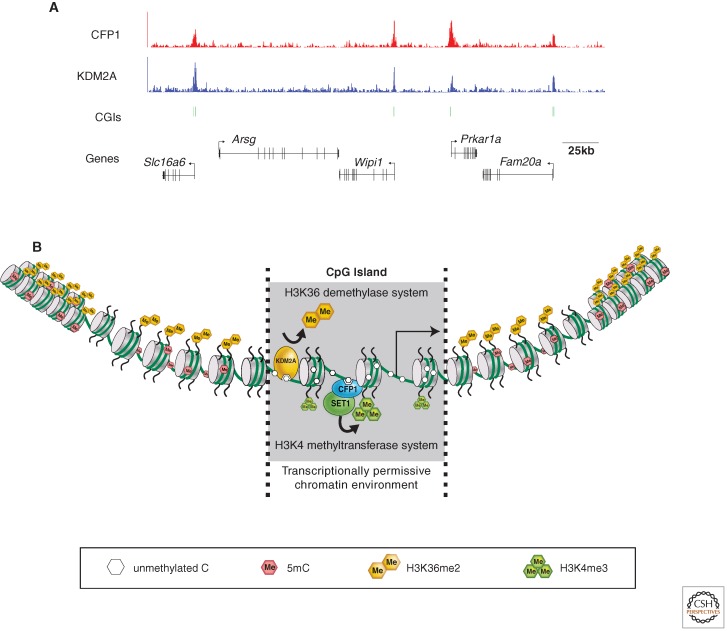
Zinc finger CxxC domain–containing proteins (e.g., CFP1 and KDM2A) bind specifically to unmethylated cytosines in CpG islands within the mammalian genome and impose a defined chromatin environment.
Abstract
Most mammalian gene promoters are embedded within genomic regions called CpG islands, characterized by elevated levels of nonmethylated CpG dinucleotides. Here, we describe recent work demonstrating that CpG islands act as specific nucleation sites for the zinc finger CxxC domain–containing proteins CFP1 and KDM2A. Importantly, both CFP1 and KDM2A are associated with enzymatic activities that modulate specific histone lysine methylation marks. The action of these zinc finger CxxC domain proteins therefore imposes a defined chromatin architecture on CpG islands that distinguishes these important regulatory elements from the surrounding genome. The functional consequence of this CpG island–directed chromatin environment is discussed.
Approximately two-thirds of mammalian gene promoters are found within genomic regions known as CpG islands (CGIs). In contrast to bulk genomic DNA in which CpG dinucleotides are underrepresented and pervasively methylated, CGIs show a high density of CpGs and are refractory to DNA methylation. Despite more than 25 years of work aimed at understanding CGI function, it remains unclear how they contribute to the activity of gene promoters. However, of notable progress in the past few years is our demonstration that CGIs, through specific recruitment of proteins that bind nonmethylated DNA, can specifically alter the local chromatin environment of gene regulatory elements (Blackledge et al. 2010; Thomson et al. 2010).
We know that methylated CpGs act as nucleation sites for methyl-CpG binding domain (MBD) proteins that are generally associated with transcriptional repression. Based on the existence of MBD proteins, Skalnik and colleagues hypothesized that proteins may exist that specifically recognize nonmethylated CpGs. Through subsequent work, they identified the CpG binding protein (CGBP), later named CxxC finger protein 1 (CFP1), as such a factor (Voo et al. 2000). Importantly, this protein was found to bind to DNA through a ZF (zinc finger)-CxxC domain that specifically recognizes CpG dinucleotides in vitro.
The question remained: Does CFP1 recognize nonmethylated CGIs in vivo and thus impact on CGI function? This possibility was particularly intriguing, as CFP1 forms part of an extended family of ZF-CxxC domain–containing proteins that include chromatin-binding factors such as DNMT1, MLL1, and MBD1. With the emergence of massively parallel sequencing technologies coupled to chromatin immunoprecipitation (ChIP-seq), an exciting opportunity was presented to formally test the relationship between ZF-CxxC proteins and CGIs. To address this possibility, we performed independent studies, focusing on the ZF-CxxC proteins CFP1 and histone lysine demethylase 2A (KDM2A, also referred to as JHDM1a, FBXL11, or CXXC8). This unbiased approach showed a remarkable genome-wide association between sites of enrichment for both of these proteins and nonmethylated CGIs (Fig. 1A) (Blackledge et al. 2010; Thomson et al. 2010). Therefore, at least in the case of CFP1 and KDM2A, the ZF-CxxC domain appears to act as a CGI-targeting module. Significantly, these studies showed for the first time that CGIs are directly interpreted through recognition of nonmethylated DNA, and that most, if not all, nonmethylated CGIs share common protein factors.
Figure 1.
ZF-CxxC proteins at CpG islands. (A) CFP1 (top panel, red) and KDM2A (bottom panel, blue) ChIP-seq analysis for an ∼300 kb region of mouse chromosome 11 (data from Blackledge et al. 2010 and Thomson et al. 2010). Annotated genes and CGIs (green bars) are illustrated below the sequence traces. (B) CFP1 and KDM2A impose a specific chromatin environment at CGIs that is distinct from the rest of the genome.
Interestingly, and of potential relevance to CGI function, most ZF-CxxC proteins are associated with chromatin-modifying activities. For example, CFP1 exists in a SETD1-containing methyltransferase complex that acts on the histone H3 lysine 4 (H3K4) residue, whereas KDM2A is a JmjC domain–containing demethylase enzyme that targets the histone H3 lysine 36 (H3K36) residue. In our studies, we observed that the histone-modifying activities associated with these ZF-CxxC proteins impose a defined chromatin environment at CGIs. Specifically, the action of CFP1 facilitates nucleation of histone H3K4 trimethylation (H3K4me3), a punctate mark generally associated with 5′ ends of genes, whereas KDM2A depletes H3K36 dimethylation (H3K36me2), a very abundant and broadly distributed modification adorning 30%–50% of total histone H3 (Blackledge et al. 2010; Thomson et al. 2010).
A surprising finding from these studies was that CFP1 and KDM2A bind at CGIs and modify CGI chromatin independently of transcriptional activity. A striking illustration of this was the demonstration that an exogenous CpG-rich sequence is sufficient to recruit CFP1 and nucleate H3K4me3 without concomitant recruitment of RNA polymerase II (RNA Pol II; Thomson et al. 2010). These bodies of work have therefore established a new paradigm, whereby the underlying DNA signal at CGIs (i.e., a high density of nonmethylated CpGs) is translated into a defined histone modification status (i.e., H3K4me3 enriched and H3K36me2 depleted). Therefore, via the ZF-CxxC system, nonmethylated CGIs have a “hard-wired” chromatin environment distinct from the rest of the genome (Fig. 1B).
The above studies prompted the question, within the context of CGI elements, what is the functional significance of ZF-CxxC protein binding and the chromatin modification states that they impose? Histone lysine methylation marks are thought to influence transcription by recruiting specific effector proteins via plant homeodomain (PHD) fingers or chromatin-modifier (chromo-) domains. In the case of H3K4me3, a number of studies suggest that this mark has the potential to recruit PHD finger proteins that support transcription, such as the core transcription factor TFIID, the NuRF chromatin remodeling complex, and ING4-containing histone acetyltransferase complexes. In contrast, studies in yeast suggest that H3K36me2 may repress transcription initiation by recruiting the chromodomain protein EAF3, a component of the RPD3S histone deacetylase complex. It is therefore possible that the combined effect of H3K4me3 enrichment and H3K36me2 depletion at CGIs creates a permissive chromatin environment that favors transcriptional initiation.
CGI promoters show a number of unique characteristics that may be attributable, at least in part, to the permissive chromatin environment created by ZF-CxxC proteins. For example, unlike classical TATA-box promoters, which use a defined transcriptional start point, CGI promoters tend to initiate transcription over a broad region of 100 bp or more. Furthermore, even in the absence of productive transcription, CGI promoters are enriched for RNA Pol II and show short, nonproductive, bidirectional transcripts (Core et al. 2008). Finally, inducible “primary response genes” that have CGI promoters can be rapidly activated by lipopolysaccharide stimulation without a requirement for chromatin-remodeling events (Ramirez-Carrozzi et al. 2009; illustrated in Fig. 9 of Busslinger and Tarakhovsky 2014). This is in contrast to primary response genes with non-CGI promoters for which productive transcriptional output requires SWI/SNF-mediated chromatin remodeling. Further studies are required to determine whether ZF-CxxC proteins contribute to some or all of these CGI characteristics and ultimately to define the precise role that ZF-CxxC proteins play in CGI function.
As a final thought, it should be emphasized that protection from DNA methylation is at the crux of CGI existence, and is essential for ZF-CxxC protein nucleation at these regions. Although the mechanisms responsible for establishing and maintaining this DNA methylation-free state are poorly defined, one alluring possibility is that ZF-CxxC proteins themselves may contribute. For example, in vitro studies suggest that H3K4me3, the mark placed by the CFP1 complex, may block de novo DNA methylation, exemplified by the inhibitory effect this mark has on DNMT3L binding, which is part of the DNMT3A/3L de novo methylating complex (described in Cheng 2014). Also, the ZF-CxxC protein TET1 is a hydroxylase enzyme able to convert methylcytosine into hydroxymethylcytosine, a reaction that has been implicated in DNA demethylation pathways (described in Li and Zhang 2014). It is therefore tempting to speculate that at CGIs, ZF-CxxC proteins provide a self-reinforcing loop of nonmethylated CpG recognition and subsequent protection from DNA methylation.
REFERENCES
*Reference is also in this collection.
- Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ 2010. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell 38: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Busslinger M, Tarakhovsky A 2014. Epigenetic control of immunity. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a19307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Cheng X 2014. Structural and functional coordination of DNA and histone methylation chromatin. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a18747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Li E, Zhang Y 2014. DNA methylation in mammals. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a19133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST 2009. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138: 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, et al. 2010. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464: 1082–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voo KS, Carlone DL, Jacobsen BM, Flodin A, Skalnik DG 2000. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol Cell Biol 20: 2108–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]