ABSTRACT
An important first line of defense against Candida albicans infections is the killing of fungal cells by professional phagocytes of the innate immune system, such as polymorphonuclear cells (PMNs) and macrophages. In this study, we employed live-cell video microscopy coupled with dynamic image analysis tools to provide insights into the complexity of C. albicans phagocytosis when macrophages and PMNs were incubated with C. albicans alone and when both phagocyte subsets were present. When C. albicans cells were incubated with only one phagocyte subtype, PMNs had a lower overall phagocytic capacity than macrophages, despite engulfing fungal cells at a higher rate once fungal cells were bound to the phagocyte surface. PMNs were more susceptible to C. albicans-mediated killing than macrophages, irrespective of the number of C. albicans cells ingested. In contrast, when both phagocyte subsets were studied in coculture, the two cell types phagocytosed and cleared C. albicans at equal rates and were equally susceptible to killing by the fungus. The increase in macrophage susceptibility to C. albicans-mediated killing was a consequence of macrophages taking up a higher proportion of hyphal cells under these conditions. In the presence of both PMNs and macrophages, C. albicans yeast cells were predominantly cleared by PMNs, which migrated at a greater speed toward fungal cells and engulfed bound cells more rapidly. These observations demonstrate that the phagocytosis of fungal pathogens depends on, and is modified by, the specific phagocyte subsets present at the site of infection.
IMPORTANCE
Extensive work investigating fungal cell phagocytosis by macrophages and PMNs of the innate immune system has been carried out. These studies have been informative but have examined this phenomenon only when one phagocyte subset is present. The current study employed live-cell video microscopy to break down C. albicans phagocytosis into its component parts and examine the effect of a single phagocyte subset, versus a mixed phagocyte population, on these individual stages. Through this approach, we identified that the rate of fungal cell engulfment and rate of phagocyte killing altered significantly when both macrophages and PMNs were incubated in coculture with C. albicans compared to the rate of either phagocyte subset incubated alone with the fungus. This research highlights the significance of studying pathogen-host cell interactions with a combination of phagocytes in order to gain a greater understanding of the interactions that occur between cells of the host immune system in response to fungal invasion.
INTRODUCTION
Candida species collectively represent the fourth leading cause of nosocomial bloodstream infections in the United States (1, 2). Within the Candida genus, Candida albicans is the major human fungal pathogen, accounting for approximately 50% of candidemia cases (3, 4). This fungus ordinarily colonizes mucosal surfaces of the skin and gastrointestinal and vulvovaginal tracts of healthy individuals. Around 75 million women suffer at least four episodes of Candida vaginitis each year, and AIDS-associated esophagitis is extremely common in untreated HIV-positive individuals (3). This opportunistic pathogen can also cause life-threatening systemic infections, with associated mortality rates of 30 to 60% in patients who have implanted medical devices, have been administered immunosuppressant drugs, or have been subjected to severe trauma (5).
An important first line of defense against Candida infections is provided by the innate immune system through the recruitment of professional phagocytes, such as polymorphonuclear cells (PMNs) and macrophages, to the site of infection (5–8). Studies to date have recognized the importance of macrophages and neutrophils in host defense against fungal pathogens and suggest a dominant role for the PMNs (9–11). This is reflected in the observation that neutropenic patients and patients with peroxidase deficiency are highly susceptible to invasive candidiasis (5, 12, 13). In particular, patients with chronic granulomatous disease, displaying impaired neutrophil function, are especially prone to systemic fungal infections (12–15). Additionally, in a mouse model where splenic macrophages were eliminated, mice demonstrated increased susceptibility to experimental disseminated candidiasis (16). Despite studies highlighting the clinical importance of phagocytes in anti-Candida infection, the process of fungal cell phagocytosis remains poorly understood at the mechanistic level.
The first step in the phagocytosis process involves macrophages and PMNs recognizing pathogen-associated molecular patterns (PAMPs) present in the fungal cell wall through pattern recognition receptors (PRRs) localized on the phagocytic cell membrane, in endosomes, and in the cytoplasm (17–19). Engagement of these receptors enables the phagocyte to directly engulf and destroy fungal cells within the phagolysosome by a number of oxidative and nonoxidative mechanisms, including the production of toxic reactive oxygen species (ROS) and reactive nitrogen species (RNS), expression of various antimicrobial peptides, and the activities of hydrolytic enzymes (5, 6, 17, 20–23). Furthermore, fungi and bacteria induce the formation of neutrophil extracellular traps (NETs) by activated PMNs, which can entrap and kill fungi and bacteria extracellularly (24). PAMP-PRR interactions also facilitate indirect killing of Candida by triggering the induction of proinflammatory cytokines and chemotactic factors which serve to activate other arms of the host immune system and aid in the clearance of Candida from the body (6, 17, 18).
Extensive work has been carried out identifying the PRRs and downstream signaling pathways that are involved in phagocyte recognition of fungal cells (6, 9, 23). These studies have identified the Toll-like receptors (TLRs) and C-type lectin receptors (CLRs) as the major PRRs involved in C. albicans PAMP recognition (6, 17, 25). Other studies have revealed how the overall phagocytic process in macrophages and PMNs is affected by fungal cell wall composition, morphogenesis, and species (5, 8, 26, 27). For example, Keppler-Ross et al. demonstrated that J774.1 macrophages preferentially phagocytosed Candida glabrata and Saccharomyces cerevisiae over C. albicans and that these macrophages displayed a strong preference for C. albicans yeast cells rather than hyphal cells (26). In addition, C. albicans mutants deficient in cell wall N- and O-linked glycans negatively affected the capacity of PMNs to phagocytose C. albicans (5). Such studies have been informative but have investigated phagocytosis under conditions where only one immune cell type and one fungal cell type were present.
By breaking down the phagocytic process into distinct stages (migration, recognition, engulfment, phagosome maturation, and killing), we have been able to determine the effect of specific factors, such as cell morphology and cell wall composition, on individual stages of the phagocytosis process (8, 28). In doing so, novel insights into the mechanisms that govern effective phagocytic clearance have been revealed. For example, dynamic imaging was used to demonstrate nonlytic expulsion of hyphal C. albicans cells from macrophages (29) and C. albicans-mediated inhibition of cell division in macrophages undergoing mitosis (30).
In this study, we assessed stage-specific C. albicans phagocytosis by human monocyte-derived macrophages and human PMNs in isolation with C. albicans and when both phagocyte subsets were present. Through this dual approach, we revealed marked differences in the behaviors of cells and in the outcomes of the interaction between PMNs and macrophages with C. albicans during phagocytosis. We show a lower overall capacity of PMNs to phagocytose fungal cells, an increased susceptibility of PMNs to C. albicans-mediated killing when phagocytes were incubated alone with C. albicans, and a marked increase in C. albicans-mediated macrophage killing when both types of phagocytes were cocultured with C. albicans.
RESULTS
Comparison of macrophage- and PMN-mediated C. albicans phagocytoses.
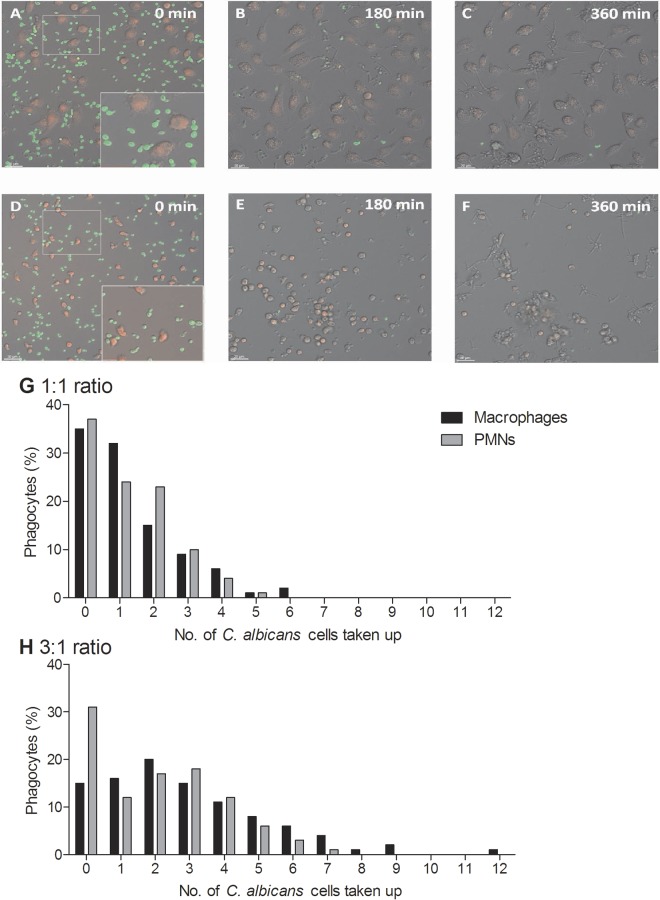
Phagocytosis of C. albicans by macrophages and PMNs is critical for elimination of fungal infections from the body; however, the relative roles that they play in this process and the interactions that occur between these two cell types remain to be elucidated. We first quantified the dynamics of C. albicans phagocytosis by PMNs or macrophages in cell cultures using pure populations of only one phagocyte type. To do this, human monocyte-derived macrophages and human PMNs were challenged with live C. albicans CAI4-CIp10. Live-cell video microscopy using our standard phagocytosis assay (28) recorded the total number of C. albicans cells taken up by individual macrophages and PMNs over a 6-h period. Video S1 in the supplemental material shows the interactions between C. albicans and macrophages, and Videos S2 to S4 show the interactions between C. albicans and PMNs. An uptake event was defined as the complete engulfment of one C. albicans cell by either a macrophage or a PMN following cell-cell contact. Figure 1A to C and D to F show snapshots of the overall clearance of C. albicans cells throughout the 6 h of the assay by macrophages and PMNs, respectively. At a multiplicity of infection (MOI) of 1, there was no significant difference between the macrophages and PMNs in terms of the average numbers of C. albicans cells taken up (Table 1) or the percentages of cells taking up at least one fungal cell (65.0 ± 3.6 and 62.7 ± 10.4, means ± standard deviations [SD] [n = 300], for macrophages and PMNs, respectively) (Fig. 1G). However, when the MOI was increased to 3, the average C. albicans cell uptake by macrophages was significantly greater than for PMNs (Table 1) (P < 0.05). Furthermore, the percentage of macrophages taking up at least one fungal cell was significantly greater than for PMNs (85.0 ± 3.6 and 68.7 ± 4.0, respectively) (Fig. 1H). Thus, at low concentrations of C. albicans cells, both phagocytes were equally as effective in taking up fungal cells; however, when C. albicans yeast cells were in excess, macrophages took up a greater number of C. albicans cells than PMNs did.
FIG 1 .
Uptake of live C. albicans cells by macrophages and PMNs. (A to F) Snapshots from live-cell video microscopy experiments tracking the phagocytosis of live wild-type C. albicans cells by human monocyte-derived macrophages (A to C) and human PMNs (D to F) over 6 h. Phagocytes (red) with live wild-type C. albicans cells (green) at 0 min, 180 min, and the end of the 6 h. Bars, 30 µm. (G and H) Numbers of C. albicans cells ingested by macrophages and PMNs at the 1:1 (G) and 3:1 (H) C. albicans cell/phagocyte ratios. Bars represent percentages of macrophages or PMNs that ingested a defined number of C. albicans cells.
TABLE 1 .
Uptake of live C. albicans cells by macrophages and PMNsa
| Phagocytes | Avg uptake (no. of C. albicans cells taken up ± SD) |
||
|---|---|---|---|
| C. albicans cell/phagocyte MOI of 1 | C. albicans cell/phagocyte MOI of 3 | Coculture assay | |
| Macrophages | 1.3 ± 0.2 | 2.9 ± 0.3 | 2.3 ± 0.6 |
| PMNs | 1.2 ± 0.1 | 2.0 ± 0.3* | 2.5 ± 0.7 |
Average number of C. albicans cells taken up ± SD by human monocyte-derived macrophages and human PMNs. n = 3; *, P < 0.05.
Human PMNs engulf C. albicans more rapidly than macrophages do.
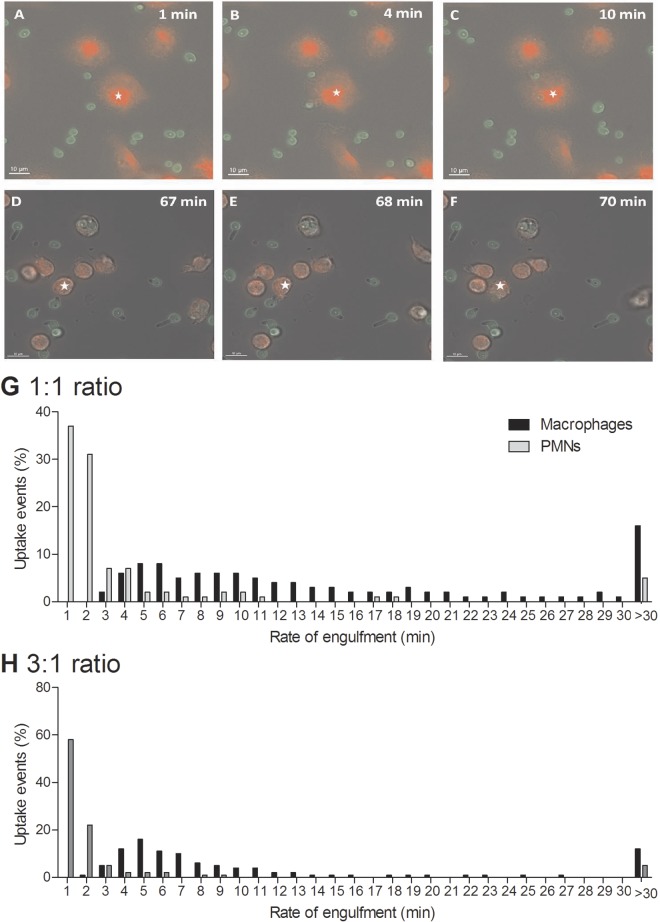
Uptake of target yeast cells is influenced by the migration of phagocytes toward the site at which fungal cells are located and the time taken from the establishment of cell-cell contact to complete uptake of C. albicans. As in our previous studies, we define the latter as the rate of engulfment (28). We examined whether there were differences in the rates of C. albicans engulfment between macrophages and PMNs by live-cell video microscopy and image analysis (Fig. 2A to C and D to F for macrophages and PMNs, respectively). First, at MOIs of both 1 and 3, PMNs engulfed C. albicans cells at a significantly higher rate than macrophages (Table 2; Fig. 2G and H). Videos S1 to S3 in the supplemental material highlight this observed difference in rates of engulfment between macrophages and PMNs. Furthermore, at both MOIs, approximately 12 to 15% of C. albicans cells bound to a macrophage took longer than 30 min to be internalized, compared with 5% of cells bound to PMNs (Fig. 2G and H). Interestingly, when macrophage activities at the different MOIs were compared, it was observed that macrophages engulfed C. albicans cells at a markedly higher rate at a MOI of 3 than at an MOI of 1 (Table 2). This accelerated uptake of yeast cells did not occur for PMNs at higher MOIs (Table 2).
FIG 2 .
Macrophage and PMN engulfment of live C. albicans cells. Shown are snapshots taken from live-cell video microscopy capturing the various stages of C. albicans engulfment by macrophages (A to C) and PMNs (D to F). (A and D) Phagocyte (red, ☆) and C. albicans cell (green) prior to cell-cell contact; (B and E) cells once cell-cell contact had been established; (C and F) C. albicans within the phagocyte following ingestion; (G and H) times taken for macrophages and PMNs to ingest live wild-type C. albicans cells at the 1:1 (G) and 3:1 (H) C. albicans cell/phagocyte ratios. The rate of engulfment was defined as the time taken from initial cell-cell contact to complete ingestion of C. albicans cells by the phagocyte. Bars represent the percentages of uptake events (n = 3).
TABLE 2 .
Rate of engulfment of live C. albicans cells by macrophages and PMNsa
| Phagocytes | Avg rate of engulfment (min ± SD) |
||
|---|---|---|---|
| C. albicans cell/phagocyte MOI of 1 | C. albicans cell/phagocyte MOI of 3 | Coculture assay | |
| Macrophages | 12.0 ± 1.3 | 8.1 ± 0.4b | 14.4 ± 0.6c |
| PMNs | 2.9 ± 0.6d | 2.1 ± 0.4d | 1.7 ± 0.2d |
Average times (min) taken ± SD for human monocyte-derived macrophages and human PMNs to engulf live wild-type C. albicans cells (n = 3).
The value for macrophages was significantly different at an MOI of 3 (P < 0.01) from the value at an MOI of 1 when macrophages were incubated alone with C. albicans.
The value for macrophages in coculture was significantly different (P < 0.0001) from that for macrophages incubated alone at an MOI of 3.
Values for PMNs were significantly different (P < 0.0001) from those for macrophages at the same MOI.
In summary, PMNs engulfed C. albicans at a significantly higher rate than macrophages did at both a low and a high MOI, and the rate of macrophage engulfment was affected by the concentration of fungal cells.
PMNs are more susceptible than macrophages to killing by C. albicans.
It has been shown previously that C. albicans can form hyphae inside phagocytes, often resulting in phagocyte rupture and death (8, 31, 32). We examined this phenomenon in more detail to determine whether there were differences between the different phagocytic cell types in this respect. It was observed that at both low and high MOIs, a significantly greater number of PMNs than macrophages were killed by C. albicans during the course of the phagocytosis assay (Fig. 3A and B). As expected, at the higher MOI, significantly more macrophages were killed than at the lower MOI (Fig. 3C). In contrast, rates of PMN killing were similar at both MOIs (Fig. 3D). The percentages of macrophages and PMNs killed increased with the number of fungal cells being taken up; however, it was observed that at both MOIs, when macrophages and PMNs had taken up the same number of C. albicans cells, a significantly greater proportion of PMNs were killed (Fig. 4A and B). This may reflect the smaller average size of PMNs (8.3 µm ± 1.1 µm) than macrophages (21.2 µm ± 3.5 µm). Furthermore, the data revealed that >90% of PMNs that took up 3 or more fungal cells were killed, whereas a high proportion (~40%) of macrophages survived even after ingesting up to 7 fungal cells. Thus, in these assays, PMNs were more susceptible to C. albicans-mediated killing than macrophages, irrespective of the number of C. albicans cells ingested.
FIG 3 .
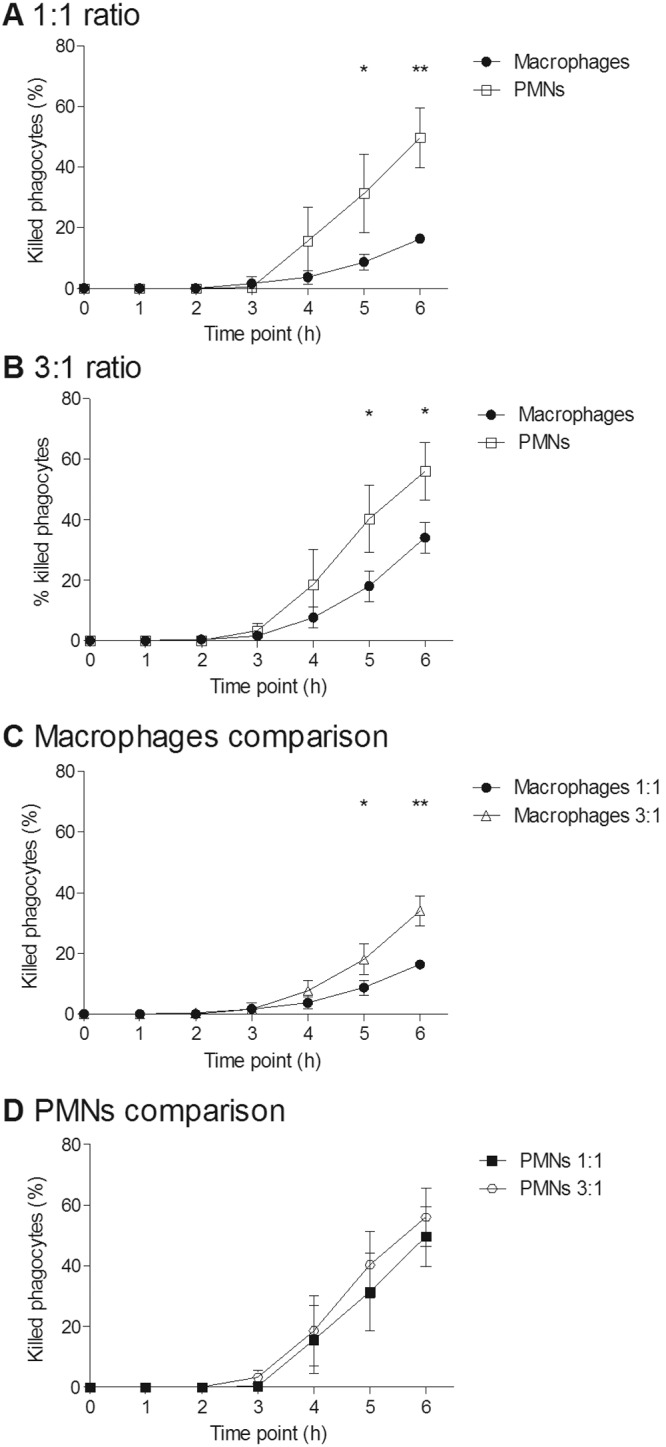
Killing of macrophages and PMNs by live C. albicans cells over a 6-h period. (A and B) Percentages of macrophages and PMNs killed by C. albicans at the 1:1 (A) and 3:1 (B) C. albicans cell/phagocyte ratios; (C and D) comparisons of C. albicans-mediated phagocyte killings at MOIs for macrophages (C) and PMNs (D). Values are means ± SD (n = 3). *, P < 0.05; **, P < 0.01.
FIG 4 .
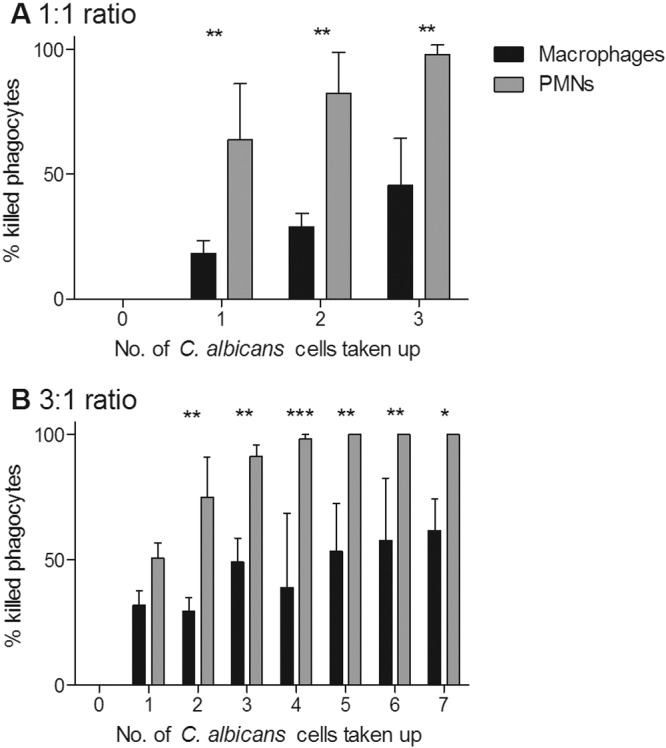
C. albicans-mediated phagocyte killing in relation to the number of C. albicans cells taken up. (A and B) Percentages of phagocytes killed after taking up a defined number of C. albicans cells at the 1:1 (A) and 3:1 (B) C. albicans cell/phagocyte ratios. Bars represent mean percentages of phagocytes killed ± SD (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Phagocytosis and killing dynamics in mixed-phagocyte cocultures.
For coculture assays, an MOI of 3 C. albicans cells for all phagocytes (macrophages plus PMNs, with macrophages and PMNs at a 1:1 ratio) was used. C. albicans uptake and engulfment by macrophages and PMNs was quantified, and C. albicans-mediated phagocyte killing was measured (see Video S5 in the supplemental material).
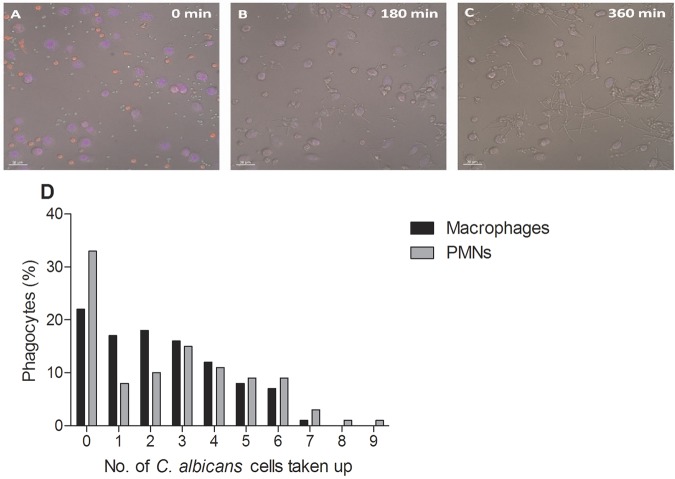
When phagocytes were cultivated with an excess of C. albicans cells, macrophages took up more fungal cells than PMNs did. We examined whether the same was true when macrophages and PMNs were present in coculture. Interestingly, no significant difference between macrophages and PMNs was observed in C. albicans uptake rates (Table 1 and Fig. 5A to C). Additionally, approximately 22% of PMNs did not take up any C. albicans cells, compared with 33% of macrophages (Fig. 5D). Thus, when both cell types were present, macrophages and PMNs contributed approximately equally to clearance of C. albicans.
FIG 5 .
Uptake of live C. albicans cells in combined macrophage-PMN cultures. (A to C) Images taken from live-cell video microscopy experiments tracking the phagocytosis of live C. albicans cells by macrophages and PMNs in the combined assay over 6 h. Shown are macrophages (blue) and PMNs (red) with live C. albicans cells (green) at 0 min, 180 min, and the end of the 6 h. Bar, 30 µm. (D) Numbers of C. albicans cells ingested by macrophages and PMNs in the combined assay. Experiments were carried out an MOI of 3. Bars represent percentages of macrophages or PMNs that ingested a defined number of C. albicans cells (n = 3).
Human PMNs engulf C. albicans more rapidly than macrophages do in mixed-phagocyte cocultures.
When both phagocyte subsets were present, PMNs engulfed C. albicans at a significantly higher rate than macrophages (Table 2 and see Video S5 in the supplemental material). Furthermore, 38% of C. albicans cells took longer than 30 min to be engulfed by macrophages, whereas only 1% of fungal cells took longer than 30 min to be ingested by PMNs (Fig. 6). The average rate of C. albicans engulfment by PMNs was similar to that observed when the PMNs were studied in isolation (Table 2 and Fig. 2H and 6). In contrast, macrophage engulfment of C. albicans was significantly delayed in the coculture assay, with macrophages taking an average of 14.4 ± 0.6 min to engulf fungal cells, compared to 8.1 ± 0.4 min in the absence of PMNs (Table 2 and Fig. 2H and 6). Thus, the rate of C. albicans engulfment by macrophages decreased when PMNs were present, whereas for the PMNs, the rate of engulfment was unaffected by the presence of macrophages and was significantly higher both in the presence and in the absence of macrophages.
FIG 6 .
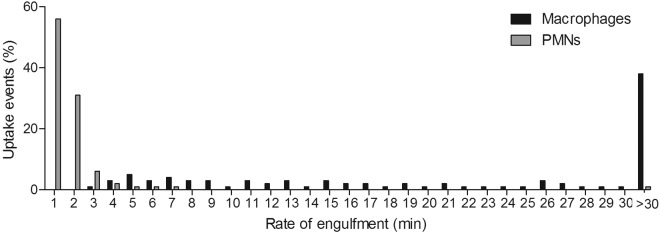
Macrophage and PMN engulfment of live C. albicans cells in combined cultures. Shown are the times taken for macrophages and PMNs to ingest live C. albicans cells in the coculture assay. Rate of engulfment was defined as the time taken from initial cell-cell contact to complete ingestion of C. albicans by the phagocyte. Experiments were carried out at a MOI of 3. Macrophages and PMNS were present at a 1:1 ratio. Bars represent the percentages of uptake events (n = 3).
C. albicans-mediated macrophage killing increases in mixed-phagocyte cocultures.
In the coculture assay, C. albicans-mediated macrophage killing was significantly increased from the level observed when macrophages were cultivated in isolation with C. albicans (Fig. 7A). Indeed, there was no difference in rates of C. albicans-mediated killing of macrophages and PMNs when both cell types were present (Fig. 7B). This contrasts with the observation that PMNs were significantly more susceptible to killing by fungal cells when PMNs and macrophages were cultured in isolation (Fig. 3B). Additionally, the percentage of macrophages killed after taking up 2 or more fungal cells in the coculture assay was >80%, compared to approximately 25% when macrophages were incubated alone with C. albicans (Fig. 4B and 7C).
FIG 7 .
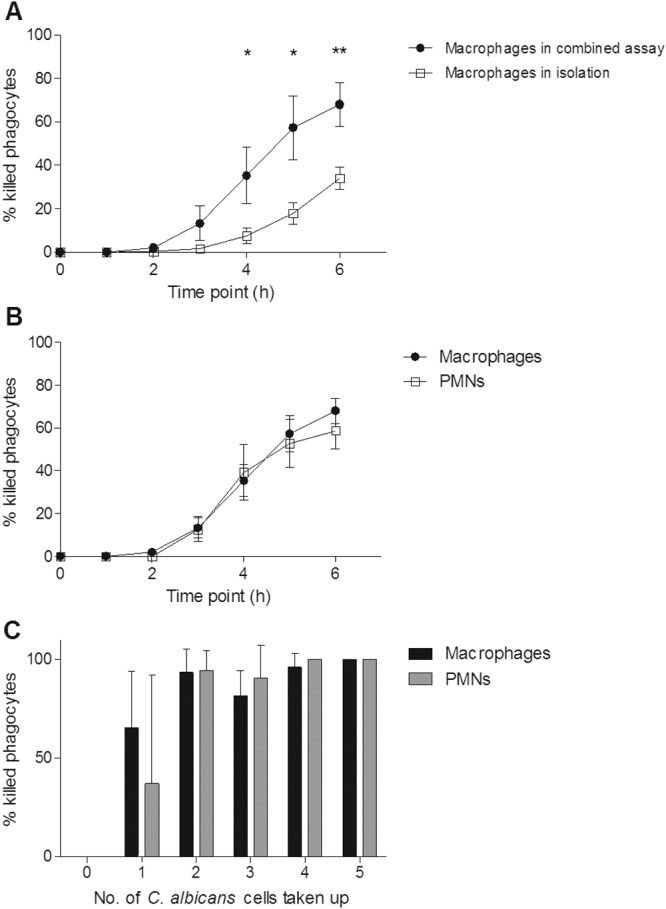
Killing of macrophages and PMNs by live C. albicans cells in the combined assay over 6 h. (A) Comparison of C. albicans-mediated killing of macrophages in the combined assay and that in isolation; (B) percentages of macrophages and PMNs killed by C. albicans in the combined culture assay; (C) percentages of phagocytes killed after taking up a defined number of C. albicans cells in the combined assay. Experiments were carried out at a 3:1 ratio of C. albicans cell to total phagocytes. Values/bars represent means ± SD (n = 3). *, P < 0.05; **, P < 0.01.
PMNs migrate more rapidly toward C. albicans cells.
To investigate this increase in C. albicans-mediated macrophage killing, we analyzed the migration of macrophages and PMNs toward fungal cells. C. albicans cells formed germ tubes rapidly under the conditions of the experiment; therefore, the time taken for a phagocyte to locate and migrate toward a fungal cell influenced whether it was taken up early in the yeast form or later on as a germ tube. It is reasonable to speculate that this may ultimately affect the susceptibility of the phagocyte to C. albicans-mediated killing. Tracking diagrams (Fig. 8A and B) illustrate the distances traveled and the directionality and velocity of PMNs and macrophages in the coculture assay in the presence of wild-type C. albicans. In these diagrams, symbols indicate the locations of macrophages or PMNs at 1-min intervals, and arrows represent directionality. Quantitative analysis of the average track revealed a 20-fold-higher average track velocity of PMNs than of macrophages toward C. albicans cells (0.007 ± 0.004 µm/s [n = 3] for macrophages versus 0.13 ± 0.07 µm/s [n = 3] for PMNs) (see Video S5 in the supplemental material). Thus, in the combined phagocyte assays, PMNs migrated at a greater velocity than macrophages toward C. albicans.
FIG 8 .
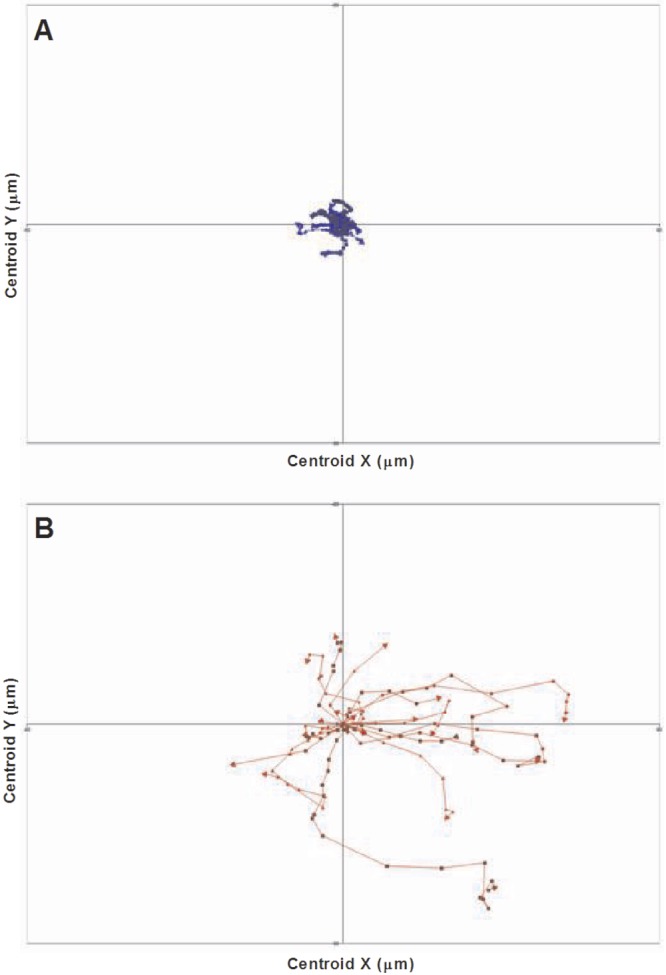
Phagocyte migration toward C. albicans. Shown are tracking diagrams illustrating the distances traveled, directionality, and velocity of macrophages (A, blue) and PMNs (B, red) in pursuit of live C. albicans cells in the combined assay. Tracks represent the movement of individual phagocytes relative to their starting position, symbols indicate the locations of phagocytes at 1-min intervals, and arrows represent directionality.
This faster migration of PMNs toward fungal cells resulted in earlier clearance of yeast cells by PMNs, leaving the remaining Candida cells time to germinate before being engulfed by macrophages as hyphae. Indeed, in the coculture assays, macrophages took up a significantly greater proportion of hyphal cells (relative to yeast cells) in the coculture assay than they did when in isolation (72.4% ± 9.6% and 59.3% ± 2.7% [means ± SD, n = 300] of hyphal cells were taken up by macrophages in coculture and isolation assays, respectively [data not shown]), and this coincided with increased C. albicans-mediated macrophage killing. These data are consistent with previous reports which indicate that uptake of hyphal cells enhances phagocyte killing (8). In addition, this increase in hyphal uptake was accompanied by a lower rate of engulfment than when macrophages were incubated alone with C. albicans (Table 2). Furthermore, in some uptake events, we observed continued extension of C. albicans hyphae during the engulfment process (see Video S6 in the supplemental material). These are all factors which are likely to have contributed to enhanced macrophage killing. Thus, in the presence of PMNs, macrophages were more susceptible to killing by the fungus.
DISCUSSION
C. albicans is a major life-threatening human fungal pathogen that causes infections associated with high morbidity and high mortality (2). Professional phagocytes, such as macrophages and PMNs, provide an important line of defense against invasive candidiasis by directly engulfing and destroying fungal cells. We conducted a detailed comparison of stage-specific phagocytosis of C. albicans by PMNs and that by macrophages in isolation and when both phagocyte subsets were present and competing for fungal cells. We observed profound differences in (i) migration, (ii) rate of engulfment, (iii) overall uptake, and (iv) phagocyte survival when C. albicans yeast cells interacted with mixtures of macrophages and PMNs rather than individual phagocyte subsets. Nonprofessional phagocytes, such as dendritic and endothelial cells, also contributed to the overall clearance of C. albicans in vivo and will be the subject of future studies.
PMNs are considered the primary mediators of phagocytic clearance (9–11, 31, 33). Indeed, neutropenia is a major predisposing factor in clinical studies for developing invasive candidiasis (21, 22). Furthermore, recent in vitro studies suggest that the onset of hyphal formation and subsequent escape of fungal cells is inhibited inside human neutrophils (33). This may suggest that PMNs not only mediate the majority of C. albicans uptake but also are less susceptible than macrophages to killing by C. albicans following phagocytosis. Surprisingly, we observed that when only one immune cell type was present, macrophages, not PMNs, had the greater capacity for C. albicans uptake despite engulfing fungal cells at a lower rate after establishing cell-cell contact. However, when both cell types were present, macrophages and PMNs contributed approximately equally to clearance of C. albicans. Our data illustrate that uptake rates are affected not only by the fungal/phagocyte ratio but also by the proportion of the phagocyte subsets encountered. This suggests that the overall phagocytic capacity in single phagocyte assays is not determined simply by the number PMNs and macrophages but is rather a consequence of the percentage of phagocytes contributing to uptake. In vivo, the contribution of phagocyte subsets to overall uptake would also be influenced further by factors such as chemokine and cytokine concentrations and spatial considerations such as location of the fungal infiltrate, the numbers of phagocytes, and accessibility to the infection sites. Interestingly, observations from a larval zebrafish model of disseminated candidiasis where phagocytosis of Candida cells was imaged in vivo showed that macrophage-like cells took up a significantly greater number of C. albicans yeast cells than neutrophils did (17).
A number of studies have previously established that C. albicans can form filaments inside macrophages and that hyphal formation is essential for C. albicans escape and phagocyte killing (8, 30–32). In this study, we examined this phenomenon further and observed that PMNs were significantly more susceptible than macrophages to C. albicans-mediated killing when incubated in isolation with C. albicans. Indeed, in vitro, nearly 50% of macrophages that took up 5 or more fungal cells survived, whereas 100% of PMNs that took up 3 or more fungal cells were killed following continued growth of hyphae inside the host cell. However, when both macrophages and PMNs were studied in coculture, macrophage survival following C. albicans phagocytosis was significantly impaired, and the rate of C. albicans-mediated killing of macrophages increased to the same level as that of PMNs incubated alone with C. albicans.
We showed here that PMNs not only migrated more rapidly than macrophages toward C. albicans cells but also engulfed C. albicans cells at a significantly higher rate once cell-cell contact was established. The differential activation and expression of PRRs displayed on macrophages and PMNs may contribute to the observed differences in rates of engulfment seen in these myeloid cells in the present study. This is in keeping with the results of a number of studies that have established that immune cell migration and fungal cell recognition is influenced by the relative levels of expression and specificities of PRRs displayed on the immune cell surface (34,35). The observations also mirror findings from our previous work showing that the glycosylation status of the C. albicans cell wall and therefore repertoire of PAMPs affects the migration, engulfment, and killing of neutrophils (5) and macrophages (28).
Taken together, the data from the current study suggest that when both macrophages and PMNs are presented to C. albicans in a coculture, PMNs mediate the majority of early C. albicans yeast cell uptake events due to their ability to migrate more rapidly toward fungal cells and then engulf bound fungal cells at a higher rate than macrophages do. As a consequence, macrophages may encounter a higher proportion of hyphal cells, which they engulf at a lower rate. Both uptake of a higher proportion of hyphal C. albicans cells and increased hyphal length at the time of uptake is reportedly associated with host cell death (8, 28, 31, 32), which may explain the increase in macrophage death seen here in the coculture assay compared to that when macrophages were incubated alone with C. albicans.
This study highlights the critical role that macrophages and PMNs play in C. albicans phagocytosis and demonstrates that major kinetic parameters related to the rate and efficiency of phagocytosis and killing are multifactorial, depending on the presence of multiple cell types and a temporal program of events that ultimately defines the outcome of the predation of fungal cells by different phagocyte cell types. The implications of this study are that extrapolations of complex interactions between pathogen cells and the myeloid cells involved in innate immunity have to be made in the knowledge that cellular interactions are significantly affected by the proximity of other cell types and dynamic changes in the predator/prey cell type ratio.
MATERIALS AND METHODS
C. albicans strains and growth conditions.
C. albicans serotype A strain CAI4 + CIp10 was used for all experiments and was the progenitor of the mutant strains used in our previous studies (28). This strain was obtained from glycerol stocks stored at −80°C and plated on synthetic complete medium lacking uracil (SC−Ura). SC−Ura plates consisted of 6.9 g yeast nitrogen base without amino acids (Formedium, Norfolk, United Kingdom), 1 ml 1 M NaOH (BDH Chemicals, VWR International, Leicestershire, United Kingdom), 10 ml 1% (wt/vol) adenine hemisulfate salt (Sigma, Dorset, United Kingdom), 50 ml 40% d-glucose (Fisher Scientific, Leicestershire, United Kingdom), 50 ml 4% SC−Ura dropout (Formedium, Norfolk, United Kingdom), and 2% (wt/vol) technical agar (Oxoid, Cambridge, United Kingdom) made up to 1,000 ml in distilled H2O.
Preparation of human monocyte-derived macrophages.
Human monocyte-derived macrophages were isolated from the blood of healthy volunteers. Peripheral venous blood was collected in EDTA-coated tubes, pooled, and diluted 1:3 in Hanks’ balanced salt solution (HBSS). Lymphoprep (Axis-Shield, Norway) kits were used to separate peripheral blood mononuclear cells (PBMCs) from whole blood. The PBMC layer was washed and resuspended in DMEM (Lonza, Slough, United Kingdom) supplemented with 200 U/ml penicillin-streptomycin antibiotics (Invitrogen, Paisley, United Kingdom) and 2 mM l-glutamine (Invitrogen, Paisley, United Kingdom). Serum was separated from blood using standard methods and heat inactivated at 56°C for 20 min before use. PBMCs were seeded at 10 × 106 in 2 ml supplemented DMEM containing 10% autologous human serum onto glass-based imaging dishes (PAA Imaging gas chromatography [GC] dishes; The Cell Culture Company, GE Healthcare, France) and incubated at 37°C with 5% CO2 for 2 h to allow for monocyte adherence to the dish. After 2 h, the supernatant containing the floating lymphocytes was removed and replaced with 2 ml warm supplemented DMEM containing 10% autologous human serum. Dishes were incubated at 37°C with 5% CO2, and medium was changed on days 3 and 6. Cells were used in imaging experiments on day 7. Immediately prior to phagocytosis experiments, supplemented DMEM was replaced with prewarmed supplemented serum-free CO2-independent medium (Gibco, Invitrogen, Paisley, United Kingdom) containing 1 µM LysoTracker red DND-99 (Invitrogen, Paisley, United Kingdom) when macrophages were the only immune cell type present or 1 µM LysoTracker blue DND-22 (Invitrogen, Paisley, United Kingdom) when both macrophages and PMNs were present to distinguish between the two immune cell types. LysoTracker red and blue DND are fluorescent dyes that stain acidic compartments in live cells, enabling tracking of these cells during phagocytosis and phagolysosome maturation.
Preparation of human PMNs.
Human PMNs were isolated from the blood of healthy volunteers and used in phagocytosis assays within 3 h of isolation. Peripheral venous blood was collected in EDTA-coated tubes, pooled, and diluted 1:2 in sterile PBS. Histopaque-1119 (Sigma, Dorset, United Kingdom) was added to a conical tube and carefully overlaid with the same volume of Histopaque-1077 (Sigma, Dorset, United Kingdom) to create a double-density gradient. An equal volume of diluted blood was carefully layered on top of the upper Histopaque gradient, and tubes were centrifuged at 700 × g for 40 min at room temperature to allow for separation of PBMCs and PMNs from erythrocytes. Serum and PBMC layers were aspirated and discarded, and the PMN layer was washed once in PBS. For hypotonic lysis of erythrocytes carried over into the PMN fraction, 3 ml of 0.2% NaCl solution was added to cells and mixed gently. After 30 to 45 s, 3 ml of 1.6% NaCl solution was added, mixed gently, and then diluted to 50 ml in PBS before centrifugation at 350 × g for 10 min at room temperature. Cells were then washed in PBS and resuspended in prewarmed supplemented serum-free CO2-independent medium containing 1 µM LysoTracker red DND‑99 (Invitrogen, Paisley, United Kingdom) and used immediately in phagocytosis assays.
Preparation of FITC-stained C. albicans.
C. albicans colonies were grown in SC−Ura medium and incubated overnight at 30°C with 200-rpm shaking. Live C. albicans cells were stained for 10 min at room temperature in the dark with 1 mg/ml fluorescein isothiocyanate (FITC) (Sigma, Dorset, United Kingdom) in 0.05 M carbonate-bicarbonate buffer (pH 9.6) (BDH Chemicals, VWR International, Leicestershire, United Kingdom). Following incubation, cells were washed three times in 1× PBS to remove any residual FITC and finally resuspended in 1× PBS.
Live-cell video microscopy phagocytosis assays.
Phagocytosis assays were performed using our standard protocol, with modifications (8, 28). Samples of live FITC-stained wild-type C. albicans (CA14-CIp10) cells were added to 1 × 106 LysoTracker red DND-99-stained human monocyte-derived macrophages or 1 × 106 LysoTracker red DND-99-stained human PMNs in glass-based imaging dishes at a multiplicity of infection (MOI) of either 1 or 3. For phagocytosis assays where both macrophages and PMNs were present at a 1:1 ratio, live, FITC-stained wild-type C. albicans (CA14-CIp10) cells were added to glass-based imaging dishes containing 5 × 105 macrophages stained with 1 µM LysoTracker blue DND-22, and 5 × 105 PMNs stained with 1 µM LysoTracker red DND-99 at an MOI of 3 (ratio of C. albicans cells to total phagocytes, 3:1). In the coculture experiments, macrophages and PMNs were not always isolated from the same donors. Video microscopy was performed using a DeltaVision Core microscope (Applied Precision, WA, USA) in a 37°C chamber, and images were captured at 1-min intervals over a 6-h period using an EMCCD camera. At least three independent experiments were performed for both macrophages and PMNs incubated alone with C. albicans, and at least 3 videos were analyzed from each experiment using softWoRx Explorer image analysis software (Applied Precision). The same setup was used for the coculture assays where both macrophages and PMNs were present. One hundred phagocytes were selected at random from each experiment and analyzed individually at 1-min intervals over a 6-h period. Measurements taken included C. albicans uptake, defined as the number of C. albicans cells taken up by an individual phagocyte over the 6-h period, the time point at which cell-cell contact was established, and the time point at which a C. albicans cell was fully engulfed; these measurements were used to determine the rate of C. albicans engulfment, defined as the time taken from establishment of cell-cell contact to complete ingestion of a C. albicans cell (a fungal cell was considered to have been fully ingested when its FITC fluorescent signal was lost, indicating that the fungal cell was now inside the phagocyte and not merely bound to the phagocyte cell surface). The viability of the phagocytes over the 6-h period was defined as the percentage of macrophages or PMNs from the total macrophage or PMN population, respectively, that had been killed by specific time points. In the coculture assays, macrophages and PMNs were also analyzed separately in this manner, so the percentage of macrophages killed by certain time points in the coculture assays represents the percentage of macrophages out of the total macrophage population sampled that had been killed. Velocity 6.2 imaging analysis software was used to track phagocyte migration at 1-min intervals throughout the 6-h phagocytosis assay. The software enabled high-throughput analysis of phagocyte migration, providing detailed information on the distances traveled, directionality, and velocity of hundreds of individual phagocytes. Data were subsequently displayed in tracking diagrams and used to calculate the mean track velocity and track lengths of phagocytes cultured with C. albicans (28).
Means values and standard deviations were calculated. Unpaired, two-tailed t tests and two-way analysis of variance (ANOVA) followed by Bonferroni multiple-comparison tests were used to determine statistical significance.
SUPPLEMENTAL MATERIAL
Macrophages incubated alone with C. albicans for 6 h (macrophages are stained red, and C. albicans cells are stained green). Download
Neutrophils incubated alone with C. albicans for 6 h (neutrophils are stained red, and C. albicans cells are stained green). Download
Neutrophils incubated alone with C. albicans over 3 h taken with a spinning-disk microscope (neutrophils are stained red, and C. albicans cells are stained green). Download
Three-dimensional video demonstrating C. albicans engulfment by neutrophils using a spinning-disk microscope (neutrophils are stained red, and C. albicans cells are stained green). Download
Macrophages and neutrophils incubated together with C. albicans in the coculture assay for 6 h (macrophages are stained blue, neutrophils are stained red, and C. albicans cells are stained green). Download
C. albicans hyphal extension during uptake by a macrophage (the macrophage is stained blue, and the C. albicans cell is stained green). Download
ACKNOWLEDGMENTS
F.M.R. was supported by a SULSA BioSkape studentship. L.P.E. and N.A.R.G. acknowledge funding by the Wellcome Trust (grants 080088, 086827, 075470, and 099215) and by an FP7-2007-2013 grant agreement (HEALTH-F2-2010-260338-ALLFUN).
Footnotes
Citation Rudkin FM, Bain JM, Walls C, Lewis LE, Gow NAR, Erwig LP. 2013. Altered dynamics of Candida albicans phagocytosis by macrophages and PMNs when both phagocyte subsets are present. mBio 4(6):e00810-13. doi:10.1128/mBio.00810-13.
REFERENCES
- 1. Spellberg B. 2011. Vaccines for invasive fungal infections. F1000 Med. Rep. 3:13. 10.3410/M3-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zaoutis TE, Argon J, Chu J, Berlin JA, Walsh TJ, Feudtner C. 2005. The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin. Infect. Dis. 41:1232–1239 [DOI] [PubMed] [Google Scholar]
- 3. Liu Y, Filler SG. 2011. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot. Cell 10:168–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fu Y, Ibrahim AS, Sheppard DC, Chen YC, French SW, Cutler JE, Filler SG, Edwards JE., Jr 2002. Candida albicans Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol. Microbiol. 44:61–72 [DOI] [PubMed] [Google Scholar]
- 5. Sheth CC, Hall R, Lewis L, Brown AJP, Odds FC, Erwig LP, Gow NAR. 2011. Glycosylation status of the C. albicans cell wall affects the efficiency of neutrophil phagocytosis and killing but not cytokine signaling. Med. Mycol. 49:513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Netea MG, Brown GD, Kullberg BJ, Gow NAR. 2008. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 6:67–78 [DOI] [PubMed] [Google Scholar]
- 7. Miramón P, Dunker C, Windecker H, Bohovych IM, Brown AJP, Kurzai O, Hube B. 2012. Cellular responses of Candida albicans to phagocytosis and the extracellular activities of neutrophils are critical to counteract carbohydrate starvation, oxidative and nitrosative stress. PLoS One 7:e52850. 10.1371/journal.pone.0052850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McKenzie CGJ, Koser U, Lewis LE, Bain JM, Mora-Montes HM, Barker RN, Gow NAR, Erwig LP. 2010. Contribution of Candida albicans cell wall components to recognition by and escape from murine macrophages. Infect. Immun. 78:1650–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng SC, Joosten LAB, Kullberg BJ, Netea MG. 2012. Interplay between Candida albicans and the mammalian innate host defense. Infect. Immun. 80:1304–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Traynor TR, Huffnagle GB. 2001. Role of chemokines in fungal infections. Med. Mycol. 39:41–50 [DOI] [PubMed] [Google Scholar]
- 11. Diamond RD. 1993. Interactions of phagocytic cells with Candida and other opportunistic fungi. Arch. Med. Res. 24:361–369 [PubMed] [Google Scholar]
- 12. Maertens J, Vrebos M, Boogaerts M. 2001. Assessing risk factors for systemic fungal infections. Eur. J. Cancer Care (Engl.) 10:56–62 [DOI] [PubMed] [Google Scholar]
- 13. Martino P, Girmenia C, Venditti M, Micozzi A, Santilli S, Burgio VL, Mandelli F. 1989. Candida colonization and systemic infection in neutropenic patients. A retrospective study. Cancer 64:2030–2034 [DOI] [PubMed] [Google Scholar]
- 14. Lehrer RI, Cline MJ. 1969. Leukocyte myeloperoxidase deficiency and disseminated candidiasis: the role of myeloperoxidase in resistance to Candida infection. J. Clin. Invest. 48:1478–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. El-Maallem H, Fletcher J. 1977. Impaired neutrophil function and myeloperoxidase deficiency in myeloid metaplasia. Br. J. Haematol. 37:323–329 [DOI] [PubMed] [Google Scholar]
- 16. Qian Q, Jutila MA, Van Rooijen N, Cutler JE. 1994. Elimination of mouse splenic macrophages correlates with increased susceptibility to experimental disseminated candidiasis. J. Immunol. 152:5000–5008 [PubMed] [Google Scholar]
- 17. Romani L. 2011. Immunity to fungal infections. Nat. Rev. Immunol. 11:275–288 [DOI] [PubMed] [Google Scholar]
- 18. Medzhitov R. 2007. Recognition of microorganisms and activation of the immune response. Nature 449:819–826 [DOI] [PubMed] [Google Scholar]
- 19. Mora-Montes HM, Netea MG, Ferwerda G, Lenardon MD, Brown GD, Mistry AR, Kullberg BJ, O’Callaghan CA, Sheth CC, Odds FC, Brown AJP, Munro CA, Gow NAR. 2011. Recognition and blocking of innate immunity cells by Candida albicans chitin. Infect. Immun. 79:1961–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seider K, Heyken A, Lüttich A, Miramón P, Hube B. 2010. Interaction of pathogenic yeasts with phagocytes: survival, persistence and escape. Curr. Opin. Microbiol. 13:392–400 [DOI] [PubMed] [Google Scholar]
- 21. Amulic B, Cazalet C, Hayes GL, Metzler KD, Zychlinsky A. 2012. Neutrophil function: from mechanisms to disease. Annu. Rev. Immunol. 30:459–489 [DOI] [PubMed] [Google Scholar]
- 22. Brothers KM, Newman ZR, Wheeler RT. 2011. Live imaging of disseminated candidiasis in zebrafish reveals role of phagocyte oxidase in limiting filamentous growth. Eukaryot. Cell 10:932–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brown GD. 2011. Innate antifungal immunity: the key role of phagocytes. Annu. Rev. Immunol. 29:1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Urban CF, Reichard U, Brinkmann V, Zychlinsky A. 2006. Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell. Microbiol. 8:668–676 [DOI] [PubMed] [Google Scholar]
- 25. Vijayan D, Radford KJ, Beckhouse AG, Ashman RB, Wells CA. 2012. Mincle polarizes human monocyte and neutrophil responses to Candida albicans. Immunol. Cell Biol. 90:889–895 [DOI] [PubMed] [Google Scholar]
- 26. Keppler-Ross S, Douglas L, Konopka JB, Dean N. 2010. Recognition of yeast by murine macrophages requires mannan but not glucan. Eukaryot. Cell 9:1776–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gow NAR, Van De Veerdonk FL, Brown AJP, Netea MG. 2012. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat. Rev. Microbiol. 10:112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lewis LE, Bain JM, Lowes C, Gillespie C, Rudkin FM, Gow NAR, Erwig LP. 2012. Stage specific assessment of Candida albicans phagocytosis by macrophages identifies cell wall composition and morphogenesis as key determinants. PLoS Pathog. 8:e1002578. 10.1371/journal.ppat.1002578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bain JM, Lewis LE, Okai B, Quinn J, Gow NAR, Erwig LP. 2012. Non-lytic expulsion/exocytosis of Candida albicans from macrophages. Fungal Genet. Biol. 49:677–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lewis LE, Bain JM, Lowes C, Gow NAR, Erwig LP. 2012. Candida albicans infection inhibits macrophage cell division and proliferation. Fungal Genet. Biol. 49:679–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ghosh S, Navarathna DHMLP, Roberts DD, Cooper JT, Atkin AL, Petro TM, Nickerson KW. 2009. Arginine-induced germ tube formation in Candida albicans is essential for escape from murine macrophage line RAW 264.7. Infect. Immun. 77:1596–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lorenz MC, Bender JA, Fink GR. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3:1076–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ermert D, Niemiec MJ, Röhm M, Glenthøj A, Borregaard N, Urban CF. 2013. Candida albicans escapes from mouse neutrophils. J. Leukoc. Biol. 94:223-236 [DOI] [PubMed] [Google Scholar]
- 34. Bain JM, Lewis LE, Erwig LP. 2012. Differential expression of macrophage and neutrophil phagocytic receptors recognising fungal pathogens in mouse and human. Immunol. Cell Biol. 90:837-838 [DOI] [PubMed] [Google Scholar]
- 35. Klaas M, Oetke C, Lewis LE, Erwig LP, Heikema AP, Easton A, Willison HJ, Crocker PR. 2012. Sialoadhesin promotes rapid proinflammatory and type I IFN responses to a sialylated pathogen, Campylobacter jejuni. J. Immunol. 189:2414-2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Macrophages incubated alone with C. albicans for 6 h (macrophages are stained red, and C. albicans cells are stained green). Download
Neutrophils incubated alone with C. albicans for 6 h (neutrophils are stained red, and C. albicans cells are stained green). Download
Neutrophils incubated alone with C. albicans over 3 h taken with a spinning-disk microscope (neutrophils are stained red, and C. albicans cells are stained green). Download
Three-dimensional video demonstrating C. albicans engulfment by neutrophils using a spinning-disk microscope (neutrophils are stained red, and C. albicans cells are stained green). Download
Macrophages and neutrophils incubated together with C. albicans in the coculture assay for 6 h (macrophages are stained blue, neutrophils are stained red, and C. albicans cells are stained green). Download
C. albicans hyphal extension during uptake by a macrophage (the macrophage is stained blue, and the C. albicans cell is stained green). Download