Abstract
An 11.7-Å-resolution cryo-EM map of the yeast 80S·eEF2 complex in the presence of the antibiotic sordarin was interpreted in molecular terms, revealing large conformational changes within eEF2 and the 80S ribosome, including a rearrangement of the functionally important ribosomal intersubunit bridges. Sordarin positions domain III of eEF2 so that it can interact with the sarcin–ricin loop of 25S rRNA and protein rpS23 (S12p). This particular conformation explains the inhibitory action of sordarin and suggests that eEF2 is stalled on the 80S ribosome in a conformation that has similarities with the GTPase activation state. A ratchet-like subunit rearrangement (RSR) occurs in the 80S·eEF2·sordarin complex that, in contrast to Escherichia coli 70S ribosomes, is also present in vacant 80S ribosomes. A model is suggested, according to which the RSR is part of a mechanism for moving the tRNAs during the translocation reaction.
Keywords: cryo-EM, eEF2, 80S ribosome, sordarin, tRNA translocation
Introduction
Proteins in all living cells are synthesized by ribosomes, which are large, RNA-based macromolecular machines (Ban et al, 2000; Wimberly et al, 2000). During protein synthesis, the macromolecular ligands of the ribosome, that is, mRNA, tRNAs, as well as the nascent peptide chain, move through the ribosome in a precise and controlled manner. These movements of the ligands are accompanied and facilitated by corresponding movements in the ribosome itself (Spahn and Nierhaus, 1998; Frank and Agrawal, 2000; Noller et al, 2002). In line with the paradigm of the ribosome as a molecular machine, structural investigations of ribosomes in defined functional states by cryo-EM and X-ray crystallography are providing increasing evidence for movements of ribosomal parts and are thereby providing a first direct look into the dynamic behavior of the ribosome (for reviews, see Ramakrishnan and Moore, 2001; Ramakrishnan, 2002; Yonath, 2002).
The translocation reaction is accompanied by large-scale movements. During this step, the ribosome changes from the pre-translocational (PRE) to the post-translocational (POST) state as the A- and P-site bound tRNAs move to the P and E sites, respectively. The translocation step is catalyzed by elongation factor EF-G in prokaryotes and eEF2 in eukaryotes. In the classical view of translocation, EF-G/eEF2 acts by a GTPase switch mechanism similar to a regulatory G protein (Kaziro, 1978). However, this view was challenged by recent time-resolved pre-steady-state experiments (Rodnina et al, 1997). These experiments indicated that GTP hydrolysis by EF-G precedes tRNA translocation and that the release of inorganic phosphate is coupled with the tRNA translocation step, suggesting that the chemical energy of GTP hydrolysis is used to perform mechanical work on the ribosome. However, it should be noted that mere binding of EF-G without GTP hydrolysis plays a major role in the catalysis of tRNA translocation and contributes more to the acceleration of translocation than GTP hydrolysis (1000-fold versus 50-fold acceleration, respectively; Rodnina et al, 2001).
The molecular mechanism of translocation and its catalysis by EF-G/eEF2 is largely unknown. However, binding of prokaryotic EF-G induces large-scale conformational changes in the ribosome that might be related to tRNA movement (Frank and Agrawal, 2000). Much less information is available about eukaryotic ribosomes, but an earlier cryo-EM structure of eEF2 bound to the yeast 80S ribosome at 17-Å resolution showed overall similarity to the prokaryotic system as well as some significant differences (Gomez-Lorenzo et al, 2000). We present here a highly improved cryo-EM map of the yeast eEF2·80S complex in the presence of the antibiotic sordarin at 11.7-Å resolution. Docking of the recent eEF2·sordarin X-ray structure (Jørgensen et al, 2003) and of a molecular model for the yeast 80S ribosome (Beckmann et al, 2001; Spahn et al, 2001a) reveals large conformational changes within eEF2 and the 80S ribosome. Interpreted in molecular terms, these indicate how the ribosome might actively facilitate tRNA translocation. In addition, the mechanism for the inhibitory action of the antifungal drug sordarin is suggested.
Results and discussion
Cryo-EM reconstruction of the ribosomal eEF2·80S complex and docking of atomic models
The reconstruction of the 80S·eEF2·sordarin complex (Figure 1) at 11.7-Å resolution (0.5 threshold; Supplementary Figure S1) was interpreted in molecular terms by docking the recently determined eEF2·sordarin (eEF2-sor) X-ray structure (Jørgensen et al, 2003) and atomic models of ribosomal components into the density map. The docking of ribosomal component models was facilitated by a previous atomic model of the POST 80S ribosome based on a 15.4-Å cryo-EM map (Spahn et al, 2001a). For this purpose, the cryo-EM map of the 80S·eEF2·sordarin complex was brought into the same orientation as the map of the POST 80S ribosome (Spahn et al, 2001a) by first aligning the 60S subunit parts of both maps. After this alignment, it became obvious that the 40S subunits of the two maps differ in position and conformation (see below). Therefore, a crosscorrelation-based 3D orientation search was performed (Spahn et al, 2001b) to find the alignment parameters for the head and body/platform domains of the 40S part of the 80S·eEF2·sordarin map relative to the map of the POST 80S ribosome (Table I). Different alignment parameters for body/platform domains and the head domain were obtained and subsequently used to transform the corresponding parts of our previous atomic model of the translating 80S ribosome (Spahn et al, 2001a). The fitting was further improved by moving nonfitting parts (e.g., RNA helices) as rigid bodies relative to the rest of the model in order to account for local conformational changes in the 40S and 60S subunits. It is estimated that atomic models of known substructures can be positioned in a cryo-EM map with an accuracy exceeding the resolution of the map several-fold (Rossmann, 2000). Indeed, estimation of the accuracy of docking for eEF2 by correlation traces (Valle et al, 2003b) indicated a positional accuracy of better than ±2 Å (Supplementary Figure S2). The resulting atomic model allows an interpretation of the interaction between eEF2 and the 80S ribosome and an analysis of conformational changes within the yeast 80S ribosome at the molecular level.
Figure 1.
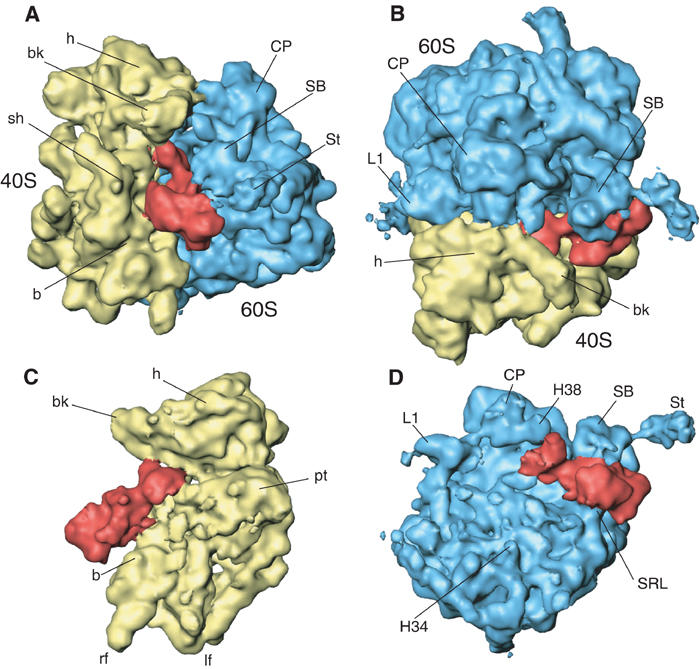
A 11.7-Å-resolution cryo-EM map of the yeast 80S·eEF2·sordarin complex. The cryo-EM map is shown (A) from the side; (B) from the top ; (C) from the 60S side, with 60S removed; and (D) from the 40S side, with 40S removed. The ribosomal 40S subunit is painted yellow, the 60S subunit blue and eEF2 red. Landmarks for the 40S subunit: b, body; bk, beak; h, head; lf, left foot; rf, right foot; pt, platform; sh, shoulder; sp, spur. Landmarks for the 60S subunit: CP, central protuberance; L1, L1 protuberance; SB, stalk base; St, stalk; H34, helix 34; H38, helix 38; SRL, sarcin–ricin loop.
Table 1.
Orientation search and alignment between different parts of the 80S ribosome
| Initial alignment at 60S subunits | Alignment at complete 40S subunit | Alignment at 40S body/platform | Alignment at 40S head | |
| Angles: | Angles: | Angles: | Angles: | |
| α=0° | α=5° | α=5° | α=4° | |
| β=0° | β=−4° | β=−3° | β=−14° | |
| γ=0° | γ=1° | γ=1° | γ=7° | |
| Crosscorrelation coefficient | ||||
| Angles α/β/γ (in case of optimal alignment) | ||||
| 60S subunit | 0.89 | 0.76 | n.d. | n.d. |
| 0/0/0 | ||||
| 40S subunit body/platform | 0.83 | 0.92 | 0.93 | 0.59 |
| 5/−3/1 | ||||
| 40S subunit head | 0.74 | 0.82 | 0.80 | 0.92 |
| |
|
|
|
4/−14/7 |
| The corresponding ribosomal parts of the POST 80S ribosome (Spahn et al, 2001a) and of the 80S·eEF2·sordarin complex are compared in different orientations with the crosscorrelation coefficient. The relative rotational orientation is given in an orthogonal coordinate system, where the x-axis is parallel to the anticodon stem helix of the P-site bound tRNA, the y-axis is parallel to the acceptor stem helix, and the z-axis points from the A-site region toward the E-site region (see Spahn et al, 2001b). The cartoon shows the outline of the 40S subunit from the 60S site and the outline of the P-site bound tRNA, together with the axes representing the coordinate system. The angles α, β and γ describe rotations around the x-, y- and z-axis, respectively. n.d., not determined. Optimal alignment for a certain ribosomal region is indicated by bold numbers and by the angles in the table. The angles describe the rotational rearrangement from the conformation of the POST 80S ribosome to the conformation of the 80S·eEF2·sordarin complex. | ||||
Large-scale conformational change in eEF2 upon binding to the 80S ribosome
The quality of the cryo-EM map allowed eEF2-sor to be docked unambiguously (Figure 2A and B). However, it was necessary to separate the eEF2 structure into two blocks corresponding to domains I/G′/II and III–V and to fit these separately as rigid bodies. The resulting model for ribosome-bound eEF2 (eEF2-cryo) is distinct from both eEF2-sor and apo-eEF2, the nucleotide-free form of eEF2 (Jørgensen et al, 2003). The rotation of domains III–V relative to domains I/G′/II in eEF2-cryo is somewhere intermediate between the two X-ray structures, and closer to that for apo-eEF2 (Figure 2A).
Figure 2.
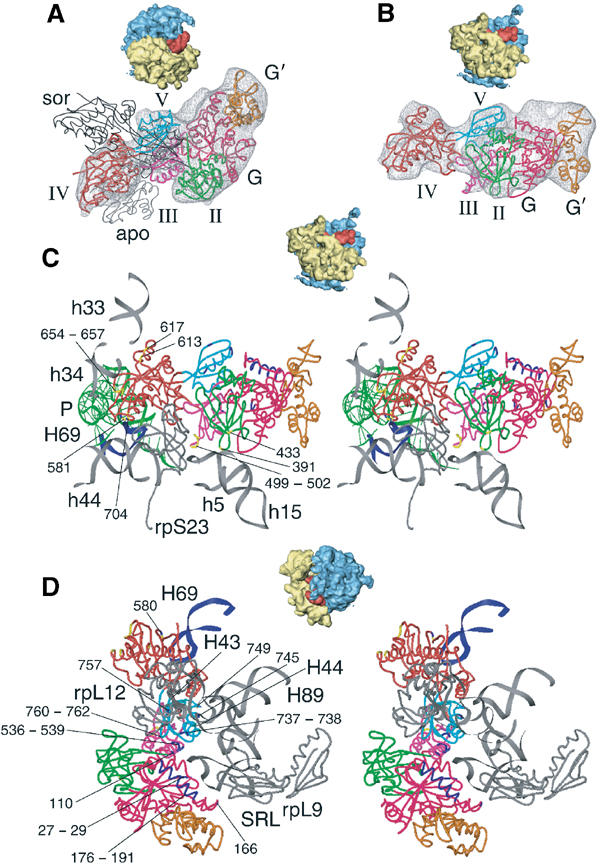
Docking of the X-ray model for eEF2 into the cryo-EM density and interactions between eEF2 and the 80S ribosome. Fitting of eEF2 (colored ribbon representation) into the corresponding cryo-EM density (A, B) and ribosomal environment of eEF2 (C, D). Thumbnails are included as an orientation aid. In (A), the X-ray structures of eEF2·sordarin (black, thin ribbon, designated sor) and the apo form of eEF2 (gray, thin ribbon, designated apo) are shown superposed onto domains I and II of the 80S bound eEF2, in order to show the conformational changes. The domains of ribosome-bound eEF2 are color-coded: G (pink), G′ (orange), II (green), III (purple), IV (red), V (cyan). Fitted atomic models of eEF2 and ribosomal components (gray ribbons) in the neighborhood of the factor in ribbon representation are displayed in stereo (C, D). The upper panel (C) focuses on the interactions between the 40S subunit and eEF2, and the lower panel (D) shows a close-up on the stalk base region of the 60S subunit. Residues of eEF2 that possibly interact with the 40S subunit are highlighted in yellow, residues that might be in contact with the 60S subunit are in blue and their numbers are indicated. (C) includes the position of the P-site bound tRNA (in green) that was derived from the POST 80S complex (Spahn et al, 2001a), in order to show the neighborhood of the tip of domain IV of eEF2 to the tRNA anticodon:codon complex.
The conformation of apo-eEF2 is thought to be closely related to both the eEF2·GDP and eEF2·GTP forms and should, therefore, represent the conformation of eEF2 in solution (Jørgensen et al, 2003). This leads us to conclude that the conformational change from apo-eEF2 to eEF2-cryo (Figure 2A) occurs upon ribosome binding and that it represents a structural transition that occurs during the translocation reaction. This conformational change is overall similar to the one that has been observed in the Escherichia coli system (Agrawal et al, 1999). However, the conformational change in the yeast system is more pronounced, and the tip of domain IV moves by some 25–30 Å between apo-eEF2 and eEF2-cryo. Additionally, a movement of domain III of eEF2 relative to domains IV and V that is present in eEF2-sor but not in apo-eEF2 (Jørgensen et al, 2003) persists in the structure of eEF2-cryo. The conformational change from apo-eEF2 to eEF2-cryo involves a rotation of domains III, IV and V relative to domains I and II and an additional rearrangement of domain III.
Molecular interactions between eEF2 and the 80S ribosome
The interaction between eEF2·sordarin and the yeast 80S ribosome involves both ribosomal subunits and all five domains of the factor (Figures 1, 2C and D). The α-sarcin-ricin loop (SRL, H95 of 25S rRNA; we will designate rRNA helices of the large subunit (LSU) by Hmn, where mn is the helix number, and helices of the small subunit by hmn), the so-called GTPase-associated center (GAC; rpL12, H43, H44), and the P proteins (P0, P1α, P1β, P2α, P2β; Ballesta and Remacha, 1996) are contact elements of the large ribosomal subunit (Table II). The latter two elements form the stalk base and stalk, respectively, of the 60S ribosomal subunit. An additional contact element on the 60S ribosomal subunit is the intersubunit bridge B2a. With respect to the 40S subunit, eEF2 interacts with the rRNA (h5, 15, 33, 34, 44) in the head and body domains, as well as the ribosomal protein rpS23 (S12p).
Table 2.
Contacts between eEF2 and the 80S ribosome from yeast
| Domain of eEF2 | eEF2 positiona | Ribosomal subunit | rRNA helixb or ribosomal protein | rRNA or protein positiona,c |
|---|---|---|---|---|
| I | H27–D29 (G1 motif, P-loop) | 60S | H95 (SRL) | 2656/2663 min |
| I | D110 | 60S | H95 (SRL) | 2660–2662 |
| I | E166 | 60S | rpL9 (L6p) | H96, G119 |
| I | Q176–T191 | 60S | P proteins | |
| II | K391 | 40S | h5 | 359 |
| II | R433 | 40S | h15 | 368 |
| III | N499–P502 | 40S | rpS23 (S12p) | N99 |
| III | L536, E538, E539 | 60S | H95 (SRL) | 2660–2662 |
| IV | P580–K582 | 60S | H69 | 1911–1913 lo |
| IV | N581, Q704 | 40S | H44 | 1492, 1493 |
| IV (inset) | K613, R617 | 40S | H33 | 1044 |
| IV | Q654–H657 | 40S | H34 | 1208 |
| V | E737, Q738 | 60S | H44 | 1095 |
| V | E757 | 60S | H43 | 1067 |
| V | S745, K749 | 60S | H89 | 2473 |
| V |
R760–G762 |
60S |
rpL12 (L11p) |
K24–K29 |
| a Amino acids and nucleotides are given that are closest to the observed contact. | ||||
| b 18S rRNA helices are designated with hmn, where mn is the helix number and 25S rRNA helices with Hmn. | ||||
| c rRNA nucleotide numbers are according to E. coli numbering. lo: loop; min: minor groove. | ||||
The highly conserved SRL of the LSU rRNA is an essential element for the binding and function of several translation factors, forming a strong interaction with the GTP-binding face of domain I (Figure 2D, Table II). The minor-groove side of the SRL stem is adjacent to His27-Asp29 of eEF2. This eEF2 sequence is part of the highly conserved G1 motif, which comprises the phosphate-binding loop (P-loop) (Jørgensen et al, 2003). A second tentative contact involves the tetraloop of the SRL and the G3 motif of eEF2 around Asp110. In addition, the interaction of domain I of eEF2 with the ribosome appears to involve rpL9 (L6p). Interestingly, the position of domain III of eEF2·sordarin appears to be such that the conserved sequence around Leu536 interacts with the tetraloop of the SRL (Figure 2D, Table II). An equivalent interaction between the SRL and domain III of EF-G has not yet been described within the bacterial system.
A second ribosomal determinant of translation factor interaction is the GAC in combination with the stalk proteins. These elements undergo a very complex interaction with eEF2 that is to some extent different from the corresponding interaction with EF-G in bacteria (Gomez-Lorenzo et al, 2000). In a separate study, we have identified an interaction between the P proteins and the α-helix D of domain I of eEF2 (Gomez et al, 2004 in preparation). The apical loops of H43 and H44 as well as rpL12 interact with domain V of eEF2 (Figure 2D, Table II). Domain V of eEF2 is further contacted by the apical loop of H89, which from its location could transmit conformational signals into the peptidyltransferase center.
The tip of domain IV of eEF2 interacts with both the 40S and 60S subunit, in the region of the decoding center. It contacts the top part of h44 and the apical loop of H69, which form the intersubunit bridge B2a (Figure 2C, Table II). rpS23 (S12p), located adjacent to the decoding center, interacts with domain III of eEF2. As domain III also interacts with the SRL (see above), this contact could be part of an information relay system between the decoding center and the SRL. A prominent difference from EF-G is the prong-like appearance of domain IV of eEF2 (Gomez-Lorenzo et al, 2000), attributed to an insertion in domain IV and a C-terminal addition (Jørgensen et al, 2003). These changes lead to a yeast-specific interaction between domain IV of eEF2 and h33 in the head of the 40S subunit. The 40S subunit also interacts with domain II of eEF2. This interaction with h5 and h15 at the shoulder of the 40S subunit (Figure 2C, Table II) appears to be similar to the corresponding interaction in the E. coli system (Frank and Agrawal, 2001).
In line with our previous suggestion (Gomez-Lorenzo et al, 2000), an interaction between the tip of domain IV of eEF2 and the P-site bound tRNA is possible according to the more recent maps, with an experimentally determined model of the P-site bound peptidyl-tRNA (Spahn et al, 2001a) and the model of ribosome-bound eEF2·sordarin. In a superposition of both models, His694-Ile698 of eEF2 proves to be close enough to interact with the codon–anticodon duplex between P-site bound tRNA and mRNA (Figure 2C). Moreover, the adjacent residue His699 is post-translationally modified to diphthamide and ADP ribosylation of diphthamide by bacterial toxins that inactivate the factor. Mutations in eEF2 that prevent diphthamide formation impair factor function (Foley et al, 1995). Taken together, this suggests an additional function for the tip of domain IV of eEF2. It is known that the accuracy of codon–anticodon interaction in the A site is enhanced by A1492 and A1493 of SSU rRNA (E. coli numbering), which interact with the minor groove of the codon–anticodon base pairs (Ogle et al, 2002). These A-minor interactions have to be disrupted for the tRNA to move from the A to the P site. The tip of domain IV of eEF2 could take over the stabilization of codon–anticodon pairing during the transition phase. In this way, eEF2 would ensure that the mRNA follows the movement of the tRNAs, thereby reducing the possibility of frameshifts.
A ratchet-like subunit rearrangement takes place in the yeast 80S ribosome
The binding of EF-G to the E. coli 70S ribosomes induces a ratchet-like subunit rearrangement (RSR) within the 70S ribosome (Frank and Agrawal, 2000). The RSR presumably corresponds to a transition state of translocation and is accompanied by a movement of a deacylated tRNA in the P site to a P/E hybrid site (Valle et al, 2003b; Zavialov and Ehrenberg, 2003). Previously, we have not detected an RSR-type conformational change upon eEF2 binding, when the 80S·eEF2·sordarin map at 17.5-Å resolution was compared to the map of the vacant 80S ribosome from yeast (Gomez-Lorenzo et al, 2000). However, when we tried to dock the atomic model of the translating yeast 80S ribosome (Spahn et al, 2001a) into the new 80S·eEF2·sordarin map at 11.7-Å resolution, it became obvious that a pronounced RSR is present in the 80S·eEF2·sordarin complex compared to the translating yeast 80S ribosome (Figure 3; Supplementary data: animation, Supplementary Figure S3).
Figure 3.
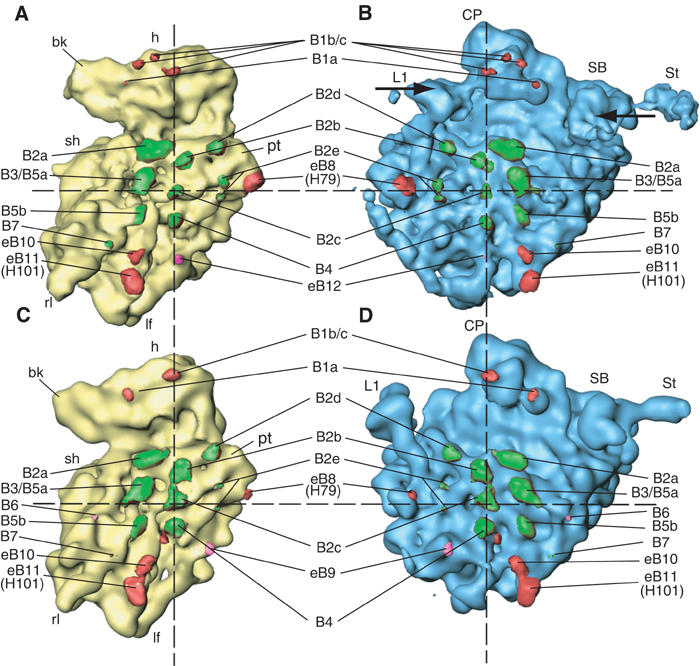
Comparison of the yeast 80S ribosome in two different conformations, and positions of the intersubunit bridges. The 80S·eEF2·sordarin complex (A, B) is compared to the POST 80S ribosome (Spahn et al, 2001a) (C, D). The two maps were computationally aligned at their respective 60S subunits (see text). The 40S subunits in yellow (A, C) and the 60S subunits in blue (B, D) are shown from their intersubunit sides. Intersubunit bridges are color-coded. Bridges that are preserved in both structures are painted green, those that are formed by different components are painted red, and those that are specific for one of the conformations are painted pink. The dashed registration lines intersect at bridge b2c, the center of rotation for the RSR. Arrows (B) indicate the inward movements of the L1 protuberance and the stalk region.
A comparison with the older maps of the vacant 80S ribosome and the 80S·eEF2·sordarin map at 17.5-Å resolution (Gomez-Lorenzo et al, 2000) shows that the reason why the RSR has not been observed previously is because the RSR state is already present in the vacant yeast 80S ribosome. The translating yeast 80S ribosome contains a peptidyl-tRNA in the P site (Beckmann et al, 2001; Spahn et al, 2001a) and is therefore in the POST state. The subunit organization of the yeast POST 80S ribosome is similar to the translating or vacant bacterial 70S ribosome, consistent with the hypothesis that the peptidyl moiety in the P site locks the prokaryotic ribosome (Valle et al, 2003b; Zavialov and Ehrenberg, 2003). The unexpected presence of the RSR in the vacant yeast ribosome shows that the conformational properties of ribosomes of yeast are different from those of E. coli. The yeast ribosome can stably adopt the RSR conformation without being stabilized by eEF2.
Normal-mode analysis is a computational approach to predict and explore global conformational changes of macromolecular assemblies. One of the motions derived from a normal-mode analysis of the X-ray structure of the Thermus thermophilus 70S ribosome (Yusupov et al, 2001) shows striking similarities to the RSR (Tama et al, 2003). The center of the rotational movement predicted by the normal-mode analysis of the bacterial ribosome was found to be h27, the same center as obtained in our experimental analysis of the RSR in the yeast 80S ribosome (see below). Therefore, the mechanical properties of the ribosome that gives rise to the RSR appear to be evolutionary conserved. This, however, leads to the question of why the conformations of the vacant 70S ribosome and the vacant 80S ribosome are different, that is, the latter shows the RSR and the former does not.
One possibility is that the two ribosomal conformations exist in an equilibrium, which is shifted toward one state in prokaryotes and toward the other in eukaryotes. In eukaryotic ribosomes, however, there might be a second factor that influences the dynamic behavior. Most of the evolutionary conserved intersubunit bridges, especially the centrally located RNA–RNA bridges, do not have to be broken for the conformation to switch (Figure 3, Table III). However, molecular interactions do have to be temporarily broken and re-formed between some components for the eukaryotic-specific outer bridges (see below), which requires a larger amount of activation energy for the transition to occur. Therefore, the vacant 80S ribosome might be kinetically trapped in the RSR conformation, and the transition between the two conformations might be impeded without proper catalysis.
Table 3.
Intersubunit bridges
| Bridge | 60S componenta | 40S componenta | Difference between eEF2·80S and POST 80Sb |
|---|---|---|---|
| B1a | H38 (886–888) | rpS15/S19p (12) | Different 40S component |
| H33 (1033) | |||
| B1b/c | rpL11/L5p (91, 92, 170) | rpS15/S19p (70) | Different 40S component |
| rpL11/L5p (84) | rpSx or | ||
| rpL11/L5p (34,35) | rpS15/S19p (N-term) | ||
| rpS18/S13p 111–118 | |||
| B2a | H69 (1910–1920) | H44 (1408/1493) | 3.5–4 Å |
| B2b | H68 (1847) | H24 (784) | 2.5 Å |
| 25S (1939–1941) | H45 (1515) | ||
| B2c | H67 (1832) | H27 (899) | 0.5–1 Å |
| H24 (771) | |||
| B2d | H68 (1848/1895) | H23 (698–703) | 5–5.5 Å |
| B2e | rpL2 (136) | H23 (712/713) | 4 Å |
| rpL43/L37ae (C-term) | H22 (671) | 3 Å | |
| B3 | H71 (1948/1960) | H44 (1418/1482) | 2–3 Å |
| B4 | H34 (716) | H20 (580/761) | 1.5 Å; no interaction between minor groove of 25S H34 and 18S H11 |
| rpS13/S15p | |||
| B5a | rpL23/L14p | H44 (1422) | 3.5 Å |
| B5b | H62 (1689/1704) | H44 (1428/1472) | 3–3.5 Å |
| B6 | rpL23/L14p (132) | H14 | No bridge |
| B7 | rpL24/L24e (47) | H44 (1446) | 6 Å |
| eB8 | H79 (exp) | rpSx | Different part of rpSx; stronger |
| eB9 | rpLx | rpSx/h21 (exp) | No bridge |
| eB10 | H63 (1713/1747) | H11 (272) | Different ribosome position |
| H101 | rpSx | ||
| eB11 | H101 (exp) | H9 (187) | Local change |
| rpSx | |||
| eB12 |
rpL19/L19e (142) |
n.d. |
New bridge |
| a Amino acids and nucleotides are given that are closest to the observed contact; rRNA nucleotide numbers are according to E. coli numbering. | |||
| b If a distance is given, the corresponding bridge appears to be formed by the same components in both states. The distance was inferred geometrically from the rigid body movement of the 40S subunit associated with the RSR. The actual relative movement might be counteracted by local changes. | |||
Local conformational changes in the 40S subunit
Part of the RSR of the 80S·eEF2·sordarin complex is a rotational movement of the head domain of the 40S subunit relative to the body/platform domains. This rotation has to occur around the neck region, which covalently connects the two parts of the 40S subunit (Supplementary Figure S3) and affects the noncovalent interactions between the head and the body/platform, which must be disrupted or accommodated by more local changes upon 40S subunit rearrangement.
Such a noncovalent head–body interaction (‘latch') that is part of the mRNA entry channel is formed between h18 in the body with h34 and rpS3 in the head (Schlünzen et al, 2000; Wimberly et al, 2000; Spahn et al, 2001a). In the 80S·eEF2·sordarin structure, the rotation of the head moves h34 inward by about 15 Å (Figure 4). This movement breaks the contact between the minor groove of h34 with h18. Instead, the interaction of h18 is predominantly with rpS3. This change of interactions seems to be facilitated by a movement of protein rpS3 itself. Interestingly, the region of h34 that forms the latch in the POST 80S ribosome is engaged in an interaction with domain IV of eEF2 (Table II). The insert in domain IV interacts with the nearby h33 and might have evolved to control and stabilize the opening of the latch during translocation.
Figure 4.
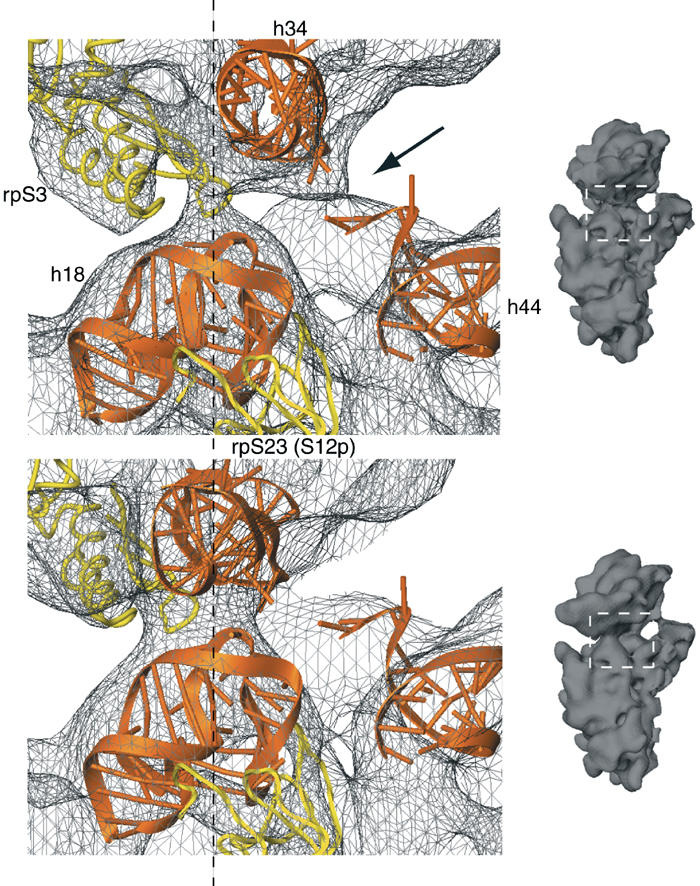
Rearrangement of the latch region of the 40S subunit. Comparison of the latch region in the 80S·eEF2·sordarin complex (upper panel) with the POST 80S ribosome (lower panel) (Spahn et al, 2001a). The corresponding cryo-EM densities are shown as a gray wire-mesh. Docked models for h18, h34 and the top of h44 are shown as orange ribbons, and models for rpS3 and rpS23 (S12p) as yellow ribbons. The dashed line goes through the center of the latch. Small inset on the right: the 40S subunits as an orientation aid. The white, dashed box indicates the latch region. The arrow in the upper panel highlights an additional connection between the head and the body of the 40S subunit that is present in the 80S·eEF2·sordarin complex.
An independent movement of the head domain of the 30S subunit relative to the body/platform domains is also part of the RSR in E. coli (Frank and Agrawal, 2000). Some movements of proteins S3, S4 and S5, involved in forming the mRNA entry channel, as well as a movement of h34 in the range of 5 Å have been reported (Gao et al, 2003). However, the magnitude of the head movement we observe in yeast (∼15 Å) and of the concurring change in the latch interaction is unprecedented as yet in prokaryotes.
Another strong network of interactions between the head and body of the 40S subunit is present at the solvent side of the 40S subunit. It involves protein rpS0A (S2p) and h36. In general, the relative movement of the head in the 80S·eEF2·sordarin complex appears to be accommodated by local conformational changes, which include a movement of the upper subdomain of the 3′ major domain of 18S rRNA (Supplementary Figure S3). This subdomain makes only a few packing contacts with the other parts of the 16S RNA in T. thermophilus (Wimberly et al, 2000). The absence of rigid contacts might be important to allow flexibility of this subdomain, which appears necessary to facilitate the head movement.
Local conformational changes in the 60S subunit
Localized changes in the 60S subunit are present mainly in its three protuberances. A conformational change takes place in the central protuberance, producing a more broadened appearance (see Figure 3). It probably facilitates the relative movement of the two ribosomal subunits, since this ribosomal region is involved in the formation of intersubunit bridges with the head of the 40S subunit (see below). Large movements can be observed at the L1 protuberance and the stalk base, which comprises the GAC.
As observed previously (Gomez-Lorenzo et al, 2000), the binding of eEF2·sordarin induces a movement of the GAC toward the central protuberance (Figure 3, Supplementary Figure S4). The inward rotation around a hinge region within H42 results in an ∼15 Å movement at the apical loops of H43 and H44, where the interaction with eEF2 takes place. A similar but smaller movement of the GAC was also observed in the E. coli 70S ribosome upon binding of EF-G (Frank and Agrawal, 2001), aa-tRNA·EF-Tu·GTP ternary complex in the presence of kirromycin (Valle et al, 2003a) and RF2 (Rawat et al, 2003). The movement of the GAC might be therefore part of a general mechanism of loading translation factors into the ribosome's factor binding site.
Another dynamic region is the L1 protuberance, the top of which is formed by rpL1, H77 and H78, while the connection to the body of the 60S subunit is formed by H76. The L1 protuberance has been observed in different positions (Harms et al, 2001; Valle et al, 2003b; Yusupov et al, 2001), which in the yeast ribosome span about 50 Å (Gomez-Lorenzo et al, 2000). Furthermore, its flexibility can be inferred from various degrees of disorder in the X-ray structures of the 50S subunit and 70S ribosomes, making an atomic interpretation of this region difficult (Ban et al, 2000; Harms et al, 2001; Yusupov et al, 2001). However, the X-ray structure of the isolated L1 protuberance at atomic resolution has been presented recently (Nikulin et al, 2003).
In the present map of the 80S·eEF2·sordarin complex, the L1 protuberance is essentially in an ‘in-position', close to the central protuberance (Figure 3), whereas in the cryo-EM map of the POST 80S ribosome (Spahn et al, 2001a) the L1 protuberance is mostly in an ‘out-position'. Even though the cause for the change in L1 position is as yet unknown, it is not related to eEF2 binding, because in vacant 80S ribosomes (Gomez-Lorenzo et al, 2000), the L1 protuberance is in the ‘in-position' as well. The L1 movement is likely to participate in the active release of the E-site bound tRNA (Gomez-Lorenzo et al, 2000; Harms et al, 2001; Valle et al, 2003b; Yusupov et al, 2001).
The conformational change at the L1 protuberance from its ‘out-position' to the ‘in-position' in the 80S·eEF2·sordarin complex appears to be rather complex (Figure 5). It entails two rotations around separate hinge points. One rotation is around a hinge point close to the H75/H76/H79 junction, and the other is around a point located within the rod-like H76. While H76 is a regular RNA double helix in prokaryotes, it contains a bulged-out nucleotide in eukaryotes exactly in the region of the second hinge. Furthermore, H76 in the ‘in-position' appears to make contact with the body of the 60S subunit at the position of this unpaired nucleotide. It is likely that the extra nucleotide provides flexibility for the second hinge movement, which would explain why much larger movements of the L1 protuberance are observed in yeast. In addition to the rotations, a third conformational change of the L1 protuberance takes place at the top of the ‘mushroom', resulting in a different domain arrangement within rpL1 (Figure 5). We note that the L1 protein can be crystallized in conformations with different arrangements of the N- and C-terminal domains (Nevskaya et al, 2000). A conformational change in rpL1 seems to be required to allow an interaction of rpL1 with the central protuberance that takes place in the ‘in-position' (Figures 1, 3 and 5). This interaction in turn might stabilize the L1 protuberance in the ‘in-position'.
Figure 5.
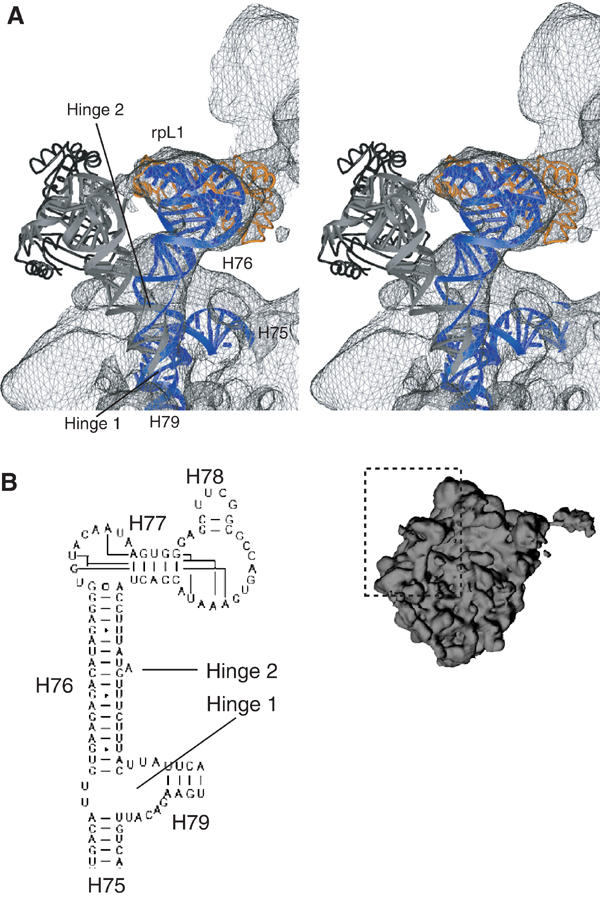
Movement of the L1 protuberance. (A) Close-up on the L1 protuberance. Elements of 25S rRNA and 60S ribosomal proteins fitted into the 80S·eEF2·sordarin complex are shown as blue and orange ribbons, respectively. The thumbnail of the 60S subunit is included as an orientation aid. The cryo-EM density of the 80S·eEF2·sordarin complex is shown as a gray wire-mesh. Docked models for the L1 protuberance in the outer position (Spahn et al, 2001a) are superposed as gray (25S rRNA) and black (rpL1) ribbons. Arrows indicate the hinge 1 and 2 regions of the conformational change. (B) Part of the secondary structure diagram of the 25S rRNA from Saccharomyces cerevisiae (http://www.rna.icmb.utexas.edu) showing the RNA corresponding to the L1 protuberance. Helices are indicated with their number. A eukaryotic-specific bulged-out nucleotide that is located at the hinge 2 region is marked by an arrow.
Dynamics of the ribosomal intersubunit bridges
The RSR of the subunits has to be regulated and made possible by conformational changes of the intersubunit bridges. The dynamic behavior of the bridges, thought to take part in the communication between the small and the large subunit, is of great functional importance. Therefore, we analyzed the intersubunit bridges of the 80S·eEF2·sordarin complex and compared the bridging regions between the subunits with the corresponding regions in our previous map of the POST 80S ribosome (Spahn et al, 2001a) (Figure 3, Table III). The RSR can be approximately described by rotational rigid-body movements of the body/platform and head domains of the 40S subunit relative to the 60S subunit. Dependent on the distance to the center of the rotation, components of both subunits that are engaged in intersubunit bridges move relative to each other, unless the movements brought about by the subunit rotation are counteracted by local changes and independent movements of the structural components forming the bridges.
Most of the central bridges are formed by RNA–RNA contacts, whereas RNA–protein and protein–protein contacts are located more toward the periphery (Spahn et al, 2001a; Yusupov et al, 2001). All central RNA–RNA intersubunit bridges (B2a, B2b, B2c, B2d, B3 and B5) are preserved in the 80S·eEF2·sordarin complex. This is very similar to the behavior observed for the bridges of the prokaryotic 70S ribosome (Gao et al, 2003; Valle et al, 2003b). As inferred from the two atomic models for the POST 80S ribosome (Spahn et al, 2001a) and the 80S·eEF2·sordarin complex, the RSR moves the corresponding RNA components relative to each other by distances ranging from 2 to 5.5 Å (Table III). Although small movements of bridge components cannot be directly observed at our resolution, it is likely that local conformational changes occur which preserve the connections despite the overall movement of the subunits. This possibility is in excellent agreement with the observed flexibility of bridge components in the X-ray structures of the ribosomal subunits (for a review, see Yonath, 2002). There is practically no relative movement at B2c, which involves the prominent switch helix h27 (Lodmell and Dahlberg, 1997). B2c can therefore be regarded as the center of the rotation of the 40S subunit relative to the 60S subunit (Figure 3; see similar observations for the 70S ribosome: Gao et al, 2003; Tama et al, 2003).
Bridges B2e and B4 involve RNA–protein contacts and are located relatively close to B2c, that is, to the center of rotation. B2e, which has been reported to be broken in E. coli ribosomes due to the RSR (Gao et al, 2003), can nevertheless be observed in both states of the yeast 80S ribosome (Table III). However, there are changes in B4. The apical loop of H34 interacts with h20 and rpS13 (S15p) in both states of the ribosome, but the second interaction, which involves the minor groove of H34 in the POST yeast 80S ribosome, appears to be absent in the 80S·eEF2·sordarin complex (Figure 3). H34 belongs to domain II of 25S rRNA, which also encompasses the GAC. Conformational signals induced at the GAC by the binding of ligands such as eEF2 might be transmitted to H34 and thereby control the subunit rearrangement.
Another candidate for an element important for controlling and regulating the intersubunit arrangement is protein rpL23 (L14p), since it is located directly at the foot of the SRL. In the POST 80S ribosome, rpL23 (L14p) participates in the formation of two bridges, B5a and B6 (Spahn et al, 2001a). Moreover, rpL23 (L14p) interacts with rpL24 (L24e), which in turn participates in B7. In support of a more active role of rpL23 (L14p) in controlling subunit rearrangement, B6 is not formed in the 80S·eEF2·sordarin complex, whereas B5a can be observed only at a low contour level (Figure 3, Table III). The L14 bridges are not changed in response to the RSR in E. coli ribosomes, but the observed conformational change of L14 (Gao et al, 2003) is in line with an active role of this protein.
In addition to those intersubunit bridges that are conserved among prokaryotes and eukaryotes, the eukaryotic 80S ribosome has extra bridges at the periphery (Spahn et al, 2001a). Bridge eB9 is disrupted in the 80S·eEF2·sordarin complex (Figure 3, Table III). However, other eukaryotic-specific bridges are present in both RSR-related conformations of the yeast 80S ribosome, apparently because intersubunit contacts are alternately formed between different parts of the subunits. This is the case for eB8. In the two states, different sites of a 40S protein density of unknown identity interact with the expansion of H79 of the 60S subunit. Different pairs of interactions for the two 80S conformations also exist for eB10, whereas the subunit movement appears to be accommodated by a local change in the case of eB11 (Table III). The new bridge eB12 involves rpL19 (L19e) and might be specific for the conformation of the RSR.
Intersubunit bridges formed by the head of the 40S subunit represent a special class. The rotation of the head, which takes place in addition to the RSR of the 40S subunit, results in a movement by about 15 Å relative to the 60S subunit when the POST 80S ribosome and the 80S·eEF2·sordarin complex are compared (see Figure 3). This movement leads to changed intersubunit bridges in the 80S·eEF2·sordarin complex, similar to the changes that have been observed for the E. coli 70S ribosome (Valle et al, 2003b). H38, the so-called A-site finger, continues to participate in bridge B1a, but the attachment site on the 40S subunit is different and appears to involve the N-terminus of rpS15 (S19p) and h33 (Table III). Moreover, the bridge can be observed only at a low contour level of the cryo-EM density map.
Protein rpL11 (L5p) of the central protuberance is also engaged in the formation of intersubunit bridges in both states. Again, bridge B1b/c is formed with differing proteins of the 40S subunit. A single-bridge interaction in the POST 80S ribosome is replaced by three interactions in the 80S·eEF2·sordarin complex that are with rpS15 (S19p) and rpS18 (S13p) (Figure 3, Table III). The set of different interactions at bridge B1b/c between the head of the 40S subunit and the central protuberance might be facilitated by concomitant conformational changes of the head proteins rpS15 (S19p) and rpS18 (S13p) and of the central protuberance.
Interaction between the 80S ribosome and eEF2 and the mechanism of sordarin
The interaction between the ribosome and the factor EF-G/eEF2 is normally transient, but the addition of nonhydrolyzable GTP analogs or certain drugs locks the system in a defined state. Fusidic acid inhibits translocation in both prokaryotic and eukaryotic systems by preventing the dissociation of EF-G/eEF2·GDP from the POST ribosome (for a review, see Spahn and Prescott, 1996). In contrast, sordarin, which has been used in this study, is highly specific for eEF2 from fungi. Similar to fusidic acid, sordarin prevents the dissociation of eEF2 from the ribosome (Justice et al, 1998), and therefore allows direct visualization of eEF2 bound to the ribosome by cryo-EM (Gomez-Lorenzo et al, 2000, and this work). However, the actual modes of inhibition of the translocation reaction by fusidic acid and sordarin appear to be different (Dominguez et al, 1999). The binding site for sordarin on eEF2 is located between domains III, IV and V (Capa et al, 1998; Jørgensen et al, 2003), whereas the fusidic acid binding site has been suggested to be located between the G domain and domains II and III (Laurberg et al, 2000).
Key to the inhibitory action of sordarin appears to be the particular conformation of domain III of eEF2 that is present in eEF2-sor (Jørgensen et al, 2003) and eEF2-cryo, which allows domain III to interact with the SRL (Figure 2). The remarkable increase in affinity of eEF2 for sordarin upon binding to the ribosome (Dominguez and Martin, 1998) is compatible with the possibility that the observed position of eEF2 domain III is also adopted on the ribosome, even in the absence of sordarin. In this case, part of the energy cost for the domain rearrangement of eEF2, which allows productive sordarin interaction with domains III, IV and V, would be paid by the energy released in the eEF2–ribosome interaction, explaining the increased sordarin affinity. Sordarin in turn might prevent domain III from moving away from the SRL, and thereby prevent the dissociation of eEF2 from the ribosome. The particular conformation of domain III of eEF2 has not yet been detected in the prokaryotic system. However, the importance of domain III for factor binding has been demonstrated for bacterial EF-G by a domain III deletion mutant (Martemyanov and Gudkov, 2000). If domain III is deleted, GTPase activity is impaired, but not the interaction of the ribosome with EF-G·GDP. As sordarin at high concentrations can stimulate the uncoupled GTPase activity when eEF2 is in excess over ribosomes (Dominguez et al, 1999), the particular conformation and ribosome interaction of domain III of eEF2 might therefore be important for GTPase activation.
Mechanism of tRNA translocation
In the course of the tRNA movement, the interactions of the tRNAs with the ribosome at the A and P sites have to be broken and new interactions have to be formed at the P and E sites, respectively. The conformational changes that the ribosome undergoes during tRNA translocation are likely to occur for the purpose of facilitating the movement of tRNA, especially since the movement of the 30S subunit during the RSR is in the expected direction of tRNA movement from A to P and P to E sites (Frank and Agrawal, 2000). Furthermore, kinetic experiments have indicated that a ribosomal conformational change precedes tRNA translocation (Savelsbergh et al, 2003). Indeed, as observed very recently, the RSR in E. coli 70S ribosomes is accompanied by the transition of a deacylated tRNA from the P site to a P/E hybrid site (Valle et al, 2003b). In an analogous way, the RSR could be involved in moving the A-site bound peptidyl-tRNA into an A/P hybrid site, directly linking the RSR to tRNA movement (Valle et al, 2003b; Zavialov and Ehrenberg, 2003). However, in contrast to the original hybrid-site model (Moazed and Noller, 1989; Noller et al, 2002), the formation of hybrid sites requires EF-G binding and does not occur spontaneously upon the peptidyl-transferase reaction.
Similar to the E. coli system (Valle et al, 2003b; Zavialov and Ehrenberg, 2003), the RSR in yeast could be directly coupled to tRNA movement. Part of the RSR in yeast is a rotational movement of the head domain of the 40S subunit that has not been observed in this form in the prokaryotic system. An intriguing possibility is that the rotation of the small subunit might occur first, prior to the additional movement of the head domain. As the tRNAs are tightly bound between body/platform and head domains, this first rotation would be forced on the tRNAs (Figure 6) and in turn break the tRNA interactions at the elbow with the large ribosomal subunit. The 3′-CCA end of the tRNAs is flexible and could move independently. It is therefore possible that this part of the RSR would lead to hybrid-site formation, as suggested for the E. coli 70S ribosome.
Figure 6.
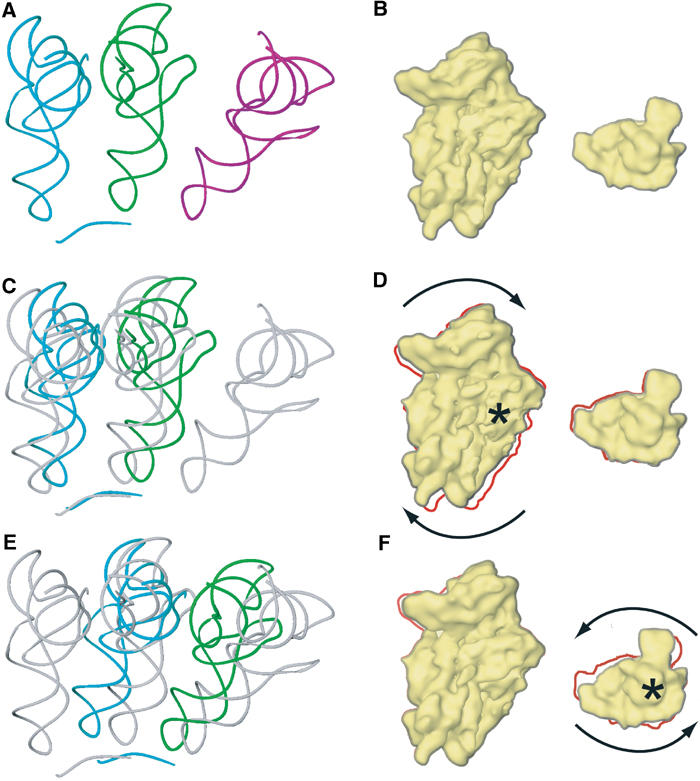
Model for tRNA movement during the translocation reaction. The coordinate transformations of the RSR, when applied to the A- and P-site bound tRNAs (A), result in hypothetical intermediates of the tRNAs during translocation (C, E) and a plausible trajectory for the path of the tRNAs from the A to the P and from the P to the E site. The corresponding state of the 40S subunit and its movements are shown in the cartoon in (B, D, F). The 40S subunit is shown on the left from the intersubunit side, and the head domain on the right from the top of the subunit. The crystallographically determined positions of the tRNAs in A (cyan), P (green) and E (purple) sites and an mRNA fragment (cyan) (Yusupov et al, 2001) are shown in (A) and are included in (C, E) as references painted in gray. (C) Shows the tRNAs in A (cyan) and P (green) sites after the coordinate transformation of the RSR for the body/platform domains of the 40S subunit (D) has been applied. The rotation of the 40S subunit is indicated by arrows, and the rotation axis at h27 of 18S rRNA by a star (D). The outline of the untransformed 40S (B) is included in (D) by the red line. The tRNA positions are further transformed (E) according to the additional movement of the head domain (F). This rotation occurs around a rotation axis through the neck of the 40S subunit (indicated by a star in (F)). The red line in (F) indicates the outline of the 40S subunit in (D).
The additional movement of the head domain that would follow next could participate in the translocation reaction also in a direct manner (Figure 6). As a result of the curvature of the head domain, the rotational movement around the neck region results in a translational movement of ribosomal residues bound to the tRNAs in the direction of the trajectory of the tRNAs. In the T. thermophilus A site, both h34 and the 965 loop of 16S rRNA in the head domain interact with the tRNA anticodon stem (Yusupov et al, 2001). In the 80S·eEF2·sordarin complex, the homologous residues are closer to the P site than to the A site. The same holds for head residues that interact with the P-site tRNA (Spahn et al, 2001a; Yusupov et al, 2001). In the 80S·eEF2·sordarin complex, these residues have moved close to the E-site region.
If the transformation parameters for the movement of the head domain from the POST 80S ribosome (Spahn et al, 2001a) to the 80S·eEF2·sordarin complex are applied to tRNAs located in A and P sites, the transformed tRNA positions are indeed close to the known P and E sites (Figure 6E and F). (We postulate in analogy to the E. coli system that in yeast the conformation of the PRE ribosome is similar to that of the POST ribosome.) It is therefore tempting to speculate that the movement of the 40S head domain occurs together with the tRNAs, such that the contacts between these moieties remain intact. The 40S head would act as a mechanical conveyer for transporting the anticodon stem loops of the tRNAs. As domain IV of eEF2 occupies the A site, it could prevent the tRNAs from moving backward when the RSR is reversed in completion of translocation. This would explain why at least an anticodon stem loop of the tRNA has to be present in the A site for translocation to occur (Joseph and Noller, 1998), as it would constitute a minimum target for interaction with domain IV. This model is reminiscent of the concept of a movable ribosomal domain (α–ɛ model; Dabrowski et al, 1998; Spahn and Nierhaus, 1998), except that in our new model the translocation reaction includes the resetting of the moveable head domain.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplemental data
Acknowledgments
We are grateful to Dr Sean Connell for help and discussion and to Michael Watters for assistance with the illustrations. This work was supported by HHMI and grants R37 GM29169 and R01 GM55440 from NIH and DBI 9871347 from NSF (to JF), R01 GM60635 (to PP), a grant of the Volkswagen Stiftung (to CS) and by grant PM99-0108 (MCyT) and Fundación Ramón Areces (to JB). The coordinates for the docked eEF2 and the ribosomal components have been deposited in the Protein Data Bank with accession numbers: 1S1H for the 40S subunit and eEF2; 1S1I for the 60S subunit. The cryo-EM map has been deposited at the 3D-EM database, EMBL-European Bioinformatics Institute, Cambridge, UK (accession number EMD-1067).
References
- Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J (1999) EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat Struct Biol 6: 643–647 [DOI] [PubMed] [Google Scholar]
- Ballesta JP, Remacha M (1996) The large ribosomal subunit stalk as a regulatory element of the eukaryotic translational machinery. Prog Nucleic Acid Res Mol Biol 55: 157–193 [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Beckmann R, Spahn CMT, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G (2001) Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107: 361–372 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54 (Part 5): 905–921 [DOI] [PubMed] [Google Scholar]
- Capa L, Mendoza A, Lavandera JL, Gomez de las Heras F, Garcia-Bustos JF (1998) Translation elongation factor 2 is part of the target for a new family of antifungals. Antimicrob Agents Chemother 42: 2694–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson M (1991) Ribbons 2.0. J Appl Cryst 24: 103–106 [Google Scholar]
- Dabrowski M, Spahn CM, Schafer MA, Patzke S, Nierhaus KH (1998) Protection patterns of tRNAs do not change during ribosomal translocation. J Biol Chem 273: 32793–32800 [DOI] [PubMed] [Google Scholar]
- Dominguez JM, Gomez-Lorenzo MG, Martin JJ (1999) Sordarin inhibits fungal protein synthesis by blocking translocation differently to fusidic acid. J Biol Chem 274: 22423–22427 [DOI] [PubMed] [Google Scholar]
- Dominguez JM, Martin JJ (1998) Identification of elongation factor 2 as the essential protein targeted by sordarins in Candida albicans. Antimicrob Agents Chemother 42: 2279–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley BT, Moehring JM, Moehring TJ (1995) Mutations in the elongation factor 2 gene which confer resistance to diphtheria toxin and Pseudomonas exotoxin A. Genetic and biochemical analyses. J Biol Chem 270: 23218–23225 [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal RK (2000) A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406: 318–322 [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal RK (2001) Ratchet-like movements between the two ribosomal subunits: their implications in elongation factor recognition and tRNA translocation. Cold Spring Harb Symp Quant Biol 66: 67–75 [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj Leith A (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116: 190–199 [DOI] [PubMed] [Google Scholar]
- Gabashvili IS, Agrawal RK, Spahn CMT, Grassucci RA, Frank J, Penczek P (2000) Solution structure of the E. coli 70S ribosome at 11.5 Å resolution. Cell 100: 537–549 [DOI] [PubMed] [Google Scholar]
- Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, Van Roey P, Agrawal RK, Harvey SC, Sali A, Chapman MS, Frank J (2003) Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell 113: 789–801 [DOI] [PubMed] [Google Scholar]
- Gomez-Lorenzo MG, Spahn CMT, Agrawal RK, Grassucci RA, Penczek P, Chakraburtty K, Ballesta JPG, Lavandera JL, Garcia-Bustos JF, Frank J (2000) Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 Å resolution. EMBO J 19: 2710–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Lorenzo MG, Spahn CMT, Linde J, Penczek PA, Ballesta JPG, Frank J (2004) Structural studies of the Saccharomyces cerevisiae ribosomal stalk, in preparation
- Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A (2001) High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107: 679–688 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zhou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A Found Crystallogr A47: 110–119 [DOI] [PubMed] [Google Scholar]
- Jørgensen R, Ortiz PA, Carr-Schmid A, Nissen P, Kinzy TG, Andersen GR (2003) Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat Struct Biol 10: 379–385 [DOI] [PubMed] [Google Scholar]
- Joseph S, Noller HF (1998) EF-G-catalyzed translocation of anticodon stem-loop analogs of transfer RNA in the ribosome. EMBO J 17: 3478–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice MC, Hsu MJ, Tse B, Ku T, Balkovec J, Schmatz D, Nielsen J (1998) Elongation factor 2 as a novel target for selective inhibition of fungal protein synthesis. J Biol Chem 273: 3148–3151 [DOI] [PubMed] [Google Scholar]
- Kaziro Y (1978) The role of guanosine 5′-triphosphate in polypeptide chain elongation. Biochim Biophys Acta 505: 95–127 [DOI] [PubMed] [Google Scholar]
- Laurberg M, Kristensen O, Martemyanov K, Gudkov AT, Nagaev I, Hughes D, Liljas A (2000) Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site. J Mol Biol 303: 593–603 [DOI] [PubMed] [Google Scholar]
- Lodmell JS, Dahlberg AE (1997) A conformational switch in Escherichia coli 16S ribosomal RNA during decoding of messenger RNA. Science 277: 1262–1267 [DOI] [PubMed] [Google Scholar]
- Martemyanov KA, Gudkov AT (2000) Domain III of elongation factor G from T. thermophilus is essential for induction of GTP hydrolysis on the ribosome. J Biol Chem 275: 35820–35824 [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF (1989) Intermediate states in the movement of transfer RNA in the ribosome. Nature 342: 142–148 [DOI] [PubMed] [Google Scholar]
- Nevskaya N, Tischenko S, Fedorov R, Al-Karadaghi S, Liljas A, Kraft A, Piendl W, Garber M, Nikonov S (2000) Archaeal ribosomal protein L1: the structure provides new insights into RNA binding of the L1 protein family. Struct Fold Des 8: 363–371 [DOI] [PubMed] [Google Scholar]
- Nikulin A, Eliseikina I, Tishchenko S, Nevskaya N, Davydova N, Platonova O, Piendl W, Selmer M, Liljas A, Drygin D, Zimmermann R, Garber M, Nikonov S (2003) Structure of the L1 protuberance in the ribosome. Nat Struct Biol 10: 104–108 [DOI] [PubMed] [Google Scholar]
- Noller HF, Yusupov MM, Yusupova GZ, Baucom A, Cate JH (2002) Translocation of tRNA during protein synthesis. FEBS Lett 514: 11–16 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V (2002) Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V (2002) Ribosome structure and the mechanism of translation. Cell 108: 557–572 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V, Moore PB (2001) Atomic structures at last: the ribosome in 2000. Curr Opin Struct Biol 11: 144–154 [DOI] [PubMed] [Google Scholar]
- Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J (2003) A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature 421: 87–90 [DOI] [PubMed] [Google Scholar]
- Rodnina M, Savelsbergh A, Katunin VI, Wintermeyer W (1997) Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 385: 37–41 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Semenkov Iu P, Savelsbergh A, Katunin VI, Peske F, Wilden B, Wintermeyer W (2001) Mechanism of tRNA translocation on the ribosome]. Mol Biol (Mosk) 35: 655–665 [PubMed] [Google Scholar]
- Rossmann MG (2000) Fitting atomic models into electron-microscopy maps. Acta Crystallogr D Biol Crystallog D56: 1341–1349 [DOI] [PubMed] [Google Scholar]
- Saad A, Ludtke SJ, Jakana J, Rixon FJ, Tsuruta H, Chiu W (2001) Fourier amplitude decay of electron cryomicroscopic images of single particles and effects on structure determination. J Struct Biol 133: 32–42 [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W (2003) An elongation factor G-induced ribosome rearrangement precedes tRNA–mRNA translocation. Mol Cell 11: 1517–1523 [DOI] [PubMed] [Google Scholar]
- Schlünzen F, Tocilj A, Zarivach R, Harms J, Glühmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A (2000) Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102: 615–623 [DOI] [PubMed] [Google Scholar]
- Spahn CMT, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J (2001a) Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA–ribosome and subunit–subunit interactions. Cell 107: 373–386 [DOI] [PubMed] [Google Scholar]
- Spahn CMT, Blaha G, Agrawal RK, Penczek P, Grassucci RA, Trieber CA, Connell SR, Taylor DE, Nierhaus KH, Frank J (2001b) Localization of the tetracycline resistance protein Tet(O) on the ribosome and the inhibition mechanism of tetracycline. Mol Cell 7: 1037–1045 [DOI] [PubMed] [Google Scholar]
- Spahn CMT, Nierhaus KH (1998) Models of the elongation cycle: an evaluation. Biol Chem 379: 753–772 [DOI] [PubMed] [Google Scholar]
- Spahn CMT, Prescott CD (1996) Throwing a spanner in the works: antibiotics and the translation apparatus. J Mol Med 74: 423–429 [DOI] [PubMed] [Google Scholar]
- Tama F, Valle M, Frank J, Brooks CL 3rd (2003) Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryo-electron microscopy. Proc Natl Acad Sci USA 100: 9319–9323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J (2003a) Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol 10: 899–906 [DOI] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J (2003b) Locking and unlocking of ribosomal motions. Cell 114: 123–134 [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V (2000) Structure of the 30S ribosomal subunit. Nature 407: 327–339 [DOI] [PubMed] [Google Scholar]
- Yonath A (2002) The search and its outcome: high-resolution structures of ribosomal particles from mesophilic, thermophilic, and halophilic bacteria at various functional states. Annu Rev Biophys Biomol Struct 31: 257–273 [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF (2001) Crystal structure of the ribosome at 5.5° resolution. Science 292: 883–896 [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Ehrenberg M (2003) Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell 114: 113–122 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplemental data


