to the community to harbor CTnDOT, as conjugative transfer bears a metabolic cost to the donor (73). While studies of CTnDOT have offered further insight as to the complex nature of conjugative elements, we still have much to learn about this element.
ACKNOWLEDGMENTS
We are grateful to Carin K. Vanderpool for feedback and discussion of this article. We thank the two anonymous reviewers for their comments on the manuscript.
Footnotes
Citation Waters JL, Salyers AA. 2013. Regulation of CTnDOT conjugative transfer is a complex and highly coordinated series of events. mBio 4(6):e00569-13. doi:10.1128/mBio.00569-13.
REFERENCES
- 1. Franks AH, Harmsen HJ, Raangs GC, Jansen GJ, Schut F, Welling GW. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moore WE, Holdeman LV. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Salyers AA. 1984. Bacteroides of the human lower intestinal tract. Annu. Rev. Microbiol. 38:293–313 [DOI] [PubMed] [Google Scholar]
- 4. Savage DC. 1977. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 31:107–133 [DOI] [PubMed] [Google Scholar]
- 5. Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C. 2012. Microbial co-occurrence relationships in the human microbiome. PLoS Comput. Biol. 8:e1002606. 10.1371/journal.pcbi.1002606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ley RE. 2010. Obesity and the human microbiome. Curr. Opin. Gastroenterol. 26:5–11 [DOI] [PubMed] [Google Scholar]
- 8. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koren O, Spor A, Felin J, Fåk F, Stombaugh J, Tremaroli V, Behre CJ, Knight R, Fagerberg B, Ley RE, Bäckhed F. 2011. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 1):4592–4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Musso G, Gambino R, Cassader M. 2011. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu. Rev. Med. 62:361–380 [DOI] [PubMed] [Google Scholar]
- 11. Hooper LV, Midtvedt T, Gordon JI. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22:283–307 [DOI] [PubMed] [Google Scholar]
- 12. Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074–2076 [DOI] [PubMed] [Google Scholar]
- 13. Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915–1920 [DOI] [PubMed] [Google Scholar]
- 14. Samuel BS, Gordon JI. 2006. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc. Natl. Acad. Sci. U. S. A. 103:10011–10016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wexler HM. 2007. Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20:593–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ottman N, Smidt H, de Vos WM, Belzer C. 2012. The function of our microbiota: who is out there and what do they do? Front. Cell. Infect. Microbiol. 2:104. 10.3389/fcimb.2012.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9:313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nguyen M, Vedantam G. 2011. Mobile genetic elements in the genus Bacteroides, and their mechanism(s) of dissemination. Mob. Genet. Elements 1:187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Speer BS, Shoemaker NB, Salyers AA. 1992. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin. Microbiol. Rev. 5:387–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sutter VL, Kwok YY, Finegold SM. 1972. Standardized antimicrobial disc susceptibility testing of anaerobic bacteria. I. Susceptibility of Bacteroides fragilis to tetracycline. Appl. Microbiol. 23:268–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bodner SJ, Koenig MG, Treanor LL, Goodman JS. 1972. Antibiotic susceptibility testing of Bacteroides. Antimicrob. Agents Chemother. 2:57–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Salyers AA, Gupta A, Wang Y. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12:412–416 [DOI] [PubMed] [Google Scholar]
- 23. Quesada-Gómez C, Rodríguez-Cavallini E, Rodríguez C. 2013. Scarce detection of mobile erm genes associated with tetQ in Bacteroides and Parabacteroides from Costa Rica. Anaerobe 21:18–21 [DOI] [PubMed] [Google Scholar]
- 24. Salyers AA, Shoemaker NB. 1995. Conjugative transposons: the force behind the spread of antibiotic resistance genes among Bacteroides clinical isolates. Anaerobe 1:143–150 [DOI] [PubMed] [Google Scholar]
- 25. Shoemaker NB, Vlamakis H, Hayes K, Salyers AA. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salyers AA, Shoemaker NB, Stevens AM, Li LY. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Burrus V, Pavlovic G, Decaris B, Guédon G. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601–610 [DOI] [PubMed] [Google Scholar]
- 28. Whittle G, Shoemaker NB, Salyers AA. 2002. The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell. Mol. Life Sci. 59:2044–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scott JR, Churchward GG. 1995. Conjugative transposition. Annu. Rev. Microbiol. 49:367–397 [DOI] [PubMed] [Google Scholar]
- 30. Whittle G, Salyers AA. 2002. Bacterial transposons—an increasingly diverse group of elements, p. 385–427 In Streips UN, Yasbin RE. (ed), Modern microbial genetics. John Wiley & Sons, Inc, New York, NY. [Google Scholar]
- 31. Salyers AA, Shoemaker NB, Li LY. 1995. In the driver’s seat: the Bacteroides conjugative transposons and the elements they mobilize. J. Bacteriol. 177:5727–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bacic MK, Smith CJ. 2008. Laboratory maintenance and cultivation of Bacteroides species. Curr. Protoc. Microbiol. Chapter 13:Unit 13C.1. 10.1002/9780471729259.mc13c01s9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rotimi VO, Duerden BI, Hafiz S. 1981. Transferable plasmid-mediated antibiotic resistance in Bacteroides. J. Med. Microbiol. 14:359–370 [DOI] [PubMed] [Google Scholar]
- 34. Franklin TJ, Snow GA. 2005. Biochemistry and molecular biology of antimicrobial drug action. Springer Verlag, New York, NY. [Google Scholar]
- 35. Whittle G, Hund BD, Shoemaker NB, Salyers AA. 2001. Characterization of the 13-kilobase ermF region of the Bacteroides conjugative transposon CTnDOT. Appl. Environ. Microbiol. 67:3488–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bacic M, Parker AC, Stagg J, Whitley HP, Wells WG, Jacob LA, Smith CJ. 2005. Genetic and structural analysis of the Bacteroides conjugative transposon CTn341. J. Bacteriol. 187:2858–2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Whittle G, Hamburger N, Shoemaker NB, Salyers AA. 2006. A Bacteroides conjugative transposon, CTnERL, can transfer a portion of itself by conjugation without excising from the chromosome. J. Bacteriol. 188:1169–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Whittle G, Shoemaker NB, Salyers AA. 2002. Characterization of genes involved in modulation of conjugal transfer of the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 184:3839–3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stevens AM, Shoemaker NB, Li LY, Salyers AA. 1993. Tetracycline regulation of genes on Bacteroides conjugative transposons. J. Bacteriol. 175:6134–6141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cheng Q, Sutanto Y, Shoemaker NB, Gardner JF, Salyers AA. 2001. Identification of genes required for excision of CTnDOT, a Bacteroides conjugative transposon. Mol. Microbiol. 41:625–632 [DOI] [PubMed] [Google Scholar]
- 41. Moon K, Shoemaker NB, Gardner JF, Salyers AA. 2005. Regulation of excision genes of the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 187:5732–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Waters JL, Salyers AA. 2012. The small RNA RteR inhibits transfer of the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 194:5228–5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Waters JL, Wang GR, Salyers AA. 27 September 2013. Tetracycline-related transcriptional regulation of the CTnDOT mobilization region. J. Bacteriol.[Epub ahead of print.] 10.1128/JB.00691-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nikolich MP, Shoemaker NB, Salyers AA. 1992. A Bacteroides tetracycline resistance gene represents a new class of ribosome protection tetracycline resistance. Antimicrob. Agents Chemother. 36:1005–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Y, Rotman ER, Shoemaker NB, Salyers AA. 2005. Translational control of tetracycline resistance and conjugation in the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 187:2673–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang Y, Shoemaker NB, Salyers AA. 2004. Regulation of a Bacteroides operon that controls excision and transfer of the conjugative transposon CTnDOT. J. Bacteriol. 186:2548–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chancey ST, Zähner D, Stephens DS. 2012. Acquired inducible antimicrobial resistance in Gram-positive bacteria. Future Microbiol. 7:959–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chopra I, Roberts M. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 56:232–260 PubMed; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Connell SR, Tracz DM, Nierhaus KH, Taylor DE. 2003. Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob. Agents Chemother. 47:3675–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stevens AM, Sanders JM, Shoemaker NB, Salyers AA. 1992. Genes involved in production of plasmidlike forms by a Bacteroides conjugal chromosomal element share amino acid homology with two-component regulatory systems. J. Bacteriol. 174:2935–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stevens AM, Shoemaker NB, Salyers AA. 1990. The region of a Bacteroides conjugal chromosomal tetracycline resistance element which is responsible for production of plasmidlike forms from unlinked chromosomal DNA might also be involved in transfer of the element. J. Bacteriol. 172:4271–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moon K, Sonnenburg J, Salyers AA. 2007. Unexpected effect of a Bacteroides conjugative transposon, CTnDOT, on chromosomal gene expression in its bacterial host. Mol. Microbiol. 64:1562–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Park J, Salyers AA. 2011. Characterization of the Bacteroides CTnDOT regulatory protein RteC. J. Bacteriol. 193:91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Keeton CM, Gardner JF. 2012. Roles of Exc protein and DNA homology in the CTnDOT excision reaction. J. Bacteriol. 194:3368–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Abbani M, Iwahara M, Clubb RT. 2005. The structure of the excisionase (Xis) protein from conjugative transposon Tn916 provides insights into the regulation of heterobivalent tyrosine recombinases. J. Mol. Biol. 347:11–25 [DOI] [PubMed] [Google Scholar]
- 56. Elantak L, Ansaldi M, Guerlesquin F, Méjean V, Morelli X. 2005. Structural and genetic analyses reveal a key role in prophage excision for the TorI response regulator inhibitor. J. Biol. Chem. 280:36802–36808 [DOI] [PubMed] [Google Scholar]
- 57. Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 58. Sutanto Y, Shoemaker NB, Gardner JF, Salyers AA. 2002. Characterization of Exc, a novel protein required for the excision of Bacteroides conjugative transposon. Mol. Microbiol. 46:1239–1246 [DOI] [PubMed] [Google Scholar]
- 59. Jeters RT, Wang GR, Moon K, Shoemaker NB, Salyers AA. 2009. Tetracycline-associated transcriptional regulation of transfer genes of the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 191:6374–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bonheyo G, Graham D, Shoemaker NB, Salyers AA. 2001. Transfer region of a Bacteroides conjugative transposon, CTnDOT. Plasmid 45:41–51 [DOI] [PubMed] [Google Scholar]
- 61. Keeton CM, Park J, Wang GR, Hopp CM, Shoemaker NB, Gardner JF, Salyers AA. 2013. The excision proteins of CTnDOT positively regulate the transfer operon. Plasmid 69:172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bonheyo GT, Hund BD, Shoemaker NB, Salyers AA. 2001. Transfer region of a Bacteroides conjugative transposon contains regulatory as well as structural genes. Plasmid 46:202–209 [DOI] [PubMed] [Google Scholar]
- 63. Peed L, Parker AC, Smith CJ. 2010. Genetic and functional analyses of the mob operon on conjugative transposon CTn341 from Bacteroides spp. J. Bacteriol. 192:4643–4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li LY, Shoemaker NB, Salyers AA. 1995. Location and characteristics of the transfer region of a Bacteroides conjugative transposon and regulation of transfer genes. J. Bacteriol. 177:4992–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wood MM, DiChiara JM, Yoneji S, Gardner JF. 2010. CTnDOT integrase interactions with attachment site DNA and control of directionality of the recombination reaction. J. Bacteriol. 192:3934–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Malanowska K, Salyers AA, Gardner JF. 2006. Characterization of a conjugative transposon integrase, IntDOT. Mol. Microbiol. 60:1228–1240 [DOI] [PubMed] [Google Scholar]
- 67. Cheng Q, Paszkiet BJ, Shoemaker NB, Gardner JF, Salyers AA. 2000. Integration and excision of a Bacteroides conjugative transposon, CTnDOT. J. Bacteriol. 182:4035–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Malanowska K, Yoneji S, Salyers AA, Gardner JF. 2007. CTnDOT integrase performs ordered homology-dependent and homology-independent strand exchanges. Nucleic Acids Res. 35:5861–5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Laprise J, Yoneji S, Gardner JF. 2013. IntDOT interactions with core sites during integrative recombination. J. Bacteriol. 195:1883–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sutanto Y, DiChiara JM, Shoemaker NB, Gardner JF, Salyers AA. 2004. Factors required in vitro for excision of the Bacteroides conjugative transposon, CTnDOT. Plasmid 52:119–130 [DOI] [PubMed] [Google Scholar]
- 71. Keeton CM, Hopp CM, Yoneji S, Gardner JF. 2013. Interactions of the excision proteins of CTnDOT in the attR intasome. Plasmid 70:190–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hinerfeld D, Churchward G. 2001. Specific binding of integrase to the origin of transfer (oriT) of the conjugative transposon Tn916. J. Bacteriol. 183:2947–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Firth N, Ippin-Ihler K, Skurray RA. 1996. Structure and function of the F factor and mechanism of conjugation, p 2377–2401 In Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 74. Ochoa-Repáraz J, Mielcarz DW, Wang Y, Begum-Haque S, Dasgupta S, Kasper DL, Kasper LH. 2010. A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol. 3:487–495 [DOI] [PubMed] [Google Scholar]